In this study, Shimazu et al. show that the histidine methyltransferase carnosine N-methyltransferase 1 (CARMT1) is the major His N1-position-specific methyltransferase and plays myriad roles in RNA metabolism. CARMT1 preferentially methylates C3H ZF-type RNA-binding proteins involved in mRNA degradation and alternative splicing and is critical for embryonic development.
Keywords: methyltransferase, CARNMT1, methylation, histidine, mRNA, splicing, degradation, zinc finger protein, U2AF1, embryonic lethality, embryonic development
Abstract
Histidine (His) residues are methylated in various proteins, but their roles and regulation mechanisms remain unknown. Here, we show that carnosine N-methyltransferase 1 (CARNMT1), a known His methyltransferase of dipeptide carnosine (βAla-His), is a major His N1-position-specific methyltransferase. We found that 52 His sites in 20 proteins underwent CARNMT1-mediated methylation. The consensus methylation site for CARNMT1 was identified as Cx(F/Y)xH, a C3H zinc finger (C3H ZF) motif. CARNMT1-deficient and catalytically inactive mutant mice showed embryonic lethality. Among the CARNMT1 target C3H ZF proteins, RNA degradation mediated by Roquin and tristetraprolin (TTP) was affected by CARNMT1 and its enzymatic activity. Furthermore, the recognition of the 3′ splice site of the CARNMT1 target C3H ZF protein U2AF1 was perturbed, and pre-mRNA alternative splicing (AS) was affected by CARNMT1 deficiency. These findings indicate that CARNMT1-mediated protein His methylation, which is essential for embryogenesis, plays roles in diverse aspects of RNA metabolism by targeting C3H ZF-type RNA-binding proteins and modulating their functions, including pre-mRNA AS and mRNA degradation regulation.
Enzymatic methylation occurs in diverse cellular molecules, including DNA, RNA, and proteins. Histidine (His) residues of cellular proteins have been reported to be methylated for more than five decades; however, the specific roles and biological functions of histidine methyltransferases (His MTases) remain largely unknown (Jakobsson 2021). In 2010, histidine protein methyltransferase 1 (HPM1) was discovered as the first His MTase methylating the N3 position of yeast ribosomal protein L3 (RPL3) (Webb et al. 2010). Methyltransferase-like 18 (METTL18), the mammalian homolog of HPM1, methylates the His of RPL3 and regulates ribosomal function (Małecki et al. 2021; Matsuura-Suzuki et al. 2022). In addition to HPM1/METTL18, SET domain-containing 3 (SETD3) was discovered as another N3-position-specific His MTase that targets β-actin (Kwiatkowski et al. 2018; Wilkinson et al. 2019). For the N1-position-specific His MTase, METTL9 was found to methylate the His residues in the alternating His repeat (HxH motif) in the mammalian proteome (Daitoku et al. 2021; Davydova et al. 2021; Lv et al. 2021).
The imidazole ring contains two nitrogen atoms, N1 (or π) and N3 (or τ), responsible for hydrophobic interactions. They also show affinity for metal cations, a feature frequently used in immobilized metal affinity chromatography. N1 and N3 atoms of the imidazole ring can be protonated, resulting in neutral and positively charged states. Methylation of the His N1 or N3 position modulates these interactions and affects various biological processes (Jakobsson 2021). For example, SETD3-mediated β-actin methylation plays a role in muscle contractility, and loss of SETD3 causes primary dystocia (Wilkinson et al. 2019). His methylation of RPL3 regulates translation elongation (Matsuura-Suzuki et al. 2022) and translation dynamics (Małecki et al. 2021) in mammals and ribosome assembly in yeast (Al-Hadid et al. 2014). His methylation by METTL9 regulates zinc ion binding and transport (Davydova et al. 2021; Lv et al. 2021) and oxidative phosphorylation (Davydova et al. 2021).
Carnosine N-methyltransferase 1 (CARNMT1; also known as UPF0586 or C9orf41) is a seven-β-strand (7BS) MTase that was originally identified as a carnosine MTase from rat skeletal muscles (Drozak et al. 2015). It catalyzes the S-adenosyl methionine (SAM/AdoMet)-dependent methylation of His on dipeptide carnosine (βAla-His). As CARNMT1 shows catalytic activity toward several His-containing dipeptides or tripeptides, this enzyme is speculated to have substrates other than carnosine (Drozak et al. 2015). Here, we investigated the properties and biological functions of CARNMT1.
Results
CARNMT1 is a His N1-position-specific protein MTase in mammals
The amino acid sequence of CARNMT1 is well conserved from yeast to mammals, though it is quite distinct from that of other 7BS MTases or SET domain MTases (Fig. 1A,B). As CARNMT1/UPF0586 acts as an N1-position-specific His MTase for dipeptide carnosine (βAla-His) and other His-containing short peptides, we tested its catalytic activity toward longer peptides (Fig. 1C). Recombinant glutathione S-transferase (GST)-tagged full-length mouse CARNMT1 protein (Supplemental Fig. S1A) as well as METTL9, SETD3, and METTL18 reacted with several peptides with known His methylation sites or poly-His (9xHis) peptides. CARNMT1 caused methylation at the C-terminal His or internal His in several peptides, suggesting that this enzyme can methylate His-containing peptides with certain sequence preferences. Notably, the peptide around H157 of rabbit myosin light chain kinase 2 (MLCK2; H148 in humans), which is an N1-methylated His-containing protein (Meyer and Mayr 1987), was highly (>95%) methylated by CARNMT1 (Fig. 1C). To confirm whether CARNMT1 introduced a methyl group into the N1 or N3 position of His (denoted as 1MH/πMH or 3MH/τMH, respectively), amino acid analysis of the methylated peptide was conducted (Fig. 1D). The methylated peptide had a single peak corresponding to the 1MH standard (t = 28.0 min) (Fig. 1D, cyan), confirming that CARNMT1 catalyzes N1-position-specific His methylation.
Figure 1.
CARNMT1 is a histidine N1-position-specific protein MTase in mammals. (A) Phylogenic analysis of human lysine MTases and His MTases. Human SET domain MTases (EHMT2: NP_006700, SUV39H1: NP_001269095, SETDB1: NP_001138887, SMYD2: NP_064582, and SETD3: NP_115609) and 7BS MTases (VCPKMT: NP_078834, CAMKMT: NP_079042, METTL21A: NP_660323, METTL9: NP_057109, METTL18: NP_219486, and CARNMT1: NP_689633) were analyzed with a neighbor-joining method using MEGA-X software (version 10.2.6). An asterisk indicates MTases that target His residues. (B) MTase domain of CARNMT1 orthologs conserved in eukaryotes (H. sapiens: NP_689633, M. musculus: NP_080396, X. tropicalis: NP_001119994, D. rerio: NP_001013361, D. melanogaster: NP_726779, C. elegans: NP_496829, S. cerevisiae: NP_014307, S. pombe: NP_596290, and A. thaliana: NP_180775). (C) Table of the in vitro peptide MTase assay. The indicated synthetic peptides were incubated with CARNMT1, METTL9, SETD3, or METTL18 in the presence of SAM for 2 h at 30°C, and the methylation was analyzed with MALDI-MS. (ND) Not detected, (—) not tested, (*) methylated at C-terminal H, (**) methylated in full-length protein. (D) CARNMT1 catalyzes N1-position-specific His methylation in vitro. The 9xHis peptide was methylated in vitro with CARNMT1 and hydrolyzed with 6 N HCl, and then the 1MH and 3MH content was analyzed with an amino acid analyzer. The methylated 9xHis peptide contained amino acids with elution time corresponding to 1MH standard. (E–H) N1-position-specific His methylation by CARNMT1 and METTL9 in HEK293T cells (E,G) or HAP1 cells (F,H). CRISPR–Cas9-mediated CARNMT1 KO, METTL9 KO (Davydova et al. 2021), and CARNMT1/METTL9 DKO cells were established in HEK293T or HAP1 cells. Total protein extracts were hydrolyzed, and their 1MH (E,F) and 3MH (G,H) content was analyzed as described in the Materials and Methods. n = 3, mean ± SD, Tukey HSD test. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (n.s.) not significant (P > 0.05). (I) Anserine production was disrupted in CARNMT1 KO cells but not in METTL9 KO HEK293T cells. Cellular carnosine (Car) and anserine content was analyzed with MRM LC-MS/MS. n = 3, mean ± SD, Tukey HSD test. (*) P < 0.05. See also Supplemental Figure S1.
CARNMT1 and/or METTL9 knockout (KO) cells were generated in human HEK293T or near-haploid HAP1 cells using the CRISPR–Cas9 system (Supplemental Fig. S1B,C), and their total protein His methylation was examined using multiple reaction monitoring (MRM) liquid chromatography–tandem mass spectrometry (LC-MS/MS) (Fig. 1E–H). CARNMT1 KO cells showed ∼50% reduction in 1MH levels (Fig. 1E,F). Consistent with our previous study, METTL9 KO cells also showed ∼50% reduction in 1MH levels (Davydova et al. 2021). CARNMT1 and METTL9 double-KO (DKO) cells showed undetectable (<0.003%) levels of 1MH, suggesting that both CARNMT1 and METTL9 are responsible for protein N1-position-specific His methylation in HEK293T and HAP1 cells. In contrast, protein 3MH levels remained unchanged in all combinations of CARNMT1 and METTL9 KO cells (Fig. 1G,H). As CARNMT1 is a carnosine MTase that produces anserine (His-methylated carnosine), the cellular anserine/carnosine content in KO cells was determined by MRM LC-MS/MS analysis (Fig. 1I). CARNMT1 KO cells had lower anserine levels than wild-type (WT) cells, whereas METTL9 KO cells had anserine levels similar to WT cells, indicating that CARNMT1, but not METTL9, contributes to anserine production. Moreover, some enzymes other than CARNMT1 also contribute to carnosine His methylation (anserine production), since anserine still presents in CARNMT1 KO cells (the amount of anserine relative to carnosine was ∼15% and ∼6% in WT and CARNMT1 KO cells, respectively). In summary, CARNMT1 is a general N1-position-specific His MTase that methylates cellular proteins and shorter peptides, such as carnosine.
Proteomic identification of substrates for CARNMT1
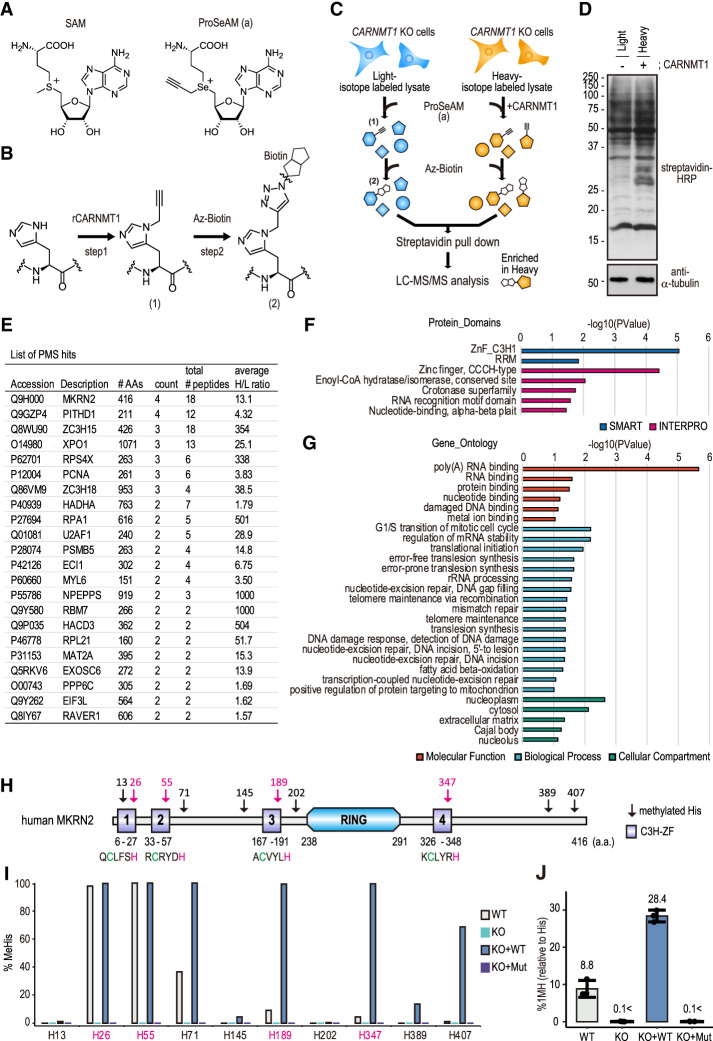
To identify the protein substrates for CARNMT1, ProSeAM–MTase–SILAC (PMS) screening was performed (Fig. 2A,B; Shimazu et al. 2014; Davydova et al. 2021; for review, see also Sohtome et al. 2021). CARNMT1 KO HEK293T cells were cultured in light isotope Lys- and Arg-containing medium or heavy isotope-labeled medium, and the cell lysates were incubated with an alkyne-substituted SAM analog, ProSeAM (Fig. 2A), with (heavy) or without (light) recombinant CARNMT1 (Supplemental Fig. S1A). After the in vitro propargylation of His residues with ProSeAM (Fig. 2B, panel 1), a biotin tag was introduced via the CuAAC reaction (Fig. 2B, panel 2), and the biotinylated proteins were purified with streptavidin beads followed by LC-MS/MS analysis (Fig. 2C,D). A total of 22 proteins was identified as CARNMT1 substrate candidates (Fig. 2E). Gene ontology (GO) analysis (DAVID) showed that several CARNMT1 substrates, such as makorin ring finger protein 2 (MKRN2), zinc finger CCCH-type containing 15 (ZC3H15), ZC3H18, and U2 small nuclear RNA auxiliary factor 1 (U2AF1), had C3H (CCCH)-type zinc finger (C3H ZF) motifs (Fig. 2F).
