Abstract
Data drift refers to differences between the data used in training a machine learning (ML) model and that applied to the model in real-world operation. Medical ML systems can be exposed to various forms of data drift, including differences between the data sampled for training and used in clinical operation, differences between medical practices or context of use between training and clinical use, and time-related changes in patient populations, disease patterns, and data acquisition, to name a few. In this article, we first review the terminology used in ML literature related to data drift, define distinct types of drift, and discuss in detail potential causes within the context of medical applications with an emphasis on medical imaging. We then review the recent literature regarding the effects of data drift on medical ML systems, which overwhelmingly show that data drift can be a major cause for performance deterioration. We then discuss methods for monitoring data drift and mitigating its effects with an emphasis on pre- and post-deployment techniques. Some of the potential methods for drift detection and issues around model retraining when drift is detected are included. Based on our review, we find that data drift is a major concern in medical ML deployment and that more research is needed so that ML models can identify drift early, incorporate effective mitigation strategies and resist performance decay.
Introduction
Machine learning (ML) is seeing an ever-expanding applicability in medical imaging and therapy. Early examples of computer vision and pattern recognition in medical imaging included models intended to help medical professionals in disease detection (computer-aided detection, or CADe), diagnosis (computer-aided diagnosis, or CADx), segmentation, and quantitative imaging. 1 These early systems often involved expert-defined features or pre-defined feature extraction methods that were combined by an ML model to provide the desired output. Current applications of ML in radiology have expanded into new areas, including image reconstruction and denoising, 2–6 triage and notification software to aid in image interpretation prioritization, 7,8 software to help guide image acquisition, 9,10 and software to contour organs at risk (OARs) in support of radiation treatment planning. 11,12 Autonomous artificial intelligence (AI), described as software that impacts treatment decisions without human input, has also been the focus of recent investigations. 13,14 More importantly, most current systems use architectures such as deep neural networks that are trained end-to-end from the images to the desired output, bypassing the laborious feature engineering step. The availability of successful ML architectures in code sharing platforms, the availability of low-cost computer hardware necessary for model development and the convenience of open-source ML frameworks make it possible to design ML solutions for imaging tasks with relative ease. ML research has also demonstrated that it is possible to train general neural network architectures using data sets from related problems (e.g. a natural image classification tasks) and then adapt the neural network to particular tasks in medical imaging using transfer learning. These and other developments increase the capabilities of ML in medical imaging and even point to the possibility of bringing ML solutions one step closer to intelligence associated with adaptation to or learning from new experiences or data.
The increased number of application types and the reliance on ML in medical imaging and therapy increases the importance for high accuracy, low bias, and robust and generalizable algorithms in order to facilitate user trust. The sizes of medical imaging data sets are typically smaller than those for natural imaging applications due to the labor-intensive data labeling process and privacy concerns in healthcare. For example, the ImageNet data set used for the Large-Scale Visual Recognition Challenge (ILSVRC) has over 1.2 million annotated training images and 50,000 annotated test images. 15 In contrast, the data set used in the 2021 SIIM-FISABIO-RSNA Machine Learning COVID-19 Challenge (one of the larger data sets used for medical imaging challenges) had 8042 COVID-19 positive and 2136 COVID-19 negative chest X-ray examinations. 16 Moreover, ML systems are often seen as black boxes, where data go in and decisions come out, but the processes between input and output are opaque. 17 Explainable ML attempts to reveal the working mechanisms of the ML model to tackle its perceived opaqueness and help build trust in the ML. 18 Finally, differences between the performance of the ML system claimed by its developer and that in clinical practice will hinder user trust. The focus of this article, data drift, is a major culprit behind these potential differences.
Due to a confluence of factors, most ML models designed for medical imaging and therapy have narrowly scoped training data such that the data may be collected from a specific clinical practice pattern, within a short time interval, with a single/limited demography, or using a limited range of acquisition systems. Differences in data characteristics between the training and clinical setting where an ML model is used can, and often does, affect performance. The term data drift in this article loosely refers to differences between the training data and the actual patient input data found in clinical operation, with a more precise definition provided in the next section. In addition to the variations in the data, deep neural networks are sensitive to small perturbations in input images 19 and prone to miscalibration, 20 a condition in which the model output is intended as a probability of a disease or condition, but in fact does not reflect the true likelihood of the event in the clinical setting. These limitations, coupled with data drift can drive down real-world ML performance compared to a setting without drift.
Types and causes data drift in medical imaging
Data drift is widely recognized as a significant concern in the real-world deployment of ML and is an active area of research in the AI/ML community. Despite its increasing importance and an early effort in unifying data drift terminology, 21 multiple terms are used in the literature for the same topic, including data set shift, domain shift, distributional shift, domain drift, data set bias, out-of-distribution generalization, and so on. Synthesizing the definitions in the literature, 21–23 data drift refers to a mismatch between the conditions for model training and clinical use. The mismatch could be in the distribution of input data or features, the clinical context of use, or the functional relationship linking the input and output in an ML model. In the definition, the reference point for the data drift is an ML model that was trained and initially tested with independent data to provide a performance estimate, and the mismatch with respect to clinical use may be due to changes of environment and/or changes over time. For example, a skin lesion characterization ML model trained and initially tested in a population consisting of light-skinned subjects will be subject to data drift when the model is clinically deployed in an environment with a different mixture of light- and dark-skinned individuals. Furthermore, even if the distribution of data in the clinic initially matches that of the training and initial test data sets, it may still change over time again leading to data drift and an unexpected loss of performance.
