Abstract
Progressive multifocal leukoencephalopathy (PML) is a severe neurological condition caused by reactivation of JC polyomavirus (JCPyV) in immunosuppression. Asymptomatic JCPyV persists in peripheral tissues. Upon reactivation, neurotropic rearrangements may emerge, and the virus gains access to the brain. To assess the mechanisms of PML pathogenesis, brain tissue material from PML patients was collected for small RNA sequencing. Upregulation of 8 microRNAs (miRNAs) in PML brain was validated using quantitative microRNA polymerase chain reaction (PCR). Bioinformatics tools were utilized to identify major associations of the upregulated miRNAs: neuroinflammation and blood-brain barrier disruption. The results indicate involvement of human miRNA regulation in PML pathogenesis.
Keywords: JC polyomavirus, JCPyV, progressive multifocal leukoencephalopathy, PML, miRNA, microRNA
To assess mechanisms of PML pathogenesis, brain tissue material from PML patients was collected for miRNA sequencing. Upregulation of 8 miRNAs in PML brain was observed. Major associations of upregulated miRNAs include neuroinflammation and blood-brain barrier permeability and damage.
Progressive multifocal leukoencephalopathy (PML), a human demyelinating neurodegenerative disease, is caused by reactivation of JC polyomavirus (JCPyV). JCPyV is ubiquitous and is carried by more than half of the global population. Upon reactivation, replication of peripheral JCPyV is enhanced, neurotropic rearrangements emerge, typically in the noncoding control region (NCCR) and the viral capsid protein (VP1) gene, and the virus gains access to the brain [1]. PML is caused by lytic JCPyV infection in glial cells, leading to destruction of brain white matter. The trigger for JCPyV reactivation is unknown but the risk is elevated in patients with weakened immune defense such as patients with HIV, patients with multiple sclerosis receiving immunomodulatory biological drugs, transplant patients receiving immunosuppressive medication, and patients with malignancies, especially lymphoma [2]. In this present work we have assessed microRNA (miRNA) expression in brain tissue samples from living and deceased PML patients to enlighten the mechanisms of brain pathogenesis.
METHODS
Study Population
The study population consisted of 9 PML patients and 2 control individuals described in detail in Supplementary Material. The study was approved by the ethics board of Helsinki University Hospital (HUS/2586/2019). Informed consent was obtained from all participants. PML diagnosis of all cases was confirmed by immunohistochemical staining for JCPyV large T antigen (Supplementary Figure 1). Six case samples were rare biopsies from living patients, while postmortem samples were acquired from 3 cases and 2 controls. Five cases had a disease or medication predisposing to PML, including chronic lymphocytic leukemia (cases 4, 5, and 9), myelofibrosis (case 3), dermatomyositis (case 7), and medications such as fludarabine (cases 4, 5, and 9), bendamustine (cases 4 and 5), rituximab (cases 4, 5, 7, and 9), as well as cyclosporine A, methotrexate, azathioprine, and prednisolone (case 7) [2] (Table 1). Interestingly, as many as 4 PML cases (cases 1, 2, 6, and 8) had no known immunosuppressive disease or medication prior to diagnosis. Cases 1–4 and 7–9 were tested human immunodeficiency virus (HIV) negative, while the HIV status of cases 5 and 6 is unknown.
Table 1.
Description of the Study Population
| Age, y | Male/Female | Biopsy/Post mortem | Location of the Lesion | Immunosuppressive Medication | Disease History | |
|---|---|---|---|---|---|---|
| Case 1 | 72 | Male | Biopsy | Parietal lobe | None | None |
| Case 2 | 70 | Male | Biopsy | Fronto-parietal lobe | None | Coronary atherosclerosis and asthma |
| Case 3 | 64 | Male | Biopsy | Parietal lobe | Hydroxyurea | Myelofibrosis |
| Case 4 | 71 | Male | Biopsy | Occipital-parietal lobe | Fludarabine, rituximab, bendamustine | Adenocarcinoma, chronic lymphocytic leukemia |
| Case 5 | 79 | Female | Biopsy | Occipital lobe | Fludarabine, rituximab, bendamustine | Chronic lymphocytic leukemia |
| Case 6 | 78 | Female | Post mortem | Fronto-parietal lobe | None | Type-2 diabetes, hypertension, hypercholesterolemia, osteoporosis, and atrial fibrillation |
| Case 7 | 46 | Female | Post mortem | Nucleus lentiformis | Prednisolone, cyclosporine A, intravenous immunoglobulin, methotrexate, azathioprine, rituximab | Dermatomyositis |
| Case 8 | 80 | Female | Biopsy | Parietal lobe | None | Breast cancer, hypertension, asthma |
| Case 9 | 83 | Female | Post mortem | Frontal lobe | Fludarabine, rituximab | Chronic lymphocytic leukemia |
| Control 1 | NA | NA | Post mortem | Frontal lobe | NA | NA |
| Control 2 | NA | NA | Post mortem | Frontal lobe | NA | NA |
Abbreviation: NA, not analyzed.