Figure 2.
Proteomic identification of substrates for CARNMT1. (A) Chemical structures of SAM and ProSeAM (a). (B) Labeling of His N1 position with ProSeAM. (Panel 1) His was alkylated with ProSeAM by recombinant CARNMT1. (Panel 2) The propargylated His was subsequently biotinylated with azide-biotin (Az-Biotin) via CuAAC reaction for the streptavidin affinity purification. (C) Schematic of the ProSeAM–MTase–SILAC (PMS) screening. CARNMT1 KO HEK293T cells were cultured with (heavy) or without (light) stable isotope-containing medium for at least six doubling times. The cells were lysed and incubated with ProSeAM alone (−) or ProSeAM plus CARNMT1 (+). Proteins were biotinylated, mixed together in one tube, and then pulled down with streptavidin beads. The bound proteins were analyzed with LC-MS/MS. (D) Western blotting of biotinylated proteins after CuAAC reaction. (Top) Streptavidin-HRP. (Bottom) Anti-α-tubulin. (E) List of PMS screening hit proteins. A total of 22 proteins was identified as screening hits. (Count) Number of peptides identified in a total of four independent screenings, (total # peptides) the total number of peptides identified in each protein, (average H/L ratio) the average heavy/light enrichment. (F,G) Summary of GO analysis. A functional gene annotation of the 22 proteins was performed with DAVID GO analysis (v6.8). (F) Functional annotation against “protein domains” (SMART and Interpro). (G) Functional annotation against “molecular function,” “biological process,” and “cellular component” gene ontologies. (H) Schematic protein domain and 1MH of human MKRN2. Amino acid sequences of four C3H zinc fingers are shown below the C3H zinc finger domain (C3H ZF). (RING) C3HC4-type zinc finger domain. An arrow indicates methylated His. (Magenta) His in C3H zinc finger, (green) Cys in C3H zinc finger. (I,J) His methylation of MKRN2-FLAG in HEK293T cells. MKRN2-FLAG was expressed in WT, KO, KO + WT rescue, and KO + Mut rescue HEK293T cells (see Supplemental Fig. S2 for stable cells). The FLAG-tagged proteins were immunoprecipitated, and their methylation was analyzed with LC-MS/MS (I) or MRM LC-MS/MS (J). n = 3, mean ± SD. See also Supplemental Figure S2 and Supplemental Tables S1 and S2.
In total, 55 and 58 C3H ZF protein genes have been identified in humans and mice, respectively. Several C3H ZF genes are enriched in macrophage-related tissues, such as the thymus, spleen, lung, intestine, and adipose tissues (Liang et al. 2008). C3H ZF proteins are involved in RNA binding and regulate RNA-related functions, such as mRNA stability, processing, or translation (Hall 2005; Liang et al. 2008). Consistent with the biological role of the C3H ZF motif, GO analysis of the identified CARNMT1 substrate candidates showed enrichment of RNA-binding proteins (RBPs), indicating that CARNMT1 preferentially targets and regulates C3H ZF proteins (Fig. 2G).
Subsequently, we examined the methylation level of each His site in MKRN2, a C3H ZF protein identified by PMS screening. CARNMT1 KO cells rescued with C-terminally HA-tagged WT CARNMT1 (WT) or a catalytically dead mutant (Mut) in which Gly (G199/201) in the catalytic domain (motif I) was substituted with Arg (G199/201R) were generated (Supplemental Fig. S2A–D). MKRN2-FLAG-expressing plasmids were transfected into each cell line to determine His methylation. As shown in Figure 2, H and I, FLAG-tagged MKRN2 was methylated at multiple His residues with higher methylation levels in the C3H ZF motifs (H26, H55, H189, and H347) (magenta in Fig. 2H,I). In contrast, no methylation was observed at His in the C3HC4-type zinc finger RING domain (H264), suggesting sequence specificity of CARNMT1 catalysis (Supplemental Table S1). Subsequent amino acid analysis of the entire MKRN2-FLAG confirmed that 1MH was present (8.8% as compared with His) in WT cells, abolished (<0.1%) in KO cells or KO cells rescued with CARNMT1 Mut-HA (KO + Mut) cells, and increased (28.4%) in KO cells rescued with WT CARNMT1-HA (KO + WT) cells (Fig. 2J). No detectable level of 3MH was observed in MKRN2-FLAG-expressing WT cells (Supplemental Table S2), confirming the N1-position-specific His methylation of MKRN2.
As CARNMT1 is a dipeptide carnosine MTase and a protein MTase, we tested whether carnosine or anserine inhibits the protein MTase function. An in vitro protein methylation inhibition assay was performed using CARNMT1 and MKRN2 as a His MTase and its substrate (Supplemental Fig. S2E,F). Carnosine inhibited MKRN2 methylation at ∼3 mM (IC50 = 3.2 mM ± 1.2 mM), whereas anserine showed no inhibitory effect up to 30 mM. This suggests that MKRN2 (protein substrate) and carnosine (dipeptide) compete as substrates for CARNMT1.
His residues in C3H ZF motifs are preferentially methylated by CARNMT1
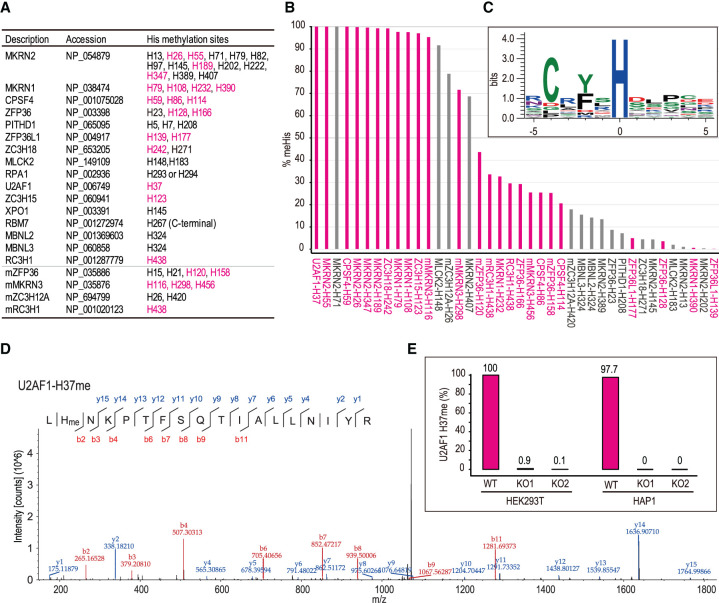
To further characterize the target His specificity of CARNMT1 catalysis, PMS screening hit proteins as well as manually selected C3H ZF proteins and MLCK2 were expressed with a FLAG tag in the CARNMT1 KO HEK293T cells or KO + WT HEK293T cells. As for MLCK2, it is not a C3H ZF protein but has been known to undergo 1MH modification in vivo (Meyer and Mayr 1987), and we observed high-level methylation of MLCK2 peptides of the surrounding sequence, including the methylation targeted His (Fig. 1C). Methylation was determined by LC-MS/MS or amino acid analysis (for RNA-binding motif protein 7 [RBP7], where the methylation site was the C-terminal His). Figure 3A summarizes the identified methylated His sites of the examined proteins. Here, we identified 52 His sites in 20 proteins that were methylated by CARNMT1. Among the 52 methylated sites, approximately half (25 out of 52) were localized in the C3H ZF motif, and the majority of the highly methylated His sites (>90% methylated) were in the C3H ZF motif regions; i.e., H37 in U2AF1, H79/H108 in MKRN1, H26/H55/H189/H347 in MKRN2, H116 in mMKRN3, H59 in CPSF4, H242 in ZC3H18, and H123 in ZC3H15 (Fig. 3B). One such highly methylated His site in the non-C3H ZF motif region was H148 in MLCK2 (Fig. 3B; Supplemental Fig. S3A,B). From the identified His sites with >50% methylation, the consensus motif of CARNMT1-mediated His methylation was determined to be Cx(F/Y)xH, which was the same consensus sequence as the C3H ZF motif (Fig. 3C). His methylation detected in the analyzed proteins was either undetectable or greatly reduced in CARNMT1 KO cells, confirming that the His methylation is catalyzed by CARNMT1 (Supplemental Table S3).
Figure 3.
The His residues in the C3H ZF motifs are preferentially methylated by CARNMT1. (A,B) List of in vivo His-methylated proteins by mammalian CARNMT1 (A) and abundance of His methylation (B). CARNMT1 KO cells rescued with overexpressed CARNMT1-HA WT were transfected with the indicated FLAG-tagged protein-expressing plasmid. IP-purified proteins were analyzed with LC-MS/MS except for the one indicated by an asterisk, the methylation site of which was determined by MRM MS. For all proteins listed, either His methylation was undetectable or trace amounts were observed when expressed in CARNMT1 KO cells (Supplemental Table S3). (Cyan) Methylated His on C3H zinc finger. (C) Consensus His methylation sites for CARNMT1. Amino acid sequences with >50% His methylation sites were analyzed with WebLogo3 (Crooks et al. 2004). (D,E) Methylation of endogenous U2AF1 in WT cells but not in CARNMT1 KO cells. (D) MS/MS spectrum of U2AF1 peptides containing methylated H37. (E) Abundance of methylated peptide and its corresponding unmethylated peptides. Methylation of U2AF1-H37 was analyzed in two independent HEK293T KO clones and two independent HAP1 KO clones. See also Supplemental Figures S2 and S3 and Supplemental Table S3.
To further evaluate the presence of newly identified His methylation in vivo, endogenous U2AF1 was immunoprecipitated with an anti-U2AF1 antibody from WT or CARNMT1 KO cells (HEK293T and HAP1 cells; two independent clones each), and their methylation was determined via LC-MS/MS analysis (Fig. 3D,E; Supplemental Fig. S3C,D). U2AF1 was almost completely (>99%) methylated at the H37 position in the first C3H ZF motif, whereas trace amounts (<0.1%) of methylation were observed in the KO cells. These results indicate that CARNMT1 is an N1-position-specific His MTase that preferentially targets the Cx(F/Y)xH motif as a consensus sequence; thus, CARNMT1 is responsible for His methylation of C3H ZF proteins.
As U2AF1 has two C3H ZF motifs, we examined whether both His residues in the zinc finger (H37 and H173) were methylated. In endogenous U2AF1, methylation of H37 but not of H173 could be detected by LC-MS/MS analysis (Supplemental Table S4). The amino acid analysis showed a significant reduction in 1MH levels in the H37A mutant, while the H173A mutation had little effect on 1MH levels (Supplemental Fig. S3E). To further investigate the specificity among ZF motifs, we constructed a swap mutant of the ZF domain in U2AF1 (ZF2/ZF1) (Supplemental Fig. S3F). Their methylation levels were determined by LC-MS/MS (Supplemental Fig. S3G). We observed a high level of methylation of ZF1 even when the position was swapped to the second ZF position (96.4% in H173 of ZF2/ZF1), whereas little methylation of ZF2 occurred regardless of the position. This suggests that there is an important sequence preference around the methylation site of the ZF domain. We found that S/L/R often presents at position −1 before the methylated His (Fig. 3C), which may contribute to the substrate specificity/preference of CARNMT1 for the C3H ZF motifs. Furthermore, the Cx(F/Y)xH sequence is critical but not sufficient for efficient His methylation by CARNMT1. High levels of methylation of several nonconsensus His residues, such as H71 and H407 in MKRN2, H148 in MLCK2, and H26 in ZC3H12A, were also observed, indicating the presence of an additional target preference for CARNMT1 other than the Cx(F/Y)xH sequence.
