Abstract
Polydopamine-assisted modification for bone substitute materials has recently shown great application potential in bone tissue engineering due to its excellent biocompatibility and adhesive properties. A scaffold material’s impact on osteoclasts is equally as important as its impact on osteoblasts when considering tissue engineering for bone defect repair, as healthy bone regeneration requires an orchestrated coupling between osteoclasts and osteoblasts. How polydopamine-functionalized bone substitute materials modulate the activity of osteoblast lineage cells has been extensively investigated, but much less is known about their impact on osteoclasts. Moreover, most of the polydopamine-functionalized materials would need to additionally load a biomolecule to exert the modulation on osteoclast activity. Herein, we demonstrated that our biomimetic polydopamine-laced hydroxyapatite collagen (PDHC) scaffold material, which does not need to load additional bioactive agent, is sufficiently able to modulate osteoclast activity in vitro. First, PDHC showed an anti-resorptive potential, characterized by decreased osteoclast differentiation and resorption capacity and changes in osteoclasts’ transcriptome profile. Next, cAMP response element-binding protein (CREB) activity was found to mediate PDHC’s anti-osteoclastogenic effect. Finally, although PDHC altered clastokines expression pattern of osteoclasts, as revealed by transcriptomic and secretomic analysis, osteoclasts’ coupling to osteoblasts was not compromised by PDHC. Collectively, this study demonstrated the PDHC material orients osteoclast behavior to an anti-resorptive pattern without compromising osteoclasts’ coupling to osteoblasts. Such a feature is favorable for the net increase of bone mass, which endows the PDHC material with great application potential in preclinical/clinical bone defect repair.
Keywords: polydopamine, osteoclasts, cell-material interaction, osteoclast-osteoblast coupling, bone defect repair
Graphical Abstract
Introduction
Critical sized bone defects that resulted from trauma, infection, or tumor, often require a bone graft to restore bone integrity and function. Innovative bone regenerative strategies through tissue engineering have garnered much attention, which utilize 3D scaffolds in combination with cells and bioactive molecules to create an “osteogenic” substitute.1 Healthy bone regeneration in bone defect repair requires an orchestrated cellular activity within the bone remodeling microenvironment. This involves osteoblasts (OBs) that form the osteoid matrix and osteoclasts (OCs) that resorb damaged bone. The interaction between bone cells and scaffold materials is critical for the optimization of bone regeneration. A great deal of work has focused on how bone substitute materials mediate the activity of osteoblastic lineage cells (OBs, mesenchymal stem cells (MSCs), etc.), but whether/how OC activity is affected by these materials is often ignored. While OCs are responsible for bone resorption, they release anabolic messages to OBs via secreted cytokines or cell-cell contact that promote OB proliferation, differentiation, and mineralization.2 This OC-derived bone formation mechanism called “coupling” is worth noting in modern development of bone substitutes.3 A material’s impact on OCs is equally as important as its effect on OBs when developing bone substitute materials for bone defect repair.
Polydopamine (PDA) has recently shown great potential in the development of bone substitutes mainly due to its biocompatibility and adhesive property.4, 5 PDA coating or cross-linkage have been demonstrated to exert anabolic effects on osteoblastic lineage cells. For example, PDA coating on TiO2 nanotubes enhances a set of activities of MC3T3-E1 pre-osteoblasts, including cell spreading, formation of focal adhesions, cell viability and proliferation.6 PDA film deposited on nanoporous titanium promotes both adhesion and proliferation of osteoblastic MG-63 cells.7 Osteogenic differentiation of MG-63 cells or MSC is enhanced by PDA coating on various bone substitutes.8, 9 Our lab recently developed a biomimetic polydopamine-laced hydroxyapatite collagen (PDHC) calcium silicate material.10–12 Through applying PDA cross-linkage to strengthen mechanical property, the material also enhanced MSC proliferation and mineralization in vitro and increased new bone formation in a calvaria critical size defect model.11
In contrast, how PDA-functionalized bone substitutes affect the behavior of OCs remains largely unknown. Moreover, most of existing PDA-functionalized materials would need to load a biomolecule (aspirin, adenosine, lactoferrin, etc.) to exert the modulation on OC activity and thus their effects actually result from the immobilized biomolecule rather than PDA functionalization.13–15 In this study, we showed that the PDHC scaffold material, which does not need to load additional bioactive agent, is enough able to modulate OC activity in vitro. Our results will form a rationale for preclinical/clinical application of PDHC material in bone defect repair.
Materials and Methods
Fabrication of PDHC disk
The fabrication of PDHC disks is performed according to the method described previously.11, 16, 17 In brief, hydroxyapatite and collagen (HC) slurry was biomimetically synthesized by a co-precipitation method using in situ hybridization of calcium silicates with HC powder.18 The HC powder, dopamine, and calcium hydroxide were mixed and cross-linked with enTMOS (bis [3-(trimethoxysilyl)-propyl] ethylenediamine) and APS on a cold stage. Then the mixture was transferred into disk molds, which were 3D-printed using a Form 2 3D printer (Formlabs) to fit 24-, 96-well tissue culture plates, for solidification and pressed with glass to smooth the surface. After air-dried and oven-dried for 7 days, the product—PDHC disks were sterilized with cold ethylene oxide gas for future cell experiment use.
OC differentiation, identification, and conditioned medium collection
RAW 264.7 (ATCC® TIB-71) cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% FBS (Gibco) and penicillin/streptomycin at 37℃, 5% CO2. Osteoclastogenic induction was performed as previously described:19 RAW cells were plated at a density of 4 × 104 cells/cm2 and treated with receptor activator of nuclear factor kappa-Β ligand (RANKL) (10 ng/mL, R&D systems) for 5d to fully differentiated to OCs. Medium was replaced every two days. For conditioned medium collection, RAW cells were plated on 24-well tissue culture plates at a density of 4 × 104 cells/cm2 and treated with 10 ng/mL RANKL. Medium (500 µl for each well) was replaced every two days. On day 5, conditioned medium from mature OCs was collected and concentrated 100-fold using 10 kDa pore size Amicon® Ultra-4 Centrifugal Filter Unit (Millipore Sigma). The concentrated OC conditioned medium was aliquoted and stored in −80℃. Upon use, it was defrosted and added to the medium for treating OBs at 1:50 ratio.
For OC identification,20 the differentiated OCs were fixed with 4% paraformaldehyde, permeabilized with Triton X-100, and stained with tartrate-resistant acid phosphatase (TRAP) staining solution. Nikon Eclipse Ti-U inverted microscope (Nikon) was used for imaging and TRAP positive, multinucleated (≥3 nuclei) cells were counted as OC.
Osteoclastic resorption capacity
For detecting resorption capacity, RAW cells were plated on bone slices (BioVendor) or PDHC disks. At indicated time points, bone slices or PDHC disks were sonicated 10 minutes for removing cells and then stained with 1% toluidine blue for 4 minutes.21 Resorption pits were visualized by Nikon SMZ18 stereo microscope (Nikon) and analyzed by ImageJ software (NIH). Additionally, since both bone slice and PDHC are composed of hydroxyapatite—a calcium phosphate, calcium can be released to the conditioned medium as soluble ions upon effective osteoclastic resorption. Thus, we also detected the calcium ions concentration in the conditioned medium using a colorimetric calcium assay kit (Abcam, #ab102505), to reflect OC resorption activity.
