Abstract
Sixty-nine Kenyan Plasmodium falciparum field isolates were tested in vitro against pyrimethamine (PM), chlorcycloguanil (CCG), sulfadoxine (SD), and dapsone (DDS), and their dihydrofolate reductase (DHFR) genotypes were determined. The in vitro data show that CCG is more potent than PM and that DDS is more potent than SD. DHFR genotype is correlated with PM and CCG drug response. Isolates can be classified into three distinct groups based on their 50% inhibitory concentrations (IC50s) for PM and CCG (P < 0.01) and their DHFR genotypes. The first group consists of wild-type isolates with mean PM and CCG IC50s of 3.71 ± 6.94 and 0.24 ± 0.21 nM, respectively. The second group includes parasites which all have mutations at codon 108 alone or also at codons 51 or 59 and represents one homogeneous group for which 25- and 6-fold increases in PM and CCG IC50s, respectively, are observed. Parasites with mutations at codons 108, 51, and 59 (triple mutants) form a third distinct group for which nine- and eightfold increases in IC50s, respectively, of PM and CCG compared to the second group are observed. Surprisingly, there is a significant decrease (P < 0.01) of SD and DDS susceptibility in these triple mutants. Our data show that more than 92% of Kenyan field isolates have undergone at least one point mutation associated with a decrease in PM activity. These findings are of great concern because they may indicate imminent PM-SD failure, and there is no affordable antimalarial drug to replace PM-SD (Fansidar).
The spread of chloroquine (CQ)-resistant Plasmodium falciparum populations in areas in Africa where malaria is endemic (5, 23, 48) has required that other drugs be introduced for malaria treatment. Pyrimethamine (PM) and proguanil (Paludrine) are specific competitive inhibitors of dihydrofolate reductase (DHFR), a key enzyme in nucleotide biosynthesis (13). Sulfadoxine (SD) is thought to inhibit dihydropteroate synthase (DHPS), an enzyme that catalyzes a reaction in the synthesis of folate in P. falciparum (13). Proguanil is metabolized to its active form, cycloguanil (CG), and has been used primarily for chemoprophylaxis. A combination of PM and SD (Fansidar) acts synergistically to inhibit the folate pathway (9).
Genetic analysis of P. falciparum isolates has demonstrated that antifolate resistance in P. falciparum is caused by point mutations in the gene that encodes the protein target of PM, DHFR, leading to amino acid changes in the active site of the enzyme. The amino acid serine at position 108 (Ser-108) is linked to sensitivity to both PM and CG. The Ser-108-to-Asn-108 mutation confers resistance to PM, and Thr-108 is associated with resistance to CG (paired with a mutation of Ala-16 to Val-16). Subsequent mutations of Asn-51 to Ile-51, Cys-59 to Arg-59, and Ile-164 to Leu-164 enhance the resistance of the isolates to PM. These findings were based on the determination of the complete DNA sequence of the DHFR-coding region in a series of P. falciparum isolates whose sensitivities to PM and CG had been previously determined by in vitro testing (10, 14, 24, 25, 55). Isolates from some sub-Saharan and Southeast Asian countries have been analyzed by DNA sequencing (3, 4). This technique is reliable but expensive and time-consuming, which limits its use in large-scale epidemiological studies in areas of endemicity. Alternatively, PCR can be adapted for the rapid detection of sequences differing by a single base pair (8, 17, 21, 32, 35). Specific primers for allele-specific PCR (ASPCR) have been designed for the detection of mutations at codons 16, 108, and 164 in the P. falciparum DHFR gene (15, 26, 27). This approach has been used to detect mutations at codon 108 in epidemiological investigations of Brazilian (26) and Malian (28) P. falciparum isolates and in a comparative study on isolates from East and West Africa and South America (29). Restriction enzyme digestion of PCR products can also be used to identify some of these point mutations in the P. falciparum DHFR gene (12, 53).
PM-SD is cheap and well tolerated and is increasingly being used as a first-line treatment against uncomplicated malaria in Kenya. As a result, there is a significant reduction in in vitro chemosensitivity to PM in Kenyan parasites. In the 1980s, an average of 20% of the samples tested were classed as PM resistant (37, 44, 46), but that average rose to about 90% between 1993 and 1995 (18). We report here the application of ASPCR and enzyme digestion methods for the detection of point mutations at codons 51, 59, 108, and 164 of the DHFR gene in in vitro-adapted Kenyan P. falciparum field isolates. The goal was to determine whether particular point mutations in the DHFR-coding region are correlated in each isolate with the isolates’ chemosensitivities to the DHFR inhibitors PM and CCG (the active metabolite of chlorproguanil [CPG] [Lapudrine]) or to the DHPS inhibitors SD and dapsone (DDS). CCG and DDS were included in the study because this combination has proved to be particularly potent against P. falciparum isolates in vitro (50) and in vivo (1).
MATERIALS AND METHODS
Parasites.
Five well-known laboratory reference strains, D6, K1, W282, V1/S, and FCR3, were used as positive and negative controls to establish the mutation-specific PCR assay and the restriction enzyme digestion. Their characteristics, drug profiles, and DHFR genotypes are published elsewhere (10, 14, 25). Field isolates were collected during a clinical trial of the antifolate combination CPG-DDS in Kilifi, on the Kenyan coast, between 1993 and 1995 (1). A total of 1 to 3 ml of venous blood was collected from each patient and cryopreserved in 10% dimethyl sulfoxide in liquid nitrogen until adaptation for in vitro culture.
In vitro adaptation and chemosensitivity assessment.