Impact of His methylation on zinc binding of the C3H ZF motif
As His methylation by METTL9 occurs on alternating His (HxH) motifs, thereby modulating their binding affinities against metal ions (Davydova et al. 2021), an isothermal titration calorimetry assay was performed to test whether His methylation of C3H ZF motifs by CARNMT1 also has an impact on their binding affinities against zinc ion (Supplemental Fig. S4A,B). A C3H ZF peptide around ZF1 of Nup475/mZFP36 (Nup475-1D) (Amann et al. 2003) was synthesized with or without N1-histidine methylation at H120. Both peptides bound to zinc ions with almost the same dissociation constant (Supplemental Fig. S4C). However, we found that the thermodynamic parameters enthalpy (ΔH) and entropy (−TΔS) changed between the two peptides (Supplemental Fig. S4D,E), suggesting that His methylation may alter the binding mode or protein conformation, since ΔH values allow characterization of proton movement linked to the association of proteins and ligands (Ward and Holdgate 2001). Thus, His methylation of C3H ZF motifs may impact the binding mode with zinc ion, which induces a conformational change around the zinc finger domain, thereby affecting its target nucleotide recognition.
His methylation by CARNMT1 regulates the RNA-binding activities of tristetraprolin (TTP) and Roquin
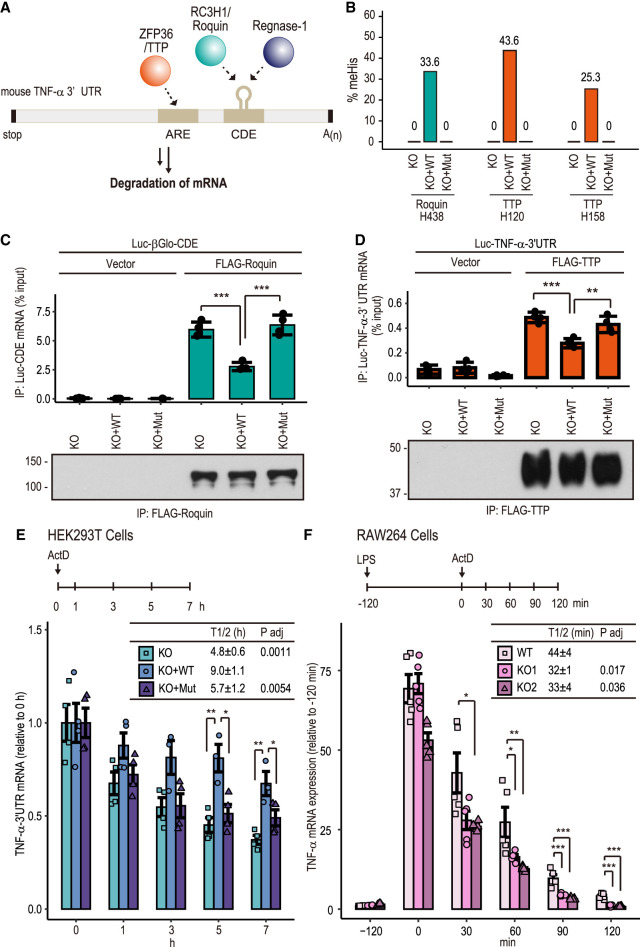
As C3H ZFs are important for RNA binding (Carballo et al. 1998; Hall 2005), we tested whether His methylation modulates RNA-binding ability. Several C3H ZF proteins, including the CARNMT1 target TTP (also known as ZFP36), Roquin (also known as RC3H1), and Regnase-1 (also known as ZC3H12A), are expressed in macrophages and regulate the mRNA stability of cytokines, such as tumor necrosis factor-α (TNF-α) (Hall 2005; Liang et al. 2008; Leppek et al. 2013; Mino et al. 2015). Expression of TNF-α, one of the most potent proinflammatory cytokines in mammals (Tracey and Cerami 1994), is tightly controlled post-transcriptionally by several regulatory elements in the 3′ untranslated region (UTR). An AU-rich element (ARE) suppresses translation and causes rapid degradation of mRNAs in the cytoplasm. It is recognized by ARE-binding proteins, such as TTP, that promote rapid mRNA decay during macrophage stimulation (Carballo et al. 1998). Roquin (Leppek et al. 2013) and Regnase-1 (Mino et al. 2015) also bind to the constitutive decay element (CDE) in the 3′ UTR and destabilize the mRNA (Fig. 4A). In all cases, the C3H ZF domain has an important role in RNA binding (Mino and Takeuchi 2018; Garg et al. 2021).
Figure 4.
His methylation by CARNMT1 regulates RNA-binding activity of TTP and Roquin. (A) Schematic of TNF-α 3′ UTR-binding proteins TTP, Roquin, and Regnase-1. TTP recognizes ARE (AU-rich element), whereas Roquin and Regnase-1 target CDE (constitutive decay element) and promote mRNA degradation. (B) LC-MS/MS analysis of His methylation of FLAG-TTP (H120 and H158) and FLAG-Roquin (H438). See Supplemental Figure S5, A–C, for LC-MS/MS spectrum. (C,D) FLAG-Roquin and FLAG-TTP bind less to TNF-a reporter mRNA in KO + WT rescue cells. Cells were transfected with the indicated Luc reporter and FLAG-Roquin (C) or FLAG-TTP (D). FLAG-tagged proteins were immunoprecipitated with M2-agarose, and then bound RNAs were isolated and analyzed by RT-qPCR. n = 3, mean ± SD, Tukey HSD test. (**) P < 0.01, (***) P < 0.001. Input RNA and input FLAG proteins are shown in Supplemental Figure S5, D and E. (E) mRNA decay assay. TNF-α 3′ UTR mRNA reporter was transfected with FLAG-TTP and FLAG-Roquin. Forty-eight hours after transfection, cells were treated with 5 mM actinomycin D at the indicated time, and the reporter mRNA was analyzed with qPCR. n ≥ 3, mean ± SEM, Tukey HSD test. (*) P < 0.05, (**) P < 01.0. (F) mRNA decay in macrophage-like RAW264 cells. Carnmt1 KO Raw264 cells (Supplemental Fig. S5F–H) were stimulated with LPS, and mRNA levels were measured after treatment with 5 µg/mL actinomycin D. n ≥ 3, mean ± SEM, Tukey HSD test. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. See also Supplemental Figures S2 and S5.
We tested whether the TNF-α mRNA-binding activities of TTP and Roquin were modulated by His methylation. FLAG-TTP or FLAG-Roquin was coexpressed with reporter constructs (Mino et al. 2015) in CARNMT1 KO HEK293T cells, KO + WT cells, or KO + Mut cells (Supplemental Fig. S2). His methylation in the C3H ZF motif of FLAG-TTP (H120 and H158) and FLAG-Roquin (H438) was determined by LC-MS/MS analysis, and methylation was undetectable in KO or KO + Mut cells and was ∼25%–40% in KO + WT cells (Fig. 4; Supplemental Fig. S5A–C). Next, the levels of TNF-α reporter mRNA coimmunoprecipitated with FLAG-tagged TTP or Roquin were measured via reverse transcriptase polymerase chain reaction (RT-PCR) (Fig. 4C,D; Supplemental Fig. S5D,E). The obtained mRNA levels decreased in KO + WT cells, suggesting that the methylation of TTP and Roquin negatively regulates TNF-α mRNA binding.
Subsequently, mRNA decay of the reporter construct (Luc-TNFα-3′ UTR) was examined in CARNMT1 KO or KO + WT cells transfected with FLAG-TTP and FLAG-Roquin (Fig. 4E). Actinomycin D (5 µg/mL) was added for the indicated time to inhibit transcription, and the mRNA level was analyzed via RT-qPCR. The reporter mRNAs were more stable in KO + WT cells (t1/2 = 9.0 h ± 1.1 h) than in KO cells (t1/2 = 4.8 h ± 0.6 h) and KO + Mut cells (t1/2 = 5.7 h ± 1.2 h).
To analyze the TNF-α mRNA decay in macrophages, Carnmt1 KO murine macrophage-like RAW264 cells were generated using the CRISPR–Cas9 method (Supplemental Fig. S5F–G). Methylation of the CARNMT1 target C3H His site (H37) of mouse U2AF1 was again almost 100% in WT RAW264 cells but was undetectable in the Carnmt1 KO cells, suggesting that CARNMT1 was not functional in the KO clones (Supplemental Fig. S5H). Lipopolysaccharide (LPS; 1 µg/mL) was added for 2 h to induce TNF-α mRNA expression, followed by 5 µg/mL actinomycin D for the indicated time. TNF-α mRNA levels (Fig. 4F) and protein levels (Supplemental Fig. S5I) were analyzed via RT-qPCR or Western blotting in WT cells and the two independent Carnmt1 KO clones. Carnmt1 KO RAW264 cells exhibited faster TNF-α mRNA decay (t1/2 = 32 min ± 1 min in KO1, and t1/2 = 33 min ± 1 min in KO2) (Fig. 4F) than WT cells (t1/2 = 44 ± 4 min) as well as rapid TNF-α protein degradation (Supplemental Fig. S5I). These results suggest that CARNMT1 regulates TNF-α mRNA stability via methylation of C3H ZFs in TTP and Roquin, which attenuates their RNA-binding activities.
CARNMT1 modulates alternative splicing (AS)
In eukaryotic cells, protein-coding genes are transcribed into precursor mRNA (pre-mRNA) transcripts, which have nontranslated intervening sequences called introns. In the nucleus, introns are excised from pre-mRNA by splicing reactions, and the mature mRNA is then exported to the cytoplasm. Introns possess consensus sequences that define the 5′ splice sites, branch sites, and 3′ splice sites, which contain a well-conserved AG dinucleotide (Will and Luhrmann 2011; Yoshida et al. 2015; Ule and Blencowe 2019). The 3′ splice site is recognized by the U2AF heterodimer, which consists of a large U2AF2 subunit and a small U2AF1 subunit (Zamore and Green 1989). The large subunit U2AF2 possesses three consecutive RNA recognition motifs (RRMs), while the small subunit U2AF1 has one RRM domain flanked by two C3H ZFs (Mollet et al. 2006).
As C3H ZFs bind to RNA and are involved in RNA regulation (Carballo et al. 1998; Hall 2005), whole-transcriptome sequencing (RNA-seq) analysis was performed using total RNA from CARNMT1 KO HEK293T cells. The sequencing reads were mapped to the human genome reference sequence, and 292 changes (false discovery rate [FDR] < 0.01; |IncLevelDifference| > 0.1) were identified in splicing from 25,212 AS events (Fig. 5A). The most observed changes in splicing in CARNMT1 KO cells were cassette exon (CE)-type events, and 133 genes were affected (Supplemental Table S5). Sashimi plots for ATP11C and MRPS18C showed differential CE-type events in CARNMT1 KO cells (Fig. 5B). CE is a splicing event in which an intervening exon between two exons from the mature mRNA sequence is either included or skipped to generate protein isoforms. CE accounts for over half of the alternatively spliced events (Cui et al. 2017).
Figure 5.
CARNMT1 modulates AS. (A) Summary of the high-throughput RNA sequencing (RNA-seq) analysis. Five representative AS patterns are depicted. The numbers of statistically significant (FDR < 0.01; |IncLevelDifference| > 0.1) splicing pattern changes compared with CARNMT1 knockout cells are indicated at the right. The numbers of total splicing events are indicated in parentheses. (B) Sashimi plot of ATP11C and MRPS18C. The X-axis indicates localization of each gene according to hg19 coordinates, and the Y-axis indicates RPKM (reads per kilobase of exon per million mapped reads). (C) LC-MS/MS analysis of endogenous H37me of U2AF1 in each cell line used for the splicing assay. (D) RT-PCR analysis of mRNAs for validation of the RNA-seq. The indicated RNAs (ATP11C, ATP13A3, FIP1L1, MRPS18C, PYCR1, STX16, TM2D3, and TPRA1) from HEK293T cells were analyzed with RT-PCR using primers targeting each CE (Supplemental Fig. S6A). n = 3, mean ± SD, Tukey HSD test. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. (E) Box plots of the strength of 3′ splice site (3′ss Maxent). All sequences of CE exons in A and human RefGene exons were analyzed. Significance was calculated by Wilcoxon rank sum test. (F) Sequence analysis on upstream 3′ss of CE exons from CARNMT1 KO cells. Motif analysis of upstream 3′ splice site CE-skipped (top), CE-included (middle), and RefGene (bottom) exons with WebLogo. The height of each letter is proportional to the probability of the corresponding base at the given position. (G, top) Schematic overview of the minigene derived from either WT or mutated (T to C) ATP11C. (Bottom) Gel-like view of capillary electrophoresis of ATP11C alternative splicing and quantification of the percentage of skipped products. (H) Schematic illustrations of RNA constructs for in vitro RNA-binding assay. (I) In vitro transcribed RNA corresponding to the 3′ half of the intron-containing branch point sequence (BPS), polypyrimidine tract (PPT), 3′ splice site, and downstream cassette exon was incubated with whole-cell extracts from either 293 control cells or CARNMT1 KO cells under splicing condition. (Left) The RNAs were immunoprecipitated with anti-U2AF1 antibody, and input RNA (2.5%) and coprecipitated RNAs were analyzed by Northern blotting. (Right) Quantifications of Northern blotting. Means ± SEM are given for three independent experiments, and two-tailed Student t-test values were calculated. (*) P < 0.01. See also Supplemental Figure S6.
