In this study, Sharma et al. describe a novel cytoplasmic poly(A) binding protein, neuPABP, specifically found in postnatal brain neurons undergoing synaptogenesis. Unlike other PABPs, neuPABP does not bind eIF4G to support mRNA translation; instead, neuPABP interacts with neuron-specific, noncoding RNA BC1, as well as select ribosomal and mitochondrial mRNAs, in free ribonucleoprotein complexes, suggesting a role for neuPABP in suppressing their translation.
Keywords: RNA binding protein, mRNA translation, poly(A) binding protein
Abstract
The mRNA 3′ poly(A) tail plays a critical role in regulating both mRNA translation and turnover. It is bound by the cytoplasmic poly(A) binding protein (PABPC), an evolutionarily conserved protein that can interact with translation factors and mRNA decay machineries to regulate gene expression. Mammalian PABPC1, the prototypical PABPC, is expressed in most tissues and interacts with eukaryotic translation initiation factor 4G (eIF4G) to stimulate translation in specific contexts. In this study, we uncovered a new mammalian PABPC, which we named neural PABP (neuPABP), as it is predominantly expressed in the brain. neuPABP maintains a unique architecture as compared with other PABPCs, containing only two RNA recognition motifs (RRMs) and maintaining a unique N-terminal domain of unknown function. neuPABP expression is activated in neurons as they mature during synaptogenesis, where neuPABP localizes to the soma and postsynaptic densities. neuPABP interacts with the noncoding RNA BC1, as well as mRNAs coding for ribosomal and mitochondrial proteins. However, in contrast to PABPC1, neuPABP does not associate with actively translating mRNAs in the brain. In keeping with this, we show that neuPABP has evolved such that it does not bind eIF4G and as a result fails to support protein synthesis in vitro. Taken together, these results indicate that mammals have expanded their PABPC repertoire in the brain and propose that neuPABP may support the translational repression of select mRNAs.
Almost all eukaryotic mRNAs contain both a 5′ m7G “cap” structure and 3′ poly(A) tail, cis-acting elements that can play an important role in regulating both mRNA translation and turnover (Gallie 1991; Tarun and Sachs 1995). The mRNA 5′ cap helps recruit eukaryotic translation initiation factor 4F (eIF4F), a protein complex that contains the cap binding protein eIF4E, the scaffold protein eIF4G, and the ATP-dependent DEAD-box helicase protein eIF4A (Grifo et al. 1983; Pelletier et al. 2015). eIF4G physically interacts with another translation initiation factor, eIF3 (LeFebvre et al. 2006), which interfaces with the smaller 40S ribosomal subunit for translation initiation (Fraser et al. 2004). In addition to these eIFs, translation initiation can also be stimulated by the cytoplasmic poly(A) binding protein (PABPC), which binds to the 3′ poly(A) tail and is highly conserved among eukaryotes (Tarun et al. 1997; Wei et al. 1998; Kahvejian et al. 2005).
PABPC contains four RNA recognition motifs (RRMs), a proline-rich linker domain, and a C-terminal mademoiselle (MLLE) domain (Fig. 1A; Gorgoni and Gray 2004). RRM1 and RRM2 preferably bind poly(A) sequences, while RRM3 and RRM4 bind both poly(A) and A/U-rich sequences equally well (Kühn and Pieler 1996; Khanam et al. 2006). A single PABPC covers ∼30 As (Baer and Kornberg 1983; Kühn and Pieler 1996), and multiple PABPC molecules can oligomerize on poly(A) sequences via interactions between the linker domain of one PABPC and RRM1 of an adjacent molecule (Schäfer et al. 2019). PABPC RRM2 can directly bind eIF4G (Safaee et al. 2012), an interaction that stimulates mRNA translation in vitro and in Xenopus oocytes (Wakiyama et al. 2000; Kahvejian et al. 2005), contexts where mRNA poly(A) tail length correlates with translational efficiency (Imataka et al. 1998; Wakiyama et al. 2000; Kahvejian et al. 2005). Notwithstanding that PABPC stimulates mRNA translation in certain systems (Wakiyama et al. 2000; Xiang and Bartel 2021), recent reports have suggested that PABPC plays a negligible role in enhancing the translation efficiency of the transcriptome in postembryonic mammalian cell lines (Xiang and Bartel 2021; Kajjo et al. 2022).
Figure 1.
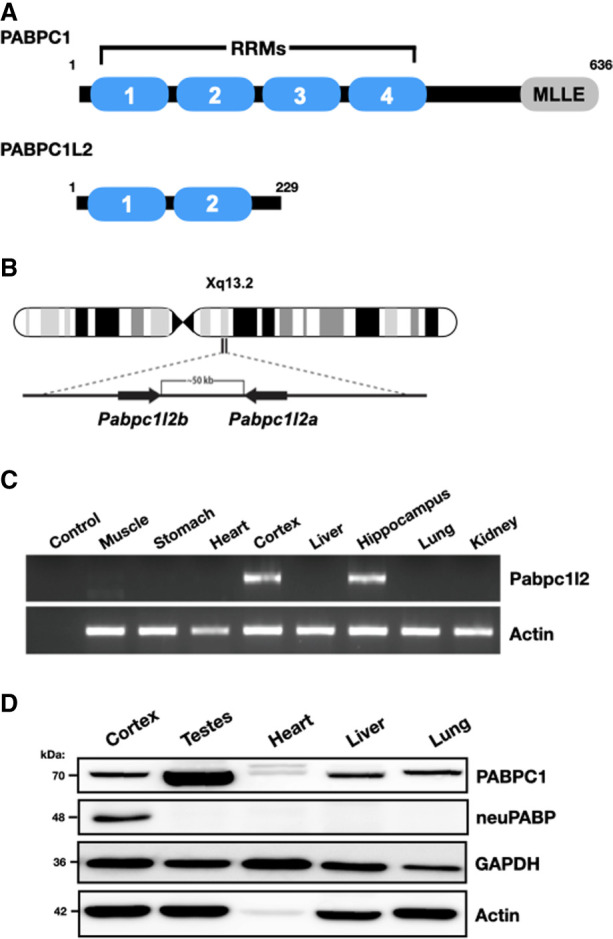
PABPC1L2 (neuPABP) displays a neural-specific expression pattern. (A) Schematic representation of PABPC1 and PABPC1L2 domain organization. (B) Schematic diagram of a human X chromosome showing the position of the Pabpc1l2 ampliconic gene. (C) Semiquantitative RT-PCR analysis of Pabpc1l2 and Actin mRNAs from multiple adult mouse tissues (C57BL/6J; age: 5 mo). (D) Western blotting of PABPC1, neuPABP, GAPDH, and Actin on lysates prepared from select adult mouse tissues (C57BL/6J; age: 5 mo).
The PABPC MLLE domain acts as a platform that directly interacts with a number of PABPC-interacting proteins. These include poly(A) binding protein-interacting proteins (PAIPs) 1 and 2 and eukaryotic release factor 3 (eRF3) (Hoshino et al. 1999; Khaleghpour et al. 2001a; Roy et al. 2002), which regulate mRNA translation. The PABPC MLLE domain also interfaces with several proteins that regulate mRNA decay, including the PAN3 subunit of the PAN2–PAN3 deadenylase complex, which trims excessively long poly(A) tails to the length of 25–40 nt (Schäfer et al. 2019), as well as proteins that recruit the CCR4–NOT deadenylase complex (e.g., Tob proteins and the miRNA-associated protein GW182/TNRC6) (Okochi et al. 2005; Fabian et al. 2011; Huntzinger et al. 2013; Jonas and Izaurralde 2015; Chen et al. 2020). PABPC has been shown to promote poly(A) tail trimming by the CCR4–NOT complex but prevents premature 3′ terminal uridylation (Yi et al. 2018) and blocks deadenylation-independent decay of normally stable transcripts (Kajjo et al. 2022).
While yeast code for only a single PABPC, Pab1, higher metazoans contain genes that encode for several PABPC paralogs with similar architectures (Gorgoni and Gray 2004). These additional PABPCs display tissue- and temporal-specific expression patterns, including testis-specific PABPC (tPABP or PABPC3), which is expressed in round spermatids (Feral et al. 2001), and embryonic PABP (ePABP), which is expressed in oocytes and early embryos but is not expressed in adult tissues (Voeltz et al. 2001; Seli et al. 2005). Here we report the identification of a new mammalian PABPC that we termed neural PABP (neuPABP). neuPABP is coded for by the X-linked Pabpc1-like gene (Pabpc1l2) and displays a neural-specific expression pattern. neuPABP is detectable in the postnatal brain, and its expression in neurons increases during neuronal maturation. neuPABP is a bona fide PABPC that localizes to postsynaptic densities and has a unique architecture as compared with PABPC1, containing only two RRMs as well as a unique N-terminal domain of unknown function. neuPABP interacts with the neuron-specific noncoding RNA BC1 and is enriched on mRNAs coding for ribosomal and mitochondrial proteins. Interestingly, in contrast to PABPC1, neuPABP-associated mRNAs are translationally dormant. In keeping with this, neuPABP has undergone evolutionary selection such that it cannot interact with eIF4G, which in turn prevents it from stimulating mRNA translation in vitro.
Results
Pabpc1l2 is predominantly expressed in neural tissues
The human X chromosome contains a two-copy ampliconic Pabp-like gene, Pabpc1-like 2 (Pabc1l2a/b), with both copies lacking introns and being >99% identical to each other at the nucleotide level (Fig. 1B). Pabc1l2a/b is conserved among mammals (Mueller et al. 2013) and is predicted to code for a truncated open reading frame as compared with PABPC1, containing only the first two RRMs (Fig. 1A; Supplemental Fig. S1). To assess Pabpc1l2 expression in adult mice, we isolated total RNA from adult mouse tissues and carried out semiquantitative RT-PCR and RT-qPCR reactions using Pabpc1l2-specific primers. We observed Pabpc1l2 mRNA expression in mouse brain tissues (e.g., the cortex and hippocampus) but could not detect its expression in any other somatic tissues (Fig. 1C; Supplemental Fig. S2). In keeping with this, Pabpc1l2 gene expression was consistently detected in the postnatal brain of P7 and 10-wk mice in neural cells as assessed by single-cell RNA sequencing analyses. The detection rate was higher in neurons, lower in glial cells (oligodendroglial cells and astrocytes), and negligible in microglia and other nonneural cell types (Supplemental Figs. S3, S4). We also analyzed Pabpc1l2 gene expression across human tissues using publicly available data from the National Institutes of Health's Genotype–Tissue Expression Project (The GTEx Consortium 2013). In keeping with data acquired from mouse tissues, Pabpc1l2 mRNA was also primarily detected in human brain tissues (Supplemental Fig. S5).