P. falciparum isolates were cultured by the methods of Haynes et al. (16) and Trager and Jensen (38) with minor modifications as previously described (44). The medium, RPMI 1640 (GIBCO BRL, Paisley, United Kingdom) containing no para-aminobenzoic acid or folic acid, was supplemented with 10% normal human serum, 25 mM bicarbonate, and 25 mM HEPES buffer. PM and CCG were obtained from Sigma Chemical, Dorset, United Kingdom. Antimalarial activity was measured by using a radioisotopic technique as previously described (44). Results were expressed as the drug concentration required for 50% inhibition of [3H]hypoxanthine incorporation into parasite nucleic acid (IC50). Before the chemosensitivity test was carried out, the following conditions had to be fulfilled: (i) a parasitemia of at least 4% and (ii) a growth rate of at least threefold per 48 h.
DNA extraction.
A total of 3 to 4 ml of an in vitro culture from a blood sample with a 6% hematocrit and 4 to 5% parasitemia was centrifuged at 3,000 rpm for 5 min. The pellet obtained was lysed with 500 μl of 0.15% saponin for 5 min, and then 10 ml of TSE buffer (20 mM Tris [pH 8.0], 100 mM NaCl, 50 mM EDTA) was added. The mixture was centrifuged at 4,000 rpm for 10 min, and the resulting pellet was resuspended in 100 to 150 μl of TSE and stored at −20°C until processed. P. falciparum DNA was purified according to a protocol described elsewhere (27). Briefly, equal volumes (50 μl) of sample and preheated 5% Chelex-100 (Bio-Rad, Richmond, Calif.) were mixed together and heated for 10 min at 95 to 100°C. The Chelex was removed by centrifuging twice for 1 min at 15,000 × g, and 1 to 2 μl of the final supernatant was used for PCR.
PCR analysis.
The method used to detect point mutations is based on nested PCR: the first round of PCR allows the amplification of the entire DHFR domain, and the second round is carried out for the allele-specific amplification. The DHFR domain was amplified with primers SP1 (26) (5′-ATGATGGAACAAGTCTGCGAC-3′ [sense]) and 86 (52) (5′-TCATATGACATGTATCTT-3′ [antisense]). Antisense primers DIA3 (5′-GAATCCTTTCCCAGC-3′), DIA9 (5′-GAATGCTTTCCCAGG-3′), and DIA12 (5′-GGAATGCTTTCCCAGT-3) were used to detect, respectively, Ser, Thr, and Asn codons at position 108 with the common sense primer SP1 (26, 27). (Two sequences have been published for DIA3, 5′-GAATGCTTTCCCAGC-3′ and 5′-GAATCCTTTCCCAGC-3′, the latter of which contained a typographical error, substituting C for G at the 5th nucleotide. We used this second sequence in the current work, resulting in the need to use a slightly lower annealing temperature [Tm] to compensate for an extra mismatched base in the middle of the primer.) Asn and Ile codons at position 51 were detected by sense oligonucleotides FR51W (5′-TTACCATGGAAATGTAA-3′) and FR51M (5′-TTACCATGGAAATGTAT-3′), respectively (29), paired with antisense oligonucleotide 86. Antisense FR59W (5′-ATGTTGTAACTGCACA-3′) and FR59M (5′-ATGTTGTAACTGCACG-3′) allow the detection of Cys and Arg codons, respectively, at position 59 with the common sense primer SP1 (29). The wild-type Ile codon at position 164 was detected by antisense FR164W (5′-CAACGGAACCTCCTAT-3′) (15) and sense SP1 primers. Amplification of the DHFR domain was carried out under standard conditions. Two hundred micromolar concentrations of each deoxynucleoside triphosphate and a one micromolar concentration of each primer were used in the presence of 3 mM MgCl2. Amplification started at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 45°C for 45 s, and extension at 72°C for 45 s with 1.5 U of DNA polymerase enzyme (Taq polymerase) (Promega, Southampton, United Kingdom) in 50 μl of the reaction mixture. A total of 10 μl of the reaction mixture was taken for electrophoresis in an ethidium bromide-stained 1.5% agarose gel. If the band was seen, 1 μl of diluted (1:10, in sterile water) PCR product was subjected to the second round of PCR. In view of the wide range of possible parameters for optimal ASPCR conditions (8, 17) and based on previous observations (26, 27), careful upward and downward adjustment of PCR Tm, MgCl2 concentration, and PCR cycles was investigated to determine discriminative conditions. DIA3 was used at 52.5°C (Tm) for 20 cycles. All other sets of primers were amplified by using 15 cycles, and the Tm was 55°C for DIA9 and DIA12, 50°C for DIA51 and DIA52, 52.5°C for DIA59 and DIA60, and 55°C for DIA164. All PCRs were performed with 0.5 U of Taq polymerase, and 1.5 mM MgCl2 was the optimal concentration. Conditions were otherwise identical to those described for the first PCR. Amplified alleles were revealed with ethidium bromide staining after migration in 2% agarose gel. DIA3, DIA9, and DIA12 amplified a fragment of 350 bp. Specific amplification at codons 51, 59, and 164 showed, respectively, fragments of 630, 190, and 540 bp.
Three restriction enzymes distinguish the alleles at position 108 (12, 53) (AluI [Ser], ScrFI [Thr], and BsrI [Asn]) by yielding 350- and 430-bp fragments. Ten microliters of a 780-bp DHFR gene fragment amplified in the first round of PCR was precipitated by overnight incubation following the addition of sodium acetate (pH 5) (final concentration, 0.3 M) and 2.5 volumes of cold absolute ethanol. The DNA was pelleted by centrifugation (5 min, 15,000 × g), washed in 70% ethanol, air dried, and resuspended in 10 μl of sterile water. The DHFR product was analyzed by digestion with 2 U each of these enzymes (New England Biolabs, Beverly, Mass.) under the conditions recommended by the manufacturer. The products were resolved on a 2% agarose gel to visualize fragments.