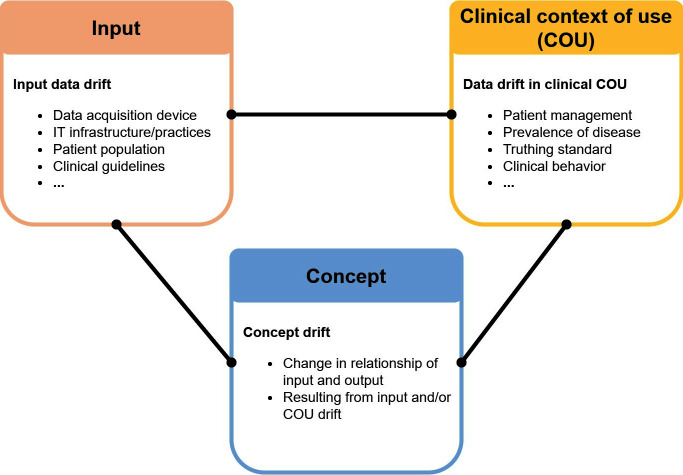
One major challenge in ML is concept drift, 24 which is a change in the relationship between the input data (e.g. images) and the target variables (e.g. classification labels, clinical outcomes, etc.) in supervised learning ML models. This definition is consistent with the definitions by Gama et al 25 and Moreno-Torres et al in which the term is called “concept shift”. 21 Note that some earlier AI literature used the term “concept drift” to refer to changes in the context of use or target variables. 26 Here, we adopted a more recent definition referring specifically to the functional relationship the ML model learned from training data to link the input to the target variable. An example of concept drift is the way in which the relationship between the input data and the classification labels changes when a new class (or comorbidity) is introduced. For example, after 2020, certain patterns of patchy ground-glass opacities in chest X-rays may no longer be labeled as bacterial pneumonia, but as COVID-19 pneumonia. 27 Because this functional relationship is learned from data, both input data drift and data drift in the clinical context of use may cause concept drift, as summarized in Figure 1.
Figure 1.
Data drift categories and associated typical causes. The two types of drift in the upper row may be caused by similar phenomena; e.g. disease prevalence may cause either or both input data drift and COU drift, depending on the context. The drift type in the lower row (concept drift) may be caused by either input data drift or COU drift. COU, clinical context of use.
The rest of this section defines and discusses the causes of input data drift and data drift in the clinical context of use. Studies that investigate the implications of data drift are then discussed in the following section.
Input data drift is a change in the characteristics of ML input data. This is sometimes referred to as “covariate shift” in the literature. 21 A common cause of input data drift is differences in the image/data acquisition devices generating the input to an ML model. As an example, there are multiple manufacturers of CT scanners and a wide array of models and acquisition protocols in clinical use, which leads to wide variations in the qualities of CT images. If an ML algorithm is trained and validated with a limited set of CT acquisition devices (or even a limited range of acquisition protocols), the algorithm may yield inferior performance or even fail after deployment when applied to images acquired from different CT scanners or using different acquisition protocols.
Another cause is differences in patient population. Models may be developed in a research environment with data samples obtained by convenience that do not represent the patient population in deployment. Inadvertent biased sampling or sampling with errors in a clinical study may result in a model that does not generalize well when deployed. Models developed in academic or specialty clinics may not generalize well when deployed to community settings. Patient populations may also change over time. For example, an urban hospital that subsequently acquires primary care practices in rural areas may end up with a substantial change in the demographics of their hospitalized population. 22 Another cause for changes is through the advent of new diseases such as the COVID-19 pandemic or other unexpected “black swan” events.
Yet another cause of input data drift is related to ML systems that utilize clinical features as input. These clinical features can change over time due to, e.g. the introduction of updated clinical guidelines. The Reporting and Data Systems (RADS) endorsed by the American College of Radiology (ACR) are guidelines for the evaluation and interpretation of disease-oriented imaging studies. Each reporting and data system is devised by a group of experts in consensus and they are periodically updated to improve diagnostic parameters. 28 If an ML algorithm is designed using an earlier version of RADS features, it may not work properly when that RADS definitions are updated. Input data drift may also be caused by changes in information technology (IT) practices, software, or infrastructure (e.g. EHR systems) on which the ML model relies. 22 Examples include changes of IT protocols such as changing from International Classification of Diseases Ninth Revision (ICD-9) to ICD-10 codes in the US, and EHR updates that change parameter definitions. Such changes can inadvertently impact ML model inputs leading to incorrect results or lower performance.
Data drift in the clinical context of use refers to changes in the clinical setting in which the model is being used. This includes how the ML system is integrated into the clinical workflow, the disease spectrum and truth-state definition that impact the clinical interpretation of the ML output and performance, and the interaction of clinicians with the ML system. Examples of how data drift in the clinical context of use can occur in practice are given below.
One typical change is an evolution in clinical patient management practice that may affect how an ML system is integrated into the clinical workflow. Taking cervical cancer as an example, an ML system may have been designed to assist clinical decision-making in combination with pap smear screening. When the HPV test is introduced for screening and integrated into clinical patient management, the role of the ML in the workflow may also have to be adjusted accordingly. Moreover, an ML model may be designed to provide its output in a standardized clinical report format but a format update within a clinical environment may make the ML outputs outdated or irrelevant.
Change of disease prevalence, e.g. proportion of cancer patients in the general population for cancer diagnosis, is an important data drift in the clinical context of use. Prevalence is also known as the a priori probability of classes in classification problems. 21 An ML model can be designed to output the a posteriori probability of disease based on the prevalence using Bayes rule at the time of development. 29,30 However, when the ML system is used in operation, if the actual prevalence in the clinic does not match that used in calibrating the algorithm output or the prevalence changes over time, this may cause erroneous interpretation of the probabilistic outputs.
