Abstract
Current pharmacological agents for human immunodeficiency virus (HIV) infection include drugs targeted against HIV reverse transcriptase and HIV protease. An understudied therapeutic target is HIV integrase, an essential enzyme that mediates integration of the HIV genome into the host chromosome. The dicaffeoylquinic acids (DCQAs) and the dicaffeoyltartaric acids (DCTAs) have potent activity against HIV integrase in vitro and prevent HIV replication in tissue culture. However, their specificity against HIV integrase in cell culture has been questioned. Thus, the ability of the DCQAs and DCTAs to inhibit binding of HIV type 1 (HIV-1) gp120 to CD4 and their activities against HIV-1 reverse transcriptase and HIV RNase H were studied. The DCQAs and DCTAs inhibited HIV-1 integrase at concentrations between 150 and 840 nM. They inhibited HIV replication at concentrations between 2 and 12 μM. Their activity against reverse transcriptase ranged from 7 μM to greater than 100 μM. Concentrations that inhibited gp120 binding to CD4 exceeded 80 μM. None of the compounds blocked HIV-1 RNase H by 50% at concentrations exceeding 80 μM. Furthermore, when the effects of the DCTAs on reverse transcription in acutely infected cells were measured, they were found to have no activity. Therefore, the DCQAs and DCTAs exhibit >10- to >100-fold specificity for HIV integrase, and their activity against integrase in biochemical assays is consistent with their observed anti-HIV activity in tissue culture. Thus, the DCQAs and DCTAs are a potentially important class of HIV inhibitors that act at a site distinct from that of current HIV therapeutic agents.
Drug therapy for AIDS is currently limited to several classes of drugs. Agents currently approved for use in the United States are targeted against the human immunodeficiency virus (HIV) reverse transcriptase (RT) and protease (PR). Such agents, including zidovudine, dideoxyinosine, and dideoxycytidine, have been the mainstay of antiretroviral therapy since the late 1980s (32, 33, 50, 51). Treatment with this class of drugs has been hampered somewhat by a relative lack of utility if used to treat patients in the asymptomatic period (1), by the emergence of drug-resistant organisms following the introduction of therapy (2, 19, 20, 30, 43, 47), and by significant toxicities. Recently, a new class of antiretroviral agent, the PR inhibitors, was approved for use in AIDS (4, 5, 10, 27). Resistance to this class of compounds has appeared following in vitro culture of HIV in the presence of some inhibitors and in patients following therapy (3, 11, 22, 24, 40, 48, 49). Recently, the use of cocktails of both RT inhibitors and PR inhibitors has looked exceptionally promising (9, 21, 23). These results suggest that potent anti-HIV therapies which greatly suppress viral replication may prevent, or at least slow significantly, both disease progression and the emergence of drug-resistant HIV.
Enzymatic targets that may be exploited in antiretroviral therapy, in addition to the RT inhibitors and PR inhibitors, have been elusive. However, one therapeutic target that has been studied recently is HIV integrase (IN). HIV, like all retroviruses, requires integration of a cDNA copy of its RNA genome into host cell chromatin for productive infection. The major classes of IN inhibitors reported to date include aurintricarboxylic acid (12) and cosalene analogs (13), caffeic acid phenylethyl ester (17, 18), DNA binding agents, (17, 18) topoisomerase inhibitors (6, 17) and bis-catechols (28). These compounds, in general, are not selective for IN. Aurintricarboxylic acid and related compounds inhibit both RT and other polynucleotidyl phosphoryltransferases (13). Inhibition of IN by DNA binding agents and topoisomerase inhibitors is relatively weak and nonselective. Importantly, most of the information on IN inhibitors has been derived from in vitro experiments using purified IN, and a protective effect of IN inhibitors against HIV infection in tissue culture is either undetectable (28) or has not been reported. Recent work from the National Cancer Institute has identified a large number of HIV IN inhibitors; however, none of the compounds reported has been demonstrated to inhibit HIV replication in tissue culture at nontoxic concentrations and few are active against HIV IN at concentrations of 1 μM or lower (25, 36, 37, 53–55).
A new class of HIV type 1 (HIV-1) IN inhibitor, the dicaffeoylquinic acids (DCQAs), that inhibits HIV-1 integration in biochemical assays and blocks viral replication at nontoxic concentrations in tissue culture, has recently been discovered (44, 46). The compounds that have been reported to date include 3,5-DCQA, 1-methoxyoxalyl-3,5-DCQA (1-MO-3,5-DCQA), 1,5-DCQA, 3,4-DCQA, and 4,5-DCQA, as well as a related dicaffeoyltartaric acid (DCTA), l-chicoric acid (l-CCA). These compounds inhibit HIV-1 integration in vitro at concentrations ranging from 150 to 840 nM and inhibit HIV-1 replication at concentrations of 2 to 12 μM. Their toxicities are all greater than 150 μM, suggesting that the compounds are relatively selective for HIV-1 IN (44, 46). As other IN inhibitors previously reported have been nonselective (7, 12, 13, 17, 18, 28), the specificity of the DCQAs for IN was investigated both to determine whether these compounds exhibited greater specificity than other candidate IN inhibitors and to better understand their mechanisms of anti-HIV activity in cell culture. The ability of these compounds to block HIV gp120 binding to CD4 and their activities against HIV-1 RT and HIV-1 RNase H are reported herein.
MATERIALS AND METHODS
Cell toxicity and anti-HIV assays.
Cell toxicity and antiviral assays were performed as reported previously (35). Briefly, samples were diluted 1:1 in growth medium, filter sterilized, and further twofold serially diluted from 1:8 to 1:1,280 in triplicate wells of a microtiter plate. To each 50 μl of diluted sample, 50 μl of growth medium was added followed by 100 μl of MT-2 cell suspension (2 × 105 cells). Cells were incubated with drug for 48 h at 37°C and then harvested for cell viability in a neutral red dye assay, as described previously (35). Cell toxicity, or the 5% lethal dose (LD5), was defined as greater than 5% inhibition of MT-2 cell growth over 48 h. This concentration is within 1 standard deviation of untreated cell controls and is a nontoxic concentration. The toxicity of several compounds (l-CCA, 3,4-DCQA, 4,5-DCQA, and 3,5-DCQA) were also tested over a period of 72 h. The LD5 of the compounds following the longer culture times were identical to the 48-h toxicities (data not shown).
Anti-HIV assays were performed as described previously (35). Based upon cell toxicity data, samples were diluted in growth medium such that a final 1:4 dilution of the sample would result in a concentration of sample equal to the LD5. The samples were then twofold serially diluted in triplicate. To each 50 μl of diluted sample, 50 μl of HIVLAI was added, and virus was incubated with the sample for 1 h at 37°C. Next, 100 μl of MT-2 cell suspension (2 × 105 cells) was added to each well, and cells were incubated for 72 h at 37°C. The final multiplicity of infection was 1 to 5. Cells were harvested for cytopathic effect in a neutral red dye assay, as described previously (35). The antiviral concentration reported is the concentration of sample necessary to protect MT-2 cells from 50% virus-induced cytopathic effect; this is referred to as the 50% effective dose (ED50).
RT assays.
For all RT assays, RT was precipitated from HIVLAI virions. This RT, rather than recombinant HIV RT, was used to better mimic the RT reactions present in cell culture during the anti-HIV assays.
(i) Homopolymeric template.