Epi Info version 6 (Centers for Disease Control, Atlanta, Ga.) and Unistat-IV (Magelon, London, United Kingdom) packages were used for statistical analyses (P < 0.01, the chi-square or Student test) and graphing, respectively.
RESULTS
Sixty-nine P. falciparum isolates were adapted for growth in vitro and their responses to PM, CCG, SD, and DDS were determined. The ranges of IC50s were 0.1 to 2408.84 nM for PM and 0.03 to 37.26 nM for CCG. The IC50s of SD and DDS were 0.36 to 74.56 nM and 0.03 to 27.260 nM, respectively. These IC50s span the range from these expected for parasites that are sensitive to these drugs to those for parasites that are highly resistant. In addition, it is clear that CCG is more potent than PM and DDS is more potent than SD against representative field isolates from this area of Kenya (Kilifi).
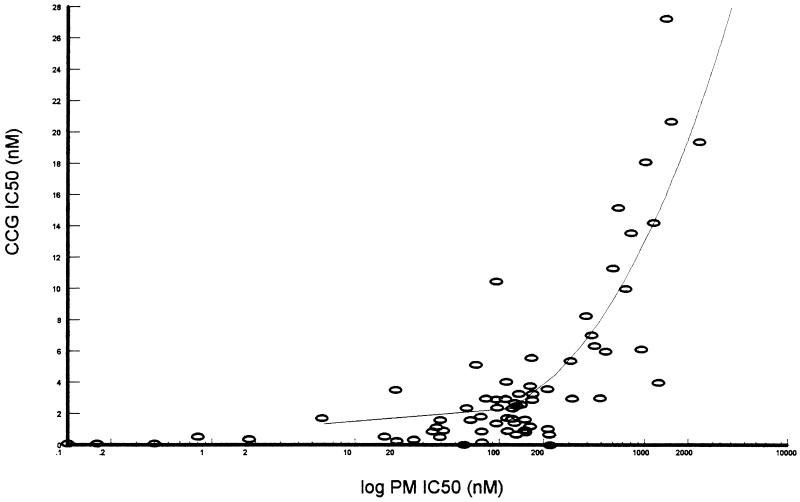
Of the 69 isolates, 37 were considered mixed isolates because amplification of two different alleles of DHFR was observed in the ASPCR. In order to address clearly the correlation between the DHFR genotype and the response to the drugs, we concentrated first on those isolates that yielded a PCR product with only one set of allele-specific oligonucleotide primers. All isolates that met this criterion were tested to determine their genotype for codons 51, 59, 108, and 164. All isolates contained the drug-sensitive Ile at position 164, and no isolates with the rather rare Thr-108 were detected. Table 1 presents the chemosensitivity profile and the DHFR genotypes at positions 51, 59, and 108 for these 32 isolates. There was a strong correlation between particular DHFR genotypes and sensitivity to PM and CCG. We observed three distinct groups based on the IC50s of PM and CCG. IC50s for the first group were within the range for drug-sensitive parasites; the mean IC50 of PM in this group was 3.71 ± 6.94 nM, and that of CCG was 0.24 ± 0.21 nM. These isolates all have the wild-type DHFR codons at positions 108, 51, and 59. The second group was composed of isolates for which the mean IC50 of PM was 92.88 ± 36.02 nM and that of CCG was 1.37 ± 0.65 nM. The mean IC50 of PM is 25-fold higher and that of CCG is 6-fold higher than the IC50 of these drugs for the sensitive group. Genetically, this group was heterogeneous. One isolate had a single change from the wild-type sequence to the Asn codon at position 108, one had an Asn codon at position 108 and the change encoding Ile-51, and seven isolates had both Asn-108 and Arg-59 codons. Parasites in which the DHFR has undergone mutations at all three positions to Ile-51, Arg-59, and Asn-108 comprise the largest group. The mean IC50 of PM was 815.25 ± 582 nM and that of CCG was 10.8 ± 7.2 nM, demonstrating the second step in resistance. These parasites were 225-fold more resistant to PM and 48-fold more resistant to CCG than the sensitive group. Despite the fairly wide range of IC50s measured within each group, the differences among the groups were statistically significant (P < 0.01). We have also established that cross-resistance between PM and CCG is characterized by a polynomial function of the form y = ax3 + bx2 + cx + d (r2 = 0.80, df = 68). The decrease in the CCG IC50s is pronounced only when the PM IC50 increases by more than three logarithmic cycles (triple mutants), thus confirming the high potency of CCG compared with PM. Finally, from the wild-type parasites to the triple mutants, the PM and CCG IC50s increase, respectively, 220- and 45-fold. All these changes in IC50s are statistically significant for both antifolates (P < 0.01) (Fig. 1).
TABLE 1.
Relationship between DHFR point mutations in Kenyan field isolates of P. falciparum and in vitro chemosensitivity to the antifolates PM and CCG
| DHFR phenotypea | IC50 (n)b
|
|
|---|---|---|
| PM | CCG | |
| Group 1 | ||
| 108(−) 51(−) 59(−) | 3.71 ± 6.94 (5) | 0.24 ± 0.21 (5) |
| Group 2 | ||
| 108(+) 51(−) 59(−) | 150.20 (1) | 0.81 (1) |
| 108(+) 51(+) 59(−) | 95.18 (1) | 1.38 (1) |
| 108(+) 51(−) 59(+) | 84.36 ± 12.5 (7) | 1.48 ± 0.94 (7) |
| Total | 92.88 ± 36.02 (9) | 1.37 ± 0.85 (9) |
| Group 3 | ||
| 108(+) 51(+) 59(+) | 815.25 ± 582 (18) | 10.80 ± 7.2 (18) |
108(−), Ser-108 (wild type); 108(+), Asn-108; 51(−), Asn-51 (wild type); 51(+), Ile-51; 59(−), Cys-59 (wild type); 59(+), Arg-59.
