Abstract
Small cell lung cancer (SCLC) exists broadly in four molecular subtypes: ASCL1, NEUROD1, POU2F3, and Inflammatory. Initially SCLC subtypes were thought to be mutually exclusive, but recent evidence shows intra-tumoral subtype heterogeneity and plasticity between subtypes. Using a CRISPR-based autochthonous SCLC GEMM to study the consequences of KDM6A/UTX inactivation, here we show that KDM6A inactivation induced plasticity from ASCL1 to NEUROD1 resulting in SCLC tumors that express both ASCL1 and NEUROD1. Mechanistically, KDM6A normally maintains an active chromatin state that favors the ASCL1 subtype with its loss decreasing H3K4me1 and increasing H3K27me3 at enhancers of neuroendocrine genes leading to a cell state that is primed for ASCL1 to NEUROD1 subtype switching. This work identifies KDM6A as an epigenetic regulator that controls ASCL1 to NEUROD1 subtype plasticity and provides an autochthonous SCLC GEMM to model ASCL1 and NEUROD1 subtype heterogeneity and plasticity, which is found in 35-40% of human SCLCs.
Keywords: KDM6A, UTX, ASCL1, NEUROD1, plasticity, subtypes, small cell lung cancer, neuroendocrine
Introduction
Small cell lung cancer (SCLC) is a high-grade neuroendocrine cancer accounting for ~15% of lung cancers1-3. Nearly all SCLCs are genetically driven by loss of function (LOF) mutations in tumor suppressor genes RB1 and TP534-6. Recent studies have uncovered SCLC molecular subtypes characterized by expression of distinct transcription factors3,7-10. These include the neuroendocrine ASCL1 and NEUROD1 subtypes (70-80%) and the non-neuroendocrine POU2F3 and inflammatory subtypes (20-30%)7,8,11.
Although SCLC subtypes were initially thought to be largely mutually exclusive3, recent immunohistochemistry (IHC) data from human SCLCs shows that most ASCL1-positive SCLCs also expressed NEUROD1, and conversely, most NEUROD1-positive SCLCs expressed ASCL19. Similar to SCLC, neuroendocrine prostate cancers contain both ASCL1 and NEUROD1 subpopulations12. A SCLC genetically-engineered mouse models (GEMM) driven by MYC, and loss of Rb1 and Trp53 (RPM model) demonstrated that MYC drives SCLC tumor evolution from ASCL1 to NEUROD1 to YAP1 demonstrating plasticity between SCLC subtypes13.
~3-4% of SCLC harbor KDM6A LOF mutations4,14. KDM6A is an H3K27 histone demethylase15,16 and also binds to KMT2D(MLL4) in the MLL3/4 COMPASS complex where KDM6A has a scaffolding role that impacts its H3K4me1 methyltransferase activity at enhancers17-20. Several KDM6A phenotypes do not require its demethylase activity suggesting a dominant functional role for KDM6A in the COMPASS complex21-24. Consistent with this, COMPASS complex members KMT2D, NCAO6, and KMT2C are the 3 top co-dependencies with KDM6A in the cancer dependency map25. KMT2C and KMT2D LOF mutations are found in ~15% of SCLCs4,14. Hence COMPASS complex mutations together comprise ~20% of SCLCs. Based on KDM6A’s role in differentiation during development and in other cancers19, we hypothesized that KDM6A inactivation in SCLC could have consequences on neuroendocrine differentiation thereby impacting SCLC subtype determination.
Results
KDM6A Loss Induces NEUROD1 Expression in SCLC Tumors
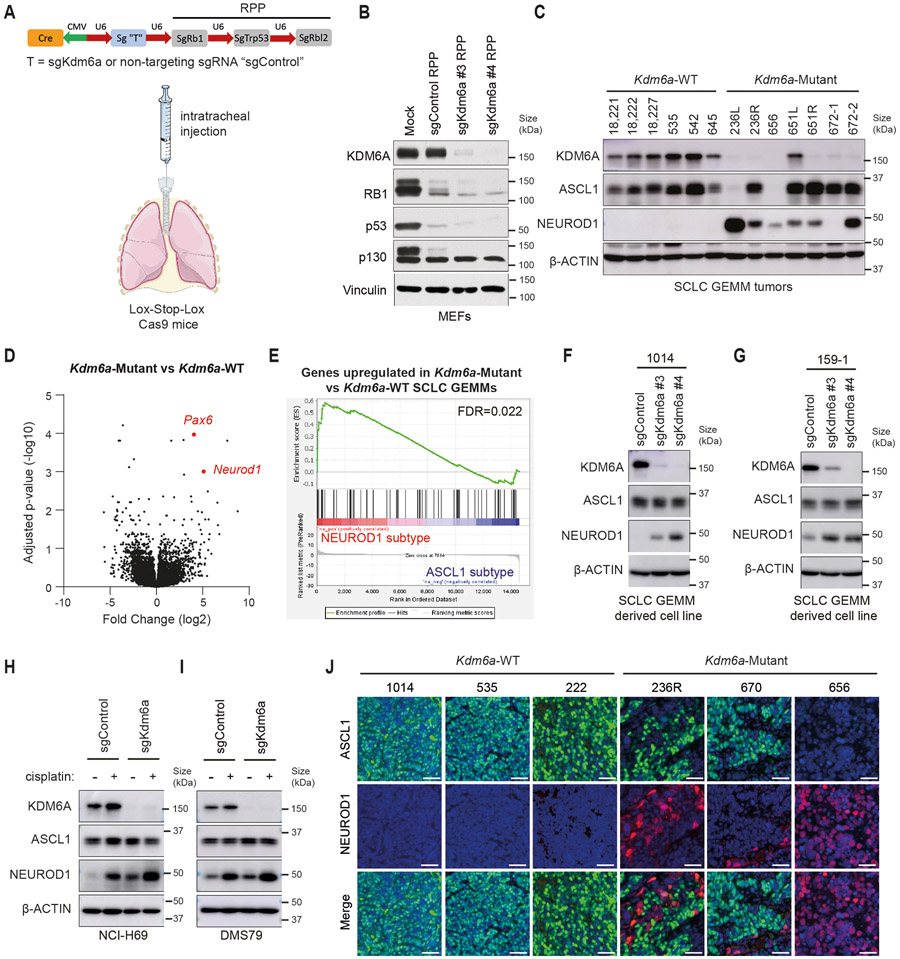
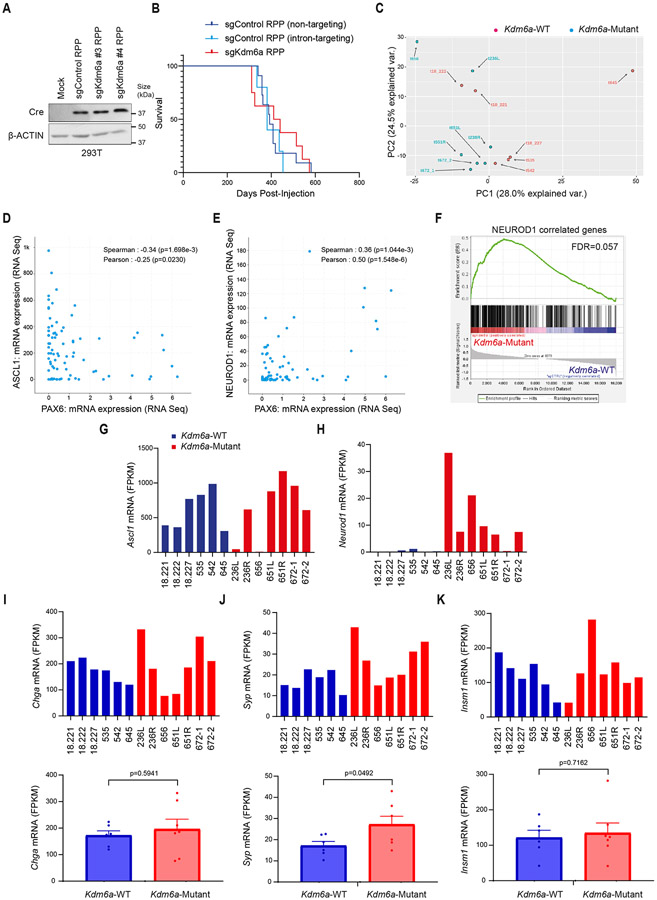
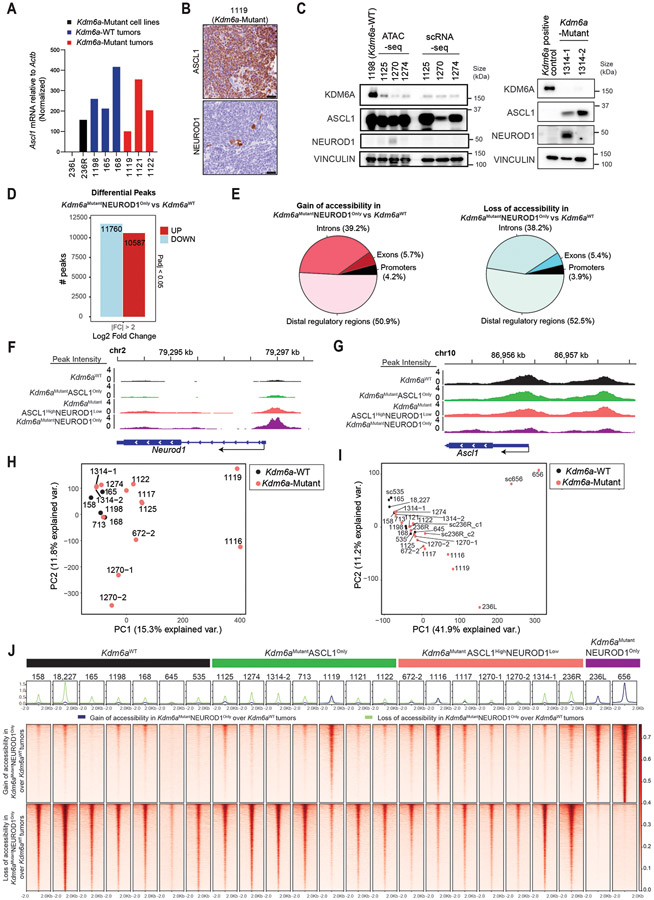
To study the consequences of KDM6A inactivation in SCLC, we used our SCLC CRISPR RPP GEMM26 to generate autochthonous SCLCs that were Kdm6a-Mutant or Kdm6a-WT (Fig. 1a). To do this, adenoviruses that encoded Cre recombinase and sgRNAs targeting Rb1, Trp53, and Rbl2 (RPP) and Kdm6a (sgKdm6a RPP) or two different sgRNA controls including a non-targeting control or intron-targeting control (sgControl RPP) were engineered and first transduced into MEFs, which confirmed simultaneously knock out of KDM6A along with RPP and Cre expression (Fig. 1b and Extended Data Fig. 1a). sgKdm6a#4 RPP or sgControl RPP adenoviruses were then introduced into the lungs of lox-stop-lox(LSL) Cas9 mice by intratracheal (IT) injection (Fig. 1a) and mice were followed until symptoms developed. KDM6A inactivation had no significant effect on overall survival (Extended Data Fig. 1b). Histopathological examination revealed that all tumors had histology consistent with SCLC (Supplementary Table 1).
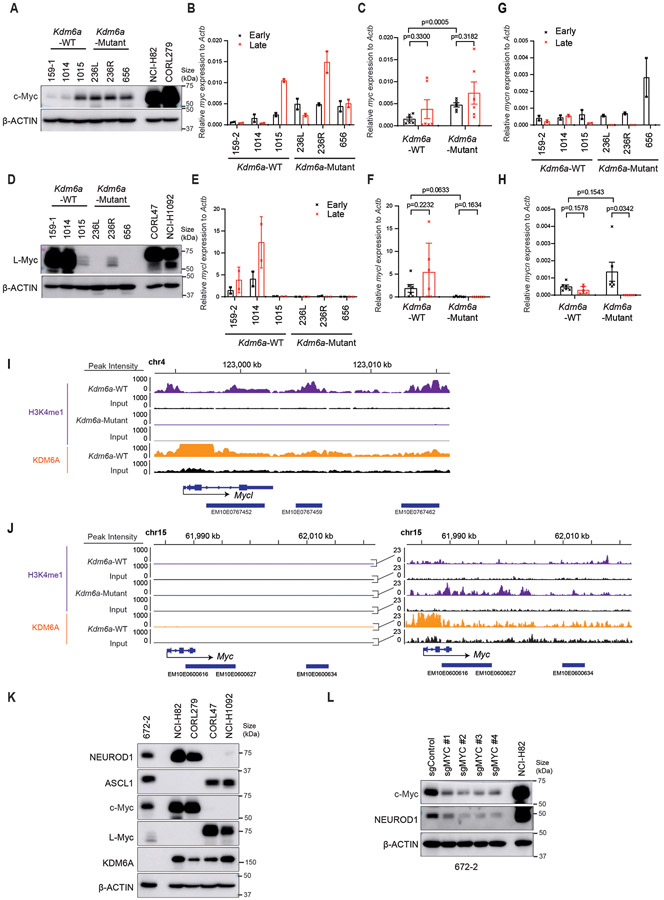
Fig. 1. KDM6A Inactivation in an Autochthonous SCLC Mouse Model Promotes NEUROD1 Expression Leading to SCLC Tumors that Express both ASCL1 and NEUROD1.
(a) Schematic of the adenovirus used for intratracheal injection (IT) into the lungs of lox-stop-lox (LSL)-Cas9 mice to generate autochthonous SCLC tumors that are Kdm6a inactivated or Kdm6a wild-type (WT). RPP=sgRb1, sgTrp53, sgRbl2; sg “T”=sgKdm6a or sgControl (non-targeting sgRNA). (b) Immunoblot analysis of MEFs expressing Cas9 infected with the sgControl RPP, sgKdm6a #3 RPP or sgKdm6a #4 RPP adenoviruses as indicated. (c) Immunoblot analysis of SCLC lung tumors formed in LSL-Cas9 mice injected with sgControl RPP (Kdm6a-WT) or sgKdm6a RPP (Kdm6a-Mutant) adenoviruses. (d) Volcano plot of differential expression analysis from RNA-seq data comparing Kdm6a-Mutant vs. Kdm6a-WT from tumors in c. n=7 Kdm6a-Mutant tumors, n=6 Kdm6a-WT tumors. FDR p-values adjusted for multiple comparisons after log transformation are shown. Neurod1 and one of its target genes Pax6 are highlighted in red. (e) Gene set enrichment analysis (GSEA) of RNA-seq data from ASCL1 and NEUROD1 human SCLC tumors34 of the upregulated genes in Kdm6a-Mutant vs Kdm6a-WT GEMMs (see Supplementary Table 2). FDR q-value adjusted for multiple comparisons is indicated. (f, g) Immunoblot analysis of two mouse SCLC cell lines derived from Kdm6a-WT mice 1014 (f) and 159-1 (g) transduced with 2 independent Kdm6a sgRNAs or a non-targeting control (sgControl) and maintained in culture for 30 days post-transduction. (h, i) Immunoblot analysis of two human ASCL1-positive SCLC cell lines, NCI-H69 (h) and DMS79 (i), nucleofected with Cas9 RNP containing a Kdm6a sgRNA or a non-targeting control (sgControl). Cells were then treated with cisplatin (1 μM) or DMSO for 3 days. (j) Multiplexed immunofluorescence for ASCL1 and NEUROD1 from 3 Kdm6a-WT and 3 Kdm6a-Mutant mouse SCLC lung tumors indicated. Scale Bar= 50 μm.
Based on KDM6A’s role in differentiation during development and in cancer17,19,27,28, we hypothesized that KDM6A was important for controlling SCLC neuroendocrine differentiation. To test this, immunoblot analysis for the neuroendocrine transcription factors ASCL1 and NEUROD1 was performed using tumors from several sgKdm6a RPP (Kdm6a-Mutant) and sgControl RPP (Kdm6a-WT) mice. Strikingly, compared to Kdm6a-WT tumors, which express only ASCL1 and not NEUROD126,29-32, Kdm6a-Mutant tumors also expressed NEUROD1 in almost all tumors with ASCL1 loss in some tumors (Fig. 1c). The magnitude of NEUROD1 expression was heterogenous across Kdm6a-Mutant tumors. Consistent with this, DEG analysis of bulk RNA-seq from Kdm6a-Mutant vs. Kdm6a-WT tumors showed that NEUROD1 and PAX6 (NEUROD1 target gene)33 as well as NEUROD1-correlated genes were highly enriched in Kdm6a-Mutant tumors (Fig. 1d, Extended Data Figs. 1c-k, and Supplementary Table 2). Upregulated genes in Kdm6a-Mutant tumors were significantly enriched in human SCLC tumors of the NEUROD1 subtype relative to the ASCL1 subtype34 suggesting that the Kdm6a-Mutant SCLC GEMM is a model relevant to the human NEUROD1 SCLC subtype (Fig. 1e). NEUROD1 induction after KDM6A loss could be recapitulated by Kdm6a CRISPR inactivation in 2 mouse SCLC cell lines established from Kdm6a-WT mice (Figs. 1f,g and Extended Data Figs. 2a,b). Similarly, KDM6A CRISPR inactivation modestly increased NEUROD1 expression in 2 human ASCL1-positive SCLC cell lines, and treatment with the front-line chemotherapy cisplatin further increased NEUROD1 expression in these KDM6A-inactivated cells (Figs. 1h,i).
IHC and multiplex immunofluorescence for ASCL1 and NEUROD1 showed that Kdm6a-Mutant tumors variably expressed ASCL1 or NEUROD1 in a mutually exclusive manner suggesting propensity to switch from ASCL1 to NEUROD1 upon Kdm6a inactivation (Fig. 1j and Extended Data Figs. 2c,d). Of note, a low percentage of tumor cells within some Kdm6a-Mutant tumors didn’t express ASCL1 nor NEUROD1 (Extended Data Fig. 2d). Both Kdm6a-Mutant and Kdm6a-WT tumors had histological features consistent with SCLC expressing neuroendocrine markers synaptophysin, chromogranin A, and INSM1 (Extended Data Figs. 1i-k, 2e-g). Interestingly, the nuclei of NEUROD1-positive cells were larger compared to ASCL1-positive cells and showed scattered pleomorphic giant cells (Extended Data Figs. 2e-g). Together, these results show that KDM6A inactivation induces heterogenous NEUROD1 expression in SCLCs of the ASCL1 subtype.
KDM6A Loss Increases Chromatin Accessibility at the Neurod1 Promoter
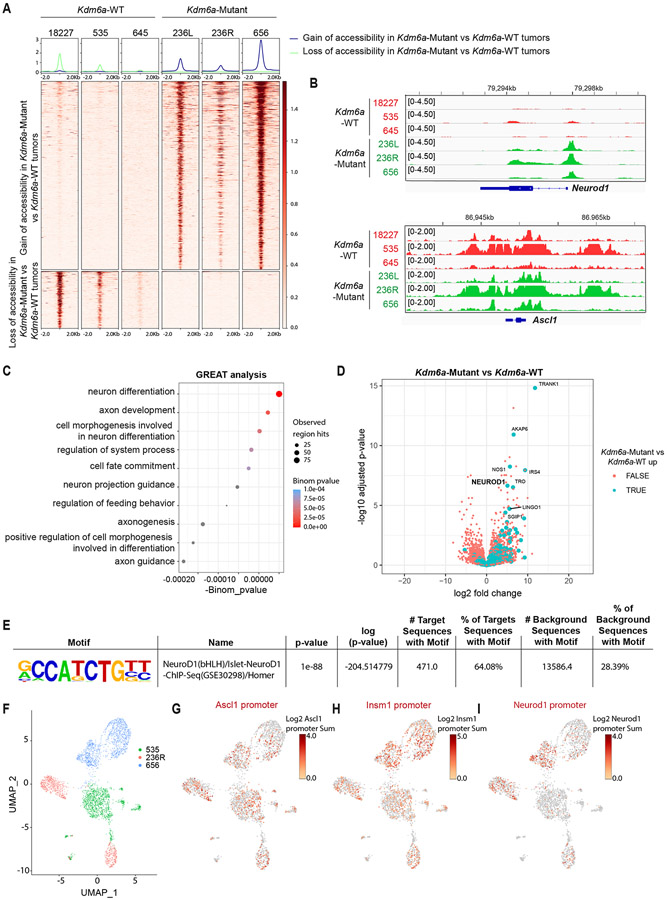
We next performed ATAC-seq to identify whether KDM6A inactivation alters chromatin accessibility at NEUROD1 or NEUROD1-related genes in SCLC mouse tumors. Bulk ATAC-seq showed both increased and decreased chromatin accessibility peaks at both promoters and enhancers in Kdm6a-Mutant tumors vs. Kdm6a-WT tumors (Fig. 2a, Extended Data Figs. 2h,i, Supplementary Table 3). Kdm6a-Mutant tumors had significantly increased chromatin accessibility at the Neurod1 promoter and largely retained chromatin accessibility at the Ascl1 promoter (Fig. 2b). Genomic regions enrichment of annotations tool (GREAT) analysis which identifies biological processes associated with differential changes in cis-regulatory regions35 showed enrichment of processes involved in axon development and neuron differentiation in Kdm6a-Mutant vs. Kdm6a-WT tumors (Fig. 2c); biological processes also enriched in NEUROD1-positive human SCLCs34. Neurod1 was a top differentially expressed gene that also significantly gained chromatin accessibility in Kdm6a-Mutant tumors (Fig. 2d). Moreover, the NEUROD1 Motif was the most significantly enriched motif in Kdm6a-Mutant tumors (Fig. 2e). Single-cell ATAC-seq in 2 Kdm6a-Mutant tumors that either heterogeneously expressed ASCL1 and NEUROD1 (236R) or expressed only NEUROD1 (656), and also on 1 Kdm6a-WT tumor (535) (Fig. 2f, also see Figs. 1c,j) confirmed these findings showing that Kdm6a-Mutant tumor cells have accessibility at both the ASCL1 and NEUROD1 promoters while Kdm6a-WT tumor cells only show accessibility for ASCL1 (Figs. 2g-i and Extended Data Fig. 2j).