Cell viability assay
For PDHC cytotoxicity assessment, RAW cells were lifted from the material surface using trypsin (Gibco) and then stained with trypan blue (Sigma-Aldrich) for living cell counting, compared to tissue culture plate group. MSC and OB viability was evaluated using tetrazolium dye MTS cell viability assay kit (Promega) following the manufacturer’s instructions. The absorbance at 490 nm was measured using the Cytation 5 imaging reader (BioTek).
RT-qPCR
Total RNA was extracted using RNAzol (Molecular Research Center) and cDNA reverse-transcription was performed using iScript™ cDNA Synthesis Kit (Bio-Rad). RT-qPCR was performed on StepOnePlus Real-time PCR system (Applied Biosystems) using the iTaq™ Universal SYBR Green Supermix reagent (Bio-Rad). ΔΔCT method was used to calculate the relative expression of target genes, which was normalized to housekeeping gene β−2-microglobulin (B2m). Primer information is listed in Supplementary Table 1.
mRNA-seq and bioinformatic analysis
Total RNA was extracted using RNeasy Mini Kit (Qiagen). A cDNA library was prepared using NEBNext Ultra DNA library prep kit (NEB) and mRNA-seq was performed on NovaSeq 6000 system (Illumina). RNAseq data of 4 samples were collected: 2 replicates in 2 sample groups (OCdisk group and OC group). Reads were mapped to Mus musculus mm10 genome by STAR software. Fragments Per Kilobase of transcript per Million mapped reads (FPKM) was calculated to estimate gene expression levels. Differential expression (DE) analysis was performed using the DESeq2 package in R software (version 2_1.6.3), to identify the significant DE genes in the comparison of OCdisk group vs. OC group. P values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate (FDR). Gene set enrichment tests were performed using the clusterProfiler package and the “camera” function in limma package in R software, to test gene sets in MsigDB collection including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG).
Western blot
Total protein was extracted by RIPA buffer plus phosphatase & protease inhibitor cocktail. The total protein was separated using XCell SureLock Mini-Cell (Invitrogen) sodium dodecyl sulfate-polyacrylamide gel electrophoresis system and transferred onto a nitrocellulose membrane by using Trans-Blot Cell system (Bio-Rad). After blocking with 5% nonfat milk, membrane was incubated with primary antibodies cAMP response element-binding protein (CREB) (Cell Signaling, #9197), phospho-CREB (Ser133) (Cell Signaling, #9198), PKA-Cα (Cell Signaling, #4782), β-Actin (Santa Cruz, #sc-47778), and corresponding peroxidase-conjugated secondary antibodies (Merck Millipore). Protein bands was visualized by ImageQuant LAS 4000 chemiluminescence camera system (GE Healthcare) and analyzed by ImageJ software (NIH).
MSCs isolation and osteogenic induction
Bone marrow-derived MSCs were obtained from 8-week-old male C57BL/6J mice, according to a protocol (No. 2020A00000064) approved by the Institutional Animal Care and Use Committee at The Ohio State University. Briefly, the femurs were removed after animal euthanasia. Bone marrow was flushed out and maintained in α-MEM (Gibco) supplemented with 10% FBS and penicillin/streptomycin at 37℃, 5% CO2. Non-adherent cells were removed after 24h and adherent cells were kept for continued culture. Growth medium was replaced every 3 days. The cells at passages 3–5 were verified by flow cytometry (positive for CD29/CD44/CD105 and negative for CD34/CD45) and used in subsequent experiments. MSCs received osteogenic medium (OM), which is complete growth medium supplemented with 10mM β-glycerophosphate, 0.2mM ascorbic acid and 10−7M dexamethasone, or OM plus specific conditioned medium for osteogenic differentiation. At indicated time points, the cells were harvested for RT-qPCR detection of osteogenic genes expression. At day 28, the cells were fixed with 70% cold ethanol and stained with 1% Alizarin Red solution (Sigma-Aldrich) for 10 min, for mineral nodule visualization. Then we extracted the stained samples using 1% (w/v) cetylpyridinium chloride solution (Sigma-Aldrich) and read the absorbance at 562 nm as quantification result.
Transwell cell migration assay
8µm pore polycarbonate membrane transwell system (Corning) was used. Before the cell migration assay, the cells were starved for 18–20h in serum-free medium.22, 23 1 X 104 MSCs or OBs in 0.2% FBS medium were seeded on the upper chamber. The lower chamber was filled with complete growth medium or conditioned medium as indicated. After 16h incubation, the cells on the upper surface of the membrane were removed and then the membranes were fixed and stained with DAPI. The number of cells migrated through the membrane was counted in 5 random areas under Nikon Eclipse Ti-U inverted fluorescence microscope (Nikon).
Antibody array
Levels of clastokines in OC conditioned medium was detected by a customized RayBio C-Series antibody array (RayBio). Total protein concentration was determined by BCA protein assay kit (Sigma-Aldrich) and samples were diluted with blocking buffer (RayBio) to final concentration of 250 μg. The antibody array membranes were incubated with sample after blocking. After washing, the membranes were incubated with biotinylated antibody cocktail. After washing, the membranes were then incubated with HRP-streptavidin. After washing, the membranes were sent for chemiluminescence detection using ImageQuant LAS 4000 camera system (GE Healthcare). Relative protein levels were analyzed using ImageJ software (NIH).
Statistical analyses
Experiments were performed in triplicate and repeated three times. Data was presented as mean ± standard error of the mean (SEM). Statistical analyses were performed using GraphPad Prism 5 (GraphPad Software), except for bioinformatic analyses. The comparisons between two groups were assessed using Student’s t test while the comparisons among multiple groups were assessed using one-way ANOVA followed by Bonferroni post-hoc test. P < 0.05 was considered significant.
Results
PDHC attenuates OC differentiation and resorption capacity
We first checked PDHC cytotoxicity on RAW 264.7 cells, a cell line commonly used as OC precursor,24 by live cell counting after trypan blue staining. As Figure 1A shows, at each time points (2d, 4d, 6d), the number of living RAW cells that grew on PDHC disks (RAWdisk group) were similar to that on tissue culture plates (RAW group). Inspired by the fact that dopamine can suppress RANKL-induced OC differentiation 19, 25, 26, we investigated whether PDHC material has similar potential. After 5 days of RANKL stimulation, the number of OCs formed on PDHC disk (OCdisk group) was much less compared to tissue culture plate (OC group) (Figure 1B and 1C). Moreover, the OCs are also smaller in size and had fewer nuclei number (Figure 1B and 1C). Decreased expression of OC specific genes (c-fos, Nfatc1, Trap, Ctsk) was also detected in OCdisk group (Figure 1D). Collectively, these data showed that OC differentiation was attenuated when cells grew on PDHC disks.
figure 1.