Changes in the definition of truth states in the clinical context of use is another cause of data drift because performance is measured by comparing the ML output with the truth. An example is the contouring definition for the rectum as an OAR in curative three-dimensional external beam radiotherapy for prostate cancer. Nitsche et al 31 discussed 13 different definitions for rectum contouring from the literature and the 3 definitions under their investigation yielded significantly different dose–volume histogram curves. When a model is trained with data from an institution with a specific OAR definition and is deployed in an institution with a different definition, its performance would suffer.
Data drift in the clinical context of use also includes changes in the behavior of physicians due to the introduction of ML systems in the clinic. 22 A typical example of physician behavior change is automation bias, i.e. overreliance on an ML system after getting used to it, which can impact the effectiveness of an ML system over time.
Implications for ML models in medicine
Data drift can have major consequences for ML models including malfunction and performance deterioration. This can be a major barrier for ML tools to generalize to a wide range of healthcare institutions, disease patterns and image acquisition technologies, especially as these factors change over time. 32 The concept of drift is familiar to clinicians, who may find their own previous experience inadequate when new clinical situations arise, leading them to work more cautiously when operating outside their clinical ‘comfort zone’. 23 Key skills in dealing with data drift are to first recognize a change has occurred and then to have enough self-awareness to know when personal intuition developed for other situations will not carry over. These skills are hard enough for clinicians to build. Since most current ML tools are not likely to have been designed to recognize a change, such a “self-awareness” is currently even harder for ML models. As a result, these models may often provide erroneous output, even with high confidence in the presence of data shift. 33
Often, medical imaging and therapy ML models in the research setting are trained on conveniently available data with images acquired from only a few scanners and with or without harmonized acquisition protocols. This type of patient cohort and controlled acquisition may represent a data drift from actual clinical populations. With this potential source of data drift in mind, Mårtensson et al 34 investigated how ML models perform with unseen clinical data sets. The authors trained and tested multiple versions of a CNN on different subsets of brain MR data collected in multiple studies with different scanners, protocols and disease populations. They found that their model generalized well to data sets acquired with similar protocols as the training data but performed substantially worse in clinical cohorts with visibly different tissue contrasts. A similar performance decline associated with data drift coming from differences in imaging systems was observed by De Fauw et al, 35 who developed an ophthalmic deep learning model using three-dimensional optical coherence tomography (OTC) scans to identify patients in need of referral for a range of 50 common sight-threatening retinal diseases. When they trained and tested with data from the same OTC scanner type (Type 1), they observed an overall error rate for referral of 5.5%. When the same model was used on test data acquired with a different type of OTC scanner (Type 2), performance fell substantially resulting in an error rate of 46.6%. These examples and other studies 36–38 have demonstrated that data drift originating from differences in acquisition can play a major role in the deterioration of ML model performance in clinical practice.
As discussed previously, distribution changes over time are another major cause for data drift. Nestor et al 39 investigated the quality of ML model prediction when the models were trained on historical data but tested on future data. Using the MIMIC-III data set, 40 the authors found that when the raw feature representation was used, models trained on historic data and tested on future data had dramatic drops of performance for both mortality prediction (e.g. up to a 0.29 drop in the area under the receiver operating characteristic curve or AUC) and long length-of-stay prediction (up to a 0.10 drop in AUC). A clinically motivated feature representation that grouped raw features into underlying concepts reduced, but did not eliminate, the drop in performance in time. The COVID-19 pandemic gave researchers multiple opportunities to investigate how ML model performance can change when data or disease distributions change in time. Duckworth et al 41 trained an ML model using pre-COVID-19 data to identify patients at high risk of admission for the emergency department. When applied during the COVID-19 pandemic, the model provided substantially lower performance (AUC = 0.826, 95% CI:[0.814,0.837]) compared to pre- COVID-19 performance (AUC = 0.856, 95% CI:[0.852,0.859]). Roland et al 42 found similar results for comparing pre-COVID and early-COVID period data to random sampling in their COVID-19 diagnosis (positive or negative) and prediction of in-hospital death models. Otles et al 43 found a drop in their model performance for predicting healthcare-associated infections over time. Their analysis revealed that most of the drop was caused by an “infrastructure shift”, i.e. changes in access, extraction and transformation of data, rather than changes in clinical workflows and patient populations.
In an effort to collect large data sets for ML training, it may be tempting to include any available data source. Due to socioeconomic, historical, or other reasons, these conveniently available data may underrepresent certain subpopulations (e.g. based on gender, race or age), which can lead to a mismatch of training and operational populations, or a population drift. 44 A mismatch between training and clinical utilization populations in ML may also be caused by differences in data collection geography, e.g. collection of data from different countries 45,46 or from urban vs rural sites. 47 Larrazabal et al 48 studied the effect of gender balance in the training data set and found a consistent decrease in performance for underrepresented genders when a minimum balance in the training data set is not fulfilled. Differences in ML model performance among different demographic subpopulations have been documented in multiple studies. 49,50 Although not all of these differences may be caused by a population drift, 51,52 data drift should be considered as a possible cause when such a difference is detected.