Fig. 2. KDM6A Inactivation Increases Chromatin Accessibility at the Neurod1 Promoter in Autochthonous SCLC Mouse Tumors.
(a) Heat maps of ATAC-seq read densities of upregulated (n=735) and downregulated (n=222) peaks near promoters in Kdm6a-Mutant (236L, 236R, 656) vs. Kdm6a-WT (18227, 535, 645) mouse SCLC lung tumors. (b) Tracks of ATAC-seq data at Neurod1 (top) and Ascl1 (bottom) promoters from the Kdm6a-WT (red) and Kdm6a-Mutant (green) mouse SCLC tumors indicated. (c) Genomic Regions Enrichment of Annotations Tool (GREAT) analysis of ATAC-seq data (from a) of the changes in Kdm6a-Mutant tumors vs. Kdm6a-WT tumors. Binomial p-values were calculated using GREAT. (d) Volcano plot of differential expression analysis from RNA-seq data from Fig. 1 comparing Kdm6a-Mutant tumors vs. Kdm6a-WT tumors. TRUE/FALSE analysis indicates if ATAC-seq differential peaks have matched nearby gene. p-values are calculated using Wald test in DEseq2 and were adjusted for multiple hypothesis testing. (e) HOMER Motif Enrichment analysis from ATAC-seq data from a showing that the Neurod1 motif is the top enriched regulatory motif in Kdm6a-Mutant tumors vs. Kdm6a-WT tumors (see Supplementary Table 3, tab 4 for complete HOMER Motif enrichment list). p-values are calculated using binomial test in HOMER2. (f) Uniform Manifold Approximation and Projection (UMAP) of single-cell ATAC-seq data of all cells from three independent tumors from 3 independent mice. n=1252 cells from Kdm6a-WT tumor in green (535), n=830 cells and n=1258 cells from 2 Kdm6a-Mutant tumors from independent mice in red (236R) and blue (656). (g-i) Chromatin accessibility at Ascl1 promoter (g), Insm1 promoter (h) and Neurod1 promoter (i) from single-cell ATAC-seq data from f.
Kdm6a Loss Alters Chromatin Accessibility for ASCL1 to NEUROD1 Subtype Switching
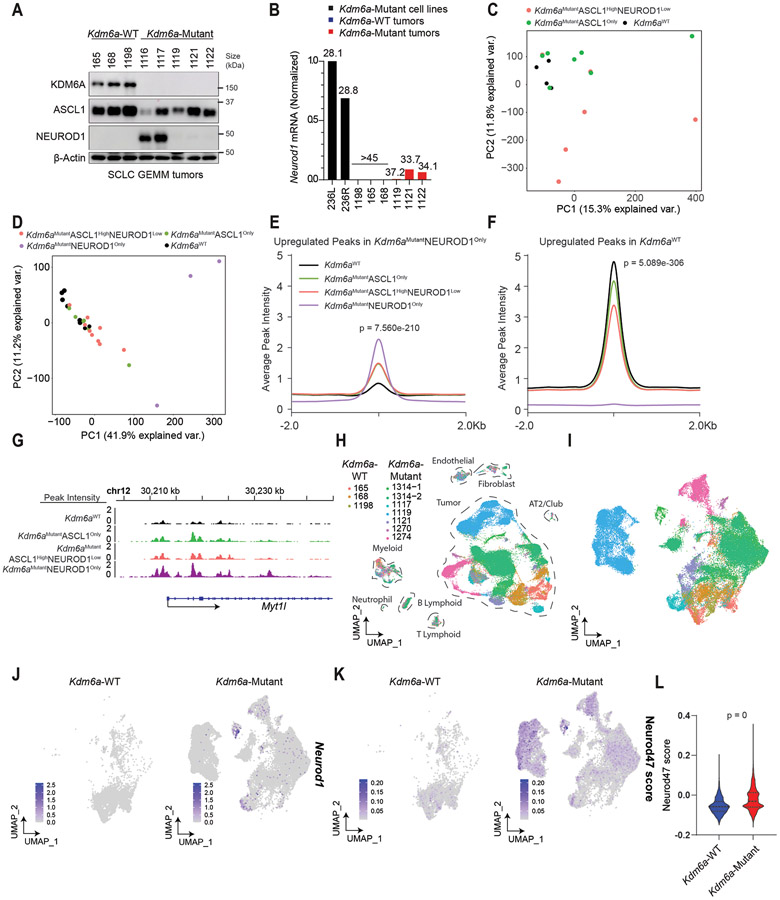
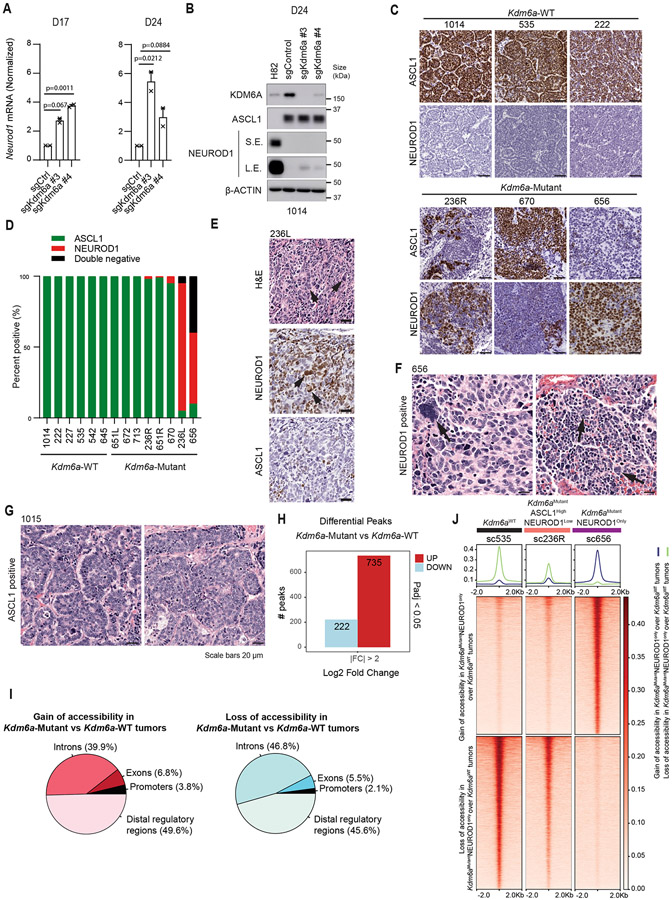
Our data shows that KDM6A loss can induce the complete transition from ASCL1 to NEUROD1, but in many tumors its loss promotes ASCL1 and NEUROD1 intra-tumoral heterogeneity (Fig. 1). We noted that one Kdm6a-Mutant tumor in our initial cohort and 3 Kdm6a-Mutant tumors in a subsequent independent cohort, all with confirmed KDM6A protein loss, grossly expressed only ASCL1 without detectable NEUROD1 protein (Figs. 1c, 3a). Neurod1 RT-qPCR showed very low but detectable Neurod1 transcripts in all these Kdm6a-Mutant “ASCL1 positive/NEUROD1 negative” tumors, but no detectable Neurod1 transcripts in any Kdm6a-WT tumor (Fig. 3b and Extended Data Fig. 3a). IHC showed very rare NEUROD1-positive cells in these Kdm6a-Mutant tumors (Extended Data Fig. 3b) suggesting these subtle increases in Neurod1 mRNA likely reflected very rare NEUROD1-positive cells within these Kdm6a-Mutant tumors.
Fig. 3. Kdm6a Inactivation Alters Chromatin Accessibility and mRNA Expression for ASCL1 to NEUROD1 Subtype Switching.
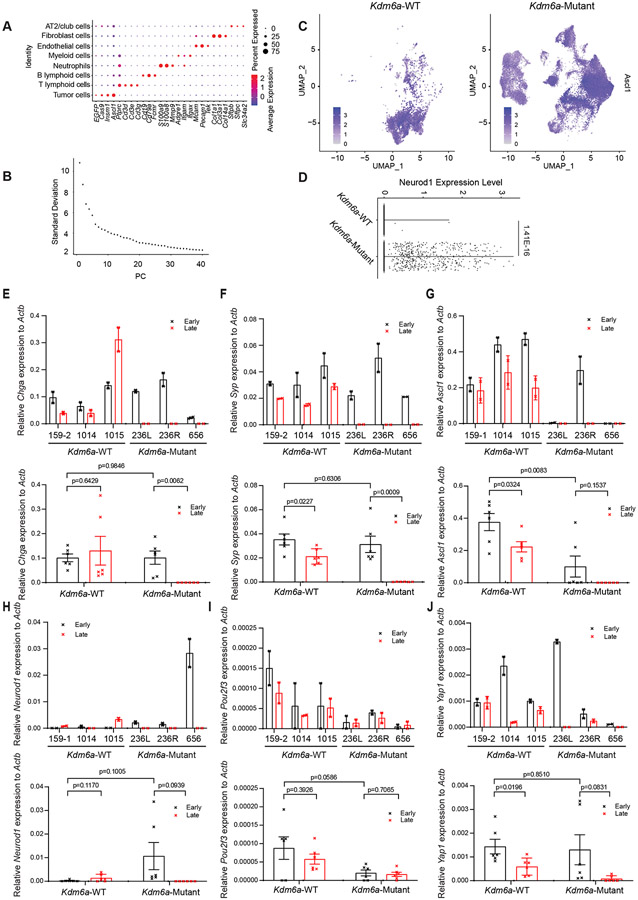
(a) Immunoblot analysis of an additional cohort of SCLC lung tumors from LSL-Cas9 mice IT injected with sgControl RPP or sgKdm6a RPP adenoviruses. (b) RT-qPCR for Neurod1 from tumors from a where NEUROD1 protein was undetectable. Ct values of each sample are indicated on top. (c) PCA of all chromatin accessibility peaks from ATAC-seq data of 4 Kdm6aWT SCLC tumors, 7 Kdm6aMutantASCL1Only SCLC tumors, and 6 Kdm6aMutantASCL1HighNEUROD1Low SCLC tumors from the 2nd cohort of mice. (d) PCA of differential chromatin accessibility peaks in Kdm6aMutantNEUROD1Only vs. Kdm6aWT SCLC tumors (see Extended Data Fig. 3d) from both bulk and pseudo-bulk sc-ATAC-seq data of all tumors: 7 Kdm6aWT SCLC tumors, 7 Kdm6aMutantASCL1Only SCLC tumors, 7 Kdm6aMutantASCL1HighNEUROD1Low SCLC tumors, and 2 Kdm6aMutantNEUROD1Only SCLC tumors. For tumors in c,d, see Supplementary Table 4. (e, f) Average peak intensity at all peaks upregulated in Kdm6aMutantNEUROD1Only vs. Kdm6aWT (e) or upregulated in Kdm6aWT vs. Kdm6aMutantNEUROD1Only (f) for all tumors in d classified by phenotype. Legend in e also applies to f. p-values for e,f indicates comparisons among the 4 groups. p-values are calculated using Anderson-Darling k-sample test in R package “kSamples”. (g) Average chromatin accessibility tracks for each phenotype at the NEUROD1 target gene Myt1l from ATAC-seq data from d. (h) UMAP of all cells from scRNA-seq of the 10 autochthonous SCLC lung tumors indicated [3 Kdm6a-WT, 7 Kdm6a-Mutant including 5 Kdm6aMutantASCL1Only and 2 Kdm6aMutantASCL1HighNEUROD1Low). n=48,651 total cells. Cell types were determined based on representative marker expression (Extended Data Fig. 4a). (I) UMAP of all tumor cells from the scRNA-seq from h. (j,k) Feature plots of Neurod1 (j) or the Neurod47_score (k). (l) Neurod47_score violin plot from k. For l, non-parametric Wilcoxon rank sum test was used to generate a two-tailed p-value. n=35,446 Kdm6a-Mutant and n=6612 Kdm6a-WT tumor cells. Minimum and maximum values define the range of violin plot. Dotted lines represent median and upper and lower quartiles.
To better understand whether these Kdm6a-Mutant “ASCL1-dominant/NEUROD1-low” tumors differed from Kdm6a-WT tumors, bulk ATAC-seq was performed on an additional cohort of Kdm6a-Mutant SCLC tumors that dominantly expressed ASCL1 with either no detectable NEUROD1 expression (Kdm6aMutantASCL1Only) or that primarily expressed ASCL1 with a minority of cells expressing NEUROD1 (Kdm6aMutantASCL1HighNEUROD1Low) vs. Kdm6a-WT tumors (Extended Data Fig. 3c and Supplementary Table 4). Chromatin accessibility signatures of Kdm6aMutantNEUROD1Only tumors (656 and 236L) vs. Kdm6a-WT tumors were used (Extended Data Figs. 3d,e) to understand whether Kdm6aMutantASCL1Only or Kdm6aMutantASCL1HighNEUROD1Low tumors had chromatin accessibility changes that resemble Kdm6aMutantNEUROD1Only tumors despite low to no NEUROD1 expression. Chromatin accessibility for ASCL1 or NEUROD1 were consistent with the expression patterns described above (Extended Data Figs. 3f,g). PCA analysis from this experiment alone or combined with the initial ATAC-seq experiment in Fig. 2 (including pseudo-bulk analysis of the scATAC-seq samples) showed that Kdm6aMutantASCL1Only or Kdm6aMutantASCL1HighNEUROD1Low tumors were different from Kdm6a-WT tumors showing an intermediate phenotype compared to the Kdm6aMutantNEUROD1Only tumors above (Figs. 3c,d and Extended Data Figs. 3h,i). Similarly, upregulated and downregulated peaks in both Kdm6aMutantASCL1Only and Kdm6aMutantASCL1HighNEUROD1Low tumors showed an intermediate phenotype compared to either Kdm6aMutantNEUROD1Only tumors or Kdm6a-WT tumors (Figs. 3e,f and Extended Data Fig. 3j). For example, Myt1l, a NEUROD1 target gene29, already displayed increased promoter accessibility in Kdm6aMutantASCL1Only or Kdm6aMutantASCL1HighNEUROD1Low tumors (Fig. 3g).
To further explore whether there are subpopulations of tumor cells within Kdm6a-Mutant “ASCL1 dominant/NEUROD1 low” tumors that show gene expression patterns observed in NEUROD1-positive SCLCs, we performed scRNA-seq on 7 Kdm6a-Mutant “ASCL1 dominant/NEUROD1 low” tumors (including 5 Kdm6aMutantASCL1only and 2 Kdm6aMutantASCL1HighNEUROD1Low tumors) vs. 3 Kdm6a-WT tumors (Fig. 3a, Extended Data Fig. 3c and Supplementary Table 5). UMAP projections showed distinct tumor cell and immune cell populations within tumors (Fig. 3h and Extended Data Fig. 4a). Consistent with our immunoblots, both Kdm6a-Mutant and Kdm6a-WT tumor cells expressed Ascl1 in most tumor cells while there were distinct small clusters of Neurod1-positive cells in the 2 Kdm6aMutantASCL1HighNEUROD1Low tumors with sporadic single NEUROD1-positive cells in other Kdm6a-Mutant tumors (Figs. 3i,j and Extended Data Figs. 4b-d). We then asked whether there were already expression changes associated with the NEUROD1 subtype despite being largely “NEUROD1-negative” by developing a NEUROD1 gene score based on genes whose expression correlated with NEUROD1-positive human SCLC tumors and cell lines29 (see Methods, referred to hereafter as Neurod47_score; Supplementary Table 5). The Neurod47_score was highly significantly enriched in Kdm6a-Mutant vs. Kdm6a-WT tumors and many Kdm6a-Mutant tumor cells had this signature upregulated despite not expressing NEUROD1 (Figs. 3k,l). Together, these data show that Kdm6a inactivation induces chromatin accessibility and mRNA expression changes correlating with NEUROD1 SCLCs even in tumors that remain ASCL1-dominant suggesting these tumors are primed for transitioning from an ASCL1-positive to an NEUROD1-positive cell state.
KDM6A Loss Accelerates Plasticity Between SCLC Subtypes
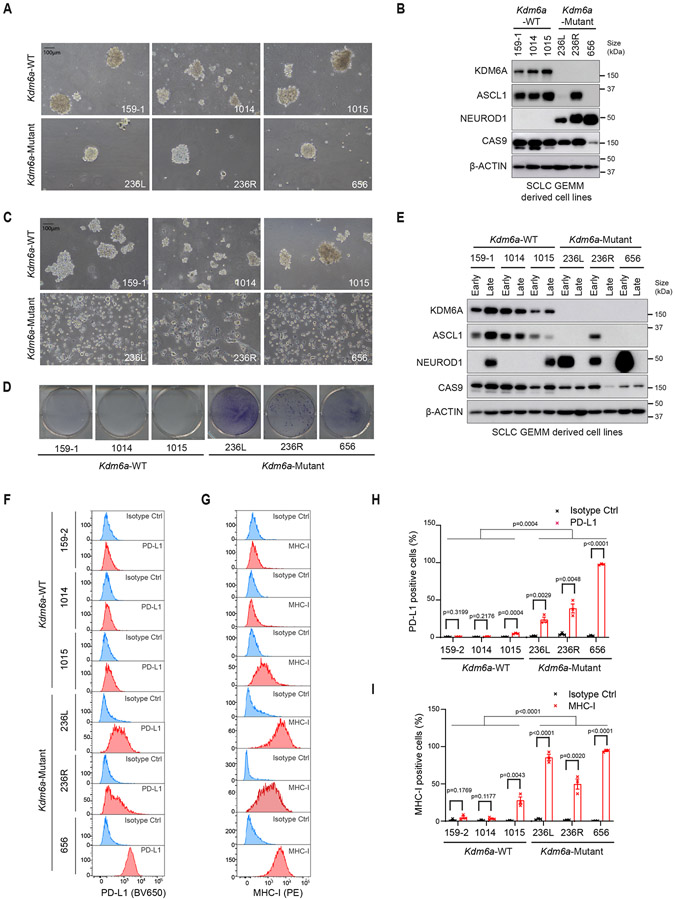
We then isolated primary cell lines from Kdm6a-Mutant and Kdm6a-WT SCLC lung tumors. Initially, cells grew as neuroendocrine spheroid aggregates similar to previous cell lines generated from SCLC GEMMs30 and both Kdm6a-Mutant and Kdm6a-WT cells highly expressed the neuroendocrine markers synaptophysin and chromogranin (Fig. 4a and Extended Data Figs. 4e,f). Consistent with our in vivo findings, early passage cell lines from Kdm6a-Mutant tumors expressed NEUROD1 and variably expressed ASCL1, while Kdm6a-WT tumors expressed only ASCL1 (Fig. 4b and Extended Data Figs. 4g,h). Surprisingly after ~1 month in culture, Kdm6a-Mutant cell lines began growing as attached cells even when plated on ultra-low attachment plates and lost chromogranin and synaptophysin, while Kdm6a-WT cell lines remained as neuroendocrine spheroid aggregates in suspension and retained chromogranin and synaptophysin (Figs. 4c,d and Extended Data Figs. 4e,f). Immunoblot analyses revealed that late passage Kdm6a-Mutant lines also lost expression of both ASCL1 and NEUROD1 (referred to hereafter as double-negative) (Fig. 4e and Extended Data Figs. 4g,h). Interestingly, over time some Kdm6a-WT cells began to acquire low NEUROD1 expression (Fig. 4e and Extended Data Figs. 4g,h) demonstrating a tendency to drift toward a NEUROD1-positive state and suggesting a temporal order of events beginning with ASCL1 followed by NEUROD1.
Fig. 4. KDM6A Inactivation Accelerates Plasticity Between SCLC Subtypes.
(a) Representative brightfield images and (b) immunoblot analysis of cell lines derived from Kdm6a-WT or Kdm6a-Mutant SCLC mouse lung tumors cultured in ultra-low attachment flasks at early times (<1 month) after the cell lines were generated (see Methods). (c) Representative brightfield images from cells in a and b after being maintained in ultra-low attachment flasks for 2 months in culture (late passage). (d) Crystal violet staining of late passage Kdm6a-WT or Kdm6a-Mutant cell lines plated on tissue culture treated 6-well plates for 48 hours. (e) Immunoblot analysis of cell lines shown in a and c comparing ASCL1 and NEUROD1 protein levels in early vs. late passage tumor-derived cell lines (see Methods). (f-i) Histograms (f,g) and quantification (h,i) of flow cytometry analysis for cell surface expression of PD-L1 (BV650) (f, h) and MHC class I (H2-Db) (PE) (g, i) in the late passage Kdm6a-WT and Kdm6a-Mutant cell lines. For f-i, n=3 biological independent experiments. Data are presented as mean values ± SEM. Statistical significance was calculated using unpaired, two-tailed students t-test and p-values are indicated on each figure panel. For all t-tests in h,i comparing all Kdm6a-Mutant vs. Kdm6a-WT cell lines, t-test compares all late passage Kdm6a-Mutant vs. all late passage Kdm6a-WT. Scale Bar=100 μm.
These attached double-negative Kdm6a-Mutant lines were reminiscent of non-neuroendocrine cells with restored antigen presentation36; the difference being that Kdm6a-Mutant cells weren’t derivative subpopulations, but comprised the entire culture. In line with this, late passage Kdm6a-Mutant lines had restored surface MHC class I and PD-L1 expression (Figs. 4f-i). They did not highly express non-neuroendocrine SCLC transcription factors including POU2F3 or YAP1 (Extended Data Figs. 4i,j). Together, these results demonstrate that KDM6A loss induces subtype plasticity from an ASCL1 to NEUROD1 cell state in vivo which transitions to a “double-negative” cell state with restored inflammatory markers when cultured ex vivo.