IC50s are expressed in nanomolar concentrations; values are means ± standard deviations; P < 0.01 for all values.
FIG. 1.
Relationship between PM and CCG in vitro chemosensitivity of Kenyan field isolates. Values are IC50s of CCG in and logarithmic cycles of IC50 of PM, both in nanomolar concentrations.
We also analyzed samples that contained more than one DHFR allele. When these were grouped according to the IC50s of PM and CCG, the same trend was observed. Isolates fell into three distinct groups: Asn-51, Cys-59, and Ser-108 (first group); Asn-108 with or without Asn-51 or Cys-59 (second group); and Ile-51, Arg-59, and Asn-108 (third group). However, in these mixed isolates, the IC50 depends both on the intrinsic sensitivity of the alleles present and the relative proportions of the alleles, so the discrimination between groups and the mean IC50s for them was less pronounced.
Because we are also interested in the responses of these isolates to sulfa drugs, we tested their sensitivity to SD and DDS (Table 2). These drugs are thought to target the DHPS enzyme in folate synthesis, not DHFR, so we were surprised to see that the sensitivity of the isolates to SD and DDS also fell into two distinct groups corresponding to the DHFR genotype of the parasites. For the triple mutants (Ile-51, Arg-59, and Asn-108) IC50s of SD were 29.77 ± 17.3 μM and those of DDS were 12.0 ± 8.5 μM. For all the isolates with DHFR alleles of the other genotypes, IC50s of SD were 11.97 ± 10.73 μM and those of DDS were 2.89 ± 3.34 μM, indicating that the isolates were four- and twofold more sensitive than the triple-mutant group. Despite the wide range of values, this difference was highly significant (P < 0.01) for both drugs. This reduction in sensitivity affected more than half of the isolates assayed, so it represents a change that may be important clinically.
TABLE 2.
Relationship between DHFR point mutations in Kenyan field isolates of P. falciparum and in vitro chemosensitivity to the antifolates SD and DDS
| DHFR phenotypea | IC50 (n)b
|
|
|---|---|---|
| SD | DDS | |
| Group 1 | ||
| 108(−) 51(−) 59(−) | 7.42 ± 6.0 (5) | 1.27 ± 2.1 (5) |
| 108(+) 51(−) 59(−) | 37.20 (1) | 2.98 (1) |
| 108(+) 51(+) 59(−) | 18.79 (1) | 10.16 (1) |
| 108(+) 51(−) 59(+) | 10.64 ± 9.6 (7) | 2.98 ± 3.0 (7) |
| Total | 11.97 ± 10.73 (14) | 2.98 ± 3.24 (14) |
| Group 2 | ||
| 108(+) 51(+) 59(+) | 29.77 ± 17.3 (18) | 12.0 ± 8.5 (18) |
108(−), Ser-108 (wild type); 108(+), Asn-108; 51(−), Asn-51 (wild type); 51(+), Ile-51; 59(−), Cys-59 (wild type); 59(+), Arg-59.
IC50s are expressed in micromolar concentrations; values are means ± standard deviations; P < 0.01 for all values.
It is important to determine the contribution of each mutation to the drug sensitivity of the enzyme the gene encodes. This has been studied by using purified enzymes in vitro but not in field samples. To begin this study, we collected the data in a different way, as shown in Table 3. Here, the IC50s of PM and of CG are correlated with the data showing whether the isolates express the codon in the drug-sensitive or mutated form. In all cases, the IC50s for isolates with the wild-type codon were lowest, the IC50s for those with mutated alleles were highest, and the IC50s for mixed populations of alleles lay between these two extremes. These differences were also highly significant statistically (P < 0.001).
TABLE 3.
Effect of each DHFR point mutation irrespective of the other two on PM and CCG IC50s for Kenyan P. falciparum field isolates
| DHFR genotype | IC50a for isolates with mutation at codon (n)
|
|||||
|---|---|---|---|---|---|---|
| 108
|
51
|
59
|
||||
| PM | CCG | PM | CCG | PM | CCG | |
| Wild type | 3.71 ± 6.94 (5) | 0.24 ± 0.20 (5) | 66.20 ± 68.60 (24) | 1.46 ± 2.30 (24) | 34.70 ± 44.21 (15) | 1.178 ± 1.43 (15) |
| Mixed alleles | 97.52 ± 99.70 (24) | 2.28 ± 2.20 (24) | 140.18 ± 83.63 (21) | 2.38 ± 1.71 (21) | 148.11 ± 96.10 (12) | 2.52 ± 2.26 (12) |
| Mutated alleles | 447.60 ± 515.09 (40) | 6.00 ± 6.57 (40) | 646.18 ± 588.98 (24) | 8.68 ± 7.23 (24) | 427.74 ± 511.42 (42) | 5.91 ± 6.47 (42) |
IC50s are in nanomolar concentrations; P < 0.01 for each group compared with the other two.
As reported in previous studies, we found that a Ser-to-Asn mutation at position 108 was a precondition for antifolate resistance and that point mutations at codons 51 and 59 occurred independently. In an attempt to address the order of appearance of point mutations at codons 51 and 59, we determined the frequencies of these mutations in populations with single alleles for DHFR. The results show that alleles that carried a mutation at codon 59 (61%) were more frequent than those that carried a mutation at codon 51 (35%). This difference was significant (P < 0.05), which may indicate the earlier appearance of the mutation at codon 59 or a stronger selection for the mutation at codon 59 than that at codon 51 if we exclude random selection.