Study Design
Brain tissue from 8 PML patients (cases 1–8) and 2 controls (controls 1 and 2) were included in miRNA sequencing. Repeated attempts to prepare a sequencing library from case 9 sample were unsuccessful. Further validation was done with 8 cases (1–3 and 5–9) and 2 controls (1 and 2). Case 4 sample was inadequate for validation. The sequencing process and validation of the findings are described in detail in Supplementary Material. Briefly, RNA was extracted using RNeasy FFPE Kit (Qiagen) to prepare libraries for small RNA sequencing. Sequencing was performed with a NextSeq500 instrument (Illumina) 75-bp single end kit. Advanced TaqMan miRNA assays (Thermo Fisher Scientific) were used to validate differential expression of 8 human miRNAs and expression of JCPyV encoded JCV-miR-J1-5p and for JCV-miR-J1-3p.
RESULTS
Small RNA Sequencing and Validation
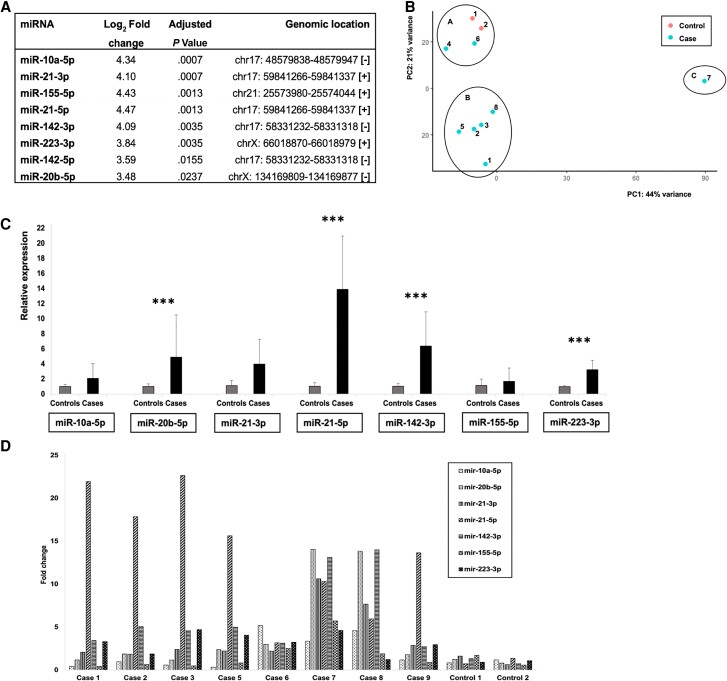
We originally identified 10 differentially expressed (DE) miRNAs using threshold values log2 fold change >2 and adjusted P value <.05 (Figure 1A). Nine of the DE miRNAs were upregulated and 1 downregulated in PML brain samples as compared to controls. The upregulated miRNAs were hsa-miR-10a-5p, hsa-miR-20b-5p, hsa-miR-21-3p, hsa-miR-21-5p, hsa-miR-142-3p, hsa-miR-142-5p, hsa-miR-155-5p, hsa-miR-223-3p, and hsa-miR-664a-3p, which all appeared to have log2 fold change as high as >3. The downregulated miRNA was hsa-miR-4800-3p. hsa-miR-664a-3p and hsa-miR-4800-3p were excluded from further analyses because they do not belong to conserved miRNA families and may represent star miRNAs or RNA fragments misannotated as miRNAs (Supplementary Material). Intriguingly, genes encoding 5 out of the 8 DE miRNAs are located in chromosome 17 (Figure 1A). Additionally, sequencing reads representing the JCPyV encoded JCV-miR-J1-5p, as well as JCV-J1-3p encoded by JCPyV and related BK polyomavirus (BKPyV), were found in all brain tissue samples, except that JCV-miR-J1-5p was not identified in case 7 (Supplementary Table 1). Also, control 1 had very low counts of reads representing both JCPyV miRNA reads. Hereafter the human miRNAs are referred to without the prefix hsa-.