We chose eight genes (ATP11C, ATP13A3, FIP1L1, MRPS18C, PYCR1, STX16, TM2D3, and TPRA1) from the genes that showed differential AS patterns of CE type (more skipped in CARNMT1 KO cells) in the RNA-seq analysis and validated such AS changes via RT-PCR. Endogenous U2AF1 methylation was determined in each cell line, confirming almost 100% methylation of H37 in WT and KO + WT rescue cells and no detectable methylation in KO and KO + Mut rescue cells (Fig. 5C). The mRNAs of all eight genes had more skipped exons in KO cells than in WT cells (Fig. 5D; Supplemental Fig. S6A). This phenotype was rescued in KO + WT cells but not in KO + Mut cells, suggesting that the catalytic activity of CARNMT1 is necessary for the regulation of AS. RNA-seq analysis revealed that 99 genes were up-regulated and 83 genes were down-regulated in CARNMT1 KO HEK293T cells (FDR < 0.01), but only three out of the 133 genes included displayed differential CE-type events (red dots vs. pink dots in Supplemental Fig. S6C; Supplemental Table S5), suggesting that this CE-type phenotype in CARNMT1 KO cells is not associated with changes in gene expression.
To further confirm the CE-type change in different cells, mRNA levels in CARNMT1 KO HAP1 cells were determined via RT-PCR (Supplemental Fig. S6B). All mRNAs affected in CARNMT1 KO HEK293T cells also showed increased CE events in KO HAP1 cells, confirming the effect of CARNMT1 depletion on CE regulation. Furthermore, the affected CE contained a weak 3′ splice site (Maxent 3′ss score based on maximum entropy model) as compared with the reference genes, but 5′ splice site strength (Maxent 5′ss), branch point sequence (BPS) strength, and exon length were not significantly different (Fig. 5E; Supplemental Fig. S6D), suggesting that 3′ splice site recognition may be important for the regulation of CE by CARNMT1. Inspecting the 23-nt 3′ intron/exon junction sequence used to calculate the 3′ splice site strength (Fig. 5F), we recognized slightly more variability in sequences between the −20 and −4 positions in alternatively spliced CE than those of consensus sequences based on the reference sequence. Furthermore, we found a mild but noticeable C-to-T shift at the −3 position (−1 position from AG site) in the CE-skipped sequences, and this difference was more obvious between CE-skipped and CE-included sequences.
C3H ZFs of U2AF1 are essential for the splicing reaction of U2AF1 (Will and Luhrmann 2011; Herdt et al. 2020), and the S34F/Y substitution near H37 in the first C3H ZF induces AS changes (Ilagan et al. 2015; Inoue et al. 2016). C versus T preference at the −3 position has been reported to be critical for differential AS patterns of CE type in S34F/Y mutant cells (Ilagan et al. 2015). We reanalyzed the AS event that occurred by the S34F substitution with our AS analysis pipelines. The sequencing reads (GSE58871) (Ilagan et al. 2015) were mapped to the human genome reference sequence, and 2163 CE-type event changes (FDR < 0.01; |IncLevelDifference| > 0.1) were identified (Supplemental Fig. S6E). A C-to-T shift at the −3 position in the CE-skipped sequence was also observed in the S34F, suggesting a similar impact on splicing between S34F and CARNMT1 KO (Supplemental Fig. S6F). CE genes were compared, and ∼10% of the differentially altered CE events detected in CARNMT1 KO HEK293T cells overlapped with those of S34F mutant cells (nine out of 89 for CE-skipped and seven out of 68 for CE-included) (Supplemental Fig. S6G). To determine whether the C-to-T shift at the −3 position is important for CARNMT1-mediated CE skip, we constructed an ATP11C minigene and its mutant minigene with a T-to-C substitution. The minigenes were expressed in WT or CARNMT1 KO HEK293T cells, and the mRNA levels were determined via RT-PCR. CE skip of the ATP11C minigene was greatly increased in KO cells, whereas that of the T-to-C mutant was significantly diminished (Fig. 5G). This indicates that T at the −3 position is important for the CE event regulated by CARNMT1. To check whether the RNA-binding activity of U2AF1 to pre-mRNA of genes exhibiting the CE phenotype is affected by His methylation, we performed an RNA-binding assay between U2AF1 and the MRPS18C pre-mRNA element transcribed in vitro (Fig. 5H). We also used human adenovirus 2 major late (AdML) pre-mRNA, which possesses a stronger 3′ splice site score than the MRPS18C 3′ splice site, as a control mRNA (Maxent scores in Fig. 5H) and C at the −3 position. The RNA-binding assay showed that U2AF1 from WT cells bound more strongly to the MRPS18C pre-mRNA-derived 3′ sequence (MRPS18C 3′ss) compared with that from KO cells, whereas they bound to the same extent toward the AdML pre-mRNA-derived 3′ sequence (AdML 3′ss) (Fig. 5I). Intriguingly, the differential interaction between MRPS18C RNA and U2AF1 between WT and KO cells was canceled by CT-(the −4 and −3 position [−2 and −1 position from the AG site])-to-AC substitution (MRPS18C CT to AC) (Fig. 5H,I), suggesting that weak 3′ splice site sequences around the −3 position are probably the major determinants of the differences between WT and KO cells.
Taken together, methylation of U2AF1 by CARNMT1 affected its 3′ss mRNA-binding activity, thereby modulating the pre-mRNA AS events in mammalian cells. In this study, we focused on CE because CE events were the most common and their analysis was simpler. However, if His methylation of C3H ZF impacts U2AF1 3′ss recognition, it would not be surprising even if CARNMT1 depletion also affected types of AS events other than CE, such as alternative 3′ splice site (A3′ss) events.
CARNMT1 is essential for mouse embryonic development
To evaluate the physiological role of CARNMT1 in mice, CRISPR–Cas9-mediated Carnmt1 KO mice were generated, and heterozygous KO (Het; +/−) male and female mice were intercrossed (Fig. 6A–C). Homozygous KO (−/−) mice were not obtained 14 d after birth (P14), indicating that Carnmt1 KO mice are embryonic or postnatally lethal (Fig. 6D). To investigate embryonic development and lethality, the genotypes and crown–rump lengths (CRLs) of embryos were determined at E11.5–E17.5. As shown in Figure 6, D–F, homozygous KO embryos showed growth retardation as early as E11.5 and died around E13.5–E15.5. This indicates that Carnmt1 KO mice are embryonically lethal.
Figure 6.
CARNMT1 is essential for mouse embryonic development. (A) Schematic of CRISPR–Cas9-mediated Carnmt1 genome editing. Two guide RNAs targeting upstream of (gRNA1) and downstream from (gRNA2) exon 4 were injected into fertilized C57/B6 eggs together with CAS9 protein. (B) Typical genotyping image of WT (+/+), Het (+/−), and KO (−/−) embryos (E11.5). (C) Total proteins from WT (+/+), Het (+/−), and KO (−/−) embryos (E11.5) were separated with SDS-PAGE and detected with anti-CARNMT1 antibody (top) or anti-α-tubulin antibody (bottom). An asterisk indicates a band corresponding to the molecular weight of CARNMT1. (D) Typical image of Carnmt1 WT, Het, and KO embryos at the indicated time point. (E,F) Crown–rump lengths (CRLs) of the embryos. Mean ± SEM, Tukey HSD test. (*) P < 0.05, (***)P < 0.001. (#) Dead pup. (G) Carnmt1 mRNA expression profile in mice. The expression level of Carnmt1 was normalized to that of Gapdh. n = 3, mean ± SD. (H) Amino acid analysis of tissues from WT (+/+) and Het (+/−) mice and E11.5 embryos from Carnmt1 (−/−) mice. Proteinous 1MH and 3MH (Supplemental Fig. S7I) were measured with MRM LC-MS/MS. n = 3, mean ± SEM, Student's t-test. (*) P < 0.05, (***) P < 0.001. (I,J) CARNMT1 protein levels in skeletal muscle from WT (+/+) and Het (+/−) adult mice were analyzed with anti-CARNMT1 antibody (top) and anti-α-tubulin antibody (bottom). (I) Quantitation of the CARNMT1 protein. Mean ± SEM, Student's t-test. (*) P < 0.05. (K) Summary of the RNA-seq analysis in mouse embryonic brains and livers. Five representative AS patterns are depicted. The numbers of statistically significant (FDR < 0.01; |IncLevelDifference| > 0.1) splicing pattern changes compared with Carnmt1 knockout embryos are indicated at the right. (L) Sequence analysis of upstream 3′ss site of CEs from Carnmt1 KO embryos. Motif analysis of upstream 3′ splice site with WebLogo (top; E13.5 brain) and RefGene (bottom; E13.5 liver). See also Supplemental Figure S7.
To investigate whether MTase activity is important for embryonic development, catalytic mutant (ΔCat) mice with G199/201R substitution in CARNMT1 were generated using the CRISPR–Cas9 system (Supplemental Fig. S7A). Heterozygous enzyme-dead (ΔCat/+) male and female mice were intercrossed to obtain homozygous enzyme-dead (ΔCat/ΔCat) mice. ΔCat/ΔCat embryos survived until E13.5 but died around E13.5–E15.5, as in the case of Carnmt1 KO (Supplemental Fig. S7A–H). These results suggest that MTase activity is essential for the critical function of CARNMT1 in mouse embryonic development.
As homozygous KO mice showed embryonic lethality, heterozygous KO mice (4 wk old, male) were used to examine the mRNA expression profile of Carnmt1 (Fig. 6G) and its protein 1MH levels (Fig. 6H) in different tissues and organs. Carnmt1 mRNA was ubiquitously expressed, especially at relatively high levels in the testis, spleen, and skeletal muscles. Total proteinaceous 1MH was remarkably enriched and 1MH level was decreased in the skeletal muscle of heterozygous mice. Skeletal muscle CARNMT1 protein levels were decreased in heterozygous KO mice compared with WT mice (Fig. 6I,J), suggesting that monoallelic expression of Carnmt1 reduced protein levels there. E11.5 embryos in the homozygous KO background but not in the heterozygous KO showed ∼50% reduction in total 1MH levels compared with WT embryos (Fig. 6H). Protein 3MH levels were unchanged in all tested tissues/organs (Supplemental Fig. S7I). These results indicate that even though CARNMT1 may be ubiquitously expressed, 1MH levels of CARNMT1 target proteins can be regulated by controlling CARNMT1 expression and activity in physiological conditions, which is one of the important issues to be addressed in the future. As homozygous KO mice are lethal, whether 1MH levels in other tissues/organs are reduced under complete KO conditions remains unknown. Further studies with tissue-specific conditional KO mice are needed to address this issue. Moreover, further analysis of His-methylated substrates and the subsequent functional analysis of their methylation are necessary to determine the essential role of CARNMT1 in embryonic development.
Finally, we performed RNA-seq analyses on total RNAs from Carnmt1 KO embryos (E13.5 brains and livers). We identified 737 and 585 changes (FDR < 0.01; |IncLevelDifference| > 0.1) in splicing events in the brains and livers, respectively (Fig. 6K). In both tissues, the most observed splicing changes in Carnmt1 KO were again CE-type events, as shown in Figure 5A. In addition, C-to-T shifts at the −3 position in the CE skip sequence were also observed in these tissues (more evident than in 293T cells) (Fig. 6L), further suggesting that 3′ splice site sequences around the −3 position are the major determinants of the differences between WT and Carnmt1 KO. Again, further analysis will be required to determine whether the AS in Carnmt1 KO accounts for the embryonic lethality.