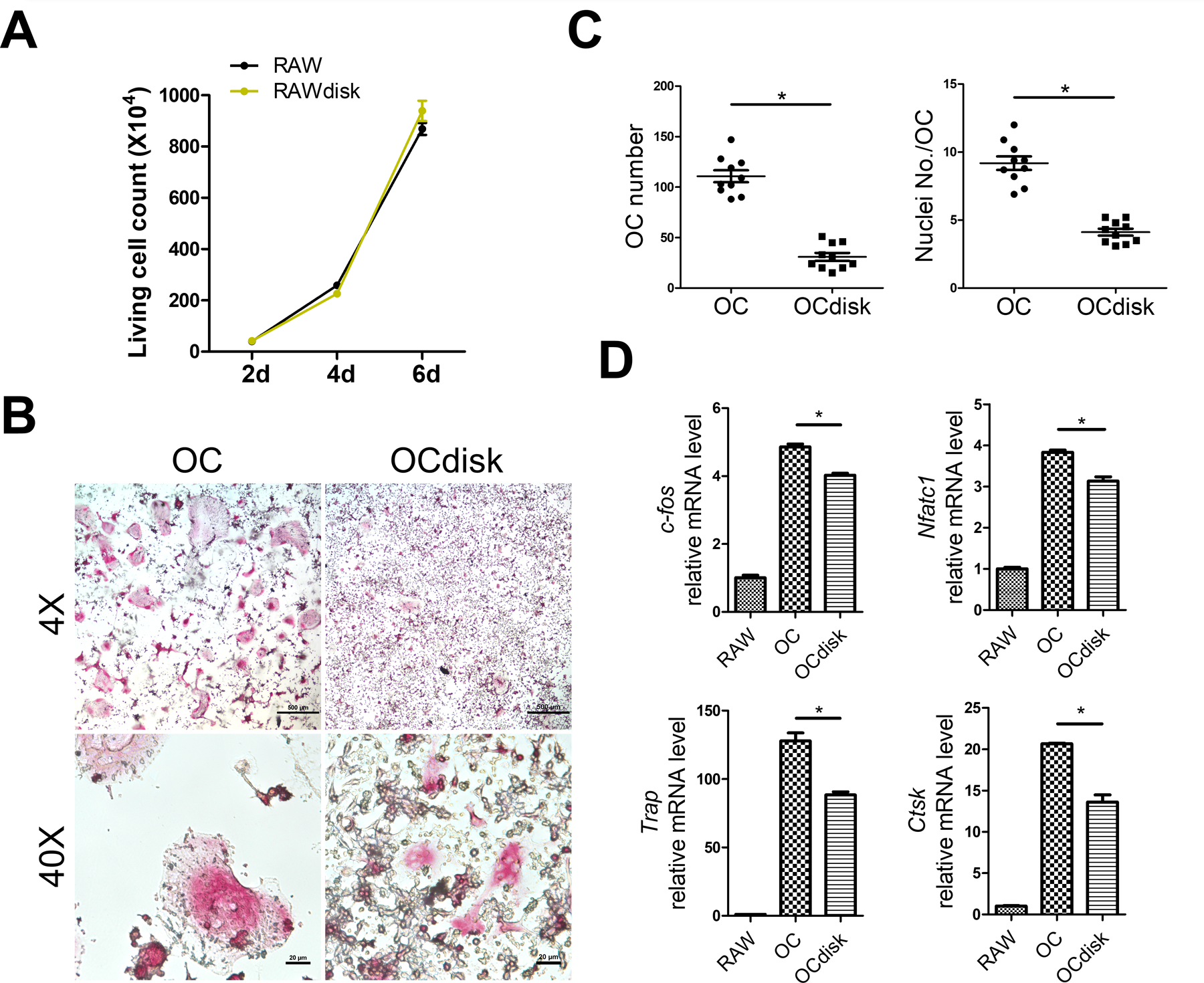
pdhc attenuates oc differentiation. (a) raw cells were seeded on tissue culture plate (raw group) or pdhc disk (rawdisk group). live cell counting was performed with trypan blue staining after lifting. for (b-d), raw cells were seeded on tissue culture plate (oc group) or pdhc disk (ocdisk group) and received 10 ng/ml rankl stimulation for 5 days to differentiate into ocs. (b) trap staining of ocs. (c) quantification of ocs: oc number and nuclei number per oc. trap positive, multinucleated (≥3 nuclei) cells were counted as oc. (d) relative expression of osteoclastic genes (c-fos, nfatc1, trap, ctsk) determined by rt-qpcr; normalized to b2m.
n = 3 for all experiments. *p < 0.05. data shown as mean ± sem.
In addition to OC differentiation, the resorption capacity of mature OCs was also suppressed when grew on PDHC disks (Figure 2). After 6 and 8 days of RANKL stimulation, RAW-cell-derived OCs generated apparent resorption pits on bone slices (OCbone group) but less resorption pits on PDHC disks (OCdisk group) (Figure 2A and 2B). In addition, since both bone slice and PDHC are composed of hydroxyapatite, which can be dissolved and release soluble calcium ions to the conditioned medium upon effective osteoclastic resorption, we further detected the calcium concentration in OC conditioned medium and observed decreased calcium level in OCdisk group (Figure 2C), which suggests an impaired OC resorption capacity.
figure 2.
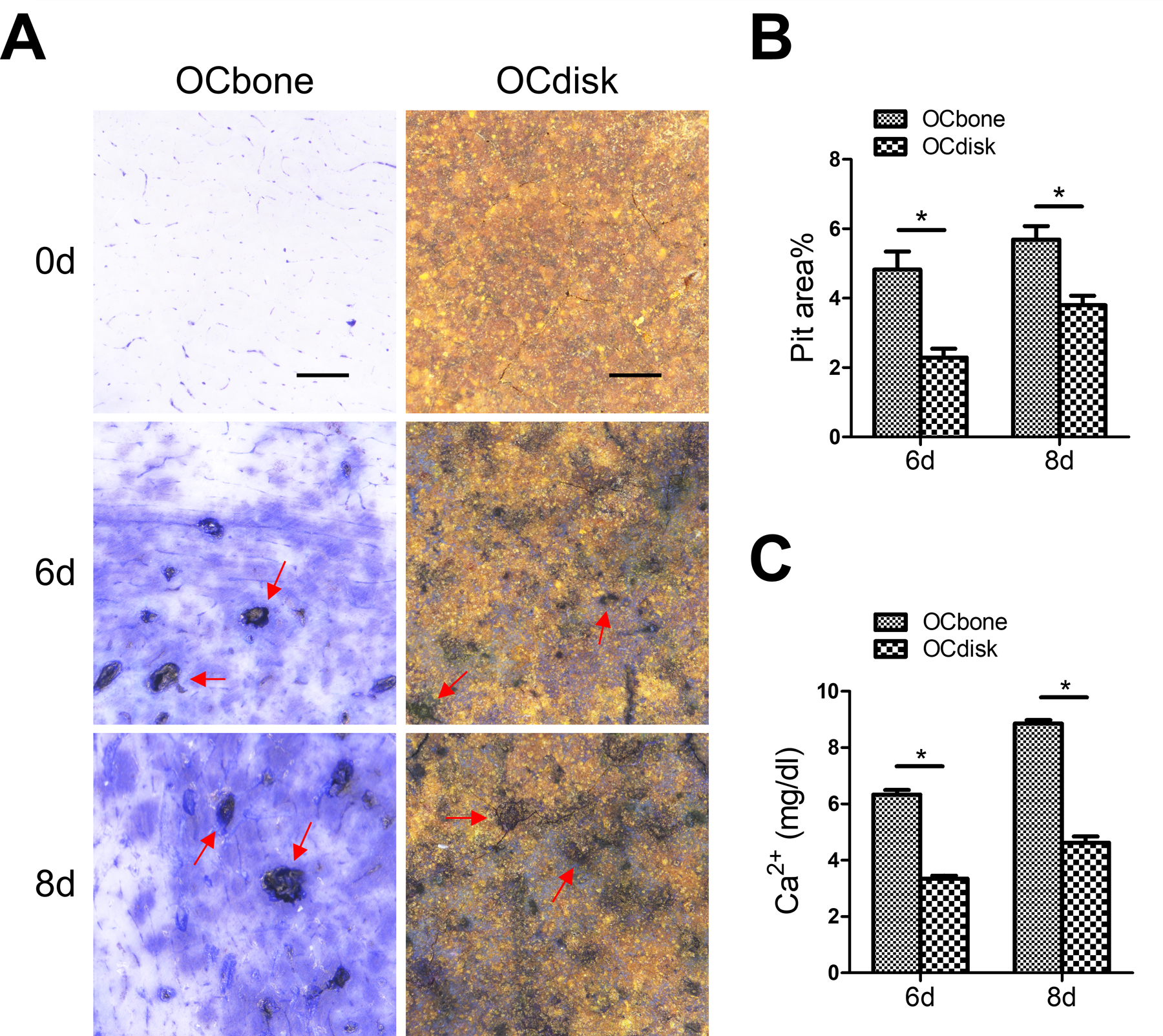
pdhc attenuates oc resorption capacity. raw cells were seeded on bone slices (ocbone group) or pdhc disks (ocdisk group) and received 10 ng/ml rankl stimulation to differentiated into mature ocs. (a) resorption pits (red arrow) were visualized by toluidine blue staining. scale bar is 1000 µm. (b) quantification of pit area. (c) calcium ions concentrations in oc conditioned medium.