DISCUSSION
Our findings provide further information on the association between DHFR genotype and parasite chemosensitivity, in addition to confirming previous reports. We found a strong correlation between antifolate drug response and DHFR genotype, as previously reported for laboratory reference strains (10, 24, 25, 55), field isolates (3), and purified enzyme measured in vitro (34). The occurrence of point mutations at codons 51 and 59 did not affect significantly the profile of chemosensitivity to PM and CCG compared to that of isolates with the codon 108 mutation alone. This observation, obtained with field isolates, is not consistent with those obtained with culture-adapted strains or studies of purified enzyme in vitro (34). In these situations, the ancillary mutations at codon 51 or 59 gave rise to a significantly higher level of PM resistance than a mutation at codon 108 alone (10, 25, 55).
When the activity of an enzyme that carried both Asn-108 and Arg-59 was measured in vitro, it had a significantly lower kcat than the wild-type enzyme or an enzyme with the Asn-108-Ile-51 combination (34). If this is the case in vivo, one would not expect the corresponding genotype to predominate in the field populations. However, 7 of 32 of our populations did show both mutations in the same isolate. Previous studies compared single laboratory reference strains, and this discrepancy could result from differences in genetic background among the isolates studied directly in the field. We have found that only when point mutations occurred at codons 51 and 59 together with the Asn-108 mutation was resistance to PM and CCG significantly increased.
In our study, no Thr-108 was detected, confirming the rarity of this phenotype in field isolates (3, 4, 26, 29). This mutation had been described only in laboratory reference strains, is always paired with a substitution of Ala for Val at codon 16, and is associated with CG resistance (14, 25). Recently, however, one field isolate from Papua New Guinea with both Thr-108 and Ala-16 was described (31). The substitution of Ile-164 for Leu-164 has been found only where resistance to PM-SD is well established, e.g., in Southeast Asia (3, 4, 54) and Bolivia (29). Our results and those reported by Plowe et al. (29) and Wang et al. (42) show that this mutation is not currently frequent in field isolates from the Kilifi region. Recently, new mutations (29, 42, 54) and a repetitive insertion (29) associated with antifolate resistance in DHFR have been described. Although isolates in this study were not screened for these new genotypes, none of the other Kenyan field isolates analyzed have these genetic modifications (29, 42).
Several earlier studies pointed out that sulfa drugs may act against P. falciparum by mechanisms other than the inhibition of DHPS (9, 11, 19, 43). Our results showing a correlation between response to the sulfa drugs and DHFR genotype is inconclusive, since we did not analyze DHPS genotypes, although the involvement of DHFR in sulfa drug activity has been reported to occur in bacteria (30) and in Plasmodium chabaudi (33). Several studies have suggested that point mutations in DHPS may underlie sulfa drug resistance in P. falciparum isolates (7, 29, 40, 41). However, our study did not address this issue because of our current working hypothesis that the primary mutations that govern parasite resistance to PM-SD occur in DHFR (47).
The high potencies of CCG compared with PM and of DDS compared with SD in vitro confirm observations from other in vitro studies (50) and of field isolates (39). We also observed cross-resistance between PM and CCG, but this phenomenon was pronounced only in isolates that carried the triple mutation in the DHFR. Even on these highly PM-resistant isolates, CCG remains effective since the change in IC50 from that for wild type is about 50-fold compared to a change of more than 225-fold in the IC50 of PM. This is comparable to results obtained with laboratory reference strains (47).
Surprisingly, only about 8% of Kenyan field isolates now have the wild-type DHFR, which indicates the operation of a powerful selective pressure over recent years. In Kenya, during the 1980s, PM-SD was rarely used for malaria treatment because it offered no advantage over CQ. Owing to the spread of CQ resistance (5, 6, 22), PM-SD has been used increasingly as the first-line treatment of uncomplicated malaria in this region. In Kilifi, this change from CQ to PM-SD was made in 1992. Based on in vitro analysis in 1986, Spencer et al. (37) found nearly 30% of the isolates to be resistant to PM in Malindi, along the Kenyan coast (this proportion was probably high since the assay employed normal medium rather than medium with no added folic acid). Two subsequent in vitro studies in the same area, in 1986 in Jilore (44) and in 1987 to 1989 in Kilifi (46), showed approximately 20% of the isolates to be resistant to PM. Parasites in our study were collected between 1993 and 1995 in Kilifi, and results show that more than 92% of isolates are resistant to PM. Thus, within a short time, there has been a major increase in the prevalence of PM-resistant isolates. In this area, there are no sources of PM-SD other than the district hospital. For home-based treatment, most patients are treated with either CQ or an antipyretic agent (20). In spite of limited availability and restricted use of PM-SD, PM chemosensitivity has decreased over a short time, demonstrating the powerful selection that results from the use of this particular antimalarial treatment. Our findings are of great concern since there is no affordable alternative antimalarial drug to replace PM-SD.