Analysis of MYC Paralogs in Kdm6a-Mutant Tumors
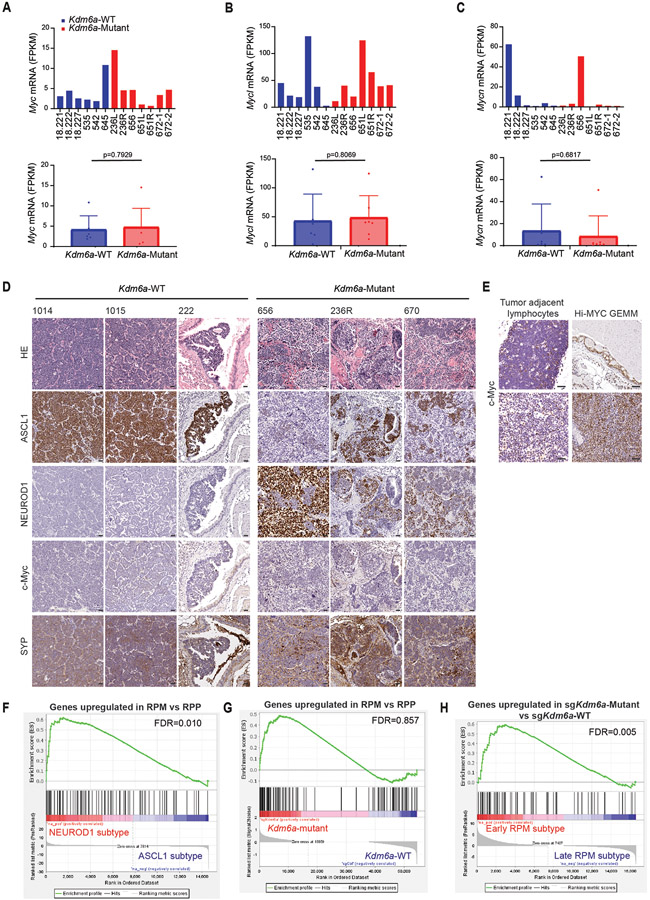
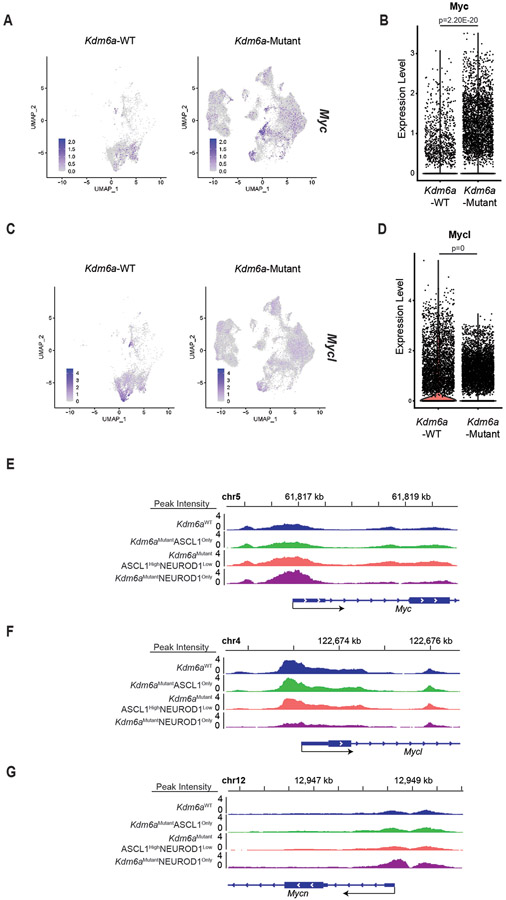
The phenotypes after Kdm6a loss draws parallels with MYC-driven SCLC subtype plasticity13. To explore whether there is a relationship between Kdm6a-Mutant and MYC-driven subtype plasticity, we analyzed MYC paralog (MYC, MYCL, MYCN) expression in Kdm6a-Mutant tumors. Bulk RNA-seq showed no significant change in MYC paralog expression in Kdm6a-Mutant vs. Kdm6a-WT tumors (Extended Data Figs. 5a-c). Both Kdm6a-Mutant and Kdm6a-WT tumors highly expressed MYCL with relatively low MYC expression. Consistent with this, IHC for c-Myc showed very low c-Myc protein levels in both Kdm6a-Mutant and Kdm6a-WT tumors (Extended Data Figs. 5d-e). Upregulated gene sets in Kdm6a-Mutant or RPM GEMMs both significantly correlated with NEUROD1-positive human SCLCs, but genes upregulated in RPM tumors did not significantly correlate with genes upregulated in Kdm6a-Mutant tumors again suggesting differences between Kdm6a-Mutant and RPM GEMMs (Extended Data Figs. 5f,g, Fig. 1e, Supplementary Table 6). A significant correlation was found between genes upregulated in Kdm6a-Mutant tumors vs. genes upregulated early on during the RPM subtype evolution (Extended Data Fig. 5h), which is when NEUROD1 is most highly expressed in RPM GEMMs13. Interestingly, higher resolution analyses from tumor cell pseudo-bulk DEG analysis showed that MYCL expression was modestly decreased and MYC expression was modestly increased in Kdm6a-Mutant vs. Kdm6a-WT tumors (Extended Data Figs. 6a-d). Moreover, MYCL promoter accessibility was decreased and MYC promoter accessibility was increased in Kdm6aMutantNEUROD1Only tumors (Extended Data Figs. 6e-g). We also further investigated MYCL/MYC in our tumor-derived cell lines. MYC levels were slightly higher; albeit at much lower levels compared to high MYC expressing human SCLC cells; and MYCL levels were lower in Kdm6a-Mutant vs. Kdm6a-WT lines (Extended Data Figs. 7a-h). Consistent with this, ChIP-seq data showed that KDM6A was enriched at the MYCL locus and that Kdm6a-Mutant cells completely lost H3K4me1 at MYCL enhancers, which was associated with a modest reciprocal gain of H3K4me1 at MYC enhancers without significant KDM6A enrichment (Extended Data Figs. 7i,j). To test whether this modest gain of c-Myc expression functionally regulates NEUROD1, MYC was CRISPR inactivated in a NEUROD1-positive Kdm6a-Mutant tumor derived cell line 672-2, which expresses ASCL1, NEUROD1, and MYC (Extended Data Fig. 7k). Acutely 672-2 cells tolerated MYC inactivation but over time all MYC inactivated cells died. At early times after infection, MYC inactivation decreased NEUROD1 levels (Extended Data Fig. 7l). Collectively, these data show both differences and similarities between Kdm6a-Mutant and RPM GEMMs and suggest that c-Myc functionally contributes to NEUROD1 expression in Kdm6a-Mutant cells ex vivo despite its very low expression levels.
KDM6A Regulates Neuroendocrine Genes to Maintain a Chromatin State Permissive for the ASCL1 Subtype
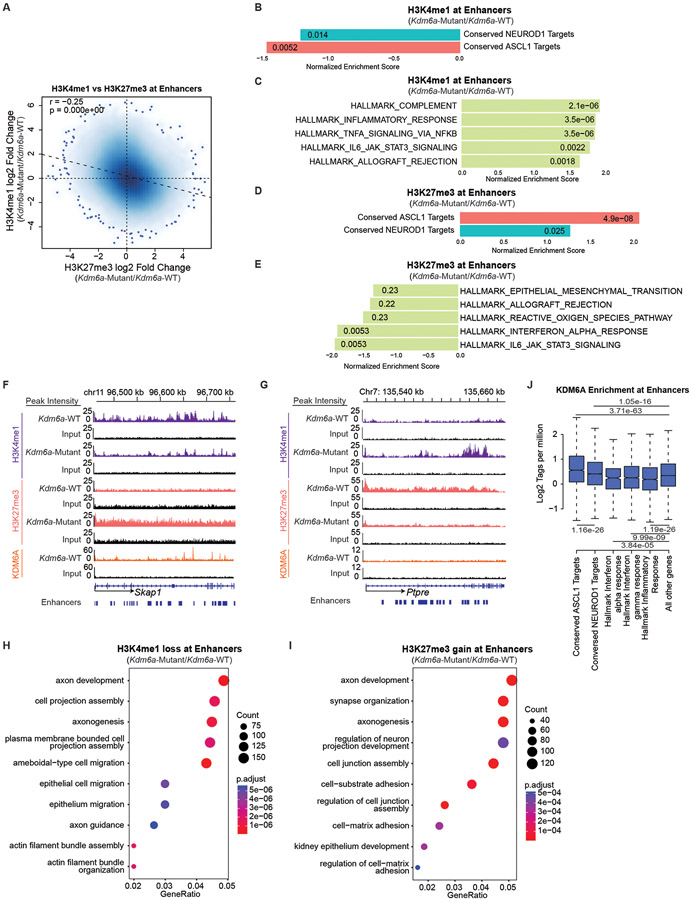
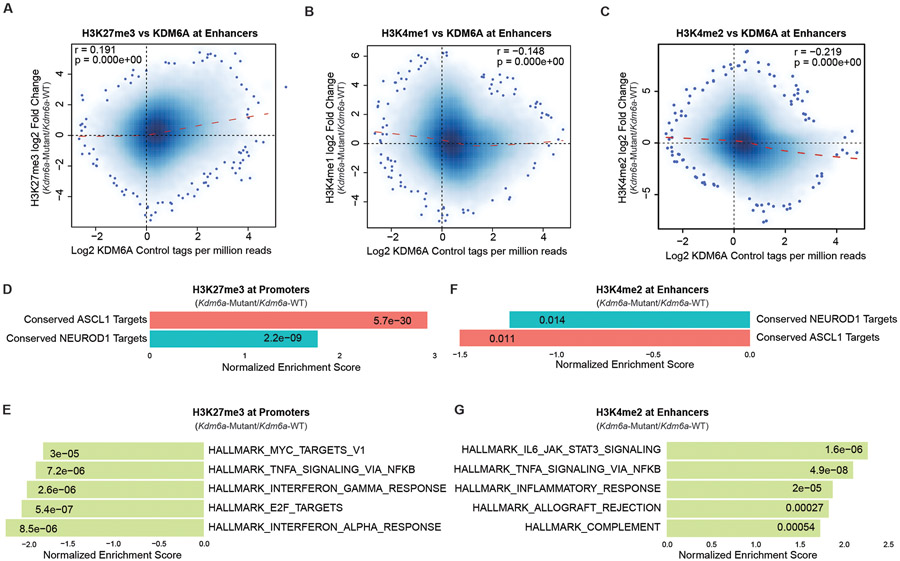
KDM6A canonically maintains a chromatin state permissive for gene activation through its H3K27me3 demethylase activity and a scaffolding role in the MLL3/4 COMPASS complex which increases H3K4me1 at enhancers19. Based on its canonical activity, we hypothesized that NEUROD1 induction after KDM6A loss is likely indirect, and that KDM6A’s direct function is to maintain a permissive chromatin state for the ASCL1 subtype. To test this, we performed ChIP-seq for KDM6A in 2 early passage Kdm6a-WT cell lines (1014, 159-1) and for the histone methylation marks that KDM6A regulates including H3K27me3, H3K4me1, and H3K4me2 in the same 2 early passage Kdm6a-WT cell lines and also in 2 early passage Kdm6a-Mutant cell lines (236L, 236R). These ChIP-seq data were analyzed for enrichment near both enhancers and promoters. As expected, KDM6A binding peaks were enriched at both enhancers and promoters. We observed expected peak patterns of H3K27me337, H3K4me1, and H3K4me2 in all samples where H3K27me3 was broadly enriched near subsets of both enhancers and promoters, H3K4me1 was highly enriched at enhancers, and H3K4me2 was enriched at both promoters and enhancers (Supplementary Figs. 1-4, Supplementary Table 7). In line with KDM6A’s canonical activity maintaining H3K4me1 and demethylating H3K27me3 at some enhancers, genome-wide correlation analysis revealed that KDM6A binding in Kdm6a-WT samples was weakly positively correlated with change in H3K27me3 at enhancers and weakly negatively correlated with change in H3K4me1 at enhancers when these marks are compared between Kdm6a-Mutant and Kdm6a-WT conditions (Extended Data Figs. 8a,b). Consistent with this, changes in H3K4me1 and H3K27me3 in Kdm6a-Mutant/Kdm6a-WT at enhancers were significantly negatively correlated (Fig. 5a). To unbiasedly ask which genes are affected by the changes in H3K27me3 or H3K4me1 in Kdm6a-Mutant vs. Kdm6a-WT cell lines, GSEA was performed using Hallmark gene sets and conserved ASCL1 and NEUROD1 target gene sets29. Consistent with our hypothesis, GSEA analysis revealed that H3K4me1 was most considerably depleted and H3K27me3 was most considerably enriched at enhancers of conserved ASCL1 target genes (Figs. 5b,d,f). Similar directional changes were observed at NEUROD1 target genes, but these changes were overall less pronounced compared to ASCL1 target genes (Figs. 5b,d). Enhancers of inflammatory/interferon genes showed opposite patterns to ASCL1 and NEUROD1 with enrichment of H3K4me1 and depletion of H3K27me3 upon KDM6A inactivation (Figs. 5c,e,g) suggesting a switch towards a chromatin state permissible for inflammatory gene expression. Similar correlations as described for H3K27me3 at enhancers were also observed for H3K27me3 at promoters (Extended Data Figs. 8d,e). Moreover, H3K4me2 ChIP-seq analysis at enhancers also yielded similar correlations as described for H3K4me1 at enhancers (Extended Data Figs. 8c,f,g). Gene ontology analyses of H3K4me1 peaks lost and H3K27me3 peaks gained at enhancers upon KDM6A inactivation revealed biological processes involved in neuron and axon development (Figs. 5h,i). In line with our H3K4me1 and H3K27me3 ChIP-seq data, KDM6A binding at enhancers was most highly enriched at ASCL1 target genes and relatively depleted at inflammatory/interferon genes suggesting that histone methylation changes at neuroendocrine genes (ASCL1>NEUROD1) were a consequence of loss of KDM6A binding at those genes (Fig. 5j). Together, these results show that KDM6A binding is enriched at enhancers of neuroendocrine genes with a preference for ASCL1 target genes.
Fig. 5. KDM6A Binds and Regulates Neuroendocrine Genes to Maintain a Chromatin State Permissive for the ASCL1 Subtype.
(a) Genome-wide correlation of log fold change in H3K4me1 ChIP-seq (Kdm6a-Mutant/Kdm6a-WT) vs. log fold change in H3K27me3 ChIP-seq (Kdm6a-Mutant/Kdm6a-WT) at enhancers. See Supplementary Figs. 2&3. For a, r=Pearson correlation coefficient. p-value was calculated using a two-sided Pearson’s correlation test. (b, c) Normalized Enrichment Score from GSEA of H3K4me1 ChIP-seq at enhancers of conserved ASCL1 target genes or conserved NEUROD1 target genes (b) or top 5 enriched Hallmarks (c) from the ChIP-seq data in a. (d, e) Normalized Enrichment Score from GSEA of H3K27me3 ChIP-seq at enhancers of conserved ASCL1 or NEUROD1 target genes (d) or top 5 depleted Hallmarks (e) from the ChIP-seq data in a. (f, g) Tracks of H3K4me1, H3K27me3, and KDM6A ChIP-seq at the ASCL1 target gene Skap1 (f) or an inflammatory gene Ptpre (g) showing changes representative of the analyses in b-e. Each track is the peak sum of: 2 Kdm6a-WT tumor-derived cell lines for Kdm6a-WT (159-1,1014) and 2 Kdm6a-Mutant tumor-derived cell lines for Kdm6a-Mutant (236L,236R) with their respective input. (h,i) Gene ontology (GO) analysis of enhancers that lose H3K4me1 (h) or gain H3K27me3 (i) in Kdm6a-Mutant/Kdm6a-WT. For h,i, p-values were generated by GSEA using a permutation test adjusted for multiple hypothesis testing using the Benjamini-Hochberg correction. Adjusted p-values are indicated. (j) Boxplot of genome-wide KDM6A ChIP-seq enrichment at enhancers of ASCL1 conserved targets, NEUROD1 conserved targets, and inflammatory genes vs. all other genes. For j, indicated p-values are calculated using unpaired, two-sided, Mann-Whitney U tests adjusted for multiple hypothesis testing. n=2,958 enhancers (ASCL1), n=9,217 enhancers (NEUROD1), n=567 enhancers (Interferon α), n=2,220 enhancers (Interferon γ), n=2,442 enhancers (Inflammatory), n=191,064 enhancers (Other) from 2 biological independent replicates. The center line is the median, the lower and upper bounds represent 25% and 75% rank and the whiskers indicate 1.5 times the interquartile range. For all experiments, H3K4me1 and H3K27me3 ChIP-seq is from 2 independent Kdm6a-Mutant cell lines (236L,236R) and 2 independent Kdm6a-WT cell lines (1014,159-1). KDM6A ChIP-seq is from 2 independent Kdm6a-WT cell lines (1014,159-1). Also see Supplementary Table 7.
NEUROD1 Expression after KDM6A Loss is Partially Mediated by KMT2A
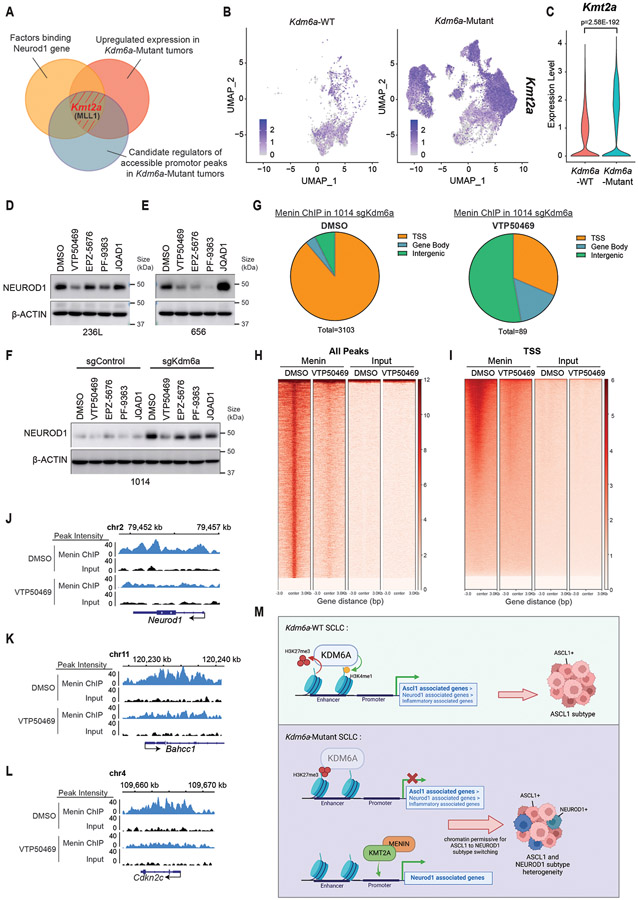
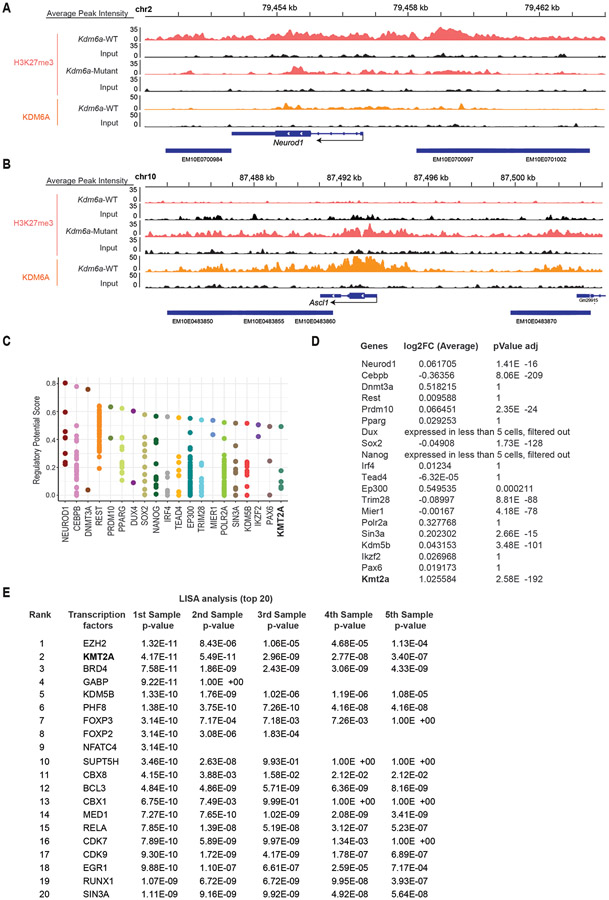
Canonically KDM6A activity maintains a chromatin state permissible for gene activation. We therefore hypothesized that NEUROD1 expression upon KDM6A loss is not a direct effect of loss of KDM6A binding to the NEUROD1 gene. Consistent with this, KDM6A is not enriched at the NEUROD1 locus itself, but is enriched at the ASCL1 locus with its loss increasing H3K27me3 (Extended Data Figs. 9a,b). To identify regulators that directly bind and promote expression of NEUROD1 correlated genes in Kdm6a-Mutant tumors, we identified candidate factors that bind NEUROD1 using public ChIP-seq data (cistrome.org) and determined which factors were also significantly upregulated in Kdm6a-Mutant tumor cells. We also performed LISA analysis38 using our ATAC-seq data from Fig. 2a of top 100 upregulated peaks near promoters in Kdm6a-Mutant NEUROD1-positive tumors (Supplementary Table 8). These analyses nominated KMT2A (MLL1) as a candidate positive regulator of NEUROD1 in Kdm6a-Mutant tumors (Figs. 6a-c, Extended Data Figs.9c-e). KMT2A(MLL1) together with Menin(MEN1) binds DNA/chromatin to maintain genes in an active chromatin state for gene expression39. To test directly whether KMT2A/Menin is necessary for NEUROD1 expression, we treated Kdm6a-Mutant tumor derived cells (236L and 656) (Fig. 4b) and 1014 Kdm6a isogenic CRISPR-inactivated cells (Fig. 1f) with VTP50469; an inhibitor that blocks the interaction of KMT2A with Menin40. Other inhibitors of epigenetic modifiers that normally maintain genes in an active state were also included. Treatment of cells with VTP50469 consistently decreased NEUROD1 protein levels across all 3 Kdm6a-Mutant cell lines (Figs. 6d-f). ChIP-seq for Menin in 1014 Kdm6a knockout cells treated with VTP50469 or DMSO showed that Menin directly binds to Neurod1 and other Menin target genes (Bahcc1 and Cdkn2c) and that VTP50469 inhibits these Menin-DNA interactions showing that VTP50469 is functional (Figs. 6g-l, Supplementary Fig. 5, Supplementary Table 8). Together this shows that NEUROD1 expression after KDM6A inactivation is at least partially mediated by increased expression and activity of KMT2A directly acting to promote NEUROD1 expression.