Potential remedies
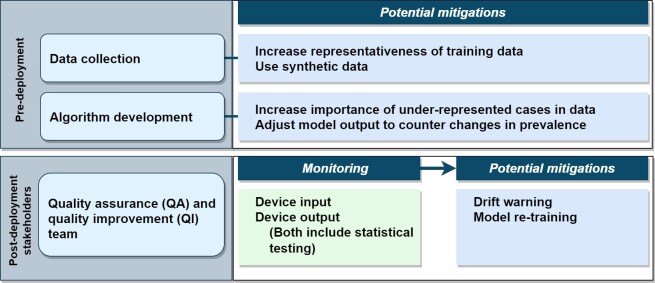
The previous section emphasized that if left unaddressed, data drift can have major consequences. In this section, we discuss how some harmful effects of data drift can be addressed. We organize our discussion into pre- and post-deployment. Pre-deployment strategies mainly focus on alleviation of the effects of mismatches between the training and deployment data. In post-deployment, one first monitors whether data drift has occurred, and if so, implements mitigation measures to reduce the impact, as depicted in Figure 2. For both pre- and post-deployment strategies, understanding the causal structure of the data 53,54 can be useful in identifying the type of data drift and for selecting the mitigations in a structured way.
Figure 2.
General description of approaches from the literature to address data drift. QA, quality assurance; QI, quality improvement.
Pre-deployment
For data drift caused by a difference in disease prevalence between the training and deployment populations, one technical approach can be to correct the probabilistic ML output using the known or estimated prevalence under the deployment condition. This correction approach, based on Bayes rule, has been shown to improve ML performance both in terms of reducing misclassification error 31 and in reducing miscalibration 30 as defined in the “Introduction”. A weakness of this approach is that the true prevalence needs to be known, or a large data set reflecting the deployment prevalence must be available. 31
Under a covariate shift, a technical approach is to perform importance weighting. 55,56 Informally, this means that cases are reweighted by their importance defined as the ratio of their likelihood in deployment over that of the training data. Examples that are rarer in the training data but more likely to occur in deployment receive higher weights, emphasizing them in the training process. 55 For example, in a CADe algorithm to detect breast masses on screening mammograms, the distribution of the mass size in a training data set may be different from the clinical distribution because of the data collection process. Knowing this difference, a developer may apply importance weighting to correct for the covariate shift. For a successful implementation of importance weighting, no part of the deployment distribution can be unseen in the training data, and relevant distributions in the clinical deployment data set must be well-described, a condition that may be difficult to satisfy for many problems. This technique is part of the larger field of domain adaptation, which utilizes data from one or more relevant source domains to execute tasks in a new target domain. 57 In the field of domain adaptation, the data drift is caused by the difference between the source and target domains. In medical imaging, a common domain adaptation technique is to apply extensive and plausible augmentations to the data available from a single source or a small number of sources with the idea that a model trained on the augmented data could generalize better on unseen domains. 57–59
Another possibility for bridging the gap between the training and deployment data distributions is to use synthetic image data. Recent work on natural imaging indicates that it is possible to generate synthetic image data for ML training with a small domain gap between the training and deployment sets even for highly demanding applications such as face analysis. 60 Pipelines for generating synthetic images using either physics-based methods applied to organ/lesion models 61 or deep-learning-based approaches 62 have been explored in recent years. A possible approach is to use synthetic images for image acquisition settings or patient demographics that are known to be inadequately represented in a training data set. However, how to do this effectively and across different domains is still a topic of research.
Post-deployment
After ML deployment, monitoring can be used to determine when data drift may pose a threat to the safety and effectiveness of the system. Monitoring can be employed at different levels including the output level (i.e. monitoring of device performance), or at the input level (i.e. monitoring drift in the input data by checking for changes in the input images or their labels).
Performance monitoring at the output is attractive because it can provide a direct measure of performance decay. Methods based on standard drift detection methods such as ADaptive WINdowing (ADWIN) 63 and statistical process control (SPC) 64 have been applied to performance monitoring and change detection for ML models in medicine. 65,66 However, rigorous performance monitoring using the same performance metrics from pre-deployment can be difficult, because the reference standard used for measuring the true performance may be difficult to collect or the time scale needed may be quite long. For example, for a mammographic CAD system, the pre-deployment reference standard may be biopsy or 1-year follow-up to establish the positive/negative status of a subject. The use of the same reference standard may necessitate a 1-year delay in monitoring, which may be unacceptable. For this reason, alternative outcomes have been suggested for monitoring. 67 For example, one could monitor the agreement of the CAD system with radiologists with a change in the agreement rate potentially indicating data drift. Another type of drift detection involves the estimation of the epistemic or model uncertainty. Changes in these uncertainty estimations can be used with ADWIN to detect drift. 68 This approach also works in scenarios where there is no access to the reference standard. Additional research is needed to evaluate the robustness of such methods, in particular their sensitivity to the size of the data, to fully realize their potential usage in drift detection.
Monitoring at the input level include monitoring changes in the target variable, ML input, relationship between these two, and detecting out-of-distribution inputs. 67,69 SPC charts, proposed for quality improvement in radiology departments, 70 can also be used for monitoring changes in the input and target variables. 67 More complex approaches such as changes in the distribution of latent features of deep-learning AI models 71 and the distance between high-dimensional feature distributions have also been proposed to monitor for a change in the relationship between the input and target variables.
Monitoring results can be used in different ways. A reasonable approach may be to use a statistical test (e.g. to determine whether a control limit has been exceeded when using a control chart 72 ) and thus to decide whether data drift has occurred. When data drift is detected, one action could be to issue a warning, with the warning content and recipients dependent on the type and risks posed by the drift. Beyond a warning, some types of data drift (e.g. a prior probability shift) can be addressed by the pre-deployment techniques discussed in Section Potential Remedies - Pre-deployment. However, model retraining on new data or even a change in ML architecture may be needed to address the drift in many situations. Several recent studies in medical imaging explored the possibility of retraining in response to data drift with minimal requirements, e.g. without access to labels for new test cases, with minimal memory requirements, and fast retraining after each new test case. 73–75 The decision to perform retraining cannot be taken lightly because retraining also brings the possibility of an unintended performance deterioration. This possibility depends on many factors, including the quality and quantity of the new training data, the magnitude of the change in the model architecture, the change of validation methods and truthing methods, and the method (or lack thereof) for keeping users informed of the change.