Discussion
CARNMT1 catalyzes N1-position-specific protein His methylation in mammals
Methylation of His residues was first discovered >50 yr ago, and it was revealed that β-actin and myosin contain 3MH (Johnson et al. 1967; Elzinga 1971). The responsible catalytic enzyme(s) remained unknown in higher eukaryotes until 2018. SETD3 is the enzyme involved in the introduction of 3MH into His (H73) of β-actin (Kwiatkowski et al. 2018; Wilkinson et al. 2019). His methylation plays an important role in the regulation of actin function and muscle contractility. METTL18, the mammalian homolog of HPM1, also catalyzes N3-position-specific His methylation of RPL3 (Małecki et al. 2021; Matsuura-Suzuki et al. 2022). METTL9 is a protein N1-position-specific His MTase that targets alternating His residues, resulting in the regulation of metal ion binding (Davydova et al. 2021). However, other than SETD3, METTL18, and METTL9, the remaining enzymes involved in His methylation of proteins in mammals remain unknown. In this study, we demonstrated that CARNMT1/UPF0586, known as the dipeptide carnosine (βAla-His) MTase, acts as the major protein N1-position-specific His MTase in mammals. Although both METTL9 and CARNMT1 methylate the N1 position of the His residue, their substrate specificities are quite distinct. METTL9 targets the alternating His (HxH) motif in proteins (Davydova et al. 2021), whereas CARNMT1 preferentially methylates the Cx(F/Y)xH motif. Kapell and Jakobsson (2021) reported that (C/H)x(M/H)xH is a consensus motif for His methylation in human cells. The (C/H)x(M/H)xH sequence may be merged with the METTL9 and CARNMT1 consensus sequences (HxHxH) and (Cx(F/Y)xH), respectively. Since the consensus motif is likely their sum, N1-position-specific His methylation by METTL9 and CARNMT1 may occur across mammalian proteomes. Indeed, both METTL9 (Davydova et al. 2021) and CARNMT1 have diverse protein substrates in mammals, whereas the N3-position-specific His MTases SETD3 and METTL18 specifically target β-actin (Kwiatkowski et al. 2018; Wilkinson et al. 2019) and RPL3 (Małecki et al. 2021; Matsuura-Suzuki et al. 2022), respectively. Notably, the Cx(F/Y)xH motif is not sufficient for methylation by CARNMT1. U2AF1 contains two C3H ZF domains, but the first ZF domain containing H37 is predominantly methylated by CARNMT1 (Supplemental Fig. S3E–G). We also observed high-level methylation on nonconsensus His residues, such as H148 of MLCK2 (Fig. 3), suggesting an additional substrate preference for CARNMT1 other than the Cx(F/Y)xH motif.
To date, no common substrate has been identified between CARNMT1 and METTL9. KO of METTL9 or CARNMT1 in HEK293T and HAP1 cells reduced approximately half of the protein 1MH level (Fig. 1E,F), indicating that both CARNMT1 and METTL9 are major His MTases that equally regulate the protein 1MH levels in the two cell lines. METTL9 may be predominant in mouse embryonic fibroblasts (MEFs), as METTL9 single KO showed ∼80% loss of 1MH in MEFs (Davydova et al. 2021), whereas CARNMT1 single KO showed no significant loss of 1MH (Supplemental Table S6). In contrast, CARNMT1-dependent 1MH may be predominant in the skeletal muscle, since the skeletal muscle from Carnmt1 heterozygous KO mice showed significant loss of 1MH (Fig. 6H).
We found that skeletal muscle is a major organ that is enriched in His-methylated proteins with both forms (Fig. 6H; Supplemental Fig. S7I). β-Actin and myosin are 3MH-containing proteins (Johnson et al. 1967; Elzinga 1971), whereas MLCK2/MYLK2 contains 1MH (Meyer and Mayr 1987; for review, see Jakobsson 2021). MLCK2 is predominantly expressed in skeletal muscle and is a Ca2+/calmodulin-dependent serine–threonine protein kinase that phosphorylates the regulatory light chain (RLC) of sarcomere myosin (Stull et al. 2011). Although the N1-position-specific H157 methylation of MLCK2 has been reported in rabbit skeletal muscle (Meyer and Mayr 1987), the responsible MTase or physiological role of this modification remains to be elucidated. In the present study, we identified CARNMT1 as a His MTase for H148 of human MLCK2, which corresponds to H157 of rabbit MLCK2 (Fig. 3; Supplemental Fig. S3A,B). As several other methylated proteins identified in this study, such as MBNL2 and MBNL3, were highly expressed in the skeletal muscle, these methylations may also contribute to the high 1MH content in this tissue. Currently, the biological functions of His methylation in skeletal muscles remain unclear. Relatedly, although we have not analyzed gene expression in KO mice, GO term analysis of RNA-seq of HEK293T KO cells showed that development-related terms, such as anatomical structure development, multicellular organism development, and system development, are enriched in the down-regulated genes (Supplemental Table S5), suggesting that CARNMT1 is involved in the regulation of the expression of development-related genes. Further analysis of conditional KO mice may reveal the biological functions of His methylation in adult mice.
CARNMT1 was originally identified as a carnosine N1-position-specific His MTase in rat skeletal muscles. Our study also demonstrated that CARNMT1, but not METTL9, contributes to carnosine methylation (anserine production) in HEK293T cells (Fig. 1I). There are several possibilities for the remaining anserine in CARNMT1 single-KO cells. Since we determined anserine by MRM LC-MS/MS, compounds with the same MRM transition were indistinguishable. If such compounds were present, the anserine level in KO cells was overestimated. Moreover, other MTases may contribute to carnosine methylation. The histamine N-methyltransferase (HNMT)-like protein acts as a carnosine methyltransferase in chickens (Drozak et al. 2013), indicating that the ortholog of HNMT-like or its related gene product(s) may function as other carnosine MTases in mammals.
CARNMT1 is a protein His methyltransferase that is essential for mouse embryonic development
Since the conventional CARNMT1 KO mice (Fig. 6) and the catalytically dead mutant mice (Supplemental Fig. S7) died around E13.5, the enzymatic activity of CARNMT1 is essential for mouse embryonic development. Wang-Eckhardt et al. (2020) showed that homozygous carnosine synthase 1 (Carns1) KO mice lacking both carnosine and anserine develop normally. This clearly indicates that CARNMT1 is crucial for embryonic development via protein His methylation but not carnosine methylation.
To our knowledge, U2AF1 is the only protein among the identified CARNMT1 targets whose homozygous knockout has been reported to cause an embryonic-lethal phenotype. U2AF1 homozygous KO shows embryonic lethality prior to organogenesis (Groza et al. 2023). Hematopoietic cell-specific conditional U2af1 KO mice also showed a later embryonic lethality (E18.5) (Wadugu et al. 2021). Although the fetal brains and livers from Carnmt1 homozygous KO embryos showed AS changes as compared with those from WT, we should carefully examine whether loss of His methylation of U2AF1 alone causes embryonic lethality. Since H37 in the U2AF1 ZF domain accommodates zinc ions, it is difficult to mutate the residue to any other amino acids that would cause disruption of zinc coordination. Further identification of protein substrates during embryogenesis will help in understanding CARNMT1 target protein His methylation that is essential for the development.
His methylation as an additional layer of epitranscriptomic regulation of mRNA
N6-methyladenosine (m6A), methylation at the N6 position of adenosine, is the most abundant epitranscriptomic modification in eukaryotic mRNA (Feng et al. 2018; Murakami and Jaffrey 2022). N6-adenosine methylation is induced by m6A MTase complexes, including METTL3 and METTL14 (Liu et al. 2014). Thousands of mRNAs in human cells are subjected to m6A modification, which affects multiple aspects of mRNA regulation, including pre-mRNA splicing, mRNA transport from the nucleus to the cytoplasm, translation, mRNA turnover, mRNA stability, and subcellular localization (Zhao et al. 2017). m6A is enriched around the stop codons in 3′ UTRs and within internal long exons (Geula et al. 2015). Wei et al. (2021) found that ∼10% of m6A peaks are located in alternative introns/exons, often close to 5′ splice sites, which modulate the splicing events.
Here, we identified His methylation of mRNA regulators, such as U2AF1, TTP, and Roquin, all of which are C3H ZF proteins. U2AF1 is a splicing factor that recognizes the 3′ splice site of pre-mRNA. We found that methylation of H37 in U2AF1 modulated the 3′ splice site recognition, thereby regulating mRNA splicing in culture cell lines (HEK293T and HAP1 cells) (Fig. 5) as well as mouse embryos (E13.5 brains and livers) (Fig. 6G,H). U2AF1-S34F mutation is frequently observed in myelodysplastic syndromes (MDSs) and MDS-related acute myeloid leukemia (AML); therefore, it has been suggested that this mutation is associated with the onset and/or pathogenesis of these hematologic diseases (Fei et al. 2018; Zhao et al. 2022). Since the effects on AS from the U2AF1-S34F mutation and the effects on AS from the deletion of CARNMT1, which eliminates U2AF1-H37 methylation, are very similar, there may be a link between reduced or defective CARNMT1 function and MDSs or MDS-related AML. As for TTP and Roquin, they both bind to the 3′ UTR of TNF-α mRNA and negatively regulate its stability. His methylation increased mRNA stability by reducing its interaction with TTP or Roquin (Fig. 4). In all cases, further structural analysis is required to determine the precise mechanism by which His methylation affects mRNA recognition.
Arg methylation is known to play a role in various cellular processes, including transcription, pre-mRNA splicing, and mRNA stability, and multiple Arg MTases target/methylate RBPs (Guccione and Richard 2019). Arg modification of splicing factors occurs in the glycine-rich domain (RGG domain) (Thandapani et al. 2013). In contrast, His methylation is often located in the C3H ZF domain (Fig. 3). We have found that 25 C3H ZFs present in 12 RBPs are subjected to His methylation by CARNMT1 (Fig. 3A, colored in magenta), and their His methylation has important effects on the ZF domain functions (Figs. 4, 5). Thus, His methylation of RBPs could be considered as an additional layer of epitranscriptomic regulation in addition to that of Arg methylation. Furthermore, methylation of RBPs, together with RNA methylation, may cooperatively regulate RNA metabolism, which contributes to proper mRNA splicing and degradation and subsequently affects proteome diversity in mammals.
Materials and methods
Reagents and antibodies
All reagents used in this study were purchased from Sigma or Nacalai Tesque unless stated otherwise. Antibodies for U2AF1, CARNMT1, and TNF-α were purchased from MBL (RN085PW), Proteintech Group (24068-1-AP), and Abcam (ab183218), respectively. All antibodies were used for Western blotting with 1:1000 dilution in 5% skim milk and PBS + 0.1% Tween-20 and incubated for 16 h at 4°C with gentle agitation.
Cloning and mutagenesis
Full-length mouse Carnmt1/UPF0586 cDNA (NM_026120) was amplified from C57BL/6J mouse cDNA and cloned into pcDNA3 vector with C-terminal FLAG or HA sequence to obtain pcDNA3-mCARNMT1-HA or pcDNA3-mCARNMT1-FLAG for mammalian expression. The cDNA was cloned into pGEX4T1 vector (pGEX4T1-mCARNMT1) or pET19b vector (pET19b-mCARNMT1) to obtain GST-CARNMT1 or 9xHis-CARNMT1 in E. coli. A catalytic mutant of CARNMT1 (Mut) in which G199 and G201 were substituted with Arg (G199/201R) was obtained with site-directed mutagenesis according to the manufacturer's protocols (Agilent). For the stable expression of CARNMT1, a cDNA fragment of WT or Mut CARNMT1 with a C-terminal HA sequence was cloned into AgeI and EcoRI sites of the pQCXIP vector (Clontech) to obtain pQCXIP-mCARNMT1-HA or pQCXIP-mCARNMT1 Mut-HA. For stable expression of SETD3-FLAG, a cDNA fragment corresponding to human SETD3 (NM_032233) was cloned into AgeI and BamHI sites of pQCXIP vector with C-terminal FLAG (pQCXIP-SETD3-FLAG). cDNAs for CARNMT1 targets were amplified from HEK293T or C57BL/6J mouse cDNA and cloned into pcDNA3 vector with a C-terminal FLAG tag (pcDNA3-X-FLAG). All sequences were confirmed by DNA sequencing. Plasmids for FLAG-tagged mouse TTP (pFLAG-mTTP-WT), mouse Roquin (pFLAG-mRoquin-WT), or TNFα luciferase reporter constructs (pGL3-βGlo-TNF-CDE and pGL3-TNF-3′ UTR) were kindly provided by Dr. Mino and Dr. Takeuchi (Mino et al. 2015). pRL-TK control vector was obtained from Promega.