Fig. 6. NEUROD1 Induction after KDM6A Inactivation is Partially Mediated by KMT2A.
(a) Venn diagram of factors binding NEUROD1 (orange), upregulated expression in Kdm6a-Mutant tumors (red), and candidate regulators of accessible peaks at promoters in Kdm6a-Mutant tumors (blue). (b,c) Kmt2a feature plot (b) and violin plot (c) using scRNA-seq data from Fig. 3i of tumor cells from Kdm6a-Mutant and Kdm6a-WT tumors. For c, non-parametric Wilcoxon rank sum test was used to generate a two-tailed p-value adjusted for multiple comparisons by Bonferroni correction. Minimum and maximum values define the range of violin plot. n=35,446 Kdm6a-Mutant and n=6612 Kdm6a-WT tumor cells. (d-f) Immunoblot analysis of 236L (d) and 656 (e) Kdm6a-Mutant tumor derived cells lines or 1014 Kdm6a isogenic cells from Fig. 1f (f) treated with inhibitors that block the function of epigenetic modifiers that normally maintain gene expression including VTP50469 (Menin/MLL1 inhibitor, 500 nM), EPZ-5676 (DOT1L inhibitor, 1 μM), PF-9363 (KAT6A/KAT6B inhibitor, 100 nM), JQAD1 (EP300 degrader, 500 nM) or DMSO for 6 days. (g) Pie charts of Menin ChIP-seq data showing peak distributions throughout the genome of 1014 cells with Kdm6a CRISPR inactivation treated with VTP50469 (500 nM) or DMSO for 5 days. (h, i) Genome wide rank-ordered heat map of Menin ChIP signal at all peaks (h) or across promoters (TSS −3kB/+3kB) (i). A second replicate of the Menin-ChIP experiment in shown in Supplementary Fig. 5. (j-l) Menin ChIP-seq tracks at Neurod1 (j) and two canonical Menin target genes Bahcc1 (k) and Cdkn2c (l). (m) Schematic of the role of KDM6A in SCLC subtype plasticity. When KDM6A is present, KDM6A binds enhancers to increase H3K4 mono-methylation (H3K4me1) and decrease H3K27 tri-methylation (H3K27me3) maintaining a chromatin state most permissive for the ASCL1 subtype. When KDM6A is inactivated, ASCL1 subtype genes lose H3K4me1 and gain H3K27me3 at enhancers and chromatin becomes less permissive for the ASCL1 subtype. Upon KDM6A inactivation, KMT2A (MLL1) expression is upregulated and KMT2A/Menin binds the NEUROD1 promoter to promote NEUROD1 expression resulting in SCLC tumors with heterogenous ASCL1 and NEUROD1 expression. This figure was created with BioRender.com and publication license has been obtained.
Discussion
Although SCLC molecular subtypes were initially thought to be largely mutually exclusive3, recent evidence supports intra-tumoral molecular subtype heterogeneity9,12,41 and the ability to dynamically evolve from one subtype to another13. Using autochthonous SCLC GEMMs generated using CRISPR/Cas926 where Kdm6a, which is mutated in human SCLCs4,14, was inactivated at tumor initiation in somatic cells in the lung, we discovered that KDM6A loss accelerates neuroendocrine subtype plasticity from a pure ASCL1 subtype to mixed SCLCs that express ASCL1 and NEUROD1. KDM6A demethylates H3K27me315,16 and binds the c-terminus of KMT2D of the MLL3/4 COMPASS complex that mono-methylates H3K4 at enhancers18,19. Our data suggests that both functions of KDM6A participates in its ability to maintain an active chromatin state most permissible for the ASCL1 subtype. Loss of KDM6A causes increased H3K27me3 and decreased H3K4me1 at enhancers of neuroendocrine genes with the strongest enrichment or depletion, respectively, at the enhancers of ASCL1 target genes ultimately resulting in a chromatin state that favors ASCL1 to NEUROD1 subtype switching. These cells evolved ex vivo to lose both neuroendocrine transcription factors (double-negative) and highly express inflammatory genes. Although we did find occasional “double-negative” tumor cells within Kdm6a-Mutant tumors, Kdm6a-Mutant tumors largely maintained their neuroendocrine identity with heterogenous expression of ASCL1 and NEUROD1 and therefore more evidence is needed to support an inflammatory subtype transition in vivo. NEUROD1 upregulation after KDM6A inactivation was not a direct effect of loss of KDM6A binding and was at least partially mediated through KMT2A, which is upregulated upon KDM6A inactivation (Fig. 6m). Given that KDM6A binds and regulates the chromatin state of many genes, there likely could be other epigenetic modifiers and transcription factors (e.g. c-Myc) that coordinately help promote subtype plasticity after KDM6A inactivation.
~35-40% of human SCLCs have expression of both ASCL1 and NEUROD19. Our autochthonous SCLC GEMM with loss of Kdm6a, Rb1, Trp53, and Rbl2 (KRPP) is, to our knowledge, the first SCLC GEMM utilizing an inactivating mutation in an epigenetic modifier to model mixed ASCL1 and NEUROD1 subtype heterogeneity. Human SCLCs have many more concurrent genomic alterations compared to SCLC GEMMs resulting in more inter-tumoral heterogeneity, which is not captured in SCLC GEMMs. GEMMs uniquely allow for the study of the functional consequences of gene inactivation during tumorigenesis and our data clearly shows that KDM6A inactivation in SCLC GEMMs causes ASCL1 and NEUROD1 intra-tumoral subtype heterogeneity. Similar to our phenotypic findings, c-Myc drives plasticity from ASCL1 positive in situ tumors to NEUROD1 positive invasive tumors in the RPM SCLC GEMM. KRPP tumors were invasive tumors from mice that were at their endpoint and showed a spectrum of ASCL1 and NEUROD1 dominance with most tumors still expressing ASCL1, while invasive RPM tumors are NEUROD1 dominant13,31. KRPP tumors had very low c-Myc expression, but did show expression changes consistent with a shift from L-Myc to c-Myc paralog expression and c-Myc was necessary for NEUROD1 expression in KDM6A inactivated cells. Hence, very low c-Myc levels could help coordinate the transcriptional program that promotes NEUROD1 expression in KRPP tumors. Clearly there are several differences between the KRPP and RPM models and both can now be utilized to better understand mechanisms of ASCL1 and NEUROD1 subtype heterogeneity and plasticity.
Although many of the KRPP tumors had mixed ASCL1 and NEUROD1 expression, KDM6A loss was not sufficient in some tumors to induce subtype switching from ASCL1 to NEUROD1. KRPP tumors of the “pure ASCL1 subtype” had evidence of chromatin accessibility and gene expression changes that correlated with the NEUROD1 subtype despite not expressing NEUROD1. This is consistent with models of epigenetic plasticity in cancer where mutations in chromatin regulators make chromatin more permissive for cell state transitions, but ultimately cell state transitions require selective pressure42,43. Since our Kdm6a-Mutant and Kdm6a-WT GEMMs both have Rb1, Trp53, and Rbl2 inactivation, which is sufficient for SCLC tumorigenesis26,32, we posit that there was not strong selective pressure for ASCL1 to NEUROD1 subtype switching in our in vivo studies. Hence tumors had variable degrees of ASCL1 to NEUROD1 subtype switching. Cisplatin induced a greater magnitude of NEUROD1 expression in SCLC cell lines after KDM6A inactivation. We hypothesize that additional selective pressures to lose ASCL1, as has been seen in human SCLCs after recurrence from front-line platinum-doublet chemotherapy7,44,45, could further promote ASCL1>NEUROD1 subtype plasticity in the absence of KDM6A and this can be explored in future studies.
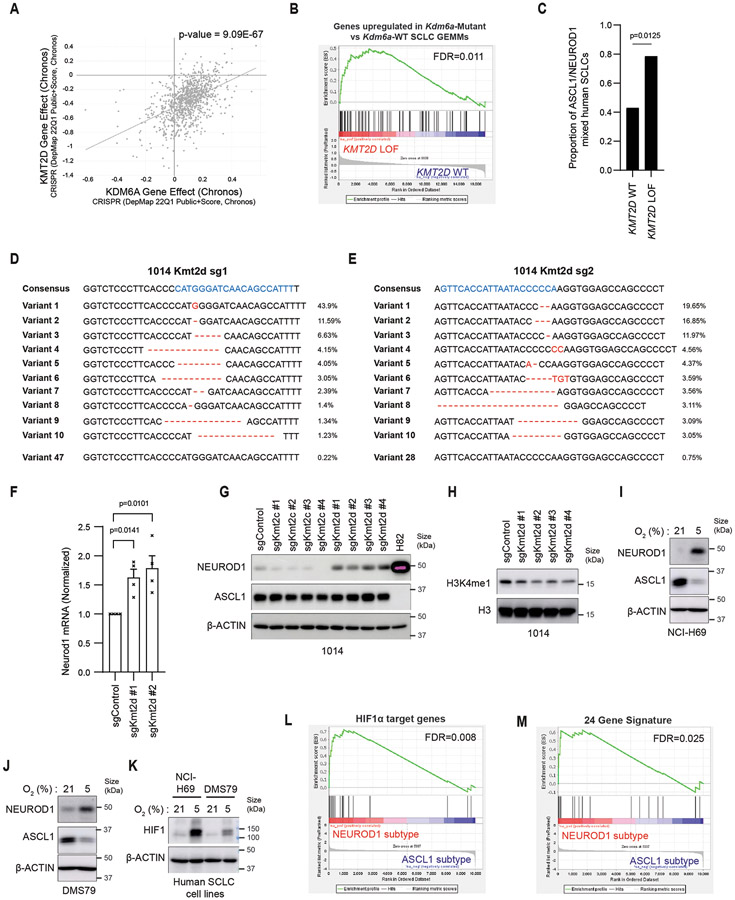
KDM6A LOF mutations are found in human SCLC, but are infrequent accounting for <5% of SCLCs4,14 and therefore do not explain the large fraction (~35-40%) of human SCLCs that express both ASCL1 and NEUROD1. KMT2D LOF mutations are more frequent in SCLC (~8%-17%)4-6,14. KDM6A and KMT2D both participate in the COMPASS complex and KDM6A loss most closely phenocopies KMT2D loss across all cancer cell lines25. Moreover, germline LOF mutations in KDM6A or KMT2D are the most frequent mutations found in patients with Kabuki syndrome46,47. Gene expression changes in Kdm6a-Mutant vs. Kdm6a-WT GEMMs correlate with gene expression changes in human SCLCs with KMT2D LOF mutations, and human SCLCs with KMT2D LOF mutations are more likely to express both ASCL1 and NEUROD1 (Extended Data Figs. 10a-c, Supplementary Table 9). Moreover, Kmt2d CRISPR inactivation, but not Kmt2c CRISPR inactivation, induced NEUROD1 expression in 1014 mouse SCLC cells (Extended Data Figs. 10d-h). Future studies can explore whether KMT2D inactivation in SCLC GEMMs phenocopies KDM6A inactivation and promotes ASCL1 and NEUROD1 intra-tumoral heterogeneity.
KDM6A can be directly inactivated by hypoxia27. Thus, KDM6A function could be lost in a hypoxic tumor microenvironment in a SCLC tumor genetically WT for KDM6A causing conversion from an ASCL1-positive to NEUROD1-positive cell state, which we have observed in human SCLC cell lines grown in hypoxia (Extended Data Figs. 10i-k). Consistent with this, scRNA-seq from human SCLCs found that hypoxia genes highly correlated with the NEUROD1 subtype34, which we validated using additional hypoxia gene sets (Extended Data Figs. 10l,m, Supplementary Table 10). Future studies will focus on better understanding what drives SCLC subtype heterogeneity through both genetic and microenvironmental mechanisms and whether these other causes are linked to KDM6A functional inactivation.
Methods
All experiments herein comply with all ethical regulations. Specifically, all mouse experiments complied with National Institutes of Health guidelines and were approved by Dana-Farber Cancer Institute Animal Care and Use Committee (DFCI, protocol 19-009). All adenoviral and lentiviral transduction experiments complied with the Biohazard Control Committee (DFCI, protocol 19-1133).
Adenoviral sgRNA Expression Vector Cloning
Effective sgRNAs targeting mouse Rb1, Trp53, and Rbl2 were first validated using lentiviral vectors as described previously26. Effective sgRNAs targeting mouse Kdm6a (sg3 and sg4) were described previously27 and validated in mouse embryonic fibroblasts expressing Cas9. The cloning method for generation of adenoviral sgRNA expression vectors encoding CMV-Cre recombinase and sgRNAs targeting Rb1, Trp53, and Rbl2 and “T” sgRNA (in this case Kdm6a sg3, Kdm6a sg4, a non-targeting sgRNA control, or an intron-targeting sgRNA control) was also described previously30. Briefly, a pENTR223-CMV-Cre-U6-sgX-U6-sgRb1-U6-sgTrp53-U6-sgRbl2 where X is sgKdm6a#3, sgKdm6a#4, sgC0111, or sgIntron was used in an LR recombination reaction to clone the 4 pENTR223-CMV-Cre-U6-sgX-U6-sgRb1-U6-sgTrp53-U6-sgRbl2 vectors described above into pAd-PL DEST (Invitrogen) according to the manufacturer’s instructions. The recombinants were transformed into HB101 cells and ampicillin-resistant colonies were screened by restriction digestion of miniprep DNA and subsequently validated by DNA sequencing. The following sgRNA oligos were used (including BsmBI sites): Rb1 mouse #11 sense (5’- CACCGCAACTAGAAAATGATACG-3’), Rb1 mouse #11 anti-sense (5’- AAACCGTATCATTTTCTAGTTGC-3’), Trp53 mouse #8 sense (5’- CACCGGTGTAATAGCTCCTGCATGG-3’), Trp53 mouse #8 anti-sense (5’- AAACCCATGCAGGAGCTATTACACC-3’), Rbl2 mouse #6 sense (5’- CACCGAGGAGGATGGCGACGCCG-3’), Rbl2 mouse #6 anti-sense (5’- AAACCGGCGTCGCCATCCTCCTC-3’), Kdm6a mouse #3 sense (5’-CACCGCCGCCTTTTCGGGTTCGTG-3’), Kdm6a mouse #3 anti-sense (5’- AAACCACGAACCCGAAAAGGCGGC-3’), Kdm6a mouse #4 sense (5’-CACCG CCTTGGCTCGACAAAAGCT-3’), Kdm6a mouse #4 anti-sense (5’-AAACAGCTTTTGTCGAGCCAAGGC-3’), C0111 (Non-targeting sgRNA, sgControl) sense (5’- CACCGGGAGGCTAAGCGTCGCAA-3’), C0111 (Non-targeting sgRNA, sgControl) anti-sense (5’- AAACTTGCGACGCTTAGCCTCCC-3’), Intron mouse sense (5’-CACCGTGTTGCAGTAATAATTAGCG-3’), Intron mouse anti-sense (5’-AAAC CGCTAATTATTACTGCAACAC-3’).
Adenovirus Production and Purification
Adenoviral production and purification were performed as described previously30. 5 μg of the adenovirus vector (pAd/PL Invitrogen #V494-20) containing the desired sgRNA sequences and Cre recombinase expression cassette (see above) was digested with PacI (New England Biolabs) for 2 hours at 37°C according to the manufacturer’s instructions and column purified using Qiagen’s gel extraction kit. 1 μg of PacI-digested pAd/PL was transfected into 1.5 X 106 293AD cells plated on a 6 cm tissue-culture dish using Lipofectamine 2000. The following day, the media was exchanged, and subsequently exchanged every 48 hours thereafter. Once 293AD cells showed evidence of adenovirus production (determined by comet formation with lysis), the cells and supernatant were harvested, which were then subjected to 4 freeze-thaw cycles by alternating between an ethanol dry ice bath and 37°C. Cell debris was removed by centrifugation and the supernatant was collected, passed through a 0.45 μm filter, aliquoted, and frozen at −80°C until use.
To generate high titer adenovirus for in vivo experiments, adenovirus was generated as described above. 50 μl of the adenovirus stock was added to each 10 cm tissue-culture dish of 293FT cells plated at 3 X 106 cells per dish (4 10 cm dishes in total for each purification). When 293FT cells showed evidence of adenovirus production, as determined by cell rounding and partial detachment (~48-72 hours after addition of adenoviral stock), the cells were collected, and adenovirus was purified using Virabind Adenovirus Purification Kit (Cell Biolabs #VPK-5112). The purified adenovirus was titered using QuickTiter Adenovirus Quantitation Kit (Cell Biolabs #VPK-106) according to the manufacturer’s instructions.
Intratracheal Injections
Intratracheal injections were performed as described previously48. Briefly, mice were anesthetized with ketamine and xylazine and pedal reflexes were monitored to ensure adequate anesthesia. Mice were maintained on a heated stage at 37° C while anesthetized. Mice were hung on stage with their top incisors and intubated with a 22-gauge 1 inch catheter (ThermoFisher Scientific #1484120). Once intubated, adenovirus (4 X 108 VP/mouse) in a total volume of 75 μl (diluted in PBS) was added to the catheter and subsequently inhaled by the mice.
sgKdm6a RPP or sgControl RPP Genetically-Engineered Mouse Models of SCLC using CRISPR/Cas9
For all experiments, pure congenic Lox-stop-lox (LSL) Cas9 BL6J mice were purchased from Jackson Labs (Jackson No. 026175) and maintained as homozygous BL6J mice. Genotyping of Cas9 and GFP at the ROSA26 were confirmed for all mice on the study (Transnetyx). Housing conditions for mice at the DFCI Vivarium include a 12 hour/12 hour day-night cycle where temperature is maintained at 72 C. Roughly equal numbers of male and female 3-4 month-old transgenic homozygous LSL-Cas9 were intratracheally injected with adenovirus (4 X 108 VP/mouse) encoding effective sgRNAs targeting Rb1, Trp53, Rbl2, and an sg “T” where T is either an sgRNA targeting Kdm6a or an sgControl (non-targeting sgRNA or sgRNA targeting an intron in the mouse genome as indicated) and CMV-Cre recombinase. For all experiments in Fig. 1, Kdm6a sg4 was used. For experiments in Fig. 3, tumors from mice 1116, 1117, 1119, 1270, 1274, and 1314 were generated with Kdm6a sg4 and tumors from mice 1121, 1122, and 1125 were generated with Kdm6a sg3. All mice used are listed in Supplementary Table 1. Mice were monitored weekly and euthanized when they became symptomatic (primarily respiratory distress), moribund, or lost 15% of their total body weight. The maximal tumor size allowed by the Dana-Farber Cancer Institute Animal Care and Use Committee is 2 cm and the maximal tumor size was not exceeded in any of our studies. Upon euthanization, half of the lung tumor specimen was immediately flash frozen on dry ice for subsequent DNA, RNA, and protein analysis, while the other half was fixed in 10% formalin for 24 hours and then stored in 70% ethanol prior to being embedded in paraffin. For some tumors, cell lines were generated (see method below). Livers were harvested and fixed and embedded as above. Slides were made for Hematoxylin and Eosin (H&E), immunohistochemistry (IHC), and immunofluorescence (IF). H&E slides were analyzed by a specialized rodent pathologist Dr. Roderick Bronson for diagnosis.
Generation of Cell Lines from Mouse SCLC Tumors and Cell Culture
sgControl RPP (159-1, 1014, 1015) and sgKdm6a RPP (236L, 236R, 656, 672-2) cell lines were generated from CRISPR-based SCLC GEMMs (see above) as described previously30. Once tumors developed, mice were euthanized with CO2 and their tumors were quickly extracted, washed in ice cold PBS, and minced several times using an ethanol sterilized razor blade. 3mL of collagenase/hyaluronidase (Stem cell biology #07912) diluted 1:10 in complete RPMI media containing [10% FBS, P/S, and HITES (10 nM hydrocortisone (Sigma Aldrich # H0135), Insulin-Transferrin-Selenium (Gemini #400-145), and 10 nM beta-estradiol (Sigma Aldrich# E2257), 100 U/mL of penicillin (P), and 100 μg/mL of streptomycin (S)], and 1mL dispase (Corning # 354235) was added to the tumor, and incubated at 37°C for 20-40 minutes with periodic pipetting ~10 every minutes (until most of the tumor cells were in suspension). The cells were then collected, centrifuged at 200 x g for 5 minutes, resuspended in complete RPMI media (see above), filtered through a 70 μm cell strainer (BD #352350), centrifuged again at 1000 rpm for 5 minutes, resuspended in fresh RPMI HITES media and placed in ultra-low adherence tissue culture dishes (Corning #3471). Media was subsequently replaced every 3 days. Histopathology on the tumors confirmed SCLC for all cell lines generated. All cell line were grown in Ultra-Low Attachment flasks (Corning™ 3814CONV) or plates (Corning™ 3471) at 37°C in the presence of 5% CO2. Once established, all cell lines were validated using immunoblot analysis for Cas9 and the SCLC neuroendocrine markers ASCL1 and NEUROD1. Early passage cell lines were tested for Mycoplasma (Lonza #LT07-218) and then were frozen using Bambanker’s freezing media (Bulldog Bio). Early passage cell lines were maintained in culture for <1.5 months and late passage cell lines were cells maintained in culture for greater than 2 months.
