Abstract
Neuroblastoma is a highly metastatic cancer, and thus is one of the leading causes of cancer-related mortalities in pediatric patients. More than 50% of NB cases exhibit 17q21-ter partial chromosomal gain, which is independently associated with poor survival, suggesting the clinical importance of genes at this locus in NB. IGF2BP1 is one such proto-oncogene located at 17q locus, and was found to be upregulated in patients with metastatic NBs. Here, utilizing multiple immunocompetent mouse models, along with our newly developed highly metastatic NB cell line, we demonstrate the role of IGF2BP1 in promoting NB metastasis. Importantly, we show the significance of small extracellular vesicles (EVs) in NB progression, and determine the pro-metastatic function of IGF2BP1 by regulating the NB-EV-protein cargo. Through unbiased proteomic analysis of EVs, we discovered two novel targets (SEMA3A and SHMT2) of IGF2BP1, and reveal the mechanism of IGF2BP1 in NB metastasis. We demonstrate that IGF2BP1 directly binds and governs the expression of SEMA3A/SHMT2 in NB cells, thereby modulating their protein levels in NB-EVs. IGF2BP1-affected levels of SEMA3A and SHMT2 in the EVs, regulate the formation of pro-metastatic microenvironment at potential metastatic organs. Finally, higher levels of SEMA3A/SHMT2 proteins in the EVs derived from NB-PDX models indicate the clinical significance of the two proteins and IGF2BP1-SEMA3A/SHMT2 axis in NB metastasis.
Keywords: neuroblastoma metastasis, IGF2BP1, extracellular vesicles, NB mouse model, pre-metastatic niche
Introduction
Neuroblastoma (NB) is the most common extra-cranial solid pediatric cancer, that arises from neural crest cells, progenitors of the sympathetic nervous system. It accounts for 7–8% of all pediatric cancers, and around 15% of cancer-related mortalities in children [1–3]. High-risk (HR) groups have increased metastasis and disease recurrence, leading to poor outcomes and lower overall survival [3]. Additionally, distant site metastasis is evident in most of the relapsed patients, reflecting the clinical aggression of the disease [1–3].
There are few well-known oncogenic drivers of NB such as MYCN amplification, 1p loss, 11q loss and 17q gain, of which, MYCN is most studied. However, MYCN amplification is detected in only about 20% of all NBs [1, 2], suggesting unknown potential oncogenic factors in MYCN non-amplified NBs. Overall, the HR NBs remain clinically aggressive despite of the most recent multimodal therapeutic plans. Identification of novel oncogenic drivers, apart from MYCN in the intermediate-risk/HR group NBs is thus essential to address this aggressive disease.
More than half of all NB cases, irrespective of MYCN status, contain a partial 17q21-ter gain, and this aberration correlates with reduced progression-free survival in MYCN amplified as well as non-amplified NB patients. Also, it has been independently associated with lower survival rates in patients with advanced-stage NBs, thus indicating the clinical significance of genes at this locus in NB pathogenesis [4, 5]. IGF2BP1 (Insulin-like growth factor 2 mRNA-binding protein 1), is one of the potential proto-oncogenes found in this locus. IGF2BP1 is found to be upregulated specifically in patients with advanced-stage metastatic NBs, and higher levels of IGF2BP1 protein was found to be associated with significantly lower survival rates in MYCN amplified and non-amplified NB patients [6].
IGF2BP1 is an RNA binding protein (RBP), that imparts a wide array of downstream functions, by regulating the stability of its target RNAs [7–9]. It is predominantly expressed in the embryonic tissues [10] and in elevated levels in multiple cancer types. IGF2BP1 imparts stability to several oncogenic mRNAs, including the MYC family in multiple cancers [6, 7, 11, 12]. We and others have shown its oncogenic potential in a variety of malignancies including colorectal cancer, leukemia, and melanoma [9, 13–17]. Also, we recently showed the role of IGF2BP1 in regulating cell proliferation and imparting chemoresistance to human NB cell lines in culture [18]. However, its importance in NB progression and metastasis is unknown. Given the association of elevated IGF2BP1 levels in patients with metastatic NBs and with poor prognosis, this study was designed to uncover the role and mechanisms of IGF2BP1 in NB metastasis with an overarching objective to establish IGF2BP1 as a potential therapeutic target for the treatment of metastatic NBs.
The importance of small extracellular vesicles (EVs) as signaling factors in multiple cancer types has been established in recent years [19–21]. EVs are defined as 30–150 nm sized vesicles, secreted by actively proliferating cells and thus by cancer cells in enormous amounts. These vesicles are generated by the inward budding of the late endosomal membrane, forming a pool of intraluminal vesicles within the multivesicular bodies (MVBs). Fusion of MVBs with the plasma membrane release these EVs into the extracellular space [22, 23].
Tumor-derived EVs (TEV) play a multitude of functions in overall cancer progression, by regulating the microenvironment, mediating immune regulation, imparting drug resistance and oncogenic properties to surrounding cancer cells in a wide range of cancers [19, 20]. Specifically, an important pro-metastatic function of EVs is to induce permissive metastatic microenvironment, including the formation of pre-metastatic niche (PMN) at the secondary sites, thus priming these organs for future metastasis. Studies have reported the involvement of EVs in the initiation and regulation of sequential events of PMN formation in various other cancers, thus promoting cancer cell-seeding and growth at these new locations [21, 24, 25]. Previous studies have reported increased levels of NB-derived EVs in the sera of patients with advanced-stage metastatic disease [26], however, the EV-mediated signaling in NB progression is poorly understood.
In this study we established the function of IGF2BP1 in promoting NB metastasis using several immunocompetent mouse models. For the first time, we demonstrate the importance of EVs in NB progression, and report the pro-metastatic function of IGF2BP1 in an EV-mediated manner. We show that IGF2BP1 promotes NB metastasis by inducing pro-metastatic microenvironment in the liver via EVs. IGF2BP1 increases fibronectin deposition and promotes the recruitment of CD45+ cells in the liver, thereby priming the livers for metastasis.
Mechanistically, we show that IGF2BP1 governs the protein cargo of these NB derived EVs by regulating its mRNA target expression in the cells. Importantly, we discovered SEMA3A and SHMT2 – two novel targets of IGF2BP1. IGF2BP1 binds and stabilizes SEMA3A and SHMT2 mRNAs thereby increasing their expression in NB cells, eventually affecting their levels in the EVs. SEMA3A and SHMT2 proteins mediate the pro-metastatic function of IGF2BP1 in NB in an EV-dependent manner.
In addition, we developed a new highly metastatic NB cell line, M1, as an efficient model system for studying NB metastasis. In vivo validation of our M1 line showed that these cells are highly penetrant, extremely metastatic and highly aggressive in immunocompetent mice as compared to the parental 9464Ds. Also, M1 cells prominently develop liver metastasis, which is one of the common sites of metastasis in NB patients, thus making it a relevant model for NB studies.
Materials and Methods
Cell lines
Mouse neuroblastoma cell line 9464D was a gift from Dr. Paul Sondel’s lab (University of Wisconsin) and Neuro2A (ATCC Cat# CCL-131, RRID:CVCL_0470) was purchased from ATCC. Both the cell lines were cultured as monolayers in Dulbecco’s modified Eagle’s medium (DMEM; VWR International), supplemented with 10% v/v FBS (Gibco by Life Technologies) and 100 units/ml penicillin and streptomycin (Corning). To generate luciferase reporter cells, parental 9464D and Neuro2A cells were transduced with pCDH-EF1-Luc-p2a-tdTomato (RRID:Addgene_72486) lentivirus and sorted for tdTomato fluorophore. Cells were then verified for luciferase activity by Promega luciferase assay system (Promega, E1500).
M1 line generation
During the process of determining a suitable cell-number for experimental-metastasis studies, standardization experiment was performed by injecting 9464D (Luc-tdT) reporter cells at different cell-titers (0.25, 0.5 and 1 million/mouse) in C57BL/6J mice (RRID:IMSR_JAX:000664). But only 2 out of 30 injected mice developed metastasis. Liver tumors from these two mice were isolated and processed as previously described protocol [27]. Briefly, isolated tumors were washed with sterile DPBS, minced and digested sequentially with collagenase/dispase/trypsin. Resuspended cell pellet was passed through 18G, 23G and 27G needles for obtaining single-cell suspensions. Different cell populations were separated out by pre-plating technique, and further homogenous purified NB cells were obtained by differential trypsinization and replating procedure. These cells were then sorted for tdTomato expression. One of the cell lines, named – M1, was validated by NMYC expression, and verified for luciferase activity. M1 line was used in the present study and was cultured using same complete DMEM medium as above.
IGF2BP1 KD and OE cell lines
IGF2BP1 KD – M1 cells were generated using doxycycline inducible SMARTvector Inducible TurboGFP shRNA Lentiviral constructs (GE Dharmacon Cat# V3SM11256-03EG140486). shRNA SMARTvector Inducible Non-targeting Control construct (GE Dharmacon Cat# VSC11654) was used to generate M1- shNT control cell line. Lentivirus particles were produced in Lenti-X 293T cells (Takara Cat# 632180) using lipofectamine 3000 transfection reagent (Invitrogen) as per manufacturer’s protocol. Cells were transduced by 8-hour infections with the lentivirus + 8 ug/ml polybrene reagent to generate stable cell lines. Cells were then subjected to puromycin selection (4 ug/ml) in growth media for a week, followed by shRNA induction with 4–6 ug/ml of doxycycline-treatment for 6 days. Induction of shIGF2BP1 showed >80% of IGF2BP1 knockdown (Supplementary Figure 3B).