n = 3 for all experiments. *p < 0.05. data shown as mean ± sem.
PDHC modifies OC transcriptome profile
To identify molecular changes that result from PDHC modification, we next investigated how PDHC changed the transcriptome profile of OCs. OCs differentiated on tissue culture plates (OC group) or on PDHC disks (OCdisk group) were harvested for mRNA-seq. mRNA-seq results showed that the transcriptome profile of PDHC-modified OCs differed from OCs (Figure 3A). 3520 genes in total were expressed differentially, according to the cut-off threshold padj < 0.05 (Figure 3B). Among them, 1776 upregulated genes and 1744 downregulated genes were identified in PDHC-modified OCs compared to OCs. Gene set enrichment test of GO (Figure 3C) and KEGG (Figure 3D) pathways showed notable enrichment of DE genes in PDHC-modified OCs. It is worth noting that “osteoclast differentiation” pathway is one of the top 20 enriched KEGG pathways (Figure 3D). On the basis of the results in Figure 1, this bioinformatic clue further suggests that affecting OC differentiation is a dominant impact of PDHC on OCs. The DE genes in “osteoclast differentiation” pathway in PDHC-modified OCs are visualized in KEGG map (Figure 3E).
figure 3.
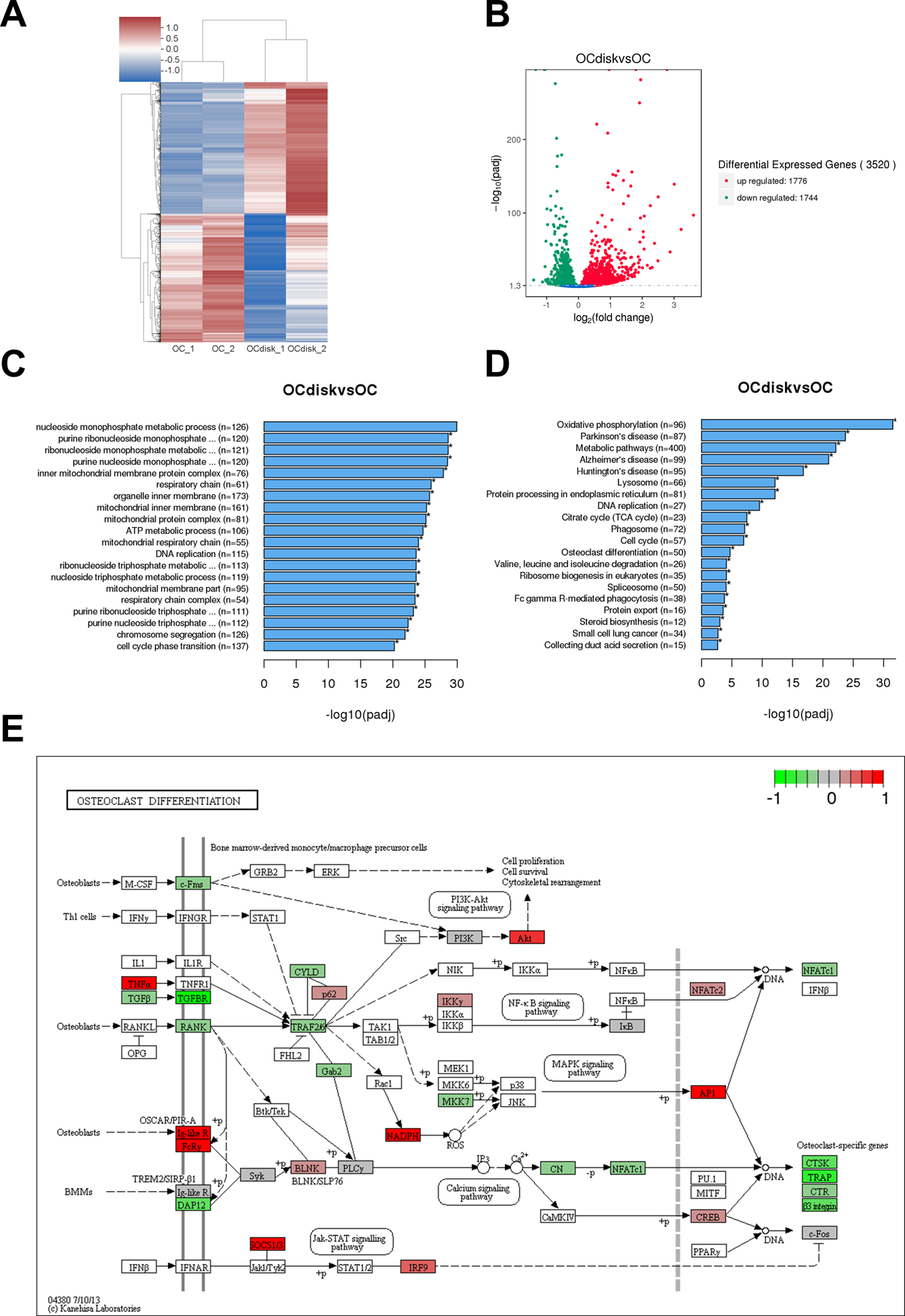
pdhc modifies oc transcriptome profile. raw cells were seeded on tissue culture plates (oc group) or pdhc disks (ocdisk group) and received 10 ng/ml rankl stimulation for 5 days to differentiate into ocs. upon differentiation, ocs were harvested for mrna-seq. (a) hierarchical clustering heatmap of differential expression (de) genes. color key represents z score. red represents high expression genes; blue represents low expression genes. (b) volcano plot of de genes in ocdisk vs. oc group. threshold: padj < 0.05. red dots represent upregulated de genes; green dots represent downregulated de genes. (c) the top 20 enriched go pathways. (d) the top 20 enriched kegg pathways. (e) kegg map (kanehisa laboratories) of de genes in “osteoclast differentiation” pathway in ocdisk vs. oc. color key represents log2fc.