Human Cell Lines
CORL47 and CORL279 were obtained from Sigma (11/2018). DMS79, NCI-H82 cells were a kind gift from Dr. Kwok-kin Wong’s laboratory (New York University) and were obtained in 8/2014 and authenticated at Genetica DNA laboratories. NCI-H1092, NCI-H69 (obtained 11/2018) and 293FT cells were originally obtained from American Type Culture Collection (ATCC). 293AD cells (AD-100) were obtained from Cell Biolabs. CORL47, CORL279, DMS79, NCI-H69, and NCI-H82 cells were maintained in RPMI-1640 media with 10% FBS and P/S. NCI-H1092 cells were maintained in DMEM/F12 media 5% FBS, P/S, and HITES. 293T, 293AD, and MEFs expressing Cas9 were maintained in DMEM media with 10% FBS and P/S. Mouse embryonic fibroblasts expressing Cas9 used to validate the adenoviruses were described previously26. Early passage cell lines were tested for Mycoplasma and were negative (Lonza #LT07-218), and then were frozen using Bambanker’s freezing media (Bulldog Bio). All experiments were performed with cell lines that were maintained in culture for <3 months at which time an early passage cell lines were thawed. No commonly misidentified cell lines were used in this study.
Pharmacological Inhibitors
The following chemicals (stored at −20°C or −80°C) were added to cell culture where indicated: Cisplatin (Selleck #S1166, stock 5 mM), VTP-50469 (MedChem Express #HY-114162, stock 10 mM in DMSO), EPZ-5676 (MedChem Express #HY-15593, stock 50 mM in DMSO), PF-9363 (MedChem Express #HY-132283, stock 10 mM in DMSO) or EP300 degrader (JQAD1; kind gift from Dr. Jun Qi, stock 10 mM in DMSO).
sgRNA Cloning to Make Lentiviruses
sgRNA sequences were designed using the Broad Institute sgRNA designer tool (http://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) and synthesized by IDT technologies. The sense and antisense oligonucleotides were mixed at equimolar ratios (0.25 nanomoles of each sense and antisense oligonucleotide) and annealed by heating to 100°C in annealing buffer (1X annealing buffer 100 mM NaCl, 10 mM Tris-HCl, pH 7.4) followed by slow cooling to 30°C for 3 hours. The annealed oligonucleotides were then diluted at 1:400 in 0.5X annealing buffer.
For CRISPR/Cas9 knockout experiments in cells, the annealed oligos were ligated into LentiGuide Puro (Addgene #52963) for experiments in mouse SCLC cell lines. Ligations were done with T4 DNA ligase for 2 hours at 25°C. The ligation mixture was transformed into HB101 competent cells. Ampicillin-resistant colonies were screened by restriction digestion of miniprep DNAs and subsequently validated by DNA sequencing.
The following sgRNA oligos were used for LentiGuide Puro vector for CRISPR knockout experiments: Kdm6a mouse #3 sense (5’-CACCGCCGCCTTTTCGGGTTCGTG-3’), Kdm6a mouse #3 anti-sense (5’- AAACCACGAACCCGAAAAGGCGGC-3’), Kdm6a mouse #4 sense (5’-CACCGCCTTGGCTCGACAAAAGCT-3’), Kdm6a mouse #4 anti-sense (5’-AAACAGCTTTTGTCGAGCCAAGGC-3’), Myc mouse #1 sense (5’- CACCGAGAGGCAAACCCCTGCCAAG-3’), Myc mouse #1 anti-sense (5’- AAACCTTGGCAGGGGTTTGCCTCTC-3’), Myc mouse #2 sense (5’- CACCGGCTGTACGGAGTCGTAGTCG-3’), Myc mouse #2 anti-sense (5’- AAACCGACTACGACTCCGTACAGCC-3’), Myc mouse #3 sense (5’- CACCGGTAGCGACCGCAACATAGGA-3’), Myc mouse #3 anti-sense (5’- AAACTCCTATGTTGCGGTCGCTACC-3’), Myc mouse #4 sense (5’- CACCGGTCAATGCACTCGGACGCGG-3’), Myc mouse #4 anti-sense (5’- AAACCCGCGTCCGAGTGCATTGACC-3’), Mouse Kmt2d #1 sense (5’-CACCGAAATGGCTGTTGATCCCATG-3’), Mouse Kmt2d #1 anti-sense (5’-AAACCATGGGATCAACAGCCATTTC-3’), Mouse Kmt2d #2 sense (5’-CACCGGTTCACCATTAATACCCCCA-3’), Mouse Kmt2d #2 anti-sense (5’-AAACTGGGGGTATTAATGGTGAACC-3’), Mouse Kmt2d #3 sense (5’- CACCGTCGGGCCGGACTAACATCCG-3’), Mouse Kmt2d #3 anti-sense (5’- AAACCGGATGTTAGTCCGGCCCGAC-3’), Mouse Kmt2d #4 sense (5’- CACCGTGGGGATGGACAGCCCGACG-3’), Mouse Kmt2d #4 anti-sense (5’- AAACCGTCGGGCTGTCCATCCCCAC-3’), Mouse Kmt2c #1 sense (5’- CACCGAAAAGGCCCATTACCCAATG-3’), Mouse Kmt2c #1 anti-sense (5’- AAACCATTGGGTAATGGGCCTTTTC-3’), Mouse Kmt2c #2 sense (5’- CACCGAGAACCATTATTAGTAAACG-3’), Mouse Kmt2c #2 anti-sense (5’- AAACCGTTTACTAATAATGGTTCTC-3’), Mouse Kmt2c #3 sense (5’- CACCGGGTGAAATGGATGATAGTCG-3’), Mouse Kmt2c #3 anti-sense (5’- AAACCGACTATCATCCATTTCACCC-3’), Mouse Kmt2c #4 sense (5’ CACCGAAGCATTACCTGAATCCATG-3’), Mouse Kmt2c #4 anti-sense (5’- AAACCATGGATTCAGGTAATGCTTC-3’), C0111 (Non-targeting sgRNA, sgControl) sense (5’- CACCGGGAGGCTAAGCGTCGCAA-3’), C0111 (Non-targeting sgRNA, sgControl) anti-sense (5’- AAACTTGCGACGCTTAGCCTCCC-3’).
Lentivirus Production
Lentiviruses were made by Lipofectamine 2000-based co-transfection of 293FT cells with the respective lentiviral expression vectors and the packaging plasmids psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) at a ratio of 4:3:1. Virus-containing supernatant was collected at 48 and 72h after transfection, pooled together (15 mL total per 10-cm tissue culture dish), passed through a 0.45-μm filter, aliquoted, and frozen at −80°C until use.
Lentiviral Infection
The cells were counted using a Vi-Cell XR Cell Counter (Beckman Coulter) and 2 X106 cells were resuspended in 1mL lentivirus with 8 μg/mL polybrene in individual wells of a 12 well plate. The plates were then centrifuged at 448 x g for 2h at 30° C. 16 hours later the virus was removed and cells were grown for 72 hours before being placed under drug selection. Cells were selected in puromycin (0.5 μg/mL).
CRISPR/Cas9 RNP Nucleofection of NCI-H69 and DMS79 Cells
Alt-R® CRISPR-Cas9 crRNA sequences were designed using the Broad Institute sgRNA designer tool (http://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) and synthesized by IDT technologies. Alt-R® CRISPR-Cas9 Negative Control crRNA #1 (IDT #1072544), Alt-R® CRISPR-Cas9 tracrRNA (IDT #1072532) and Alt-R™ S.p. Cas9 Nuclease V3 (IDT #1081058) were ordered from IDT technologies. Preparation of the RNAs and the nucleofection were performed following IDT’s protocol about the Alt-R® CRISPR-Cas9 System. Briefly, Alt-R® CRISPR-Cas9 crRNAs and tracrRNA were resuspend at 200 μM in IDTE Buffer, were mixed at equimolar ratios to a final duplex concentration of 100 μM and annealed by heating to 95°C for 5 min to form the gRNA complex. The gRNA complexes (120 pmol) were then mixed with Cas9 enzyme (104 pmol), diluted in PBS for a final volume of 5 μL and incubated for 20 min at room temperature to form the RNP complex. In parallel, 1x106 cells per reaction were prepared, washed in sterile PBS, and resuspend in 20 μL of SF Cell Line Nucleofector™ Solution supplemented (Lonza V4XC-2032). The cells and the RNP complex were carefully mixed and transferred to the wells of the 16-well Nucleocuvette™. Nucleofection were performed using CM-137 program on 4D-Nucleofector™ (Lonza). Cells were carefully resuspended with pre-warmed media, transferred into a 12 well plate and incubated at 37°C, 5% CO2 for 72 hours at which time an initial immunoblot was performed to test knockout efficiency. The following crRNA oligo was used for CRISPR/Cas9 RNP Nucleofection: Kdm6a human #1 sense (5’-GATTTGGTCTGGATAAATGC-3’). Cisplatin experiments in Figs. 1h,i were performed after >3 weeks in culture after generation of KDM6A isogenic cell lines by nucleofection. NCI-H69 and DMS79 cells were plated at 100,000 cells/mL and then treated with 1 μM cisplatin or DMSO for 72 hours at which time cell lysates were harvested for immunoblot analysis.
Immunoblotting
Cell pellets were lysed in a modified EBC lysis buffer (50mM Tris-Cl pH 8.0, 250 mM NaCl, 0.5% NP-40, 5 mM EDTA) supplemented with a protease inhibitor cocktail (Complete, Roche Applied Science, #11836153001) and phosphatase inhibitors (PhosSTOP Sigma #04906837001). Soluble cell extracts were quantified using the Bradford Protein Assay. 20 μg of protein per sample was boiled after adding 3X sample buffer (6.7% SDS, 33% Glycerol, 300 mM DTT, and Bromophenol Blue) to a final concentration of 1X, resolved by SDS-PAGE using either 10% or 8% SDS-PAGE, semi-dry transferred onto nitrocellulose membranes, blocked in 5% milk in Tris-Buffered Saline with 0.1%Tween 20 (TBS-T) for 1h, and probed with the indicated primary antibodies overnight at 4°C. Membranes were then washed three times in TBS-T, probed with the indicated horseradish peroxidase conjugated (HRP) secondary antibodies for 1h at room temperature, and washed three times in TBS-T. Bound antibodies were detected with enhanced chemiluminescence (ECL) western blotting detection reagents (Immobilon, Thermo Fisher Scientific, #WBKLS0500) or Supersignal West Pico (Thermo Fisher Scientific, #PI34078). The primary antibodies and dilutions used were: Rabbit Anti-KDM6A (UTX) (Cell Signaling #33510, 1:1000), Rabbit Anti-ASCL1 (Abcam #Ab211327, 1:1000), Rabbit Anti-NEUROD1 [EPR4008] (Abcam #Ab109224, 1:1000), Rabbit anti-c-Myc (Y69) (Abcam #32072, 1:1000), Rabbit anti-L-Myc (E3M5P) (Cell Signaling Technology #76266S, 1:1000), rabbit α-Rb1 (Abcam #181616 used at 1:2000), rabbit rodent specific anti-p53 (Cell Signaling D2H9O #32532, 1:1000), rabbit anti-p130 (Abcam #Ab76234, 1:1000), rabbit anti-Cre Recombinase (D7L7L) (Cell Signaling #15036, 1:1000), Mouse Anti-Cas9 (Cell Signaling Technology #14697, 1:1000), Rabbit anti-HIF1α (Cell Signaling #14179, 1:1000), Mouse Anti-β-actin (Sigma, clone AC-15, #A3854, 1:25,000), mouse α-Vinculin (Sigma, hVIN-1, # V9131 used at 1:10000). Histone extractions were performed as described previously26 with the following primary antibodies: Rabbit anti-Histone H3 (D1H2) (Cell Signaling Technology #4499S, 1:1000) and Rabbit anti-Mono-Methyl-Histone H3 (Lys4) (D1A9) (Cell Signaling Technology #5326T, 1:1000). The secondary antibodies and dilutions used were: Goat Anti-Mouse (Jackson ImmunoResearch #115-035-003) and Goat anti-Rabbit (Jackson ImmunoResearch #111-035-003) and used at 1:5000.
Bulk RNA-Sequencing and Analysis
SCLC tumors were harvested at necropsy and were flash-frozen. RNA was extracted using RNeasy mini kit (Qiagen #74106) including a DNase digestion step according to the manufacturer’s instructions and RNA sequencing was performed as described below.
Total RNA samples in each experiment were submitted to Novogene Inc. The libraries for RNA-seq are prepared using NEBNext Ultra II non-stranded kit. Paired end 150bp sequencing was performed on Novaseq6000 sequencer using S4 flow cell. Sequencing reads were mapped to the mm10 genome by STAR. Statistics for differentially expressed genes were calculated by DESeq2 (1.36.0).
Gene Set Enrichment Analysis of RNA-seq Data
GSEA software was downloaded from the Gene Set Enrichment Analysis website [http://www.broad.mit.edu/gsea/downloads.jsp]49. GSEA was performed using ‘NEUROD1 correlated genes’ of 401 genes (see Supplemental Table 2, tab 7)29 in Extended Data Fig. 1f. Upregulated genes in Kdm6a-Mutant vs Kdm6a-WT in the SCLC GEMM (see Supplemental Table 2, tab 5) were used to perform GSEA in Fig. 1e and Extended Data Figs. 5h,10b.
Pre-ranked GSEA was performed on pseudo-bulk RNAseq data from Chan et al., Cancer Cell, 202134. R package DESeq2 (1.36.0) was used to perform differential analysis on the pseudo-bulk sample-level read counts aggregates (Supplementary Table 2, tab 6) in Fig. 1e and Extended Data Fig. 5f. Differential gene expression analysis between RPM and RPP tumor was performed on read counts with R package DESeq2 following gene exclusion rules in the original paper (Ireland, Cancer Cell, 2020)13 (Supplementary Table 6 for Extended Data Fig. 5f-h) where variation from tumor RNA-seq sample preparation was adjusted by removing all non-coding features, histones, and ribosomal RNAs from count matrices. The top 100 genes with largest fold changes were used to perform GSEA in Extended Data Fig. 5. Pre-ranked GSEA was performed on KMT2D loss of function mutations vs. KMT2D WT human tumor samples that are neuroendocrine (express either ASCL1 and/or NEUROD1) from George et al., Nature, 20154 in Extended Data Fig. 10. Specifically, FPKM values were first log transformed with a pseudo-count of 1, then R package Limma (3.52.4) was used to calculate differential expression profile with trend=TRUE in the eBayes function (Supplementary Table 9 for Extended Data Fig. 10b). For Extended Data Figs. 10l,m, 24 Hypoxia genes50 and HIF1 target genes51 were used (Supplementary Table 10).
Reverse-Transcriptase Quantitative PCR (RT-qPCR)
RNA was extracted using Quick-RNA™ Miniprep kit (Zymo Research, CA, USA) according to the manufacturer’s instructions. RNA concentration was determined using the Nanodrop 8000 (Thermofisher Scientific). A cDNA library was synthesized using iScript Reverse Transcription Supermix for RT-qPCR (Biorad #1708841) according to the manufacturer’s instructions. qPCR for Ascl1 and Neurod1 were performed using the SsoAdvanced Universal SYBR Green Supermix (Biorad #1725271) according to the manufacturer’s instructions. qPCR for Chromogranin A, Synaptophysin, Pou2f3, yap1, myc, mycl and mycn were performed using the LightCycler 480 (Roche) with the LightCycler 480 Probes Master Kit (Roche) and Taqman probes (ThermoFisher Scientific) according to the manufacturer’s instructions. The ΔΔCT Method was used to analyze data. The CT values for each probe were then normalized to the CT value of ActB. The data from Fig. 3b and Extended Data Fig. 2a, 3a, and 10f were then normalized to the control to determine the relative fold change in mRNA expression. The following probes were used for qPCR with SYBR Green: Mouse Ascl1 Forward (5’-AGGGATCCTACGACCCTCTTAG-3’), Mouse Ascl1 Reverse (5’-GTCGTTGGCGAGAAACACTAAAG-3’), Mouse Neurod1 Forward (5’-AGGCTCCAGGGTTATGAGATCG-3’), Mouse Neurod1 Reverse (5’-TGAGAACTGAGACACTCATCTG-3’), Mouse ActB Forward (5’-CATTGCTGACAGGATGCAGAAGG-3’), Mouse ActB Reverse (5’-TGCTGGAAGGTGGACAGTGAGG-3’). The following TaqMan probes were used: Actb mouse (Mm00607939_s1), Chga (Mm00514341_m1), Syp (Mm00436850_m1), Pou2f3 (Mm01179139_g1), Yap1 (Mm01143263_m1), Myc (Mm00487804_m1), Mycl (Mm00493155_m1) and Mycn (Mm00476449_m1).
Immunohistochemistry and Multiplexed Immunofluorescence
For in situ immunolabeling studies, ASCL1 (clone EPR19840, ab211327, Abcam), NEUROD1 (clone EPR17084, ab205300, Abcam), Synaptophysin (clone D4, SC-17750, Santa Cruz Biotech) and c-Myc (clone Y69, ab32072, Abcam) specific antibodies were used. In brief, 5-micron paraffin sections were de-waxed and rehydrated following standard protocols. Antigen retrieval consisted of steaming for 40 min in Target Retrieval Solution (S1700, Agilent, Santa Clara, CA) for NEUROD1 and steaming in citrate buffer (Vector Laboratories, Burlingame, CA) for 20 min for ASCL1, Synaptophysin and c-Myc. Slides were then washed and equilibrated in TBS-Tween buffer (Sigma, St. Louis, MO) for 10 min. For immunohistochemistry, primary antibodies were applied at 37°C for 60 min at dilutions of 1:100 (ASCL1), 1:25 (NEUROD1), 1:100 (c-Myc) and 1:200 (Synaptophysin). Immunocomplexes were visualized using PowerVision Poly-HRP Anti-Rabbit (Leica, Buffalo Grove, IL) secondary antibodies with DAB (3,3′-Diaminobenzidine, Sigma) as the chromogen. For dual-immunofluorescence labeling studies, tyramide-based signal amplification was carried out by first incubating slides with NEUROD1 specific antibodies at 1:25 for 60 min at 37°C. Next, slides were incubated with PowerVision Poly-HRP Anti-Rabbit (Leica) secondary antibodies followed by Tyramide568 (Tyramide SuperBoost kit, Thermo Fisher) according to manufacturer’s protocols. Slides were then stripped by steaming in citrate buffer (Vector) for 20 min and subsequently incubated with ASCL1 specific antibodies (1:100, 60 min, 37°C) followed by PowerVision Poly-HRP Anti-mouse (Leica) secondary antibodies and Tyramide488 (Tyramide SuperBoost kit, Thermo Fisher). Slides were counterstained with DAPI (Thermo Fisher) and mounted with Prolong (Thermo Fisher). All bright field slides were imaged using a Ventana DP200 slide scanned (Roche Diagnostics, Indianapolis, IN). Fluorescence images were taken a Nikon Eclipse E800 (Nikon, Melville, NY) microscope. All analyses were carried out using QuPath (v0.3.0)52.
Nuclei isolation for ATAC-sequencing
Fragments of the SCLC frozen tissues were cut and resuspended in 300 μl of cold 3-detergent ATAC Resuspension Buffer (RSB) containing 0.1% NP40, 0.1% Tween-20, and 0.01% Digitonin. Tissues were dounced ten times each with a loose and a tight pestle each until homogenization was complete. The homogenate was then transferred to a 1.5 ml pre-chilled microfuge tube and incubated on ice for 10 min. After lysis, 300 μl of ATAC-RSB containing 0.1% Tween-20 was added and the tubes were inverted to mix. Lysates were filtered through a 40 μm cell strainer and nuclei were centrifuged for 10 min at 1500 relative centrifugal force (RCF) in a pre-chilled (4°C) fixed-angle centrifuge. Nuclei were resuspended with 300 μl of ATAC-RSB containing 0.1% Tween-20 and counted with a hemocytometer using Trypan blue stain.
Bulk ATAC-Sequencing
100,000 nuclei were resuspended in 50 μl of transposition mix (25 μl 2x TD buffer, 2.5 μl transposase (100 nM final), 16.5 ul PBS, 0.5 ul 1% Digitonin, 0.5 μl 10% Tween-20, 5 μl H2O). Transposition reactions were incubated at 37 °C and shaken at 1000 RPM for 30 minutes on a thermomixer. Transposed DNA was purified using Qiagen columns. Libraries were amplified as described previously53. Thirtyfive basepair paired-end reads were sequenced on a NextSeq instrument (Illumina).
Single Cell ATAC-Sequencing
Nuclei were prepared as described above. For scATAC-seq, nuclei were transposed according to 10XGenomics specifications. Approximately 10,000 cells were targeted for each tissue analyzed and pooled and processed according to the 10x Genomics scATAC-seq sample preparation protocol (Chromium Single Cell ATAC Library & Gel Bead Kit, 10x Genomics) to obtain a single sequencing library (containing both barcoded cell populations) that was sequenced on a NovaSeq6000 using a 100 cycle kit with a 66bp read one, 8bp index read, 16bp index two read, and 47bp read two.
Bulk ATAC-seq Data Analysis
Quality control
All samples were processed through the computational pipeline developed at the Dana-Farber Cancer Institute Center for Functional Cancer Epigenetics (CFCE) using primarily open-source programs54,55. Sequence tags were aligned with Burrows-Wheeler Aligner (BWA)56 to build mm9 and uniquely mapped, non-redundant reads were retained. These reads were used to generate binding sites with Model-Based Analysis of ChIP-Seq 2 (MACS v2.1.1.20160309), with a q-value (FDR) threshold of 0.0157. We evaluated multiple quality control criteria based on alignment information and peak quality: (i) sequence quality score; (ii) uniquely mappable reads (reads that can only map to one location in the genome); (iii) uniquely mappable locations (locations that can only be mapped by at least one read); (iv) peak overlap with Velcro regions, a comprehensive set of locations – also called consensus signal artifact regions – in the genome that have anomalous, unstructured high signal or read counts in next-generation sequencing experiments independent of cell line and of type of experiment; (v) number of total peaks (the minimum required was 8,000); (vi) high-confidence peaks (the number of peaks that are tenfold enriched over background); (vii) percentage overlap with known DHS sites derived from the ENCODE Project (the minimum required to meet the threshold was 80%); and (viii) peak conservation (a measure of sequence similarity across species based on the hypothesis that conserved sequences are more likely to be functional). Genome tracks were visualized by IGV (v2.14.1)58.