RT activities in culture supernatant fluids were determined by the method of Poiesz et al., with poly(rA-dT)15 as a template primer and 25 μCi of [methyl-3H]dTTP per reaction (specific activity, 80.4 Ci/mmol; NEN) (41). In other assays, poly(rC-dG)15 was used as a primer and [8-3H]dGTP (specific activity, 13 Ci/mmol; ICN) was substituted for the dTTP in each reaction. Inhibition assays were performed by replacing the autoclaved deionized water used as a solvent in the reaction mix with an equal volume of autoclaved deionized water containing the compound. Reactions were performed at 37°C for 1 h. The reaction mixtures were precipitated with ice-cold 10% trichloroacetic acid and vacuum filtered through DEAE paper with a Bio-Rad slot blotter. Slots were cut out, placed in 3 ml of ScintiVerse II aqueous scintillant (Fisher), and allowed to incubate at room temperature for 3 h, and then counts per minute were determined with a Beckman β-scintillation counter.
(ii) Heteropolymeric template.
Virus supernatant fluids were precipitated in 30% polyethylene glycol as described by Poiesz et al. (41). Viral pellets were lysed with reagents available in a nonradioactive RT detection kit (DuPont). In preliminary experiments, assays were performed by diluting standard heteropolymeric template solution with an equal volume of autoclaved, deionized water. It was found that the assay results were comparable to those with undiluted template solution (data not shown). For inhibition assays, the template solution was diluted with water or compound in water. Virus lysate was added and reactions were allowed to proceed for 1 h, according to the manufacturer’s instructions. Completed reactions were captured in a sandwich oligonucleotide capture assay with a streptavidin-coated RT capture plate and biotinylated capture probe and a horseradish peroxidase-conjugated detection probe supplied with the kit. A final colorimetric detection step was performed with a 3,3′,5,5′-tetramethylbenzidine solution supplied with the kit. After a 60-min enzymatic step, reactions were stopped with an equal volume of stop solution; 1 h later, optical density was measured at 450 nm on a TiterTek II microcolorimeter.
CD4-gp120 binding assay.
For CD4-gp120 binding assays, a commercially available assay was obtained (DuPont) and the standard protocol was applied. Briefly, a standard curve was generated by diluting gp120 from 0 to 80 ng/ml. The gp120 solution was added to a CD4-coated microtiter plate. Test wells contained 20 ng of gp120 and compounds in a final volume equivalent to the standard curve. Plates were incubated overnight and washed extensively with wash buffer. Next, a murine anti-gp120 monoclonal antibody conjugated with horseradish peroxidase was added to each well and incubated at room temperature for 1 h. After extensive washing, hydrogen peroxide and a color indicator, O-phenylaminediamine, were added to each well and a blank well; the reactions proceeded at room temperature. Reactions were stopped by the addition of 100 μl of 1 N sulfuric acid, and absorption at 490 nm was measured on a TiterTek II microcolorimeter. All reactions were performed in duplicate.
RNase H assays. (i) SPA.
Recombinant HIV-1 RT was obtained from Agouron Pharmaceuticals. This RT contains an active HIV-1 RNase H domain (14). RNase H activity was measured in a scintillation proximity assay (SPA) for RNase H according to the manufacturer’s instructions (Amersham). The protein was diluted in RNase H assay buffer (50 mM Tris-HCl [pH 8.0], 50 mM KCl, 8 mM MgCl, 2 mM dithiothreitol, 50 μg of bovine serum albumin (BSA) per ml, 0.05% [wt/vol] sodium azide, 0.05% [vol/vol] Tween 20). In a microcentrifuge tube, inhibitor was added to the assay buffer followed by a [3H]RNase H substrate, and diluted enzyme was added to each tube. The final reaction conditions were inhibitor (50 μg/ml), 20 ng of RT per ml, 35 mM Tris-HCl, 35 mM KCl, 5.6 mM MgCl, 1.4 mM dithiothreitol, 35 μg of BSA per ml, 0.035% sodium azide, and 0.035% Tween 20. Reactions were allowed to proceed for 15 min at 25°C. The reactions were terminated by addition of a streptavidin-SPA bead solution and allowed to equilibrate for 5 min at 25°C, and then the tubes were counted on a Beckman scintillation counter with the window set for 3H. For those reactions in which inhibitor was allowed to preincubate with the enzyme for 1 h, the same protocol was followed except that the substrate was added to the reaction mixtures 1 h after the enzyme was added to the reaction buffer containing inhibitor.
(ii) Gel cleavage assay.
Inhibition of specific RNase H activity was based on experiments first described by Hostomsky et al. (26). A portion of the gag region of HIV-1 (nucleotides 629 to 714 from the BH-10 clone of HIV-1 [34]) that contained six RNase H cleavage sites was cloned into pTZ18R (Pharmacia). The gene fragment was 10 20 30 40 pppGGGAAUUCGAGCUCAAGCUUUAGACAAGAUAGAGGAAGAGC 50 60 70 80 AAAACAAAAGUAAGAAAAAAGCACAGCAAGCAGCAGCUGGGAUC BamHI
The RNase H cleavage sites, numbered I to VI (42), are located between nucleotides 28 and 29 (VI), 30 and 31 (V), 31 and 32 (IV), 36 and 37 (III), 39 and 40 (II), and 48 and 49 (I). Uniformly labeled runoff transcripts of this region were prepared from the plasmid by using an RNA synthesis kit (Stratagene) and [γ-32P]ATP. The 3′ ends of these transcripts were generated by BamHI digestion of the template DNA. The transcripts were gel purified and hybridized with an oligodeoxyribonucleotide complementary to nucleotides 17 to 55. Five nanograms of hybrid substrate (20,000 cpm) were used for each assay. The substrate, in the presence or absence of inhibitors, was incubated with 0.1 μM HIV-1 RT in 10 μl of buffer containing 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 8 mM MgCl2, and 5 mM dithiothreitol at 37°C for 15 min. Inhibitor concentrations tested were between 5 and 100 μg/ml. Reactions were then terminated by boiling for 3 min in 2 μl of formamide containing bromophenol blue. Samples were analyzed by polyacrylamide gel electrophoresis through 10% polyacrylamide containing 6 M urea. Gels were soaked in 10% acetic acid for 15 min, dried, and exposed on PhosphorImager plates.
RT processivity by PCR.
Work by Zack et al. in quiescent primary lymphocytes demonstrated that specific oligonucleotides and PCR could be employed to determine both how far proviral synthesis has progressed and whether mRNA splicing has occurred within HIV-infected cells (52). All PCRs were performed on a titration curve (25, 28, and 30 cycles) and at different time points after virus inoculation to ensure that relative levels of spliced RNA between drug-treated and untreated cells could be compared.
MT-2 cells were inoculated with HIVLAI at a multiplicity of infection of >1 in the presence of either H2O, zidovudine, l-CCA, or chlorogenic acid. Concentrations of each inhibitor were chosen based on previously determined ED50. Thus, each concentration of drug was greater than, but near, the ED50 to eliminate any low-level toxicity (except the chlorogenic acid, which was chosen to be the same concentration, 5 μg/ml, as l-CCA). At 24 and 48 h after infection, cells were washed to remove virus and DNA was isolated by dissolving the cells in lysis buffer (10 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2, 0.5% Nonidet P-40 and 0.5% Tween 20 [pH 8.3]) and then extracted with phenol-chloroform and nucleic acids precipitated with ethanol. This is a modification of the original protocol, leading to better and more consistent recovery of nucleic acids than the original urea lysis buffer conditions reported previously (52). The ethanol precipitate was dissolved in Tris-HCl and used in PCRs.
