Abstract
It is discovered that activated caspase-3 tends to induce apoptosis in gasdermin E (GSDME)-deficient cells, but pyroptosis in GSDME-sufficient cells. The high GSDME expression and apoptosis resistance of pancreatic ductal adenocarcinoma (PDAC) cells shed light on another attractive strategy for PDAC treatment by promoting pyroptosis. Here we report a hGLuc-hGSDME-PCA system for high-throughput screening of potential GSDME activators against PDAC. This screening system neatly quantifies the oligomerization of GSDME-N to characterize whether pyroptosis occurs under the stimulation of chemotherapy drugs. Based on this system, ponatinib and perifosine are screened out from the FDA-approved anti-cancer drug library containing 106 compounds. Concretely, they exhibit the most potent luminescent activity and cause drastic pyroptosis in PDAC cells. Further, we demonstrate that perifosine suppresses pancreatic cancer by promoting pyroptosis via caspase-3/GSDME pathway both in vitro and in vivo. Collectively, this study reveals the great significance of hGLuc-hGSDME-PCA in identifying compounds triggering GSDME-dependent pyroptosis and developing promising therapeutic agents for PDAC.
Key words: Pyroptosis, Caspase-3, Gasdermin E, Pancreatic ductal adenocarcinoma, hGLuc-hGSDME-PCA, High-throughput screening, Ponatinib, Perifosine
Graphical abstract
Based on the developed hGLuc-hGSDME-PCA system, ponatinib and perifosine were screened out from drug library. The results demonstrated that they can trigger pyroptosis in PDAC cells through caspase-3/GSDME pathway to inhibit tumor growth.
1. Introduction
Pancreatic cancer is a malignancy gastrointestinal cancer characterized by late diagnosis, limited treatment success and dismal prognosis1. Pancreatic ductal adenocarcinoma (PDAC), which originates from the exocrine portion of the pancreas, accounts for 95% of pancreatic cancers2. Despite numerous researches, PDAC remains a major medical challenge with a 5-year survival rate below 10%, an increasing global incidence and a high mortality rate3. Both tumorigenesis and pathogenesis of PDAC remain extremely elusive4. Furthermore, all available treatments, including chemotherapy, radiotherapy, and immunotherapy, have limited clinical benefits for PDAC patients5. For now, the main treatment against PDAC relies on gemcitabine. The poor therapeutic response to PDAC is possibly due to the unique genetic makeup and tumor microenvironment. Moreover, the capacity of PDAC cells to evade death also plays a vital role in this process6,7. As resistance to apoptosis is a general hallmark of cancer, non-apoptotic cell death, like pyroptosis, provide alternative strategies for killing PDAC cells8, 9, 10.
Pyroptosis is a lytic pro-inflammatory type of programmed cell death (PCD), characterized by cellular swelling, large bubbles blowing from the plasma membrane, and rapid cell lysis11,12. It is triggered mainly by caspase-1, -4, -5 and -1113,14. Once activated, these inflammatory caspases cleave gasdermin D (GSDMD) to liberate cytotoxic N-terminal fragments, which integrate into the plasma membrane and form large oligomeric pores, ultimately causing pyroptosis15. Apart from this, pyroptosis can also be caused by gasdermin E (GSDME), which is specifically cleaved by pro-apoptotic caspase-316,17. Recent studies unveiled that GSDME could switch caspase-3-mediated apoptosis to pyroptosis after certain chemotherapy drug treatments18. Henceforth, GSDME-mediated pyroptosis has drawn significant attention in oncology areas19, 20, 21. Unexpectedly, GSDME is silenced in most tumor cells due to epigenetic suppression, meaning that these cells can hardly be killed though pyroptosis21,22. In contrast to this, GSDME is highly expressed in PDAC23. Overall, cell pyroptosis triggered by GSDME is a potentially innovative strategy against PDAC.
Protein–protein interactions (PPIs) are crucial for signal transduction, protein trafficking and movement. For instance, GSDME N-terminal proteins interact with each other and oligomerize to form pores in cell membrane during pyroptosis. Numerous approaches have been developed to study PPIs in vivo and in vitro, including the protein-fragment complementation assay (PCA)24. In the PCA strategy, two proteins of interest (proteins A and B) are fused to an inactive complementary fragment of a reporter protein, individually. When proteins A and B interact, the split reporter fragments are brought together and fold into the functional conformation, enabling reconstitution of the reporter activity25. The reporter protein was usually chosen from dihydrofolate reductase, β-lactamase, luciferase or fluorescent proteins24. Luciferase PCAs have been developed, mainly including three luciferases from the firefly (Photinus pyralis), the sea pansy (Renilla reniformis), and the marine copepod (Gaussia princeps)26,27. The humanized form of Gaussia luciferase (hGLuc) generates a bioluminescent signal that is over 100-fold higher than the humanized forms of firefly luciferases (hFLuc) or Renilla luciferases (hRLuc) in cell lysates28. On account of its small size, strong luminescent activity, low reaction background, simple reaction condition, and high stability, hGLuc has become a widely used reporter for high-throughput assays29,30.
To our knowledge, no practicable assay has been developed to evaluate pyroptosis-inducing activity quantitatively. We created a luminescent PCA to quantify GSDME-N oligomerization using complementary hGLuc(N) and hGLuc(C) fused separately to full-length GSDME. Fusion fragments were co-expressed by dual promoter expression vector pBudCE4.1. When GSDME-N oligomerizes biologically, the luminescent activity that come from the complementation of hGLuc(N) and hGLuc(C) will be altered. In this study, screening results of the FDA-approved anti-cancer drug library indicated that compounds ponatinib and perifosine exhibited the most robust luminescent activity and caused drastic pyroptosis in PDAC cells. Subsequent experiments also revealed that perifosine inhibited pancreatic cancer by promoting pyroptosis via caspase-3/GSDME pathway both in vitro and in vivo. Taken together, we believe that hGLuc-hGSDME-PCA is a great high-throughput screening model that can identify compounds triggering pyroptosis in PDAC cells. Our work underscores the importance of hGLuc-hGSDME-PCA in developing promising drugs against PDAC.
2. Materials and methods
2.1. Reagents
Ponatinib (HY-12047), perifosine (HY-50909), sorafenib (HY-10201), gemcitabine hydrochloride (HY-B0003), Z-DEVD-FMK (HY-12466), GSK-872 (hydrochloride) (HY-101872A) and necrostatin-1 (HY-15760) were purchased from MedChemExpress (Shanghai, China). A customized FDA-approved Anti-Cancer Drug Library was purchased from SelleckChem (Houston, TX, USA). Recombinant murine TNF-α (315-01A) was purchased from PeproTech (Rocky Hill, NJ, USA). jetOPTIMUS® reagent (117-15) was purchased from Polyplus Transfection® (Illkirch, France). Lipofectamine™ RNAiMAX transfection reagent (13778075) and Zeocin™ Selection Reagent (Bleomycin) (R25001) were purchased from Invitrogen (Carlsbad, CA, USA). M-PER™ Mammalian Protein Extraction Reagent (78501) was purchased from Thermo Scientific (Boston, MA, USA). Renilla Luciferase Assay System (E2820) was purchased from Promega Corporation (Madison, WI, USA). Lipopolysaccharides (O55:B5) (L6529), etoposide (E1383) and β-mercaptoethanol (2-mercaptoethanol, 2ME, M6250) were purchased from Sigma–Aldrich (St. Louis, MO, USA). PhoSTOP phosphatase inhibitor cocktail (04906837001) and cOmplete Table-EDTA free protease inhibitor cocktail (04693132001) were purchased from Roche (Mannheim, Germany). Enhanced BCA Protein Assay Kit (P0009), Crystal Violet Staining Solution (C0121), CellTiter-Lumi™ Plus Luminescent Cell Viability Assay Kit (C0068S), Apoptosis and Necrosis Assay Kit (C1056) and Reactive Oxygen Species Assay Kit (S0033S) were from Beyotime Biotechnology (Shanghai, China). Cell Counting Kit-8 (KGA317), kFluor488 Click-iT EdU Imaging Kit (KGA331), Annexin V-FITC/PI Double-stained Apoptosis Detection Kit (KGA108) and Cell Membrane Red Fluorescent Dye DiD (KGMP0025) were from KeyGEN BioTECH (Nanjing, China). LDH Cytotoxicity Assay Kit (40209ES76) was from Yeasen Biotechnology (Shanghai, China) for in vitro studies. CheKine™ Micro Lactate Dehydrogenase (LDH) Assay Kit (KTB1110) was from Abbkine (Wuhan, China) in vivo studies. Alanine aminotransferase Assay Kit (C009-2-1) and aspartate aminotransferase Assay Kit (C010-2-1) were from Nanjing Jiancheng Bioengineering Institute (Nanjing, China).