9464D and Neuro2A Luc-tdT reporter cells were used for ectopic expression of IGF2BP1. GFP-tagged IGF2BP1 overexpression and EGFP-control vector cell lines were generated using LV-GFP-hIMP1 (pLKO. hIMP1_3G from Dr. Joel Yisraeli’s Lab) and EGFP-pReceiver LV105 (Genecopoeia Cat# EX-GFP-Lv105-B) constructs, respectively. Stable cell lines were generated by lentiviral transduction as mentioned above. Positive cells with efficient GFP expression were obtained by cell sorting, and IGF2BP1 overexpression was verified by western (Supplementary Figure 3C, D). IGF2BP1 rescue M1 line was obtained using the same GFP-tagged IGF2BP1 construct and verified by western (Supplementary Figure 3E).
SEMA3A and SHMT2 OE/KD cell lines
SEMA3A_pDONR221 and SHMT2_pLX304 plasmids were obtained from DNASU plasmid repository at Arizona State University. pDONR221_EGFP (RRID:Addgene_25899) and pLX304 empty vector (RRID:Addgene_25890) were obtained from Addgene. SEMA3A and EGFP were then cloned into pLX304 by LR recombination reaction (Gateway™ LR Clonase™ II Enzyme mix, Invitrogen) as per Invitrogen’s Gateway cloning protocol. Stable 9464D-SEMA3A OE, 9464D-SHMT2 OE and 9464D-EGFP cell lines were generated by lentiviral transduction as mentioned above. Transduced cells were selected by 4–5 ug/ml of Blasticidin selection (3–4 days), followed by verification of SEMA3A/SHMT2 overexpression by western (Supplementary Figure 8B). 9464D-EGFP/IGF2BP1 OE + SEMA3A/SHMT2 KD stable cell lines were generated using doxycycline inducible SMARTvector Inducible TurboRFP shRNA Lentiviral constructs (GE Dharmacon Cat# V3SM11253-231319154, V3SM11253-231281864), as above-mentioned procedure. All the above parental and generated cell lines were regularly tested for Mycoplasma contamination (MycoAlert plus mycoplasma detection kit, Lonza), and were cultured for less than a month for experiments.
NB patient data analysis
To determine if IGF2BP1 was independently associated with poor survival in NB, we performed a statistical analysis using the genomic data from two large national NB databases – TARGET (https://ocg.cancer.gov/programs/target) and GSE62564 (GEO). Clinical and gene expression data was first downloaded, then preprocessed with log2 transformation and quantile normalization. Kaplan-Meier survival curves were generated for overall and event-free survival with the online R2 genome tool (https://hgserver1.amc.nl/). No patient data samples were excluded from the analysis. The best cutoff points were selected with the minimal P-value of the log-rank test.
Cell-based in vitro functional assays
Incucyte cell proliferation and migration assays:
M1-shIGF2BP1 and shNT cells were seeded at 5000 cells/well in 96 well plates on day 5 post dox-induction for proliferation assay. Live-cell phase contrast images were obtained using a 10× objective lens (three images per well), and cell confluence was analyzed using IncuCyte Live Cell Analysis (v2019B) software. For migration assay, 30,000 cells/well (100ul/well) were seeded on day 5 (post-dox induction) in IncuCyte ImageLock 96-well plates (Sartorius 4379), and after 12–14 hours, scratches were made using IncuCyte 96-well WoundMaker Tool (Sartorius 4563) as per manufacturer’s protocol. Images were obtained every 2 hours for 48 hours, and relative wound density was quantified using the Incucyte Cell Migration Analysis software (Sartorius 9600-0012).
Invasion assay:
Invasion assay was performed using the Corning Biocoat Matrigel invasion chambers (Corning 354480) as per manufacturer’s protocol. Briefly, 25000 cells in 500 ul of serum free media were seeded in the inserts, and complete media (+10% FBS) was added to the wells of the companion plate as a chemoattractant. After 24 hours, inserts were fixed and permeabilized by subjecting them to 4% formaldehyde (in PBS), followed by 100% methanol. The inserts were then stained with 0.25% crystal violet (in 20% methanol), rinsed, dried and analyzed for invaded cell numbers under 20x phase contrast microscope. Images from 4–5 random areas acquired for quantification. Invasion assay with EV-treatments was performed in a similar manner, except for NIH3T3 (ATCC Cat# CRL-1658, RRID:CVCL_0594) cells treated with 25 ug/ml of EVs were plated in serum-free media in the inserts at a density of 25000 cells/insert. Invaded cells were then analyzed after 18–24 hours, as described above.
Cell viability assay:
Cell viability assay was performed using CyQUANT™ XTT Cell Viability Assay Kit (Invitrogen X12223) as per manufacturer’s protocol. M1-shNT and M1-shIGF2BP1 cells were induced with doxycycline for IGF2BP1 knockdown for 6 days in culture. On day 6 post-dox induction, cells were seeded in 96 well plates as 5000 cells/well, and after 24 hours, 70 uls of XTT working solution was added to each well, and absorbance was measured as per manufacturer’s protocol. M1 cells without any treatment were used as a negative control, and M1 cells treated with 1, 2 uM Doxorubicin chemotherapeutic, (a topoisomerase inhibitor) were used as a positive control.
Experimental metastasis model – Intravenous (IV) injections
All animal procedures were performed in accordance with the conditions approved by the Institutional Animal Care and Use Committee (IACUC) at Pennsylvania State University College of Medicine. For in vivo validation of M1, 1x106 M1 or 9464D cells were IV-injected in C57BL/6J mice, followed by monitoring these mice for metastasis growth by in vivo bioluminescence imaging system and survival. Weights (gm) of metastatic tumors were quantified at experimental endpoint of recommended humane euthanasia.
For studying the effect of IGF2BP1, 1x106 IGF2BP1 KD-M1 cells, 0.5x106 IGF2BP1 OE-9464D cells and 0.25x106 IGF2BP1 OE-Neuro 2A cells expressing luciferase construct were injected intravenously, and mice were monitored for the development of metastases using in vivo bioluminescence imaging system. M1 and 9464D cells were injected in 6–8 weeks old C57BL/6J mice, and Neuro2A cells were injected in 6–8 weeks old A/J mice (RRID:IMSR_JAX:000646). Mice were anesthetized using 2.5% isoflurane (IsoSol Isoflurane, USP, VEDCO) and injected with 0.15 mg/g body weight of D-Luciferin (Goldbio # LUCK-100) dissolved in PBS, 10 minutes prior to imaging. Mice were imaged once or twice a week. Luminescence (radiance) was calculated using region of interest (ROI) measurements and was used to quantify the metastatic tumor growth. Animals were followed for survival analysis, humane euthanasia/death was the experimental endpoint for these experiments. Male and female mice were equally distributed and randomly assigned to each group (n≥5 animals/group) for all animal experiments. Mice to be injected with shIGF2BP1 or shNT-M1 cells were switched to 200 mg/kg doxycycline feed (Bio-Serv # S3888), two-days prior to cell-injections.
Primary tumor growth studies
1x106 9464D or M1 cells were injected subcutaneously, and mice were monitored for primary tumor growth. Tumor sizes (in mm) were measured twice a week, until they reached the experimental endpoint size of ≥20mm. n≥8 animals per group used for the study.
Western blot
Western blotting was performed as previously described [18] with minor modifications. 60µg of total protein was separated on 10% polyacrylamide/SDS gel, transferred to PVDF membranes, and membranes were blocked with 5% BSA in TBS (+0.1% Tween-20). Primary antibodies include: IGF2BP1 (Cell Signaling Technology Cat# 8482, RRID:AB_11179079)), β-Actin (Cell Signaling Technology Cat# 4970, RRID:AB_2223172), SEMA3A (Thermo Fisher Scientific Cat# PA5-67972, RRID:AB_2691930), SHMT2 (Cell Signaling #33443), β-tubulin (Cell Signaling Technology Cat# 2146, RRID:AB_2210545), Caspase 3 (Proteintech Cat# 19677-1-AP, RRID:AB_10733244) and Cleaved Caspase-3 (Cell Signaling Technology Cat# 9661, RRID:AB_2341188). Horseradish peroxidase-conjugated secondary antibody: anti-rabbit IgG (Cell Signaling Technology Cat# 7074, RRID:AB_2099233)
Small extracellular vesicle isolation and characterization
EV-depleted FBS was prepared by centrifuging heat-inactivated FBS at 120,000g for 18 hours at 4°C, followed by filtering the supernatant through 0.22µM filter.
IGF2BP1 OE or EGFP control-9464D cells were plated in complete media with 10% FBS in 150 mm culture plates, 24 hours after plating, cells were then rinsed with PBS and maintained in DMEM with EV-depleted serum-media for 48 hours. While, shIGF2BP1 and shNT-M1 cells were cultured alongwith doxycycline for 4 days, and were changed to EV-depleted media on day 5 of dox-induction. Conditioned media was collected and centrifuged at 2000g for 20 mins at 4°C for removing dead cells and debris. Supernatant was then subjected to differential steps of ultracentrifugation: 10,000g for 30 minutes, followed by 100,000g for 3 hours at 4°C. At the end of this centrifugation step, supernatant was carefully removed, and the pellet was washed once with PBS and centrifuged at 120,000g for 3 hours at 4°C. The resulting EV-pellet was resuspended in 350–500 uls of PBS and stored at −80°C for further in vivo and in vitro experiments. Purified EVs were characterized by transmission electron microscopy and nanoparticle tracking analysis.
For transmission electron microscopy (TEM), 30 µL of the resuspended EV-sample was pipetted onto a 400-mesh copper grid with carbon-coated formvar film and incubated for 2 min. Excess liquid was removed by blotting. The grid was briefly placed on 30 µL of 1% uranyl acetate for 2 min, followed by blotting to remove the excess liquid. The grid was then allowed to dry and was examined using the JEOL JEM1400 Transmission Electron Microscope (JEOL USA Inc., Peabody, MA, USA).