Generation of CARNMT1 KO cells
CARNMT1 KO human cells were generated with CRISPR–Cas9-mediated gene editing. Guide RNA for introns upstream of (5′-ACATATGCACCAATTAGAAG-3′) and downstream from (5′-GTTATATTAGTACTTAAAAA-3′) exon 2 were cloned into pL-CRISPR.EFS.GFP vector (Addgene 57818) and pL-CRISPR.EFS.tRFP vector (Addgene 57819), respectively. The plasmids (pL.CRISPR.GFP-gUPF0586 and pL.CRISPR.tRFP-gUPF0586) were transfected into HEK293T or HAP1 cells using PEI transfection reagent (Polysciences, Inc.), and GFP/RFP-double-positive cells were sorted and plated onto a 96-well plate. The KO clones were screened by genomic PCR. CARNMT1 and Mettl9 DKO HEK293T and HAP1 cells were generated essentially as described by Davydova et al. (2021). Briefly, guide RNAs targeted to introns upstream of (5′-TAATAAGTGATTATGGTTGT-3′) and downstream from (5′-GCAGTATTTTTCTGGAGCGG-3′) exon 3 in METTL9 were cloned into the BbsI and BsaI sites, respectively, of the PX330-B/B vector. The gRNA vectors were transfected into CARNMT1 KO cells with pEGFP-C1 vector, and GFP-positive cells were sorted and cloned. For generation of CARNMT1 KO murine RAW264 cells (RIKEN BRC RCB0535), the two gRNA plasmids for targeting exon 3 in CARNMT1 (upstream in exon junction [GFP]: 5′-AATTGCCTTGTTTCATTTCA-3′; downstream in intron [rRFP]: 5′-GATTCATAATGCTTATACCGA-3′) were transfected with Fugene HD transfection reagent (Promega), and GFP/RFP-double-positive cells were sorted and cloned as described above.
For generation of the CARNMT1 KO cells rescued with C-terminally HA-tagged WT CARNMT1 (KO + WT rescue) or mutant CARNMT1 with G199/201R substitutions (KO + Mut rescue), retrovirus packaging cells were transfected with the plasmids pQCXIP-mCARNMT1-HA or pQCXIP-mCARNMT1 Mut-HA using PEI transfection reagent (Polysciences, Inc.) and cultured for 24 h. The virus containing culture supernatant was then transferred to CARNMT1 KO HEK293T cells with 4 µg/mL polybrene. Twenty-four hours after infection, cells were selected with 1 µg/mL puromycin and cultured for an additional 2 wk with the drug.
Recombinant proteins
Recombinant METTL9 (Davydova et al. 2021) and METTL18 (Matsuura-Suzuki et al. 2022) were described previously. To obtain GST-CARNMT1, 9xHis-CARNMT1, or salmonella MTAN recombinant protein (a gift from Vern Schramm; Addgene plasmid 64041, http://n2t.net/addgene:64041, RRID: Addgene_64041), BL21 (pLysS) strains were transformed with the pGEX4T plasmids or pET19 plasmids, and the bacteria were cultured in 2× YT medium with 100 µg/mL ampicillin and 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 18 h at 18°C. The cells were pelleted and lysed with 1× PBS/0.5% NP-40 by sonication with a Branson Sonifier (Branson Ultrasonics Corp. S-250D) for 5 min on ice. The lysates were centrifuged at 15,000g for 10 min, and the supernatants were incubated with glutathione sepharose 4B (GE Healthcare) or Ni-NTA agarose (Qiagen) for 1 h at 4°C with gentle agitation. The agarose beads were washed five times with GST wash buffer (1× PBS at pH 7.4) or His wash buffer (50 mM Tris-HCl at pH 7.4, 25 mM imidazole) and eluted with GST elution buffer (50 mM Tris-HCl at pH 8.0, 50 mM glutathione) or His elution buffer (50 mM Tris-HCl at pH 7.4, 250 mM imidazole). The purified proteins were dialyzed with dialysis buffer (50 mM Tris-HCl at pH 8.0, 100 mM NaCl, 0.2 mM DTT, 10% glycerol) using Slide-A-Lyzer dialysis cassettes (10 kDa MWCO; Thermo Fisher Scientific) or an Amicon Ultra-15 centrifugal filter unit (10 kDa MWCO; Millipore), and their concentration was measured using the Bradford protein assay kit (Bio-Rad Laboratories). SETD3-FLAG was stably expressed in HEK293T cells with retrovirus infection. The FLAG-tagged SETD3 protein was purified with M2-agarose beads (Sigma-Aldrich), and the concentration of bound protein was determined by SDS-PAGE with BSA as the protein standard.
Peptide methylation assay
Peptides around His methylation sites were synthesized with a peptide synthesizer (ABI 433A) and purified with HPLC. Each peptide (100 µM) was incubated with 1 µg of MTases for 2 h at 30°C in 10 µL of a reaction buffer (50 mM Tris-HCl at pH 8.0). The reaction was stopped by adding 1% TFA, and the peptides were analyzed with a MALDI mass spectrometer (Microflex, Bruker Daltonics).
Methyl His content analysis
Methyl His content analysis was performed as described previously (Davydova et al. 2021). Briefly, cell pellets precipitated with ice-cold acetone were hydrolyzed with 6 N HCl for 24 h at 110°C, and the amino acids were dissolved in 25 µL of 5 mM ammonium formate with 0.001% formic acid. These were applied to a Vanquish UHPLC liquid chromatograph (Thermo Fisher Scientific) coupled to a TSQ Vantage or TSQ Altis triple quadrupole mass spectrometer (Thermo Fisher Scientific). The amino acids were separated on a C18 column (YMC-Triart C18, 2.0-mm × 100-mm length, 1.9-µm particle size; YMC Co., Ltd.). Mobile phase A was comprised of 5 mM ammonium formate with 0.001% formic acid, and mobile phase B was comprised of acetonitrile. The following slopes were used for a gradient elution at a flow rate of 0.3 mL/min: 0 min/0% B–1.5 min/0% B–2 min/95% B–4 min/95% B–4.1 min/0% B–7 min/0% B. The effluent from the column was directed to an HESI-II electrospray ion source (Thermo Fisher Scientific) connected to a triple quadrupole mass spectrometer operating in the positive ion multiple reaction monitoring mode, and the intensities of specific MH+ → fragment ion transitions were recorded (His m/z: 156.1 → 83.3, 93.2, 110.2; 1MH m/z: 170.1 → 95.3, 97.3, 109.2; and 3MH m/z: 170.1 → 81.3, 83.3, 124.2). The electrospray conditions were spray voltage: 3000 V, vaporizer temperature: 450°C, sheath gas pressure: 50 arbitrary units, auxiliary gas pressure: 15 arbitrary units, and collision gas pressure: 1.0 mTorr. With each batch of experimental samples, a series of standard samples was simultaneously prepared and run. Calibration curves were constructed for 1MH, 3MH, and His from the data obtained from the standard samples with a range of 1–250 nM. The measured concentrations and percentages of 1MH and 3MH in each experimental sample were calculated from the calibration curves.
Carnosine and anserine content analysis
HEK293T cells (1 × 107) were collected in a 1.5-mL tube. One-hundred microliters of 1× PBS was added to the cell pellets and then sonicated for 10 sec on ice. Protein concentrations were determined with a Bradford protein assay (Bio-Rad Laboratories). Three-hundred micrograms of the cell lysates was precipitated with 4 vol of ice-cold acetonitrile. Precipitated protein was removed by centrifugation at 14,000g for 10 min, and the cleared supernatants were applied to a Vanquish UHPLC liquid chromatograph (Thermo Fisher Scientific) coupled to a TSQ Altis triple quadrupole mass spectrometer. Cleared supernatants were separated on a Diol-HILIC column (2.0-mm × 50-mm length; YMC Co., Ltd.). Mobile phase A was comprised of 7.5 mM ammonium formate/0.001% FA, and mobile phase B was comprised of 90% ACN/7.5 mM ammonium formate/0.001% FA. The following slopes were used for a gradient elution at a flow rate of 0.4 mL/min: 0 min/99% B–3.5 min/53.5% B–3.6 min/10% B–4.0 min/10% B–4.1 min/99% B–6.0 min/99% B. The effluent from the column was directed to an HESI-II electrospray ion source (Thermo Fisher Scientific) connected to the triple quadrupole mass spectrometer operating in the positive ion multiple reaction monitoring mode, and the intensities of specific MH+→ fragment ion transitions were recorded (carnosine m/z 227.1 → 110.2, 83.2; anserine m/z 241.1 → 109.2, 95.2). The electrospray conditions were spray voltage: 3000 V, vaporizer temperature: 450°C, sheath gas pressure: 50 arbitrary units, auxiliary gas pressure: 15 arbitrary units, and collision gas pressure: 1.5 mTorr. Calibration curves were constructed from the data obtained from the standard samples with a range of 0.06–8 nM. The measured concentrations and percentages in each experimental sample were calculated from the calibration curves.
ProSeAM–MTase–SILAC (PMS) screening
ProSeAM was prepared as reported by Sohtome et al. (2018). PMS screening analysis was performed as described by Davydova et al. (2021) with some modifications. Briefly, CARNMT1 KO HEK293T cells were cultured in DMEM containing either light Arg and Lys or heavy isotope-labeled Arg (13C6 15N4 L-arginine) and Lys (13C6 15N2 L-lysine) (Thermo Scientific) for at least six doubling times. The cells were harvested and lysed in a lysis buffer (50 mM Tris-HCl at pH 8.0, 50 mM KCl, 10% glycerol, 0.5% NP-40). The cell lysates containing 150 μg of proteins were incubated for 2 h at 20°C with 150 μM ProSeAM with (heavy) or without (light) 10 μg of recombinant CARNMT1 (protein:enzyme ratio 15:1) in MTase reaction buffer (50 mM Tris-HCl at pH 8.0) supplemented with 1 μM salmonella MTAN to remove by-product (SeAH) after MTase reaction. The reaction was stopped by adding 4 vol of ice-cold acetone. After centrifugation at 15,000g for 5 min, precipitates were washed once with ice-cold acetone. The pellet was then dissolved in 39 µL of 1× PBS/0.2% SDS, after which 10 µL of 5× click reaction buffer and 1 µL of 10 mM azide-PEG4-Biotin (Click Chemistry Tools) were added, and the reaction mixture was incubated for 60 min at 20°C. The click reaction was stopped with 4 vol of ice-cold acetone. The pellet was resuspended in 75 µL of binding buffer (1× PBS, 0.1% Tween-20, 2% SDS, 20 mM dithiothreitol [DTT]) at 50°C. The light and heavy samples were mixed in a tube, 300 µL of IP buffer (TBS; 0.1% Tween-20) containing 3 µg of Dynabeads M-280 Streptavidin (Life Technologies Japan Ltd.) was added to the tube, and the tube was incubated for 30 min at room temperature (the final SDS concentration in the reaction was 0.5%). The protein-bound beads were washed three times with wash buffer (1× PBS, 0.1% Tween-20, 0.5% SDS) and twice with 100 mM ammonium bicarbonate (ABC) buffer and then analyzed by mass spectrometry. DTT (20 mM) was added to protein-bound Dynabeads in 100 mM ABC buffer, and the mixture was incubated for 30 min at 56°C. Next, iodoacetamide was added, and the mixture was incubated for 30 min at 37°C in the dark. The protein samples were then digested with 1 µg of trypsin (Promega). The protein fragments were applied to an EASY-nLC 1000 liquid chromatograph (Thermo Fisher Scientific) coupled to a Q Exactive hybrid quadrupole orbitrap mass spectrometer (Thermo Fisher Scientific) with a nanospray ion source in positive mode. The peptides derived from the protein fragments were separated on a Nano-HPLC C18 capillary column (0.075-mm inner diameter × 150-mm length, 3-mm particle size; Nikkyo Technos). Mobile phase A was comprised of water with 0.1% formic acid, and mobile phase B was comprised of acetonitrile with 0.1% formic acid. Two different slopes were used for a gradient elution for 120 min at a flow rate of 300 nL/min: 0%–30% of phase B for 100 min and 30%–65% of phase B for 20 min. The mass spectrometer was operated in the top 10 data-dependent scan mode. The parameters for operating the mass spectrometer were as follows: spray voltage: 2.3 kV, capillary temperature: 275°C, mass to charge ratio: 350–1800, and normalized collision energy: 28%. Raw data were acquired using Xcalibur software (Thermo Fisher Scientific). The MS and MS/MS data were searched against the Swiss-Prot database using Proteome Discoverer 2.2 (Thermo Fisher Scientific) using Mascot search engine software version 2.6.0 (Matrix Science). The peptides were considered identified when their false discovery rates (FDRs) were <5%. For substrate identification, proteins exhibiting at least a 1.5-fold enrichment in more than two out of four replicative experiments were defined as positive hit proteins.