2.2. Cell lines and cell culture
All cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and incubated at 37 °C in a humidified atmosphere (5% CO2 and 95% air). The cell culture medium was changed every 1–2 days depending on cell density. When cells reach 80%–90% confluence, routine passaging was performed at a ratio of 1:2 to 1:6. The PANC-1, MIA PaCa-2 and HPDE6-C7 (H6c7) cells were cultured in Dulbecco's Modified Eagle Medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco) and 100× Penicillin‒Streptomycin (KeyGEN BioTECH). The AsPC-1 and BxPC-3 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 Medium (Gibco) supplemented with 10% FBS and 100× Penicillin‒Streptomycin.
2.3. Plasmid construction
To construct the recombinant vector, the sequences of amino acids 18–109 of hGLuc(N) and 110–185 of hGLuc(C) were synthesized and fused to the 5′ end of GSDME, respectively. The secretion signal sequence of GLuc was removed to avoid secretion outside the cell. 5′ ends of the two fusions were further modified to facilitate immunodetection by adding the Flag-tag or HA-tag. The coding sequence of a flexible polypeptide linker (Gly-Gly-Gly-Gly-Ser)2 was inserted between the tag and hGLuc fragments26. The same linker was also inserted between the hGLuc and hGSDME fragments.
Two target fragments were amplified by PCR and ligated sequentially with T4 DNA ligase into dual promoter expression vector pBudCE4.1. Specifically, HA-hGLuc(C)-hGSDME segment was cloned into CMV MCS using HindIII and BamHI restriction enzyme, while Flag-hGLuc(N)-hGSDME segment was cloned into EF-1α MCS using NotI and KpnI restriction enzyme. Ligations were transformed into competent E. coli, and individual colonies on plates containing zeocin were cultured in Luria–Bertani medium for DNA extraction. The restriction digest and Sanger sequencing at AZENTA (Suzhou, China) confirmed the DNA sequences. Eventually, the pBudCE4.1-HA-hGLuc(C)-hGSDME-Flag-hGLuc(N)-hGSDME recombinant reporter plasmid (called pBudCE4.1-coexpression for short) was obtained for expressing two target genes in mammalian cell lines simultaneously.
2.4. Plasmid and siRNA transfection
For the comparison of 12 commercially available transfection reagents, PANC-1 was seeded in 48-well plates overnight and then transfected using 12 types of reagents (Supporting Information Table S1). The transfected plasmid was a GFP-expressing plasmid (500 ng per well). The DNA amount to reagent volume ratio was 1:1–1:4. Cells were fluorescently imaged 24 h after transfection. The transfection efficiency and cell damage were considered to determine the best transfection reagent suitable for PANC-1 cells.
For subsequent plasmid transfection to PANC-1, jetOPTIMUS reagent was employed according to the manufacturer's protocol. Transfections were performed with 2 μg of DNA/4 μL jetOPTIMUS reagent per well of 6-well plate and 0.2 μg of DNA/0.4 μL jetOPTIMUS reagent per well of 96-well plate. After transfection for 24 h, cells were cultured in fresh medium for further experiments.
The small interfering RNA (siRNA) duplexes were purchased from GenePharma (Suzhou, China). siRNAs were transfected into cells using Lipofectamine™ RNAiMAX transfection reagent. Cells were harvested 48 h later to evaluate knock-down efficiency via Western blotting. The siRNA sequences of control, GSDME, GSDMD or CASP-3 were as follows:
NC 5′-UUCUCCGAACGUGUCACGUTT-3′,
GSDME 5′-GCAGCAAGCAGCUGUUUAUTT-3′,
GSDMD 5′-GCAGGAGCUUCCACUUCUATT-3′,
CASP-3 5′-AUGCCGACAAGCUUGAAUUTT-3′.
2.5. Luciferase activity assay
PANC-1 cells cultured in 96-well plates were transiently transfected with 0.2 μg plasmid per well. Chemical treatments were carried out 24 h post-transfection. Cells were incubated with 50 μmol/L compounds of our FDA-approved Anti-Cancer Drug Library and positive drug sorafenib for the indicated time. To detect the hGLuc PCA signal, cells were lysed with M-PER Mammalian Protein Extraction Reagent (100 μL/well) according to the manufacturer's instructions. The plate was gently shaken for 30 min. The supernatants of cell lysates were collected, and each sample's total protein concentration was determined by BCA protein assays. In addition, after a 30 μL sample and 100 μL Renilla Luciferase Assay Substrate (1X) were transferred into 96-well white microplate (Corning, NY, USA), the luminescence signal was immediately measured on the multimode plate reader (EnVision 2105, PerkinElmer, USA) at 37 °C. Light emission was integrated over 5 s after an initial 1-s preread delay. Finally, luciferase activity was presented as either relative luminescence unit per μg protein (RLU/μg) or further calculated comparative values according to Eq. (1):
| (1) |
where FB is the RLU/μg value of the blank control well transfected with GSDME plasmid and treated with DMSO; FC is the RLU/μg value of the negative control well transfected with reporter plasmid pBudCE4.1 and treated with DMSO; FT is the RLU/μg value of the treatment well transfected with reporter plasmid pBudCE4.1 and treated with drugs.
For co-transfected system assays, 0.2 μg each of pcDNA3.1-HA-hGLuc(C)-hGSDME and pcDNA3.1-Flag-hGLuc(N)-hGSDME were co-transfected together into PANC-1 cells. When transfection finished, cells were treated with DMSO or 50 μmol/L sorafenib for 24 h. Then luciferase activity was measured as described above.
2.6. Cell viability assay
Cell viability was performed using Cell Counting Kit-8 (CCK-8) following the manufacturer's instructions. AsPC-1, BxPC-3, PANC-1, MIA PaCa-2 or H6c7 cells were seeded in 96-well plates (5 × 104 cells per well) and cultured at 37 °C overnight. Ponatinib and sorafenib were dissolved in DMSO, while perifosine was dissolved in sterile water. Afterward, five different cell types were treated with ponatinib, perifosine, or sorafenib for the indicated time and concentrations. Subsequently, 10 μL of CCK-8 reagent was added to each well, and the cells were incubated for 2 h at 37 °C. The absorbance was measured at 450 nm using a microplate reader (Biotek, USA) to estimate cell viability.
2.7. LDH release assay
The LDH released into the medium was used to analyze chemical-mediated cytotoxicity and membrane integrity. Briefly, cell culture supernatants were collected after various treatments and detected by LDH Cytotoxicity Assay Kit. 120 μL supernatant of each sample was transferred into one well of a fresh 96-well plate, then 60 μL of LDH detection reagent was added to the same well. The plates were then incubated for 30 min protected from light at room temperature. The absorbance was measured at 490 nm on a microplate reader (Biotek). Relative LDH release was calculated as Eq. (2):
| (2) |
2.8. Clonogenic assay
We assessed the clonogenicity of the PANC-1 cells using 6-well plates. We seeded individually 1000 cells/well of PANC-1 and cultivated them overnight. Then, the cells were treated with drugs as indicated for 24 h followed by replacement with fresh medium. Two weeks after drug treatments, the proliferating cells were washed with PBS, fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet dyes for 30 min. After washing with water several times, the number of colonies was counted for statistical analysis.
2.9. EdU incorporation assay
PANC-1 cells were planted in 35 mm glass-bottom dishes (Cellvis, D35-20-1-N) and subjected to the indicated treatments. kFluor488 Click-iT EdU Imaging Kit was utilized to measure cell proliferation based on the manufacturer's instructions. Briefly, EdU (20 μmol/L) was added to the medium for 2.5 h. After fixation and permeabilization, cells were stained by the Click-iT reaction cocktail for 30 min and 5 μg/mL Hoechst 33342 for 20 min. From this step onwards, the samples should be protected from light. After nuclear staining, images were captured with a confocal microscope (Olympus FV3000). The proliferating cell nuclei incorporated with EdU were marked by green fluorescence. The number of EdU+ cells was quantified using ImageJ.
