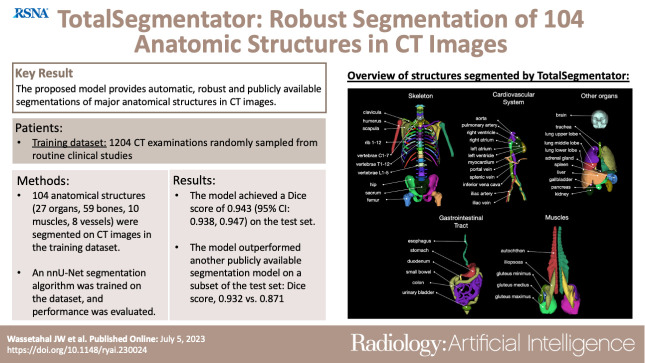
Abstract
Purpose
To present a deep learning segmentation model that can automatically and robustly segment all major anatomic structures on body CT images.
Materials and Methods
In this retrospective study, 1204 CT examinations (from 2012, 2016, and 2020) were used to segment 104 anatomic structures (27 organs, 59 bones, 10 muscles, and eight vessels) relevant for use cases such as organ volumetry, disease characterization, and surgical or radiation therapy planning. The CT images were randomly sampled from routine clinical studies and thus represent a real-world dataset (different ages, abnormalities, scanners, body parts, sequences, and sites). The authors trained an nnU-Net segmentation algorithm on this dataset and calculated Dice similarity coefficients to evaluate the model’s performance. The trained algorithm was applied to a second dataset of 4004 whole-body CT examinations to investigate age-dependent volume and attenuation changes.
Results
The proposed model showed a high Dice score (0.943) on the test set, which included a wide range of clinical data with major abnormalities. The model significantly outperformed another publicly available segmentation model on a separate dataset (Dice score, 0.932 vs 0.871; P < .001). The aging study demonstrated significant correlations between age and volume and mean attenuation for a variety of organ groups (eg, age and aortic volume [rs = 0.64; P < .001]; age and mean attenuation of the autochthonous dorsal musculature [rs = −0.74; P < .001]).
Conclusion
The developed model enables robust and accurate segmentation of 104 anatomic structures. The annotated dataset (https://doi.org/10.5281/zenodo.6802613) and toolkit (https://www.github.com/wasserth/TotalSegmentator) are publicly available.
Keywords: CT, Segmentation, Neural Networks
Supplemental material is available for this article.
© RSNA, 2023
See also commentary by Sebro and Mongan in this issue.
Keywords: CT, Segmentation, Neural Networks
Summary
TotalSegmentator provides automatic, easily accessible segmentations of major anatomic structures on CT images.
Key Points
■ The proposed model was trained on a diverse dataset of 1204 CT examinations randomly sampled from routine clinical studies; the dataset contained segmentations of 104 anatomic structures (27 organs, 59 bones, 10 muscles, and eight vessels) that are relevant for use cases such as organ volumetry, disease characterization, and surgical or radiation therapy planning.
■ The model achieved a high Dice similarity coefficient (0.943; 95% CI: 0.938, 0.947) on the test set encompassing a wide range of clinical data, including major abnormalities, and outperformed other publicly available segmentation models on a separate dataset (Dice score, 0.932 vs 0.871; P < .001).
■ Both the training dataset (https://doi.org/10.5281/zenodo.6802613) and developed model (https://www.github.com/wasserth/TotalSegmentator) are publicly available.
Introduction
In the past few years, both the number of CT examinations performed and the available computing power have steadily increased (1,2). Moreover, the capability of image analysis algorithms has vastly improved given advances in deep learning techniques (3,4). The resulting increases in data, computational power, and algorithm quality have enabled radiologic studies using large sample sizes. For many of these studies, segmentation of anatomic structures plays an important role. Segmentation is useful for extracting advanced biomarkers based on radiologic images, automatically detecting abnormalities, or quantifying tumor load (5). In routine clinical analysis, segmentation is already used for applications such as surgical and radiation therapy planning (6). Thus, the associated algorithms could ultimately enter routine clinical use to improve the quality of radiologic reports and reduce radiologist workload.
For most applications, segmentation of the relevant anatomic structure is the first step. Building and training a segmentation algorithm, however, are complex because they require tedious manual annotation of training data and technical expertise for training the algorithm. Providing a ready-to-use segmentation toolkit that enables automatic segmentation of most of the major anatomic structures on CT images would considerably simplify many radiology studies, thereby accelerating research in the field.
Several publicly available segmentation models are currently available. However, these models are generally specific for a single organ (eg, the pancreas, spleen, colon, or lung) (7–11). They cover only a small subset of relevant anatomic structures and are trained on relatively small datasets that are not representative of routine clinical imaging, which is characterized by differences in contrast phases, acquisition settings, and diverse abnormalities. Thus, researchers must often build and train their own segmentation models, which can be costly.