Figure 1.
MicroRNA (miRNA) expression in progressive multifocal leukoencephalopathy (PML) and control brain samples. A, Log2 fold changes and adjusted P values of differentially expressed miRNAs in brain tissue of PML patients versus controls. Genomic location of each differentially expressed miRNA gene is also listed. B, Principal component analysis (PCA) based on normal transformation. Clustering of PML brain tissue samples and controls to 3 clusters, A, B, and C, based on miRNA expression patterns. C, Relative expression of each miRNA in cases versus controls, average, and standard deviation, normalized to the mean expression of miR-191-5p. miR-20b-5p, miR-21-5p, miR-142-3p, and miR-223-3p with significant P value, *** P < .05. D, Fold change of each miRNA in each individual case and control as compared to mean of controls.
Based on miRNA sequencing results, the cases and controls form 3 clusters in principal component analysis (PCA; Figure 1B). Cluster A consists of controls 1 and 2 and cases 4 and 6, cluster B consists of cases 1, 2, 3, 5, and 8, and cluster C consists of case 7. No biological or medical parameter defining the clusters could be identified, except that the brain tissue sample of case 7 was taken from a specific location, nucleus lentiformis, which may explain some of the differences.
The expression of 8 DE miRNAs was validated by quantitative polymerase chain reaction (qPCR). Relative expression of miR-10a-5p, miR-20b-5p, miR-21–3p, miR-21-5p, miR-142-3p, miR-155-5p, and miR-223-3p in cases versus controls was calculated using miR-191-5p for normalization (Figure 1C). Significant change (P < .05) in relative expression could be shown for miR-20b-5p, miR-21-5p, miR-142-3p, and miR-223-3p. The mean relative expression of miR-21-5p was more than 10 times higher in cases versus controls. Also, the mean relative expression of miR-142-3p was over 5 times higher in cases versus controls. Strong expression of miR-142-5p was measured in cases 1–5 and 9, but not in cases 6–8 or controls, and thus its relative expression in cases versus controls could not be calculated or indicated by fold change. The absence of qPCR signals in controls, despite the presence of sequencing reads, may be explained by Advanced TaqMan assay characteristics producing very low amplification curve profiles possibly affecting the calculations.
Expression of each miRNA varied among cases (Figure 1D). miR-21-5p expression was strongly upregulated, with fold change >10, in all cases except case 6. In addition, strong upregulation of miR-20b-5p, miR-21-3p, and miR-142-3p was detected in cases 7 and 8. miR-10a-5p was upregulated in cases 6, 7, and 8. Strongest upregulation of miR-155-5p was seen in case 7. Less variation was seen in miR-223-3p expression between the samples. The largest number of upregulations was seen in case 7, which may indeed explain different clustering in PCA.
In qPCR validation, JCPyV-specific JCV-miR-J1-5p was identified in brain tissues from all cases except case 7, again pointing out that this particular case is different. JCV-miR-J1-3p, common to JCPyV and BKPyV, was identified in all PML cases used for validation. Neither JCV-miR-J1-3p nor JCV-miR-J1-5p was identified in control samples by qPCR.
Data Mining
Among statistically significantly (false discovery rate, < 0.05) enriched genes associated with more than 2 of the DE miRNAs, a total of 326 genes were identified, including 12 genes associated with 4 or more DE miRNAs analyzed by the Mienturnet analysis tool. The 10 most significantly enriched genes and their functions are presented in Supplementary Figure 2A and 2B. Interestingly the 10 most significant miRNA-function associations in PML brain include several functions related to immune response or inflammation, as well as latent virus replication analyzed by Tam 2.0 web server (Supplementary Figure 2C). Additionally, interesting association of the upregulated miRNAs based on the literature are listed in Supplementary Table 2.
DISCUSSION
To study the mechanisms of brain damage in PML, we assessed miRNA expression in brain tissue samples and identified 8 upregulated human miRNAs in brain tissue samples from PML patients by sequencing: miR-10a-5p, miR-20b-5p, miR-21-3p, miR-21-5p, miR-142-3p, miR-142-5p, miR-155-5p, and miR-223-3p. Interestingly, the genes encoding 5 of the 8 upregulated miRNAs appear to be located in chromosome 17.