2.10. Immunoblot analysis
Cells or frozen tumors (20–40 mg) were lysed in RIPA buffer (50 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, and 2 mmol/L EDTA (pH 8.0)) supplemented with protease inhibitor and phosphatase inhibitor. Protein concentration was measured using the BCA protein assay kit. Equal amounts of protein were separated with SDS-PAGE gels and transferred to nitrocellulose membranes. The membrane was blocked using 5% skim milk for 1 h at room temperature and incubated with primary antibodies at 4 °C overnight, followed by secondary antibodies for 2 h at room temperature. The antibodies used were as follows: anti-DFNA5/GSDME-N-terminal (1:1000, ab215191, Abcam), anti-GSDMD (1:2000, 20770-1-AP, Proteintech), anti-Caspase-3 (1:1000, 9662S, Cell Signaling Technology), anti-β-tubulin (1:2500, 10094-1-AP, Proteintech), anti-HA-tag (1:4000, ab9110, Abcam), anti-DYKDDDDK Tag (1:1000, 14793S, Cell Signaling Technology), HRP-labeled goat anti-rabbit IgG(H + L) (1:2000, A0208, Beyotime), and HRP-labeled goat anti-mouse IgG(H + L) (1:2000, A0216, Beyotime). Immunoreactive bands were visualized with High-sig ECL Western Blotting Substrate and Tanon Imaging System (Tanon, Shanghai, China). Tubulin was used as a loading control. The images were analyzed in a semi-quantitative manner through ImageJ software.
For the oligomerization analysis, the cells were lysed in buffer containing 50 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 1% Triton X-100 and 1 mmol/L PMSF, then disrupted by a sonicator in an ice-water bath. The harvested lysates were prepared with nonreduced loading buffer, then electrophoresed through a 6% NativePAGE Tris-Gly gel (Beyotime) in NativePAGE running buffer at 100 V for 15 min and 150 V for 65 min. Proteins were then transferred to a polyvinyl difluoride (PVDF) membrane at 380 mA for 1.5 h in NativePAGE transfer buffer for immunoblotting.
2.11. Immunoprecipitation
After transfection and treatment of sorafenib, the cells were lysed in buffer containing 50 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 1% Triton X-100 and 1 mmol/L PMSF, then disrupted by a sonicator in an ice-water bath. For the oligomerization analysis, the nonreduced cell samples were incubated with or without 2ME, respectively. The proteins were immunoprecipitated from the cell lysates by incubating with the relevant antibody at 4 °C overnight and subsequently with protein A/G agarose beads for another 2 h at 4 °C. Then, the immunoprecipitates were collected and washed with lysis buffer. The samples were subjected to Native-PAGE and transferred to PVDF membranes, which were probed with indicated antibodies and detected.
2.12. Hoechst 33342/PI fluorescent staining
The treated PANC-1 cells were stained with Apoptosis and Necrosis Assay Kit according to the manufacturer's instructions. Briefly, cells were treated as indicated. After washing with PBS, 800 μL staining buffer, 5 μL Hoechst 33342 solution and 5 μL PI solution were sequentially added to each well for 30 min at 4 °C. The cell images were obtained from a confocal microscope.
2.13. Immunofluorescence analysis
PANC-1 cells were seeded on confocal dishes. After treatment, cells were fixed in 4% paraformaldehyde before permeabilization with 0.5% Triton X-100. Intracellular GSDME and cleaved caspase-3 were detected using the anti-GSDME polyclonal antibody (1:250, 13075-1-AP, Proteintech) and anti-cleaved caspase-3 (Asp175) (1:400, 9661T, Cell Signaling Technology) overnight at 4 °C. The secondary antibody Alexa Fluor 488-labeled goat anti-rabbit IgG(H + L) (1:1000, P0176-1, Beyotime) was incubated for 1 h in the dark at room temperature. Finally, the counterstain of DiD or DAPI was operated at room temperature in the dark for 10 min. Fluorescent images were captured using Olympus FV3000 confocal microscope with the software FV31S-SW (Olympus Corporation, Japan).
Paraffin-embedded mice tumor sections (5 μm) were incubated with anti-cleaved caspase-3 (Asp175) (1:200, 9661T, Cell Signaling Technology) overnight at 4 °C. Then, the secondary antibody, Cy3 conjugated goat anti-rabbit IgG(H + L) (1:300, GB21303, Servicebio), was incubated for 1 h in the dark at room temperature. After the counterstain of DAPI, tissue images were obtained from the Olympus FV3000 system.
2.14. Flow cytometry
PANC-1 cells or BxPC-3 cells were treated with ponatinib, perifosine and sorafenib according to the indicated treatments. For flow cytometry detection, 5 × 105 cells per group in 6-well plates were collected and washed with PBS twice. Cells were resuspended with 500 μL Binding Buffer and stained using 5 μL annexin V-fluorescein isothiocyanate (FITC) and 5 μL propidium iodide (PI) from KeyGEN BioTECH for 10–15 min in the dark. The pyroptosis was determined by BD Accuri C6 Flow Cytometer (BD Biosciences, San Jose, USA) over FL1 and FL3 channels, and data analysis was performed with FlowJo v10 software.
2.15. Intracellular ROS detection
The intracellular ROS generation was determined by DCFH-DA. PANC-1 cells were seeded on confocal dishes, cultured for 12 h, and then incubated with ponatinib or perifosine plus 5 mmol/L NAC for another 6 h. After being rinsed with PBS, the cells were incubated with a serum-free culture medium containing 10 μmol/L DCFH-DA for 20 min at 37 °C. Washed with PBS three times, the fluorescence images of the cells were captured using a confocal microscope at 488 nm excitation. The semi-quantitative analysis of ROS staining images was determined by Image J.
2.16. Xenograft tumor model
BALB/c Nude (CAnN.Cg-Foxn1nu/Crl) female mice were purchased from Beijing Vital River Laboratory Animal Technology (Beijing, China), with licensing number SCXK-2019-0001. Mice were maintained in specific pathogen-free isolator units and with a 12-h light–dark cycle, temperature 22 ± 2 °C and humidity 50 ± 10%. The animals received standard sterilized food and water ad libitum. All animal experiments were conducted in accordance with institutional guidelines following approval by the China Pharmaceutical University Animal Care and Use Committee.
For tumor suppression assay, 5 × 106 PANC-1 cells were resuspended in 0.1 mL PBS and subcutaneously implanted into the dorsal part (above the right axilla) of the mice at 6-week-old31,32. Only single-cell suspensions with >90% viability were used for injection. Tumor volume was determined based on caliper measurements using the formula (length × width2)/2 (mm3). Treatment was initiated when the tumors became palpable (∼100 mm3). Tumor-bearing mice were randomized to six groups (n = 6/group) with the same tumor volume calculated: (i) vehicle; (ii) perifosine (10 mg/kg); (iii) perifosine (20 mg/kg); (iv) perifosine (30 mg/kg); (v) gemcitabine (30 mg/kg); (vi) sorafenib (20 mg/kg). Perifosine and sorafenib were administered intraperitoneally five times per week, whereas gemcitabine was administered intraperitoneally once every two days. Perifosine and gemcitabine were dissolved in sterile saline solution, and sorafenib was suspended in 5% DMSO/40% PEG 300/5% Tween 80/50% saline solution. Tumor size was monitored every three days over 24 days until reaching the maximum allowable size. When the treatment was terminated, the tumors, livers, kidneys and serums were collected from mice after euthanasia. The treated over control tumor volume ratio (T/C%) was calculated by Eq. (3):
| (3) |
where RV = relative volume33,34.
For intratumoral GSDME knckdown experiment, AAV (AAV-shC or AAV-shGSDME) produced by ViGene Biosciences (Jinan, China) was intratumorally injected twice in 3-day intervals with 5.0 × 1011 vg/mouse when the subcutaneous tumor becomes palpable35, 36, 37. Treatment was initiated when the tumors volume up to 100–120 mm3. Tumor-bearing mice were randomized four groups (n = 6/group) with the same tumor volume: (i) AAV-shC + Vehicle; (ii) AAV-shC + Perifosine (30 mg/kg); (iii) AAV-shGSDME + Vehicle; (iv) AAV-shGSDME + Perifosine (30 mg/kg). Perifosine were administered intraperitoneally five times per week. Tumor size was monitored every three days over 21 days.