For Nanoparticle tracking analysis, EV samples were diluted from 1:100 to 1:500 dilution to a final volume of 1ml in sterile particle free water. Each sample was loaded by syringe pump into the NanoSight NS300 (Malvern Instruments Ltd, Malvern, Worcestershire, UK) set in scatter mode, and four 10-second videos were generated at 24.98 frames per second, and Particle size distribution and concentration were calculated using the NTA 3.2 software (Malvern Instruments Ltd).
EVs were examined for the presence of EV-markers by western blotting (30 µg total protein loaded), using CD81 (Santa Cruz Biotechnology Cat# sc-166029, RRID:AB_2275892), Alix (Cell Signaling Technology Cat# 2171, RRID:AB_2299455), and TSG101 (Santa Cruz Biotechnology Cat# sc-7964, RRID:AB_671392) as well as for EV-negative marker GM130 (Novus Cat# NBP2-53420, RRID:AB_2916095) antibodies. HRP-conjugated secondary antibodies from Cell signaling used: anti-mouse IgG (Cell Signaling Technology Cat# 7076, RRID:AB_330924), anti-rabbit IgG (Cell Signaling Technology Cat# 7074, RRID:AB_2099233).
EV-mediated effect in vivo
Mice were IV-injected with 10µg of EVs in 100 µl PBS or 100 µl plain PBS, every alternate day for a course of three weeks. For experimental metastasis studies, two days post last EV-injection, mice were injected with 0.5x106 M1 cells and were monitored for the growth of metastasis. At experimental endpoint of 7 weeks post cell-injections or at recommended humane euthanasia, metastatic tumors were extracted and examined by weight. For PMN studies, mice were injected with EVs for three weeks as described above. Two days post last EV-injection, mice were euthanized and potential organs – liver, femur bone and lungs were harvested and formalin-fixed for ex-vivo analysis.
Tissue processing and Immunofluorescence (IF) staining
Liver, lung and bone tissues were fixed in 4% formalin or formical-4 Decalcifier for 24–48 hours, followed by 24 hours in 70% ethanol. Tissues were processed through graded alcohols, cleared in xylene and infiltrated with paraffin using a Sakura VIP5 Tissue processor on a 12-hour overnight cycle. Following overnight processing, tissues were embedded in paraffin using a Tissue Tek embedding console. Blocks were then sectioned into 6-micron thickness using a Leitz 1512 microtome, and mounted on charged slides and dried prior to staining.
IF staining was performed by deparaffinizing and hydrating the tissues in xylene and decreasing concentrations of ethanol. Antigen retrieval was done by adding pre-heated antigen retrieval buffer (100mM Tris-EDTA buffer (pH:9) for fibronectin; 10mM sodium citrate buffer (pH:6) for CD45) on the tissue sections and incubating the slides at 95° C in a heater-hybridizer for 15 minutes. Slides were then cooled by adding a few drops of room temperature (RT) buffer on bench-top for 5 minutes, followed by again heat (95°C) incubation with pre-heated buffer for 5 minutes. This 5-minute heat-cold cycle was repeated 3 times. At the end of this cycle, slides were cooled at RT for 30 minutes, followed by 2x washes with 1x TBS (+0.025% Triton- X-100), and blocking the tissue with 5% donkey serum (in 1% BSA) for 2 hours at RT. Tissues were then incubated with primary antibodies against fibronectin (Abcam Cat# ab2413, RRID:AB_2262874) or CD45 (R and D Systems Cat# AF114, RRID:AB_442146) overnight at 4°C. Next day, slides were washed twice with 1x TBS (+0.1% Tween 20), followed by fluorophore-conjugated secondary antibody incubation (anti-goat Alexa Fluor 488, (Jackson ImmunoResearch Labs Cat# 705-545-003, RRID:AB_2340428); anti-rabbit Rhodamine red, (Jackson ImmunoResearch Labs Cat# 711-295-152, RRID:AB_2340613)) in dark, for 1 hour at RT. Slides were washed twice with 1xTBS (+0.1% Tween 20), and mounted using VectaShield antifade with Dapi (Vector labs #H-1500-10) medium. Images, 8–10 random fields/slide from different areas were acquired on Biorad ZOE Fluorescent cell Imager.
Proteomic analysis of EVs
TMT-labelling and mass spectrometry:
EVs from shIGF2BP1/shNT-M1, IGF2BP1OE/EGFP control-9464D cells were quantified for protein concentration using the RC DC™ Protein Assay (Bio-Rad Laboratories). Mass spectrometry analysis of EV-proteins was performed at the University of Michigan. EV-samples (50ug each), 4 biological replicates per condition were proteolyzed and labelled with TMT 16-plex kit (Thermo Fisher, A44521) as per the manufacturer’s protocol. Briefly, upon reduction (5 mM DTT, 30 min at 45°C) and alkylation (15 mM 2-chloroacetamide, 30 min at RT) of cysteines, proteins were precipitated by adding 6 volumes of ice-cold acetone followed by overnight incubation at −20°C. The precipitate was spun and the pellet was air dried, prior to resuspension in 0.1M TEAB. Resuspension was digested with trypsin/Lys-C mix (1:25 protease: protein, Promega) by constant shaking on thermomixer at 37°C, overnight. Labelling was performed by incubating the entire digest with TMT reagent (dissolved in 41uls anhydrous acetonitrile) for 1 hour at RT, followed by quenching the reaction with 5% hydroxyl amine, 15 mins. Labeled samples were then mixed together and dried using a vacufuge, followed by reconstitution in 800 uls of 0.1% formic acid/2% acetonitrile and LC-MS/MS analysis. Orbitrap Fusion (Thermo Fisher Scientific) and RSLC Ultimate 3000 nano-UPLC (Dionex) was used to acquire the data [28]. 200 uls of combined sample was taken for offline fractionation using high pH reversed-phase peptide fractionation kit according to manufacturer’s protocol (Pierce; Cat #84868).
Data processing:
Proteome Discoverer (v2.4; Thermo Fisher) was used for data processing. MS2 spectra were searched against M musculus uniport protein database (17054 entries; reviewed entries; downloaded on 08/06/2021) using the following search parameters: MS1 and MS2 tolerance were set to 10 ppm and 0.6 Da, respectively; carbamidomethylation of cysteines (57.02146 Da) and TMT labeling of lysine and N-termini of peptides (304.207 Da) were considered static modifications; oxidation of methionine (15.9949 Da) and deamidation of asparagine and glutamine (0.98401 Da) were considered variable. Identified proteins and peptides were filtered to retain only those that passed ≤1% FDR threshold. Quantitation was performed using high-quality MS3 spectra (Average signal-to-noise ratio of 16 and <50% isolation interference).
Bioinformatic data analysis:
Protein abundances reported by Proteome Discoverer were taken for bioinformatic analysis. Exosome quality was assessed by visualizing abundances of known exosome markers [29] and by comparison of our data to known high-quality exosome samples [30]. Protein abundances were normalized so that each sample had the same median abundance and then analyzed by limma [31] for differential abundance with q-value correction for multiple testing. A threshold of q < 0.3 (30% False Discovery Rate, FDR) was used to identify differentially abundant proteins. t-statistics from the limma analysis were used to identify the proteins with opposite regulation in the two comparisons. An o-index is defined as the product of the two t-statistics: OE vs EGFP and shIGF2BP1 vs shNT. The more negative it is, the more likely that the protein is oppositely regulated. A threshold o-index of −2 was set to filter the candidates for opposing regulation.
mRNA stability, Pre-mRNA quantification and binding (CLIP) assays
The effect of IGF2BP1 on mRNA stability, was determined by actinomycin D chase assay. IGF2BP1-OE/EGFP control-9464D cells plated in 150 mm tissue culture dish, were treated with 10 µg/ml Actinomycin D (Invitrogen, A7592). Plate area was divided in approximately equal areas, and cells were scraped after 1, 3, 5 and 7-hours post-treatment from the same plate. Total RNA was isolated using TRIzol reagent (Invitrogen) as per manufacturer’s protocol. Equal amount (ng) of total RNA was used for further cDNA synthesis (Bio-Rad iScript cDNA synthesis kit) as per manufacturer’s protocol. Then real-time PCR analysis was done using iTaq Universal SYBR Green Supermix (Bio-Rad) on a CFX96 Touch™ Real-Time PCR Detection System with SEMA3A and SHMT2 primers (Supplementary Table1). Relative transcript levels were calculated by comparative Ct method, normalized to RPS18.
CLIP was performed as previously described [17]. Briefly, 9464D-IGF2BP1-OE cells upon reaching 75–80% confluency, were fixed with 3% formaldehyde in PBS for 10 minutes and lysed using sonication (10 pulses for 10 s). Anti-IGF2BP1 antibody (MBL International Cat# RN007P, RRID:AB_1570640) linked to Dynabeads (Thermo Fisher Scientific Cat# 11203D, RRID:AB_2783009) were used for overnight immunoprecipitation at 4°C. RNA-protein complexes precipitated by magnetic separation, were reverse cross-linked and washed five times with lysis buffer, prior to RNA extraction (TRIzol reagent, Invitrogen). To remove traces of genomic DNA, isolated RNA was treated with RNase-free DNase I (Thermo Scientific). First-strand cDNA synthesis was performed using Bio-Rad iScript cDNA synthesis kit according to manufacturer’s protocol, followed by real-time PCR with an iTaq Universal SYBR Green kit (Bio-Rad). PCR amplification conditions were as follows: initial denaturation for 3 minutes at 95°C, PCR amplification for 45 cycles with denaturation for 20 seconds at 95°C, annealing for 20 seconds at 58°C, and elongation for 20 seconds at 72°C. Fold change in gene expression was determined using Biorad’s CFX software. The expression of BTRC/SHMT2/SEMA3A genes was normalized to their respective input samples.