Several of the upregulated miRNAs have been associated with neurological conditions, neuroinflammation, the blood-brain barrier (BBB), or polyomavirus before. Frequent dysregulation of miR-21-5p, miR-155-5p, and miR-223-3p in neurodegenerative diseases has been reported [3]. Regulation of miR-21-5p in brain tissue has been previously associated with PML [4]. miR-21-5p is involved in astrocyte activation and remyelination [5]. Upregulated miR-155-5p again has been associated with demyelination [6], which is a crucial event in PML pathogenesis.
Immunosuppressive and immunomodulatory drugs involve PML risk [2]. Upregulation of miR-142-5p and miR-20b-5p upon natalizumab treatment has been shown [7, 8], suggesting that the mechanisms by which biological drugs elevate the risk of JCPyV reactivation and PML may thus be linked to upregulation of specific miRNAs in infected glial cells.
Association with neuroinflammation has been shown for miR-21, miR-142-3p, miR-155-5p, and miR-223-3p [9, 10]. Among them, miR-155-5p and miR-142-3p are proinflammatory whereas miR-21-5p and miR-223-3p are anti-inflammatory modulators. Neuroinflammation is a protective response in the brain, but uncontrolled neuroinflammation may lead to tissue damage mediated by elevated glial cell activation with significant innate immune mediator secretion, BBB permeability, and infiltration of peripheral immune cells, such as in multiple sclerosis [9]. The mechanisms of damage in JCPyV-infected brain may involve regulation miRNAs implicated in neuroinflammation.
Reduced barrier function of the BBB might enable JCPyV access from the periphery to the brain. miR-155 has been shown to contribute to BBB disruption under inflammatory conditions by downregulating key junctional proteins [11]. miR-21-3p may aggravate BBB damage by promoting apoptosis and inflammation, whereas miR-21-5p protects the BBB by suppressing apoptosis and inflammation [12]. Here, we observed concordant upregulation of both arms of miR-21 and also of miR-142, although the frequency or significance of such a phenomenon is not completely understood.
miRNAs may also modify viral infection, as exemplified by miR-155, whose irregular expression has been suggested to support viral persistence in general and pathologic reactions via immune response modulation [13]. Importantly, miR-223-3p regulates JCPyV infection through downregulation of NF-1A protein expression, a negative regulator of JCPyV replication [14].
All 8 upregulated miRNAs have been identified in extracellular vesicles in different disease conditions, which is potentially interesting in JCPyV pathogenesis as JCPyV may utilize extracellular vesicles to infect target cells in a receptor-independent manner [15].
In addition to JCPyV, miRNAs upregulated in PML patients may be implicated in BKPyV disease. Upregulation of miR-10a-5p, miR-21-3p, miR-21-5p, and miR-142-5p has been reported in nephropathy, a severe condition of kidney transplant recipients caused by BKPyV but occasionally also by JCPyV [4]. Upregulation of miR-21-5p in PML, nephropathy, and in cells infected with the rearranged Mad-1 JCPyV strain has been shown [4].
In conclusion, we have identified and validated upregulation of 8 human miRNAs in PML brain. Whether regulation of these miRNAs is directly due to JCPyV remains to be studied. The reported functional associations of the upregulated miRNAs include neuroinflammation and BBB disruption. Future studies will assess the role of these miRNAs in PML pathogenesis, as well as their value as prognostic and diagnostic biomarkers of PML.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Anni Honkimaa, Department of Virology, Helsinki University Hospital and University of Helsinki, Helsinki, Finland.
Joni Suppula, Institute of Biotechnology, DNA Sequencing and Genomics Laboratory, University of Helsinki, Helsinki, Finland.
Olli Tynninen, Department of Pathology, Helsinki University Hospital and University of Helsinki, Helsinki, Finland.
Mika Saarela, Department of Neurology, Helsinki University Hospital and University of Helsinki, Helsinki, Finland.
Hanna Liimatainen, Department of Virology, Helsinki University Hospital and University of Helsinki, Helsinki, Finland.
Pia Laine, Institute of Biotechnology, DNA Sequencing and Genomics Laboratory, University of Helsinki, Helsinki, Finland.
Petri Auvinen, Institute of Biotechnology, DNA Sequencing and Genomics Laboratory, University of Helsinki, Helsinki, Finland.