PM-SD treatment was not associated with clinical failure in the Kilifi trial from which our isolates were obtained. However, return of parasitemia within 21 days is now common, in contrast to the situation in the early 1980s (36), indicating low-level drug resistance (2). This early return of parasitemia after PM-SD treatment is likely to be associated with a high prevalence of parasites carrying the triple mutations in DHFR, which represent nearly 60% of our isolates. In Tanzania, infections which exhibit an R1 response to PM-SD remain sensitive to the CPG-DDS combination (39), a fact which is supported by pharmacodynamic and pharmacokinetic theory (47). Since CPG-DDS is a short-half-life combination (45, 51), immediate introduction into operational use in East Africa may delay further selection of antifolate resistance factors. Studies by us and others in East Africa indicate that the mutation at codon 164, which is associated with high-level resistance to all antifolate treatment drugs (47), at present must occur at a very low frequency, since it has not been detected (29). Continued use of PM-SD with the associated strong resistance-selective pressure (46) is likely to quickly select for the mutation at codon 164, as has happened in Southeast Asia. Once this occurs, CPG-DDS will probably also become ineffective as a short-course treatment of malaria (49). An important opportunity to extend the useful therapeutic life of antifolate antimalarial drugs in Africa exists.
ACKNOWLEDGMENTS
We thank the Director of KEMRI, for permission to publish these results.
The work was supported by the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR) and the Wellcome Trust. A.M.N. is grateful to the University of Washington, Seattle, for technical support. W.M.W. and P.A.W. are grateful to Smithkline Beecham Pharmaceuticals for financial support. W.M.W. and E.K.M. are grateful to the Wellcome Trust of Great Britain for personal and project support (WT grant reference 045010).
REFERENCES
- 1.Amukoye E, Winstanley P A, Watkins W M, Snow R W, Hatcher J, Mosobo M, Ngumbao E, Lowe B, Ton M, Minyiri G, Marsh K. Chlorproguanil-dapsone: effective treatment for uncomplicated falciparum malaria. Antimicrob Agents Chemother. 1997;41:2261–2264. doi: 10.1128/aac.41.10.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anabwani G M, Esamai F O, Menya D A. A randomised controlled trial to assess the relative efficacy of chloroquine, amodiaquine, halofantrine and Fansidar® in the treatment of uncomplicated malaria in children. East Afr Med J. 1996;73:155–158. [PubMed] [Google Scholar]
- 3.Basco L K, Eldin de Pecoulas P, Wilson C M, Le Bras J, Mazabraud A. Point mutations in the dihydrofolate reductase-thymidylate synthase gene and pyrimethamine and cycloguanil resistance in Plasmodium falciparum. Mol Biochem Parasitol. 1995;69:135–138. doi: 10.1016/0166-6851(94)00207-4. [DOI] [PubMed] [Google Scholar]
- 4.Basco L K, Eldin de Pecoulas P, Le Bras J, Wilson C M. Plasmodium falciparum: molecular characterization of multidrug-resistant Cambodian isolates. Exp Parasitol. 1996;82:97–103. doi: 10.1006/expr.1996.0013. [DOI] [PubMed] [Google Scholar]
- 5.Bloland P B, Lackritz E M, Kazembe P N, Were J B, Steketee R, Campbell C C. Beyond chloroquine: implications of drug resistance for evaluating malaria therapy efficacy and treatment policy in Africa. J Infect Dis. 1993;167:932–937. doi: 10.1093/infdis/167.4.932. [DOI] [PubMed] [Google Scholar]
- 6.Brandling Bennett A D, Oloo A J, Watkins W M, Boriga D A, Kariuki D M, Collins W E. Chloroquine treatment of falciparum malaria in an area of Kenya of intermediate chloroquine resistance. Trans R Soc Trop Med Hyg. 1988;82:833–837. doi: 10.1016/0035-9203(88)90009-0. [DOI] [PubMed] [Google Scholar]
- 7.Brooks D R, Wang P, Read M, Watkins W M, Sims P F, Hyde J E. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur J Biochem. 1994;224:397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- 8.Cha R S, Zarlb H, Keohavong P, Thilly W G. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Applications. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 9.Chulay J D, Watkins W M, Sixsmith D G. Synergistic antimalarial activity of pyrimethamine and sulfadoxine against Plasmodium falciparum in vitro. Am J Trop Med Hyg. 1984;33:325–330. doi: 10.4269/ajtmh.1984.33.325. [DOI] [PubMed] [Google Scholar]
- 10.Cowman A F, Morry M J, Biggs B A, Cross G A, Foote S J. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dieckmann A, Jung A. Mechanisms of sulfadoxine resistance in Plasmodium falciparum. Mol Biochem Parasitol. 1986;19:143–147. doi: 10.1016/0166-6851(86)90119-2. [DOI] [PubMed] [Google Scholar]
- 12.Eldin de Pecoulas P, Basco L K, Abdallah B, Dje M K, Le Bras J, Mazabraud A. Plasmodium falciparum: detection of antifolate resistance by mutation-specific restriction enzyme digestion. Exp Parasitol. 1995;80:483–487. doi: 10.1006/expr.1995.1060. [DOI] [PubMed] [Google Scholar]
- 13.Ferone R. Folate metabolism in malaria. Bull W H O. 1977;55:291–298. [PMC free article] [PubMed] [Google Scholar]
- 14.Foote S J, Galatis D, Cowman A F. Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc Natl Acad Sci USA. 1990;87:3014–3017. doi: 10.1073/pnas.87.8.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gyang F N, Peterson D S, Wellems T E. Plasmodium falciparum: rapid detection of dihydrofolate reductase mutations that confer resistance to cycloguanil and pyrimethamine. Exp Parasitol. 1992;74:470–472. doi: 10.1016/0014-4894(92)90209-s. [DOI] [PubMed] [Google Scholar]
- 16.Haynes J D, Diggs C L, Hines F A, Desjardins R E. Culture of human malaria parasite Plasmodium falciparum. Nature. 1976;263:767–769. doi: 10.1038/263767a0. [DOI] [PubMed] [Google Scholar]
- 17.Huang M M, Arhneim N, Goodman M F. Extension of base mispairs by Taq polymerase: implication for single nucleotide discrimination in PCR. Nucleic Acids Res. 1992;20:4567–4573. doi: 10.1093/nar/20.17.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mberu, E. K., and W. M. Watkins. Unpublished data.