While mouse Pabpc1l2 is predicted to code for a protein that contains only two RRMs and is classified by the National Center for Biotechnology Information as a putative pseudogene (NM_001384267.1), the predicted amino acid sequence across these motifs is highly conserved in PABPC1L2 homologs, sharing a high degree of identity and homology with the first two RRMs of PABPC1 (Supplemental Fig. S1). This suggests that as opposed to Pabpc1l2 being a pseudogene, there has been evolutionary pressure on it to maintain an open reading frame. To determine whether Pabpc1l2 is expressed at the protein level, we generated a polyclonal antibody that recognizes a peptide corresponding to the predicted mouse PABPC1L2 C terminus (ERGAWARQSTSADFKDFD), a unique sequence that is not present in other proteins, including other PABPCs (Supplemental Fig. S1). To evaluate the specificity of our antibody, we transiently transfected HeLa cells with a plasmid coding for PABPC1L2 and carried out Western blotting on lysates (Supplemental Fig. S6A). Importantly, our antibody identified ectopic PABPC1L2 in transfected cell lysates, but no corresponding band was observed in lysates derived from nontransfected cells. In keeping with Pabpc1l2 mRNA expression patterns, Western blotting analysis of mouse tissues using our antibody only detected a protein in neural tissues (Fig. 1D; Supplemental Figs. S6B, S7). In contrast, PABPC1 was detected in all somatic tissues, albeit at different levels of expression (Fig. 1D). Thus, these data suggest that PABPC1L2 displays a neural-specific expression pattern; hence, we termed it neural PABP (neuPABP).
neuPABP contains a unique N-terminal domain of unknown function
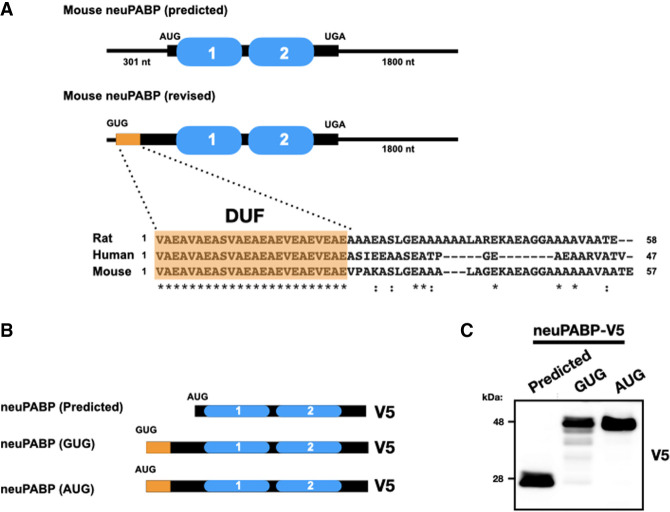
Mouse Pabpc1l2 mRNA is predicted to contain a 301-nt 5′ UTR and an open reading frame (ORF) encoding a short protein (229 amino acids): neuPABP. However, endogenous neuPABP migrates at a higher position on SDS-PAGE (∼48 kDa) than what would be predicted by its ORF. Using 5′RACE, we verified that the predicted Pabpc1l2 mRNA 5′ terminus is accurate (Supplemental Fig. S8). While it is possible that neuPABP maintains post-translational modifications that may alter its molecular weight, another explanation for this discrepancy is that the predicted ORF encoding neuPABP is incomplete. Thus, we set out to verify the sequence of full-length neuPABP. To this end, endogenous neuPABP was immunoprecipitated from adult mouse cortex lysate and subjected to mass spectrometry analysis to determine whether neuPABP peptide coverage extends beyond its predicted N and C termini. While we were unable to detect additional peptides C-terminal to the neuPABP stop codon, our analysis identified significant peptide coverage corresponding to the Pabpc1l2 mRNA 5′ UTR that is in-frame with the predicted neuPABP open reading frame (Supplemental Fig. S9). This additional N-terminal sequence suggests that mouse neuPABP is 312 amino acids in length with a short (24-amino-acid) conserved N-terminal domain of unknown function (DUF) comprised almost exclusively of valine, glutamate, and alanine amino acids (Fig. 2A). This region has no initiator ATG codon; however, GTG at positions 53–55 (Supplemental Fig. S9) would code for the N-terminal valine identified by mass spectrometry analysis. Moreover, the sequence flanking this codon (gcggcgaccGUGgcg) is very similar to the Kozak consensus sequence for non-AUG initiators (gccgcca/gcc(non-AUG)ga/cu) (Boeck and Kolakofsky 1994; Grünert and Jackson 1994). To test this, a modified neuPABP ORF, along with all 5′-terminal nucleotides, was fused to a C-terminal V5 tag and subsequently transfected into HeLa cells (Fig. 2B). Western blotting with a V5 antibody demonstrated that this construct produced an ∼48-kDa protein, similar to the size of endogenous neuPABP (Fig. 2C). Moreover, mutating the initiator GTG in our construct to ATG generated a protein of similar size. This is in contrast to a construct with the predicted neuPABP ORF, which generated a significantly smaller protein (∼28 kDa). Collectively, these data indicate that Pabpc1l2 encodes a GUG-initiated ORF and that neuPABP contains a unique N-terminal domain that is not found in other PABPCs.
Figure 2.
neuPABP is a GUG-initiated protein with a misannotated N-terminal extension. (A) Schematic diagram of the predicted and revised neuPABP open reading frame, along with the predicted AUG and validated GUG initiator codons, respectively. The N-terminal region (highlighted in orange) corresponds to the domain of unknown function (DUF) that is predicted to be conserved between human, mouse, and rat neuPABP. (B) Schematic diagram of mouse neuPABP expression constructs containing C-terminal V5 tags. (C) Western blot analysis of HeLa cells transfected with plasmids encoding V5-tagged predicted neuPABP or containing the Pabpc1l2 5′ UTR containing the GTG codon or ATG codon.
neuPABP is a bona fide PABP that is expressed during neuronal maturation
neuPABP is predicted to contain two RRMs that maintain a high degree of identity to RRM1 and RRM2 of PABPC1 (Supplemental Fig. S1). To determine whether neuPABP can bind to RNA, we purified recombinant GST-tagged neuPABP and carried out an in vitro selection—called RNAcompete (Ray et al. 2013)—by incubating GST-neuPABP with a complex collection of short RNAs. RNAcompete analysis identified AAAAAA as the consensus binding motif for neuPABP, indicating that neuPABP is a true poly(A) binding protein (Fig. 3A; Supplemental Table S1). To determine the affinity of neuPABP for poly(A) RNA, we purified recombinant PABPC1 and neuPABP (Fig. 3B) and carried out electrophoretic mobility shift assays using a 32P-end-labeled (A)25 oligoribonucleotide (Fig. 3C). In keeping with the RNAcompete data, we observed that neuPABP bound (A)25 RNA with an affinity similar to that of PABPC1. Moreover, a secondary shift in neuPABP binding suggests that two neuPABP proteins can bind 25 As, as compared with PABPC1, where only a single protein can bind (Baer and Kornberg 1983; Kühn and Pieler 1996).
Figure 3.
neuPABP specificity for poly(A) RNA. (A) Summary of RNAcompete experiments for GST-neuPABP. The sequence logo of the neuPABP RNA binding motif is shown, along with a scatter plot displaying the Z scores and motifs for the two halves of the RNA pool (set A and set B). (B) Recombinant PABPC1 and neuPABP were analyzed by SDS-PAGE and Coomassie blue staining. (C) High-affinity binding of neuPABP to oligo(A) RNA. EMSA was carried out as described in the Materials and Methods. A constant amount of 32P-oligo(A)25 RNA was incubated with specific concentrations of neuPABP or PABPC1. The KD value of ∼50 nM was calculated from three biological experiments for both PABPC1 and neuPABP. Recombinant GST (control) did not lead to a gel shift of radiolabeled oligo.
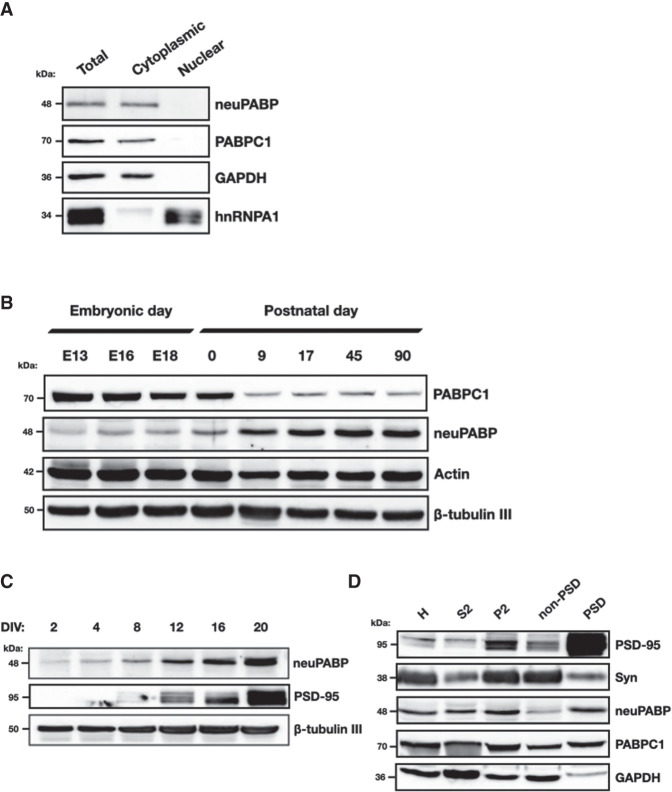
Nuclear/cytoplasmic fractionation experiments on lysates derived from adult cortex tissue indicate that like PABPC1, neuPABP is a cytoplasmic PABP (Fig. 4A). We next set out to determine the temporal expression of neuPABP during mouse brain development. To this end, we isolated the brains of mice at ages E13, E16, and E18 as well as several postnatal ages. Lysates generated from isolated tissues were then resolved by SDS-PAGE and analyzed using antibodies against PABPC1, neuPABP, actin, and β-tubulin III (controls) (Fig. 4B). Strikingly, we observed that PABPC1 and neuPABP displayed opposing temporal expression patterns during brain development. PABPC1 was highly expressed in embryonic tissues, but its levels were significantly lower in postnatal brain tissues. In contrast, neuPABP was barely detectable in embryonic brain tissue. However, its expression steadily increased during postnatal brain development, reaching a maximum at around P17 and remaining at this level into adulthood (Fig. 4B). Interestingly, this period of expression coincided with synaptogenesis, where synapses are formed between neurons, which is one of the key events that takes place in rodents during the first few postnatal weeks of life (Li et al. 2010). To determine whether neuPABP displays a similar expression pattern during neuronal maturation, we isolated mouse primary cortical neurons from P0 pups and cultured them to promote their maturation in vitro. Lysates were then generated from cultured neurons and resolved by SDS-PAGE. Similar to what we observed over the course of mouse brain development, neuPABP levels were barely detectable in newly cultured neurons. However, neuPABP expression rapidly increased over time, with its expression pattern overlapping with that of the synaptic marker PSD-95 (Fig. 4C). Moreover, neuPABP levels in mature neurons approached those of PABPC1 as assessed by Western blots using recombinant protein ladders for direct comparisons (Supplemental Fig. S10). Interestingly, while neuPABP was easily detected in mature neurons, we could not detect neuPABP expression in two neuroblastoma cell lines (SH-SY5Y and Neuro2a) even after differentiating them into neuronal-like cells (data not shown). In keeping with its expression during synaptogenesis, we also detected the subcellular localization of neuPABP in synaptosomes following synaptosome fractionation of the mouse cortex. neuPABP was detected by Western blotting in the PSD-95-enriched postsynaptic density (PSD) fraction (Fig. 4D), whereas neuPABP was barely detectable in the non-PSD fraction, which was enriched in synaptophysin. This is in contrast to PABPC1, which was equally detectable in both PSD and non-PSD fractions. In keeping with these data, proteomic analyses of isolated human neural tissues identified neuPABP peptides in synaptosomes and postsynaptic densities (Föcking et al. 2016; Hesse et al. 2019). Taken together, these data indicate that neuPABP is a bona fide poly(A) binding protein whose expression coincides with synaptogenesis.