Differential Binding Analyses
Peaks from all samples were merged to create a union set of sites for each transcription factor and histone mark using bedops59. Sample-sample correlation and differential peaks analysis were performed by the CoBRA pipeline55. Read densities were calculated for each peak for each sample and used for the comparison of cistromes across samples. Sample similarity was determined by hierarchical clustering using the Spearman correlation between samples. Differential peaks were identified by DEseq2 with adjusted p<0.05 and ∣log2FoldChange∣ > 2. A total number of reads in each sample was applied to the size factor in DEseq2, which can normalize the sequencing depth between samples. Peaks from each group were used for motif analysis by the motif search findMotifsGenome.pl in HOMER (v3.0.0)60, with cutoff q-value≤ 1e-10. The signals of each sample on differential binding sites were visualized by Deeptools61. For the PCA of chromatin accessibility in Fig. 3c and Extended Data Fig. 3h, all peaks from ATAC-seq data from the 2nd cohort of mice (see Supplementary Table 4, tab 1) were used. For the PCA of chromatin accessibility in Fig. 3d and Extended Data Fig. 3i, differential peaks in Kdm6aMutantNEUROD1Only vs. Kdm6aWT SCLC tumors were used (see Extended Data Fig. 3d and Supplementary Table 4, tabs 2,3).
GREAT Analysis
Kdm6a-WT versus Kdm6a-Mutant differential peaks (KDM6A enriched regions) were put into GREAT web tool (http://great.stanford.edu/public/html/) to predict functions35. We set the threshold for single nearest genes within 400kb. The GO Biological Process prediction results were visualized using ggplot2 (H. Wickham. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, 2016.)
Correlation of RNA-Seq and ATAC-Seq Analysis
RNA-seq data reads alignment, quality control and analysis were performed using Visualization Pipeline for RNA-seq data(VIPER)62. Alignment to the mm9 genome was done using STAR v2.7.0f63 followed by transcript assembly using cufflinks v2.2.164 and RseQC v2.6.265. Differential gene expression analyses were performed on absolute gene counts for RNA-Seq data and raw read counts for transcriptomic profiling data using DESeq2 v1.18.166. RNA-seq differential analysis result was visualized by volcano plot. Points in plots were highlighted by Kdm6a-Mutant vs Kdm6a-WT differential peaks (KDM6A enriched regions) in ATAC-seq that have matched nearby genes in RNA-seq. The nearby genes of ATAC-seq differential peaks were found by bedtools67. Volcano plot was made by ggplot2.
Profile plot on differential binding regions
We first calculated the average signal of 7 Kdm6aWT SCLC tumors (158, 18227, 165, 1198, 168, 645, 535, sc535), 7 Kdm6aMutantASCL1Only SCLC tumors (1125, 1274, 1314-2, 713, 1119, 1121, 1122), 7 Kdm6aMutantASCL1HighNEUROD1Low SCLC tumors (672-2, 1116, 1117, 1270-1, 1270-2, 1314-2, 236R, sc236R_c1, sc236R_c2) and 2 Kdm6aMutantNEUROD1Only (236L, 656, sc656) tumors using WiggleTools68. Profile plots were based on Kdm6aMutantNEUROD1Only vs. Kdm6aWT differential binding regions created by Deeptools.
Single-cell ATAC-seq data analysis
Single cell ATAC-seq data of 3 samples (535=Kdm6a-WT, 236R= Kdm6aMutantASCL1HighNEUROD1Low, 656=Kdm6aMutantNEUROD1Only) were run by cellranger-atac(v2.0.0)69 pipeline with default parameters70. R package Seurat71 and Signac(v1.6.0)72 were used to perform QC filtering of low-quality cells. Cells were filtered based on strength of nucleosome-binding pattern, transcription start site enrichment score, number of fragments in peaks and fraction of fragments in peaks. Samples were then integrated by a common peak set derived from peaks of each sample with 20< peakwidths < 10000. Integrated data normalization and dimensionality reduction were performed using Signac with latent semantic indexing (LSI), consisting of term frequency-inverse document frequency (TF-IDF) normalization and singular-value decomposition (SVD) for dimensionality reduction, using the top 20% of peaks in terms of their variability across the samples. The first LSI component reflected sequencing depth across the samples and was not utilized in downstream analyses. Pseudo-bulk analysis of scATAC-seq samples was performed by pooling all reads from good quality cells in each sample.
Single Cell RNA-Sequencing Sample Preparation
The single-cell RNA sequencing (scRNA-seq) experiments in autochthonous CRISPR-based SCLC GEMM models (see description above) were performed as follows: 3-4 month old male and female homozygous BL6J LSL-Cas9 mice (Jackson No. 026175) were intratracheally injected with Kdm6a-Mutant or Kdm6a-WT adenovirus (see adenovirus method above). Once mice became symptomatic from their tumors (see method above), 6 independent Kdm6a-Mutant and 3 independent Kdm6a-WT mice were euthanized and lung tumors dissected and finely minced mechanically using a razor blade and then enzymatically digested with Mouse Tumor Dissociation Kit (Miltenyi Biotec, #130-096-730) following the manufacturer’s instruction. Briefly, minced tumor tissue was transferred to a gentleMACS C Tube containing enzyme mix prepared with 20% of Enzyme R option to preserve cell surface epitopes. Dissociation using the gentleMACS Octo Dissociator with Heaters (Miltenyi Biotec, #130-096-427) was performed using the 37C_m_TDK_2 gentleMACS Program. The single cell suspensions were resuspended in RPMI containing 10% FBS and subsequently passed through a 70 μm Cell Strainer (Greiner, #542070) and centrifuged at 300 x g for 3 min followed by 2 washes with 0.04% UltraPure Bovine Serum Albumin (Invitrogen, AM2616) in DPBS. Finally, dissociated cells were resuspended in DPBS with 0.04% UltraPure BSA and cell counts were measured with a Vi-CELL XR Cell Viability Analyzer (Beckman Coulter). Cells were then diluted in 0.04% BSA/DPBS at a cell concentration of 1000 cells/μL. About 16,000 cells were loaded onto a 10× Genomics Chromium™ instrument (10× Genomics) according to the manufacturer’s instructions. The scRNA-seq libraries were processed using Chromium Next GEM Single Cell 5' Kit v2 kit (10× Genomics). Quality controls for amplified cDNA libraries and final sequencing libraries were performed using Bioanalyzer High Sensitivity DNA Kit (Agilent). The sequencing libraries for scRNAseq were normalized to 4 nM concentration and pooled. The pooled sequencing libraries were sequenced on Illumina NovaSeq S4 300 cycle platform. The sequencing parameters used were: Read 1 of 26bp, Read 2 of 90bp, Index 1 of 10bp and Index 2 of 10bp.
Single-Cell RNA Sequencing Data Analysis
Custom reference genome was established by adding EGFP and Cas9 sequence73 to the mouse genome (mm10-2020-A). Cell ranger version 6.0.2 pipeline (10× Genomics) was used to align sequencing data to the custom mouse reference genome and generate the gene-level counts matrix. Unfiltered raw counts data was imported into Seurat v4 R package (version 4.1.0)74 for downstream processing and analysis. Low quality cells were filtered out using following thresholds: total UMI counts < 500, number of transcripts < 350, log10TranscriptsPerUMI <= 0.8, and cells with more than 15% transcripts mapping to mitochondrial genes. In addition, genes expressed in less than ten cells were removed. The UMI counts matrices were then natural-log normalized and scaled with Seurat’s ‘NormalizeData’ and ‘ScaleData’ functions.
Dimension Reduction, Cluster Analysis and Visualization of scRNA-Seq Data
The Seurat v4 R package was used for dimension reduction and clustering. Top 2,000 genes with the highest variance were selected using the ‘FindVariableFeatures’ function with ‘vst’ method to perform linear dimensional reduction (principal component analysis) using the ‘RunPCA’ function, and top 40 principal components were used to perform graph-based unsupervised clustering with the ‘FindNeighbors’ and ‘FindClusters’ functions and Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) (arXiv:1802.03426) for data visualization in two-dimensional space. Automatic cluster annotation using the R package SingleR (version 1.8.1)75 with ImmGenData76,77 and manual tumor cluster annotation with markers of EGFP, Cas9, Ascl1, and Insm1 were performed to select the tumor population. Then, the tumor subpopulations were reanalyzed. Seurat’s ‘CellCycleScoring’ function and S phase and G2/M phase specific gene lists (Supplementary Table 5, tabs 4, 5) were used to calculate the G2M and S phase gene expression scores and to be regressed out using the ‘var.to.regress’ option in the ‘SCTransform’ function with default parameters. Top 3,000 genes excluding mitochondrial and ribosomal genes with the highest variance were selected using the ‘FindVariableFeatures’ function with ‘vst’ method to perform linear dimensional reduction (principal component analysis) using the ‘RunPCA’ function, and top 40 principal components were used to plot UMAP for data visualization in two-dimensional space. Differential expression profile were obtained with Seurat’s ‘FindMarkers’ function using test.use = “wilcox”, logfc.threshold = 0, min.pct=0, to compare differentially expressed genes between Kdm6a-Mutant and Kdm6a-WT (Supplementary Table 5, tab 2).
A Neurod1 gene score (Neurod47_score) was calculated based on overlapping genes between NEUROD1-positive primary human SCLC tumors and NEUROD1-positive SCLC cell lines29, which yielded a gene list of 47 genes that are correlated with the NEUROD1 subtype (Supplementary Table 5, tab 3).
Adherence Assays
Late passage Kdm6a-WT (159-1, 1014, 1015) or Kdm6a-Mutant (236L, 236R, 656) tumor-derived SCLC mouse cells were counted on day 0 using a Vi-Cell XR Cell Counter and plated in tissue culture-treated 6 well plates at 100,000 cells/mL in 2 mLs of complete media. 48 hours later the cells were washed twice in PBS to remove non-adherent cells. Representative images were acquired using brightfield microscopy with a 10X objective and then stained with crystal violet for visualization of the entire well.
Flow Cytometry
Late passage Kdm6a-WT (159-1, 1014, 1015) or Kdm6a-Mutant (236L, 236R, 656) tumor-derived SCLC mouse cells were collected, washed twice in PBS, resuspended in FACS buffer (D-PBS containing 2% FBS) and transferred in 1.5 mL Eppendorf tubes. After washing, cells were incubated with fluorophore conjugated anti-PD-L1/CD274 (Biolegend #124307, B7-H1, clone 10F.9G2), anti-H-2Ld/H-2Db (Biolegend #114507, clone 28-14-8) or the isotype control (Biolegend #400212, IgG2a, κ isotype Ctrl) at 1:100 dilution in the dark for 30 minutes at room temperature. After washing, cells were resuspended in FACS buffer, transferred to flow cytometry tubes containing a 70 μm filter and analyzed on a LSR Fortessa flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Data analyses were performed with FlowJo software. A gating strategy for flow cytometry experiments is shown in Supplementary Fig. 6.
H3K4me1, H3K4me2, H3K27me3, and KDM6A ChIP-Sequencing Experiments
Early passage 236L and 236R cell lines derived from Kdm6a-Mutant tumors and early passage 1014 and 159-1 cell lines derived from Kdm6a-WT tumors (see Figs. 4a,b) were plated in ultra-low attachments plates at 0.4 X 106 cells/mL for all H3K4me1, H3K4me2, H3K27me3, and KDM6A ChIP-seq experiments; in 8 mLs (3.2 X 106 cells total) of complete media for H3K4me1, in 12 mLs (4.8 X 106 cells total) of complete media for both H3K4me2 and H3K27me3 ChIP-seq or 20 mLs (8 X 106 cells total) of complete media for KDM6A ChIP. 24 hours later, cells were collected, and centrifuged. The supernatant was aspirated and the cells were washed once in PBS. For the KDM6A ChIP experiments, cells were cross-linked by incubation with 2 mM disuccinimidyl glutarate (DSG) with rocking for 30 minutes at room temperature prior to fixation step. Then, for all ChIP experiments, cells were fixed in 1% paraformaldehyde diluted in PBS for 10 minutes at room temperature. Excess formaldehyde was quenched by dropwise addition of freshly made Glycine at a final concentration 1.25 M followed by rocking the cells for an additional 5 minutes at room temperature. The paraformaldehyde/glycine was then removed by spinning at 400 x g for 5 minutes at 4°C and the cells were washed twice with ice cold PBS.
The cells were then resuspended in 1 mL of lysis buffer (50 mM Hepes-NaOH pH 8, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% TX-100 supplemented with a protease inhibitors and phosphatase inhibitors) and incubated by rotating for 10 minutes at 4°C. Intact nuclei were then collected by centrifugation at 1900 x g for 5 minutes at 4°C. The supernatant was then gently aspirated and the pellet was resuspended in 1 mL of wash buffer (10mM Tris-HCl pH 8.0, 200mM NaCl, 1mM EDTA, supplemented with a protease inhibitor cocktail and phosphatase inhibitors) and incubated by rotating for 10 minutes at 4°C. The nuclei were again collected by centrifugation at 1900 x g for 5 minutes at 4°C. The supernatant was then gently aspirated and the tube was rinsed with 500 μL of shearing buffer (0.1% SDS, 1 mM EDTA, 10 mM Tris-HCl pH 8.0 supplemented with protease inhibitors and phosphatase inhibitors). The nuclei were again collected by centrifugation at 1900 x g for 5 minutes at 4°C, the supernatant was carefully removed, and the pellet was gently resuspended in 1mL shearing buffer and transferred to a 1 mL AFA fiber tube (Covaris #520130).
The chromatin was then sonicated using a Covaris E220 sonicator at 100 peak incident power, 10% duty cycles, 200 cycles per burst, water level 15 (left indicator)/10 (right indicator) for 900 seconds. The samples were then transferred to 1.5 mL Eppendorf tubes and centrifuged in an Eppendorf microcentrifuge at 15,900 x g for 5 minutes at 4°C. 900 μL of the supernatants were transferred to a new Eppendorf tube, to which 1% Triton X-100 and 150 mM NaCl were added. 20μl of Dynabeads Protein G (ThermoFisher #10004D), prewashed with Shearing Buffer, was then added and samples were rotated for 1 hour at 4°C to pre-clear the sonicated chromatin. The Dynabeads were then removed using a magnetic stand and the supernatants were transferred to new tubes. 5% of the total lysate was removed to make genomic DNA to be used for the ChIP-sequencing input DNA. For the H3K27me3 and H3K4me2 ChIP, the remaining lysate (95% of total) were equally distributed into two Eppendorf tubes to perform the H3K27me3 and H3K4me2 ChIPs. 3 μg of the rabbit anti-H3k27me3 (C36B11) (Cell signaling, #9733) or 3 μg of the rabbit anti-H3K4me2 (Abcam #Ab32356) primary antibodies were added. The KDM6A and H3K4me1 ChIPs were performed separately also using 5% input and 95% for ChIP. 10 μg of the rabbit anti-KDM6A [Bethyl lab, A302-374A (UTX antibody)] or 1 μg of the rabbit anti-H3K4me1 (D1A9) (CST #5326) primary antibodies were added. All samples were incubated overnight while rotating at 4°C.
The following morning, 50 μL of Dynabeads, prewashed with Shearing buffer, were added and the samples were incubated for another 2 hours while rotating at 4°C. A magnetic stand was used to isolate the Dynabeads. The supernatant was removed and the Dynabeads were washed once in 1 mL of low salt immune complex wash buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% SDS, 1% TX-100, 2 mM EDTA) and incubated for 5 minutes while rotating at 4°C. The beads were again isolated using a magnetic stand. Following removal of the supernatant the Dynabeads were washed once in 1 mL of high salt immune complex wash buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 0.1% SDS, 1% TX-100, 2 mM EDTA) and incubated for 5 minutes while rotating at 4°C. The beads were again pelleted with a magnetic stand. Following removal of the supernatant, the Dynabeads were washed once in 1 mL of Lithium Chloride immune complex wash buffer (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 1% SDS, 1% NP-40, 1 mM EDTA) and incubated for 5 minutes while rotating at 4°C. The beads were then washed twice in TE pH8 at room temperature and the beads were then resuspended in 100μL TE pH8.
RNase A (Qiagen) was then added to a final concentration of 0.2 mg/mL and the samples were incubated for 30 minutes at 37°C. To reverse the formaldehyde crosslinking and remove protein, proteinase K was then added to a final concentration of 0.8 mg/mL and the samples were incubated for 30 minutes at 42°C and then an additional 6 hours at 65°C. 300 μL QG buffer and 100 μL of isopropanol was then added to each sample and a magnetic stand was used to pellet the Dynabeads. The supernatant was then collected and DNA was isolated using QIAquick gel extraction kit (Qiagen) according to the manufacturer’s instructions. DNA concentration was determined using the Nanodrop 8000 (Thermofisher Scientific) and sample were stored at −80°C. For all experiments, an isotype matched control IgG ChIP (Cell Signaling #3900 used at total amount in μg as the experimental ChIP antibody described above) was performed with an equal number of cells as a control for ChIP pulldown signal, which yielded no measurable DNA concentration and hence only experimental ChIP samples and inputs were sequenced.
ChIP-Sequencing Library Preparation, Sequencing, and Analysis
The ends of the purified DNA fragments were repaired and ligated to loop adapter sequences and then amplified by PCR. A 200-400bp insert DNA library was constructed using NEB Next Ultra II DNA Library Prep Kit tailored for Novogene. The DNA were then sequenced on the Illumina Novaseq 6000 S4 platform using PE150 strategy (Novogene, Beijing).
Sequenced reads generated from KDM6A, H3K27me3, H3K4me1 and H3K4me2 ChIP-Seq and respective input libraries were aligned to the mouse genome (mm10) using Bowtie278. The R package SPP79 was used to calculate tag densities using a Gaussian kernel bandwidth of 35bp, and BigWig format files were generated for both tag enrichment and tag density for visualization in the genome browser80,81. ChIP-Seq peaks were called using MACS37 version 3.0.0a7 with parameters -f BAM -g mm -B -q 0.01 –broad. Mouse genome reference transcript annotations were obtained from the University of California Santa Cruz Genomics Institute (UCSC)82. The mouse ENCODE Candidate Cis-Regulatory Elements (cCREs) data set (ENCODE Project Consortium)83 were used to identify putative enhancers. For the final analysis we retained only those cCREs that were annotated as “dELS” (distal enhancer-like signature) and at least 2,000 bp from an annotated transcription start site (see Supplementary Table 7, tab 2). Enhancers were labeled with the nearest annotated gene. Histone mark and KDM6A enrichment for enhancers (within +/− 1Kb of enhancer center point) or transcription start site (within +/− 1Kb of start site) was calculated as ChIP tags per million divided by input tags per million.
GSEA analysis and Gene Ontology analyses were performed using ClusterProfiler version 4.2.28. For GSEA analysis, log2 fold changes of mark enrichment (Kdm6a-Mutant vs Kdm6a-WT) at enhancers or transcription start site proximal regions were used with GSEA function options “nPermSimple”=10000, “eps”=0, and “maxGSSize”=1100 to produce normalized enrichment scores and adjusted p-values. Because multiple enhancers may be annotated with the same gene, the enhancer with the highest absolute value log2 fold change for each gene was chosen as input for GSEA analysis. Gene ontology (GO) enrichment was performed with the function enrichGO with options “pvalueCutoff” = 0.05, “qvalueCutoff” = 0.1, “ont” = "BP", “maxGSSize” = 500, and the “universe” set to the subset of genes with one or more annotated enhancers. All GO analyses were performed on genes with enhancers that overlapped replicating (Kdm6a-WT_1014 and Kdm6a-WT_159-1) KDM6A ChIP-Seq MACS3 peaks. For H3K4me1, replicating (Kdm6a-WT_1014 and Kdm6a-WT_159-1) H3K4me1 ChIP-Seq MACS3 peaks were required and log2 fold changes <=−1 were required for H3K4me1 Kdm6a-Mutant_236R vs Kdm6a-WT (mean of Kdm6a-WT_1014 and Kdm6a-WT_159-1) and Kdm6a-Mutant_236L vs Kdm6a-WT. For H3K27me3, above median H3K27me3 enrichment at enhancers was required in both Kdm6a-Mutant_236L and Kdm6a-Mutant_236R and log2 fold changes >= 1 were required for H3K27me3 Kdm6a-Mutant_236R vs Kdm6a-WT (mean of Kdm6a-WT_1014 and Kdm6a-WT_159–1) and Kdm6a-Mutant_236L vs Kdm6a-WT. For Fig. 5a and Extended Data Figs. 8a-c, Pearson’s correlations and p-values were calculated using the R function cor.test. For Fig. 5a, the trend line was generated using the lm function while for Extended Data Figs. 8a-c, the trend line was generated using the function lowess.
LISA Analysis
Lisa Analysis38 was performed to find candidate epigenetic/transcription positive regulators of the accessible promoter peaks found in Kdm6a-Mutant vs Kdm6a-WT tumors from the ATAC-seq data using LISA website (http://lisa.cistrome.org/). For this analysis, the top 100 genes, based on the p-value < or equal to 0.05, a log fold change > or equal to 1, and then sorted by ascending p-value, were as the input and LISA was run for human orthologs (Supplementary Table 8, tabs 1,2). The complete analysis results can be found in Supplementary Table 8, Tab 3.