Pre-mRNA levels of SEMA3A and SHMT2 were analyzed by real-time PCR as described above, using pre-mRNA primers (Supplementary Table1).
EV isolations from PDX-serum samples
Patient-derived xenograft (PDX) cells (COG-N-415x, COG-N-453x, COG-N-496x, COG-N-424x, and COG -N-603x) were obtained from the Childhood Cancer Repository maintained by the Children’s Oncology Group (Pediatric Preclinical Tumor Consortium, https://www.pdxfinder.org/source/pptc/). Cells were suspended in Matrigel diluted 1:2 with PBS, and 1–3× 106 cells (200 µL) were injected subcutaneously into the left flank of NSG mice (NOD SCID gamma, Jackson Laboratory). Once the tumors reached approximately 1500 mm3, mice were euthanized, and tumors were collected, processed, and cryopreserved. For EV isolation, blood was collected via intra-cardiac puncture from these PDX-bearing or control (without tumors) mice. The collected blood was then left undisturbed for 1 hour at RT, followed by a 15-minute centrifugation at 3000 rpm. Supernatant (serum) was transferred to a fresh tube and stored at −80°C until EV-isolation. EVs were isolated from serum samples using the ExoQuick® ULTRA EV Isolation Kit for Serum and Plasma (SBI system Biosciences # EQULTRA-20A-1) following the manufacturer’s instructions. Isolated EV-samples were subjected to protein estimation (RC DC™ Protein Assay), and western blotting as described above.
Statistical analysis
GraphPad Prism 9 was used for statistical analysis. Unpaired two tailed-t tests were used for two group comparisons, One-way ANNOVA was used for comparing more than 2 groups, and ordinary Two-way ANNOVA was used for multiple comparisons. Data are presented as mean ± SEM. Effect on tumor growth-rate was monitored using the nonlinear regression model. Survival curves were generated using the Kaplan-Meier survival analysis. Statistically significant values are indicated on the graphs as well as in the figure legends (significance level: α = 0.05).
Results
Validation and characterization of a novel highly metastatic transplantable neuroblastoma cell line
Due to the limitations of current models to study spontaneous NB metastasis, we used the model of experimental metastasis in immunocompetent mice for investigating the function of IGF2BP1. 9464D is a widely used NB mouse cell line for primary tumor studies, however it shows poor metastatic efficiency in experimental metastasis model. Thus, in order to establish a more efficient metastatic model, we developed a novel transplantable NB cell line (Figure 1A). 9464D cells were injected intravenously in C57Bl6 immunocompetent mice, of the 30 injected mice only two developed metastatic tumors. Liver tumor from one of these mice was isolated and processed to generate an established line - named as M1. In vivo validation of the newly generated M1 cell line showed a remarkably high (100%) metastatic penetrance in the model of experimental metastasis, relative to the 9464Ds that had only 18% penetrance rate (Figure 1B). Moreover, mice injected with M1 cells showed increased growth of metastasis and reduced survival (Figure 1C, D). Importantly, mice injected with M1 cells develop tumors specifically in the livers, their site of origin and a prominent site of metastasis in NB patients. M1 cells did not show a differential primary tumor growth as compared to 9464Ds (Figure 1E).
Figure 1. Generation and characterization of a novel highly metastatic transplantable mouse NB cell line - M1.

A) schematic of M1 cell line generation; In vivo characterization of M1 cells - B) tumor penetrance (Bioluminescence in-vivo imaging, statistical analysis – unpaired t-test, two-tailed, *** p-value − 0.0003. Data representation from two independent experiments, n=8 animals per group/experiment); C) metastatic tumor weights at experimental endpoint (statistical analysis – unpaired t-test, two-tailed, mean ± SEM, ** p-value − 0.0011); D) effect on mouse survival (Kaplan-Meier analysis, Log-rank test, p-value − 0.0007) n=6 animals/group for C) & D); E) Primary tumor growth rate, 1 million cells/mouse subcutaneously-injected, tumor sizes (in mm) measured until experimental endpoint, statistical analysis - non-linear regression model, F-test, p-value − 0.82); F) effect of 9464D/M1 cell-derived EVs on recipient NIH3T3 cell-invasion (using Biocoat Matrigel invasion inserts, EV-treatment: 25ug/ml); G) EV-treated NIH3T3 Invasion assay quantification (statistical analysis - one-way ANNOVA, * p-value <0.02, ** p-value <0.005); H) Fibronectin and CD45 expression in the livers of EV-treated mice (10ug/100ul EV-treatments), IHC/IF staining, n=4 animals/group; I,J) FN and CD45 expression quantification, corrected total fluorescence (CTF) calculated for >6 areas/slide (statistical analysis - one-way ANNOVA, mean ± SEM, ** p-value <0.005 *** p-value <0.0003, **** p-value <0.0001).
Additionally, in vitro characterization of M1 line revealed a surprisingly distinct phenotype as compared to 9464Ds. M1 cells showed reduced cell proliferation, migration and invasion in culture (Supplementary figure 1). Since M1 cells did not possess pro-metastatic phenotypes in culture, we hypothesized that its high metastatic potential in vivo could be associated with secreted factors such as extracellular vesicles. TEV or cancer-cell secreted EVs have known to alter the surrounding cancer and non-cancer cells within the primary tumor, and at secondary locations upon cancer cell homing at these new sites. TEVs can promote metastasis by regulating the metastatic microenvironment of secondary locations through processes such as affecting the resident fibroblasts and inducing other pre-metastatic factors. Interestingly, mouse fibroblast NIH3T3 cells treated with M1-derived EVs showed a significantly higher invasion rate as compared to the 9464D-derived EV-treated cells and untreated cells. 9464D-derived EV-treatment did have a notable increase in cell invasion relative to the untreated cells (Figure 1F). Additionally, M1-EV treated mice showed significantly increased Fibronectin (FN) expression and CD45+ cell accumulation in the livers, as compared to 9464D-EV treated and PBS-treated groups, indicative of inducing a pro-metastatic environment (Figure 1H). This data suggests EVs as one of the potential mechanisms responsible for the metastatic nature of M1 cells.
Overall, the in vivo characterization confirms that our novel M1 cell line is highly metastatic in nature, and thus is a more efficient model system for studying NB metastasis in syngeneic C57Bl6 mice. We thus utilized M1 cells along with other previously established syngeneic NB mouse models to understand the role of IGF2BP1 in NB progression.
IGF2BP1 promotes NB metastasis
In addition to its previously reported prognostic value [6], our patient data analysis from two national neuroblastoma databases show IGF2BP1 to be independently associated with overall poor and disease-free survival (Supplementary figure 2). Given the association between elevated levels of IGF2BP1 expression in patients with advanced-stages and poor prognosis of metastatic NB, we hypothesized its role in NB progression specifically via promoting metastasis. We found that down-regulation of IGF2BP1 in the highly metastatic M1 cells resulted in significant reduction of their metastatic potential (Figure 2A, B), thereby prolonging survival of these cell-injected mice (Figure 2C). Conversely, mice injected with IGF2BP1 overexpressing (IGF2BP1-OE-9464D) cells showed increased growth of experimental metastasis (Figure 2D, E) at week 5, and significantly lowered survival (Figure 2F) as compared to the EGFP expressing control-cell injected mice. Re-expressing IGF2BP1 in IGF2BP1-knockdown M1 cells substantially restored the metastatic potential of these cells (Supplementary Fig 3A), further indicating that observed metastatic phenotype was specifically governed by IGF2BP1.
Figure 2. IGF2BP1 promotes metastasis of NB and reduces survival in mouse models.

A), B) Effect of IGF2BP1 knockdown on NB metastasis using M1 model, metastasis monitored by bioluminescence imaging, luminescence (radiance) used for quantification; C) survival of cell-injected mice (in-vivo bioluminescence image – week 5, n=7 animals/group, statistical analysis – Two-tailed t-test, mean ± SEM, ** p-value − 0.0069; survival analysis - Kaplan-Meier analysis, Log-rank test, p-value < 0.001); D), E) Effect of IGF2BP1 overexpression on NB metastasis using 9464Ds; F) survival of mice (in-vivo bioluminescence image – week4, n=5 animals/group, survival analysis - Kaplan-Meier analysis, Log-rank test, p-value − 0.045); IGF2BP1 function validation using A/J mouse model - G), H) metastasis growth and bioluminescence quantification; and I) survival of mice (n=7 animals/group, tumor growth monitored using bioluminescence imaging, statistical analysis - non-linear regression model, F-test, *** p-value − 0.0001; survival analysis - Kaplan-Meier analysis, Log-rank test, p-value − 0.025).
We further validated the function of IGF2BP1 using a different (A/J) NB mouse model along with its syngeneic Neuro2A cells, to rule out the possibility of model-specific artefact. Neuro2A cells were chosen here as this cell line expresses low levels of endogenous IGF2BP1, and thus makes it a suitable model for ectopic expression of IGF2BP1. Consistently, Neuro2A cells overexpressing IGF2BP1 showed increased metastatic burden (Figure 2G, H) and reduced survival (Figure 2I) in mice. Taken together, our data demonstrates the role of IGF2BP1 in promoting NB metastasis.