LC-MS/MS analysis of His methylation sites
The plasmids for C-terminally FLAG-tagged proteins (pcDNA3-X-FLAG) were transfected into CARNMT1 KO cells and KO + WT rescue cells. Forty-eight hours after transfection, the cells were harvested and lysed in IP buffer (50 mM Tris-HCl at pH 7.5, 400 mM NaCl, 0.5% NP-40, protease inhibitors). M2-agarose beads (Sigma-Aldrich) were added and incubated with the lysate for 1 h at 4°C and then washed three times with IP buffer. The bound proteins were separated with SDS-PAGE, and the gel band corresponding to FLAG-tagged proteins was excised and in-gel-digested with trypsin unless described otherwise. The trypsinized protein fragments were applied to a liquid chromatograph (Easy-nLC 1000) coupled to a Q Exactive hybrid quadrupole orbitrap mass spectrometer as described previously (Nguyen-Tien et al. 2022). The acquired data were processed using Mascot 2.8 and Proteome Discoverer 2.4. The search parameters for MS and MS/MS data were as follows: enzyme: trypsin; static modifications: none; dynamic modifications: oxidation (Met), methylation (His), propionamide (Cys) acetyl (protein N-term), and Gln>; precursor mass tolerance: ±15 ppm; fragment mass tolerance: ±30 mmu; maximum missed cleavages: three; and instrument type: ESI-TRAP. The proteins were considered identified when their false discovery rates (FDRs) were <1%. Each His methylation level was calculated from the intensity of the methylated peptides divided by the intensity of the none methylated peptides.
RNA-seq analysis
High-throughput RNA sequencing (RNA-seq) analyses were performed in three independent total RNAs derived from HEK293T cells. Mouse brains and livers were prepared by RNeasy mini kit (Qiagen), and correlation among replicates are described in Supplemental Table S5. The rRNA depletion was performed with NEBNext rRNA depletion kit v2 (NEB). RNA libraries were prepared using the NEBNext Ultra RNA library preparation kit for Illumina (NEB). These samples were sequenced on a HiSeq 2500 high-throughput sequencing platform (Illumina). The sequencing data were analyzed as previously described (Fukumura et al. 2021). Obtained sequence reads were mapped onto the human genome reference sequences (hg19) using HISAT2 version 2.2.1 (Kim et al. 2019). Sequence reads with low-quality scores were eliminated using Trimmomatic (Bolger et al. 2014) with the “leading:20 trailing:20 slidingwindow:4:15 minlen:36” option. Transcript assembly based on mapped sequence reads was performed using StringTie version 2.1.7 (Pertea et al. 2016). Using the obtained GTF files as a reference, the BAM files were analyzed using rMATS version 4.1.1 to examine the changes of AS isoforms (Shen et al. 2014). Significant changes of splicing events were defined as when the false discovery rate (FDR) was calculated to be <0.01. Sashimi plot was depicted using rmats2sashimiplot (https://github.com/Xinglab/rmats2sashimiplot). The strengths of the 5′ and 3′ splice sites were calculated using MaxentScan (Yeo and Burge 2004), and branch point strength was calculated by SVM-BPfinder (Corvelo et al. 2010). The raw data from the RNA-seq analysis have been deposited in the NCBI Gene Expression Omnibus under accession numbers GSE217990 and GSE228894.
RT-PCR analysis
Total RNAs were isolated from HEK293T cells, and splicing efficiency was analyzed by RT-PCR using a primer set targeting skipped exon (Supplemental Table S7). The PCR products were separated on a 2.5% agarose gel in 0.5× TBE buffer (44.5 mM Tris-borate, 1 mM EDTA) and visualized with a FAS-V gel imaging system (Nippon Genetics Co., Ltd.). The unspliced pre-mRNA and spliced mRNA were quantified using NIH ImageJ software and normalized according to their product lengths.
RNAIP and qPCR analysis
RNAIP was performed as described by Mino et al. (2015). Briefly, HEK293T cells transfected with the indicated expression plasmids were lysed in RNAIP lysis buffer (0.5% NP-40, 150 mM NaCl, 20 mM Tris-HCl at pH 7.5, Complete mini protease inhibitor cocktail without EDTA [Roche], 0.2 U/mL RNasin [Promega]). Anti-FLAG M2 affinity gel (Merck) was incubated for 1 h at 4°C with lysates, and the beads were washed three times with RNAIP lysis buffer. Bound RNAs were eluted from the beads using Sepasol-RNA I Super G (Nacalai Tesque) and analyzed by RT-qPCR. Reverse transcription was performed using an Omniscript RT kit (Qiagen) according to the manufacturer's instructions. For quantitative PCR, cDNA fragments were amplified with Power SYBR Green master mix (Thermo Fisher Scientific) and detected with a StepOne real-time PCR system (Applied Biosystems). Primers used for qPCR are listed in Supplemental Table S7. Proteins were eluted from the beads using SDS sample buffer and analyzed by Western blot with the indicated antibodies. For mRNA decay experiments, 5 µg/mL actinomycin D was treated for the indicated times, total RNA was isolated using Sepazol, and mRNA levels were determined by RT-qPCR analysis.
For in vitro experiments, whole-cell extracts from either control or CARNMT1 KO 293 cells were prepared according to a previous study (Kataoka and Dreyfuss 2004). Briefly, ∼1.0 × 107 cells were rinsed twice with ice-cold PBS and collected into 1.5-mL tubes. Cells were suspended with 200 µL of buffer E (20 mM HEPES-KCl at pH 7.9, 100 mM KCl, 0.2 mM EDTA, 10% glycerol, 1× protein inhibitor cocktail [Merck P1860], 1 mM dithiothreitol) and sonicated three times for 5 sec with 30-sec intervals at 4°C using a QSonica Q125 at 20% amplitude. The lysates were centrifuged at 13,500 rpm for 10 min, and supernatants were transferred to a new tube. RNA probes were generated with in vitro transcription using CUGA 7 in vitro transcription kit (Nippon Gene 307-13531) according to the manufacturer's instructions. The reaction mixture was then treated with DNase I for 30 min at 37°C and purified with a G-50 column (Cytiva 28903408) to remove nucleotides. The binding was performed in a 50-µL mixture containing 1 µL of 16 mM MgCl2, 15 µL of cell extracts, 15 µL of D'K-100 buffer (20 mM HEPES-KOH at pH 8.0, 100 mM KCl, 0.2 mM EDTA, 1 mM dithiothreitol), and 1 pmol of in vitro transcribed RNA for 10 min at 30°C. The mixture was incubated with 1 µg of either anti-U2AF1 (MBL RN085PW) or FLAG (Merck F7425) conjugated with 20 µL of protein-G beads (Nacalai 02198-64) and incubated for 1 h at 4°C with continuous rotation. The beads were washed six times with wash buffer (20 mM HEPES-NaOH at pH 7.9, 150 mM NaCl, 0.05% Triton X-100). The beads were suspended in Homomix (50 mM Tris-HCl at pH 7.4, 5 mM EDTA, 1.5% SDS, 300 mM NaCl, 1.5 mg/mL proteinase K) for 60 min at 50°C, extracted with phenol-chloroform, precipitated with ethanol, and subjected to subsequent Northern blot analyses.
Total RNAs (∼1.0 µg) were separated with denaturing PAGE containing 8 M urea. RNAs were then transferred to Hybond-N+ (Merck RPN203B) using semidry transfer platform with 300 mA of constant current for 40 min. The membrane was cross-linked and hybridized overnight with DIG-labeled DNA in PerfectHyb Plus (Merck H7033) at 60°C. After being washed in 2× SSC followed by a stringent wash in 0.2× SSC, hybridized probes were detected with an alkaline phosphatase-conjugated anti-DIG antibody (Merck 11093274910) and CDP-Star (Merck 11685627001) following the manufacturer's instructions. The chemiluminescent signals were detected using ImageQuant 800 (Cytiva). The acquired bands were quantified with NIH Image. RNA substrates and probes used for Northern analysis are listed in Supplemental Table S8.
Mouse experiments
Animals were maintained on a 12-h light/dark cycle with access to food (CLEA Japan, Inc., CE-2) and water ad libitum. The temperature and humidity were maintained at 22°C–23°C and 50%–60%, respectively. Animal health was checked by the animal facility staff five times per week. All experiments involving mice complied with all relevant ethical regulations for animal testing and research and were carried out according to protocols approved by the Animal Experiment Committee of the RIKEN Center for Brain Science.
Generation of Carnmt1 KO mice
The Carnmt1 KO mice were generated by the CRISPR–Cas9 system. Guide RNAs that target exon 4 of mouse Carnmt1 were screened with CRISPR design tools (http://crispr.mit.edu) and CRISPRdirect (Naito et al. 2015). CRISPR RNA (crRNA) oligos for introns upstream of (5′-AACTTGAGTTTTAAGAAGTC-3′) and downstream from (5′-ATTCATAATGCTTATACCGA-3′) exon 4 were purchased from Fasmac. The crRNAs, Cas9 protein (NEB M0646T), and tracrRNA (Fasmac) were injected into fertilized eggs (mouse strain C57BL/6J), and the mutated Carnmt1 gene versions were screened by PCR. Two out of 30 founder mice carried the expected heterozygous deletion of exon 4. These heterozygous mice were crossed with C57BL/6J mice, and the male (+/−) and female (+/−) mouse colonies were maintained.
Quantification and statistical analysis
Mean values, SD values, SEM values, and Student's t-test (two-tailed unpaired) were calculated using Excel (Microsoft). One-way ANOVA and Tukey–Kramer post-hoc test were performed in R. P-values <0.05 were considered significant (not significant [n.s.], P < 0.05 [*], P < 0.01 [**], and P < 0.001 [***]).
Data availability
Source data for Figures 1–6 are in Supplemental Tables S1–S8. All reads from the RNA-seq experiments generated in this study have been submitted to Gene Expression Omnibus under accession numbers GSE217990 and GSE228894. Read numbers of NGS data are listed in Supplemental Table S5. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the data set identifier PXD037965.
Supplementary Material
Acknowledgments
We are grateful to Takashi Mino and Osamu Takeuchi for kind support on TNFα experiments, Daisuke Kaida for important advice related to AS, and Takashi Kondo and Haruhiko Koseki for teaching experimental techniques on dissection of mouse embryos. We thank Ayane Kataoka for technical support of mouse genotyping and Western blot analysis; the staff of Research Resources Division, RIKEN Center for Brain Science, for generation of CARNMT1 heterozygous KO mice, peptide synthesis, and LC-MS/MS analysis; and especially Masaya Usui and Hiromasa Morishita for MRM quantitation. This work was supported by the “Epigenome Manipulation Project” of the All-RIKEN Projects (to Y. Shinkai, M.S., T. Shimazu, Y. Sohtome, and A.O.), Japan Ministry of Education, Culture, Sports, Science, and Technology Grant-in-Aid for Scientific Research 20K06497 (to T. Shimazu) and 22K05565 (to R.Y.), Grant-in-Aid for Scientific Research (B) 22H02214 (to Y. Sohtome and T. Shimazu), and Grant-in-Aid for Transformative Research Areas (B) 22H05019 (to Y. Sohtome, and 22H05020 (to T. Shimazu), and the Takeda Science Foundation (to T. Shimazu).
Author contributions: T. Shimazu designed and performed experiments, analyzed the data, and prepared the manuscript. R.Y. analyzed RNA-seq and performed data analysis and RNAIP experiments. K.K. performed experiments. T. Suzuki and N.D. performed the mass spectrometry and analyzed the data. S.M., M.H., and A.O. generated Carnmt1 catalytically dead mice. M.A., Y. Sohtome, and M.S. prepared ProSeAM. Y. Shinkai supervised the study, planned the experiments, interpreted the data, and prepared the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350755.123.
Competing interest statement
The authors declare no competing interests.