Menin ChIP-Seq Experiments and Analysis
For the Menin ChiP-seq experiments, 1014 cells with Kdm6a CRISPR inactivation (see Fig. 6f) cells were counted and plated at 0.1x106 per mL in 100 mLs (10 X 107 cells total) for each condition and treated with VTP-50469 (500 nM) or DMSO for 5 days per replicate. Crosslinking was performed with 10 × 107 cells per immunoprecipitation. After drug (or vehicle) treatment, cells were collected, washed once with ice-cold PBS, and flash-frozen. Cells were resuspended in ice-cold PBS and cross-linked using 1% paraformaldehyde (PFA; Electron Microscopy Sciences) for 10 minutes at room temperature with gentle rotation. Unreacted PFA was quenched with glycine (final concentration 125 mmol/L) for 5 minutes at room temperature with gentle rotation. Cells were washed once with ice-cold PBS and pelleted by centrifugation (800 × g for 5 minutes). To obtain a soluble chromatin extract, cells were resuspended in 1 mL of LB1 (50 mmol/L HEPES pH 7.5, 140 mmol/L NaCl, 1 mmol/L EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, and 1× complete protease inhibitor cocktail) and incubated at 4°C for 10 minutes while rotating. Samples were centrifuged (1,400 × g for 5 minutes), resuspended in 1 mL of LB2 (10 mmol/L Tris-HCl pH 8.0, 200 mmol/L NaCl, 1 mmol/L EDTA, 0.5 mmol/L EGTA, and 1× complete protease inhibitor cocktail), and incubated at 4°C for 10 minutes while rotating. Finally, samples were centrifuged (1,400 × g for 5 minutes) and resuspended in 1 mL of LB3 (10 mmol/L Tris-HCl pH 8.0, 100 mmol/L NaCl, 1 mmol/L EDTA, 0.5 mmol/L EGTA, 0.1% sodium deoxycholate, 0.5% N-Lauroylsarcosine, and 1× complete protease inhibitor cocktail). Triton X-100 was added to a final concentration of 1%. Chromatin extracts were sonicated for 30 minutes using a Covaris E220-focused ultrasonicator. Lysates were centrifuged at maximum speed for 10 minutes at 4°C, and 5% of supernatant was saved as input DNA. Beads were prepared by incubating them in 0.5% BSA in PBS and antibodies overnight [100 μL of Dynabeads Protein A plus 15 μL of anti-Menin (Bethyl, A300-105A) antibody. Antibody–bead mixes were washed with 0.5% BSA in PBS and then added to the lysates overnight while rotating at 4°C. Beads were then washed six times with RIPA buffer (50 mmol/L HEPES pH 7.5, 500 mmol/L LiCl, 1 mmol/L EDTA, 0.7% sodium deoxycholate, and 1% NP-40) and once with TE-NaCl buffer (10 mmol/L Tris-HCl pH 8.0, 50 mmol/L NaCl, and 1 mmol/L EDTA). Chromatin was eluted from beads in elution buffer (50 mmol/L Tris-HCl pH 8.0, 10 mmol/L EDTA, and 1% SDS) by incubating at 65°C for 30 minutes while shaking, supernatant was removed by centrifugation, and crosslinking was reversed by further incubating chromatin overnight at 65°C. The eluted chromatin was then treated with RNaseA (10 mg/mL) for 1 hour at 37°C and with Proteinase K (Roche) for 2 hours at 55°C. Following de-crosslinking, DNA was purified with AMPure XP beads (Beckman Coulter). DNA fragments were quantified by TapeStation 4200 (Agilent) using HSD1000 Tape and Reagent (Agilent) and Qubit (ThermoFisher). 1-10 ng of DNA was used in preparation of Illumina compatible libraries using SMARTer ThruPLEX DNA-Seq Kit (Takara) followed by sequencing using NextSeq550 (Illumina) to obtain 20-30 million 37bp, paired-end reads.
For Menin ChIP-seq analysis, raw Illumina sequencer output was converted to FASTQ format using bcl2fastq (v2.20.0.422). A summary of all total and mapped reads of ChIP-seq samples used in this study (Supplementary Table 8, tab 4). Reads (paired-end 37mers) were aligned to the mouse (Gencode M24/mm10) genome using STAR (v2.7.5a; params –alignIntronMax 1 –alignEndsType EndToEnd – alignMatesGapMax 2000 for ChIP-seq analysis), sorted, and duplicates were marked/removed with picard pipeline tools (v2.9.4). Final “deduped” .BAM files were indexed using SAMtools (v1.95). ChIP-seq data visualizations were produced using IGVtools (TDF signal pileups; v2.3.75) and deeptools (tornado plots; v3.1.3; regions around central points of control sample MACS2 peaks plotted using referencePoint option and k = 1). Deeptools TSS-based signal plots were visualized using 4kb intervals centered around annotated protein-coding transcript transcription start sites. ChIP-seq peaks were called using MACS2 with appropriate input samples used as controls. Peak overlaps with genes, TSS regions, known non-specific high-signal blacklist regions and peaks were determined using the bedtools (v2.28.0) intersect function to compare genomic coordinates in pairwise fashion.
CRISPR-Amplicon Sequencing
Genomic DNA from the cell lines indicated was isolated using the QIAamp DNA Blood Mini Kit (Qiagen #51106) according to the manufacturer’s instructions. PCR was done by using KOD Xtreme polymerase (EMD Millipore #71975) and following set of primers to amplify the genomic region of Kmt2d targeted by sgKmt2d #1 and #2: forward Kmt2d #1 (5’- GACAGGCACATTTGTGGGCAC −3’), reverse Kmt2d #1 (5’- CGTAGTGGGGACAGCCCATAG −3’), and forward Kmt2d #2 (5’- AAATGAGAAGGCTGAGCGAGAAG −3’), reverse Kmt2d #2 (5’- ACTGCTGGGGAACGATCCTGTAC −3’). The final PCR product was then column purified using Qiagen’s gel extraction kit and sequenced using next-generation sequencing by the MGH DNA Core Facility.
Hypoxia Experiments
Cells were plated at 0.05x106 per mL in 2 10 cm tissue-culture dish. One of the plates was kept in a culture incubator in the presence of 21% O2 (37°C, 5% CO2) and the other plate was incubated in a hypoxia chamber in the presence of 5% O2 (37°C, 5% CO2) for 7 days. Cells were then placed on ice and lysed immediately as described above.
Human ASCL1/NEUROD1 Subtype Analysis from RNA-seq Data
Correlations between FPKM values of ASCL1 and NEUROD1 and IHC H-score were determined using data reported by Caeser et al.84. ASCL1 IHC H-scores of >8 from this study reflected ASCL1 positive tumors which correlated with ASCL1 FPKM values of greater than 55. NEUROD1 IHC H-scores of >76 from this study reflected NEUROD1 positive tumors which correlated with NEUROD1 FPKM values greater than 1.5. Thus, FPKM values of 55 for ASCL1 and 1.5 for NEUROD1 were used as threshold cut-offs to call ASCL1 and NEUROD1 positive tumors, respectively using other publicly available RNA-sequencing studies of human neuroendocrine lung tumors4,41.
Statistics and Reproducibility
For all GSEA analysis from RNA-sequencing and ChIP-sequencing data, statistical significance was calculated using FDR corrected for multiple hypothesis testing where FDR is indicated on each figure. For the GREAT analysis of ATAC-sequencing data in Fig. 2, a binomial p-value was calculated as described previously35. For genome-wide ATAC-seq peak comparison in Figs. 3e,f, an Anderson-Darling k-sample test in R package “kSamples” was used to calculate p-values. For the ChIP-sequencing analysis, the correlation p-values are based on Pearson's product moment correlation coefficient and t distribution with n-2 degrees of freedom. p-values for the boxplot in 5j were calculated using unpaired, two-sided, Mann-Whitney U tests adjusted for multiple hypothesis. For the single-cell RNA expression data in Figs. 3l, 6c and Extended Data Figs. 4d, 6b, 6d, statistical significance of gene expression at a single cell level was calculated using non-parametric Wilcoxon rank sum test. For all other experiments, statistical significance was calculated using unpaired, two-tailed Students t-test. p-values were considered statistically significant if the p-value was <0.05. Error bars represent S.E.M. unless otherwise indicated.
For all experiments with statistical data, the number of independent biological experiments are described in each figure legend. For immunoblot analyses in Figs. 1b, 1f-i, 4b, 4e, 6d-f, and extended data figs. 1a, 2b, 7a, 7d, 7k, 7l, and 10g-k, at least 3 biological independent experiments were performed and representative immunoblots are shown. For immunoblot analyses in Figs. 1c, 3a, and Extended Data Figs. 3c, immunoblots contains multiple independent tumors from independent mice. For immunohistochemistry and immunofluorescence experiments in Figs. 1j and Extended Data Figs. 2c,2e-g, representative micrographs are shown from 6 independent Kdm6a-WT tumors and 8 independent Kdm6a-Mutant tumors and the data are quantified in Extended Data Fig. 2d. Micrographs in Figs. 4a, 4c, and 4d are representative from 3 biological independent experiments. Micrograph in Extended Data Fig. 3b is representative of IHC from 3 independent Kdm6a-Mutant tumors. Extended Data Fig. 5d shows 3 independent tumors from independent mice with the positive control in Extended Data Fig. 5e.
For the initial mouse experiments in Fig. 1, at least 8 mice per arm were included and mice were completely randomized to receive sgKdm6a RPP or sgControl RPP adenoviruses. For all sequencing experiments (RNA-seq, ATAC-seq, scRNA-seq, and ChIP-seq) the investigators that processed and sequenced the samples [DFCI’s CFCE core (ATAC-seq), DFCI TIGL core (scRNA-seq), or Novogene (RNA-seq and ChIP-seq)] were blinded to the identity of the samples. For all cell culture experiments, each experiment was repeated in at least 3 biological independent experiments as specified in legends. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications26,30. Data distribution was assumed to be a normal distribution, but this was not formally tested. For cell culture experiments, blinding was not possible. For all experiments, no data were excluded from any analyses.
Extended Data
Extended Data Fig. 1. KDM6A Inactivation in an Autochthonous SCLC Mouse Model Promotes NEUROD1 Expression Leading to SCLC Tumors that Express both ASCL1 and NEUROD1.
(a) Immunoblot analysis of 293T cells infected with adenoviruses that encoded Cre recombinase and the indicated sgRNAs. (b) Kaplan-Meier survival analysis of LSL-Cas9 mice IT injected with the indicated adenoviruses. p=0.3447 for sgKdm6a RPP vs. sgControl RPP (non-targeting), p=0.2781 for sgKdm6a RPP vs. sgControl RPP (intron-targeting), p=0.3481 for sgControl RPP (non-targeting) vs. sgControl RPP (intron-targeting). n=11 mice sgControl RPP (non-targeting), n=5 mice sgControl RPP (intron-targeting), and n=8 mice (sgKdm6a#4 RPP). (c) Principal component analysis (PCA) of gene expression from RNA-seq data of 6 Kdm6a-WT SCLC mice tumors (18,221, 18,222, 18,227, 535, 542, 645) and 7 Kdm6a-Mutant SCLC mice tumors (236L, 236R, 656, 651L, 651R, 672-1, 672-2) in Fig. 1d. (d, e) mRNA expression of ASCL1 vs. PAX6 (D) or NEUROD1 vs. PAX6 (e) from publicly available RNA-seq data set of 81 human SCLC samples from George et al. Nature 20154. p-values are generated from cbioportal.org. (f) Gene set enrichment analysis (GSEA) of RNA-seq data (from Fig. 1d, Extended Data Fig. 1c) of NEUROD1 correlated genes (401 genes; see Borromeo et al. Cell Reports 201629, see Supplemental Table 2)29. FDR q-value calculated using GSEA is indicated. (g-k) mRNA expression from the RNA-seq data (from Fig. 1d) for Ascl1 (g), Neurod1 (h), Chromogranin A (i), Synaptophysin (j) and Insm1 (k) of individual Kdm6a-WT and Kdm6a-Mutant mouse SCLC tumors. For i, j, and k, lower graphs show average mRNA expression in Kdm6a-WT vs. Kdm6a-Mutant lung tumors (see Supplementary Table 2). For i,j,k, data are presented as mean values ± SEM. Statistical significance was calculated using unpaired, two-tailed students t-test and p-values are indicated on each figure panel. n=6 Kdm6a-WT tumors from independent mice and n=7 Kdm6a-Mutant tumors.
Extended Data Fig. 2. IHC and ATAC-seq Data from Kdm6a-Mutant vs. Kdm6a-WT Autochthonous SCLC tumors from Figs. 1&2.
(a) RT-qPCR for Neurod1 at the times indicated after transduction of mouse SCLC cell lines derived from Kdm6a-WT mice (1014) with 2 independent Kdm6a sgRNAs or a non-targeting sgRNA (sgControl). Data are relative to Actb and then normalized to Neurod1 expression in sgControl sample. n=2 biological independent experiments. Data are presented as mean values ± SEM. Statistical significance was calculated using unpaired, two-tailed students t-test and p-values are indicated. (b) Immunoblot analysis of the cells from a 24 days after transduction. NCI-H82 cells, a human NEUROD1-high SCLC cell line, is used a positive control for NEUROD1 expression. (c) Immunohistochemistry (IHC) for ASCL1 and NEUROD1 from 3 Kdm6a-WT and 3 Kdm6a-Mutant mouse SCLC lung tumors. Scale Bar=50 μm. (d) Quantification of ASCL1-positive, NEUROD1-positive and ASCL1/NEUROD1-negative (double-negative) cells in Kdm6a-WT tumors (n=6 tumors) and Kdm6a-Mutant tumors (n=7 tumors) from the multiplex-IF data in Fig. 1j. (e) Representative H&E and IHC staining for NEUROD1 and ASCL1 of a Kdm6a-Mutant tumor (236L). (f,g) Representative H&E of a Kdm6a-Mutant tumor (656) (f) and a Kdm6a-WT tumor (1015) (g). For e and f, black arrows show enlarged nuclei of NEUROD1-positive cells with occasional multi-nucleated giant cells. Scale Bar=20 μm. (h) Bar graph of upregulated (red) and downregulated (blue) differential accessible peaks with LFC>2, pAdj<0.05 from ATAC-seq data in Fig. 2a in Kdm6a-Mutant vs Kdm6a-WT tumors. p-values are calculated using Wald test in DEseq2 and adjusted for multiple hypothesis testing. (i) Pie charts of genomic location of differential accessible peaks from a. (j) Heat maps of ATAC-seq read densities from pseudo-bulk analysis of scATAC-seq data from Fig. 2f showing gain (n=11,760 peaks) or loss (n=10,587) of accessibility of Kdm6aMutantNEUROD1Only vs Kdm6aWT SCLC tumors (see Extended Data Fig. 3d).
Extended Data Fig. 3. Analysis of Bulk ATAC-seq from Kdm6a-Mutant vs. Kdm6a-WT Tumors from Fig. 3.
(a) RT-qPCR for Ascl1 mRNA expression relative to Actb and then normalized to Ascl1 expression in the 236L cell line of mouse SCLC lung tumors from Fig. 3a. (b) IHC for ASCL1 and NEUROD1 from the 1119 Kdm6a-Mutant mouse SCLC tumor showing rare NEUROD1-positive cells. Scale Bar= 50 μm. (c) Immunoblot analysis of SCLC lung tumors formed in an additional cohort of LSL-Cas9 mice injected with sgControl RPP (Kdm6a-WT) or sgKdm6a RPP (Kdm6a-Mutant) adenoviruses. For the immunoblot on the left, a separate piece of each tumor was used for bulk ATAC-seq and scRNA-seq experiments and hence immunoblots were run for each sample. Note that the piece of tumor 1270 used for ATAC-seq did show faint NEUROD1 expression while the 1270 sample used for scRNA-seq did not show any detectable NEUROD1 expression. This was likely related to heterogeneity within the tumor sample. (d) Bar graph of upregulated (red) and downregulated (blue) differential accessible peaks with LFC>2, pAdj<0.05 from ATAC-seq data used to identify accessibility changes in Kdm6aMutantNEUROD1Only tumors for analyses in Figs. 3e,f and hence differential accessibility analysis was performed comparing Kdm6aMutantNEUROD1Only tumors (236, 656) vs. Kdm6aWT SCLC tumors (158, 18227, 165, 1198, 168, 645, 535, sc535). p-values are calculated using Wald test in DEseq2 and adjusted for multiple hypothesis testing. (e) Pie charts of genomic location of differential accessible peaks from d. (f,g) Chromatin accessibility tracks for the average of each phenotype indicated (see Figs. 3c-g) at Neurod1 (f) or Ascl1 (g) from all ATAC-seq data from Fig. 3d. (h, i) PCA of all ATAC-seq data from Fig. 3c (h) and Fig. 3d (i) now classified by genotype (Kdm6a-Mutant vs. Kdm6a-WT) rather than phenotype. (j) Heat maps of ATAC-seq read densities from all bulk ATAC-seq data from Figs. 3d-f showing gain (n=11,760 peaks) or loss (n=10,587) of accessibility in Kdm6aMutantNEUROD1Only vs Kdm6aWT SCLC tumors (see Extended Data Fig. 3d) for all phenotypes indicated.
Extended Data Fig. 4. Additional scRNA-seq Data from Kdm6a-Mutant vs. Kdm6a-WT Tumors from Fig. 3 and Additional RT-qPCR Data from Tumor Derived Cell Lines from Fig. 4.
(a) Dot plot showing the average expression of marker genes to identify immune cell subpopulations of all cells in Fig 3h. The size of the dot represents the percentage of cells expressing a particular gene while color represents the mean gene expression levels (blue is low and red is high). (b) Elbow plot showing the standard deviation associated with the top 40 PCs in the tumor population. (c) Feature plot of Ascl1 of all tumor cells from scRNA-seq data in Kdm6a-Mutant vs. Kdm6a-WT (see Fig. 3i). (d) Neurod1 violin plot using scRNA-seq data from Fig. 3j of tumor cells from Kdm6a-Mutant and Kdm6a-WT tumors. For d, non-parametric Wilcoxon rank sum test was used to generate a two-tailed p-value adjusted for multiple comparisons by Bonferroni correction. n=35,446 Kdm6a-Mutant and n=6,612 Kdm6a-WT tumor cells. Each individual dot represents a cell. (e-j) RT-qPCR for Chromogranin A (e), Synaptophysin (f), Ascl1 (g), Neurod1 (h) Pou2f3 (i) and Yap1 (j) mRNA expression in the early vs. late passage tumor-derived cell lines of individual Kdm6a-WT and Kdm6a-Mutant (upper graphs) or average mRNA expression in Kdm6a-WT vs. Kdm6a-Mutant cell lines (lower graphs). n=6 biological independent experiments for each genotype (2 biological independent experiments for each cell line). Data are presented as mean values ± SEM. Statistical significance was calculated using unpaired, two-tailed students t-test and p-values are indicated on each figure panel. For all t-tests comparing Kdm6a-Mutant vs. Kdm6a-WT cell lines, all early passage Kdm6a-Mutant were compared to all early passage Kdm6a-WT cell lines.
Extended Data Fig. 5. Comparison of Kdm6a-Mutant SCLC GEMM with the Myc-driven RPM SCLC GEMM using RNA-seq and IHC from tumors.
(a-c) mRNA expression from the RNA-seq data (from Fig. 1d) for Myc (a), Mycl (b) and Mycn (c) of individual Kdm6a-WT and Kdm6a-Mutant mouse SCLC tumors (top) or combined tumors by genotype (bottom). See Supplementary Table 2. For a,b,c, data are presented as mean values ± SEM. Statistical significance was calculated using unpaired, two-tailed students t-test and p-values are indicated on each figure panel. n=6 Kdm6a-WT tumors from independent mice and n=7 Kdm6a-Mutant tumors. (d) H&E and IHC staining for ASCL1, NEUROD1, c-Myc and Synaptophysin from 3 Kdm6a-WT (1014, 1015, 222) and 3 Kdm6a-Mutant (656, 236R, 670) mouse SCLC lung tumors. Scale Bar=50 μm. (e) IHC staining for c-Myc from tumor adjacent lymphocytes and high c-Myc prostate cancer GEMM as positive control of c-Myc staining. Scale Bar=50 μm. (f-h) GSEA of genes upregulated in the RPM vs. RPP GEMM13 with publicly available scRNA-seq data from human SCLCs of the ASCL1 and NEUROD1 subtypes34 (see Methods) (f), or with RNA-seq data from Kdm6a-Mutant vs. Kdm6a-WT tumors from Fig. 1d (g), or using GSEA of genes upregulated in Kdm6a-Mutant tumors (see Fig. 1e) with late RPM vs. early RPM subtypes13. FDR q-values from GSEA adjusted for multiple hypothesis testing are indicated.
Extended Data Fig. 6. Myc and Mycl mRNA Expression and Chromatin Accessibility in Kdm6a-Mutant vs. Kdm6a-WT Tumors.
(a-d) Feature plots with violin plot of single cells from tumors for Myc (a,b) and Mycl (c,d) expression from scRNA-seq data from Fig. 3i. For b,d, non-parametric Wilcoxon rank sum test was used to generate two-tailed p-values adjusted for multiple comparisons by Bonferroni correction. n=35,446 Kdm6a-Mutant and n=6,612 Kdm6a-WT tumor cells. Each individual dot represents a cell. Minimum and maximum values defined the range of violin plot. (e-g) Chromatin accessibility tracks for the average of each phenotype indicated (see Figs. 3c-g) at Myc (e), Mycl (f) and Mycn (g) from all ATAC-seq data from Fig. 3d.
Extended Data Fig. 7. Analysis of c-Myc and L-Myc in Kdm6a-Mutant vs. Kdm6a-WT Tumor Derived Cell Lines.