IGF2BP1 modulation does not impact the phenotype of M1-cells in vitro and does not affect primary tumor growth
IGF2BP1 has been previously shown to play a pleotropic role in promoting development and progression of multiple malignancies [8, 11, 13–16, 32]. To determine if IGF2BP1 regulates various pro-tumorigenic phenotypes in M1 cells in culture, we examined the effect of IGF2BP1 knockdown (KD) on cell survival, proliferation, migration and invasion of these cells. Surprisingly, despite of highly metastatic in nature, M1 cell line failed to show any difference in cell viability and the pro-metastatic cellular processes such as cell proliferation, migration or invasion in vitro, upon IGF2BP1 modulation (Figure 3, Supplementary figure 4). This prompted us to explore the extracellular mechanisms potentially involved in regulating the function of IGF2BP1 in promoting NB metastasis. In addition to its role in promoting NB metastasis, we wanted to determine if IGF2BP1 was essential for NB tumor initiation and primary tumor growth. Mice were subcutaneously injected with either IGF2BP1 KD/OE cells and their respective control cells, followed by time-dependent tumor monitoring. We saw no difference in the rate of tumor growth (Supplementary figure 5A) or tumor size at the experimental endpoint (Supplementary figure 5B, C) upon IGF2BP1 modulation. Likewise, there was no difference in the survival of these mice due to primary tumor burden (Supplementary figure 5D), thus indicating the function of IGF2BP1 specifically in NB-metastasis.
Figure 3. IGF2BP1 does not impact the pro-metastatic properties of highly metastatic M1 cells in vitro.
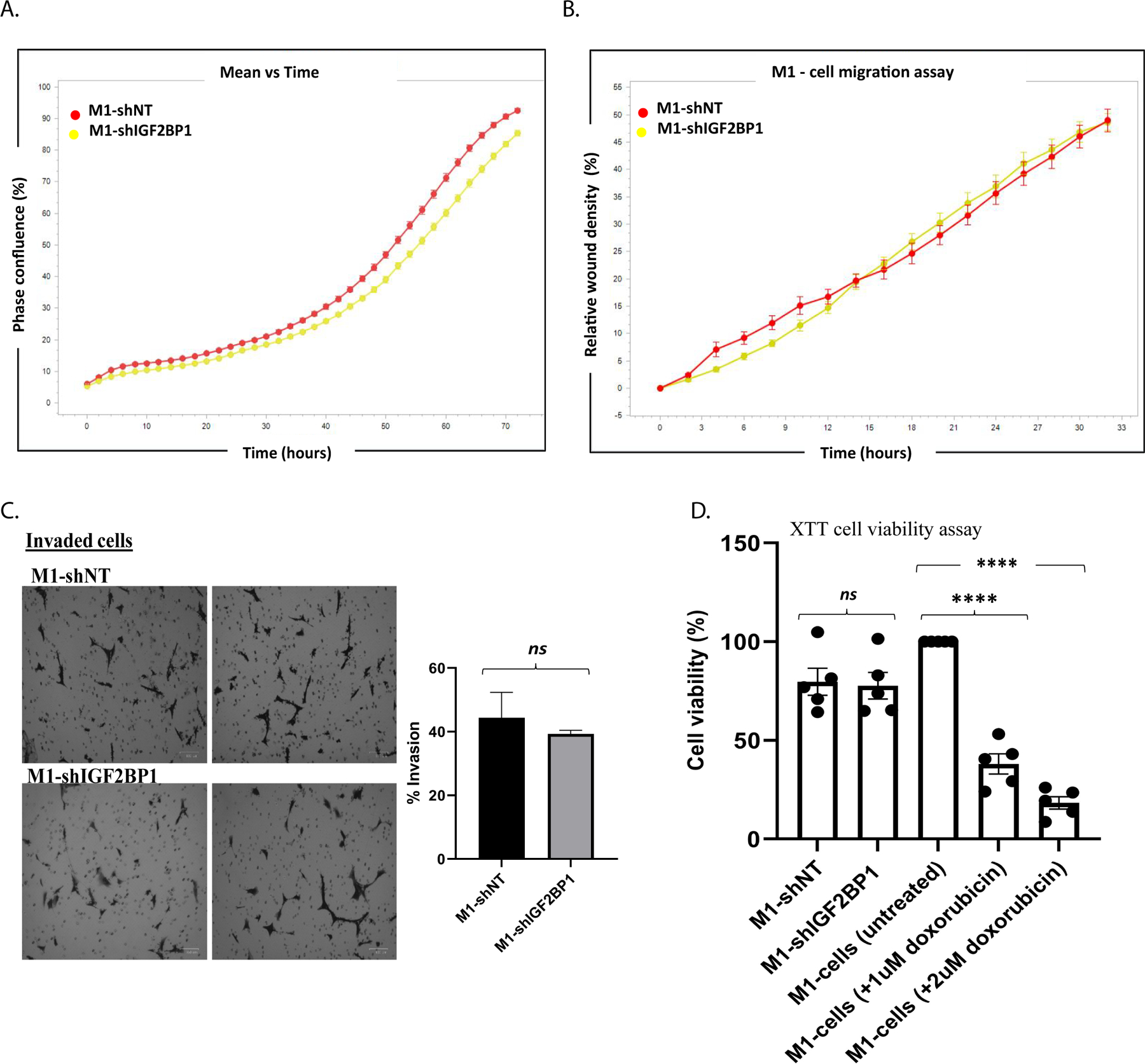
Effect of IGF2BP1 knockdown on A) cell proliferation; B) migration (IncuCyte live-cell analysis system); C) invasion (Biocoat Matrigel invasion inserts, statistical test- unpaired Two-tailed t-test, p-value: non-significant (0.592)); D) survival (XTT cell viability assay, statistical test- one-way ANNOVA, mean ± SEM, M1-shNT vs M1-shIGF2BP1: p-value − ns (0.998), **** p-value <0.0001). (each assay repeated with n>3 independent experiments).
IGF2BP1 promotes metastasis in NB via EVs
One of the salient features of metastatic tumors is secretion of several tumor derived factors and EVs as pro-metastatic signals. Previous studies have shown the importance of EVs in metastasis in other cancer types. With clinical reports showing the presence of EVs in the sera of patients with advanced-staged NBs [26], we hypothesized the importance of EVs in NB metastasis.
To determine the involvement of EVs and if the pro-metastatic function of IGF2BP1 is EV-dependent, we examined the role of EVs isolated from our IGF2BP1 modulated NB cells. EVs were isolated from IGF2BP1-KD/OE or their respective control NB cells in culture using differential ultracentrifugation method. Vesicles were analyzed and validated by Transmission electron microscopy (TEM) and Nanoparticle tracking analysis (NTA) for particle size, morphology and concentration (Supplementary figure 6A, B), and for the presence of EV-specific surface marker proteins (Supplementary figure 6C). Our extracellular vesicles are of 100–120 nm in size, falling within the defined size range for small EVs and meeting the specifications set by International society for extracellular vesicles (ISEV) [33].
To examine the importance of EVs in regulating NB metastasis, mice were treated with either IGF2BP1-KD-M1 or non-targeting (NT) shRNA control-M1 cell derived EVs for three weeks prior to parental M1 cell line injection. These mice were monitored for the formation of metastatic tumors, tumor weights were analyzed at experimental endpoint. We found that pre-treatment with EVs derived from NT shRNA control cells promoted the growth of liver metastasis in mice, demonstrating the significance of EVs in promoting NB metastasis. However, IGF2BP1-KD-M1 cell derived EVs failed to promote metastasis as compared to NT-M1 cell derived EVs, indicating the EV-mediated pro-metastatic function of IGF2BP1 in NB (Figure 4A). These results thus show the significance of EVs in promoting NB metastasis, that is impacted by IGF2BP1 modulation.
Figure 4. IGF2BP1 promotes metastasis in NB via small extracellular vesicles (EVs).

A) Effect of M1-shNT and M1-shIGF2BP1 cell-derived EVs on experimental metastasis (10 ug/100ul EV-treatments, tumor weights examined at experimental endpoint, n=5 animals/group, statistical analysis: one-way ANNOVA, mean ± SEM, * p-value − 0.03, *** p-value − 0.0009); Effect of NB-cell derived EVs on PMN formation - B) fibronectin (FN) and CD45+ expression in mouse livers treated with M1-shNT/M1-shIGF2BP1 cell-derived EVs; C), D) IF data quantified by calculating corrected total fluorescence (CTF) of >8 areas/slide (10 ug/100ul EV-treatments, n=4 animals/group, statistical analysis - One way ANOVA, mean ± SEM, * p-value <0.03, *** p-value <0.0003, **** p-value <0.0001); E) FN and CD45+ expression in the livers of 9464D-IGF2BP1OE/9464D-EGFP derived EV-treated mice; F), G) quantification of E) (n=3 animals/group, statistical analysis - One way ANOVA, mean ± SEM, * p-value <0.03).
To further determine how these EVs promote metastasis in NB, we examined their role in regulating PMN formation. Based on the intrinsic feature of cancer derived EVs to prime the potential secondary organs for metastasis, we explored several markers of PMN in the livers, bones and lungs of EV-treated mice. We found increased levels of PMN marker expression specifically in the livers of the EV-treated mice, corelating with the preferred metastatic site that we have previously observed in this mouse model. EVs derived from NT shRNA control-M1 cells increased the expression of PMN markers such as fibronectin (FN) expression (Figure 4B, C) and CD45+ cell accumulation (Figure 4B, D) in mouse livers. Importantly, EVs derived from IGF2BP1-KD-M1 cells failed to induce these markers of PMN formation. Conversely, EVs derived from IGF2BP1-OE-9464D cells increased the expression of PMN markers (Figure 4E–G).
We found no significant difference in the PMN formation in the lungs and bone of these mice (data not shown), indicating the specificity of EVs in formation of PMN at the potential metastatic sites. Overall, our findings demonstrate the importance of EVs and EV-mediated function of IGF2BP1 in promoting metastasis of NB via induction of PMN formation.