- 19.Milhous W K, Weatherly N F, Bowdre J H, Desjardins R E. In vitro activities of and mechanisms of resistance to antifol antimalarial drugs. Antimicrob Agents Chemother. 1985;27:525–530. doi: 10.1128/aac.27.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mwenesi H, Harpham T, Marsh K, Snow R W. Child malaria treatment among mothers in Kenya. Soc Sci Med. 1995;49:1271–1277. doi: 10.1016/0277-9536(94)00250-w. [DOI] [PubMed] [Google Scholar]
- 21.Newton C R, Heptinstall L E, Summers C, Super M, Schwarz M, Anwar R, Graham A, Smith J C, Markham A F. Amplification refractory mutation system for prenatal diagnosis and carrier assessment in cystic fibrosis. Lancet. 1989;ii:1481–1483. doi: 10.1016/s0140-6736(89)92931-0. [DOI] [PubMed] [Google Scholar]
- 22.Pasvol G, Newton C R, Winstanley P A, Watkins W M, Peshu N M, Were J B, Marsh K, Warrell D A. Quinine treatment of severe falciparum malaria in African children: a randomized comparison of three regimens. Am J Trop Med Hyg. 1991;45:702–713. doi: 10.4269/ajtmh.1991.45.702. [DOI] [PubMed] [Google Scholar]
- 23.Payne D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol Today. 1987;3:241–246. doi: 10.1016/0169-4758(87)90147-5. [DOI] [PubMed] [Google Scholar]
- 24.Peterson D S, Walliker D, Wellems T E. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson D S, Milhous W K, Wellems T E. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci USA. 1990;87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson D S, Di Santi S M, Povoa M, Calvosa V S, do Rosario V E, Wellems T E. Prevalence of the dihydrofolate reductase Asn-108 mutation as the basis for pyrimethamine-resistant falciparum malaria in the Brazilian Amazon. Am J Trop Med Hyg. 1991;45:492–497. doi: 10.4269/ajtmh.1991.45.492. [DOI] [PubMed] [Google Scholar]
- 27.Plowe C V, Djimde A, Bouare M, Doumbo O, Wellems T E. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- 28.Plowe C V, Djimde A, Wellems T E, Kouriba B, Doumbo O K. Community pyrimethamine use and prevalence of resistant Plasmodium falciparum genotypes in Mali: a model for deterring resistance. Am J Trop Med Hyg. 1996;55:467–471. doi: 10.4269/ajtmh.1996.55.467. [DOI] [PubMed] [Google Scholar]
- 29.Plowe C V, Cortese J F, Djimde A, Nwanyanwu O C, Watkins W M, Winstanley P A, Estrada-Franco J G, Mollinedo R E, Avila J C, Cespedes J L, Carter D, Doumbo O K. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis. 1997;176:1590–1596. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- 30.Poe M. Antibacterial synergism: a proposal for chemotherapeutic potentiation between trimethoprim and sulfamethoxazole. Science. 1976;194:533–535. doi: 10.1126/science.788154. [DOI] [PubMed] [Google Scholar]
- 31.Reeder J C, Rieckmann K H, Genton B L, Lorry K, Wines B, Cowman A F. Point mutation in the dihydrofolate reductase and dihydropteroate synthase genes and in vitro susceptibility to pyrimethamine and cycloguanil of Plasmodium falciparum isolates from Papua New Guinea. Am J Trop Med Hyg. 1996;55:209–213. doi: 10.4269/ajtmh.1996.55.209. [DOI] [PubMed] [Google Scholar]
- 32.Sarkar G, Cassady J, Bottema C D, Sommer S S. Characterization of polymerase chain reaction amplification of specific alleles. Anal Biochem. 1990;186:64–68. doi: 10.1016/0003-2697(90)90573-r. [DOI] [PubMed] [Google Scholar]
- 33.Sirawaraporn W, Yuthavong Y. Potentiating effect of pyrimethamine and sulfadoxine against dihydrofolate reductase from pyrimethamine-sensitive and pyrimethamine-resistant Plasmodium chabaudi. Antimicrob Agents Chemother. 1986;29:899–905. doi: 10.1128/aac.29.5.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirawaraporn W, Sathitkul T, Sirawaraporn R, Yuthavong Y, Santi D V. Antifolates-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc Natl Acad Sci USA. 1997;94:1124–1129. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sommer S S, Groszbach A R, Bottema C D. PCR amplification of specific alleles (PASA) is a general method for rapidly detecting known single-base changes. BioTechniques. 1992;12:82–87. [PubMed] [Google Scholar]
- 36.Spencer H C, Watkins W M, Sixsmith D G, Koech D K, Chulay J D. A new in vitro test for pyrimethamine/sulfadoxine susceptibility of Plasmodium falciparum and its correlation with in vivo resistance in Kenya. Bull W H O. 1984;62:615–621. [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer H C, Watkins W M, Sixsmith D G, Koech D K. Response of Plasmodium falciparum to dihydrofolate reductase inhibitors in Malindi, Kenya. Trans R Soc Trop Med Hyg. 1986;80:201–203. doi: 10.1016/0035-9203(86)90009-x. [DOI] [PubMed] [Google Scholar]