Eeva Auvinen, Department of Virology, Helsinki University Hospital and University of Helsinki, Helsinki, Finland.
Notes
Author contributions . E. A. contributed conceptualization. M. S. performed clinical characterization of patients. J. S. and O. T. contributed methodology. A. H., J. S., O. T., and P. L. performed formal analysis. A. H. and J. S. performed investigation. E. A. and A. H. acquired funding. E. A. and P. A. performed project administration and supervision. A. H. wrote the original draft. All authors contributed to writing, reviewing, and editing.
Acknowledgments . We thank Ms Anu Kaitonen and the personnel of the DNA sequencing and genomics laboratory for valuable technical assistance. We appreciate the help of Dr Aila Särkkä in statistical analyses.
Financial support . This work was supported by the Emil Aaltonen Foundation; and the Finnish Society for the Study of Infectious Diseases. Funding to pay the Open Access publication charges for this article was provided by FinELib.
Data availability . The data for this study have been deposited in the European Nucleotide Archive at EMBL-EBI under accession number PRJEB59548 (https://www.ebi.ac.uk/ena/browser/view/PRJEB59548).
References
- 1. Wollebo HS, White MK, Gordon J, Berger JR, Khalili K. Persistence and pathogenesis of the neurotropic polyomavirus JC. Ann Neurol 2015; 77:560–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kartau M, Sipilä JO, Auvinen E, Palomäki M, Verkkoniemi-Ahola A. Progressive multifocal leukoencephalopathy: current insights. Degener Neurol Neuromuscul Dis 2019; 9:109–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Juźwik CA, Drake S, Zhang Y, et al. microRNA dysregulation in neurodegenerative diseases: a systematic review. Prog Neurobiol 2019; 182:101664. [DOI] [PubMed] [Google Scholar]
- 4. Takahashi K, Sato Y, Sekizuka T, et al. High expression of JC polyomavirus-encoded microRNAs in progressive multifocal leukoencephalopathy tissues and its repressive role in virus replication. PLoS Pathog 2020; 16:e1008523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bai X, Bian Z. MicroRNA-21 is a versatile regulator and potential treatment target in central nervous system disorders. Front Mol Neurosci 2022; 15:842288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maciak K, Dziedzic A, Miller E, Saluk-Bijak J. miR-155 as an important regulator of multiple sclerosis pathogenesis. A review. Int J Mol Sci 2021; 22:4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sievers C, Meira M, Hoffmann F, Fontoura P, Kappos L, Lindberg RL. Altered microRNA expression in B lymphocytes in multiple sclerosis: towards a better understanding of treatment effects. Clin Immunol 2012; 144:70–9. [DOI] [PubMed] [Google Scholar]
- 8. Ingwersen J, Menge T, Wingerath B, et al. Natalizumab restores aberrant miRNA expression profile in multiple sclerosis and reveals a critical role for miR-20b. Ann Clin Transl Neurol 2015; 2:43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaudet AD, Fonken LK, Watkins LR, Nelson RJ, Popovich PG. MicroRNAs: roles in regulating neuroinflammation. Neuroscientist 2018; 24:221–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mandolesi G, De Vito F, Musella A, et al. miR-142-3p is a key regulator of IL-1β-dependent synaptopathy in neuroinflammation. J Neurosci 2017; 37:546–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lopez-Ramirez MA, Wu D, Pryce G, et al. MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J 2014; 28:2551–65. [DOI] [PubMed] [Google Scholar]
- 12. Ge X, Li W, Huang S, et al. Increased miR-21-3p in injured brain microvascular endothelial cells after traumatic brain injury aggravates blood-brain barrier damage by promoting cellular apoptosis and inflammation through targeting MAT2B. J Neurotrauma 2019; 36:1291–305. [DOI] [PubMed] [Google Scholar]
- 13. Jafarzadeh A, Naseri A, Shojaie L, et al. MicroRNA-155 and antiviral immune responses. Int Immunopharmacol 2021; 101:108188. [DOI] [PubMed] [Google Scholar]
- 14. Ravichandran V, Major EO. DNA-binding transcription factor NF-1A negatively regulates JC virus multiplication. J Gen Virol 2008; 89:1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morris-Love J, Gee GV, O'Hara BA, et al. JC polyomavirus uses extracellular vesicles to infect target cells. mBio 2019; 10:e00379-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.