Figure 4.
neuPABP is expressed during neuronal maturation. (A) Subcellular fractionation of an adult mouse brain cortex (C57BL/6J; age: 2 mo) shows cytoplasmic localization of both neuPABP and PABPC1. GAPDH and hnRNPA1 were used as markers for cytoplasmic and nuclear fractions, respectively. (B) Western blot analysis of PABPC1, neuPABP, Actin, and β-tubulin III on lysates prepared from mouse brain cortices isolated at different stages of embryonic and postnatal development. (C) Western blot analysis of neuPABP, PSD-95, and β-tubulin III on lysates prepared from mouse primary cortical neurons. Neurons were isolated from P0 pups and cultured for defined days in vitro (DIV). (D) Western blot analysis of subcellular fractions of an adult mouse cortex (C57BL/6J; age: 6 mo) prepared by synaptosome fractionation. Lysates were probed with the postsynaptic (PSD) marker PSD-95 and the presynaptic marker synaptophysin (Syn), as well as neuPABP, PABPC1, and GAPDH. Cortex homogenates (H) were generated, and supernatant (S2) and the crude synaptosomal pellet (P2) were acquired after high-speed centrifugation of the S1 supernatant. The crude synaptosomal fraction was further fractionated into a Triton X-100-soluble non-PSD fraction (extrasynaptic) and a Triton X-100-insoluble PSD-containing fraction (synaptic).
neuPABP interacts with BC1 RNA and select nontranslating mRNAs
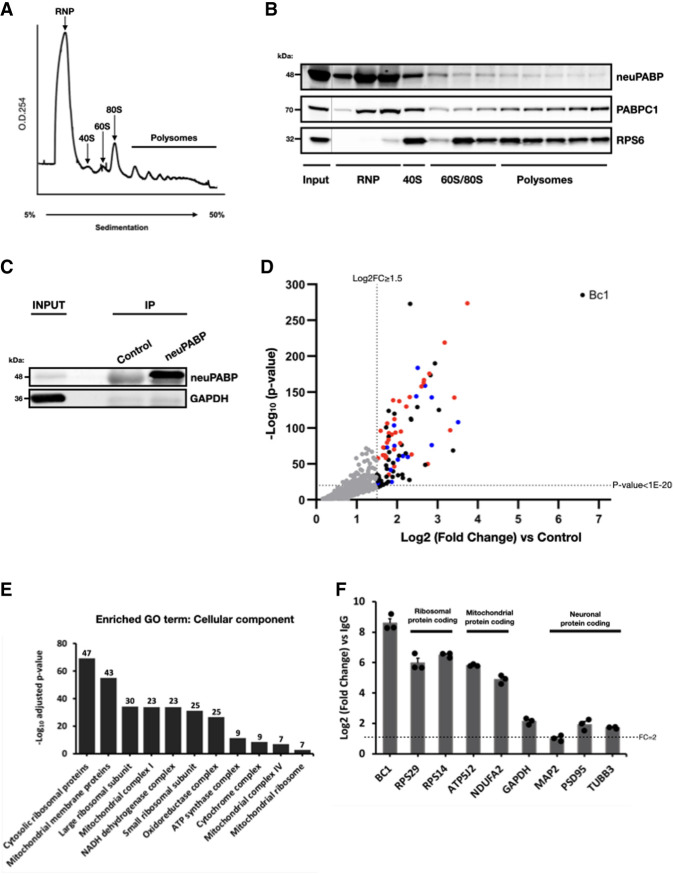
As our data indicate that neuPABP can bind poly(A) RNA with similar affinity to PABPC1, we next wished to determine whether neuPABP is associated with actively translating mRNAs. To this end, we isolated polysome profile fractions derived from P9 mouse cortex lysate (Fig. 5A) and assessed the distributions of PABPC1 and neuPABP by Western blotting (Fig. 5B). Consistent with many studies, we observed PABPC1 throughout the polysome gradient, including in heavy polysome fractions that contain highly translated mRNAs (Tcherkezian et al. 2014; Fonseca et al. 2015). In stark contrast, the vast majority of neuPABP did not sediment in heavy polysome fractions. Instead, neuPABP sedimented in early fractions, including those that contain free ribonucleoprotein (RNP) complexes, with a small amount of neuPABP in fractions that contain 40S subunits (Fig. 5B). These data suggest that unlike PABPC1, neuPABP is not associated with actively translating mRNAs.
Figure 5.
neuPABP localizes with early RNP fractions on polysome gradients and interacts with specific RNAs. (A) Polysome profile traces of lysates prepared from mouse cortices (C57BL/6J; age: P9). (B) Lysates were fractionated by sucrose gradient centrifugation. Fractions were subsequently collected, TCA-precipitated, and resolved by SDS-PAGE, and Western blotting was subsequently performed using antibodies against neuPABP, PABPC1, and a ribosomal protein marker (RPS6). (C) Immunoprecipitation of neuPABP from an adult mouse hippocampus (C57BL/6J; age: 6 mo). Immunoprecipitated complexes were subjected to SDS-PAGE, and Western blotting was performed using anti-neuPABP and anti-GAPDH antibodies. neuPABP-enriched RNAs were isolated using RNA purification kit (Qiagen) and identified by RNA-seq. (D) Volcano scatter plot showing most significantly enriched RNAs with neuPABP (threshold set at log2 FC ≥ 1.5 and P-value set at <1 × 10−20). BC1 RNA and mRNAs encoding ribosomal proteins (red) and nuclear-encoded mitochondrial proteins (blue) were enriched. (E) Top Wikipathway (WP) and associated gene ontology (GO) terms (cellular component) significantly enriched among proteins coded for by neuPABP-enriched mRNAs (FC ≥ 2). The number above each column represents the number of genes associated with its corresponding term. (F) RT-qPCR analyses of neuPABP-enriched transcripts identified by RNA-seq. neuPABP was immunoprecipitated from adult mouse hippocampi (C57BL/6J; age: 6 mo), and associated RNAs were Trizol-extracted. Error bars represent SEM from biological replicates (n = 3). Data points for biological replicates are shown as solid circles. Data were normalized to an in vitro transcribed RLuc spiked-in RNA.
We next set out to identify RNAs associated with neuPABP by carrying out RNA immunoprecipitation sequencing (RIP-seq). Briefly, neuPABP-interacting RNAs were immunopurified with anti-neuPABP antibody from adult mouse hippocampal lysates in three biological replicates and sent for deep sequencing to identify neuPABP-enriched RNAs (Fig. 5C-E; Supplemental Table S2). The most highly enriched RNA associated with neuPABP was BC1 (brain cytoplasmic 1) (Fig. 5D), a neuron-specific noncoding RNA that contains an internal stretch of adenosines (Supplemental Fig. S11A; Martignetti and Brosius 1995; Rozhdestvensky et al. 2001). BC1 has been reported to play a role in translational repression and, like neuPABP, also sediments in early polysome gradient fractions containing free RNP complexes (Supplemental Fig. S11B–D; Wang et al. 2002; Zalfa et al. 2003). To verify that neuPABP can interact with BC1 RNA, HeLa cells were transfected with plasmids encoding V5-tagged neuPABP or V5-tagged PABPC1 and a plasmid that expresses the BC1 RNA. Ectopic PABPC1 and neuPABP, which were expressed at similar levels, were immunoprecipitated with V5 antibody, and BC1 RNA association was assessed by RT-qPCR (Supplemental Fig. S11E,F). We also tested whether V5-tagged neuPABP could interact with BC200 RNA, a primate- and neuron-specific noncoding RNA that also contains a stretch of internal adenosines and is expressed in HeLa cells (Supplemental Fig. S11A; Martignetti and Brosius 1993; Tiedge et al. 1993; Shin et al. 2017). While neither V5-neuPABP nor V5-PABPC1 coprecipitated a histone mRNA (1H4H) lacking a poly(A) tail (control), BC1 and BC200 RNAs were equally enriched with both poly(A) binding proteins (Supplemental Fig. S11E,F). In addition to interacting with BC1, our gene set enrichment analyses of neuPABP-interacting RNAs displayed an enrichment of mRNAs coding for ribosomal proteins and proteins with mitochondrial functions (Fig. 5D,E), which were subsequently validated by RT-qPCR analysis from hippocampal lysates (Fig. 5F). These included ribosomal protein-encoding mRNAs (RPS29 and RPS14) and mitochondrial protein-encoding mRNAs (ATP5J2 and NDUFA2). In contrast, other mRNAs coding for neuron-specific proteins (MAP2, PSD-95, and β-tubulin III) displayed significantly lower enrichment with neuPABP.
As our data show that neuPABP sediments in early RNP fractions that contain untranslated RNAs, we next set out to determine whether neuPABP directly binds the identified target RNAs in RNP fractions. As native RNA–protein interactions can be preserved by cross-linking, we formaldehyde-cross-linked adult mouse cortex tissue prior to generating lysates and carrying out polysome profiling. Early RNP fractions were isolated from polysome gradients (Figs. 6A,B) and immunoprecipitated with IgG (control) or neuPABP antibody to isolate neuPABP-associated RNAs (Fig. 6C). Importantly, while early RNP fractions contained both neuPABP and PABPC1, PABPC1 did not coprecipitate with neuPABP (Fig. 6C). Nevertheless, BC1 and mRNAs coding for ribosomal and mitochondrial proteins were enriched with neuPABP as assessed by RT-qPCR analyses (Fig. 6D). In contrast, neuPABP pulled down significantly lower levels of two mRNAs coding for neuronal-specific proteins (PSD-95 and MAP2) and failed to interact with a mitochondrially encoded ND1 mRNA (control). Taken together, these data indicate that neuPABP interacts with BC1 noncoding RNA and select translationally suppressed mRNA populations.
Figure 6.

neuPABP associates with untranslated mRNAs present in early RNP fraction. Cortices of adult mice (C57BL/6J; age: 6 mo) were triturated and formaldehyde-cross-linked. Lysates were prepared and fractionated by sucrose gradient centrifugation. (A) Ribosome traces of lysates prepared from formaldehyde-cross-linked adult mouse cortices (C57BL/6J; age: 6 mo). (B) Free RNP fractions (depleted of ribosomal subunits) were collected from the polysome gradient and resolved by SDS-PAGE, and Western blotting was performed using antibodies against RPS6, neuPABP, PABPC1, and GAPDH. (C) Immunoprecipitation of neuPABP from free RNP fractions from B. Immunoprecipitated complexes were resolved by SDS-PAGE, and Western blotting was performed using antibodies against neuPABP, PABPC1, and GAPDH. (D) RT-qPCR analysis of neuPABP-associated RNAs isolated from C. Error bars represent SEM from biological replicates (n = 3), which are shown as solid circles. A mitochondrial mRNA (mt.ND1) was used as a negative control. Data were normalized to an in vitro transcribed RLuc spiked-in RNA.
neuPABP has lost its ability to interact with eIF4G and represses mRNA translation in vitro
Our data suggest that neuPABP associates with an abundant noncoding RNA, BC1, as well as specific mRNAs that are translationally dormant. We therefore next assessed the impact of neuPABP on protein synthesis using a Krebs cell-free in vitro translation (IVT) system. This system was used previously to biochemically determine that PABPC1 can function as a translation factor (Kahvejian et al. 2005). To this end, we generated recombinant glutathione S-transferase (GST)-tagged neuPABP (Fig. 7A) and added it to our IVT system. In contrast to recombinant glutathione S-transferase (GST), which did not affect protein synthesis, GST-tagged neuPABP inhibited the expression of a firefly luciferase (FL)-encoding polyadenylated mRNA in a dose-dependent manner (Fig. 7B). In contrast, addition of neuPABP had no observable impact on the expression of an unadenylated FL reporter mRNA (Supplemental Fig. S12A). We also assessed the impact of neuPABP on mRNA translation in a SH-SY5Y neuroblastoma cell line using a bicistronic reporter in which translation initiation of the first open reading frame (Renilla luciferase) is cap-dependent, while cap-independent translation of the second open reading frame (firefly luciferase) is mediated by the hepatitis C virus internal ribosome entry site (IRES) (Supplemental Fig. S12B). Ectopic expression of FLAG-tagged neuPABP inhibited cap-dependent translation in a dose-dependent manner while having no observable impact on the translation of the HCV IRES-driven open reading frame (Supplemental Fig. S12C,D). Collectively, these data suggest that neuPABP can repress cap-dependent translation of polyadenylated mRNAs.
Figure 7.