2.17. Immunohistochemistry (IHC)
Mice tumors were fixed immediately after excision in 4% paraformaldehyde in PBS for 24 h at room temperature before being dehydrated and paraffin-embedded. IHC staining was performed on 5 μm paraffin-embedded sections. The primary antibody against Ki67 (1:2000, 27309-1-AP, Proteintech) was diluted at 1:250 and then incubated overnight at 4 °C in a humidified container. After three washes with PBST, the tissue slides were treated with the secondary antibody (1:500, SA00004-2, Proteintech) for 1 h at room temperature. Sections were washed in PBST three times, stained with DAB according to the manufacturer's protocol, mounted on slides, and photographed using a digital microscope camera (Nikon, Japan).
2.18. Statistical analysis
Statistical analyses were performed with GraphPad Prism 8.0 (GraphPad Software Inc., La Jolla, USA). All data represented at least three independent experiments and were presented as means ± SEM. Two-group comparisons were made using a two-tailed Student's t-test, while one-way ANOVA followed by Tukey's post hoc test was used to assess differences among three groups or more. P < 0.05 was considered statistically significant; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001.
For high-throughput analysis, the screening quality can be assessed using the Z′ factor, calculated as Eq. (4):
| (4) |
where μ was the mean value of the signal (RLU/μg), and σ was the standard deviation of that. The signal-to-background (S/B) ratio was calculated using the μsora/μdmso ratio, and the coefficient of variation was calculated as Eq. (5):
| (5) |
3. Results
3.1. Construction of recombinant reporter plasmid
The GSDME protein has no cell death-inducing activity in the cytoplasm under normal conditions. However, full length GSDME can be cleaved by caspase-3 into GSDME-N and GSDME-C, when caspase-3 is triggered in response to particular stimulation18. The subsequent interaction between GSDME-N represents the occurrence of pyroptosis, which is crucial to the quantification of pyroptosis in our study. Using the simple and sensitive GLuc reporter, we constructed the hGLuc-hGSDME-PCA system to search for compounds that can specifically activate GSDME and lead to pyroptosis. When GSDME-N fragments oligomerized together, hGLuc(N) and hGLuc(C) folded into hGLuc with measurable bioluminescence activity (Fig. 1A). In our design, hGLuc(N) corresponds to the residues 18–109 of hGLuc, while hGLuc(C) corresponds to the residues 110–185 of hGLuc. Notably, the 16 amino acids N-terminal secretory signal sequence of GLuc was eliminated to prevent its natural secretion26 (Fig. 1B). Both fusion fragments, HA-hGLuc(C)-hGSDME and Flag-hGLuc(N)-hGSDME, were subcloned into the mammalian expression vector pBudCE4.1 (Fig. 1C). The production of restriction digest on agarose gel electrophoresis, along with Sanger sequencing, verified the accuracy of the DNA sequences (Fig. 1D). Due to the fact that pBudCE4.1 is a dual promoter expression vector, two fused target proteins can be expressed in the host cell both independently and concurrently39. We used the GSDME and tag antibodies to perform Western blot analysis after transiently transfecting the recombinant plasmid into 293T cells. The expression levels of two target proteins rose in tandem with the amount of DNA, demonstrating that the reporter plasmid constructed successfully (Fig. 1E).
Figure 1.
The construction process of the recombinant reporter plasmid. (A) The schematic diagram of the hGLuc-hGSDME-PCA screening system. (B) The insertion sites and amino acid sequences of the two fusion proteins. (C) The chart of the pBudCE4.1-HA-hGLuc(C)-hGSDME-Flag-hGLuc(N)-hGSDME recombinant reporter plasmid (called pBudCE4.1-coexpression for short). (D) Plasmid products after restriction digestion were analyzed by agarose gel electrophoresis. (E) The correctness of fusion protein expression was verified in 293T cells using antibodies against HA, Flag, GSDME and β-tubulin.
3.2. Fusion-type GSDME were cleaved and oligomerized in response to pyroptosis elicited by sorafenib in PANC-1 cells
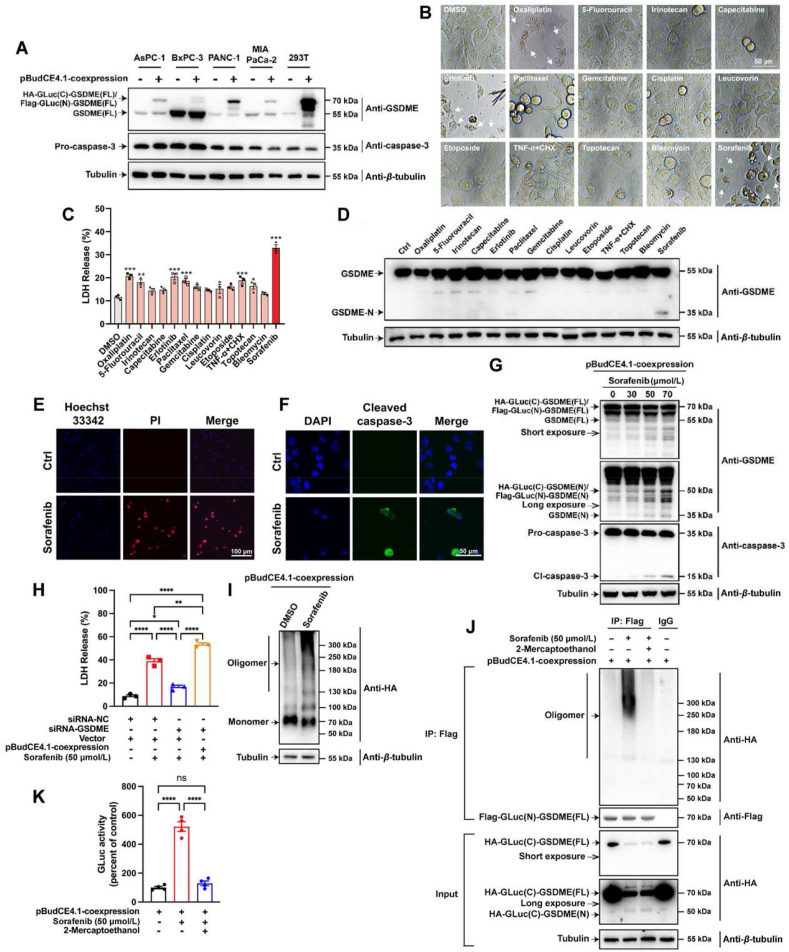
Since GSDME has the capacity to switch apoptosis into pyroptosis in a cell type-specific manner17, we evaluated the GSDME expression utilizing TCGA and GTEx project through the GEPIA2 web server (http://gepia2.cancer-pku.cn/#index). In ten leading cancer types with the highest mortality rates3, data showed that the expression of the GSDME gene was considerably only increased in PDAC tumor tissues compared to normal counterpart tissue (Supporting Information Fig. S1A and B). Because of the greater transfection efficiency, lower endogenous wild-type GSDME level, and higher exogenous fusion-type GSDME level, PANC-1 was chosen out of the four PDAC cells as a tool cell for the screening system (Fig. 2A; Fig. S1C and D). In order to screen out the best positive control suitable for our system, multiple compounds from clinical treatments of pancreatic cancer or published pyroptosis inducers were administrated in PANC-1 cells. Out of the 14 compounds, sorafenib exhibited the best effects on inducing pyroptosis which was much beyond our expectations. Sorafenib not only promoted the highest LDH release and GSDME-N protein levels but also induced obvious pyrolytic bubbles morphology in PANC-1 cells (Fig. 2B‒D). Since the pyroptosis executed by GSDME is accompanied by cell membrane rupture and caspase-3 cleavage11, we confirmed the occurrence of these processes using PI staining and immunofluorescence (Fig. 2E and F). Like endogenous GSDME protein, exogenous fusion-type GSDME protein were also cleaved by sorafenib to produce the N-terminal domain (Fig. 2G; Fig. S1E‒G). Therefore, the luciferase fragment fused to the N-terminus of GSDME did not interfere with the cleavage of GSDME. With the ability of inducing pyroptosis by cleaving GSDME and further releasing LDH, sorafenib also inhibited the viability and colony formation of PANC-1 cells in a dose-dependent manner (Fig. S1H‒L).