The DNA was amplified in PCRs (as described above) in the presence of unlabeled primers (a modification of the original protocol that leads to semiquantitative results). For determination of provirus synthesis, DNA was amplified with HIV-1 long terminal repeat-gag primers (complete proviral DNA synthesis) that lead to a 200-bp product. The resultant products were separated on 1.5% agarose gels, and the products were visualized by ethidium bromide staining and UV illumination. The primers for these reactions were M667 and M661, yielding a 200-bp product representative of complete reverse transcription (double-stranded proviral DNA).
Immunofluorescence assay.
For immunofluorescence assays, approximately 106 cells were washed with phosphate-buffered saline (PBS), resuspended in 100 μl of PBS, and spotted onto a glass slide. Cells were air-dried and then fixed with a 1:1 solution of acetone-methanol. Fixed cells were incubated with an HIV-immune globulin fraction (45) diluted 1:100 in PBS containing 1% BSA. After 30 min at 37°C, the cells were washed in PBS containing 0.1% Tween 80. Cells were next incubated for 30 min at 37°C with goat anti-human immunoglobulin G conjugated with fluorescein (Cappell) diluted 1:200 in PBS containing Evan’s blue counterstain. Slides were washed extensively with PBS containing 0.1% Tween 80, and coverslips were mounted. Cells were observed under a fluorescent microscope, and epifluorescent positive cells were enumerated.
RESULTS
Activity against HIV-1 gp120.
The DCQAs were assayed for the ability to block gp120 binding to CD4 with a commercially available assay. As illustrated in Table 1, several of the compounds were active in this assay. l-CCA inhibited gp120 binding to CD4 by nearly 50% at a concentration of 50 μg/ml (105 μM). Weak activity in this reaction was also observed for the other DCQAs, except 4,5-DCQA. However, the maximum effect for the other DCQAs was 5 to 9% inhibition of gp120 binding to CD4. In addition, the results were inconsistent with this being a mechanism of action for the DCQAs, as none of the compounds, including the best inhibitor of CD4 binding, l-CCA, blocked HIV-1-induced syncytium formation (data not shown).
TABLE 1.
Inhibition of gp120 binding to CD4 by DCQAsa
| Compound | Bound gp120 (ng) | % Inhibition of binding |
|---|---|---|
| H2O | 20 | 0 |
| Quinic acid | 20 | 0 |
| Caffeic acid | 25 | −20 |
| Chlorogenic acid | 20 | 0 |
| 1,5-DCQA | 19.1 | 5 |
| 1-MO-3,5-DCQA | 18.9 | 5 |
| 3,5-DCQA | 18.9 | 5 |
| 4,5-DCQA | 20.9 | −5 |
| 3,4-DCQA | 18.1 | 9 |
| l-CCA | 10.3 | 48 |
| OKT4ab | 1 | 95 |
| HIV-immune globulinc | 0 | 99 |
gp120 binding to CD4 was measured in a commercially available enzyme-linked immunosorbent assay kit developed for HIV drug discovery (DuPont). Reactions were performed according to the manufacturer’s instructions. The concentration of gp120 added to each reaction mix was 20 ng. The final concentration of drug added to each reaction mix was 50 μg/ml.
OKT4a is an HIV-neutralizing murine monoclonal antibody directed against CD4 and was a kind gift from Ortho Diagnostics.
HIV-immune globulin is a polyclonal human gamma globulin fraction from HIV-seropositive individuals and was obtained from the AIDS Reference Reagent Repository. This reagent contains antibodies directed against HIV-1 gp120.
Activity against HIV-1 RT.
All of the DCQAs were tested for activity against HIV-1 RT by the method of Poiesz et al. (41). In these assays, HIVLAI RT was precipitated from culture supernatants and incubated with compounds in distilled water at doses exceeding that which protected MT-2 cells from HIV or with distilled water alone for 1 h. Next, RT activity was determined against a poly(rA)-oligo(dT) template. In one representative assay, there was no inhibition of RT activity: l-CCA (500 μg/ml; 1.05 mM)-treated RT was 291,000 ± 13,000 cpm/ml (mean ± standard deviation), and solvent-treated RT was 296,000 ± 7,800 cpm/ml. In other experiments, neither 1-MO-3,5-DCQA, caffeic acid, nor chlorogenic acid inhibited [3H]thymidine uptake into the poly(rA)-oligo(dT) template (not shown).
Similar results were obtained when a poly(rC)-oligo(dG) template was used, although several DCQAs did weakly inhibit HIV-1 RT against this template. In these experiments, using HIV-1LAI RT with poly(rC)-oligo(dG) and [3H]guanosine-triphosphate, 1-MO-3,5-DCQA inhibited HIV-1 RT by 27% at a concentration of 250 μg/ml (415 μM) and by 23% at a concentration of 100 μg/ml (166 μM); caffeic acid, which had no anti-HIV activity in tissue culture, inhibited HIV-1 RT by 0% (HIV-1, 113,160 ± 403 cpm/ml; caffeic acid, 125,000 ± 7,816 cpm/ml [250 μg/ml; 1.39 mM] and 113,448 ± 3,784 cpm/ml [100 μg/ml; 556 μM]; 1-MO-3,5-DCQA, 82,168 ± 6,088 cpm/ml [250 μg/ml; 415 μM] and 87,720 ± 2,184 [100 μg/ml; 166 μM]). A representative experiment yielding similar results for 3,5-DCQA and quinic acid is shown in Table 2. The data for all of the DCQAs are summarized in Table 5.
TABLE 2.
Representative experiment: inhibition of HIV-1 RT in vitro by DCQAs with a poly(rC)-oligo(dG) homopolymeric template
| Compound | Concn (μg/ml) | RT (cpm/ml)a | % Inhibition |
|---|---|---|---|
| Virus control | NAb | 118,720 (13,952) | NA |
| Quinic acid | 250 | 93,584 (8,288) | 21 |
| 100 | 98,135 (13,040) | 17 | |
| 50 | 89,216 (8,232) | 25 | |
| 3,5-DCQA | 250 | 112,456 (25,360) | 5 |
| 100 | 110,576 (14,864) | 7 | |
| 50 | 119,416 (5,376) | 1 |
RT was determined by the method of Poiesz et al. (41), with a poly(rC)-oligo(dG) template. All reactions were performed in triplicate. Values are mean cpm/ml of [8-3H]dGTP incorporation over a 2-h time period at 37°C. Values in parentheses are standard deviations of the triplicate experiments. This experiment is representative of five experiments.
NA, not applicable.
TABLE 5.