CREB activity is involved in PDHC-attenuated OC differentiation
Gene set enrichment testing result indicated that several GO pathways, which are cAMP/PKA/CREB-relevant, showed significant enrichment in PDHC-modified OCs (Figure 4A), which arouses our interest in checking CREB activity. In addition, since modulation of cAMP/PKA/CREB activity is one of the molecular mechanisms by which dopamine suppresses OC differentiation, as reported in our previous work,19 we next investigated if CREB activity is involved in PDHC-modified OC differentiation. RAW cells were seeded on tissue culture plates (OC group) or PDHC disks (OCdisk group) and received RANKL stimulation for osteoclastogenesis induction. At different stages of osteoclastogenesis (24h and 48h after RANKL exposure), decreased levels of CREB phosphorylation and PKA expression were observed in OCdisk group (Figure 4B and 4C), which corresponds with a decrease in osteoclastic gene (c-fos, Nfatc1, Trap, Ctsk) expression (Figure 4D). Taken together, our data suggest that PDHC modulates CREB activity, which is a potential mechanism by which PDHC attenuates OC differentiation.
figure 4.
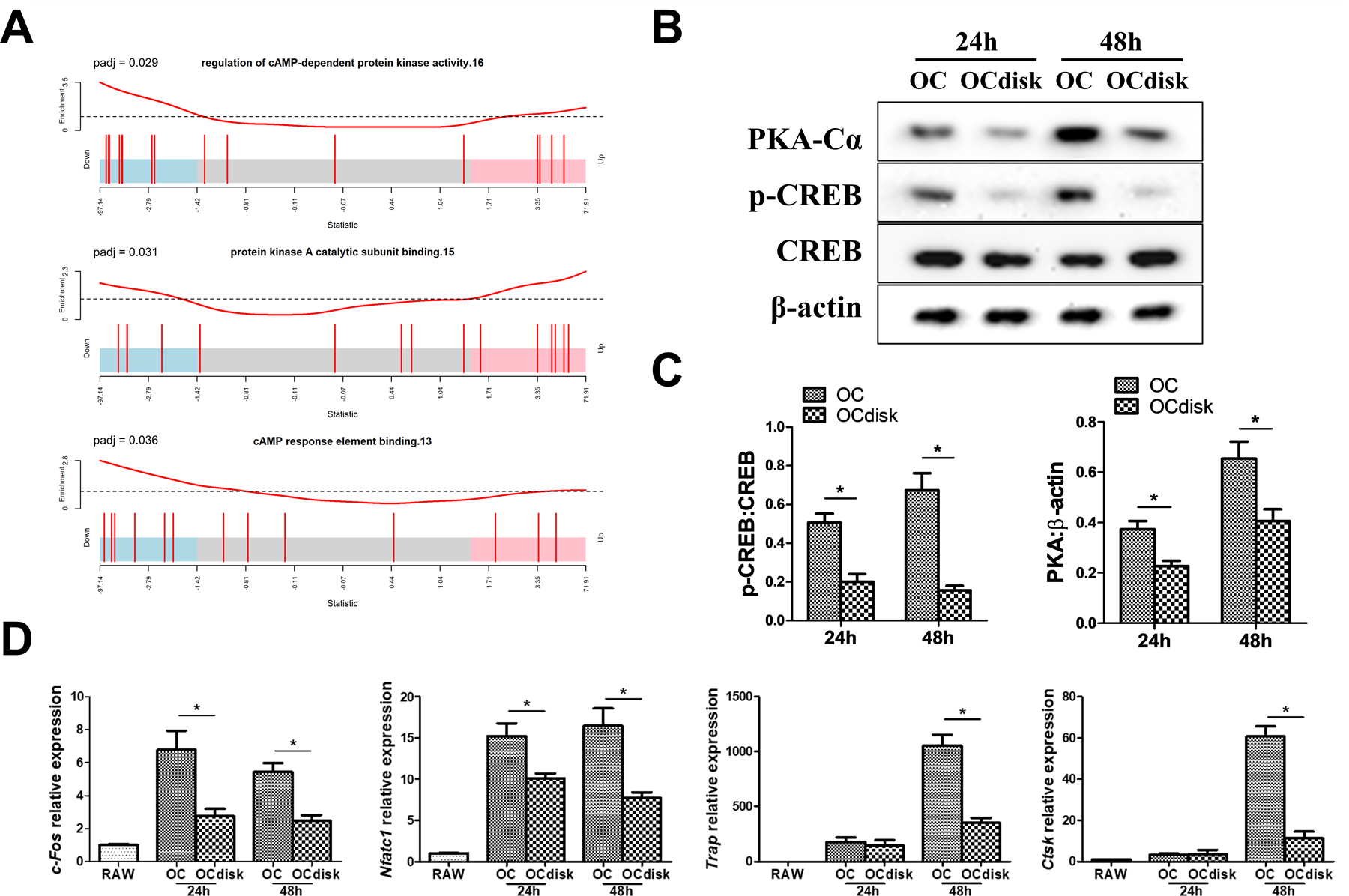
creb activity is involved in pdhc-attenuated oc differentiation. (a) gene set enrichment testing result of camp/pka/creb-relevant go pathways in pdhc-modified ocs (ocdisk group). for (b-d), raw cells were seeded on tissue culture plates (oc group) or pdhc disks (ocdisk group) and received 10 ng/ml rankl stimulation. cells were harvested at different time points during osteoclastogenesis. (b, c) western blot detection of p-creb/creb and pka. (d) relative expression of osteoclastic genes (c-fos, nfatc1, trap, ctsk) determined by rt-qpcr; normalized to b2m.
n = 3 for all experiments. *p < 0.05. data shown as mean ± sem.
PDHC does not compromise OC’s coupling to OBs
Based on the above results, PDHC exhibited significant anti-osteoclastogenic effects in vitro. Such an exciting anti-resorptive potential may be beneficial during bone defect regeneration. However, although OCs are responsible for bone resorption, they also stimulate osteoblastic bone formation through a “coupling” mechanism.3 Ideally, bone substitutes should be able to maintain the crosstalk between bone cells to promote a microenvironment appropriate for bone regeneration. Thus, we next investigated whether PDHC impairs OC’s coupling to OBs using conditioned medium system. Osteoprogenitors—bone marrow-derived MSCs or OBs (14d-osteogenically-differentiated MSCs) were treated with the conditioned medium harvested from PDHC disk alone (Disk group), OCs (OC group), or PDHC-modified OCs (OCdisk group). A control group without any treatment was also included (NT group). As Figure 5A shows, both OC’s and PDHC-modified OC’s conditioned medium enhanced MSC (on day 3 and 5) or OB (on day 5) viability, and there is no difference between OC’s and PDHC-modified OC’s conditioned medium treatment. In a Transwell cell migration system (Figure 5B), OC’s and PDHC-modified OC’s conditioned medium equally enhanced MSC or OB migration capacity.
figure 5.