To overcome this problem, we aimed to develop a model with the following characteristics: (a) publicly available (including its training data), (b) easy to use, (c) segments most anatomically relevant structures throughout the body, and (d) exhibits robust performance in any clinical setting. As an example application, we applied our segmentation model to a large dataset of 4004 patients with whole-body CT scans collected in a polytrauma setting and analyzed age-dependent changes of the volume and attenuation of different structures.
Materials and Methods
The Ethics Committee Northwest and Central Switzerland approved the ethics waiver for this retrospective study (EKNZ BASEC Req-2022–00495).
Datasets
Two datasets were aggregated for this study: one dataset for training the proposed model (training dataset) and a second dataset for the aging study example application (aging study dataset).
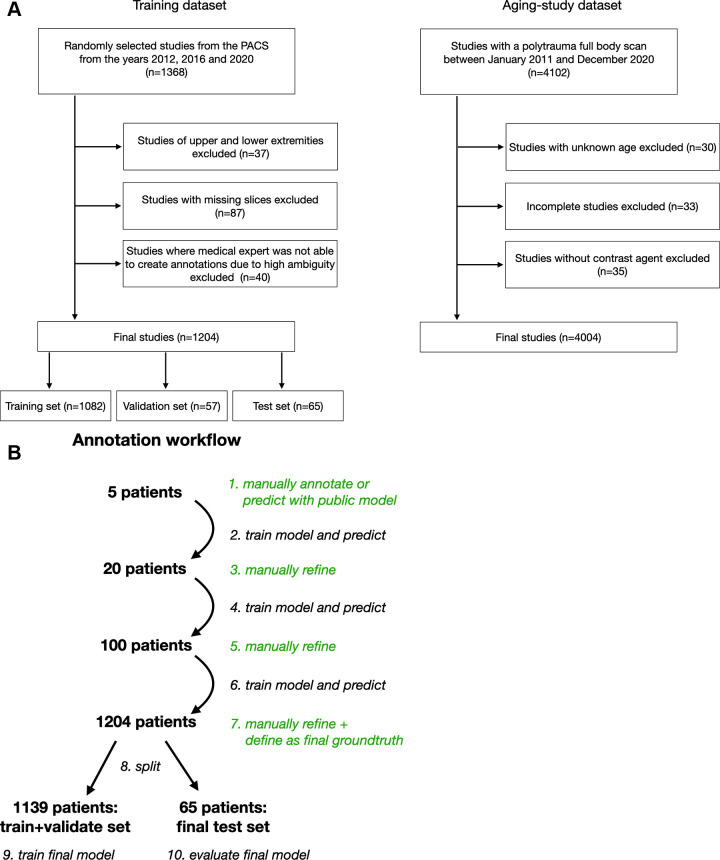
Training dataset.—To generate a comprehensive and highly variant dataset, 1368 CT examinations were randomly sampled from 2012, 2016, and 2020 from the University Hospital Basel picture archiving and communication system. CT series of upper and lower extremities (n = 37), CT series with missing slices (n = 87), and CT series for which the human annotator could not segment certain structures because of high ambiguity (eg, structures highly distorted as a result of abnormality) (n = 40) were excluded. The CT series were sampled randomly from each examination to obtain a wide variety of data. All images were resampled to 1.5-mm isotropic resolution. The final dataset of 1204 CT series was divided into a training dataset of 1082 patients (90%), a validation dataset of 57 patients (5%), and a test dataset of 65 patients (5%) (Fig 1A).
Figure 1:
(A) Diagram shows the inclusion of patients into the study. (B) Diagram shows the iterative annotation workflow of the training dataset. Steps involving manual annotation are shown in green. In step 9, a completely new model was trained independently of the intermediate models (steps 2, 4, and 6). This avoids leakage of information from the test set into the training set. PACS = picture archiving and communication system.
Aging study dataset.—All patients with polytrauma who underwent whole-body CT between 2011 and 2020 at the University Hospital Basel were initially included (n = 4102). Patients with unknown age (n = 30) or incomplete images (n = 33) or who underwent studies without administration of a contrast agent (n = 35) were excluded. In all patients, the same examination protocol was applied: contrast-enhanced, whole-body CT in an arteriovenous split-bolus phase, with a similar amount of contrast agent. Thus, we assumed that variation of attenuation (in Hounsfield units) due to the contrast agent was minimal for all images and that the attenuation values were comparable.