Figure 2.
PANC-1 cells and sorafenib were adopted to the system, and wild-type and fusion-type GSDME were cleaved and oligomerized in response to pyroptosis elicited by sorafenib. (A) The protein levels of endogenous wild-type GSDME and exogenous fusion-type GSDME in four kinds of PDAC cells (AsPC-1, BxPC-3, PANC-1 and MIA PaCa-2). 293T cells served as an expression control. (B) The morphology of PANC-1 cells treated with more than a dozen commonly used therapeutic agents for pancreatic cancer and reported pyroptosis inducers. (C) The LDH release of PANC-1 cells treated with the drugs above-mentioned. (D) The activation of GSDME protein in PANC-1 cells treated with the drugs above-mentioned. (E) Hoechst 33342/PI fluorescent staining assay of PANC-1 cells treated with sorafenib or vehicle. Scale bar = 100 μm. (F) Immunofluorescence analysis about the activation of caspase-3 in PANC-1 cells. Scale bars = 50 μm. (G) PANC-1 cells were transfected with the recombinant plasmid for 24 h and treated with sorafenib (0–70 μmol/L) for another 24 h, and then total cellular extracts were subjected to Western blotting using antibodies against caspase-3, GSDME and β-tubulin. (H) PANC-1 cells were transfected with siRNA-NC or siRNA-GSDME then transfected with empty vector or the recombinant plasmid, and then treated with or without sorafenib. The LDH release was analyzed. (I) PANC-1 cells, transfected with the recombinant plasmid, treated with or without sorafenib and lysed under non-reducing conditions, were resolved on a native gel, immunoblotted for HA. (J) Flag immunoprecipitation of lysates of PANC-1 cells, transfected with the recombinant plasmid and treated with sorafenib, were lysed with or without 2ME and resolved on a native gel. (K) PANC-1 cells, transfected with the recombinant plasmid and treated with sorafenib, were lysed with or without 2ME and then analyzed by luminescence detection. Data were presented as mean ± standard error of mean (SEM), n = 3. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
After that, it needs to be proved that the fusion-type GSDME is still biologically active after being cleaved. The results in Fig. 2H showed that the LDH release was strongly raised under alone sorafenib treatment; the LDH release was greatly reduced when endogenous GSDME was knocked down first and then sorafenib treatment. Nevertheless, supplementing exogenous fusion GSDME could completely reverse the above decreasing trend. Hence, the expressed fusion-type GSDME protein still has pore-forming activity. Afterward, the level of GSDME oligomerization in cells was detected by native gel immunoblot (Fig. 2I). The transfected samples both showed the expression of fusion GSDME monomer, while the protein levels of dimers and oligomers in the high molecular weight range were significantly increased after sorafenib treatment, demonstrating that fusion-type GSDME protein can oligomerize in host cells. Meanwhile, Flag immunoprecipitation was conducted to prove the binding between HA-GLuc(C)-GSDME(N) and Flag-GLuc(N)-GSDME(N) protein (Fig. 2J). Because the combination of these two stands for the occurrence of dimerization and oligomerization, and then promising restored luminescent activity of GLuc. We detected HA-related proteins in the form of oligomers in samples transfected with pBudCE4.1-coexpression plasmid and treated with sorafenib, proving that exogenously expressed HA-GLuc(C)-GSDME protein and Flag-GLuc(N)-GSDME proteins can oligomerize together. However, after the paralleled sample was treated with reducing agent 2ME, the HA-related protein almost disappeared, because of that 2ME inhibited the oligomerization state. Finally, using the hGLuc-hGSDME-PCA system, we preliminarily performed a luciferase activity detection (Fig. 2K). The luciferase activity after transfection and administration was significantly higher than transfection alone, which indicates the exogenously expressed fusion GSDME could interact to restore the activity of GLuc. When the cell lysate was pre-treated with 2ME, the luciferase activity decreased to the base value, also confirming that the increase of GLuc signal resulted from the oligomerization between the fusion GSDME. Based on the above results, we believe that the luciferase fragment fused to the N-terminus of GSDME does not interfere with the oligomerization of GSDME-NT. In conclusion, PANC-1 cells and sorafenib are suitable for the screening system.
3.3. Condition optimization of the hGLuc-hGSDME-PCA screening system
Although the screening system was established successfully (Fig. 3A), there were still some parts that need to be optimized. During the experiments, it was discovered that the transfection efficiency in PANC-1 cells was unsatisfactory. In order to find the best transfection reagents in PANC-1 cells, 12 commercially available transfection reagents were compared. Polyplus jetOPTIMUS, which had the highest transfection efficiency and the least degree of cell damage, was chosen for the subsequent plasmid transfection (Fig. 3B). Sorafenib was administrated in PANC-1 cells which were transfected with 0.2 μg of control plasmid or pBudCE4.1-HA-hGLuc(C)-hGSDME-Flag-hGLuc(N)-hGSDME recombinant reporter plasmid. The hGLuc-hGSDME-PCA system worked successfully in reconstituting the GLuc protein and exerting enzymatic activity after pyroptosis stimulation. Concurrent with increasing doses or time, a discernible rise in GLuc activity happened in cell lysates, reaching its peak at 24 h (Fig. 3C and D). The amount of transfected DNA and the ratio of DNA to the transfection agent were then optimized (Fig. 3E and F). We also discovered that the efficiency of transfection and drug delivery could be impacted by cell inoculation density. Then, the GLuc activity was compared under densities of 2.0 × 104–3.2 × 104 cells/well, and 2.4 × 104 cells/well was chosen as cell seeding density (Fig. 3G). These assays above optimized our hGLuc-hGSDME-PCA system. In conclusion, 2.4 × 104 cells/well of inoculation density, 200 ng of DNA amount, two-fold reagent volume and 50 μmol/L sorafenib treatment for 24 h were chosen for the following experiments.
Figure 3.
The experimental procedure was optimized for the hGLuc-hGSDME-PCA screening system. (A) The schematic diagram for protocol of hGLuc-hGSDME-PCA screening system. (B) The transfection effect on PANC-1 cells using 12 commercially available transfection reagents. (C) The dose-dependence of GLuc activity in PANC-1 cells. (D) The changes of GLuc activity in PANC-1 cells as time goes on after sorafenib treatment. (E) GLuc activity of cells transfected with different amounts of DNA. (F) GLuc activity of cells transfected with different ratios of DNA to transfection reagent. (G) GLuc activity of cells inoculated at a different density. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.4. Methodological validation and evaluation of the hGLuc-hGSDME-PCA screening system
In our screening system, two complementary GLuc proteins were expressed simultaneously by the same vector. In order to reveal whether the co-expression vector benefit our screening system better than two vectors transfecting concurrently, a test with 16 replicated wells each group was arranged40. The co-expression group was managed to transfect with the single plasmid pBudCE4.1-HA-hGLuc(C)-hGSDME-Flag-hGLuc(N)-hGSDME, while the co-transfection group was managed to transfect with plasmids pcDNA3.1-HA-hGLuc(C)-hGSDME and pcDNA3.1-Flag-hGLuc(N)-hGSDME at the same amount. Compared with co-transfection group, the co-expression group showed more consistent RLU values and reduced variance among wells (Fig. 4A). Similarly, it was discovered that the co-expression group had lower RSD values after six replicated administrations (Fig. 4B). The co-expression mode exhibited superior temporal stability and decreased inter-group variance in the two proteins’ expression levels, explaining why the luminescence signal was more steady in the co-expression mode (Fig. 4C and D; Supporting Information Fig. S2A). Particularly, higher levels of GSDME after transfection for 36 h, benefited compound-induced protein cleavage and subsequent luminescence activity assessment (Fig. 4D). Taken together, we confirmed that the co-expression mode is more suitable for our screening system.
Figure 4.