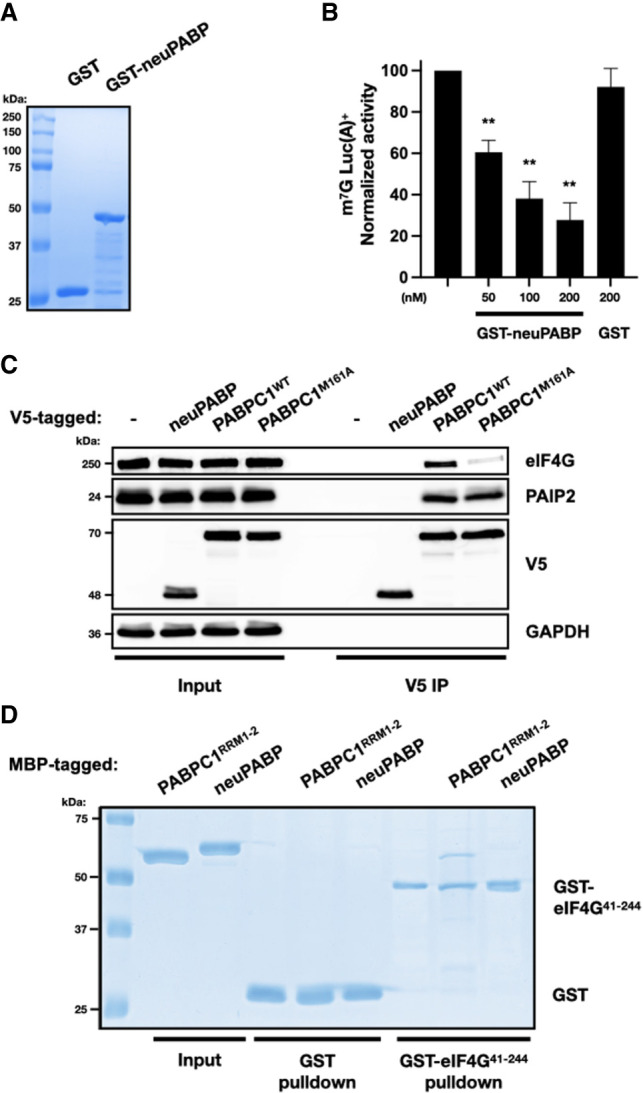
neuPABP represses translation in vitro and does not interact with eIF4G. (A) Recombinant GST and GST-tagged neuPABP were prepared and then analyzed by SDS-PAGE and Coomassie blue staining. (B) Capped poly(A)+ luciferase reporter RNA was incubated in Krebs-2 extract. Reactions were supplemented with either buffer alone (control), recombinant GST-neuPABP, or GST alone, as indicated. Normalized luciferase activity was measured relative to control. Error bars represent SEM from biological replicates (n = 3). A two-tailed Student's t-test (equal variance) was conducted (vs. control) to assess significance. (**) P-values <0.003 were calculated in GST-neuPABP treatment groups. (C) Immunoprecipitation of V5-tagged neuPABP, PABPC1WT, or PABPC1M161A from HeLa cells. Immunoprecipitated complexes were subjected to SDS-PAGE, and Western blot analysis was performed using anti-V5, anti-eIF4G, anti-PAIP2, and anti-GAPDH antibodies. (D) Recombinant glutathione-S-transferase (GST) or GST-tagged eIF4G41–244 was incubated with maltose-binding protein (MBP)-tagged PABPC1RRM1+2 or neuPABP. Precipitated proteins were separated by SDS-PAGE and visualized by Coomassie blue staining.
PABPC1 stimulates mRNA translation by directly binding to eIF4G via RRM2 (Kahvejian et al. 2005; Safaee et al. 2012). neuPABP also contains RRM2 yet paradoxically represses protein synthesis in vitro. To test whether neuPABP can bind eIF4G, HeLa cells were transfected with plasmids encoding V5-tagged neuPABP, wild-type PABPC1, or a PABPC1 mutant (M161A) that disrupts its interaction with eIF4G (control) (Kahvejian et al. 2005). V5-PABPC1WT, V5-PABPC1M161A, and V5-neuPABP were affinity-purified with V5 antibody, and coimmunoprecipitating proteins were resolved by SDS-PAGE followed by Western blotting (Fig. 7C). As expected, V5-PABPC1WT coprecipitated both eIF4G and PAIP2, whereas V5-PABPC1M161A failed to efficiently interact with eIF4G. As neuPABP lacks a C-terminal MLLE domain, it was not surprising that it did not interact with PAIP2, which uses this domain to interact with PABPC1 (Khaleghpour et al. 2001a). However, even though neuPABP contains RRM2, it failed to associate with eIF4G. To determine whether neuPABP can directly contact eIF4G, we performed in vitro pull-down assays using a recombinant GST-tagged fragment of eIF4G (amino acids 41–244) that directly binds PABPC1 (Kahvejian et al. 2005), as well as maltose-binding protein (MBP)-fused neuPABP or a PABPC1 fragment containing RRM1 and RRM2 (Fig. 7D). In keeping with previous reports, GST-eIF4G41–244 efficiently bound MBP-PABPC1RRM1+2. However, MBP-neuPABP failed to bind to this eIF4G fragment.
PABPC1 uses several amino acids in RRM2 to directly contact eIF4G (Fig. 8A; Safaee et al. 2012). While neuPABP also contains RRM2 and binds poly(A) RNA, a comparative sequence analysis of the neuPABP RRM2 revealed nonconservative substitutions in two of the amino acids that PABPC1 uses to interact with eIF4G. In addition, while neuPABP maintains a methionine corresponding to M161 in PABPC1, it also contains an adjacent phenylalanine substitution. To determine whether neuPABP fails to bind eIF4G due to substitutions in these key amino acids, we mutated these corresponding amino acids in tandem in MBP-neuPABP to those in PABPC1 (MBP-neuPABPMUT [Ile221Thr, Phe265Leu, and Tyr268Asp]). While GST-eIF4G41–244 did not interact with MBP-neuPABPWT, it efficiently bound MBP-neuPABPMUT (Fig. 8B). Moreover, while GST-neuPABPWT repressed protein synthesis in vitro, GST-neuPABPMUT incubated in Krebs extract did not (Fig. 8C,D). Taken together, these data suggest that neuPABP does not support mRNA translation due to its inability to interact with eIF4G.
Figure 8.
neuPABP has been selected to not bind eIF4G. (A) Schematic diagrams of PABPC1 and neuPABP domain organization, along with a comparative sequence analysis of human (Hs) and mouse (Mm) PABPC1 with human, mouse, and bat (Pk) neuPABP RRM2. Amino acids that play a role in PABPC1 binding to eIF4G are denoted by a green dot. Corresponding amino acids or those in proximity to eIF4G-intearcting residues are red. (B) Recombinant glutathione-S-transferase (GST)-tagged eIF4G41–244 was incubated with maltose-binding protein (MBP)-tagged PABPC1RRM1+2, neuPABPWT, or neuPABPMUT (Ile221Thr, Phe265Leu, and Tyr268Asp). Precipitated proteins were separated by SDS-PAGE and visualized by Coomassie staining. (C) Recombinant GST, GST-tagged neuPABPWT, and neuPABPMUT proteins were analyzed by SDS-PAGE and Coomassie blue staining. (D) Capped poly(A)+ luciferase reporter RNA was incubated in Krebs-2 extract. Reactions were supplemented with either recombinant GST, GST-neuPABPWT, or GST-neuPABPMUT, as indicated. Normalized luciferase activity was measured relative to buffer alone (control). Error bars represent SEM from biological replicates (n = 3). A two-tailed Student t-test (equal variance) was conducted (vs. GST) to access significance. (**) P-value <0.004 was calculated in GST-neuPABPWT treatment.
Discussion
We have identified a tissue-specific mammalian poly(A) binding protein, neuPABP, which is expressed in the brain and whose levels rapidly increase in neurons as they mature. neuPABP is homologous to the first two RRMs of PABPC1 but lacks RRM3, RRM4, the linker region, and the MLLE domain. In addition, neuPABP contains a unique N-terminal extension not seen in other PABPCs, the function of which is not known. As expected from sequence homology, neuPABP binds to poly(A) RNA but cannot bind PAIP2. Moreover, neuPABP has evolved to maintain amino acid substitutions in RRM2 that prevent it from interacting with eIF4G and stimulating translation in vitro. In accordance with these observations, polysome profiling and RNA sequencing data indicate that neuPABP associates with the neuron-specific noncoding RNA BC1, as well as select mRNAs that are translationally dormant.
While PABPCs are evolutionarily conserved from yeast to humans, higher-order metazoans have acquired several additional PABPC genes that often display tissue- and temporal-specific expression patterns (Gorgoni and Gray 2004). These include a testis-specific PABP (tPABP) that is expressed in round spermatids (Feral et al. 2001); inducible PABP (iPABP, also known as PABPC4), which has been reported to be expressed in activated T-cells (Yang et al. 1995); ovary-specific PABP (PABPC5) (Blanco et al. 2001); and embryonic PABP (ePABP), which is expressed in oocytes and early embryos (Seli et al. 2005). neuPABP therefore represents a new PABPC that mammals have added to their genetic repertoire. Like other newly acquired PABPCs during metazoan evolution, neuPABP also displays a unique temporal- and tissue-specific expression pattern. neuPABP is barely detectable in embryonic brain tissues but becomes robustly expressed in the postnatal brain. In contrast, PABPC1 levels are significantly lower in the postnatal brain as compared with PABPC1 levels in the embryonic brain. This is reminiscent of ePABP, which is expressed in early embryos and oocytes when PABPC1 is barely detectable (Voeltz et al. 2001; Cosson et al. 2002). Cytoplasmic PABPs, including PABPC1 and yeast Pab1, multimerize on mRNA poly(A) tails through PABPC–PABPC oligomerization (Yao et al. 2007; Schäfer et al. 2019). Specifically, the linker region between RRM4 and the C-terminal MLLE plays an important role in this process by contacting RRM1 of an adjacent PABPC on the poly(A) tail (Baer and Kornberg 1980; Deo et al. 1999; Sawazaki et al. 2018; Schäfer et al. 2019). neuPABP lacks the PABPC1 linker stretch but does contain a unique N-terminal extension that is not present in canonical PABPCs. Together, this unique architecture may prevent neuPABP from multimerizing with other canonical PABPCs on mRNA poly(A) tails. In keeping with this, our data suggest that neuPABP and PABPC1 do not concurrently bind target RNAs. Nevertheless, more research is required to test this hypothesis and determine the function of the conserved N-terminal DUF.
Many factors that regulate mRNA stability directly interact with the PABPC1 C-terminal MLLE domain. These include the PAN3 subunit of the PAN2–PAN3 deadenylase complex (Schäfer et al. 2019) as well as Tob and GW182, both of which recruit the CCR4–NOT deadenylase complex to targeted mRNAs (Okochi et al. 2005; Fabian et al. 2011; Huntzinger et al. 2013; Jonas and Izaurralde 2015; Chen et al. 2020). In contrast to PABPC1, neuPABP lacks a MLLE domain. This may prevent neuPABP from recruiting mRNA decay factors to neuPABP-interacting RNAs (Fig. 9). PABPC1 also uses its MLLE domain to interact with PAIP2, which when bound prevents PABPC1 from binding to RNA (Khaleghpour et al. 2001a,b). Interestingly, PAIP2 has been reported to regulate synaptic plasticity by binding to PABPC1 to inhibit protein synthesis (Khoutorsky et al. 2013). PAIP2 is then degraded following activity-dependent stimulation, which may allow PABPC1 to stimulate translation (Khoutorsky et al. 2013). As neuPABP does not interact with PAIP2 (Fig. 7C), this may allow it to bind RNAs even when PAIP2 levels are elevated.
Figure 9.
Model for the biological role of neuPABP. (Panel i) neuPABP binds to BC1 RNA and select translationally dormant mRNAs that may be transported to postsynaptic compartments. As neuPABP also lacks the PABPC1 MLLE domain, it may protect mRNAs from mRNA decay factors that can interact with this domain, including the PAN2–PAN3 complex and Tob, which interacts with the CCR4–NOT deadenylase complex. (Panel ii) It is possible that in specific contexts (depicted as a question mark), PABPC1 may displace neuPABP from mRNA poly(A) tails, bind eIF4G, and stimulate their mRNA translation.
PABPC1 can stimulate translation via its contact with eIF4G (Kahvejian et al. 2005; Safaee et al. 2012). While our data suggest that neuPABP is a bona fide PABPC, it does not stimulate translation and may even function as a translational repressor. This is based on the observations that (1) neuPABP has evolved such that it does not interact with eIF4G and as a result represses mRNA translation in vitro, (2) neuPABP-associated mRNAs are not associated with polysomes, and (3) neuPABP and PABPC1 do not co-occupy neuPABP-interacting mRNAs. neuPABP may associate with mRNAs that are already translationally repressed or potentially help maintain these mRNAs in a translationally repressed state by binding to their poly(A) tails and not supporting PABPC–eIF4G contact. Nevertheless, the exact role of neuPABP when bound to these RNAs remains to be established.