Summary of anti-HIV activities of dicaffeoylquinic acids, precursors, and active analogs
| Compound | LD5 (μM) | ED50 (μM) | IC50 (μM) against HIV-1:
|
||||
|---|---|---|---|---|---|---|---|
| gp120a | RTb | RTc | RNase Hd | INe | |||
| Quinic acid | 911 | >1,822 | NTf | NT | >>2,604 | >260 | NT |
| Caffeic acid | 1,389 | >1,389 | >278 | >278 | >>2,780 | >278 | >278 |
| Chlorogenic acid | 250 | >499 | NA | >142 | >1,420 | >142 | >142 |
| 1-MO-3,5-DCQA | 372 | 7 | >83 | 7 | >830 | >83 | 0.47 |
| 1,5-DCQA | 145 | 4 | >97 | >97 | >970 | >>97 | 0.84 |
| 4,5-DCQA | 145 | 4 | >97 | >97 | >>970 | >>97 | 0.30 |
| 3,5-DCQA | 290 | 2 | >97 | 107 | >>970 | >>97 | 0.66 |
| 3,4-DCQA (synthetic) | 116 | 12 | >97 | 103 | NT | >>97 | 0.71 |
| l-CCA | 264 | 4 | 105 | 17 | 422 | >105 | 0.15 |
gp120-CD4 inhibition was measured with a commercially available enzyme immunoassay kit (DuPont). NA, no activity.
RT activity against a heteropolymeric template was measured with a commercially available, nonradioactive kit (DuPont). IC50 are approximate, as several compounds never reached 50% inhibition.
RT activity against a poly(rC)-oligo(dT) template. >>, <10% inhibition at concentrations of 500 μg/ml or greater. None of the compounds had activity against a poly(rA)-oligo(dT) homopolymeric template.
IC50 of the compounds against HIV-1 RNase H was determined with an Amersham SPA kit according to the manufacturer’s instructions. >, some inhibition of the reaction was observed in the experiment; >>, no inhibition of RNase H activity was observed at 50 μg/ml (see Table 3).
IC50 of HIVLAI IN activity in disintegration assays as described by Chow et al. (8). Percent inhibition of other IN activities (3′-end processing and 3′ strand transfer) were determined for all compounds; the IC50 for those reactions tended to be approximately twofold less than in the disintegration assays. Bands were quantitated by densitometry and percent inhibition was calculated from the solvent and test bands. Results have been previously reported (44).
NT, not tested.
To better mimic a natural template reaction, all of the DCQAs were tested with a commercially available nonradioactive RT kit. As shown in Table 3, all of the DCQAs blocked RT activity in this heteropolymeric assay; however, caffeic acid, which had no anti-HIV activity in tissue culture, blocked HIV-1 RT to a level comparable to one of the active compounds, 1,5-DCQA. Only two of the compounds, l-CCA and the 1-MO-3,5-DCQA, had 50% inhibitory concentrations (IC50) of 5 μg/ml or less in this assay.
TABLE 3.
Inhibition of HIV-1 RT in vitro by DCQAs with a heteropolymeric templatea
| Compound | % Inhibition of HIV-1 RT with a heteropolymeric template at compound concentrations of:
|
||
|---|---|---|---|
| 50 μg/ml | 5 μg/ml | 0.5 μg/ml | |
| H2O | 0 | 0 | 0 |
| Caffeic acid | 25 | NTb | NT |
| Chlorogenic acid | −9 | NT | NT |
| 1,5-DCQA | 26 | 13 | −15 |
| 1-MO-3,5-DCQA | 88 | 56 | −17 |
| 3,5-DCQA | 45 | 18 | −1 |
| 4,5-DCQA | 50 | 17 | 13 |
| 3,4-DCQA | 49 | 21 | −3 |
| l-CCA | 89 | 45 | 0 |
Inhibition of HIV-1 RT activity was measured in a nonradioactive reaction against a heteropolymeric RNA template with a kit available from DuPont and tested according to the manufacturer’s instructions. Each value represents the mean inhibition of triplicate reactions.
NT, not tested.
Activity against HIV-1 RNase H.
The DCQAs had been previously described as inhibitors of HIV-1 IN and had weak activity against HIV-1 RT (Tables 2 and 3) in vitro. Previous data on the crystallographic structure of HIV-1 IN demonstrated that the core domain of IN had a remarkable degree of structural homology to RNase H of HIV-1 RT (15). It was hypothesized that such structural homology might explain why the DCQAs weakly inhibit HIV-1 RT in vitro. To test this hypothesis, the ability of the DCQAs to inhibit HIV-1 RT in a commercially available assay for HIV RNase H activity was assessed. The results of these studies are shown in Table 4.
TABLE 4.
Percent inhibition by DCQAs of HIV-1 RNase H-catalyzed RNA-DNA hybrid cleavage
| Compound | No preincubationa
|
Preincubationb | ||
|---|---|---|---|---|
| Expt 1 | Expt 2 | Avg | ||
| Quinic acid | 7 | 18 | 13 | 17 |
| Caffeic acid | 6 | 2 | 4 | 1 |
| Chlorogenic acid | −9 | 9 | 0 | 4 |
| 1-MO-3,5-DCQA | 37 | 18 | 28 | 41 |
| 1,5-DCQA | −7 | −15 | −11 | 5 |
| 4,5-DCQA | −27 | −19 | −23 | 12 |
| 3,5-DCQA | −29 | −24 | −26 | −11 |
| 3,4-DCQA (synthetic) | −26 | −22 | −24 | −10 |
| l-CCA | 14 | −5 | 5 | 22 |
Compounds were tested with the Amersham HIV-1 IN SPA kit according to the manufacturer’s instructions. Values are percent inhibitions of reactions after 15 min. Compounds were added to the reaction mix at the same time as the full-length RT proteins. The final concentration of each compound was 50 μg/ml. Values for each experiment are means of three separate reactions.
Compounds were preincubated with HIV-1 RT for 1 h at room temperature prior to testing in the Amersham HIV-1 RNase H SPA kit. Values are percent inhibitions of reactions after 15 min. The final concentration of each compound was 50 μg/ml. Results are means of triplicate samples in one experiment.
Only two of the DCQAs inhibited HIV-1 RNase H to any degree at 50 μg/ml. These two compounds, 1-MO-3,5-DCQA and l-CCA, inhibited RNase H by less than 50% even at these high concentrations. The remaining DCQAs had no activity against HIV-1 RNase H. All three of the compounds without anti-HIV activity in tissue culture—quinic acid, caffeic acid, and chlorogenic acid—inhibited HIV-1 RNase H better than the DCQAs. To ensure that a competitive reaction did not favor an RNase H-RNA interaction over an RNase H inhibitor complex, all of the DCQAs were allowed to preincubate with the RNase H for 1 h prior to the assay. As illustrated, this preincubation increased the activity of the DCQAs against HIV-1 RNase H, but none of the compounds inhibited the reaction by 50% (Table 4).
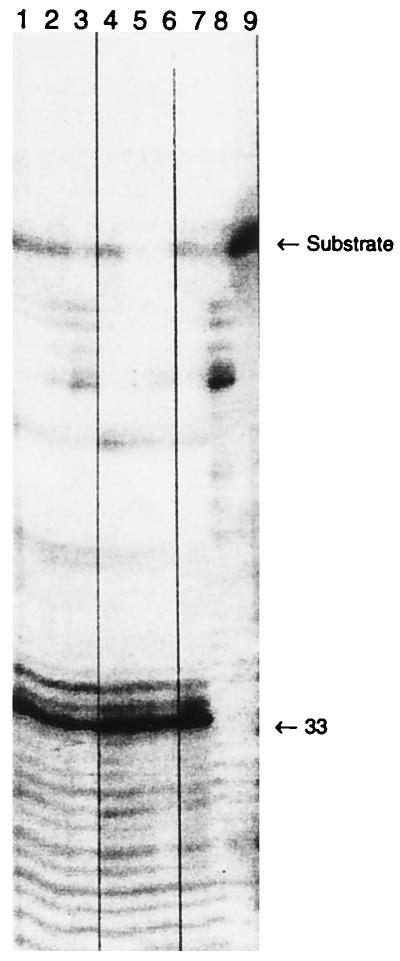
To determine whether the activity of l-CCA in the SPA was specific or nonspecific, a gel cleavage assay for RNase H activity was used (26). As illustrated in Fig. 1, neither the most potent inhibitor of HIV IN, l-CCA, nor the inactive compound, chlorogenic acid, demonstrated any specific inhibition of HIV RNase H. At concentrations ranging from over 200 μM to approximately 10 μM, there was no inhibition of RNase H-specific cleavage of an RNA-DNA hybrid substrate. Chlorogenic acid, however, did demonstrate some nonspecific degradation of the substrate in the absence of RNase H (Fig. 1, lane 8).