The hGLuc-hGSDME-PCA screening system is accurate and reliable for HTS assays of pyroptosis inducers. (A) The luminescence values of different groups transfected with only one plasmid pBudCE4.1-HA-hGLuc(C)-hGSDME-Flag-hGLuc(N)-hGSDME or transfected with two plasmids pcDNA3.1-HA-hGLucC-hGSDME and pcDNA3.1-Flag-hGLucN-hGSDME simultaneously. (B) GLuc activity in six times experiments using co-transfection mode or co-expression mode. (C) The protein expression of HA-GLuc(C)-GSDME and Flag-GLuc(N)-GSDME in six PANC-1 samples using co-transfection mode or co-expression mode. (D) The protein expression changes of HA-GLuc(C)-GSDME and Flag-GLuc(N)-GSDME in PANC-1 cells as time goes on after transfection. (E) The luminescence values of cells treated with sorafenib were then collected from different rows on the same culture plate. (F) GLuc activity of cells which expressed two fusion proteins independently or simultaneously and then treated with vehicle or sorafenib. (G) The linear relationship between the number of lysed cells and the luminescence values. (H) The RLU/μg protein values in a large number of samples treated with vehicle or sorafenib. (I) GLuc activity of cells treated with drugs known as caspase-3/GSDME pathway activators or involved in irrelevant pathways. FMK refers to Z-DEVD-FMK. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
Meantime, the intra-row and intra-column stability of the luminescence values in the present system was excellent41 (Fig. 4E and Fig. S2B). Two fusion proteins were single- or co-expressed in PANC-1 cells and stimulated by sorafenib, to verify the specificity of the PCA signal. It was found that GLuc activity significantly increased only when sorafenib was administrated after both proteins were simultaneously expressed (Fig. 4F). Additionally, the luminescence readings were linearly related to the number of lysed cells (Fig. 4G). After the lysed cell samples were kept at room temperature for up to 8 h, the GLuc activity remained largely stable, allowing for a smooth high-throughput analysis (Fig. S2D). Z′ factor, reflecting the signal variation and dynamic range, is a crucial parameter for evaluating the quality of HTS assays42. The Z′ factor value for sorafenib of this system was calculated to be 0.72, which satisfied the HTS requirements, over 0.5 (Fig. 4H)40. Finally, we used positive and negative compounds to validate the screening system's sensitivity and specificity38. GLuc activity was considerably elevated when drugs known to activate the caspase-3/GSDME pathway were administrated, while the activity stayed the same when drugs unrelated to the pathway were delivered (Fig. 4I). It was implied that our screening system had a low false positive rate in identifying GSDME activators. In general, our system for HTS assays of pyroptosis inducers was accurate and reliable.
3.5. High-throughput screening for pyroptosis inducers from FDA-approved anti-cancer drug library
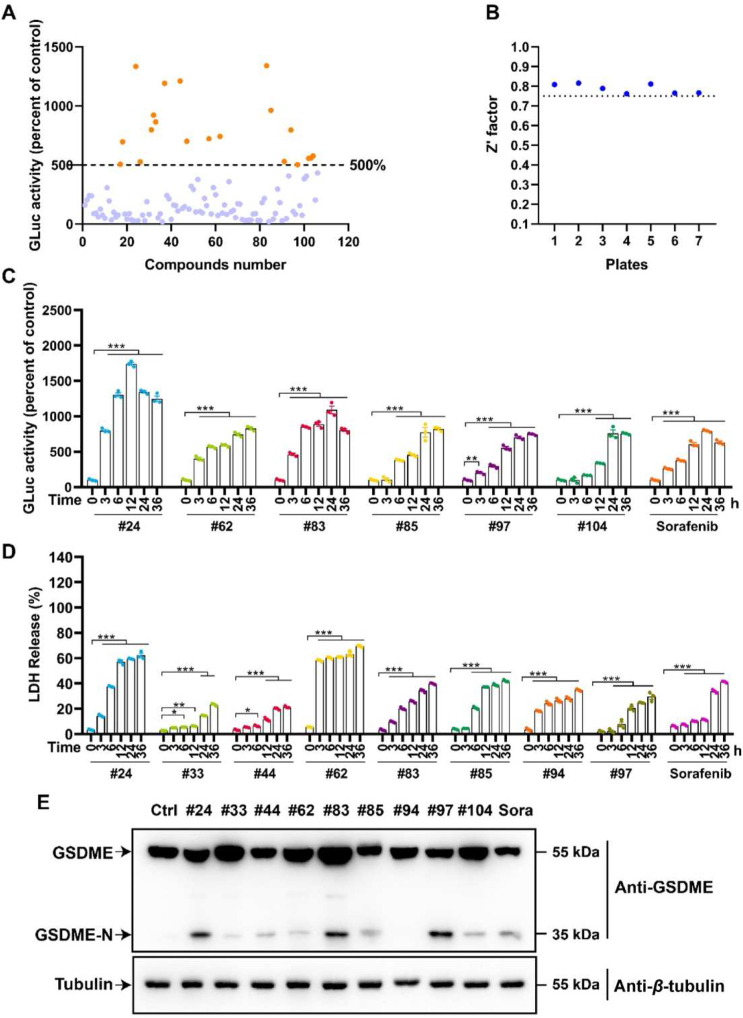
With the help of hGLuc-hGSDME-PCA screening system, we were committed to discovering compounds that could activate the caspase-3/GSDME pathway and trigger pyroptosis. After the screening of the FDA-approved anti-cancer drug library, it was found that twenty compounds exhibited more than 500% GLuc activity (Fig. 5A). Besides, Z′ factors of the seven plates for HTS all exceeded 0.75, demonstrating that our screening results was credible (Fig. 5B). For further screening, these 20 compounds were administrated in a time-gradient way. Noticeably, the six compounds significantly increased GLuc activities over time (Fig. 5C and Supporting Information Fig. S3A), while eight compounds caused intense LDH releases over time (Fig. 5D and Fig. S3B). All compounds above that boosted either GLuc activities or LDH release time-dependently were selected for further confirmation by Western blot. Encouragingly, compounds 24, 83 and 97 were found strongly activate GSDME to produce the N-terminal fragment (Fig. 5E). Interestingly, the screened component 97, doxorubicin, is a commonly used pyroptosis inducer in GSDME-expressing cells, demonstrating the reliability of our screening system18,43. Meantime, compounds 24 ponatinib and 83 perifosine, were first discovered to induce the cleavage of GSDME.
Figure 5.
Ponatinib and perifosine were screened out as pyroptosis inducers performing HTS from FDA-approved anti-cancer drug library. (A) The GLuc activity acquired from our hGLuc-hGSDME-PCA screening system of these 106 compounds in the FDA-approved anti-cancer drug library. Where the orange dots represented compounds with GLuc activity greater than 500%, the blue dots represented compounds with GLuc activity less than 500%. (B) Z′ values of seven plates used for this HTS. (C) The compounds had noticeable time-dependent GLuc activity in the top 20 GLuc activity. (D) The compounds with noticeable time-dependent LDH release in the top 20 GLuc activity. (E) The GSDME-N expression levels of the compounds screened by GLuc activity and LDH release. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.6. Ponatinib and perifosine inhibited PDAC cells growth
To explore the anti-cancer effects of ponatinib and perifosine on pancreatic cancer, we employed the PDAC cell lines (AsPC-1, BxPC-3, PANC-1, MIA PaCa-2) as well as human normal pancreatic duct epithelial cell HPDE6-C7 (H6c7). Ponatinib and perifosine treatments both significantly suppressed the cell viability of BxPC-3 and PANC-1 cells in a dose- and time-dependent manner. However, the growth of H6c7 cells was almost unaffected by ponatinib and perifosine (Fig. 6A and B). The clone formation assay visually demonstrated the suppression effects of ponatinib and perifosine on PANC-1 cell proliferation (Fig. 6C). Likewise, the EdU insertion assay also showed that ponatinib and perifosine affected the proliferative process of PANC-1 cells (Fig. 6D). LDH release which suggested plasma membrane rupture and leakage, was dose-dependently elevated in BxPC-3 and PANC-1 cells rather than H6c7 cells after ponatinib and perifosine administration (Fig. 6E). In addition to this, the treated BxPC-3 and PANC-1 cells both exhibited large bubbles emerging from the plasma membrane and cell swelling, which were characteristic pyroptotic cell morphology12 (Fig. 6F). Ponatinib and perifosine also produced similar proliferation inhibition and LDH release promotion on AsPC-1 and MIA PaCa-2 (Supporting Information Fig. S4A–D). In conclusion, ponatinib and perifosine inhibited the growth of pancreatic cancer cells in vitro, indicating the possibility of being anti-PDAC drug candidates.
Figure 6.