In synaptic compartments, mRNAs are actively translated and contribute to synaptic plasticity (Hafner et al. 2019; Biever et al. 2020). Many studies have reported that ribosomal protein-coding mRNAs localize into dendrites, and their local translation in neurites play an important role in ribosome recycling and repair independent of canonical ribosome biogenesis in the nucleolus and cytoplasm (Gumy et al. 2011; Cajigas et al. 2012; Middleton et al. 2019; Biever et al. 2020; Fusco et al. 2021; Perez et al. 2021). Similarly, nuclear-encoded mitochondrial protein-coding mRNAs are also localized into axons and presynaptic terminals and are locally translated to provide proteins for healthy mitochondrial function (Kaplan et al. 2009). We show that neuPABP localizes to dendritic postsynaptic compartments and associates with a subpopulation of ribosomal and mitochondrial protein-coding mRNAs in their untranslated state. It is possible that neuPABP plays an active role in maintaining these mRNAs and in alleviating their translation repression in a context-dependent manner after stimulation.
Brain cytoplasmic (BC) RNAs are neuronal-specific RNAs that localize to synapses and have been reported to control local protein synthesis by acting as translational repressors (Tiedge et al. 1991; Wang et al. 2002; Zalfa et al. 2003; Cristofanilli et al. 2006; Robeck et al. 2016). BC RNAs in rodents (BC1) and primates (BC200) maintain internal stretches of adenosines that have been previously reported to interact with PABPC1 (Muddashetty et al. 2002). We show here that neuPABP interacts with BC1 RNA in vivo and both BC RNAs in vitro. As levels of both BC1 and neuPABP increase during neuronal maturation and mouse brain development (Supplemental Fig. S13; Muslimov et al. 1998), it is possible that neuPABP binding to BC1 ensures that PABPC1 is not completely sequestered by this highly abundant noncoding RNA, thus allowing PABPC1 to focus on interacting with the mRNA transcriptome. Knocking out BC1 in a mouse model has been reported to lead to cognitive dysfunction and epileptogenic susceptibility (Zhong et al. 2009; Briz et al. 2017). Thus, it will be interesting to determine whether neuPABP plays a role in supporting BC1 functionality.
Materials and methods
Details of the reagents used in this study are shown in Supplemental Table S3.
Antibodies
Antibodies were purchased from Abcam (PABPC1, hnRNPA1, PSD-95, RPS6, and synaptophysin), Cell Signaling Technologies (PABPC1, V5, eIF4G, and β-Actin), BioLegend (β3tubulin), Invitrogen (V5 tag), Santa Cruz Biotechnology (GAPDH), and Sigma-Aldrich (PAIP2 and FLAG). A peptide encompassing the C-terminal end of mouse neuPABP (ERGAWARQSTSADFKDFD), which is not conserved in other mammalian PABPC proteins, was injected into rabbits for neuPABP antibody production (Thermo Fisher).
DNA constructs, cell lines, and primary cultures
Bacterial expression vectors
N-terminal GST-tagged neuPABP and eIF4G (41–244) expression clones were generated by cloning into pGEX-6P1 plasmid (Addgene). neuPABP and eIF4G (41–244) coding sequences were PCR-amplified; restriction-digested with enzymes BamHI, and SalI for neuPABP and with NotI and SalI for eIF4G (41–244); and ligated in-frame with the N-terminal GST tag in pGEX-6P1 plasmid. N-terminal His-tagged PABPC1-expressing pET-28b-PABPC1 plasmid was a gift from Dr. Sonenberg at McGill University. N-terminal malE-tagged neuPABP and PABPC1 (RRM1+2) expression clones were generated by cloning into pMAL-c5X plasmid (NEB) using restriction enzyme sites NotI and SacI. neuPABP and PABPC1(RRM1+2) coding sequences were PCR-amplified, restriction-digested with enzymes NotI and SacI, and ligated in-frame with the N-terminal malE tag in pMAL-c5X plasmid.
Mammalian expression vectors
C-terminal V5-tagged neuPABP and PABPC1 expression clones were generated by gateway cloning (Thermo Fisher). Coding sequences were PCR-amplified using gene-specific gateway primers that contained flanking attB sites for recombination with attP sites in the donor pDONR221 plasmid to generate entry clones with attL sites. Furthermore, neuPABP and PABPC1 expression clones were generated by recombining the attL sites in the pDONR221-neuPABP and PABPC1 entry clones and the attR sites in the pLEX-307 destination vector (Addgene). The lncRNA BC1 gene sequence was cloned into PLKO.1-puro vector (Sigma). The BC1 gene sequence was PCR-amplified using primers containing restriction enzyme sites for EcoRI and XmaI, restriction-digested, and ligated into PLKO.1-puro plasmid that was digested with EcoRI and AgeI (BshTI). The bicistronic luciferase reporter system was a generous gift from Dr. Jerry Pelletier. The N-terminal FLAG-tagged neuPABP construct was generated by cloning the coding sequence into pBABE-puro vector (Addgene) using restriction enzyme sites BamHI and SalI.
Cell lines
Human epithelial carcinoma HeLa cells, human embryonic kidney (HEK) 293T cells, and human neuroblastoma SH-SY5Y cells were purchased from ATCC. Cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) or DMEM/F12 (for SH-SY5Y cells) supplemented with 10% fetal bovine serum, 50 U/mL penicillin, and 50 μg/mL streptomycin.
Primary neuronal cultures
Mouse primary cortical neuron cultures were prepared from P0 pup cortices. Mouse pups were collected soon after birth and decapitated according to animal-handling protocol. Brains were immediately harvested into cold Dulbecco's modified Eagle's medium–F12 media (DMEM/F12), and the cerebellum and olfactory bulbs were removed. Cortices were gently triturated in prewarmed DMEM/F12 medium by pipetting with a Pasteur pipette to get homogenous cell suspension and centrifuged at 700 rpm for 2 min. Only half of the supernatant was removed from the top, replaced with PPD saline (0.1% papain, 0.01% deoxyribonuclease-I, 0.1% neutral protease-dispase-II, 10 mM MgCl2 in HBSS [without Ca2+ and Mg2+]), mixed with a Pasteur pipette, and incubated for 30 min at 37°C, pipetting gently every 10 min. Cells were centrifuged at 800 rpm for 10 min, resuspended in DNase-I saline (0.1% deoxyribonuclease-I in DMEM), and incubated for 15 min at 37°C. Cells were then collected by centrifugation at 800 rpm for 10 min, resuspended in complete neuronal medium (neurobasal medium containing 1× B27 supplement, 2 mM L-glutamine, 0.5× pen/strep), and plated on poly-L-lysine-coated culture flasks. Seventy-two hours after plating, 3 µM Ara-C (arabinosylcytosine) drug was added to control glial cell overgrowth. Sterile conditions were maintained throughout the procedure.
5′RACE (rapid amplification of cDNA ends)
RNAs were isolated from mouse primary neuronal cultures at 7 d in vitro using Trizol reagent. RNAs were dephosphorylated using FastAP (alkaline phosphatase) kit (Thermo Fisher) and decapped using RppH (RNA 5′ pyrophosphohydrolase) kit (NEB). RNA adapter (5′-GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA-3′) was ligated to 5′ monophosphate ends of the decapped RNAs by using T4 RNA ligase 1 (NEB). RNAs were reverse-transcribed using random hexamer priming and an AffinityScript kit (Agilent). PCR amplification using KAPA HotStart kit (Millipore Sigma) was carried out using an adapter-specific forward primer (5′-GCTGATGGCGATGAATGAACACTG-3′) and a pabpc1l2 sequence-specific reverse primer (5′-CACCGGTTGCTGGTAGTTGA-3′). A fraction of PCR reaction was further amplified using adapter-specific forward and pabpc1l2 sequence-specific reverse gateway primers containing attB sites (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTACCGCTGATGGCGATGAATGAACACTG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCCAGGCTGGCCTCCTCAA-3′, respectively) and cloned into a gateway donor plasmid, pDONR221 (Thermo Fisher). Plasmid constructs were sequenced to identify the pabpc1l2 mRNA 5′ end.
Recombinant protein purification
GST-tagged recombinant proteins were expressed in Rosetta-2(DE3) Escherichia coli cells (Millipore) and purified by using glutathione agarose resin (Thermo Fisher). GST cleavage was performed using HRV-3C protease (Thermo Fisher). His-tagged recombinant proteins were purified by using Ni-NTA beads (Qiagen). MBP-tagged recombinant proteins were purified by using amylose resin (NEB).
GST pull-down assays
GST or GST-eIF4G (1–244) recombinant bait proteins (200 pmol) were allowed to bind to 25 µL of packed glutathione beads in binding buffer (50 mM Tris, 150 mM NaCl, 0.5% NP-40, 5% glycerol, 1.5 mM DTT) for 2 h at 4°C with gentle rotation. MBP-tagged neuPABP or PABPC1 (RRM1+2) prey proteins (160 pmol) were then added, and the reactions were incubated overnight at 4°C with gentle rotation. Beads were washed to remove unbound proteins, boiled in Laemmli buffer to release the bound proteins, and resolved on an SDS-PAGE gel. Resolved proteins were visualized by Coomassie blue staining.
RNAcompete
RNA pool generation, RNAcompete pull-down assays, and microarray hybridizations were performed as previously described (Ray et al. 2009, 2013, 2017). Briefly, RNAcompete experiments used defined RNA pools that were generated from 244,000 Agilent custom DNA microarrays. Pool design was based on a de Bruijn sequence of order 11 that was subsequently modified to minimize secondary structure in the designed sequences and minimize intramolecular RNA cross-hybridization. After these modifications, not every 11-mer was represented, but each 9-mer was represented at least 16 times. To facilitate internal data comparisons, the pool was split computationally into two sets: set A and set B. Each set contained at least 155 copies of all 7-mers except GCTCTTC and CGAGAAG, which were removed because they corresponded to the SapI/BspQI restriction site used during DNA template pool generation. A φ2.5 bacteriophage T7 promoter initiating with an AGA or AGG sequence was added at the beginning of each probe sequence in the DNA template pool to enable RNA synthesis. The final RNA pool consisted of 241,399 individual sequences up to 41 nt in length (Ray et al. 2013). The microarray design is detailed in Ray et al. (2013) and can be ordered from Agilent Technologies using AMADID 024519. In RNAcompete assays, 20 pmol of GST-tagged neuPABP and 1.5 nmol of RNA pool were incubated for 30 min at 4°C in 1 mL of binding buffer (20 mM HEPES at pH 7.8, 80 mM KCl, 20 mM NaCl, 10% glycerol, 2 mM DTT, 0.1 mg/mL BSA) containing 20 µL of glutathione Sepharose 4B beads (GE Healthcare) prewashed three times in binding buffer and subsequently washed four times for 2 min with binding buffer at 4°C. One-sided Z scores were calculated for the motifs as described previously (Ray et al. 2013).
Electrophoretic mobility shift assay
RNA oligo(A)25 (150 pmol) was 5′ end-radiolabeled with 10 µCi of [γ32-P]-ATP (Perkin Elmer) using the enzyme T4 polynucleotide kinase (Thermo Fisher) for 1 h at 37°C. The end-labeled oligoribonucleotide was diluted to 2 pmol/µL with double-distilled water and column-purified (Roche), and 2 pmol of RNA oligonucleotide was used for each EMSA reaction. RNA–protein EMSA binding reactions as well as protein dilutions were made in standard phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) supplemented with 160 ng/mL double-stranded DNA and 40 U/mL RNase inhibitor (Promega). Radiolabeled RNA oligonucleotide (2 pmol per reaction) was mixed with different amounts of recombinant protein (neuPABP or PABPC1) in a final reaction volume of 20 µL and incubated for 1 h at 30°C. Binding reactions were supplemented with glycerol containing bromophenol blue dye (to 5% glycerol concentration) and electrophoresed on a 10% (w/w) nondenaturing polyacrylamide/bis-acrylamide gel at 29:1 in Tris-borate-EDTA running buffer at 120 V for 60–120 min on ice. Gels were analyzed using a PhosphorImager (GE Healthcare). Free RNA as well as gel-shifted RNA bands were quantified for each gel lane by ImageJ. The fraction of bound RNA in each lane was calculated by using the expression bound/(bound + unbound). The dissociation constant (KD) values were calculated by GraphPad Prism as the protein concentration (in nanomolar) at which only 50% of free RNA remained unbound.
In vitro translation experiments
Krebs-2 cell-free lysates were prepared as described previously for in vitro translation experiments (Svitkin and Agol 1978; Svitkin and Sonenberg 2004). Capped poly(A)+ and poly(A)− luciferase RNAs were incubated in Krebs-2 extract, in vitro translation reactions were incubated for 1 h at 30°C, and translation output from the reporter mRNAs was accessed by a luciferase assay (Promega).