Data Annotation
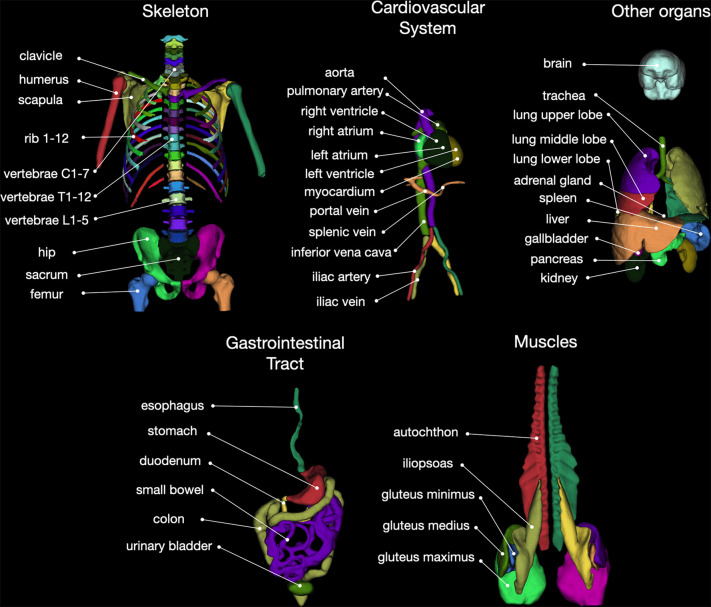
We identified 104 anatomic structures for segmentation (Fig 2, Appendix S1). The Nora Imaging Platform was used for manual segmentation or further refinement of generated segmentations (12). Segmentation was supervised by two physicians with 3 (M.S.) and 6 (H.C.B.) years of experience in body imaging, respectively. The work was split between them.
Figure 2:
Overview of all 104 anatomic structures segmented by the TotalSegmentator. autochthon = autochthonous dorsal musculature.
If an existing model for a given structure was publicly available (Appendix S2), that model was used to create a first segmentation, which was then validated and refined manually (10,13–16).
To speed the process further, we used an iterative learning approach, as follows. After manual segmentation of the first five patients was completed, a preliminary nnU-Net was trained, and its predictions were manually refined, if necessary. The nnU-Net was retrained after review and refinement of five patients, 20 patients, and 100 patients (Fig 1B).
In the end, all 1204 CT examinations had annotations that were manually reviewed and corrected whenever necessary. These final annotations served as the ground truth for training and testing. The model was trained on the dataset of 1082 patients, validated on the dataset of 57 patients, and tested on the dataset of 65 patients. This final model was independent of the intermediate models trained during the annotation workflow, which reduced bias in the test set to a minimum. Using completely manual annotations in the test set would have introduced a distribution shift and thus greater bias.
Model
We used the model from the nnU-Net framework, which is a U-Net–based implementation that automatically configures all hyperparameters based on the dataset characteristics (17,18). One model was trained on CT scans with 1.5-mm isotropic resolution. To allow for lower technical requirements (RAM and GPU memory), we also trained a second model on 3-mm isotropic resolution (for more details on the training, see Appendix S4). The runtime for the prediction of one case was measured on a local workstation with an Intel Core i9 3.5-GHz CPU and NVIDIA GeForce RTX 3090 GPU.
Statistical Analysis
Training dataset.—As evaluation metrics, the Dice similarity coefficient, a commonly used spatial overlap index, and the normalized surface distance (NSD), which measures how often the surface distance is less than 3 mm, were calculated between the predicted segmentations and the human approved ground truth segmentations. Both metrics range between 0 (worst) and 1 (best) and were calculated on the test set.
For additional evaluation, we compared our model to a nnU-Net trained on the dataset from the Multi-Atlas Labeling Beyond the Cranial Vault – Workshop and Challenge (https://www.synapse.org/#!Synapse:syn3193805/wiki/217780) (BTCV dataset) acquired at Vanderbilt University Medical Center. Because that dataset provided labels for only 13 structures (Appendix S5), the comparison was limited to those 13 structures. We ran two comparisons: one on our test set and one on the BTCV dataset. The 95% CIs were calculated using nonparametric percentile bootstrapping with 10 000 iterations. A Wilcoxon signed rank test was used to compare the Dice and NSD metrics between our model and the BTCV model. P values less than .05 were considered to indicate statistically significant differences.
Aging study dataset.—To evaluate the effect of age on different structures, the correlation between age and volume and the mean attenuation in Hounsfield units were calculated for all structures, excluding structures with failed segmentations. The segmentation of a body structure was assumed as failed if the volume of the respective body structure was too small to be anatomically plausible given by a lower bound (Table S1). The Kolmogorov-Smirnov test was used to evaluate whether continuous variables were normally distributed. The association between continuous variables was examined using Spearman rank correlation coefficient. Patients were grouped into four age quartiles and compared using the Kruskal-Wallis test. Post hoc analysis was performed using the Wilcoxon rank sum test. Bonferroni correction was performed, and P values less than .0001 were considered to indicate statistically significant differences. Outliers are not shown in the figures to maintain scaling.