Ponatinib and perifosine inhibited cell proliferation and induced cell death of PDAC cells. (A) BxPC-3, PANC-1 and H6c7 cells were treated with ponatinib (0–20 μmol/L) or perifosine (0–90 μmol/L) for 24 h, and then cell viability was analyzed by CCK-8 assay. (B) BxPC-3 and PANC-1 cells were treated with ponatinib (10 μmol/L) or perifosine (50 μmol/L) for the indicated time, and then cell viability was analyzed by CCK-8 assay. (C) PANC-1 cells were treated with ponatinib (0–5 μmol/L) or perifosine (0–30 μmol/L) for 24 h. After two weeks, cells were stained with crystal violet and pictured. (D) Confocal analysis of EdU incorporation in PANC-1 cells treated with ponatinib or perifosine. (E) BxPC-3, PANC-1 and H6c7 cells were treated with the two compounds at indicated concentrations for 24 h, and LDH-release was analyzed using an LDH assay kit. (F) BxPC-3 and PANC-1 cells were treated with ponatinib, perifosine or sorafenib for 24 h, and then microscopic imaging was performed. Arrowheads indicated ballooned cell membrane characteristics of pyroptotic cells, scale bar = 50 μm, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.7. Ponatinib and perifosine triggered PDAC cell pyroptosis via the caspase-3/GSDME pathway
As ponatinib and perifosine caused cell swelling with large bubbles, characteristics of pyroptosis (Fig. 6F), the following experiments were performed to verify if ponatinib and perifosine inhibited PDAC cell growth though pyroptosis. It was obvious that ponatinib and perifosine induced cleavage of caspase-3 in a dose-dependent manner whether in BxPC-3 or PANC-1 cells (Fig. 7A and B). More importantly, treatment with ponatinib and perifosine also promoted the cleavage of GSDME, a pyroptosis hallmark, in BxPC-3 and PANC-1 cells (Fig. 7A and B). Consistent results were also observed in AsPC-1 and MIA PaCa-2 cells (Supporting Information Fig. S5A). Similarly, ponatinib and perifosine induced cleavage of caspase-3 and GSDME over time in PDAC cells (Fig. 7A and B). As expected, cleaved caspase-3 was observed in PANC-1 cells after ponatinib or perifosine administration (Fig. 7C). Subsequently, we found that intracellular GSDME tended to localize to the cell membrane rather than the cytoplasm. It can be speculated that this dose-dependent aggregation was derived from the oligomerization of active fragment GSDME-N on plasma membranes (Fig. 7D). To confirm whether intracellular GSDME-N protein finally oligomerized after administration of ponatinib or perifosine, native gel immunoblot was performed (Fig. 7E). Compared with the control group, the GSDME dimer and oligomer bands were significantly increased after drug treatment, and more evident after overexpressing GSDME in advance. After the pre-knockdown of GSDME, the high molecular weight protein band basically disappeared. The data confirmed that ponatinib and perifosine could not only induce the cleavage of GSDME in cells, but also finally induce the oligomerization of GSDME. Thereby, the plasma membrane permeability was altered after ponatinib or perifosine stimulation, proved by the dose-dependent increases of PI-positive PANC-1 cells (Fig. 7F). As cell swelling and membrane rupture were also displayed in cells undergoing necroptosis14, the RIPK1 inhibitor necrostatin-1 and RIPK3 inhibitor GSK-872 were utilized to distinguish pyroptosis from necroptosis. As expected, necroptosis inhibitors exhibited no significant effects on cell death and LDH release induced by ponatinib or perifosine (Fig. 7G and Fig. S5B). Apart from this, PANC-1 and BxPC-3 cells proceeded directly to the annexin V and PI double-positive stage treated by ponatinib or perifosine, also proving that the two compounds induced pyroptosis instead of apoptosis (Fig. 7H and Fig. S5C).
Figure 7.
Ponatinib and perifosine triggered PDAC cell pyroptosis via the caspase-3/GSDME pathway. (A) BxPC-3 cells were treated with ponatinib (0–10 μmol/L) for 24 h or with ponatinib (10 μmol/L) for 6, 12 and 24 h. PANC-1 cells needed to be treated with two-fold concentration of ponatinib or for a longer time. Total cellular extracts were subjected to Western blot analyses using antibodies against caspase-3, GSDME and β-tubulin. (B) BxPC-3 and PANC-1 cells were treated with perifosine (0–70 μmol/L) for 24 h or with perifosine (70 μmol/L) for 12, 18 and 24 h. (C) Immunofluorescence analysis about the activation of caspase-3 in PANC-1 cells. Scale bars = 50 μm. (D) Membrane localization analysis of GSDME was performed in PANC-1 cells with anti-GSDME antibody, membrane dye DiD and nuclear dye DAPI. Scale bars = 20 μm. (E) PANC-1 cells were transfected with siRNA-GSDME or overexpression plasmid-GSDME, and then treated with ponatinib or perifosine. Cell samples lysed under non-reducing conditions and resolved on a native gel, immunoblotted for GSDME. (F) Hoechst 33342/PI fluorescent staining assay of PANC-1 cells treated with ponatinib, perifosine or vehicle. Scale bar = 100 μm. (G) After pre-treated with DMSO, necrostatin-1 or GSK-872, PANC-1 cells were treated with ponatinib or perifosine. The LDH release was analyzed. (H) Flow cytometry detection of the percentage of annexin V+/PI+ PANC-1 cells treated with ponatinib, perifosine or vehicle. (I) PANC-1 cells were treated with ponatinib or perifosine for 24 h, and total cellular extracts were subjected to Western blot analyses using antibodies against GSDMD and β-tubulin. (J) PANC-1 were pre-treated with or without Z-DEVD-FMK (20 μmol/L) for 2 h, then treated with ponatinib (10 μmol/L) or perifosine (50 μmol/L) for 24 h, and total cellular extracts were subjected to Western blot analyses using antibodies against caspase-3, GSDME, and β-tubulin. (K) BxPC-3 and PANC-1 cells were treated with ponatinib or perifosine in the absence or presence of Z-DEVD-FMK (20 μmol/L) for 6, 12 and 24 h, then LDH-release was analyzed. ∗∗∗P < 0.001, vs. drug treatment alone at each indicated time. (L) and (M) PANC-1 cells were transfected with siRNA-NC, siRNA-CASP3 or siRNA-GSDME and then treated with ponatinib or perifosine; total cellular extracts were subjected to Western blot analyses using antibodies against caspase-3, GSDME and β-tubulin. (N) PANC-1 cells were incubated with or without NAC and treated with ponatinib or perifosine. Microscopic imaging was performed. Arrowheads indicated ballooned cell membrane characteristics of pyroptotic cells, scale bar = 50 μm ns, no significance, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
After that, we sought to verify whether GSDME and caspase-3 is indispensable for ponatinib-induced or perifosine-induced pyroptosis in PDAC cells. Since both GSDME and GSDMD can induce pyroptosis, it was necessary to find out if ponatinib and perifosine activate GSDMD protein. However, ponatinib scarcely induced cleavage of GSDMD, whereas perifosine induced a small amount of cleavage of GSDMD. In other words, ponatinib and perifosine induced pyroptosis mainly though GSDME (Fig. 7I). To confirm that caspase-3 is specifically required for the cleavage of GSDME and release of LDH, cells were pre-treated with caspase-3-specific inhibitor Z-DEVD-FMK. As a result, not only the cell morphology but also GSDME cleavage and LDH extracellular release were recovered after the pre-treatment with Z-DEVD-FMK, compared with ponatinib or perifosine administration only (Fig. 7J and K; Fig. S5D and E). In addition to the caspase-3 inhibitors, the knockdown of CASP3 was also applied to demonstrate that caspase-3 and GSDME were important for ponatinib or perifosine-induced pyroptosis. Likewise, the knockdown of CASP3 resulted in a visible decrease in GSDME-N generation and the improvement of pyroptotic cell morphology (Fig. 7L and Fig. S5F). The knockdown of GSDME led to decreased LDH release and improved pyroptotic cell morphology without affecting the activation of caspase-3 (Fig. 7M; Fig. S5F and G). Moreover, the knockdown of GSDMD did not affect the cell morphology and LDH release, indicating that GSDMD was not engaged in ponatinib or perifosine-induced pyroptosis (Fig. S5F and H). In conclusion, we concluded that ponatinib and perifosine provoked pyroptosis in PDAC cells via activating the caspase-3/GSDME pathway.