(a) Immunoblot analysis for c-Myc with a c-Myc antibody (Y69) in Kdm6a-WT and Kdm6a-Mutant SCLC cell lines cultured in ultra-low attachment flasks at late times (>2 months) after cell line generation. NCI-H82 and CORL279 human SCLC cell lines are included as benchmark controls for SCLCs with high c-Myc expression. (b, c) RT-qPCR for Myc mRNA expression in the early vs. late passage Kdm6a-WT and Kdm6a-Mutant tumor-derived cell lines shown as individual lines (b) or grouped by genotype (c). (d) Immunoblot analysis for L-Myc with a specific L-Myc antibody (Cell Signaling E3M5P) in Kdm6a-WT and Kdm6a-Mutant SCLC cell lines cultured in ultra-low attachment flasks at late times (>2 months) after cell line generation. CORL47 and NCI-H1092 human SCLC cell lines are included as benchmark controls for SCLC cell lines with high L-Myc expression. CORL47 and NCI-H1092 express the RLF-MYCL fusion and hence MYCL migrates at higher molecular weights compared to WT L-Myc. (e-h) RT-qPCR for Mycl (e,f) or Mycn (g,h) mRNA expression in the early vs. late passage Kdm6a-WT and Kdm6a-Mutant lines shown as individual lines (e,g) or grouped by genotype (f,h). For all RT-qPCR experiments, n=6 biological independent experiments for each genotype (2 biological independent experiments for each cell line). Data are presented as mean values ± SEM. Statistical significance was calculated using unpaired, two-tailed students t-test and p-values are indicated. For all t-tests comparing Kdm6a-Mutant vs. Kdm6a-WT cell lines, early passage Kdm6a-Mutant vs. Kdm6a-WT were compared. (I,j) Tracks of H3K4me1, and KDM6A ChIP-seq for Mycl (i), and Myc (j) from ChIP-seq data from Fig. 5. Each track is the sum of 2 Kdm6a-WT cell lines (1014, 159-1) and 2 Kdm6a-Mutant cell lines (236L,236R) peaks with their respective input. (k) Immunoblot analysis for c-Myc and L-Myc in a Kdm6a-Mutant SCLC cell line (672-2) using highly specific antibodies with benchmark controls above. (l) Immunoblot analysis of 672-2 cells transduced with 4 independent Myc sgRNAs or a non-targeting control (sgControl) acutely after transduction and puromycin selection.
Extended Data Fig. 8. Correlation and GSEA Analyses of KDM6A, H3K27me3, H3K4me1, and H3K4me2 ChIP-seq Data in Kdm6a-WT and Kdm6a-Mutant SCLC Primary Cell Lines.
(a) Genome-wide correlation of log fold change in H3K27me3 ChIP-seq (Kdm6a-Mutant/Kdm6a-WT) vs. log fold change in KDM6A ChIP-seq at enhancers. (b) Genome-wide correlation of log fold change in H3K4me1 ChIP-seq (Kdm6a-Mutant/Kdm6a-WT) vs. log fold change in KDM6A ChIP-seq at enhancers. (c) Genome-wide correlation of log fold change in H3K4me2 ChIP-seq (Kdm6a-Mutant/Kdm6a-WT) vs. log fold change in KDM6A ChIP-seq at enhancers. For a-c, r Pearson correlation coefficient and p-value are indicated. p-values were calculated using a two-sided Pearson’s correlation test and are indicated. (d, e) Normalized Enrichment Score from GSEA of H3K27me3 ChIP-seq at transcription start sites of conserved ASCL1 target genes or conserved NEUROD1 target genes (d) or top 5 enriched Hallmarks (e) from the ChIP-seq data in Fig. 5. (f, g) Normalized Enrichment Score from GSEA of H3K4me2 ChIP-seq at enhancers of conserved ASCL1 target genes or conserved NEUROD1 target genes (f) or top 5 enriched Hallmarks (g) from the ChIP-seq data in Fig. 5. For d-g, p-values were generated by GSEA using a permutation test adjusted for multiple hypothesis testing using the Benjamini-Hochberg correction. Adjusted p-values are indicated.
Extended Data Fig. 9. NEUROD1 Expression after KDM6A Inactivation is Partially Mediated by KMT2A.
(a,b) Tracks of H3K27me3 and KDM6A ChIP-seq of Neurod1 (a) and Ascl1 (b) from ChIP-seq data from Fig. 5. Each track is the sum of the peaks of: 2 Kdm6a-WT tumor-derived cell lines for Kdm6a-WT (1014, 159-1) and 2 Kdm6a-Mutant tumor-derived cell lines for Kdm6a-Mutant (236L, 236R) with their respective input. (c) Cistrome analysis of transcription factors and chromatin regulators of Neurod1 gene in human hg38 (http://dbtoolkit.cistrome.org/) within 10 kilobases of the Neurod1 gene. (d) Pseudo-bulk differentially expressed gene (DEG) analysis from the scRNA-seq data in Fig. 3i in Kdm6a-Mutant vs Kdm6a-WT tumors cells of the genes in c. Non-parametric Wilcoxon rank sum test was used and p-values adjusted for multiple hypothesis testing are shown. (e) List of the top 20 candidate regulators of accessible peaks determined by LISA analysis of the 100 top differentially accessible peaks at TSSs with LFC>1 sorted by ascending p-value in Kdm6a-Mutant vs. Kdm6a-WT using ATAC-seq data from Fig. 2a (see Supplementary Table 8, tab 1 for complete list).
Extended Data Fig. 10. Analyses of KMT2D Loss or Hypoxia with NEUROD1 Expression in SCLC.
(a) Correlation of KDM6A dependency vs. KMT2D depedency across hundreds of cancer cell lines25. KMT2D is the #1 co-dependency with KDM6A (p-value=9.09x10−67 calculated using dependency map25). (b) GSEA using publicly available datasets of human SCLCs with KMT2D LOF mutations or KMT2D WT4 using genes upregulated in Kdm6a-Mutant vs. Kdm6a-WT SCLC GEMMs (see Fig. 1d). FDR q-value generated using GSEA, adjusted for multiple-hypothesis testing, is indicated. (c) Proportion of mixed ASCL1 and NEUROD1 human SCLCs from RNA-seq data from 3 independent data sets4,41,84 (see methods) in SCLCs with KMT2D LOF mutations vs. KMT2D WT. Fisher’s exact test was used to generate p-value. For b,c, n=14 KMT2D LOF, n=142 KMT2D WT. See also Supplementary Table 9. (d, e) CRISPR amplicon sequencing of 1014 Kdm6a-WT cells transduced with 2 independent Kmt2d sgRNAs. sgRNA sequences are in blue and gene editing is in red. (f) RT-qPCR for Neurod1 in 1014 Kmt2d knockout cells in d,e 3 weeks after transduction. Data are relative to Actb and then normalized to Neurod1 expression in the sgControl (non-targeting) cell line. n=4 biological replicates and error bars represent mean ± SEM. Statistical significance was calculated using unpaired, two-tailed students t-test and p-values are indicated. (g,h) Immunoblot (g) and histone blot (h) analyses in 1014 cells transduced with 4 independent Kmt2c and Kmt2d sgRNAs or a non-targeting control (sgControl) 30 days post-transduction. (i,j) Immunoblot analysis of NEUROD1 and ASCL1 expression of (i) NCI-H69 and (j) DMS79 ASCL1-positive human SCLC cells cultured under 5% or 21% O2 for 7 days. (k) Immunoblot analysis for HIF1α protein of cells lines in i,j. (l, m) GSEA analysis of scRNA-seq from human ASCL1 or NEUROD1 SCLC tumors34 using (l) HIF1α target gene list51 or (m) 24 genes induced by hypoxia50 (see methods) (see Supplemental Table 10). FDR q-value adjusted for multiple hypothesis testing is indicated. Model-based Analysis of Single Cell Transcriptomics (MAST) was used to generate the GSEA expression profile.
Supplementary Material
Acknowledgements:
We thank Dr. Abhishek Chakraborty for insightful discussions and members of the Oser, Barbie, and Janne laboratories for thoughtful comments during our lab meetings. M.G.O. is a William Raveis Charitable Fund Damon Runyon Clinical Investigator. This works was supported by the Damon Runyon Cancer Research Foundation (CI-101-19, M.G.O.), an NCI/NIH K08 grant (no. K08CA222657, M.G.O.), NCI R37 grant (no. R37CA269990, M.G.O.), and the Kaplan Family Fund (M.G.O.). S.A.A. was supported by NIH grants CA176745, CA259273 and CA066996. S.L. was supported by the NCI Research Specialist Award (R50CA251956).
Footnotes
Competing Interests: M.G.O. has (currently or previously) sponsored research agreements (SRAs) with Takeda, Eli Lilly, Novartis, Circle Pharma, and Bristol Myers Squibb, and is a consultant for Daiichi Sankyo and Ananke Therapeutics. None of these SRAs were used to fund this work. S.A.A. has been a consultant and/or shareholder for Neomorph Inc, Imago Biosciences, Cyteir Therapeutics, C4 Therapeutics, Nimbus Therapeutics and Accent Therapeutics. S.A.A. has received research support from Janssen and Syndax. S.A.A. is named as an inventor on a patent application related to MENIN inhibition WO/2017/132398A1. The remaining authors declare no competing interests.
Data Availability
Data from bulk RNA-sequencing experiments in Fig. 1 are included as Supplementary Table 2. Data from bulk ATAC-sequencing experiments from Fig. 2 or Fig. 3 are included as Supplementary Tables 3 and 4. Data from single-cell RNA-seq experiments in Fig. 3 are included as Supplementary Table 5. Data from Extended Data Figs. 5-7 are included as Supplementary Table 6. Data from ChIP-sequencing experiments in Fig. 5 are included as Supplementary Table 7. Data from Menin ChIP-sequencing and LISA analysis in Fig. 6 are included as Supplementary Table 8. Data from Extended Data Fig. 10 are included as Supplementary Tables 9,10. All source data are included within the supplementary information associated with this article. RNA–seq, ATAC-seq and ChIP-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE228347 (RNA-seq Fig. 1, Menin-ChiP seq Fig. 6), 2. GSE215829 (ATAC-seq Figs. 2,3) 3. GSE211242 (scRNA-seq Fig. 3), 4. ChIP-seq Data: GSE211165 (ChIP-seq Fig. 5). Any other data and materials can be requested from the corresponding author upon reasonable request.
Code Availability
Custom code for ATAC-seq analysis in Figs. 2,3 is here: https://bitbucket.org/cfce/viper/src/master/, https://bitbucket.org/cfce/cobra/src/master/ (ATAC-seq experiments). Custom R code for ChIP-seq analysis in Fig. 5 is here: DOI 10.5281/zenodo.7786472.
References
- 1.Kalemkerian GP et al. Small cell lung cancer. J Natl Compr Canc Netw 11, 78–98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudin CM, Brambilla E, Faivre-Finn C & Sage J Small-cell lung cancer. Nat Rev Dis Primers 7, 3, doi: 10.1038/s41572-020-00235-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudin CM et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 19, 289–297, doi: 10.1038/s41568-019-0133-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George J. et al. Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53, doi: 10.1038/nature14664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peifer M. et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44, 1104–1110, doi: 10.1038/ng.2396 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudin CM et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44, 1111–1116, doi: 10.1038/ng.2405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gay CM et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 39, 346–360 e347, doi: 10.1016/j.ccell.2020.12.014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baine MK et al. POU2F3 in SCLC: Clinicopathologic and Genomic Analysis With a Focus on Its Diagnostic Utility in Neuroendocrine-Low SCLC. J Thorac Oncol 17, 1109–1121, doi: 10.1016/j.jtho.2022.06.004 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baine MK et al. SCLC Subtypes Defined by ASCL1, NEUROD1, POU2F3, and YAP1: A Comprehensive Immunohistochemical and Histopathologic Characterization. J Thorac Oncol 15, 1823–1835, doi: 10.1016/j.jtho.2020.09.009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qu S. et al. Molecular Subtypes of Primary SCLC Tumors and Their Associations With Neuroendocrine and Therapeutic Markers. J Thorac Oncol 17, 141–153, doi: 10.1016/j.jtho.2021.08.763 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang YH et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev 32, 915–928, doi: 10.1101/gad.314815.118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cejas P. et al. Subtype heterogeneity and epigenetic convergence in neuroendocrine prostate cancer. Nat Commun 12, 5775, doi: 10.1038/s41467-021-26042-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ireland AS et al. MYC Drives Temporal Evolution of Small Cell Lung Cancer Subtypes by Reprogramming Neuroendocrine Fate. Cancer Cell 38, 60–78 e12, doi: 10.1016/j.ccell.2020.05.001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Augert A. et al. Small Cell Lung Cancer Exhibits Frequent Inactivating Mutations in the Histone Methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance). J Thorac Oncol 12, 704–713, doi: 10.1016/j.jtho.2016.12.011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agger K. et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734, doi: 10.1038/nature06145 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Lee MG et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318, 447–450, doi: 10.1126/science.1149042 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Tran N, Broun A & Ge K Lysine Demethylase KDM6A in Differentiation, Development, and Cancer. Mol Cell Biol 40, doi: 10.1128/MCB.00341-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JH et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res 74, 1705–1717, doi: 10.1158/0008-5472.CAN-13-1896 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L & Shilatifard A UTX Mutations in Human Cancer. Cancer Cell 35, 168–176, doi: 10.1016/j.ccell.2019.01.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang SP et al. A UTX-MLL4-p300 Transcriptional Regulatory Network Coordinately Shapes Active Enhancer Landscapes for Eliciting Transcription. Mol Cell 67, 308–321 e306, doi: 10.1016/j.molcel.2017.06.028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andricovich J. et al. Loss of KDM6A Activates Super-Enhancers to Induce Gender-Specific Squamous-like Pancreatic Cancer and Confers Sensitivity to BET Inhibitors. Cancer Cell 33, 512–526 e518, doi: 10.1016/j.ccell.2018.02.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faralli H. et al. UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J Clin Invest 126, 1555–1565, doi: 10.1172/JCI83239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gozdecka M. et al. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat Genet 50, 883–894, doi: 10.1038/s41588-018-0114-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morales Torres C, Laugesen A & Helin K Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS One 8, e60020, doi: 10.1371/journal.pone.0060020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsherniak A. et al. Defining a Cancer Dependency Map. Cell 170, 564–576 e516, doi: 10.1016/j.cell.2017.06.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oser MG et al. The KDM5A/RBP2 histone demethylase represses NOTCH signaling to sustain neuroendocrine differentiation and promote small cell lung cancer tumorigenesis. Genes Dev 33, 1718–1738, doi: 10.1101/gad.328336.119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty AA et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363, 1217–1222, doi: 10.1126/science.aaw1026 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalisz M. et al. HNF1A recruits KDM6A to activate differentiated acinar cell programs that suppress pancreatic cancer. EMBO J 39, e102808, doi: 10.15252/embj.2019102808 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borromeo MD et al. ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell Rep 16, 1259–1272, doi: 10.1016/j.celrep.2016.06.081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong D. et al. Plasticity in the Absence of NOTCH Uncovers a RUNX2-Dependent Pathway in Small Cell Lung Cancer. Cancer Res, doi: 10.1158/0008-5472.CAN-21-1991 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mollaoglu G. et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 31, 270–285, doi: 10.1016/j.ccell.2016.12.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaffer BE et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res 70, 3877–3883, doi: 10.1158/0008-5472.CAN-09-4228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marsich E, Vetere A, Di Piazza M, Tell G & Paoletti S The PAX6 gene is activated by the basic helix-loop-helix transcription factor NeuroD/BETA2. Biochem J 376, 707–715, doi: 10.1042/BJ20031021 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan JM et al. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell 39, 1479–1496 e1418, doi: 10.1016/j.ccell.2021.09.008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLean CY et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28, 495–501, doi: 10.1038/nbt.1630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahadevan NR et al. Intrinsic immunogenicity of small cell lung carcinoma revealed by its cellular plasticity. Cancer Discov, doi: 10.1158/2159-8290.CD-20-0913 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young MD et al. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res 39, 7415–7427, doi: 10.1093/nar/gkr416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin Q. et al. Lisa: inferring transcriptional regulators through integrative modeling of public chromatin accessibility and ChIP-seq data. Genome Biol 21, 32, doi: 10.1186/s13059-020-1934-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krivtsov AV & Armstrong SA MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 7, 823–833, doi: 10.1038/nrc2253 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Krivtsov AV et al. A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL-Rearranged Leukemia. Cancer Cell 36, 660–673 e611, doi: 10.1016/j.ccell.2019.11.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lissa D. et al. Heterogeneity of neuroendocrine transcriptional states in metastatic small cell lung cancers and patient-derived models. Nat Commun 13, 2023, doi: 10.1038/s41467-022-29517-9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Easwaran H, Tsai HC & Baylin SB Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell 54, 716–727, doi: 10.1016/j.molcel.2014.05.015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flavahan WA, Gaskell E & Bernstein BE Epigenetic plasticity and the hallmarks of cancer. Science 357, doi: 10.1126/science.aal2380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart CA et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat Cancer 1, 423–436, doi: 10.1038/s43018-019-0020-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simpson KL et al. A biobank of small cell lung cancer CDX models elucidates inter- and intratumoral phenotypic heterogeneity. Nat Cancer 1, 437–451, doi: 10.1038/s43018-020-0046-2 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Adam MP et al. Kabuki syndrome: international consensus diagnostic criteria. J Med Genet 56, 89–95, doi: 10.1136/jmedgenet-2018-105625 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Cheon CK et al. Identification of KMT2D and KDM6A mutations by exome sequencing in Korean patients with Kabuki syndrome. J Hum Genet 59, 321–325, doi: 10.1038/jhg.2014.25 (2014). [DOI] [PubMed] [Google Scholar]
References for Methods
- 48.DuPage M, Dooley AL & Jacks T Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4, 1064–1072, doi: 10.1038/nprot.2009.95 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L. et al. Validation of a hypoxia related gene signature in multiple soft tissue sarcoma cohorts. Oncotarget 9, 3946–3955, doi: 10.18632/oncotarget.23280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benita Y. et al. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res 37, 4587–4602, doi: 10.1093/nar/gkp425 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bankhead P. et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878, doi: 10.1038/s41598-017-17204-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buenrostro JD et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490, doi: 10.1038/nature14590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taing L. et al. CHIPS: A Snakemake pipeline for quality control and reproducible processing of chromatin profiling data [version 1; peer review: 1 not approved]. F1000Research 10, doi: 10.12688/f1000research.52878.1 (2021). [DOI] [Google Scholar]
- 55.Qiu X. et al. CoBRA: Containerized Bioinformatics Workflow for Reproducible ChIP/ATAC-seq Analysis. Genomics Proteomics Bioinformatics 19, 652–661, doi: 10.1016/j.gpb.2020.11.007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, doi: 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137–R137, doi: 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson JT et al. Integrative genomics viewer. Nat Biotechnol 29, 24–26, doi: 10.1038/nbt.1754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neph S. et al. BEDOPS: high-performance genomic feature operations. Bioinformatics 28, 1919–1920, doi: 10.1093/bioinformatics/bts277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinz S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589, doi: 10.1016/j.molcel.2010.05.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramirez F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165, doi: 10.1093/nar/gkw257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cornwell M. et al. VIPER: Visualization Pipeline for RNA-seq, a Snakemake workflow for efficient and complete RNA-seq analysis. BMC Bioinformatics 19, 135–135, doi: 10.1186/s12859-018-2139-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trapnell C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 511–515, doi: 10.1038/nbt.1621 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L, Wang S & Li W RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185, doi: 10.1093/bioinformatics/bts356 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550–550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842, doi: 10.1093/bioinformatics/btq033 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zerbino DR, Johnson N, Juettemann T, Wilder SP & Flicek P WiggleTools: parallel processing of large collections of genome-wide datasets for visualization and statistical analysis. Bioinformatics 30, 1008–1009, doi: 10.1093/bioinformatics/btt737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Satpathy AT et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol 37, 925–936, doi: 10.1038/s41587-019-0206-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Satpathy AT et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol 37, 925–936, doi: 10.1038/s41587-019-0206-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stuart T. et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821, doi: 10.1016/j.cell.2019.05.031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stuart T, Srivastava A, Madad S, Lareau CA & Satija R Single-cell chromatin state analysis with Signac. Nat Methods 18, 1333–1341, doi: 10.1038/s41592-021-01282-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Platt RJ et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455, doi: 10.1016/j.cell.2014.09.014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hao Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529, doi: 10.1016/j.cell.2021.04.048 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aran D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20, 163–172, doi: 10.1038/s41590-018-0276-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heng TS, Painter MW & Immunological Genome Project C The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9, 1091–1094, doi: 10.1038/ni1008-1091 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Benayoun BA et al. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res 29, 697–709, doi: 10.1101/gr.240093.118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kharchenko PV, Tolstorukov MY & Park PJ Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26, 1351–1359, doi: 10.1038/nbt.1508 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robinson JT et al. Integrative genomics viewer. Nat. Biotechnol 29, 24–26, doi: 10.1038/nbt.1754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thorvaldsdóttir H, Robinson JT & Mesirov JP Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics 14, 178–192, doi: 10.1093/bib/bbs017 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Casper J. et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res 46, D762–D769, doi: 10.1093/nar/gkx1020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moore JE et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710, doi: 10.1038/s41586-020-2493-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caeser R. et al. Genomic and transcriptomic analysis of a library of small cell lung cancer patient-derived xenografts. Nat Commun 13, 2144, doi: 10.1038/s41467-022-29794-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from bulk RNA-sequencing experiments in Fig. 1 are included as Supplementary Table 2. Data from bulk ATAC-sequencing experiments from Fig. 2 or Fig. 3 are included as Supplementary Tables 3 and 4. Data from single-cell RNA-seq experiments in Fig. 3 are included as Supplementary Table 5. Data from Extended Data Figs. 5-7 are included as Supplementary Table 6. Data from ChIP-sequencing experiments in Fig. 5 are included as Supplementary Table 7. Data from Menin ChIP-sequencing and LISA analysis in Fig. 6 are included as Supplementary Table 8. Data from Extended Data Fig. 10 are included as Supplementary Tables 9,10. All source data are included within the supplementary information associated with this article. RNA–seq, ATAC-seq and ChIP-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE228347 (RNA-seq Fig. 1, Menin-ChiP seq Fig. 6), 2. GSE215829 (ATAC-seq Figs. 2,3) 3. GSE211242 (scRNA-seq Fig. 3), 4. ChIP-seq Data: GSE211165 (ChIP-seq Fig. 5). Any other data and materials can be requested from the corresponding author upon reasonable request.
Custom code for ATAC-seq analysis in Figs. 2,3 is here: https://bitbucket.org/cfce/viper/src/master/, https://bitbucket.org/cfce/cobra/src/master/ (ATAC-seq experiments). Custom R code for ChIP-seq analysis in Fig. 5 is here: DOI 10.5281/zenodo.7786472.