FIG. 1.
Inhibition of specific RNA-DNA hybrid cleavage by HIV-1 RNase H. RNase H was incubated for 15 min with a radiolabelled RNA-DNA hybrid in the presence of either chlorogenic acid or l-CCA. Reactions were stopped and products were separated on a 10% polyacrylamide gel containing 6 M urea. All lanes contained substrate. Lanes 1 to 7 contained 0.1 μM HIV-1 RT. Lanes 1 to 3 contained l-CCA at concentrations of 100 (lane 1), 20 (lane 2), and 5 (lane 3) μg/ml; lanes 4 to 6 contained chlorogenic acid at concentrations of 100 (lane 4), 20 (lane 5), and 5 (lane 6) μg/ml. Lane 7 contained RT plus substrate. Lane 8 contained substrate and chlorogenic acid (100 μg/ml), while lane 9 contained substrate alone.
Activity of the DCQAs against HIV IN.
As described previously (44, 46), the DCQAs are potent inhibitors of HIV-1 IN. Their IC50 against HIV-1 IN are summarized in Table 5. Comparisons of the data in Table 5 demonstrate that all of the DCQAs which are active against HIV-1 in tissue culture also inhibit HIV-1 IN at concentrations of less than 900 ng/ml. The most active compound is l-CCA (IC50, 150 ng/ml; 316 nM) while the IC50 of the remaining DCQAs range from approximately 300 to 840 ng/ml. These IC50 are for the disintegration reaction, the reaction for which the compounds are the least active. The IC50 for these compounds in 3′-end processing and 3′ strand transfer reactions ranged from 60 to 300 ng/ml (44).
Effect on HIV-1 production from productively-infected cells.
To determine whether these compounds might have activity in steps of viral replication that occur after integration, the DCQAs were tested for their effects on productively infected cells. H9 cells chronically infected with HIVLAI were treated with 50 μg/ml of either 1-MO-3,5-DCQA (83 μM), chlorogenic acid (142 μM), or l-CCA (105 μM). All three cell lines produced equivalent amounts of polyethylene glycol-precipitable RT activity. Furthermore, the number of infectious virions produced as measured by endpoint dilution analysis on MT-2 cells was equivalent (data not shown). These results are inconsistent with these compounds having an effect on steps in the viral life cycle subsequent to integration (e.g., PR, viral assembly, or budding).
Effects of DCTAs on acute infection.
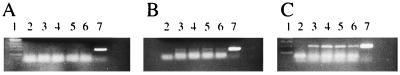
To confirm that in vitro assays of DCQA and DCTA activity were consistent with their activity in tissue culture, an RT processivity assay was used to determine whether l-CCA had any effect on HIV RT processivity in acutely infected cells. Because RT processivity reactions (52) measure the relative amounts of cDNA synthesis, inhibition at any step in viral replication prior to cDNA synthesis should result in decreased synthesis of cDNA. For example, if an inhibitor blocks gp120 binding to CD4, there will be less RNA in cells and thus less cDNA to integrate. Cells were treated with zidovudine (1 μM), chlorogenic acid (5 μg/ml; 14 μM), or l-CCA (5 μg/ml; 10 μM), HIVLAI was added, cells were washed, and DNA was isolated 4 and 24 h later. Next, 10 μl of an equivalent amount of DNA from each sample was added to PCRs with M661 and M667 primers (complete cDNA synthesis) (52). PCRs were run for 25, 28, and 30 cycles. Figure 2 illustrates the PCR products from 24-h-postinoculation DNA. Pretreatment with zidovudine led to a decrease in the relative amount of HIV DNA compared to control infections, l-CCA-treated virus, or chlorogenic acid-treated virus (Fig. 2B). There was no amplification in the cell control. Results were in the linear range, as 25-cycle PCR led to no visible bands and 30-cycle PCR resulted in reaction plateaus. The 4-h DNA was not visible following these conditions, but there was evidence of cDNA if twice the amount of DNA was used and the PCR cycle was allowed to proceed for 30 cycles (data not shown). In addition, when DNA from an equivalent number of cells (106) was added rather than the same amount of DNA, a similar result was obtained (data not shown). Thus, one cannot argue that these results are representative of toxicity, which might have limited the total amount of DNA in each sample and caused a relative increase in viral DNA when equivalent amounts of DNA were added to each PCR mixture. Chlorogenic acid, which is inactive against HIV in tissue culture, weakly inhibited DNA synthesis (whereas active l-CCA did not). Similar inhibition of RT was seen in biochemical assays for RT inhibition (Table 1). Clearly, steps through cDNA synthesis are not involved in the observed anti-HIV activity of l-CCA, and weak inhibition of RT by chlorogenic acid is not sufficient to produce an observable antiviral effect in tissue culture. These data confirm that l-CCA, when used alone, has no effect on the early stages of HIV infection through synthesis of cDNA.
FIG. 2.
HIV cDNA synthesis is impaired by treatment with zidovudine but not with l-CCA. Total DNA isolated 24 h after HIV infection of MT-2 cells was subjected to 25 cycles (A), 28 cycles (B), or 30 cycles (C) of PCR amplification in the presence of the M661 and M667 primer pair (complete cDNA synthesis). Lane 1, size markers (each band is 100 bases); lane 2, mock-infected control DNA; lane 3, mock-treated (water) HIV infection; lane 4, HIV treated with 1 μM zidovudine; lane 5, HIV treated with l-CCA (5 μg/ml); lane 6, HIV treated with chlorogenic acid (5 μg/ml); lane 7, NL4-3 plasmid DNA amplified with the M661 and M667 primer pair (product size marker).
DISCUSSION
Previous studies had demonstrated that the DCQAs and l-CCA (a DCTA) are potent inhibitors of HIV-1 IN in vitro and also inhibit HIV-1 in tissue culture. Other HIV-1 IN inhibitors are relatively nonspecific for HIV-1 IN; therefore, to determine whether the DCQAs were good lead compounds for the development of clinically useful IN inhibitors, the activity of the DCQAs against other viral enzymes and proteins was assessed. As illustrated in Table 1, only l-CCA and 3,4-DCQA had any activity in blocking gp120 binding to CD4. The degree of inhibition for all of the DCQAs and their analogs ranged from a low of −5% inhibition for 4,5-DCQA to nearly 50% for l-CCA at concentrations fully 10-fold greater than the concentrations that inhibit HIV replication in tissue culture. In addition, neither 1-MO-3,5-DCQA nor l-CCA blocked HIV-induced syncytium formation, which would be expected if the compounds blocked gp120 binding to CD4 (data not shown). The compounds were at least 100-fold more potent inhibitors of HIV IN than of gp120 binding to CD4. Furthermore, there was no correlation between inhibition of gp120 binding to CD4 and anti-HIV effect in tissue culture. These data are consistent with those of Mahmood et al. (29), who reported that 3,4-DCQA irreversibly inhibited gp120 binding to CD4. Taken in toto, however, our findings do not support a role for inhibition of gp120 binding to CD4 in the mechanism of action for the DCQAs as a class, or 3,4-DCQA as a member of that class, since inhibition of gp120 binding to CD4 was weak (Table 1) and there was no inhibition of cell fusion (not shown) or evidence for blockade of virus entry in RT processivity reactions (Fig. 2).