References
- Al-Hadid Q, Roy K, Munroe W, Dzialo MC, Chanfreau GF, Clarke SG. 2014. Histidine methylation of yeast ribosomal protein Rpl3p is required for proper 60S subunit assembly. Mol Cell Biol 34: 2903–2916. 10.1128/MCB.01634-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann BT, Worthington MT, Berg JM. 2003. A Cys3His zinc-binding domain from Nup475/tristetraprolin: a novel fold with a disklike structure. Biochemistry 42: 217–221. 10.1021/bi026988m [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281: 1001–1005. 10.1126/science.281.5379.1001 [DOI] [PubMed] [Google Scholar]
- Corvelo A, Hallegger M, Smith CW, Eyras E. 2010. Genome-wide association between branch point properties and alternative splicing. PLoS Comput Biol 6: e1001016. 10.1371/journal.pcbi.1001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Cai M, Stanley HE. 2017. Comparative analysis and classification of cassette exons and constitutive exons. Biomed Res Int 2017: 7323508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H, Someya M, Kako K, Hayashi T, Tajima T, Haruki H, Sekiguchi N, Uetake T, Akimoto Y, Fukamizu A. 2021. siRNA screening identifies METTL9 as a histidine Nπ-methyltransferase that targets the proinflammatory protein S100A9. J Biol Chem 297: 101230. 10.1016/j.jbc.2021.101230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydova E, Shimazu T, Schuhmacher MK, Jakobsson ME, Willemen H, Liu T, Moen A, Ho AYY, Małecki J, Schroer L, et al. 2021. The methyltransferase METTL9 mediates pervasive 1-methylhistidine modification in mammalian proteomes. Nat Commun 12: 891. 10.1038/s41467-020-20670-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozak J, Chrobok L, Poleszak O, Jagielski AK, Derlacz R. 2013. Molecular identification of carnosine N-methyltransferase as chicken histamine N-methyltransferase-like protein (HNMT-like). PLoS One 8: e64805. 10.1371/journal.pone.0064805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozak J, Piecuch M, Poleszak O, Kozlowski P, Chrobok L, Baelde HJ, de Heer E. 2015. UPF0586 protein C9orf41 homolog is anserine-producing methyltransferase. J Biol Chem 290: 17190–17205. 10.1074/jbc.M115.640037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga M. 1971. Amino acid sequence aroung 3-methylhistidine in rabbit skeletal muscle actin. Biochemistry 10: 224–229. 10.1021/bi00778a005 [DOI] [PubMed] [Google Scholar]
- Fei DL, Zhen T, Durham B, Ferrarone J, Zhang T, Garrett L, Yoshimi A, Abdel-Wahab O, Bradley RK, Liu P, et al. 2018. Impaired hematopoiesis and leukemia development in mice with a conditional knock-in allele of a mutant splicing factor gene U2af1. Proc Natl Acad Sci 115: E10437–E10446. 10.1073/pnas.1812669115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Li Q, Meng R, Yi B, Xu Q. 2018. METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J Cell Mol Med 22: 2558–2568. 10.1111/jcmm.13491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumura K, Yoshimoto R, Sperotto L, Kang H-S, Hirose T, Inoue K, Sattler M, Mayeda A. 2021. SPF45/RBM17-dependent, but not U2AF-dependent, splicing in a distinct subset of human short introns. Nat Commun 12: 4910. 10.1038/s41467-021-24879-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, Roske Y, Yamada S, Uehata T, Takeuchi O, Heinemann U. 2021. PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases. Nucleic Acids Res 49: 5369–5381. 10.1093/nar/gkab316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. 2015. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347: 1002–1006. 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- Groza T, Gomez FL, Mashhadi HH, Muñoz-Fuentes V, Gunes O, Wilson R, Cacheiro P, Frost A, Keskivali-Bond P, Vardal B, et al. 2023. The International Mouse Phenotyping Consortium: comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res 51: D1038–D1045. 10.1093/nar/gkac972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Richard S. 2019. The regulation, functions and clinical relevance of arginine methylation. Nat Rev Mol Cell Biol 20: 642–657. 10.1038/s41580-019-0155-x [DOI] [PubMed] [Google Scholar]
- Hall TM. 2005. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 15: 367–373. 10.1016/j.sbi.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Herdt O, Reich S, Medenbach J, Timmermann B, Olofsson D, Preußner M, Heyd F. 2020. The zinc finger domains in U2AF26 and U2AF35 have diverse functionalities including a role in controlling translation. RNA Biol 17: 843–856. 10.1080/15476286.2020.1732701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, Bradley P, Bradley RK. 2015. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res 25: 14–26. 10.1101/gr.181016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Bradley RK, Abdel-Wahab O. 2016. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev 30: 989–1001. 10.1101/gad.278424.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson ME. 2021. Enzymology and significance of protein histidine methylation. J Biol Chem 297: 101130. 10.1016/j.jbc.2021.101130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P, Harris CI, Perry SV. 1967. 3-methylhistidine in actin and other muscle proteins. Biochem J 105: 361–370. 10.1042/bj1050361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapell S, Jakobsson ME. 2021. Large-scale identification of protein histidine methylation in human cells. NAR Genom Bioinform 3: lqab045. 10.1093/nargab/lqab045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Dreyfuss G. 2004. A simple whole cell lysate system for in vitro splicing reveals a stepwise assembly of the exon-exon junction complex. J Biol Chem 279: 7009–7013. 10.1074/jbc.M307692200 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski S, Seliga AK, Vertommen D, Terreri M, Ishikawa T, Grabowska I, Tiebe M, Teleman AA, Jagielski AK, Veiga-da-Cunha M, et al. 2018. SETD3 protein is the actin-specific histidine N-methyltransferase. Elife 7: 37921. 10.7554/eLife.37921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G. 2013. Roquin promotes constitutive mRNA decay via a conserved class of stem–loop recognition motifs. Cell 153: 869–881. 10.1016/j.cell.2013.04.016 [DOI] [PubMed] [Google Scholar]
- Liang J, Song W, Tromp G, Kolattukudy PE, Fu M. 2008. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One 3: e2880. 10.1371/journal.pone.0002880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. 2014. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv M, Cao D, Zhang L, Hu C, Li S, Zhang P, Zhu L, Yi X, Li C, Yang A, et al. 2021. METTL9 mediated N1-histidine methylation of zinc transporters is required for tumor growth. Protein Cell 12: 965–970. 10.1007/s13238-021-00857-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Małecki JM, Odonohue MF, Kim Y, Jakobsson ME, Gessa L, Pinto R, Wu J, Davydova E, Moen A, Olsen JV, et al. 2021. Human METTL18 is a histidine-specific methyltransferase that targets RPL3 and affects ribosome biogenesis and function. Nucleic Acids Res 49: 3185–3203. 10.1093/nar/gkab088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura-Suzuki E, Shimazu T, Takahashi M, Kotoshiba K, Suzuki T, Kashiwagi K, Sohtome Y, Akakabe M, Sodeoka M, Dohmae N, et al. 2022. METTL18-mediated histidine methylation of RPL3 modulates translation elongation for proteostasis maintenance. Elife 11: e72780. 10.7554/eLife.72780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HE, Mayr GW. 1987. Nπ-methylhistidine in myosin-light-chain kinase. Biol Chem Hoppe-Seyler 368: 1607–1612. 10.1515/bchm3.1987.368.2.1607 [DOI] [PubMed] [Google Scholar]
- Mino T, Takeuchi O. 2018. Post-transcriptional regulation of immune responses by RNA binding proteins. Proc Jpn Acad Ser B Phys Biol Sci 94: 248–258. 10.2183/pjab.94.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino T, Murakawa Y, Fukao A, Vandenbon A, Wessels HH, Ori D, Uehata T, Tartey S, Akira S, Suzuki Y, et al. 2015. Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 161: 1058–1073. 10.1016/j.cell.2015.04.029 [DOI] [PubMed] [Google Scholar]
- Mollet I, Barbosa-Morais NL, Andrade J, Carmo-Fonseca M. 2006. Diversity of human U2AF splicing factors. FEBS J 273: 4807–4816. 10.1111/j.1742-4658.2006.05502.x [DOI] [PubMed] [Google Scholar]
- Murakami S, Jaffrey SR. 2022. Hidden codes in mRNA: control of gene expression by m6A. Mol Cell 82: 2236–2251. 10.1016/j.molcel.2022.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Hino K, Bono H, Ui-Tei K. 2015. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31: 1120–1123. 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Tien D, Suzuki T, Kobayashi T, Toyama-Sorimachi N, Dohmae N. 2022. Identification of the interacting partners of a lysosomal membrane protein in living cells by BioID technique. STAR Protoc 3: 101263. 10.1016/j.xpro.2022.101263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc Natl Acad Sci 111: E5593–E5601. 10.1073/pnas.1419161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Barjau J, Sohtome Y, Sodeoka M, Shinkai Y. 2014. Selenium-based S-adenosylmethionine analog reveals the mammalian seven-β-strand methyltransferase METTL10 to be an EF1A1 lysine methyltransferase. PLoS One 9: e105394. 10.1371/journal.pone.0105394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohtome Y, Shimazu T, Barjau J, Fujishiro S, Akakabe M, Terayama N, Dodo K, Ito A, Yoshida M, Shinkai Y, et al. 2018. Unveiling epidithiodiketopiperazine as a non-histone arginine methyltransferase inhibitor by chemical protein methylome analyses. Chem Commun 54: 9202–9205. 10.1039/C8CC03907K [DOI] [PubMed] [Google Scholar]
- Sohtome Y, Shimazu T, Shinkai Y, Sodeoka M. 2021. Propargylic Se-adenosyl-l-selenomethionine: a chemical tool for methylome analysis. Acc Chem Res 54: 3818–3827. 10.1021/acs.accounts.1c00395 [DOI] [PubMed] [Google Scholar]
- Stull JT, Kamm KE, Vandenboom R. 2011. Myosin light chain kinase and the role of myosin light chain phosphorylation in skeletal muscle. Arch Biochem Biophys 510: 120–128. 10.1016/j.abb.2011.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thandapani P, O'Connor TR, Bailey TL, Richard S. 2013. Defining the RGG/RG motif. Mol Cell 50: 613–623. 10.1016/j.molcel.2013.05.021 [DOI] [PubMed] [Google Scholar]
- Tracey KJ, Cerami A. 1994. Tumor necrosis factor: a pleiotropic cytokine and theraputic target. Annu Rev Med 45: 491–503. 10.1146/annurev.med.45.1.491 [DOI] [PubMed] [Google Scholar]
- Ule J, Blencowe BJ. 2019. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol Cell 76: 329–345. 10.1016/j.molcel.2019.09.017 [DOI] [PubMed] [Google Scholar]
- Wadugu BA, Nonavinkere Srivatsan S, Heard A, Alberti MO, Ndonwi M, Liu J, Grieb S, Bradley J, Shao J, Ahmed T, et al. 2021. U2af1 is a haplo-essential gene required for hematopoietic cancer cell survival in mice. J Clin Invest 131: e141401. 10.1172/JCI141401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Eckhardt L, Bastian A, Bruegmann T, Sasse P, Eckhardt M. 2020. Carnosine synthase deficiency is compatible with normal skeletal muscle and olfactory function but causes reduced olfactory sensitivity in aging mice. J Biol Chem 295: 17100–17113. 10.1074/jbc.RA120.014188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward WH, Holdgate GA. 2001. Isothermal titration calorimetry in drug discovery. Prog Med Chem 38: 309–376. 10.1016/S0079-6468(08)70097-3 [DOI] [PubMed] [Google Scholar]
- Webb KJ, Zurita-Lopez CI, Al-Hadid Q, Laganowsky A, Young BD, Lipson RS, Souda P, Faull KF, Whitelegge JP, Clarke SG. 2010. A novel 3-methylhistidine modification of yeast ribosomal protein Rpl3 is dependent upon the YIL110W methyltransferase. J Biol Chem 285: 37598–37606. 10.1074/jbc.M110.170787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Almeida M, Pintacuda G, Coker H, Bowness JS, Ule J, Brockdorff N. 2021. Acute depletion of METTL3 implicates N6-methyladenosine in alternative intron/exon inclusion in the nascent transcriptome. Genome Res 31: 1395–1408. 10.1101/gr.271635.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson AW, Diep J, Dai S, Liu S, Ooi YS, Song D, Li TM, Horton JR, Zhang X, Liu C, et al. 2019. SETD3 is an actin histidine methyltransferase that prevents primary dystocia. Nature 565: 372–376. 10.1038/s41586-018-0821-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394. 10.1089/1066527041410418 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Park SY, Oda T, Akiyoshi T, Sato M, Shirouzu M, Tsuda K, Kuwasako K, Unzai S, Muto Y, et al. 2015. A novel 3′ splice site recognition by the two zinc fingers in the U2AF small subunit. Genes Dev 29: 1649–1660. 10.1101/gad.267104.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Green MR. 1989. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci 86: 9243–9247. 10.1073/pnas.86.23.9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, He C. 2017. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18: 31–42. 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Cai W, Hua Y, Yang X, Zhou J. 2022. The biological and clinical consequences of RNA splicing factor U2AF1 mutation in myeloid malignancies. Cancers 14: 4406. 10.3390/cancers14184406 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Figures 1–6 are in Supplemental Tables S1–S8. All reads from the RNA-seq experiments generated in this study have been submitted to Gene Expression Omnibus under accession numbers GSE217990 and GSE228894. Read numbers of NGS data are listed in Supplemental Table S5. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the data set identifier PXD037965.