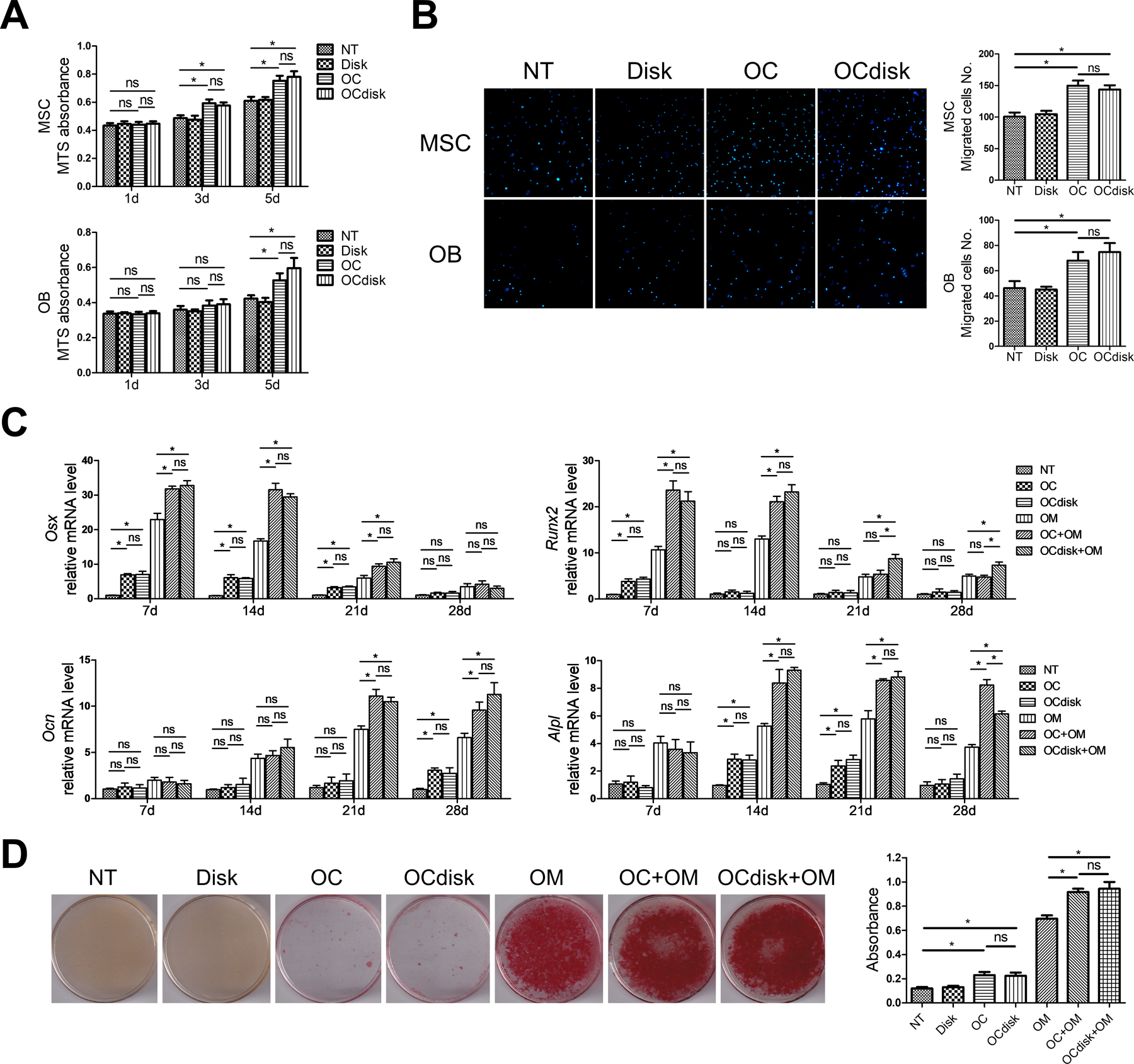
pdhc does not compromise oc’s coupling to obs. for (a, b), mscs and obs (14d-osteogenically-differentiated mscs) were treated with the conditioned medium harvested from pdhc disk alone (disk group), ocs (oc group), or pdhc-modified ocs (ocdisk group). a control group without any treatment was also included (nt group). (a) cell viability determined by mts absorbance. (b) dapi staining results of 16h transwell cell migration assay. for (c, d), indicated conditioned medium, with or without osteogenic medium (om), was used for osteogenic induction of mscs. (c) relative expression of osteogenic genes on day 7, 14, 21, and 28 determined by rt-qpcr; normalized to b2m. (d) mineral nodule visualization and quantification on day 28 by alizarin red staining.
n = 3 for all experiments. *p < 0.05, ns: p > 0.05. data shown as mean ± sem.
Next, OC’s (OC group) or PDHC-modified OC’s (OCdisk group) conditioned medium, either with or without osteogenic medium (OM), was used for osteogenic induction of MSCs. When in the absence of OM, while both OC’s and PDHC-modified OC’s conditioned medium showed very limited pro-osteogenic potential, no difference was found between these two (Figure 5C). When in the presence of OM, both OC’s and PDHC-modified OC’s conditioned medium enhanced Osx and Runx2 expression generally at early & medium time points, and enhanced Ocn and Alpl expression generally at medium & late time points (Figure 5C). In general, no difference was found between OC’s and PDHC-modified OC’s conditioned medium treatment, in spite of some exceptions (Runx2 on day 21 and 28, and Alpl on day 28). For mineralization capacity (Figure 5D), both OC’s and PDHC-modified OC’s conditioned medium equally increased mineral deposition in the presence of OM. When in the absence of OM, while both OC’s and PDHC-modified OC’s conditioned medium increased mineral deposition just a little, no difference was found between these two. Taken together, our data confirm the role of OCs in promoting osteoblastic bone formation and that PDHC does not impair OC’s coupling to OBs.
PDHC modifies OC clastokines expression pattern
We sought to further confirmed our finding that PDHC does not impair OC’s coupling to OBs through another experimental approach. OCs are known to modulate OB activity via a specific set of secretory proteins termed “clastokines”.27, 28 The OC conditioned medium used in our experiment system is expected to contain OC-secreted clastokines, which play a critical role in modulating OB activity. We first found PDHC altered the expression level of several known clastokine genes in our mRNA-seq data set (Figure 6A) and verified with RT-qPCR (Figure 6B). Apparently, the expression pattern of clastokine genes looks different in PDHC-modified OCs (OCdisk group) compared to OCs (OC group), characterized by some genes were upregulated while other genes were downregulated (Figure 6A and 6B). We next checked the secretome profile of these OCs. The levels of these clastokines in the conditioned medium harvested from OCs (OC group) and PDHC-modified OCs (OCdisk group) were detected by an antibody array. Generally consistent with the transcriptomic findings (Figure 6A and 6B), PDHC-modified OCs displayed a distinct clastokine expression pattern, characterized as C3a/Ctf1 upregulated while BMP6/Cthrc1 downregulated (Figure 6C and 6D). It is speculated that the actions of upregulated clastokines and downregulated clastokines on OBs get “neutralized” so the overall OC’s coupling effect has little changes, which potentially explains why PDHC did not alter OC’s coupling to OBs in spite of this modified clastokines expression pattern.
figure 6.