To further confirm the functional relevance of our IGF2BP1-modulated cell-derived EVs on the recipient cells, we examined their effect on the recipient cell-phenotype. NIH3T3 mouse fibroblast cells were treated with either IGF2BP1-OE or EGFP control cell-derived EVs followed by seeding them in the Matrigel invasion assay inserts. We found that treatment with IGF2BP1 OE-EVs significantly increased the invasive ability of 3T3 cells, while control-EVs just slightly increased cell invasion as compared to the PBS treated 3T3 cells (Supplementary figure 7).
IGF2BP1 imparts pro-metastatic phenotype via regulating the EV-protein cargo
To determine the EV-mediated mechanism of IGF2BP1 function, we performed an unbiased proteomic analysis on the EVs derived from IGF2BP1-modulated M1 and 9464D cells. Since our EVs were not different in particle concentration (NTA analysis) or in overall protein levels, we ruled out the possibility of IGF2BP1 affecting the vesicular release or non-specific protein loading. Proteomic characterization of EV samples showed the abundance of specifically cellular origin proteins (Supplementary figure 8B), while only negligible amounts of serum or culture media contaminant proteins were detected (Supplementary figure 8C). Additionally, the samples were enriched in EV-marker proteins (Supplementary figure 8D), thus validating the purity of the isolated vesicles and the cellular origin of the EV content. This analysis revealed a differentially regulated protein cargo, with 48 proteins significantly up or down-regulated in IGF2BP1 KD-EVs as compared to the NT shRNA control-EVs (Supplementary figure 8A). Collagen alpha-1(1) (Col1A) (p-value: 0.01) and Ephrin B1 (p-value: 0.003) were picked for further analysis based on their known function.
Additionally, using t-statistics, we identified the proteins that were oppositely regulated in IGF2BP1-KD vs NT shRNA EVs and IGF2BP1-OE vs EGFP control EVs (Figure 5A). An o-index of −2 was set as a threshold cutoff to identify the top most oppositely regulated proteins. Of the top oppositely regulated proteins, we were interested in examining the candidates that were highly relevant in terms of predicted/previously known function, and were detected in higher intensities in our EVs. We chose Semaphorin 3A (SEMA3A) with an o-index −5.90 and mitochondrial serine hydroxymethyltransferase 2 (SHMT2) with an o-index of −4.97, along with Ephrin-B1 and Col1A1 as candidates for our further analysis.
Figure 5. IGF2BP1-regulated proteins SEMA3A and SHMT2 induce pre-metastatic niche in an EV-mediated manner.

A) Proteomic analysis of EVs derived from IGF2BP1 KD/OE cells showing oppositely regulated proteins, red points represent the top candidates for opposing regulation at set threshold o-index of −2; B) Invasion assay of NIH3T3 cells treated with PBS, EGFP/IGF2BP1 OE/SHMT2 OE/SEMA3A OE derived EVs; C) NIH3T3 cell invasion assay after treatments with EGFP/IGF2BP1 OE/IGF2BP1 OE+shSEMA3A/IGF2BP1 OE+shSHMT2 cell-derived EVs (25ug/ml EV-treatments, statistical analysis - One-way ANOVA, mean ± SEM, ** p-value <0.005, *** p-value <0.0005, **** p-value <0.0001); D) CD45 expression in the livers of SEMA3AOE and SHMT2OE derived EV-treated mice; E) quantification of CD45 expression, corrected total fluorescence (CTF) for >8 areas/slide (l0ug/100ul EV-treatments, n =4 animals/group, statistical analysis - One-way ANNOVA, mean ± SEM, **** p-value <0.0001).
We confirmed the differential expression of these proteins by western blotting in our IGF2BP1-modulated cell lysates and EVs (Supplementary figure 9A).
IGF2BP1 regulated-SHMT2 and SEMA3A proteins affect the recipient cell phenotype and promote PMN formation through EVs
Initially, to ascertain the independent functions of these identified proteins, we overexpressed them individually in 9464D cells (Supplementary figure 9B) and isolated the EVs from these cells. NIH3T3 cell-invasion assay with EV treatments, was used to screen the most efficient candidates for in vivo studies. We found specifically SHMT2 and SEMA3A (but not Col1A or Ephrin B1) to be functionally active, and phenocopied the EV-mediated effect of IGF2BP1 on recipient cells. Col1A or Ephrin B1, that were not among the oppositely regulated candidates failed to show an EV-mediated functional effect (Supplementary figure 10D–F), thus indicating the specificity of the identified IGF2BP1-regulated targets. Reduced levels of these two (Col1A and EphrinB1) proteins in the M1-shIGF2BP1-EVs could possibly be a result of an alternative mechanism. NIH3T3 cells treated with either SHMT2 or SEMA3A overexpressing cell-derived EVs showed a significantly higher cell invasion rate as compared to Control-EV treated cells (Supplementary figure 9C, Figure 5B). While EVs derived from IGF2BP1 OE+shSEMA3A or IGF2BP1 OE+shSHMT2 cells reduced this EV-mediated effect of IGF2BP1 on recipient NIH3T3 cell invasion (Supplementary figure 9D, Figure 5C). However, EVs from IGF2BP1 OE+shSEMA3A&shSHMT2 double knockdown cells did not further reduce NIH3T3 cell invasion (Supplementary figure 10A–C). To further validate the function of SHMT2 and SEMA3A, we examined their individual effect via EVs on PMN formation. Mice were treated with either SHMT2 OE/SEMA 3A OE/EGFP control cell derived EVs followed by harvesting the livers for immunostaining, as described previously. Consistent with in vitro assays, we found increased expression of PMN marker (CD45) in the livers of the mice treated with SEMA OE/SHMT2 OE EVs (Figure 5D, E).
Overall these results demonstrate that IGF2BP1-regulated SEMA3A and SHMT2 phenocopy the effects of IGF2BP1 on PMN formation in an EV-mediated manner, and mediate the pro-metastatic function of IGF2BP1 in the recipient cells.
IGF2BP1 increases the expression of SEMA3A and SHMT2 by stabilizing their mRNAs
To determine the mechanisms of SHMT2 and SEMA3A regulation by IGF2BP1, we first examined the mRNA expression levels of SHMT2 and SEMA3A by qPCR, in response to IGF2BP1 modulation in NB cells. We found increased levels of both SHMT2 and SEMA3A mRNA expression upon overexpressing IGF2BP1 (Figure 6A). IGF2BP1 is an RBP that was shown to bind and stabilize a variety of its target RNAs [7, 9, 12, 17, 34–36]. Our results demonstrate that overexpression of IGF2BP1 stabilized both SHMT2 and SEMA3A mRNAs (Figure 6B). BTRC, a known target of IGF2BP1 is used as a positive control. Cross-linking and immunoprecipitation (CLIP) assay determined the direct interaction between IGF2BP1 protein and SHMT2 and SEMA3A mRNAs, similar to BTRC. (Figure 6C). We found no difference in the pre-mRNA transcript levels upon IGF2BP1 OE (Figure 6D), thus indicating that IGF2BP1 does not regulate SEMA3A and SHMT2 transcriptionally. Taken together, these results demonstrate that IGF2BP1 increases the expression of SEMA3A and SHMT2 in the cells by stabilizing their mRNAs, which eventually leads to the elevated levels of these proteins in the cells and EVs.
Figure 6. IGF2BP1 increases the expression of SEMA3A and SHMT2 by stabilizing their mRNAs.

A) mRNA expression levels of SEMA3A and SHMT2 in 9464D-EGFP and 9464D-IGF2BP1 OE cells (relative normalized expression, normalized to internal reference RPS18, n=4 independent experiments, statistical analysis – Two-way ANNOVA (ordinary, multiple comparisons with Sidaks’s correction), mean ± SEM, ** p-value − 0.0036, *** p-value − 0.008, **** p-value<0.0001); B) mRNA stability of SEMA3A, SHMT2 and BTRC transcripts accessed by actinomycin D assay in 9464D-EGFP/IGF2BP1 OE cells (samples collected until 7 hours post-treatment and mRNA expression quantified by qPCR, n=3 experiments); C) Cross-linking and immunoprecipitation (CLIP) assay for IGF2BP1 binding to SEMA3A, SHMT2, and BTRC mRNAs in 9464D-IGF2BP1 OE cells, n=3 experiments (statistical analysis – unpaired t-test, two-tailed, mean ± SEM, BTRC p-value − 0.02, SEMA3A p-value − 0.016, SHMT2 p-value − 0.016), index figure - western blot for immunoprecipitation of IGF2BP1 using anti-IGF2BP1-IgG (1.5% Input and Flow through (FT), and 3% bead-bound samples loaded); D) pre-mRNA levels of SEMA3A and SHMT2 in 9464D-EGFP or IGF2BP1 OE cells quantified by qPCR (n=5 independent experiments, statistical analysis – unpaired t-test with Welch’s correction, p-value - ns), Overexpression of IGF2BP1 confirmed in the same experimental samples (statistical analysis : unpaired two-tailed t-test, mean ± SEM, p-value <0.0001).
Serum derived EVs of NB PDX bearing mice have elevated levels of SEMA3A and SHMT2
With the limitation of accessibility to NB patient serum samples, we utilized multiple NB PDX models derived from stage 4 tumors, consisting of both primary and relapsed disease-types. We found increased levels of SEMA3A and SHMT2 protein in the serum derived EVs of PDX-bearing mice as compared to control-serum derived EVs (Supplementary figure 11A, B), indicating the potential clinical relevance of these two proteins in NB. In particular, we detected SEMA3A and SHMT2 proteins explicitly in the purified EVs (Supplementary figure 11C), confirming EVs as their source in the serum.