Results
Characteristics of the Study Sample
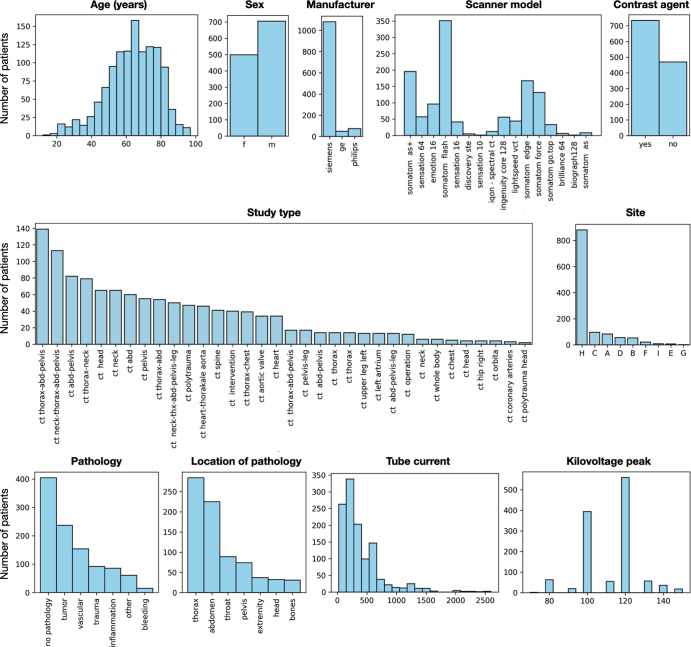
Data on basic demographic characteristics of patients included in the training dataset of 1204 CT images are shown in Figure 3. The dataset contained a wide variety of CT images, with differences in slice thickness, resolution, and contrast phase (native, arterial, portal venous, late phase, and others). Dual-energy CT images obtained using different tube voltages were also included. Different kernels (soft-tissue kernel, bone kernel), as well as CT images from eight different sites and 16 different scanners, were included in the dataset; however, most images were acquired using a Siemens manufacturer. A total of 404 patients showed no signs of abnormality, whereas 645 showed different types of abnormality (tumor, vascular, trauma, inflammation, bleeding, and other). Information on presence of abnormalities was not available for 155 patients because of missing radiology reports (Fig 3).
Figure 3:
Graphs show the distribution of different parameters of the training dataset, demonstrating the dataset’s high diversity.
The aging study dataset of CT scans from 4004 patients showed uniform age distribution, ranging from 18 to 100 years (Fig S2). The sex distribution was less balanced (2543 men [63.5%] and 1461 women [36.5%]).
Segmentation Evaluation
The model trained on CT images with a resolution of 1.5 mm showed high accuracy. The Dice score was 0.943 (95% CI: 0.938, 0.947), and the NSD was 0.966 (95% CI: 0.962, 0.971). The 3-mm model showed a lower Dice score of 0.840 (95% CI: 0.836, 0.844), but the NSD was 0.966 (95% CI: 0.962, 0.969) because minor inaccuracies introduced by the lower resolution were still within the bounds of the 3-mm distance threshold. Thus, the 3-mm model still delivered correct results, but the borders were less precise. Results for each structure independently are shown in Figure S1 and at https://github.com/wasserth/TotalSegmentator/blob/master/resources/results_all_classes.json.
In a direct comparison of our 1.5-mm model to an nnU-Net trained on the BTCV dataset, our model achieved a significantly higher Dice coefficient (0.932 [95% CI: 0.920, 0.942] vs 0.871 [95% CI: 0.855, 0.887], respectively; P < .001) and NSD score (0.971 [95% CI: 0.961, 0.979] vs 0.921 [95% CI: 0.907, 0.936]; P < .001) on our test set. When we tested our 1.5-mm model on the BTCV dataset, it achieved a Dice coefficient of 0.849 (95% CI: 0.833, 0.862) and NSD score of 0.932 (95% CI : 0.920, 0.943), showing generalizability to CT studies from a different continent. Our model achieved higher values (P < .001) than the nnU-Net trained on the BTCV dataset itself (Dice coefficient, 0.839 [95% CI: 0.821, 0.856]; NSD score, 0.915 [95% CI: 0.900, 0.930]) (for more information, see Appendix S7).
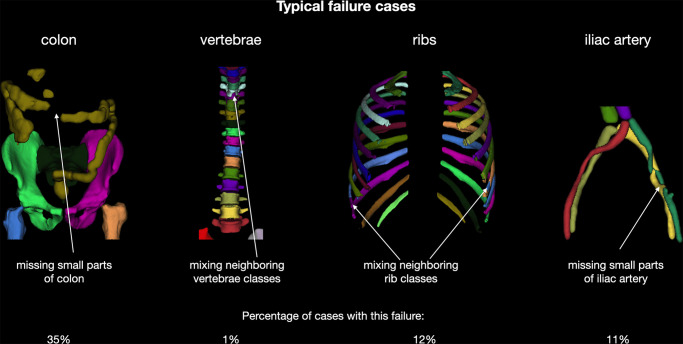
Typical failure cases.—Despite the high Dice coefficient and NSD score, our model failed in some cases. Figure 4 shows the most typical failure cases, such as missing small parts of the colon or iliac arteries and mixing up neighboring vertebrae and ribs.