The DCQAs had mixed results in assays measuring HIV-1 RT. First, they had no activity against HIV-1 RNase H (Table 4; Fig. 1). Furthermore, they had no activity against polyethylene glycol-precipitated HIV-1 when poly(rA)-oligo(dT) was used and only weak activity when poly(rC)-oligo(dG) was used (Table 2). Several of the compounds were active against a heteropolymeric template (Table 3). The IC50 for l-CCA and 1-MO-3,5-DCQA were 5 μg/ml, a concentration equal to their ED50 against HIV-1 in tissue culture. The other DCQAs, however, were active only at concentrations 10-fold greater than their ED50 in tissue culture. Therefore, for these compounds to be acting at the level of RT in tissue culture, they would need to be concentrated at least 10-fold within the cell. To confirm that the DCQAs and DCTAs do not inhibit HIV reverse transcription in cells, RT processivity was assessed in tissue culture. As shown in Fig. 2, there was no inhibition of cDNA synthesis in cells treated with l-CCA, a DCTA. Therefore, these data not only confirm that the DCTAs do not inhibit HIV RT inside cells, but they also rule out any effect on the virus life cycle up through cDNA synthesis. In enzymatic assays, the compounds demonstrate at least a 10-fold-greater preference for inhibition of IN than of RT (1-MO-3,5-DCQA); this preference may exceed 100-fold (other DCQAs). Although the data reported herein for the DCQAs clearly demonstrate a lack of inhibition of RT as a likely mechanism of action for the DCQAs and DCTAs, work by others has shown that a related (but different) class of compounds, the digalloylquinic acids, do have inhibitory activity against HIV-1 RT in vitro (38, 39). The mechanism for this difference in RT inhibition between the DCQAs and DCTAs and the digalloylquinic acids remains to be determined.
The activity of the DCQAs against HIV-1 IN, in contrast to the other biochemical assays described, was quite potent (Table 5). l-CCA was the most active, while the remaining DCQAs had relatively similar activities in vitro. The IC50 of these compounds in the disintegration reaction ranged from 150 to 840 ng/ml. These concentrations are consistent with their observed anti-HIV activities in tissue culture. Furthermore, the DCQAs demonstrate at least 100-fold selectivity for HIV IN over HIV RNase, HIV RT, or gp120 binding to CD4.
Other IN inhibitors reported previously have either been ineffective at blocking HIV replication or have not been tested for activity in tissue culture. Only two other small, nonnucleotide compounds which block HIV replication and HIV IN have been described. The first, curcumin, is a naturally occurring compound that blocks HIV replication at a concentration of 5 μg/ml yet blocks HIV integration at concentrations of 50 μg/ml (31), a finding incompatible with inhibition of integration by curcumin as a mechanism of action. The second compound, suramin, is a large, polyanionic compound (net negative charge of 6) that mediates its effects in tissue culture through a blockade of binding of gp120 to CD4 (7). The DCQAs do not act through such a mechanism. Furthermore, the net negative charge (between 1 and 2) of the DCQAs does not appear to be important in their activity, as both the chlorogenic and caffeic acids studied have the same net charge as the DCQAs yet have no activity against HIV IN or in tissue culture.
The DCQAs and DCTAs are bis-catechols, like many of the other small-molecule IN inhibitors reported to date (summarized in reference 16). Bis-catechols may chelate metal ions; therefore, it has been suggested that DCQAs and DCTAs may inhibit HIV IN nonspecifically via chelation of Mg2+ or Mn2+. However, such a mechanism seems unlikely, as the compounds exhibit little activity against either HIV-1 RT or HIV-1 RNase H, both metalloenzymes. The data may be interpreted to suggest that the DCQAs act through a variety of mechanisms in tissue culture. Indeed, it is possible that the compounds exert their activities at multiple sites within the virus life cycle. Such multiple sites of action might exert a kind of synergy that is represented by greater activity against HIV-1 than that reported for other IN inhibitors. However, the data are not supportive of a mechanism whereby the DCQAs block binding of HIV to the cell surface. They do not appear to inhibit HIV-1 RT within the cell (Fig. 2). In short, the data suggest that the compounds are acting directly through inhibition of IN.
Although mechanistic data within the cell are still lacking, the data strongly support the interpretation that the DCQAs act at the level of HIV integration. Thus, the DCQAs mark important lead compounds that can be utilized to study IN and the mechanism of integration within the cell. The DCQAs and DCTAs exhibit anti-HIV activity in tissue culture only if preincubated with virus or if added to target cells at the time of HIV addition. Why such a preincubation of inhibitor is required is unclear. Future experiments using radiolabelled l-CCA or radiolabelled DCQAs may help determine whether DCQAs must enter the cell with the virus or whether IN must be inhibited prior to reverse transcription. Through the isolation of mutant viruses that are resistant to the action of the inhibitors, it will be possible to better determine the exact site of action of the compounds and to study the effects of such mutations on HIV replication and, more specifically, the mechanics of viral integration. Finally, these compounds may prove useful in structure-activity relationship studies on HIV IN and may ultimately lead to new classes of potent HIV therapeutics that act at this unique site. The long-term goal of such studies would be the use of HIV IN inhibitors in combination with existing antiviral agents, the PR and RT inhibitors.
ACKNOWLEDGMENTS
We thank Elena Levin and Michael Aye for assistance on the RT processivity experiments.
This work was supported in part by Public Health Service grants 1RO1-AI41360 (W.E.R.), from the National Institute of Allergy and Infectious Diseases, and 5T32-GM-07311 (P.J.K.), as well as a generous gift from Agouron Pharmaceuticals, Inc.