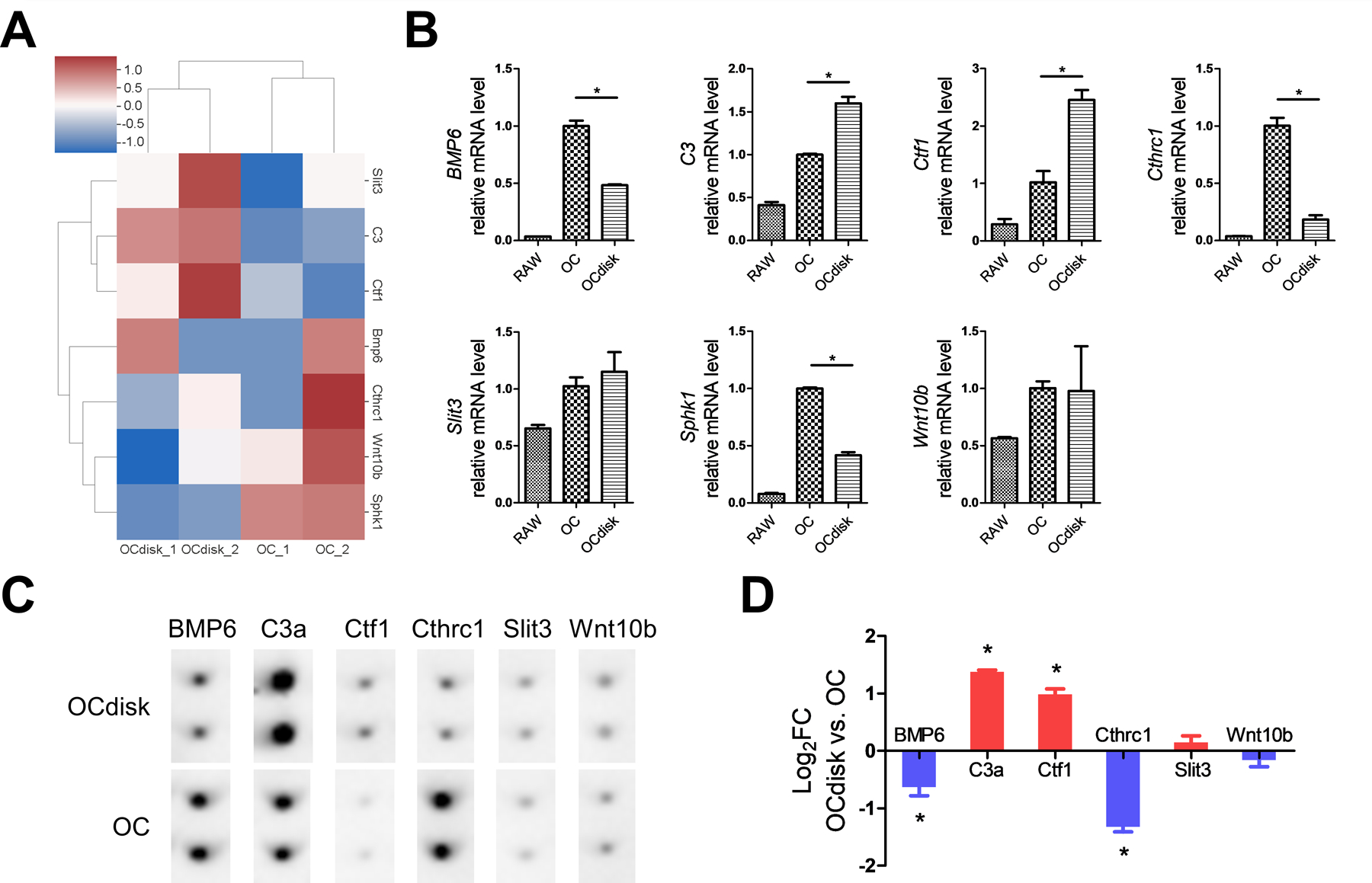
pdhc modifies oc clastokines expression pattern. raw cells were seeded on tissue culture plates (oc group) or pdhc disks (ocdisk group) and received 10 ng/ml rankl stimulation for 5 days to differentiate into oc. (a) hierarchical clustering heatmap of clastokine genes expression; based on mrna-seq data. color key represents z score. red represents high expression genes; blue represents low expression genes. (b) relative expression of clastokine genes determined by rt-qpcr; normalized to b2m. *p < 0.05. (c) antibody array detection of clastokines at the protein level in conditioned medium. (d) quantification result of antibody array. *p < 0.05 ocdisk vs. oc.
n = 2 for all experiments. data shown as mean ± sem.
Discussion
In this study, we reported that our PDHC scaffold material, which does not need to load additional bioactive agent, is enough able to modulate OC activity in vitro. First, PDHC showed an anti-resorptive potential, characterized by decreased OC differentiation and resorption capacity and changes in OC transcriptome profile. Next, we found that PDHC’s anti-osteoclastogenic effect was mediated by modulation of CREB activity. Finally, despite an altered clastokines expression pattern revealed by transcriptomic and secretomic analysis, OCs’ coupling to OBs was not impaired by PDHC. Given that bone is a dynamic tissue that is continuously being broken down and constructed, such material characteristics are favorable for the net increase of bone mass. Additionally, considering PDHC’s good biocompatibility and osteoconductivity in vivo demonstrated in our previous studies,11, 17 the new anti-OC function/mechanism revealed in the present study further supports PDHC’s application potential for bone defect repair.
Both OBs and OCs are essential for the bone regenerative process following bone defect. Healthy bone regeneration requires the activities of both OBs and OCs and relies on an orchestrated cell-cell communication.29 Osteoblastic lineage cells (OBs, MSCs, etc.) are usually the focus of interest when considering tissue engineering for bone defect repair. Therefore, how osteoblastic lineage cells interact with scaffold materials has been extensively investigated. In contrast, much less is known regarding how OC behavior is affected by bone substitute materials.30 OC-derived bone formation mechanism is worth noting in modern development of bone substitutes.3 Several types of scaffold materials, such as decellularized natural tissue extracellular matrices or bone type resorbable mineral matrices, can simultaneously induce bone resorption and formation.3 In addition, it has been proposed that OC introduction seems like a promising strategy for the structural and functional improvement of tissue-engineered bone because the absence of OCs leads to abnormalities of bone formation.31 This OC introduction idea was later tested in vivo, where iPSC-derived-OBs and OCs together promoted bone regeneration in a subcutaneous transplantation model.32 Collectively, a bone substitute material’s impact on OBs and OCs are equally important when developing bone substitute materials for bone defect repair.
PDA-assisted modification has emerged as a popular strategy for optimizing bone substitutes properties in bone tissue engineering.33, 34 A lot of evidence has suggested that PDA-functionalized bone substitute materials orient osteoblastic lineage cells to a pro-bone-formation pattern, characterized by enhanced cell adhesion, proliferation, or osteogenic differentiation.6–9 In contrast, much less is known about their effects on OCs. A few studies have explored the effects of PDA-assisted, biomolecule-immobilized materials on OCs, but these effects actually result from the immobilized biomolecule (aspirin, adenosine, lactoferrin, etc.) rather than PDA coating.13–15 In this study, we showed that the PDHC material, which does not load additional bioactive agent, is able to modify cellular activity and transcriptome profile of OCs. According to our results, PDHC orients OC behavior to an anti-resorptive pattern without impairing OC’s coupling to OBs.
It seems that PDHC’s impact on OC behavior is similar to what dopamine does: suppressing OC differentiation via modulation of CREB activity.19 The anti-osteoclastogenic effect of PDHC may be attributed to the bioactivity of dopamine, PDA-altered material properties, or both. Genetic approach can be considered to further explore this question. For example, deleting dopamine receptors in OC precursor cells and see if it can abolish PDHC’s anti-osteoclastogenic effect.
Although PDHC did not compromise OC’s coupling to OBs, clastokines expression pattern did change in PDHC-modified OCs. In the present study, we only focused on clastokines because we utilized a conditioned medium culture system. Beyond clastokines, OCs can also modulate OB activity via cell-cell direct contact and the release of matrix-embedded growth factors.27, 28, 35 In addition, it has also been indicated that OC-derived microRNAs-containing exosomes are involved in the regulation of osteoblastic activity.36, 37 Thus, future studies can consider exploring these additional possible mechanisms of OC-OB coupling in direct co-culture systems or in vivo animal models.
In conclusion, this study demonstrated that PDHC material orients OC behavior to an anti-resorptive pattern without compromising OC’s coupling to OBs. Such a material property is favorable for the net increase of bone mass, which endows PDHC material with great application potential in preclinical/clinical reconstruction of bone defects.