Figure 4:
Overview of typical failure cases of the proposed model. Users should be aware that these problems may occur.
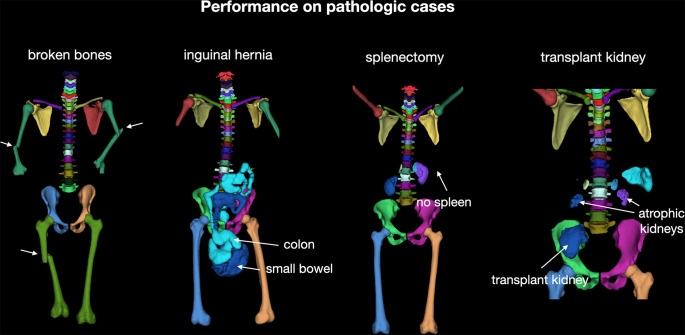
Performance on pathologic cases.—Because our model was trained on a diverse dataset, it generated robust results on patients with major abnormalities. Figure 5 shows qualitative results for several abnormalities.
Figure 5:
Overview of performance of the proposed model on different abnormalities on the test set. Our model showed robust, accurate results even when structures were distorted (broken bones), displaced (bowels displaced by inguinal hernia), completely missing (splenectomy), or duplicated (transplant kidney).
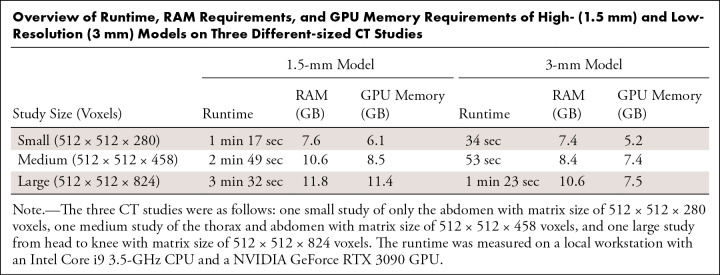
Runtime.—The Table shows an overview of the runtime, RAM requirements, and GPU memory requirements of the high-resolution (1.5 mm) and low-resolution (3 mm) models for three CT studies with different dimensions: a small study of an abdomen with matrix size of 512 × 512 × 280 voxels, a medium-sized study of the thorax and abdomen with matrix size of 512 × 512 × 458 voxels, and a large study from head to knee with matrix size of 512 × 512 × 824 voxels.
Overview of Runtime, RAM Requirements, and GPU Memory Requirements of High- (1.5 mm) and Low-Resolution (3 mm) Models on Three Different-sized CT Studies
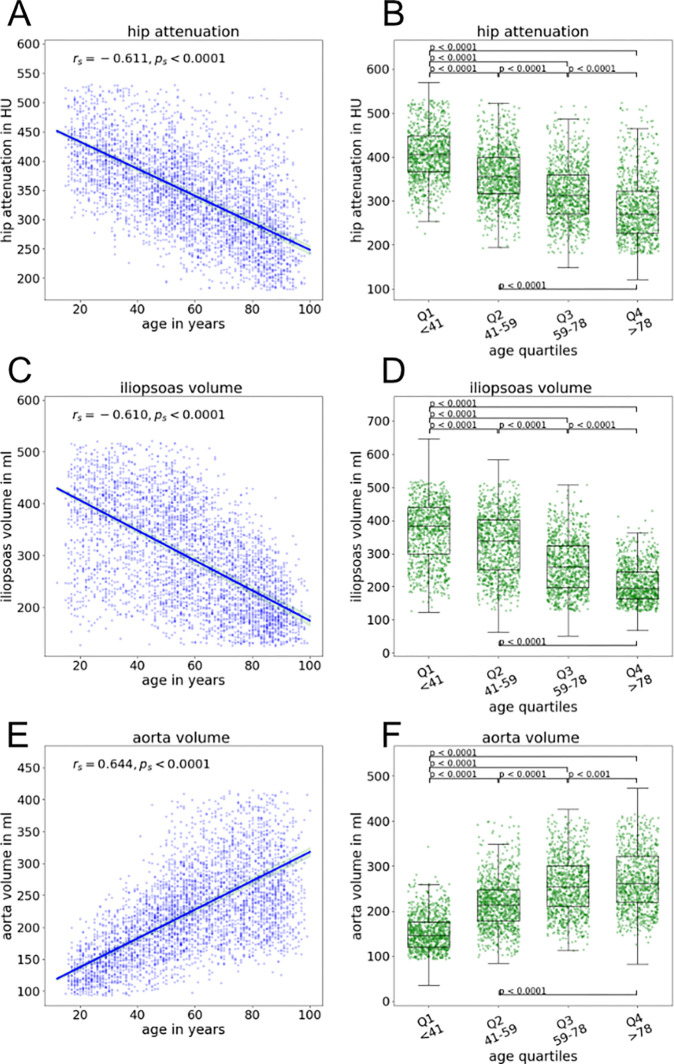
Evaluation of age-related differences.—For the aging study dataset, we observed a negative correlation between age and attenuation in the clavicle (rs = −0.53; P < .0001), hips (rs = −0.61; P < .0001 [Fig 6]), and all ribs and the scapula (Fig S3). A moderately negative correlation was observed between age and mean attenuation of the lumbar vertebrae (lumbar vertebra 4, rs = −0.55; P < .0001).
