Abstract
Summary
We present the phippery software suite for analyzing data from phage display methods that use immunoprecipitation and deep sequencing to capture antibody binding to peptides, often referred to as PhIP-Seq. It has three main components that can be used separately or in conjunction: (i) a Nextflow pipeline, phip-flow, to process raw sequencing data into a compact, multidimensional dataset format and allows for end-to-end automation of reproducible workflows. (ii) a Python API, phippery, which provides interfaces for tasks such as count normalization, enrichment calculation, multidimensional scaling, and more, and (iii) a Streamlit application, phip-viz, as an interactive interface for visualizing the data as a heatmap in a flexible manner.
Availability and implementation
All software packages are publicly available under the MIT License. The phip-flow pipeline: https://github.com/matsengrp/phip-flow. The phippery library: https://github.com/matsengrp/phippery. The phip-viz Streamlit application: https://github.com/matsengrp/phip-viz.
1 Introduction
Modern oligonucleotide synthesis allows researchers to generate highly multiplexed assays such as Phage Immunoprecipitation Sequencing (PhIP-Seq) (Mohan et al. 2018). This protocol is used to investigate linear epitopes by quantifying antibody-peptide interactions with phage-display libraries. These libraries may be general purpose, such as VirScan (Xu et al. 2015), or highly specialized as in Phage-DMS (Garrett et al. 2020). A typical PhIP-Seq dataset includes empirical serological or monoclonal antibody samples, negative controls by performing the protocol without antibodies present, and sequencing of the phage library to determine the input abundance of each peptide. The goal is to identify peptides in empirical samples that show robust enrichment due to antibody binding, relative to their expected abundances in the input phage library or no-antibody control.
Much of the published code for PhIP-Seq data analysis are specific to an experiment or task, forcing new researchers to piece together snippets from others or developing from scratch. Recently, Chen et al. (2022) published a R-based pipeline that includes enrichment quantification and plotting. The focus of our tool is to provide general infrastructure to standardize workflows and an API library applicable for any study design.
2 Design and usage
Here, we give brief descriptions of phip-flow, phippery, and phip-viz. The web documentation https://matsengrp.github.io/phippery/ provides more details, as well as instructions on how to contribute.
Nextflow pipeline: The phip-flow software is built using Nextflow (Di Tommaso et al. 2017) and presents end-to-end automation of reproducible workflows for PhIP-Seq data. The primary task of this workflow is quantification, normalization, and organized aggregation of results for any PhIP-Seq experiment starting from raw reads. Nextflow makes it easy to modify the pipeline, such as the use of alternate software for read alignment via editing template shell scripts with pre-defined inputs. Each step in the pipeline is run in a containerized environment, making it straightforward to add dependencies while maintaining workflow portability across computing platforms. By default normalization is done via edgeR (Robinson et al. 2010) although more sophisticated methods can be used (Chen et al. 2022). The inputs to the pipeline are comma-separated values (CSV) files specifying the sequencing data fastq files, the oligonucleotide sequences encoded in the phage library, and annotations for supplementary sample and peptide information. By default, alignment is performed with Bowtie2 (Langmead and Salzberg 2012), and the peptide counts are collated with SAMtools (Danecek et al. 2021). The workflow can easily scale to large sample sizes and handle any peptide library design.
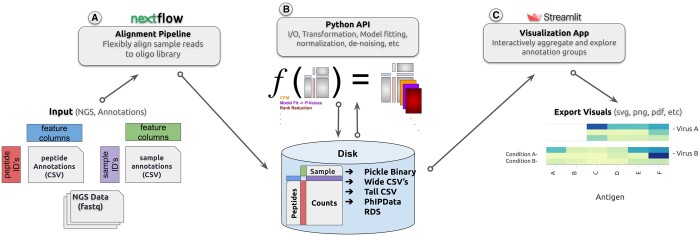
Python API: The core data structure is a formatted xarray object (Hoyer and Hamman 2017), a multidimensional array well-suited for compiling the PhIP-Seq dataset into a single object to facilitate queries on the data. The phippery API imports commonly used data science dependencies and is designed to operate with a PhIP-Seq data xarray object. It is straightforward to build up an analysis with the phippery library of functions. Functions provided by the API include querying samples or peptides to a specific subset before exporting to CSV, essential tasks such as enrichment and Z-score calculations, as well as higher level analysis such as principal component analysis (Fig. 1). Several PhIP-Seq results have been published using phippery (Stoddard et al. 2021, Garrett et al. 2022, Willcox et al. 2022).