- 38.Trager W, Jensen J B. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 39.Trigg J K, Mbwana H, Chambo O, Hills E, Watkins W M, Curtis C F. Resistance to pyrimathamine/sulfadoxine in Plasmodium falciparum in 12 villages in north east Tanzania and a test of chlorproguanil/dapsone. Acta Trop. 1997;63:185–189. doi: 10.1016/s0001-706x(96)00617-1. [DOI] [PubMed] [Google Scholar]
- 40.Triglia T, Cowman A F. Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1994;91:7149–7153. doi: 10.1073/pnas.91.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P, Read M, Sims P F G, Hyde J E. Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutation in dihydropteroate synthethase and an additional factor associated with folate utilization. Mol Microbiol. 1997;23:979–986. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang P, Lee C S, Bayoumi R, Djimde A, Doumbo O, Swedberg G, Dao L D, Mshinda H, Tanner M, Watkins W M, Sims P F G, Hyde J E. Resistance to antifolates in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthase and dihydrofolate reductase alleles in a large number of field isolates of diverse origins. Mol Biochem Parasitol. 1997;89:161–177. doi: 10.1016/s0166-6851(97)00114-x. [DOI] [PubMed] [Google Scholar]
- 43.Watkins W M, Sixsmith D G, Chulay J D, Spencer H C. Antagonism of sulfadoxine and pyrimethamine antimalarial activity in vitro by p-aminobenzoic acid, p-aminobenzoylglutamic acid and folic acid. Mol Biochem Parasitol. 1985;14:55–61. doi: 10.1016/0166-6851(85)90105-7. [DOI] [PubMed] [Google Scholar]
- 44.Watkins W M, Howells R E, Brandling Bennett A D, Koech D K. In vitro susceptibility of Plasmodium falciparum isolates from Jilore, Kenya, to antimalarial drugs. Am J Trop Med Hyg. 1987;37:445–451. doi: 10.4269/ajtmh.1987.37.445. [DOI] [PubMed] [Google Scholar]
- 45.Watkins W M, Chulay J D, Sixsmith D G, Spencer H C, Howells R E. A preliminary pharmacokinetic study of the antimalarial drugs, proguanil and chlorproguanil. J Pharm Pharmacol. 1987;39:261–265. doi: 10.1111/j.2042-7158.1987.tb06263.x. [DOI] [PubMed] [Google Scholar]
- 46.Watkins W M, Mosobo M. Treatment of Plasmodium falciparum malaria with pyrimethamine-sulfadoxine: selective pressure for resistance is a function of long elimination half-life. Trans R Soc Trop Med Hyg. 1993;87:75–78. doi: 10.1016/0035-9203(93)90431-o. [DOI] [PubMed] [Google Scholar]
- 47.Watkins W M, Mberu E K, Winstanley P A, Plowe C. The efficacy of antifolate antimalarial combination in Africa: a predictive model based on pharmacodynamic and pharmacokinetic analyses. Parasitol Today. 1997;13:459–464. doi: 10.1016/s0169-4758(97)01124-1. [DOI] [PubMed] [Google Scholar]
- 48.Wernsdorfer W, Payne D. The dynamics of drug resistance in Plasmodium falciparum. Pharmacol Ther. 1991;50:95–121. doi: 10.1016/0163-7258(91)90074-v. [DOI] [PubMed] [Google Scholar]
- 49.Wilairatana D E, Kyle P, Looaresuwan S, Chingwongprom K, Amradee S, White N J, Watkins W M. Efficacy of combinations of dapsone with proguanil and dapsone with chlorproguanil for acute uncomplicated falciparum malaria in Thailand. Ann Trop Med Parasitol. 1996;91:125–132. [PubMed] [Google Scholar]
- 50.Winstanley P A, Mberu E K, Szwandt I S F, Breckenridge A M, Watkins W M. In vitro activities of novel antifolate drug combinations against Plasmodium falciparum and human granulocyte CFUs. Antimicrob Agents Chemother. 1995;39:948–952. doi: 10.1128/aac.39.4.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winstanley P A, Watkins W M, Muhia D, Szwandt I S F, Amukoye E, Marsh K. Chlorcycloguanil-dapsone for uncomplicated falciparum malaria in young children: pharmacokinetics and therapeutic range. Trans R Soc Trop Med Hyg. 1997;91:322–327. doi: 10.1016/s0035-9203(97)90093-6. [DOI] [PubMed] [Google Scholar]
- 52.Wooden J M, Hartwell L H, Vasquez B, Sibley C H. Analysis of antimalarial drugs that target the dihydrofolate reductase of Plasmodium falciparum. Mol Biochem Parasitol. 1997;85:24–40. doi: 10.1016/s0166-6851(96)02808-3. [DOI] [PubMed] [Google Scholar]
- 53.Zindrou S, Dao L D, Xuyen P T, Dung N P, Sy N D, Skold O, Swedberg G. Rapid detection of pyrimethamine susceptibility of Plasmodium falciparum by restriction endonuclease digestion of the dihydrofolate reductase gene. Am J Trop Med Hyg. 1996;5:185–188. doi: 10.4269/ajtmh.1996.54.185. [DOI] [PubMed] [Google Scholar]
- 54.Zindrou S, Dung N P, Sy N D, Skold O, Swedberg G. Plasmodium falciparum: mutation pattern in the dihydrofolate reductase-thymidylate synthase genes of Vietnamese isolates, a novel mutation, and coexistence of two clones in a Thai patient. Exp Parasitol. 1996;84:56–64. doi: 10.1006/expr.1996.0089. [DOI] [PubMed] [Google Scholar]
- 55.Zolg J W, Plitt J R, Chen G X, Palmer S. Point mutations in the dihydrofolate reductase-thymidylate synthase gene as the molecular basis for pyrimethamine resistance in Plasmodium falciparum. Mol Biochem Parasitol. 1989;36:253–262. doi: 10.1016/0166-6851(89)90173-4. [DOI] [PubMed] [Google Scholar]