Discussion
Due to the lack of a reliable model of spontaneous NB metastasis in immunocompetent environment, experimental metastasis model is the best currently available approach to study later stages of NB metastasis. A limitation of this model is, bypassing early events of the metastatic cascade such as cell migration & invasion at primary location and intravasation. 9464D cell line, derived from the spontaneous NB tumors of TH-MYCN mice [37], is a well-known and most suitable model due to its gene expression profile and tumor-phenotype similar to human NBs [38]. However, 9464D cells are poorly metastatic even in the model of experimental metastasis. Here we developed a new mouse NB cell line – M1. M1 cells show 100% metastatic penetrance and aggressive disease progression, compared to parental 9464Ds. We also found EVs to be one of the potential mechanisms responsible for the metastatic nature of M1 cells. Possibly, EVs secreted by homed M1 cells induce a pro-metastatic environment at the new sites, thereby promoting the establishment and growth of metastases. Notably, M1 cells do not show a difference in primary tumor growth, relative to 9464D, indicating their specifically acquired metastatic ability. High metastatic potential with prominent liver metastasis, which is one of the prevalent metastatic sites in human NBs, makes M1 an efficient and clinically relevant model system for NB-metastasis studies.
Neuroblastoma being a highly metastatic cancer, is one of the leading causes of cancer-related deaths in pediatric patients. IGF2BP1 is located on most commonly amplified region (17q) in NB, its expression was shown to be associated with poor prognosis [4, 5], and its downregulation in vitro sensitizes NB cells to chemotherapeutics [18]. However, the role of IGF2BP1 in NB progression, and metastasis remained to be elucidated. In this study we have shown its importance for NB metastasis, especially its pro-metastatic function in an EV-mediated manner. We demonstrated the function of IGF2BP1 in promoting NB metastasis using both genetic knockdown and overexpression studies, utilizing several models of experimental metastasis. Highly metastatic M1 cells were used for genetic knockdown of IGF2BP1, while less metastatic 9464D was used for overexpressing IGF2BP1. Neuro2A cells express miniscule levels of endogenous IGF2BP1, and thus were also used for IGF2BP1 overexpression studies.
IGF2BP1 did not affect the cellular phenotype of the highly aggressive M1 cells in culture, suggestive of its potential role via extracellular mechanisms in NB. Although known for their multiple roles in metastasis and cancer progression in other cancer-types [19, 20, 39], the involvement of EVs in NB progression is unclear. Here we reveal the functional importance of EVs in NB progression in general, and demonstrate the EV-mediated pro-metastatic effect of IGF2BP1, that is novel for NB, and is in line with our previous observations in melanoma [39]. NB-derived EVs augmented liver metastasis, while EVs from IGF2BP1-KD NB cells failed to accelerate the metastatic process. Importantly, we show that IGF2BP1 promotes the formation of pre-metastatic niche (PMN) by inducing fibronectin deposition and CD45+ cell recruitment in the liver via EVs. Consistent with the previous observations [21] of FN upregulation in the hepatic tissues upon EV treatments, our data indicates IGF2BP1 as a pro-metastatic factor in NB.
Unbiased proteomic analysis of IGF2BP1-modulated NB-derived EV cargo led us to determine the mechanism of IGF2BP1 in NB. We identified COL1A, EPHRIN-B1, SEMA3A and SHMT2 among the top most differentially regulated proteins in the EVs, in response to cellular IGF2BP1 modulation. Initial testing of these proteins by their individual EV-mediated effects using the most reliable in vitro system in our hands – NIH3T3 cell-invasion assay, identified SEMA3A and SHMT2 (that were among the oppositely regulated candidates), as most functionally effective. NIH3T3 is a mouse fibroblast cell line, used here as a model for normal fibroblasts, with a rationale to examine the effect of TEVs on resident fibroblasts. Cancer associated fibroblasts (CAFs) are one of the major components of the tumor microenvironment (TME) that have multiple cancer-supportive functions, thereby promoting tumor growth and metastasis. Cancer cells instruct the transition of normal stromal fibroblasts to CAFs by either direct signaling or via signals from other cells within the TME. The major function of CAFs is remodeling of the ECM, thereby promoting local invasion and distal metastasis [40, 41]. Moreover, tumor-derived EVs have been found to activate normal fibroblasts to CAFs in several cancer models [19, 42, 43]. The EV-mediated effects of SEMA3A and SHMT2 were further validated in vivo demonstrating their ability to induce PMN formation, phenocopying the IGF2BP1 induced phenotype, indicating an IGF2BP1-associated function of SEMA3A and SHMT2 in NB. Moreover, downregulation of SEMA3A/SHMT2 in IGF2BP1 OE cells reduced the effect of these cell-derived EVs on recipient cell invasion, thus confirming SEMA3A and SHMT2 to be mediating the pro-metastatic function of IGF2BP1 via EVs. We chose SEMA3A and SHMT2 from the top oppositely regulated candidates based on their intensity and known functions, although there could be other potential targets of IGF2BP1 among the identified list of proteins. Some of the other top hits were integrins, extracellular matrix receptors, and endocytosis-promoting proteins that could potentially have a role in regulating the EV-uptake in recipient tissues, and are of interest for future investigation.
SEMA3A was recently shown to promote immune cell infiltration, influence the cancer-stromal cell communications in multiple cancer types [44, 45], and to regulate vascular permeability via EVs in glioblastoma patients [46]. SEMA3A-receptor, Nrp1 was found to be expressed on T-regs, tumor infiltrating PD1+ lymphocytes in non-small-cell lung cancer, and its activation by SEMA3A was shown to negatively regulate antitumor immunity[47]. Its previously known functions and our data showing its EV-mediated effects, indicate a potential role for SEMA3A in NB progression. Likewise, SHMT2 – a crucial metabolic enzyme in cancers, has been positively correlated with immune checkpoint marker-PD-L1 and VISTA expression, and is found to regulate immune signaling in oral carcinomas [48]. This correlates to the observed PMN marker (CD45+) expression in our study, suggestive of its importance in immune cell recruitment in NB. A recent study found increased expression of SHMT2 due to IGF2BP2-mediated stabilization of its mRNA in glioblastoma [49]. Here we propose a novel role for SEMA3A and SHMT2 proteins in NB metastasis and describe the associated mechanism. Importantly, we found high levels of SEMA3A and SHMT2 proteins were present in the serum-derived EVs from NB-PDX bearing mice, indicating their potential diagnostic and prognostic value in NB. Due to the inaccessibility to NB patient serum and with limited access to PDXs, we utilized a few PDX models here, derived from stage 4 patient tumors. Our finding serves as a preliminary basis for further investigation of these proteins in NB patient sera.
IGF2BP1 is a known post-transcriptional regulator that binds and stabilizes several oncogenic transcripts [7–9, 17, 34]. We show that IGF2BP1 directly interacts and stabilizes SHMT2 and SEMA3A mRNAs, thereby resulting in elevated levels of these proteins in the NB cells and secreted EVs. EV-mediated delivery of these proteins to the potential metastatic sites prime these organs for future metastases. Notably, no change observed in the pre-mRNA levels of SHMT2 and SEMA3A in response to IGF2BP1 modulation validates their specific post-transcriptional mode of regulation. The IGF2BP1-SEMA3A/SHMT2 axis could be a potential therapeutic target for metastatic NBs. Previous studies in NB have largely focused on more conventional pathways and oncogenic drivers, whereas our study establishes IGF2BP1 as a novel driver of metastasis. Additionally, with observed IGF2BP1 enrichment at advanced stages in MYCN amplified and non-amplified NBs, targeting IGF2BP1 could be a potentially efficient strategy for high risk NBs irrespective of MYCN status. Newly developed pharmacological inhibitors of IGF2BP1 [50] could hold promise for future treatment of metastatic NB.
Taken together, we developed a novel model of NB metastasis, established an important role of IGF2BP1 in NB metastasis, identified its two novel targets and propose the associated mechanisms. To the best of our knowledge, our study is the first to show the significance of EVs in NB progression.
Supplementary Material
Statement of Significance.
The uncovered here EV-mediated role of IGF2BP1 in neuroblastoma metastasis establishes IGF2BP1 as a novel therapeutic target. It identifies EV protein cargo, regulated by IGF2BP1, that confer pro-metastatic functions of neuroblastoma-derived EVs.
Acknowledgements
This study was supported by the NIH grant R01 CA243167 (V.S.S.), and Four Diamonds Fund (V.S.S.). The authors are thankful to Dr. Paul Sondel’s lab for the generous gift of reagents, Venkatesha Basrur and University of Michigan mass-spectrometry core for help with EV-mass spec. We also thank Joe Bednarczyk and Flow-Cytometry Core, Han Chen and TEM Facility, and Molecular & Histopathology Core services at Penn State College of Medicine for help with cell-sorting, TEM and tissue mounting, respectively.
Footnotes
Disclosure of competing interests: The authors declare no competing financial interests.
Data availability
Data generated in the study are available within the article and its supplementary data files. Raw EV-mass spectrometry data are available upon request from the corresponding author.