Figure 1.
(A) The processing of sequencing data from a PhIP-Seq experiment begins with the alignment pipeline, which generates either a CSV file or xarray object containing the peptide counts across samples and associated annotations. (B) Custom analysis is performed with phippery library function calls, for example via the command line interface, and (C) visualization of results can be presented in an interactive manner using our Streamlit app.
Streamlit visualization: A large majority of PhIP-Seq analysis tasks are simply aggregating samples from various treatment groups, and comparing their binding responses across peptide groups of interest in the phage library. For this, phip-viz may be used to explore subsets and group aggregations using widgets that provide insight into the dataset. This application takes as input only the binary formatted xarray dataset described above, and allows the user to subset data groups, aggregate, visualize, and export the resulting heatmaps.
Contributor Information
Jared G Galloway, Computational Biology, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA.
Kevin Sung, Computational Biology, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA.
Samuel S Minot, Data Core, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA.
Meghan E Garrett, Human Biology Division, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA; Molecular and Cellular Biology Program, University of Washington, Seattle, WA 98195, USA.
Caitlin I Stoddard, Human Biology Division, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA.
Alexandra C Willcox, Human Biology Division, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA; Molecular and Cellular Biology Program, University of Washington, Seattle, WA 98195, USA; Medical Scientist Training Program, University of Washington, Seattle, WA 98195, USA.
Zak A Yaffe, Human Biology Division, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA; Molecular and Cellular Biology Program, University of Washington, Seattle, WA 98195, USA; Medical Scientist Training Program, University of Washington, Seattle, WA 98195, USA.
Ryan Yucha, Human Biology Division, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA; Department of Microbiology, University of Washington School of Medicine, Seattle, WA 98195, USA.
Julie Overbaugh, Computational Biology, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA; Human Biology Division, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA.
Frederick A Matsen, IV, Computational Biology, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA; Howard Hughes Medical Institute, Seattle, WA, 98109, USA.
Conflict of interest
None declared.
Funding
This work was supported by the National Institutes of Health [R01 AI146028, R01 AI138709, F30 AI165112, T32 AI083203] and the Howard Hughes Medical Institute. F.A.M. is an Investigator at the Howard Hughes Medical Institute.
References
- Chen A, Kammers K, Larman HB. et al. Detecting antibody reactivities in phage ImmunoPrecipitation sequencing data. BMC Genomics 2022;23:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J. et al. Twelve years of SAMtools and BCFtools. Gigascience 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P, Chatzou M, Floden EW. et al. Nextflow enables reproducible computational workflows. Nat Biotechnol 2017;35:316–9. [DOI] [PubMed] [Google Scholar]
- Garrett ME, Galloway JG, Wolf C. et al. Comprehensive characterization of the antibody responses to SARS-CoV-2 spike protein finds additional vaccine-induced epitopes beyond those for mild infection. Elife 2022;11:73490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett ME, Itell HL, Crawford KHD. et al. Phage-DMS: a comprehensive method for fine mapping of antibody epitopes. iScience 2020;23:101622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer S, Hamman J.. Xarray: N-D labeled arrays and datasets in python. JORS 2017;5:10. [Google Scholar]
- Langmead B, Salzberg S.. Fast gapped-read alignment with bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan D, Wansley DL, Sie BM. et al. PhIP-Seq characterization of serum antibodies using oligonucelotide-encoded peptidomes. Nat Protoc 2018;13:1958–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. et al. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CI, Galloway J, Chu HY. et al. Epitope profiling reveals binding signatures of SARS-CoV-2 immune response in natural infection and cross-reactivity with endemic human CoVs. Cell Rep 2021;35:109164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox AC, Sung K, Garrett ME. et al. Detailed analysis of antibody responses to SARS-CoV-2 vaccination and infection in macaques. PLoS Pathog 2022;18:e1010155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GJ, Kula T, Xu Q. et al. Comprehensive serological profiling of human populations using a synthetic human virome. Science 2015;348:aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]