The approaches described above require collaboration and co-ordination among many stakeholders. For monitoring to be successfully implemented, clinics and radiology departments may need to rely on dedicated quality assurance (QA) and quality improvement (QI) teams made up of technologists, clinicians, administrators, software experts, and biostatisticians. Device manufacturers, realizing that the success of ML in medical imaging and therapy partly depends on mitigating the effect of data drift, should be expected to provide tools and procedures to facilitate monitoring. When retraining is necessary, the roles and responsibilities of the different stakeholders should be clearly defined. Regulatory agencies can be expected to provide frameworks to allow stakeholders address the effects of data drift while assuring device safety and effectiveness. Research into better ways to detect data drift, to decide whether to retrain a model, and most effective ways to implement retraining are also essential.
Conclusion
Data drift can significantly affect the performance of ML-based software in medical imaging and therapy. Data drift is due to a host of reasons, including sampling biases at the design stage, changes in image acquisitions and patient populations, and changes or differences in the clinical context of use across time or clinical sites. All stakeholders, including developers, medical professionals, administrators and regulators should be vigilant about recognizing the risks of data drift because it can seriously impact ML performance and impede the clinical acceptance and long-term success of ML-based software in medical imaging and therapy. Several methods have been proposed in the ML literature to mitigate the effects of data drift. However, these methods have not been widely applied in medical applications in general, and in imaging/therapy systems in particular. While monitoring medical imaging ML systems for data drift and implementing retraining when substantiative drift is detected can be effective remedies, they have not been widely implemented. More research is needed into best practices for detecting and mitigating the impact of data drift, along with strategies to make solutions practical and cost effective in the clinical setting. Advances in these areas are necessary to fully realize the potential of medical ML for improving public health.
Footnotes
Conflicts of interest: None.
Contributor Information
Berkman Sahiner, Email: Berkman.Sahiner@fda.hhs.gov.
Weijie Chen, Email: weijie.chen@fda.hhs.gov.
Ravi K. Samala, Email: ravi.samala@fda.hhs.gov.
Nicholas Petrick, Email: nicholas.petrick@fda.hhs.gov.
REFERENCES
- 1. Giger ML, Chan HP, Boone J. Anniversary paper: history and status of CAD and quantitative image analysis: the role of medical physics and AAPM. Med Phys 2008; 35: 5799–5820. doi: 10.1118/1.3013555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Qin C, Schlemper J, Caballero J, Price AN, Hajnal JV, Rueckert D. Convolutional recurrent neural networks for dynamic Mr image reconstruction. IEEE Trans Med Imaging 2019; 38: 280–90. doi: 10.1109/TMI.2018.2863670 [DOI] [PubMed] [Google Scholar]
- 3. Ravishankar S, Ye JC, Fessler JA. Image reconstruction:from sparsity to data-adaptive methods and machine learning. Proc IEEE Inst Electr Electron Eng 2020; 108: 86–109. doi: 10.1109/JPROC.2019.2936204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang G, Ye JC, De Man B. Deep learning for tomographic image reconstruction. Nat Mach Intell 2020; 2: 737–48. doi: 10.1038/s42256-020-00273-z [DOI] [Google Scholar]
- 5. Akagi M, Nakamura Y, Higaki T, Narita K, Honda Y, Zhou J, et al. Deep learning reconstruction improves image quality of abdominal ultra-high-resolution CT. Eur Radiol 2019; 29: 6163–71. doi: 10.1007/s00330-019-06170-3 [DOI] [PubMed] [Google Scholar]
- 6. Higaki T, Nakamura Y, Zhou J, Yu Z, Nemoto T, Tatsugami F, et al. Deep learning reconstruction at CT: phantom study of the image characteristics. Acad Radiol 2020; 27: 82–87. doi: 10.1016/j.acra.2019.09.008 [DOI] [PubMed] [Google Scholar]
- 7. Yahav-Dovrat A, Saban M, Merhav G, Lankri I, Abergel E, Eran A, et al. Evaluation of artificial intelligence-powered identification of large-vessel occlusions in a comprehensive stroke center. AJNR Am J Neuroradiol 2021; 42: 247–54. doi: 10.3174/ajnr.A6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elijovich L, Dornbos Iii D, Nickele C, Alexandrov A, Inoa-Acosta V, Arthur AS, et al. Automated emergent large vessel occlusion detection by artificial intelligence improves stroke workflow in a hub and spoke stroke system of care. J Neurointerv Surg 2022; 14: 704–8. doi: 10.1136/neurintsurg-2021-017714 [DOI] [PubMed] [Google Scholar]