REFERENCES
- 1.Aboulker J, Swart A M. Preliminary analysis of the Concorde trial. Lancet. 1993;341:889–890. doi: 10.1016/0140-6736(93)93096-j. [DOI] [PubMed] [Google Scholar]
- 2.Albert J, Wahlberg J, Lundeberg J, Cox S, Sandstrom E, Wahren B, Uhlen M. Persistence of azidothymidine-resistant human immunodeficiency virus type 1 RNA genotypes in posttreatment sera. J Virol. 1992;66:5627–5630. doi: 10.1128/jvi.66.9.5627-5630.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin E T, Bhat T N, Liu B, Pattabiraman N, Erickson J W. Structural basis of drug resistance for the V82A mutant of HIV-1 proteinase. Nat Struct Biol. 1995;2:244–249. doi: 10.1038/nsb0395-244. [DOI] [PubMed] [Google Scholar]
- 4.Brinkworth R I, Fairlie D P. Non-peptidic anti-AIDS agents: inhibition of HIV-1 proteinase by disulfonates. Biochem Biophys Res Commun. 1992;188:624–630. doi: 10.1016/0006-291x(92)91102-v. [DOI] [PubMed] [Google Scholar]
- 5.Brinkworth R I, Woon T C, Fairlie D P. Inhibition of HIV-1 proteinase by non-peptide carboxylates. Biochem Biophys Res Commun. 1991;176:241–246. doi: 10.1016/0006-291x(91)90915-t. [DOI] [PubMed] [Google Scholar]
- 6.Carteau S, Mouscadet J F, Goulaouic H, Subra F, Auclair C. Effect of topoisomerase inhibitors on the in vitro HIV DNA integration reaction. Biochem Biophys Res Commun. 1993;192:1409–1414. doi: 10.1006/bbrc.1993.1573. [DOI] [PubMed] [Google Scholar]
- 7.Carteau S, Mouscadet J F, Goulaouic H, Subra F, Auclair C. Inhibitory effect of the polyanionic drug suramin on the in vitro HIV DNA integration reaction. Arch Biochem Biophys. 1993;305:606–610. doi: 10.1006/abbi.1993.1468. [DOI] [PubMed] [Google Scholar]
- 8.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 9.Collier A C, Coombs R W, Schoenfeld D A, Bassett R L, Timpone J, Baruch A, Jones M, Facey K, Whitacre C, McAuliffe V J, Friedman H M, Merigan T C, Reichman R C, Hooper C, Corey L. Treatment of human immunodeficiency virus infection with saquinavir, zidovudine, and zalcitabine. N Engl J Med. 1996;334:1011–1017. doi: 10.1056/NEJM199604183341602. [DOI] [PubMed] [Google Scholar]
- 10.Craig J C, Duncan I B, Hockley D, Grief C, Roberts N A, Mills J S. Antiviral properties of Ro 31-8959, an inhibitor of human immunodeficiency virus (HIV) proteinase. Antivir Res. 1991;16:295–305. doi: 10.1016/0166-3542(91)90045-s. [DOI] [PubMed] [Google Scholar]
- 11.Culberson J C, Bush B L, Sardana V V. Qualitative study of drug resistance in retroviral protease using structural modeling and site-directed mutagenesis. Methods Enzymol. 1994;241:385–394. doi: 10.1016/0076-6879(94)41075-5. [DOI] [PubMed] [Google Scholar]
- 12.Cushman M, Golebiewski W M, Pommier Y, Mazumder A, Reymen D, De Clercq E, Graham L, Rice W G. Cosalane analogues with enhanced potencies as inhibitors of HIV-1 protease and integrase. J Med Chem. 1995;38:443–452. doi: 10.1021/jm00003a007. [DOI] [PubMed] [Google Scholar]
- 13.Cushman M, Sherman P. Inhibition of HIV-1 integration protein by aurintricarboxylic acid monomers, monomer analogs, and polymer fractions. Biochem Biophys Res Commun. 1992;185:85–90. doi: 10.1016/s0006-291x(05)80958-1. [DOI] [PubMed] [Google Scholar]
- 14.Davies J F, II, Hostomska Z, Hostomsky Z, Jordan S R, Matthews D A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 15.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 16.Farnet C, Wang B, Lipford J R, Bushman F D. Differential inhibition of HIV-1 preintegration complexes and purified integrase protein by small molecules. Proc Natl Acad Sci USA. 1996;93:9742–9747. doi: 10.1073/pnas.93.18.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fesen M R, Kohn K W, Leteurtre F, Pommier Y. Inhibitors of human immunodeficiency virus integrase. Proc Natl Acad Sci USA. 1993;90:2399–2403. doi: 10.1073/pnas.90.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fesen M R, Pommier Y, Leteurtre F, Hiroguchi S, Yung J, Kohn K W. Inhibition of HIV-1 integrase by flavones, caffeic acid phenethyl ester (CAPE) and related compounds. Biochem Pharmacol. 1994;48:595–608. doi: 10.1016/0006-2952(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 19.Gao Q, Gu Z, Parniak M A, Li X, Wainberg M A. In vitro selection of variants of human immunodeficiency virus type 1 resistant to 3′-azido-3′-deoxythymidine and 2′,3′-dideoxyinosine. J Virol. 1992;66:12–19. doi: 10.1128/jvi.66.1.12-19.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu, Z., Q. Gao, H. Fang, M. A. Parniak, B. G. Brenner, and M. A. Wainberg. 1994. Identification of novel mutations that confer drug resistance in the human immunodeficiency virus polymerase gene. Leukemia 8(Suppl. 1):S166–S169. [PubMed]
- 21.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 22.Gulnik S V, Suvorov L I, Liu B, Yu B, Anderson B, Mitsuya H, Erickson J W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 23.Hammer S M, Squires K E, Hughes M D, Grimes J M, Demeter L M, Currier J S, Eron J J, Feinberg J E, Balfour H H, Jr, Deyton L R, Chodakewitz J A, Fischl M A ACTG. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 24.Ho D D, Toyoshima T, Mo H, Kempf D J, Norbeck D, Chen C-M, Wideburg N E, Burt S K, Erickson J W, Singh M K. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong H, Neamati N, Wang S, Nicklaus M C, Mazumder A, Zhao H, Burke T R, Jr, Pommier Y, Milne G W A. Discovery of HIV-1 integrase inhibitors by pharmacophore searching. J Med Chem. 1997;40:930–936. doi: 10.1021/jm960754h. [DOI] [PubMed] [Google Scholar]
- 26.Hostomsky Z, Hostomska Z, Hudson G O, Moomaw E W, Nodes B O. Reconstitution in vitro of RNase H activity by using purified N-terminal and C-terminal domains of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci USA. 1991;88:1148–1152. doi: 10.1073/pnas.88.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kageyama S, Weinstein J N, Shirasaka T, Kempf D J, Norbeck D W, Plattner J J, Erickson J, Mitsuya H. In vitro inhibition of human immunodeficiency virus (HIV) type 1 replication by C2 symmetry-based HIV protease inhibitors as single agents or in combinations. Antimicrob Agents Chemother. 1992;36:926–933. doi: 10.1128/aac.36.5.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaFemina R L, Graham P L, LeGrow K, Hastings J C, Wolfe A, Young S D, Emini E A, Hazuda D J. Inhibition of human immunodeficiency virus integrase by bis-catechols. Antimicrob Agents Chemother. 1995;39:320–324. doi: 10.1128/aac.39.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahmood N, Moore P S, De Tommasi N, De Simone F, Colman S, Hay A J, Pizza C. Inhibition of HIV infection by caffeoylquinic acid derivatives. Antivir Chem Chemother. 1993;4:235–240. [Google Scholar]