Tissue expression analysis
Western blotting
Adult mice (C57BL/6J; age 5 mo) were sacrificed according to the animal euthanization protocol, different tissues were dissected out including different brain regions, and lysates were prepared in a lysis buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.5% NP-40, 2 mM EDTA, 1 mM DTT). Western blotting was performed to assess the expression of neuPABP PABPC1 and other proteins.
RT-PCR and RT-qPCR analyses
Adult mice (C57BL/6J; age 5 mo) were sacrificed according to the animal euthanization protocol. Different tissues/brain regions were dissected out, and total RNA was isolated using Trizol reagent. RNAs were reverse-transcribed (Thermo Fisher), and semiquantitative PCR and qPCR analyses were performed to assess the differential expression of pabpc1l2 among mouse tissues.
Single-nucleus RNA-seq data processing
The 10x multiome RNA + ATAC data for healthy mouse cortices at postnatal time points P7 and 10 wk were obtained from GSE199885. Sequencing reads were reprocessed to allow the quantification of Pabpc1l2, as the genomic annotations for Pabpc1l2a/b were absent in the mm10 reference genome build used in the original report (Khazaei et al. 2023). A custom reference was built, adding the coordinates for Pabpc1l2b (chromosome X: 103,013,563–103,016,208) to the gene annotation. In addition, since there was high homology in Pabpc1l2a/b genes resulting in multimapping reads, the sequence for Pabpc1l2a (chromosome X: 103,064,742–103,067,538) was masked using bedtools maskfasta. Thus, the expression of the two Pabpc1l2a/b genes was profiled as a single feature (Pabpc1l2) in downstream analysis, and no attempt was made to distinguish expression of Pabpc1l2a and Pabpc1l2b separately. Sequencing reads were then aligned and demultiplexed using CellRanger-arc v2.0.0 (10x Genomics) using this modified mm10 reference genome build coupled with the modified Ensembl 98 gene annotation. Quality control (QC) and data processing steps were performed using Signac v1.3.0 (Stuart et al. 2021) and Seurat v4.0.0 (Hao et al. 2021) as described in the original report (Khazaei et al. 2023). Briefly, QC metrics for RNA and ATAC modalities were jointly used to filter cells. In the RNA modality, cells were filtered on the number of genes, unique molecular identifiers (UMIs), and mitochondrial content. In the ATAC modality, cells were filtered on the number of peaks detected, transcription start site enrichment, and nucleosome signal. Next, RNA libraries were scaled to 10,000 UMIs per cell and log-normalized, and UMI counts and mitochondrial content were regressed out. Ten-week samples were integrated by merging the samples without batch correction, followed by scaling and normalization. Dimensionality reduction was performed using PCA on the top 2000 most variable features. The first 25 principal components were used as input for projection into two dimensions (uniform manifold approximation and projection) (McInnes et al. 2020) and for clustering (shared nearest-neighbor algorithm) (Hao et al. 2021). Doublets were identified using scDblFinder (Germain et al. 2021) with the recommended cluster-based approach, and subsequently the doublets were filtered. Last, annotations of cell types was performed using four machine learning-based prediction methods (SciBet, SingleCellNet, SingleR, and Support Vector Machines) (Aran et al. 2019; Tan and Cahan 2019; Li et al. 2020). For this, two murine brain cell type atlases were used. Nonneuronal cell types were annotated using a developmental murine atlas (Jessa et al. 2019), and cells predicted as neurons were subsequently annotated using a more detailed neuronal-enriched atlas (Yao et al. 2021). A consensus cell type annotation was assigned when at least two methods agreed. Finally, cell type labels were aggregated into broad cell type classes.
Subcellular fractionation
Whole-cell, cytoplasmic, and nuclear fractions
The protocol for cell fractionation of mouse brain cortices (C57BL/6J; age: 2 mo) was adapted and optimized from a previously published study describing a rapid and efficient method to subfractionate human cell lines (Suzuki et al. 2010). Briefly, an adult mouse was euthanized according to animal-handling protocols, the brain was dissected out, and the cerebellum and olfactory bulbs were removed. The brain hemispheres were separated, and the cerebral cortex was dissected out from one hemisphere. The cortex tissue was triturated in phosphate-buffered saline (PBS) by pipetting and centrifuged at 200g for 3 min. The pellet was again triturated in PBS and incubated for 3 min on ice to allow the tissue chunks to settle down. Cell suspension was obtained from the top by avoiding the tissue chunks. A part of this cell suspension was then centrifuged at 2500g for 3 min. The cell pellet thus obtained was triturated in lysis buffer (0.1% NP-40 in PBS) by pipetting six to seven times using a p1000 micropipette. One-third of this homogenous cell suspension was kept as the “whole-cell fraction,” and the remaining homogenate was pipetted three more times and centrifuged at 7000g for 30 sec. One-half of the supernatant was kept as the “cytoplasmic fraction,” while the remaining supernatant was discarded. The pellet was again triturated in the lysis buffer by pipetting five times and centrifuged at 7000g for 30 sec, and the supernatant was discarded. The residual pellet was kept as the “nuclear fraction.” Laemmli buffer containing benzonase (Millipore) was added to each collected fraction, and Western blot analysis was performed for nuclear and cytosolic proteins.
Crude synaptosome preparation
Subcellular fractionation of adult mouse brain cortices (C57BL/6J; age: 6 mo) was carried out using a protocol adapted from Qiu et al. (2013), Briz et al. (2017), and Wang et al. (2018). Briefly, an adult mouse brain was dissected out, and the cerebellum and olfactory bulbs were removed. The cortex tissue was homogenized in homogenization buffer (320 mM sucrose, 5 mM HEPES-NaOH at pH 7.4, 2 mg/mL BSA, 1 mM EDTA) by 15 strokes using a Dounce homogenizer to obtain brain homogenate fraction (H), which was centrifuged at 1000g for 10 min at 4°C to obtain supernatant S1. The S1 supernatant was centrifuged at 14,000g for 20 min at 4°C to obtain the cytosolic supernatant S2 fraction, and a crude synaptosome-containing pellet P2 fraction. We further fractionated the crude synaptosomes into a Triton X-100 detergent-soluble “non-PSD” fraction and a detergent-insoluble “PSD-enriched” fraction. The P2 fraction was resuspended once in resuspension buffer (5 mM HEPES-NaOH at pH 7.4, 1 mM EDTA) and centrifuged at 14,000g for 15 min at 4°C. The washed pellet thus obtained was resuspended in buffer A (5 mM HEPES-NaOH at pH 7.4, 100 mM NaCl, 0.5% Triton X-100), rotated for 15 min at 4°C, and centrifuged at 14,000g for 15 min at 4°C to obtain a Triton X-100-soluble “non-PSD” fraction as the supernatant. The remaining pellet was resuspended in buffer B (5 mM HEPES NaOH at pH 7.4, 0.15 mM NaCl, 1% Triton X-100, 1% SDS, 1% deoxycholic acid, 1 mM DTT), rotated for 75 min at 4°C, and centrifuged at 14,000g for 15 min at 4°C to obtain a Triton X-100-insoluble “PSD-enriched” fraction as the supernatant. The brain homogenate, cytosolic, and crude synaptosomal fractions were lysed completely by supplementing with RIPA buffer (10 mM Tris-HCl at pH 8.0, 140 mM NaCl, 1% Triton X-100, 0.1% deoxycholic acid, 0.1% SDS, 1 mM EDTA). The buffers used in crude synaptosomal preparation were supplemented with protease inhibitor cocktail and 1 mM phenylmethylsulfonyl fluoride (PMSF).
Mass spectrometry analysis
An adult mouse brain (C57BL/6J; age: 2 mo) was dissected out, the cerebellum and olfactory bulbs were removed, and the cerebral cortex was isolated. Briefly, fresh cortex tissue was triturated in cold phosphate-buffered saline by pipetting and centrifuged at 200g for 3 min. The tissue pellet was lysed in a lysis buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.5% NP-40, 2 mM EDTA, 1 mM DTT) and centrifuged at 20,000g for 15 min at 4°C to obtain the lysate. The tissue lysate was clarified with Protein G agarose beads (Millipore), and total protein was quantified and immunoprecipitated with neuPABP antibody. Protein complexes were eluted with neuPABP-specific peptide (ERGAWARQSTSADFKDFD) and resolved by SDS-PAGE. Gels were subsequently stained with colloidal stain, and neuPABP protein was excised, digested with trypsin or subtilisin, and analyzed by the Lady Davis Institute Proteomics Centre (Montreal, Canada).
Polysome profiling
A mouse brain (C57BL/6J; P9 mouse pup) was dissected out, and the cerebellum and olfactory bulbs were removed. The remaining brain tissue was triturated by pipetting in dissection buffer (10 mM HEPES-NaOH at pH 7.5, 0.1 mg/mL cycloheximide) and centrifuged at 300g for 3 min. The tissue pellet was lysed in polysome lysis buffer (10 mM HEPES-NaOH at pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, 1% NP-40, 0.1 mg/mL cycloheximide) and centrifuged twice at 20,000g for 10 minat 4°C. Lysates were loaded onto a 5%–50% linear sucrose gradient and centrifuged at 130,000g for 2 h at 4°C. Four to 16 gradient fractions were collected as described previously (Gandin et al. 2014), and proteins were TCA-precipitated from each fraction and dissolved in Laemmli buffer to perform Western blot analysis.
For lncRNA BC1, polysome fractionation was done from adult mice (C57BL/6J; age: 3 mo). RNA from each polysome gradient fraction was Trizol-extracted and reverse-transcribed, and semiquantitative-PCR analysis was carried out on lncRNA BC1 and Actin. In parallel, qPCR analysis was carried out on lncRNA BC1, and data were normalized to the total cortex RNA (input) to calculate the percent distribution across fractions.
RIP RNA sequencing (RIP-seq)
Mouse hippocampi (C57BL/6J; age: 6 mo; Pabpc1l2+/+ and Pabpc1l2−/−) were dissected out and flash-frozen on dry ice. Tissue was lysed in a lysis buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.5% NP-40, 2 mM EDTA, 1 mM DTT) and centrifuged at 20,000g for 15 min at 4°C to obtain the lysate. Tissue lysates were clarified with Protein G agarose beads (Millipore), total proteins were quantified, and immunoprecipitation was carried out by first incubating with neuPABP antibody, followed by a pull-down of immunoprecipitants with Protein-G agarose beads. Immunoprecipitation of neuPABP was confirmed by Western blotting. neuPABP-bound RNAs were extracted directly from the beads by using an RNA purification kit (Qiagen). Biological triplicate libraries were prepared from immunoprecipitated RNAs. RNA was depleted of ribosomal RNA, and libraries were prepared using the KAPA stranded RNA-seq kit with RiboErase (Roche). Sequencing reactions were carried out by paired-end 150-bp sequencing on a NextSeq500 platform (Genomics Platform at the Institute for Research in Immunology and Cancer, Montreal). Sequences were trimmed for sequencing adapters and low-quality 3′ bases using Trimmomatic version 0.35 (Bolger et al. 2014) and aligned to the reference mouse genome version GRCm38 (gene annotation from Gencode version M25, based on Ensembl 100) using STAR version 2.7.1a (Wang et al. 2013). Gene expressions were obtained as read count directly from STAR as well as computed using RSEM (Li and Dewey 2011) in order to obtain normalized gene- and transcript-level expression in TPM values for stranded RNA libraries. DEseq2 version 1.22.2 was then used to normalize gene read counts (Love et al. 2014).
Volcano plot and gene ontology analysis
Volcano plot was generated using a list of transcripts with fold change ≥ 1.10 (∼2200 transcripts that had P-value < 0.05). A log2FC ≥ 1.5 cutoff was further used in the volcano plot to highlight highly enriched transcripts. Gene set enrichment analyses for Wikipathway (WP) terms enriched among neuPABP-enriched transcripts (FC ≥ 2) were performed using the g:Profiler online platform (Raudvere et al. 2019).