- 9. Narang A, Bae R, Hong H, Thomas Y, Surette S, Cadieu C, et al. Utility of a deep-learning algorithm to guide novices to acquire echocardiograms for limited diagnostic use. JAMA Cardiol 2021; 6: 624–32. doi: 10.1001/jamacardio.2021.0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schneider M, Bartko P, Geller W, Dannenberg V, König A, Binder C, et al. A machine learning algorithm supports ultrasound-naïve novices in the acquisition of diagnostic echocardiography loops and provides accurate estimation of LVEF. Int J Cardiovasc Imaging 2021; 37: 577–86. doi: 10.1007/s10554-020-02046-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ibragimov B, Xing L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med Phys 2017; 44: 547–57. doi: 10.1002/mp.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oktay O, Nanavati J, Schwaighofer A, Carter D, Bristow M, Tanno R, et al. Evaluation of deep learning to augment image-guided radiotherapy for head and neck and prostate cancers. JAMA Netw Open 2020; 3(): e2027426. doi: 10.1001/jamanetworkopen.2020.27426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raya-Povedano JL, Romero-Martín S, Elías-Cabot E, Gubern-Mérida A, Rodríguez-Ruiz A, Álvarez-Benito M. AI-based strategies to reduce workload in breast cancer screening with mammography and tomosynthesis: A retrospective evaluation. Radiology 2021; 300: 57–65. doi: 10.1148/radiol.2021203555 [DOI] [PubMed] [Google Scholar]
- 14. Shoshan Y, Bakalo R, Gilboa-Solomon F, Ratner V, Barkan E, Ozery-Flato M, et al. Artificial intelligence for reducing workload in breast cancer screening with digital breast tomosynthesis. Radiology 2022; 303: 69–77. doi: 10.1148/radiol.211105 [DOI] [PubMed] [Google Scholar]
- 15. Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis 2015; 115: 211–52. doi: 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- 16. Lakhani P, Mongan J, Singhal C, Zhou Q, Andriole KP, Auffermann WF, et al. The 2021 SIIM-FISABIO-RSNA machine learning COVID-19 challenge: annotation and standard exam classification of COVID-19 chest radiographs. J Digit Imaging 2022; 1–8. doi: 10.1007/s10278-022-00706-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. The Lancet Respiratory M . Opening the black box of machine learning. The Lancet Respiratory Medicine 2018; 6: 801. doi: 10.1016/S2213-2600(18)30425-9 [DOI] [PubMed] [Google Scholar]
- 18. Chen H, Gomez C, Huang C-M, Unberath M. Explainable medical imaging AI needs human-centered design: Guidelines and evidence from a systematic review. NPJ Digit Med 2022; 5(): 156. doi: 10.1038/s41746-022-00699-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Szegedy C, Zaremba W, Sutskever I, Bruna J, Erhan D, Goodfellow I, et al. Intriguing properties of neural networks. 2013. Available from: arXiv preprint arXiv:13126199
- 20. Guo C, Pleiss G, Sun Y, Weinberger KQ, editors. On calibration of modern neural networks. International conference on machine learning; 2017: PMLR. [Google Scholar]
- 21. Moreno-Torres JG, Raeder T, Alaiz-Rodríguez R, Chawla NV, Herrera F. A unifying view on dataset shift in classification. Pattern Recognition 2012; 45: 521–30. doi: 10.1016/j.patcog.2011.06.019 [DOI] [Google Scholar]
- 22. Finlayson SG, Subbaswamy A, Singh K, Bowers J, Kupke A, Zittrain J, et al. The clinician and dataset shift in artificial intelligence. N Engl J Med 2021; 385: 283–86. doi: 10.1056/NEJMc2104626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Challen R, Denny J, Pitt M, Gompels L, Edwards T, Tsaneva-Atanasova K. Artificial intelligence, bias and clinical safety. BMJ Qual Saf 2019; 28: 231–37. doi: 10.1136/bmjqs-2018-008370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bayram F, Ahmed BS, Kassler A. From concept drift to model degradation: an overview on performance-aware drift detectors. Knowledge-Based Systems 2022; 245: 108632. doi: 10.1016/j.knosys.2022.108632 [DOI] [Google Scholar]
- 25. Gama J, Žliobaitė I, Bifet A, Pechenizkiy M, Bouchachia A. A survey on concept drift adaptation. ACM Comput Surv 2014; 46: 1–37. doi: 10.1145/2523813 [DOI] [Google Scholar]
- 26. Widmer G, Kubat M. Learning in the presence of concept drift and hidden contexts. Mach Learn 1996; 23: 69–101. doi: 10.1007/BF00116900 [DOI] [Google Scholar]
- 27. Huang SC, Chaudhari AS, Langlotz CP, Shah N, Yeung S, Lungren MP. Developing medical imaging AI for emerging infectious diseases. Nat Commun 2022; 13(): 7060. doi: 10.1038/s41467-022-34234-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. An JY, Unsdorfer KML, Weinreb JC. TI-RADS: reporting and data systems. Radiographics 2019; 39: 1435–36. doi: 10.1148/rg.2019190087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horsch K, Giger ML, Metz CE. Prevalence scaling: applications to an intelligent workstation for the diagnosis of breast cancer. Acad Radiol 2008; 15: 1446–57. doi: 10.1016/j.acra.2008.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Latinne P, Saerens M, Decaestecker C. Adjusting the outputs of a classifier to new a priori probabilities may significantly improve classification accuracy: Evidence from a multi-class problem in remote sensing. In: Paper presented at the ICML ’01: Proceedings of the Eighteenth International Conference on Machine Learning2001. [Google Scholar]
- 31. Nitsche M, Brannath W, Brückner M, Wagner D, Kaltenborn A, Temme N, et al. Comparison of different contouring definitions of the rectum as organ at risk (OAR) and dose-volume parameters predicting rectal inflammation in radiotherapy of prostate cancer: which definition to use? Br J Radiol 2017; 90(): 20160370. doi: 10.1259/bjr.20160370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo LL, Pfohl SR, Fries J, Posada J, Fleming SL, Aftandilian C, et al. Systematic review of approaches to preserve machine learning performance in the presence of temporal dataset shift in clinical medicine. Appl Clin Inform 2021; 12: 808–15. doi: 10.1055/s-0041-1735184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amodei D, Olah C, Steinhardt J, Christiano P, Schulman J, Mané D. Concrete problems in AI safety. 2016. Available from: arXiv preprint arXiv:160606565