References
- 1.Nakagawara A, Li YY, Izumi H, Muramori K, Inada H, Nishi M. Neuroblastoma. Japanese Journal of Clinical Oncology 2018; 48: 214–241. [DOI] [PubMed] [Google Scholar]
- 2.Irwin MS, Park JR. Neuroblastoma: paradigm for precision medicine. Pediatr Clin North Am 2015; 62: 225–256. [DOI] [PubMed] [Google Scholar]
- 3.Cohn SL, Pearson ADJ, London WB, Monclair T, Ambros PF, Brodeur GM et al. The International Neuroblastoma Risk Group (INRG) Classification System: An INRG Task Force Report. Journal of Clinical Oncology 2009; 27: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bown N, Cotterill S, Lastowska M, O’Neill S, Pearson ADJ, Plantaz D et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. New England Journal of Medicine 1999; 340: 1954–1961. [DOI] [PubMed] [Google Scholar]
- 5.Bown N, Lastowska M, Cotterill S, O’Neill S, Ellershaw C, Roberts P et al. 17q gain in neuroblastoma predicts adverse clinical outcome. Medical and Pediatric Oncology 2001; 36: 14–19. [DOI] [PubMed] [Google Scholar]
- 6.Bell JL, Turlapati R, Liu T, Schulte JH, Huttelmaier S. IGF2BP1 Harbors Prognostic Significance by Gene Gain and Diverse Expression in Neuroblastoma. Journal of Clinical Oncology 2015; 33: 1285–+. [DOI] [PubMed] [Google Scholar]
- 7.Noubissi FK, Elcheva I, Bhatia N, Shakoori A, Ougolkov A, Liu JH et al. CRD-BP mediates stabilization of beta TrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature 2006; 441: 898–901. [DOI] [PubMed] [Google Scholar]
- 8.Zirkel A, Lederer M, Stohr N, Pazaitis N, Huttelmaier S. IGF2BP1 promotes mesenchymal cell properties and migration of tumor-derived cells by enhancing the expression of LEF1 and SNAI2 (SLUG). Nucleic Acids Research 2013; 41: 6618–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elcheva IA, Wood T, Chiarolanzio K, Chim B, Wong MD, Singh V et al. RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia 2020; 34: 1354–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaniv K, Fainsod A, Kalcheim C, Yisraeli JK. The RNA binding protein Vg1 RBP is required for cell migrations during early neural development. Developmental Biology 2003; 259: 483–483. [DOI] [PubMed] [Google Scholar]
- 11.Bell JL, Wachter K, Muleck B, Pazaitis N, Kohn M, Lederer M et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cellular and Molecular Life Sciences 2013; 70: 2657–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goswami S, Tarapore RS, Poenitzsch Strong AM, TeSlaa JJ, Grinblat Y, Setaluri V et al. MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor (MITF) mRNA is inhibited by coding region determinant-binding protein (CRD-BP). J Biol Chem 2015; 290: 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elcheva I, Tarapore RS, Bhatia N, Spiegelman VS. Overexpression of mRNA-binding protein CRD-BP in malignant melanomas. Oncogene 2008; 27: 5069–5074. [DOI] [PubMed] [Google Scholar]
- 14.Vainer G, Vainer-Mossel E, Pkarsky A, Shenoy SM, Oberman F, Yeffet A et al. A role for VICKZ proteins in the progression of colorectal carcinomas: regulating lamellipodia formation. Journal of Pathology 2008; 215: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stohr N, Huttelmaier S. IGF2BP1 A post-transcriptional “driver” of tumor cell migration. Cell Adhesion & Migration 2012; 6: 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton KE, Noubissi FK, Katti PS, Hahn CM, Davey SR, Lundsmith ET et al. IMP1 promotes tumor growth, dissemination and a tumor-initiating cell phenotype in colorectal cancer cell xenografts. Carcinogenesis 2013; 34: 2647–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh V, Gowda CP, Singh V, Ganapathy AS, Karamchandani DM, Eshelman MA et al. The mRNA-binding protein IGF2BP1 maintains intestinal barrier function by up-regulating occludin expression. J Biol Chem 2020; 295: 8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biegel JM, Dhamdhere M, Gao S, Gowda CP, Kawasawa YI, Spiegelman VS. Inhibition of the mRNA-Binding Protein IGF2BP1 Suppresses Proliferation and Sensitizes Neuroblastoma Cells to Chemotherapeutic Agents. Frontiers in Oncology 2021; 11 (608816). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-Mediated Metastasis: Communication from a Distance. Developmental Cell 2019; 49: 347–360. [DOI] [PubMed] [Google Scholar]
- 20.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer and Metastasis Reviews 2013; 32: 623–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang HY, Thakur BK et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nature Cell Biology 2015; 17: 816–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020; 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McAndrews KM, Kalluri R. Mechanisms associated with biogenesis of exosomes in cancer. Mol Cancer 2019; 18: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peinado H, Kovic MA, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET (vol 18, pg 883, 2012). Nature Medicine 2016; 22: 1502–1502. [DOI] [PubMed] [Google Scholar]
- 25.Guo YX, Ji X, Liu JB, Fan DD, Zhou QB, Chen C et al. Effects of exosomes on pre-metastatic niche formation in tumors. Molecular Cancer 2019; 18 (39). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morini M, Cangelosi D, Segalerba D, Marimpietri D, Raggi F, Castellano A et al. Exosomal microRNAs from Longitudinal Liquid Biopsies for the Prediction of Response to Induction Chemotherapy in High-Risk Neuroblastoma Patients: A Proof of Concept SIOPEN Study. Cancers 2019; 11 (1476). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gharaibeh B, Lu A, Tebbets J, Zheng B, Feduska J, Crisan M et al. Isolation of a slowly adhering cell fraction containing stem cells from murine skeletal muscle by the preplate technique. Nat Protoc 2008; 3: 1501–1509. [DOI] [PubMed] [Google Scholar]
- 28.McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem 2014; 86: 7150–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Askeland A, Borup A, Ostergaard O, Olsen JV, Lund SM, Christiansen G et al. Mass-Spectrometry Based Proteome Comparison of Extracellular Vesicle Isolation Methods: Comparison of ME-kit, Size-Exclusion Chromatography, and High-Speed Centrifugation. Biomedicines 2020; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burton JB, Carruthers NJ, Hou Z, Matherly LH, Stemmer PM. Pattern Analysis of Organellar Maps for Interpretation of Proteomic Data. Proteomes 2022; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vikesaa J, Hansen TVO, Jonson L, Borup R, Wewer UM, Christiansen J et al. RNA-binding IMPs promote cell adhesion and invadopodia formation. Embo Journal 2006; 25: 1456–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles 2018; 7: 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glass M, Misiak D, Bley N, Muller S, Hagemann S, Busch B et al. IGF2BP1, a Conserved Regulator of RNA Turnover in Cancer. Frontiers in Molecular Biosciences 2021; 8 (632219). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elcheva I, Goswami S, Noubissi FK, Spiegelman VS. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell 2009; 35: 240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noubissi FK, Goswami S, Sanek NA, Kawakami K, Minamoto T, Moser A et al. Wnt signaling stimulates transcriptional outcome of the Hedgehog pathway by stabilizing GLI1 mRNA. Cancer Res 2009; 69: 8572–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss WA, Aldape K, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. European Journal of Cancer 1997; 33: 2137–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stauffer JK, Lincoln E, Greer B, Khan T, Salcedo R, Hixon JA et al. Molecular profiling of a novel transplantable model of murine neuroblastoma. Journal of Immunotherapy 2006; 29: 682–683. [Google Scholar]
- 39.Ghoshal A, Rodrigues LC, Gowda CP, Elcheva IA, Liu ZQ, Abraham T et al. Extracellular vesicle-dependent effect of RNA-binding protein IGF2BP1 on melanoma metastasis. Oncogene 2019; 38: 4182–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nature Reviews Cancer 2020; 20: 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Libring S, Shinde A, Chanda MK, Nuru M, George H, Saleh AM et al. The Dynamic Relationship of Breast Cancer Cells and Fibroblasts in Fibronectin Accumulation at Primary and Metastatic Tumor Sites. Cancers 2020; 12 (1270). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He CS, Wang LN, Li L, Zhu GQ. Extracellular vesicle-orchestrated crosstalk between cancer-associated fibroblasts and tumors. Translational Oncology 2021; 14 (101231). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giusti I, Di Francesco M, Poppa G, Esposito L, D’Ascenzo S, Dolo V. Tumor-Derived Extracellular Vesicles Activate Normal Human Fibroblasts to a Cancer-Associated Fibroblast-Like Phenotype, Sustaining a Pro-Tumorigenic Microenvironment. Frontiers in Oncology 2022; 12 (839880). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang XL, Klamer B, Li J, Fernandez S, Li L. A pan-cancer study of class-3 semaphorins as therapeutic targets in cancer. Bmc Medical Genomics 2020; 13 (45). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yazdani U, Terman JR. The semaphorins. Genome Biology 2006; 7 (211). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treps L, Edmond S, Harford-Wright E, Galan-Moya EM, Schmitt A, Azzi S et al. Extracellular vesicle-transported Semaphorin3A promotes vascular permeability in glioblastoma. Oncogene 2016; 35: 2615–2623. [DOI] [PubMed] [Google Scholar]
- 47.Leclerc M, Voilin E, Gros G, Corgnac S, de Montpreville V, Validire P et al. Regulation of antitumour CD8 T-cell immunity and checkpoint blockade immunotherapy by Neuropilin-1. Nat Commun 2019; 10: 3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu ZZ, Wang S, Yang QC, Wang XL, Yang LL, Liu B et al. Increased Expression of SHMT2 Is Associated With Poor Prognosis and Advanced Pathological Grade in Oral Squamous Cell Carcinoma. Frontiers in Oncology 2020; 10 (588530). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han W, Wang S, Qi Y, Wu F, Tian N, Qiang B et al. Targeting HOTAIRM1 ameliorates glioblastoma by disrupting mitochondrial oxidative phosphorylation and serine metabolism. iScience 2022; 25: 104823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallis N, Oberman F, Shurrush K, Germain N, Greenwald G, Gershon T et al. Small molecule inhibitor of Igf2bp1 represses Kras and a pro-oncogenic phenotype in cancer cells. Rna Biology 2022; 19: 26–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated in the study are available within the article and its supplementary data files. Raw EV-mass spectrometry data are available upon request from the corresponding author.