RNA immunoprecipitation with V5 antibody
HeLa cells were cotransfected with V5-tagged neuPABP- or PABPC1-expressing constructs and a lncRNA BC1-expressing plasmid. Forty-eight hours after transfection, lysates were prepared in a lysis buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.5% NP-40, 2 mM EDTA, 1 mM DTT) and precleared using Protein-G agarose beads (Millipore). Lysates were incubated with rabbit antibodies IgG (control) or V5 tag antibody (Cell Signaling Technologies), followed by a pull-down of immunoprecipitants with Protein-G agarose beads. Immunoprecipitation of V5-tagged proteins was confirmed by Western blotting. Coimmunoprecipitated RNA was extracted using Trizol reagent and reverse-transcribed. qPCR analysis was carried out to assess fold enrichments (vs. IgG control) of lncRNAs BC1 and BC200 with V5-tagged proteins. A nonpolyadenylated histone (1H4H) mRNA was used as a negative control.
Formaldehyde-cross-linked RNA immunoprecipitation
Formaldehyde cross-linking of mouse cortices
Brain cortices of adult mice (C57BL/6J; age: 6 mo) were dissected out and gently triturated in dissection buffer (Hank's balanced salt solution containing 10 mM HEPES at pH 7.5) and spun at 1000g to collect the triturated tissue as a pellet. The pellet was then gently resuspended in dissection buffer containing 0.1% formaldehyde and incubated for 10 min at room temperature. Formaldehyde was quenched by adding glycine to a final concentration of 200 mM for 5 min. The tissue suspension was chilled on ice and pelleted by spinning at 1000g. The tissue pellet was washed twice (without resuspending) with dissection buffer containing glycine at a final concentration of 200 mM.
Tissue lysis and polysome RNP fractionation
The cross-linked tissue pellet was lysed in polysome lysis buffer (10 mM HEPES-NaOH at pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, 1% NP-40, 0.1 mg/mL cycloheximide) and centrifuged twice at 20,000g for 10 min at 4°C. Lysates were loaded on a 5%–50% linear sucrose gradient and centrifuged at 130,000g for 2 h at 4°C. Gradient fractions (1 and 2) corresponding to the RNP fraction were collected and combined. Proteins from a part of the RNP fraction were TCA-precipitated and dissolved in Laemmli buffer to perform Western blot analysis to assess the depletion of ribosomal subunits in comparison with the total cortex lysate (marker: RPS6).
Cross-link RNP RNA immunoprecipitation
The RNP fraction was diluted 1:2 in RIPA buffer (25 mM Tris-HCl at pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM DTT), and immunoprecipitation was carried out by first incubating with antibodies IgG (control) or neuPABP, followed by a pull-down of immunoprecipitants with Protein-G agarose beads (Millipore). Immunoprecipitation of neuPABP was confirmed by Western blotting. Coimmunoprecipitated RNAs were eluted from the beads in elution buffer (50 mM Tris-HCl at pH 8.0, 2 mM EDTA, 1% SDS, 10 mM DTT) containing 100 U/mL Proteinase-K (NEB). Reverse cross-linking was performed on a thermomixer at 1200 rpm, for 30 min at 60°C and for 15 min at 70°C. Eluted RNAs were further isolated using Trizol reagent (Thermo Fisher). qPCR analysis was carried out to assess fold enrichments (vs. IgG control) of lncRNA BC1 and other neuPABP target and nontarget mRNAs. A mitochondrial genome-encoded mRNA (mt.ND1) was used as a control to negate postlysis reassociation artifacts.
Bicistronic luciferase assay
SH-SY5Y cells were cotransfected with the bicistronic luciferase reporter and N-terminally FLAG-tagged neuPABP construct or a plasmid expressing GFP (control). Twenty-four hours after transfection, cells were collected and lysed in passive lysis buffer (Promega), and RL and FL activities were measured using dual-luciferase assay (Promega). In parallel, the expression of FLAG-tagged proteins was validated by Western blot analyses. RL to FL ratios were calculated and plotted as normalized luciferase activity. A GFP-expressing construct was used as a control for normalization. Experiments were conducted in biological replicates (n = 4).
Statistics
All experiments were carried out at least in triplicates. Graphs for in vitro translation assays were generated using GraphPad Prism software and Excel. Means and standard error of the mean (SEM) from biological replicates (n = 3) were calculated. For in vitro and in cellulo translation assays, a two-tailed Student's t-test (equal variance) was carried out to assess the significance of the data in Excel. Statistical significances are as follows: P > 0.05 (n.s.), P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
Supplementary Material
Acknowledgments
We are grateful to Jerry Pelletier for providing the bicistronic reporter system and Krebs extract, Vincent Richard and Rene Zahedi at the Jewish General Hospital Proteomics Centre, Matthew Leibovitch and Ivan Topisirovic for assistance with polysome profiling, and Ji-Young Youn, Anne-Claude Gingras, Siraj Zahr, and Freda Miller for invaluable assistance at the early point of this project. This work was supported by Canadian Institutes of Health Research (CIHR) grants (PJT-156356 to M.R.F., PJT-162255 to T.R.H. and Q.M., and FDN-148403 to T.R.H.), a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant (RGPIN-2015-03712 to M.R.F.), a National Institutes of Health grant (R01HG008613 to T.H and Q.M.), and a Fonds de Recherche du Québec-Santé (FRQS) Chercheur-Boursier Senior award to M.R.F. Graduate student support was by FRQS to S.S. and by NSERC to S.K., and Z.H. was supported by a NSERC Undergraduate Student Research Award.
Author contributions: M.R.F. and S.S. designed the research. S.S., Z.H., S.K., D.R., and R.L.G. carried out experiments. All authors contributed to writing and/or editing the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350597.123.
Competing interest statement
The authors declare no competing interests.
References
- Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, et al. 2019. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20: 163–172. 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer BW, Kornberg RD. 1980. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc Natl Acad Sci 77: 1890–1892. 10.1073/pnas.77.4.1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer BW, Kornberg RD. 1983. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol 96: 717–721. 10.1083/jcb.96.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biever A, Glock C, Tushev G, Ciirdaeva E, Dalmay T, Langer JD, Schuman EM. 2020. Monosomes actively translate synaptic mRNAs in neuronal processes. Science 367: eaay4991. 10.1126/science.aay4991 [DOI] [PubMed] [Google Scholar]
- Blanco P, Sargent CA, Boucher CA, Howell G, Ross M, Affara NA. 2001. A novel poly(A)-binding protein gene (PABPC5) maps to an X-specific subinterval in the Xq21.3/Yp11.2 homology block of the human sex chromosomes. Genomics 74: 1–11. 10.1006/geno.2001.6530 [DOI] [PubMed] [Google Scholar]
- Boeck R, Kolakofsky D. 1994. Positions +5 and +6 can be major determinants of the efficiency of non-AUG initiation codons for protein synthesis. EMBO J 13: 3608–3617. 10.1002/j.1460-2075.1994.tb06668.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briz V, Restivo L, Pasciuto E, Juczewski K, Mercaldo V, Lo AC, Baatsen P, Gounko NV, Borreca A, Girardi T, et al. 2017. The non-coding RNA BC1 regulates experience-dependent structural plasticity and learning. Nat Commun 8: 293. 10.1038/s41467-017-00311-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. 2012. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74: 453–466. 10.1016/j.neuron.2012.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CA, Strouz K, Huang KL, Shyu AB. 2020. Tob2 phosphorylation regulates global mRNA turnover to reshape transcriptome and impact cell proliferation. RNA 26: 1143–1159. 10.1261/rna.073528.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B, Couturier A, Le Guellec R, Moreau J, Chabelskaya S, Zhouravleva G, Philippe M. 2002. Characterization of the poly(A) binding proteins expressed during oogenesis and early development of Xenopus laevis. Biol Cell 94: 217–231. 10.1016/S0248-4900(02)01195-4 [DOI] [PubMed] [Google Scholar]
- Cristofanilli M, Iacoangeli A, Muslimov IA, Tiedge H. 2006. Neuronal BC1 RNA: microtubule-dependent dendritic delivery. J Mol Biol 356: 1118–1123. 10.1016/j.jmb.2005.11.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo RC, Bonanno JB, Sonenberg N, Burley SK. 1999. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98: 835–845. 10.1016/S0092-8674(00)81517-2 [DOI] [PubMed] [Google Scholar]
- Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, et al. 2011. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol 18: 1211–1217. 10.1038/nsmb.2149 [DOI] [PubMed] [Google Scholar]
- Feral C, Guellaen G, Pawlak A. 2001. Human testis expresses a specific poly(A)-binding protein. Nucleic Acids Res 29: 1872–1883. 10.1093/nar/29.9.1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Föcking M, Dicker P, Lopez LM, Hryniewiecka M, Wynne K, English JA, Cagney G, Cotter DR. 2016. Proteomic analysis of the postsynaptic density implicates synaptic function and energy pathways in bipolar disorder. Transl Psychiatry 6: e959. 10.1038/tp.2016.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca BD, Zakaria C, Jia JJ, Graber TE, Svitkin Y, Tahmasebi S, Healy D, Hoang HD, Jensen JM, Diao IT, et al. 2015. La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J Biol Chem 290: 15996–16020. 10.1074/jbc.M114.621730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Lee JY, Mayeur GL, Bushell M, Doudna JA, Hershey JW. 2004. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J Biol Chem 279: 8946–8956. 10.1074/jbc.M312745200 [DOI] [PubMed] [Google Scholar]
- Fusco CM, Desch K, Dörrbaum AR, Wang M, Staab A, Chan ICW, Vail E, Villeri V, Langer JD, Schuman EM. 2021. Neuronal ribosomes exhibit dynamic and context-dependent exchange of ribosomal proteins. Nat Commun 12: 6127. 10.1038/s41467-021-26365-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 5: 2108–2116. 10.1101/gad.5.11.2108 [DOI] [PubMed] [Google Scholar]
- Gandin V, Sikstrom K, Alain T, Morita M, McLaughlan S, Larsson O, Topisirovic I. 2014. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp 2014: 51455. 10.3791/51455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain PL, Lun A, Garcia Meixide C, Macnair W, Robinson MD. 2021. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res 10: 979. 10.12688/f1000research.73600.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B, Gray NK. 2004. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief Funct Genomic Proteomic 3: 125–141. 10.1093/bfgp/3.2.125 [DOI] [PubMed] [Google Scholar]
- Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC. 1983. New initiation factor activity required for globin mRNA translation. J Biol Chem 258: 5804–5810. 10.1016/S0021-9258(20)81965-6 [DOI] [PubMed] [Google Scholar]
- Grünert S, Jackson RJ. 1994. The immediate downstream codon strongly influences the efficiency of utilization of eukaryotic translation initiation codons. EMBO J 13: 3618–3630. 10.1002/j.1460-2075.1994.tb06669.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2013. The Genotype–Tissue Expression (GTEx) Project. Nat Genet 45: 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Yeo GS, Tung YC, Zivraj KH, Willis D, Coppola G, Lam BY, Twiss JL, Holt CE, Fawcett JW. 2011. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA 17: 85–98. 10.1261/rna.2386111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner AS, Donlin-Asp PG, Leitch B, Herzog E, Schuman EM. 2019. Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science 364: eaau3644. 10.1126/science.aau3644 [DOI] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. 2021. Integrated analysis of multimodal single-cell data. Cell 184: 3573–3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse R, Hurtado ML, Jackson RJ, Eaton SL, Herrmann AG, Colom-Cadena M, Tzioras M, King D, Rose J, Tulloch J, et al. 2019. Comparative profiling of the synaptic proteome from Alzheimer's disease patients with focus on the APOE genotype. Acta Neuropathol Commun 7: 214. 10.1186/s40478-019-0847-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem 274: 16677–16680. 10.1074/jbc.274.24.16677 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Kuzuoğlu-Öztürk D, Braun JE, Eulalio A, Wohlbold L, Izaurralde E. 2013. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res 41: 978–994. 10.1093/nar/gks1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Gradi A, Sonenberg N. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 17: 7480–7489. 10.1093/emboj/17.24.7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessa S, Blanchet-Cohen A, Krug B, Vladoiu M, Coutelier M, Faury D, Poreau B, De Jay N, Hébert S, Monlong J, et al. 2019. Stalled developmental programs at the root of pediatric brain tumors. Nat Genet 51: 1702–1713. 10.1038/s41588-019-0531-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16: 421–433. 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19: 104–113. 10.1101/gad.1262905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajjo S, Sharma S, Chen S, Brothers WR, Cott M, Hasaj B, Jovanovic P, Larsson O, Fabian MR. 2022. PABP prevents the untimely decay of select mRNA populations in human cells. EMBO J 41: e108650. 10.15252/embj.2021108650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan BB, Gioio AE, Hillefors M, Aschrafi A. 2009. Axonal protein synthesis and the regulation of local mitochondrial function. Results Probl Cell Differ 48: 225–242. 10.1007/400_2009_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Kahvejian A, De Crescenzo G, Roy G, Svitkin YV, Imataka H, O'Connor-McCourt M, Sonenberg N. 2001a. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol 21: 5200–5213. 10.1128/MCB.21.15.5200-5213.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. 2001b. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 7: 205–216. 10.1016/S1097-2765(01)00168-X [DOI] [PubMed] [Google Scholar]