Figure 6:
Example correlations of CT attenuation and volume with patient age. (A) Graph shows negative correlation between hip attenuation and patient age. (B) Box plots of hip attenuation for age quartiles show a decrease with increasing age. (C) Graph shows negative correlation between iliopsoas muscle volume and patient age. (D) Box plots of iliopsoas muscle volume for age quartiles show a decrease with increasing age. (E) Graph shows positive correlation between aortic volume and patient age. (F) Box plots of aortic volume for age quartiles show an increase with increasing age. For box plots, the central mark indicates the median, the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. Whiskers extend to the most extreme data points not considering outliers and are defined by the 25th percentile subtracted by 1.5 times the IQR or the 75th percentile added by 1.5 times the IQR, respectively. Outliers are not displayed.
Negative correlations between age and attenuation were also observed for the musculature, with moderate to strong correlations for the autochthonous dorsal musculature (rs = −0.74; P < .0001) and gluteal musculature (gluteus maximus: rs = −0.51 [P < .0001]; gluteus medius: rs = −0.61 [P < .0001]; gluteus minimus: rs = −0.79 [P < .0001]). Age was moderately to strongly negatively correlated with CT attenuation and volume of the iliopsoas muscle, respectively (rs = −0.57 [P < .0001] and rs = −0.61 [P < .0001]) (Fig 6).
A positive correlation was observed between the volume of the aorta and patient age (rs = 0.64; P < .0001) (Fig 6), most likely due to aneurysm development. The correlation was less positive for the iliac arteries (rs = 0.33; P < .0001).
Regarding organ volumetry, age was negatively correlated to kidney volume (rs = −0.49; P < .0001) and pancreas volume (rs = −0.49; P < .0001).
The same analysis was done for all 104 classes (Fig S3).
Discussion
In this study, we developed a tool for segmentation of 104 anatomic structures on 1204 CT datasets obtained using different CT scanners, acquisition settings, and contrast phases. The tool demonstrated high accuracy (Dice coefficient of 0.943) and works robustly on a wide range of clinical data, outperforming other freely available segmentation tools. Furthermore, we evaluated and reported age-related changes in volume and attenuation in multiple organs using a large dataset of more than 4000 CT examinations.
Numerous models are available for segmentation of single or several organs on CT images (eg, the pancreas, spleen, colon, or lung), and work has also been conducted on segmenting several anatomic structures in one dataset and model (7–11). All previous models cover only a small subset of relevant anatomic structures and are trained on small datasets that are not representative of routine clinical imaging, which involves different contrast phases, acquisition settings, and diverse abnormalities (18).
To our knowledge, only three other studies have examined segmenting a larger number of structures on CT images. First, the algorithm reported by Chen et al (6) segments 50 different structures. Apart from the fact that many organs and anatomic structures are still not represented in that model, neither the dataset nor the model is publicly available, and the dataset is relatively homogeneous (most of the training data came from the same scanner using the same CT sequence). Second, the algorithm developed by Shiyam Sundar et al (19) segments 120 structures, and their model is publicly available. However, the model requires 256 GB of RAM, making it difficult to apply. Moreover, the training data consist of fewer than 100 individuals, making the model less robust for broad application to any CT data. Third, the algorithm developed by Trägårdh et al (20) segments 100 structures. However, because the 339 training samples are homogeneous, the model does not perform well on images with diverse slice or body orientations or involving different contrast phases.
Many segmentation models and datasets are not publicly available, which strongly reduces their benefit to the scientific community (6,21–23). Datasets that are made available often require time-consuming paperwork to request access (eg, UK Biobank, National Institutes of Health National Institute of Mental Health Data Archive) or are uploaded to data providers that are difficult to use (eg, The Cancer Imaging Archive, which requires a third-party download manager) or rate limited (eg, Google drive). We made our model easily accessible by providing it as a pretrained Python package. Our model requires less than 12 GB of RAM and does not require a GPU. Thus, it can be run on a normal laptop. In addition, our dataset is freely available to download; it does not require any access requests and can be downloaded with one click.