Supplementary Material
Funding
This work was supported by The Ohio State University College of Dentistry and the NIH/NIDCR grant (R01DE022816 to CCK).
Footnotes
Declaration of conflicting interests
The authors declare no potential conflicts of interest.
Supplemental material
Supplemental material for this article is available online.
REFERENCES
- 1.Fernandez de Grado G, Keller L, Idoux-Gillet Y, Wagner Q, Musset AM, Benkirane-Jessel N, Bornert F and Offner D, J Tissue Eng, 2018, 9, 2041731418776819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borciani G, Montalbano G, Baldini N, Cerqueni G, Vitale-Brovarone C and Ciapetti G, Acta Biomater, 2020, 108, 22–45. [DOI] [PubMed] [Google Scholar]
- 3.Kylmaoja E, Nakamura M and Tuukkanen J, Curr Stem Cell Res Ther, 2016, 11, 626–633. [DOI] [PubMed] [Google Scholar]
- 4.Batul R, Tamanna T, Khaliq A and Yu A, Biomater Sci, 2017, 5, 1204–1229. [DOI] [PubMed] [Google Scholar]
- 5.Jia L, Han F, Wang H, Zhu C, Guo Q, Li J, Zhao Z, Zhang Q, Zhu X and Li B, J Orthop Translat, 2019, 17, 82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albu AM, Draghicescu W, Munteanu T, Ion R, Mitran V, Cimpean A, Popescu S and Pirvu C, Mater Sci Eng C Mater Biol Appl, 2019, 98, 461–471. [DOI] [PubMed] [Google Scholar]
- 7.Steeves AJ, Atwal A, Schock SC and Variola F, J Mater Chem B, 2016, 4, 3145–3156. [DOI] [PubMed] [Google Scholar]
- 8.Sun Y, Deng Y, Ye Z, Liang S, Tang Z and Wei S, Colloids Surf B Biointerfaces, 2013, 111, 107–116. [DOI] [PubMed] [Google Scholar]
- 9.Rim NG, Kim SJ, Shin YM, Jun I, Lim DW, Park JH and Shin H, Colloids Surf B Biointerfaces, 2012, 91, 189–197. [DOI] [PubMed] [Google Scholar]
- 10.Dyke JC, Hu H, Lee DJ, Ko CC and You W, J Mater Chem B, 2014, 2, 7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee DJ, Lee YT, Zou R, Daniel R and Ko CC, Sci Rep, 2017, 7, 12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu H, Dyke JC, Bowman BA, Ko CC and You W, Langmuir, 2016, 32, 9873–9882. [DOI] [PubMed] [Google Scholar]
- 13.Wei Y, Liu Z, Zhu X, Jiang L, Shi W, Wang Y, Xu N, Gang F, Wang X, Zhao L, Lin J and Sun X, Biomaterials, 2020, 257, 120237. [DOI] [PubMed] [Google Scholar]
- 14.Ahmad T, Byun H, Shin HJ, Lee J, Madhurakkat Perikamana SK, Kim EM, Shin YM and Shin H, Biomater Sci, 2020, 8, 2825–2839. [DOI] [PubMed] [Google Scholar]
- 15.Shen T, Yang W, Shen X, Chen W, Tao B, Yang X, Yuan J, Liu P and Cai K, ACS Biomater Sci Eng, 2018, 4, 3211–3223. [DOI] [PubMed] [Google Scholar]
- 16.Lee DJ, Kwon J, Kim YI, Wang X, Wu TJ, Lee YT, Kim S, Miguez P and Ko CC, Orthod Craniofac Res, 2019, 22 Suppl 1, 127–133. [DOI] [PMC free article] [PubMed]
- 17.Wang L, Lee DJ, Han H, Zhao L, Tsukamoto H, Kim YI, Musicant AM, Parag-Sharma K, Hu X, Tseng HC, Chi JT, Wang Z, Amelio AL and Ko CC, J Tissue Eng, 2021, 12, 2041731421995465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang MC, Ko CC and Douglas WH, Biomaterials, 2003, 24, 2853–2862. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Han L, Xue P, Hu X, Wong SW, Deng M, Tseng HC, Huang BW and Ko CC, Cell Signal, 2021, 78, 109847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue P, Hu X, Chang E, Wang L, Chen M, Wu TH, Lee DJ, Foster BL, Tseng HC and Ko CC, Exp Mol Med, 2021. [DOI] [PMC free article] [PubMed]
- 21.Vesprey A and Yang W, Bio Protoc, 2016, 6. [DOI] [PMC free article] [PubMed]
- 22.Chen L, Shi K, Andersen TL, Qiu W and Kassem M, Cell Death Dis, 2019, 10, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pijuan J, Barceló C, Moreno DF, Maiques O, Sisó P, Marti RM, Macià A and Panosa A, Front Cell Dev Biol, 2019, 7, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collin-Osdoby P and Osdoby P, Methods Mol Biol, 2012, 816, 187–202. [DOI] [PubMed] [Google Scholar]
- 25.Hanami K, Nakano K, Saito K, Okada Y, Yamaoka K, Kubo S, Kondo M and Tanaka Y, Bone, 2013, 56, 1–8. [DOI] [PubMed] [Google Scholar]
- 26.Yang H, Xu Y, Zhu M, Gu Y, Zhang W, Shao H, Wang Y, Ping Z, Hu X, Wang L and Geng D, Biomaterials, 2016, 80, 1–10. [DOI] [PubMed] [Google Scholar]
- 27.Sims NA and Martin TJ, Front Endocrinol (Lausanne), 2015, 6, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim BJ and Koh JM, Cell Mol Life Sci, 2019, 76, 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Wang Z, Duan N, Zhu G, Schwarz EM and Xie C, Connect Tissue Res, 2018, 59, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Detsch R and Boccaccini AR, J Tissue Eng Regen Med, 2015, 9, 1133–1149. [DOI] [PubMed] [Google Scholar]
- 31.Han D and Zhang Q, Med Hypotheses, 2006, 67, 75–78. [DOI] [PubMed] [Google Scholar]
- 32.Jeon OH, Panicker LM, Lu Q, Chae JJ, Feldman RA and Elisseeff JH, Sci Rep, 2016, 6, 26761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madhurakkat Perikamana SK, Lee J, Lee YB, Shin YM, Lee EJ, Mikos AG and Shin H, Biomacromolecules, 2015, 16, 2541–2555. [DOI] [PubMed] [Google Scholar]
- 34.Kaushik N, Nhat Nguyen L, Kim JH, Choi EH and Kumar Kaushik N, Int J Mol Sci, 2020, 21. [DOI] [PMC free article] [PubMed]
- 35.Sims NA and Martin TJ, Annu Rev Physiol, 2020, 82, 507–529. [DOI] [PubMed] [Google Scholar]
- 36.Li D, Liu J, Guo B, Liang C, Dang L, Lu C, He X, Cheung HY, Xu L, He B, Liu B, Shaikh AB, Li F, Wang L, Yang Z, Au DW, Peng S, Zhang Z, Zhang BT, Pan X, Qian A, Shang P, Xiao L, Jiang B, Wong CK, Xu J, Bian Z, Liang Z, Guo DA, Zhu H, Tan W, Lu A and Zhang G, Nat Commun, 2016, 7, 10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang JX, Xie P, Li YS, Wen T and Yang XC, Cell Signal, 2020, 70, 109504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