- 30.Martin J L, Wilson J E, Haynes R L, Furman P A. Mechanism of resistance of human immunodeficiency virus type 1 to 2′,3′-dideoxyinosine. Proc Natl Acad Sci USA. 1993;90:6135–6139. doi: 10.1073/pnas.90.13.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazumder A, Raghavan K, Weinstein J, Kohn K W, Pommier Y. Inhibition of human immunodeficiency virus type-1 integrase by curcumin. Biochem Pharmacol. 1995;49:1165–1170. doi: 10.1016/0006-2952(95)98514-a. [DOI] [PubMed] [Google Scholar]
- 32.Merigan T C, Skowron G, ACTG-NIAID SG- Safety and tolerance of dideoxycytidine as a single agent. Results of early phase studies in patients with acquired immunodeficiency syndrome (AIDS) or advanced AIDS-related complex. Am J Med. 1990;88:11S–15S. doi: 10.1016/0002-9343(90)90415-a. [DOI] [PubMed] [Google Scholar]
- 33.Mitsuya H, Weinhold K J, Furman P A, St. Clair M H, Nusinoff-Lehrman S, Gallo R C, Bolognesi D, Barry D W, Broder S. 3′-Azido-3-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc Natl Acad Sci USA. 1985;82:7096–7100. doi: 10.1073/pnas.82.20.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizrahi V. Analysis of ribonuclease H activity of HIV-1 reverse transcriptase using RNA-DNA hybrid substrates derived from the gag region of HIV-1. Biochemistry. 1989;28:9088–9094. doi: 10.1021/bi00449a020. [DOI] [PubMed] [Google Scholar]
- 35.Montefiori D C, Robinson W E, Jr, Schuffman S S, Mitchell W M. Evaluation of antiviral drugs and neutralizing antibodies against human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J Clin Microbiol. 1988;26:231–235. doi: 10.1128/jcm.26.2.231-235.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neamati N, Hong H, Mazumder A, Wang S, Sunder S, Nicklaus M C, Milne G W A, Proksa B, Pommier Y. Depsides and depsidones as inhibitors of HIV-1 integrase: discovery of novel inhibitors through 3D database searching. J Med Chem. 1997;40:942–951. doi: 10.1021/jm960759e. [DOI] [PubMed] [Google Scholar]
- 37.Nicklaus M C, Neamati N, Hong H, Mazumder A, Sunder S, Chen J, Milne G W A, Pommier Y. HIV-1 integrase pharmacophore: discovery of inhibitors through three-dimensional database searching. J Med Chem. 1997;40:920–929. doi: 10.1021/jm960596u. [DOI] [PubMed] [Google Scholar]
- 38.Nishizawa M, Yamagishi T, Dutschman G E, Parker W B, Bonder A J, Kilkuskie R E, Cheng Y-C, Lee K-H. Anti-AIDS agents. 1. Isolation and characterization of four new tetragalloylquinic acids as a new class of HIV reverse transcriptase inhibitors from tannic acid. J Nat Prod. 1989;52:762–768. doi: 10.1021/np50064a016. [DOI] [PubMed] [Google Scholar]
- 39.Nonaka G-I, Nishioka I, Nishizawa M, Yamagishi T, Kashiwada Y, Dutschman G E, Bonder A, Kilkuskie R E, Cheng Y-C, Lee K-H. Anti-AIDS agents. 2. Inhibitory effect of tannins on HIV reverse transcriptase and HIV replication in H9 lymphocyte cells. J Nat Prod. 1990;53:589–595. doi: 10.1021/np50069a008. [DOI] [PubMed] [Google Scholar]
- 40.Otto M J, Garber S, Winslow D L, Reid C D, Aldrich P, Jadhav P K, Patterson C E, Hodge C N, Cheng Y S. In vitro isolation and identification of human immunodeficiency virus (HIV) variants with reduced sensitivity to C-2 symmetrical inhibitors of HIV type 1 protease. Proc Natl Acad Sci USA. 1993;90:7543–7547. doi: 10.1073/pnas.90.16.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poiesz B J, Ruscetti F W, Gazder A F, Bunn B A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Person M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 43.Richman D D. HIV drug resistance. AIDS Res Hum Retroviruses. 1992;8:1065–1071. doi: 10.1089/aid.1992.8.1065. [DOI] [PubMed] [Google Scholar]
- 44.Robinson W E, Jr, Cordeiro M, Abdel-Malek S, Jia Q, Chow S A, Reinecke M G, Mitchell W M. Dicaffeoylquinic acid inhibitors of human immunodeficiency virus (HIV) integrase: inhibition of the core catalytic domain of HIV integrase. Mol Pharmacol. 1996;50:846–855. [PubMed] [Google Scholar]
- 45.Robinson W E, Jr, Montefiori D C, Gillespie D H, Mitchell W M. Complement-mediated, antibody-dependent enhancement of human immunodeficiency virus type 1 (HIV-1) infection in vitro increases HIV-1 RNA and protein synthesis and infectious virus production. J Acquired Immune Defic Syndr. 1989;2:33–42. [PubMed] [Google Scholar]
- 46.Robinson W E, Jr, Reinecke M G, Abdel-Malek S, Jia Q, Chow S A. Inhibitors of HIV-1 replication that inhibit HIV integrase. Proc Natl Acad Sci USA. 1996;93:6326–6331. doi: 10.1073/pnas.93.13.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rooke R, Parniak M A, Tremblay M, Soudeyns H, Li X, Gao Q, Yao X-J, Wainberg M A. Biological comparison of wild-type and zidovudine-resistant isolates of human immunodeficiency virus type 1 from the same subjects: susceptibility and resistance to other drugs. Antimicrob Agents Chemother. 1991;35:988–991. doi: 10.1128/aac.35.5.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanstrom R. Characterization of HIV-1 protease mutants: random, directed, selected. Curr Opin Biotechnol. 1994;5:409–413. doi: 10.1016/0958-1669(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 49.Tisdale M, Myers R E, Maschera B, Parry N R, Oliver N M, Blair E D. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob Agents Chemother. 1995;39:1704–1710. doi: 10.1128/aac.39.8.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yarchoan R, Mitsuya H, Thomas R V, Pluda J M, Hartman N R, Perno C-F, Marczyk K S, Allain J-P, Johns D G, Broder S. In vivo activity against HIV and favorable toxicity profile of 2′,3′-dideoxyinosine. Science. 1989;245:412–415. doi: 10.1126/science.2502840. [DOI] [PubMed] [Google Scholar]
- 51.Yarchoan R, Perno C F, Thomas R V, Klecker R W, Allain J-P, Wills R J, McAtee N, Fischl M A, Dubinsky R, McNeely M C, Mitsuya H, Pluda J M, Lawley T J, Leuther M, Safai B, Collins J M, Myers C E, Broder S. Phase I studies of 2′,3′-dideoxycytidine in severe human immunodeficiency virus infection as a single agent and alternating with zidovudine (AZT) Lancet. 1988;i:76–81. doi: 10.1016/s0140-6736(88)90283-8. [DOI] [PubMed] [Google Scholar]
- 52.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 53.Zhao H, Neamati N, Hong H, Mazumder A, Wang S, Sunder S, Milne G W A, Pommier Y, Burke T R., Jr Coumarin-based inhibitors of HIV integrase. J Med Chem. 1997;40:242–249. doi: 10.1021/jm960450v. [DOI] [PubMed] [Google Scholar]
- 54.Zhao H, Neamati N, Mazumder A, Sunder S, Pommier Y, Burke T R., Jr Arylamide inhibitors of HIV-1 integrase. J Med Chem. 1997;40:1186–1194. doi: 10.1021/jm960449w. [DOI] [PubMed] [Google Scholar]
- 55.Zhao H, Neamati N, Sunder S, Hong H, Wang S, Milne G W A, Pommier Y, Burke T R., Jr Hydrazide-containing inhibitors of HIV-1 integrase. J Med Chem. 1997;40:937–941. doi: 10.1021/jm960755+. [DOI] [PubMed] [Google Scholar]