- 34. Mårtensson G, Ferreira D, Granberg T, Cavallin L, Oppedal K, Padovani A, et al. The reliability of A deep learning model in clinical out-of-distribution MRI data: A multicohort study. Med Image Anal 2020; 66: S1361-8415(20)30078-5. doi: 10.1016/j.media.2020.101714 [DOI] [PubMed] [Google Scholar]
- 35. De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med 2018; 24: 1342–50. doi: 10.1038/s41591-018-0107-6 [DOI] [PubMed] [Google Scholar]
- 36. Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of A deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med 2018; 15(): e1002683. doi: 10.1371/journal.pmed.1002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pooch EHP, Ballester P, Barros RC. Can We Trust Deep Learning Based Diagnosis? The Impact of Domain Shift in Chest Radiograph Classification. Springer International Publishing; 2020, pp.74–83. [Google Scholar]
- 38. AlBadawy EA, Saha A, Mazurowski MA. Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing. Med Phys 2018; 45: 1150–58. doi: 10.1002/mp.12752 [DOI] [PubMed] [Google Scholar]
- 39. Nestor B, McDermott MBA, Boag W, Berner G, Naumann T, Hughes MC. Feature robustness in non-stationary health records: Caveats to deployable model performance in common clinical machine learning tasks. Machine Learning for Healthcare Conference PMLR. ; 2019. [Google Scholar]
- 40. Johnson AEW, Pollard TJ, Shen L, Lehman L-WH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3(): 160035. doi: 10.1038/sdata.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Duckworth C, Chmiel FP, Burns DK, Zlatev ZD, White NM, Daniels TWV, et al. Using explainable machine learning to characterise data drift and detect emergent health risks for emergency department admissions during COVID-19. Sci Rep 2021; 11(): 23017. doi: 10.1038/s41598-021-02481-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roland T, Böck C, Tschoellitsch T, Maletzky A, Hochreiter S, Meier J, et al. Domain shifts in machine learning based covid-19 diagnosis from blood tests. J Med Syst 2022; 46(): 23. doi: 10.1007/s10916-022-01807-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Otles E, Oh J, Li B, Bochinski M, Joo H, Ortwine J, et al. Mind the Performance Gap: Examining Dataset Shift During Prospective Validation. In: Ken J, Serena Y, Mark S, eds. Proceedings of the 6th Machine Learning for Healthcare Conference; Proceedings of Machine Learning Research: PMLR. ; 2021. pp. 506–34. [Google Scholar]
- 44. Bernhardt M, Jones C, Glocker B. Potential sources of dataset bias complicate investigation of underdiagnosis by machine learning algorithms. Nat Med 2022; 28: 1157–58. doi: 10.1038/s41591-022-01846-8 [DOI] [PubMed] [Google Scholar]
- 45. Nagaraj Y, de Jonge G, Andreychenko A, Presti G, Fink MA, Pavlov N, et al. Facilitating standardized COVID-19 suspicion prediction based on computed tomography radiomics in a multi-demographic setting. Eur Radiol 2022; 32: 6384–96. doi: 10.1007/s00330-022-08730-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sáez C, Romero N, Conejero JA, García-Gómez JM. Potential limitations in COVID-19 machine learning due to data source variability: A case study in the ncov2019 dataset. J Am Med Inform Assoc 2021; 28: 360–64. doi: 10.1093/jamia/ocaa258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Celi LA, Cellini J, Charpignon M-L, Dee EC, Dernoncourt F, Eber R, et al. Sources of bias in artificial intelligence that perpetuate healthcare disparities-A global review. PLOS Digit Health 2022; 1(): e0000022. doi: 10.1371/journal.pdig.0000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Larrazabal AJ, Nieto N, Peterson V, Milone DH, Ferrante E. Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc Natl Acad Sci U S A 2020; 117: 12592–94. doi: 10.1073/pnas.1919012117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cirillo D, Catuara-Solarz S, Morey C, Guney E, Subirats L, Mellino S, et al. Sex and gender differences and biases in artificial intelligence for biomedicine and healthcare. NPJ Digit Med 2020; 3(): 81. doi: 10.1038/s41746-020-0288-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang J, Galal G, Etemadi M, Vaidyanathan M. Evaluation and mitigation of racial bias in clinical machine learning models: Scoping review. JMIR Med Inform 2022; 10(): e36388. doi: 10.2196/36388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Abbasi-Sureshjani S, Raumanns R, Michels BEJ, Schouten G, Cheplygina V. In: Risk of Training Diagnostic Algorithms on Data with Demographic Bias2020. Cham :Springer International Publishing; [Google Scholar]
- 52. Seyyed-Kalantari L, Zhang H, McDermott MBA, Chen IY, Ghassemi M. Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nat Med 2021; 27: 2176–82. doi: 10.1038/s41591-021-01595-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Castro DC, Walker I, Glocker B. Causality matters in medical imaging. Nat Commun 2020; 11(): 3673. doi: 10.1038/s41467-020-17478-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dockès J, Varoquaux G, Poline J-B. Preventing dataset shift from breaking machine-learning biomarkers. Gigascience 2021; 10(): giab055. doi: 10.1093/gigascience/giab055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shimodaira H. Improving predictive inference under covariate shift by weighting the log-likelihood function. Journal of Statistical Planning and Inference 2000; 90: 227–44. doi: 10.1016/S0378-3758(00)00115-4 [DOI] [Google Scholar]