An nnU-Net based model was used for the present study because it was shown to deliver accurate results across a wide range of tasks and has been established as the standard for medical image segmentation, outperforming most other methods (19). It might be possible to improve on the default nnU-Net through more hyperparameter optimization and exploration of newer models, such as transformers (24).
Our model has multiple potential applications. Besides its use for surgery, rapid and readily available organ segmentation also allows for individual dosimetry, as shown for the liver and kidneys (6). Automated segmentation may also enhance research and provide normal or even age-dependent values (eg, Hounsfield units and volume) and biomarkers for clinicians. Combined with a lesion-detection model, our model could be used to estimate body part–specific tumor load. Moreover, our model can be used as a first step in building models to detect specific abnormalities. More than 4500 researchers have already downloaded our model (https://zenodo.org/record/6802342), using it for a wide range of applications.
With our aging study, we demonstrated a sample application for our comprehensive segmentation model that could provide insights into the age dependency of organ volumes and attenuation. Such big-data evaluations were previously not feasible or required substantial time by expert researchers. Using a dataset of more than 4000 patients who underwent polytrauma CT, we showed correlations between age and volume of many segmented organs. Common literature values for normal organ sizes and age-dependent organ development are typically based on sample sizes of a few hundred patients. The number of diverse application examples, such as the evaluation of organ size or density as a function of age, sex, ethnicity, disease, medication intake, or drug use, is almost limitless and can provide a new (radiologic) approach for evaluating the physiology of disease processes.
A limitation of our study was that male patients were overrepresented in the study dataset, possibly because more men are part of the overall hospital population (25). We consider our model to serve as the basis for large radiologic population studies. For example, the model can be used to obtain new reference values for organ volumes or to create a new approach for evaluating different diseases using segmentation. In future research, we plan to add more anatomic structures to our dataset and model. Furthermore, we are preparing a more detailed aging study by using more patients, correcting for confounders, and analyzing more correlations.
In conclusion, we developed a CT segmentation model that is (a) publicly available (https://github.com/wasserth/TotalSegmentator), including training data (https://doi.org/10.5281/zenodo.6802613); (b) is easy to use; (c) segments most anatomically relevant structures in the whole body; and (d) works robustly in any clinical setting.
Authors declared no funding for this work.
Data sharing: Data generated by the authors or analyzed during the study are available at: https://doi.org/10.5281/zenodo.6802613 and https://github.com/wasserth/TotalSegmentator.
Disclosures of conflicts of interest: J.W. No relevant relationships. H.C.B. No relevant relationships. M.T.M. No relevant relationships. M.P. No relevant relationships. D.H. No relevant relationships. A.W.S. No relevant relationships. T.H. No relevant relationships. D.T.B. No relevant relationships. J.C. No relevant relationships. S.Y. No relevant relationships. M.B. No relevant relationships. M.S. No relevant relationships.
Abbreviations:
- BTCV
- Multi-Atlas Labeling Beyond the Cranial Vault – Workshop and Challenge
- NSD
- normalized surface distance
References
- 1. Power SP , Moloney F , Twomey M , James K , O’Connor OJ , Maher MM . Computed tomography and patient risk: facts, perceptions and uncertainties . World J Radiol 2016. ; 8 ( 12 ): 902 – 915 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sevilla J , Heim L , Ho A , Besiroglu T , Hobbhahn M , Villalobos P . Compute trends across three eras of machine learning . arXiv 2202.05924 [preprint] https://arxiv.org/abs/2202.05924. Posted February 11, 2022. Accessed November 2022 . [Google Scholar]
- 3. Suzuki K . Overview of deep learning in medical imaging . Radiol Phys Technol 2017. ; 10 ( 3 ): 257 – 273 . [DOI] [PubMed] [Google Scholar]
- 4. McBee MP , Awan OA , Colucci AT , et al . Deep learning in radiology . Acad Radiol 2018. ; 25 ( 11 ): 1472 – 1480 . [DOI] [PubMed] [Google Scholar]
- 5. Almeida SD , Santinha J , Oliveira FPM , et al . Quantification of tumor burden in multiple myeloma by atlas-based semi-automatic segmentation of WB-DWI . Cancer Imaging 2020. ; 20 ( 1 ): 6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X , Sun S , Bai N , et al . A deep learning-based auto-segmentation system for organs-at-risk on whole-body computed tomography images for radiation therapy . Radiother Oncol 2021. ; 160 : 175 – 184 . [DOI] [PubMed] [Google Scholar]