- Khanam T, Muddashetty RS, Kahvejian A, Sonenberg N, Brosius J. 2006. Poly(A)-binding protein binds to A-rich sequences via RNA-binding domains 1+2 and 3+4. RNA Biol 3: 170–177. 10.4161/rna.3.4.4075 [DOI] [PubMed] [Google Scholar]
- Khazaei S, Chen CCL, Andrade AF, Kabir N, Azarafshar P, Morcos SM, Franca JA, Lopes M, Lund PJ, Danieau G, et al. 2023. Single substitution in H3.3G34 alters DNMT3A recruitment to cause progressive neurodegeneration. Cell 186: 1162–1178 e1120. 10.1016/j.cell.2023.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoutorsky A, Yanagiya A, Gkogkas CG, Fabian MR, Prager-Khoutorsky M, Cao R, Gamache K, Bouthiette F, Parsyan A, Sorge RE, et al. 2013. Control of synaptic plasticity and memory via suppression of poly(A)-binding protein. Neuron 78: 298–311. 10.1016/j.neuron.2013.02.025 [DOI] [PubMed] [Google Scholar]
- Kühn U, Pieler T. 1996. Xenopus poly(A) binding protein: functional domains in RNA binding and protein – protein interaction. J Mol Biol 256: 20–30. 10.1006/jmbi.1996.0065 [DOI] [PubMed] [Google Scholar]
- LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE. 2006. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem 281: 22917–22932. 10.1074/jbc.M605418200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Cui Z, Niu Y, Liu B, Fan W, Yu D, Deng J. 2010. Synaptogenesis in the developing mouse visual cortex. Brain Res Bull 81: 107–113. 10.1016/j.brainresbull.2009.08.028 [DOI] [PubMed] [Google Scholar]
- Li C, Liu B, Kang B, Liu Z, Liu Y, Chen C, Ren X, Zhang Z. 2020. Scibet as a portable and fast single cell type identifier. Nat Commun 11: 1818. 10.1038/s41467-020-15523-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignetti JA, Brosius J. 1993. BC200 RNA: a neural RNA polymerase III product encoded by a monomeric Alu element. Proc Natl Acad Sci 90: 11563–11567. 10.1073/pnas.90.24.11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignetti JA, Brosius J. 1995. BC1 RNA: transcriptional analysis of a neural cell-specific RNA polymerase III transcript. Mol Cell Biol 15: 1642–1650. 10.1128/MCB.15.3.1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J. 2020. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv 10.48550/arXiv.1802.03426 [DOI] [Google Scholar]
- Middleton SA, Eberwine J, Kim J. 2019. Comprehensive catalog of dendritically localized mRNA isoforms from sub-cellular sequencing of single mouse neurons. BMC Biol 17: 5. 10.1186/s12915-019-0630-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty R, Khanam T, Kondrashov A, Bundman M, Iacoangeli A, Kremerskothen J, Duning K, Barnekow A, Hüttenhofer A, Tiedge H, et al. 2002. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J Mol Biol 321: 433–445. 10.1016/S0022-2836(02)00655-1 [DOI] [PubMed] [Google Scholar]
- Mueller JL, Skaletsky H, Brown LG, Zaghlul S, Rock S, Graves T, Auger K, Warren WC, Wilson RK, Page DC. 2013. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat Genet 45: 1083–1087. 10.1038/ng.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov IA, Banker G, Brosius J, Tiedge H. 1998. Activity-dependent regulation of dendritic BC1 RNA in hippocampal neurons in culture. J Cell Biol 141: 1601–1611. 10.1083/jcb.141.7.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okochi K, Suzuki T, Inoue J, Matsuda S, Yamamoto T. 2005. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes Cells 10: 151–163. 10.1111/j.1365-2443.2005.00826.x [DOI] [PubMed] [Google Scholar]
- Pelletier J, Graff J, Ruggero D, Sonenberg N. 2015. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res 75: 250–263. 10.1158/0008-5472.CAN-14-2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JD, Dieck ST, Alvarez-Castelao B, Tushev G, Chan IC, Schuman EM. 2021. Subcellular sequencing of single neurons reveals the dendritic transcriptome of GABAergic interneurons. Elife 10: e63092. 10.7554/eLife.63092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Chen T, Koga K, Guo YY, Xu H, Song Q, Wang JJ, Descalzi G, Kaang BK, Luo JH, et al. 2013. An increase in synaptic NMDA receptors in the insular cortex contributes to neuropathic pain. Sci Signal 6: ra34. 10.1126/scisignal.2003778 [DOI] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. 2019. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47: W191–W198. 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Chan ET, Pena Castillo L, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR. 2009. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol 27: 667–670. 10.1038/nbt.1550 [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Ha KCH, Nie K, Zheng H, Hughes TR, Morris QD. 2017. RNAcompete methodology and application to determine sequence preferences of unconventional RNA-binding proteins. Methods 118-119: 3–15. 10.1016/j.ymeth.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robeck T, Skryabin BV, Rozhdestvensky TS, Skryabin AB, Brosius J. 2016. BC1 RNA motifs required for dendritic transport in vivo. Sci Rep 6: 28300. 10.1038/srep28300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy G, De Crescenzo G, Khaleghpour K, Kahvejian A, O'Connor-McCourt M, Sonenberg N. 2002. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol Cell Biol 22: 3769–3782. 10.1128/MCB.22.11.3769-3782.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Kopylov AM, Brosius J, Hüttenhofer A. 2001. Neuronal BC1 RNA structure: evolutionary conversion of a tRNAAla domain into an extended stem–loop structure. RNA 7: 722–730. 10.1017/S1355838201002485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaee N, Kozlov G, Noronha AM, Xie J, Wilds CJ, Gehring K. 2012. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol Cell 48: 375–386. 10.1016/j.molcel.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Sawazaki R, Imai S, Yokogawa M, Hosoda N, Hoshino SI, Mio M, Mio K, Shimada I, Osawa M. 2018. Characterization of the multimeric structure of poly(A)-binding protein on a poly(A) tail. Sci Rep 8: 1455. 10.1038/s41598-018-19659-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer IB, Yamashita M, Schuller JM, Schüssler S, Reichelt P, Strauss M, Conti E. 2019. Molecular basis for poly(A) RNP architecture and recognition by the Pan2-Pan3 deadenylase. Cell 177: 1619–1631 e1621. 10.1016/j.cell.2019.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seli E, Lalioti MD, Flaherty SM, Sakkas D, Terzi N, Steitz JA. 2005. An embryonic poly(A)-binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc Natl Acad Sci 102: 367–372. 10.1073/pnas.0408378102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Lee J, Kim Y, Jang S, Lee Y, Kim S, Lee Y. 2017. Knockdown of BC200 RNA expression reduces cell migration and invasion by destabilizing mRNA for calcium-binding protein S100A11. RNA Biol 14: 1418–1430. 10.1080/15476286.2017.1297913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Srivastava A, Madad S, Lareau CA, Satija R. 2021. Single-cell chromatin state analysis with signac. Nat Methods 18: 1333–1341. 10.1038/s41592-021-01282-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Bose P, Leong-Quong RY, Fujita DJ, Riabowol K. 2010. REAP: a two minute cell fractionation method. BMC Res Notes 3: 294. 10.1186/1756-0500-3-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Agol VI. 1978. Complete translation of encephalomyocarditis virus RNA and faithful cleavage of virus-specific proteins in a cell-free system from Krebs-2 cells. FEBS Lett 87: 7–11. 10.1016/0014-5793(78)80121-5 [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Sonenberg N. 2004. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol Biol 257: 155–170. 10.1385/1-59259-750-5:155 [DOI] [PubMed] [Google Scholar]
- Tan Y, Cahan P. 2019. Singlecellnet: a computational tool to classify single cell RNA-seq data across platforms and across species. Cell Syst 9: 207–213.e2. 10.1016/j.cels.2019.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ Jr, Sachs AB. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev 9: 2997–3007. 10.1101/gad.9.23.2997 [DOI] [PubMed] [Google Scholar]
- Tarun SZ Jr, Wells SE, Deardorff JA, Sachs AB. 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci 94: 9046–9051. 10.1073/pnas.94.17.9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP. 2014. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev 28: 357–371. 10.1101/gad.231407.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge H, Fremeau RT Jr, Weinstock PH, Arancio O, Brosius J. 1991. Dendritic location of neural BC1 RNA. Proc Natl Acad Sci 88: 2093–2097. 10.1073/pnas.88.6.2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge H, Chen W, Brosius J. 1993. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci 13: 2382–2390. 10.1523/JNEUROSCI.13-06-02382.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Ongkasuwan J, Standart N, Steitz JA. 2001. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev 15: 774–788. 10.1101/gad.872201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Imataka H, Sonenberg N. 2000. Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr Biol 10: 1147–1150. 10.1016/S0960-9822(00)00701-6 [DOI] [PubMed] [Google Scholar]
- Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sonenberg N, Lomakin IB, Tiedge H. 2002. Dendritic BC1 RNA: functional role in regulation of translation initiation. J Neurosci 22: 10232–10241. 10.1523/JNEUROSCI.22-23-10232.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Liu J, Shen L, Tonti-Filippini J, Zhu Y, Jia H, Lister R, Whitaker JW, Ecker JR, Millar AH, et al. 2013. STAR: an integrated solution to management and visualization of sequencing data. Bioinformatics 29: 3204–3210. 10.1093/bioinformatics/btt558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lv X, Wu Y, Xu T, Jiao M, Yang R, Li X, Chen M, Yan Y, Chen C, et al. 2018. Postsynaptic RIM1 modulates synaptic function by facilitating membrane delivery of recycling NMDARs in hippocampal neurons. Nat Commun 9: 2267. 10.1038/s41467-018-04672-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CC, Balasta ML, Ren J, Goss DJ. 1998. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 37: 1910–1916. 10.1021/bi9724570 [DOI] [PubMed] [Google Scholar]
- Xiang K, Bartel DP. 2021. The molecular basis of coupling between poly(A)-tail length and translational efficiency. Elife 10: e66493. 10.7554/eLife.66493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Duckett CS, Lindsten T. 1995. iPABP, an inducible poly(A)-binding protein detected in activated human T cells. Mol Cell Biol 15: 6770–6776. 10.1128/MCB.15.12.6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Chiang YC, Zhang C, Lee DJ, Laue TM, Denis CL. 2007. PAB1 self-association precludes its binding to poly(A), thereby accelerating CCR4 deadenylation in vivo. Mol Cell Biol 27: 6243–6253. 10.1128/MCB.00734-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Liu H, Xie F, Fischer S, Adkins RS, Aldridge AI, Ament SA, Bartlett A, Behrens MM, Van den Berge K, et al. 2021. A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex. Nature 598: 103–110. 10.1038/s41586-021-03500-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Park J, Ha M, Lim J, Chang H, Kim VN. 2018. PABP cooperates with the CCR4–NOT complex to promote mRNA deadenylation and block precocious decay. Mol Cell 70: 1081–1088.e5. 10.1016/j.molcel.2018.05.009 [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. 2003. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112: 317–327. 10.1016/S0092-8674(03)00079-5 [DOI] [PubMed] [Google Scholar]
- Zhong J, Chuang SC, Bianchi R, Zhao W, Lee H, Fenton AA, Wong RK, Tiedge H. 2009. BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J Neurosci 29: 9977–9986. 10.1523/JNEUROSCI.3893-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.