- 56. Wang M, Deng W. Deep visual domain adaptation: A survey. Neurocomputing 2018; 312: 135–53. doi: 10.1016/j.neucom.2018.05.083 [DOI] [Google Scholar]
- 57. Zhang L, Wang X, Yang D, Sanford T, Harmon S, Turkbey B, et al. Generalizing deep learning for medical image segmentation to unseen domains via deep stacked transformation. IEEE Trans Med Imaging 2020; 39: 2531–40. doi: 10.1109/TMI.2020.2973595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hesse LS, Kuling G, Veta M, Martel AL. Intensity augmentation to improve generalizability of breast segmentation across different MRI scan protocols. IEEE Trans Biomed Eng 2021; 68: 759–70. doi: 10.1109/TBME.2020.3016602 [DOI] [PubMed] [Google Scholar]
- 59. Ouyang C, Chen C, Li S, Li Z, Qin C, Bai W, et al. Causality-inspired single-source domain generalization for medical image segmentation. IEEE Trans Med Imaging 2022. doi: 10.1109/TMI.2022.3224067 [DOI] [PubMed] [Google Scholar]
- 60. Wood E, Baltrusaitis T, Hewitt C, Dziadzio S, Cashman TJ, Shotton J. Fake it till you make it: Face analysis in the wild using synthetic data alone. In: Paper presented at the In: Paper presented at the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada. doi: 10.1109/ICCV48922.2021.00366 [DOI] [Google Scholar]
- 61. Badano A, Graff CG, Badal A, Sharma D, Zeng R, Samuelson FW, et al. Evaluation of digital breast tomosynthesis as replacement of full-field digital mammography using an in silico imaging trial. JAMA Netw Open 2018; 1(): e185474. doi: 10.1001/jamanetworkopen.2018.5474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Han C, Hayashi H, Rundo L, Araki R, Shimoda W, Muramatsu S, et al. GAN-based synthetic brain MR image generation. In: Paper presented at the In: Paper presented at the In: Paper presented at the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC. doi: 10.1109/ISBI.2018.8363678 [DOI] [Google Scholar]
- 63. Bifet A, Gavaldà R. Learning from Time-Changing Data with Adaptive Windowing. Proceedings of the 2007 SIAM International Conference on Data Mining. Philadelphia, PA; 26 April 2007. pp. 443–48. doi: 10.1137/1.9781611972771.42 [DOI] [Google Scholar]
- 64. Benneyan JC, Lloyd RC, Plsek PE. Statistical process control as a tool for research and healthcare improvement. Qual Saf Health Care 2003; 12: 458–64. doi: 10.1136/qhc.12.6.458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Davis SE, Greevy RA, Lasko TA, Walsh CG, Matheny ME. Detection of calibration drift in clinical prediction models to inform model updating. J Biomed Inform 2020; 112: S1532-0464(20)30239-2. doi: 10.1016/j.jbi.2020.103611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Minne L, Eslami S, de Keizer N, de Jonge E, de Rooij SE, Abu-Hanna A. Statistical process control for monitoring standardized mortality ratios of a classification tree model. Methods Inf Med 2012; 51: 353–58. doi: 10.3414/ME11-02-0044 [DOI] [PubMed] [Google Scholar]
- 67. Feng J, Phillips RV, Malenica I, Bishara A, Hubbard AE, Celi LA, et al. Clinical artificial intelligence quality improvement: towards continual monitoring and updating of AI algorithms in healthcare. NPJ Digit Med 2022; 5(): 66. doi: 10.1038/s41746-022-00611-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baier L, Schlör T, Schöffer J, Kühl N. Detecting Concept Drift With Neural Network Model Uncertainty. Available from: https://arxiv.org/abs/2107.01873
- 69. Park C, Awadalla A, Kohno T, Patel S. Reliable and Trustworthy Machine Learning for Health Using Dataset Shift Detection. In: Ranzato M, Beygelzimer A, Dauphin Y, et al., eds. Advances in Neural Information Processing Systems: Curran Associates, Inc. ; 2021, pp. 3043–56. [Google Scholar]
- 70. Cheung YY, Jung B, Sohn JH, Ogrinc G. Quality initiatives: statistical control charts: simplifying the analysis of data for quality improvement. Radiographics 2012; 32: 2113–26. doi: 10.1148/rg.327125713 [DOI] [PubMed] [Google Scholar]
- 71. Stacke K, Eilertsen G, Unger J, Lundstrom C. Measuring domain shift for deep learning in histopathology. IEEE J Biomed Health Inform 2021; 25: 325–36. doi: 10.1109/JBHI.2020.3032060 [DOI] [PubMed] [Google Scholar]
- 72. Mohammed MA, Worthington P, Woodall WH. Plotting basic control charts: tutorial notes for healthcare practitioners. Qual Saf Health Care 2008; 17: 137–45. doi: 10.1136/qshc.2004.012047 [DOI] [PubMed] [Google Scholar]
- 73. Karani N, Erdil E, Chaitanya K, Konukoglu E. Test-time adaptable neural networks for robust medical image segmentation. Medical Image Analysis 2021; 68: 101907. doi: 10.1016/j.media.2020.101907 [DOI] [PubMed] [Google Scholar]
- 74. Wang R, Chaudhari P, Davatzikos C. Embracing the disharmony in medical imaging: A simple and effective framework for domain adaptation. Med Image Anal 2022; 76: S1361-8415(21)00354-6. doi: 10.1016/j.media.2021.102309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. He Y, Carass A, Zuo L, Dewey BE, Prince JL. Autoencoder based self-supervised test-time adaptation for medical image analysis. Medical Image Analysis 2021; 72: 102136. doi: 10.1016/j.media.2021.102136 [DOI] [PMC free article] [PubMed] [Google Scholar]