- 7. Simpson AL , Antonelli M , Bakas S , et al . A large annotated medical image dataset for the development and evaluation of segmentation algorithms . arXiv 1902.09063 [preprint] https://arxiv.org/abs/1902.09063. Posted February 25, 2019. Accessed November 2022 . [Google Scholar]
- 8. Okada T , Linguraru MG , Hori M , et al . Multi-organ segmentation in abdominal CT images . In: 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE , 2012. , 3986 – 3989 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gayathri Devi K , Radhakrishnan R . Automatic segmentation of colon in 3D CT images and removal of opacified fluid using cascade feed forward neural network . Comput Math Methods Med 2015. ; 2015 : 670739 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hofmanninger J , Prayer F , Pan J , Röhrich S , Prosch H , Langs G . Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem . Eur Radiol Exp 2020. ; 4 ( 1 ): 50 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rister B , Yi D , Shivakumar K , Nobashi T , Rubin DLCT-ORG . CT-ORG, a new dataset for multiple organ segmentation in computed tomography . Sci Data 2020. ; 7 ( 1 ): 381 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anastasopoulos C , Reisert M , Kellner E . “Nora Imaging”: a web-based platform for medical imaging . Neuropediatrics 2017. ; 48 ( S 01 ): S1 – S45 . [Google Scholar]
- 13. Landman B , Xu Z , Igelsias J , Styner M , Langerak T , Klein A . Multi-atlas labeling beyond the cranial vault - workshop and challenge . In: Proc MICCAI Multi-Atlas Labeling Beyond Cranial Vault—Workshop Challenge . 2015. ; 12 . [Google Scholar]
- 14. Sekuboyina A , Husseini ME , Bayat A , et al . VerSe: a vertebrae labelling and segmentation benchmark for multi-detector CT images . Med Image Anal 2021. ; 73 : 102166 . [DOI] [PubMed] [Google Scholar]
- 15. Jin L , Yang J , Kuang K , et al . Deep-learning-assisted detection and segmentation of rib fractures from CT scans: development and validation of FracNet . EBioMedicine 2020. ; 62 : 103106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng S , Zhao H , Shi F , et al . CPFNet: Context pyramid fusion network for medical image segmentation . IEEE Trans Med Imaging 2020. ; 39 ( 10 ): 3008 – 3018 . [DOI] [PubMed] [Google Scholar]
- 17. Isensee F , Petersen J , Klein A , et al . nnU-Net: Self-adapting framework for U-Net-based medical image segmentation . arXiv 1809.10486 [preprint] https://arxiv.org/abs/1809.10486. Posted September 27, 2018. Accessed July 2022 .
- 18. Isensee F , Jaeger PF , Kohl SAA , Petersen J , Maier-Hein KH . nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation . Nat Methods 2021. ; 18 ( 2 ): 203 – 211 . [DOI] [PubMed] [Google Scholar]
- 19. Shiyam Sundar LK , Yu J , Muzik O , et al . Fully automated, semantic segmentation of whole-body 18F-FDG PET/CT images based on data-centric artificial intelligence . J Nucl Med 2022. ; 63 ( 12 ): 1941 – 1948 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trägårdh E , Borrelli P , Kaboteh R , et al . RECOMIA-a cloud-based platform for artificial intelligence research in nuclear medicine and radiology . EJNMMI Phys 2020. ; 7 ( 1 ): 51 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xia Y , Yu Q , Chu L , et al . The FELIX Project: deep networks to detect pancreatic neoplasms . medRxiv 2022.09.24.22280071 [preprint]. Posted February 27, 2023. Accessed November 2022 . [Google Scholar]
- 22. Chen Y , Ruan D , Xiao J , et al . Fully automated multiorgan segmentation in abdominal magnetic resonance imaging with deep neural networks . Med Phys 2020. ; 47 ( 10 ): 4971 – 4982 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Okada T , Linguraru MG , Hori M , Summers RM , Tomiyama N , Sato Y . Abdominal multi-organ CT segmentation using organ correlation graph and prediction-based shape and location priors . In: Mori K , Sakuma I , Sato Y , Barillot C , Navab N , eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. MICCAI 2013. Lecture Notes in Computer Science , vol 8151 . Springer; , 2013. , 275 – 282 . [DOI] [PubMed] [Google Scholar]
- 24. Hatamizadeh A , Nath V , Tang Y , Yang D , Roth HR , Xu D . Swin UNETR: Swin transformers for semantic segmentation of brain tumors in MRI images . In: Crimi A , Bakas S , eds. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2021. Lecture Notes in Computer Science , vol 12962 . Springer; , 2022. ; 272 – 284 . [Google Scholar]
- 25. Cullen P , Möller H , Woodward M , et al . Are there sex differences in crash and crash-related injury between men and women? A 13-year cohort study of young drivers in Australia . SSM Popul Health 2021. ; 14 : 100816 . [DOI] [PMC free article] [PubMed] [Google Scholar]