Finally, we found that ponatinib and perifosine elicited the overproduction of ROS in PANC-1 cells, which was markedly abrogated by the ROS scavenger NAC (Fig. S5I). To further demonstrate the involvement of ROS in ponatinib- and perifosine-induced cell death, we performed the following experiment. The pyroptotic cell morphology was significantly inhibited by NAC (Fig. 7N) and the almost entirely reversed activation level of GSDME by NAC (Fig. S5J and K) demonstrated that ROS overproduction is an upstream event of GSDME activation in this process. Since ROS is a regulator of caspase-3 activation43, it was quite possible that ponatinib and perifosine induced PDAC cell pyroptosis through the ROS-dependent pathway.
3.8. Perifosine and sorafenib inhibited the xenograft tumor growth and induced PDAC cells pyroptosis in vivo
To verity the anti-PDAC activity of perifosine in vivo, BALB/c nude mice were used to establish the PANC-1 xenograft tumor model before drug administration. As a positive drug, gemcitabine strongly inhibited tumor growth, which was consistent with the earlier researches18,44. PANC-1 solid tumor growth was greatly slowed down by perifosine at all three doses. The effects of perifosine at the high dose (30 mg/kg) on tumor volume and weight were comparable to those of gemcitabine (Fig. 8A and B). Images of the tumors confirmed that perifosine reduced tumor burden in a dose-dependent manner in the PANC-1 tumor model (Fig. 8C). Further findings from tumor proliferation rates revealed that high-dose perifosine dramatically inhibited the tumor development with comparable efficacy to gemcitabine (Fig. 8D). Histological analysis of tumors from the vehicle group presented hypercellularity and hyperchromatic, in particular asymmetrically dividing nuclei. Treatment with perifosine (20 and 30 mg/kg) displayed obviously cell cavitation, decreased tumor cells and deep-stained nuclei in tumor mass (Fig. 8E). Perifosine administration declined Ki67 levels, which was the indicator of cell proliferation (Fig. 8E). The average body weight and survival state of perifosine-treated mice did not decidedly change compared with control mice (Supporting Information Fig. S6B and C). In addition, there were no apparent morphological changes in the livers and kidneys and no significant enhancement in ALT/AST levels, suggesting that perifosine had no obvious toxicity to the xenograft mice (Fig. S6D‒F).
Figure 8.
Perifosine and sorafenib inhibited the xenograft tumor growth and induced PDAC cells pyroptosis in vivo. PANC-1 cells were inoculated into mice to establish the xenograft tumor model as described in Methods. (A) The tumor volume measurement proceeded every three days. (B) The subcutaneous tumors were dissected and weighed. (C) Representative photographs of isolated tumors on Day 24. (D) The tumor proliferation rate (%) was calculated using the initial and final tumor volume. (E) Histopathological and immunohistochemical analysis of subcutaneous tumors at the end of the experiment. Scale bars were 200 μm (upper) or 50 μm (lower). (F) The release of serum LDH in the mice was measured by LDH assay. (G) Western blot analyses of GSDME, GSDME-N, caspase-3 and cleaved caspase-3 expression in harvest tumor tissues. (H) Immunofluorescence analysis of cleaved caspase-3 in subcutaneous tumors at the end of the experiment. Scale bars = 50 μm. (I) The expressions of GSDME proteins were tested by Western blot using one tumor sample isolated on Days 7 and 14, respectively. (J) Representative photographs of isolated tumors on Day 21 in intratumoral GSDME knockdown experiment. (K) The tumor volume measurement proceeded every three days in intratumoral GSDME knockdown experiment. (L) The release of serum LDH was measured in intratumoral GSDME knockdown experiment. (M) Western blot analyses of GSDME, GSDME-N, caspase-3 and cleaved caspase-3 expression in harvest tumor tissues in intratumoral GSDME knockdown experiment. Data were presented as mean ± SEM, n = 6. ns, no significance. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
To find out whether the tumor inhibition was link to perifosine-induced pyroptosis, the following experiments were conducted. In contrast with control mice, the serum LDH was increased in the perifosine treatment group but not in the gemcitabine treatment group (Fig. 8F). Moreover, we analyzed the pyroptosis-related proteins in tumor tissues. The outcomes showed that perifosine therapy upregulated the activation of caspase-3 and GSDME (Fig. 8G). Besides, perifosine boosted the protein levels of cleaved caspase-3 in tumor tissue cells, according to immunofluorescence images (Fig. 8H). Interestingly, sorafenib usually used in HCC, exhibited similar inhibition effects on pancreatic cancer through pyroptosis, which was found at the first time. Sorafenib reduced the tumor size and weight, promoted the LDH release and activated GSDME protein (Fig. 8A, B, F and G). In summary, these results suggested that perifosine and sorafenib might inhibit the growth of PDAC tumors in vivo by inducing GSDME-mediated pyroptosis.
To prove the effect of perifosine comes from GSDME-dependent pyroptosis, we constructed the adeno-associated virus (AAV) packaging GSDME interference plasmid, namely AAV-shGSDME. The AAV-shGSDME or AAV-shC was injected into subcutaneous tumors in situ formed by PANC-1 GSDME WT cells to reduce the expression of GSDME in tumor tissues (Fig. 8I). It was found that the tumor volume and tumor weight of the AAV-shC + Perifosine group were significantly lower than those of the AAV-shC + Vehicle group, while the therapeutic effect of perifosine was almost reversed by AAV-shGSDME (Fig. 8J and K, Fig. S6I), indicating that the inhibitory effect of perifosine on the growth of PANC-1 xenograft tumors was greatly weakened after intratumoral knockdown of GSDME. The trends in tumor proliferation rates were similar (Fig. S6J). In addition, the serum LDH level, GSDME-N, and cleaved caspase-3 level of the AAV-shC + Perifosine group were significantly higher than those of the AAV-shC + Vehicle group (Fig. 8L and M). By contrast, the trend of elevated LDH level by perifosine was reversed in the AAV-shGSDME + Perifosine group. It implied that GSDME could not perform the function of pyroptosis executor when administrated due to knockdown, making it difficult for tumor cells to undergo cell rupture and pyroptosis. Taken together, we believe that GSDME-dependent pyroptosis plays a critical role in treating PDAC by perifosine.
4. Discussion
Plenty of studies have highlighted the pivotal role of the gasdermin family members in mediating inflammatory cell death through their pore-forming activity45, 46, 47. Activated by chemotherapeutic agents, GSDME can trigger cell pyroptosis and enhance antitumor immunity. Thus, GSDME has been implicated in cancer as a tumor suppressor protein21,48. Adversely, GSDME is generally silenced due to high methylation of the promoter region23. In most types of cancers, such as acute myeloid leukemia, colon adenocarcinoma, breast invasive carcinoma, prostate adenocarcinoma, cervical carcinoma, bladder urothelial carcinoma, and rectum adenocarcinoma, the expression of GSDME is decreased18. Whereas the expression of GSDME is increased in some other types of tumors, such as pancreatic adenocarcinoma, glioblastoma multiforme, skin cutaneous melanoma, and head and neck squamous cell carcinoma19,23,49. It is hard to explain why some tumor cells would highly express GSDME. Among cancers with high GSDME expression, pancreatic adenocarcinoma belongs to the ten leading cancer types with the highest mortality rates3. Gemcitabine is the standard therapy for PDCA patients at all stages, but it only provides a limited survival benefit6. One of the reasons is that 50% of PDAC cases have TP53 mutations, which leads to the decreased pro-apoptosis ability of TP53 and increased chemoresistance of PDAC cells to gemcitabine1,8. Therefore, it is urgent to identify more promising PDAC therapeutics that can play a role through non-apoptotic cell death pathways. It has been proposed that upregulated GSDME in PDAC cells can be used to mediate the resistance to pancreatic enzymatic digestion through a GSDME-YBX1-mucin pathway23. Thus, we must utilize the pore-forming activity of GSDME in PDAC cells to trigger pyroptosis, and more importantly, reverse its possible tumor-promoting role.
Monitoring cell pyroptosis probably plays a vital role in developing potential therapeutics for many cancers, including PDAC50, 51, 52. Currently, the commonly used method for monitoring pyroptosis relies on detecting caspase cleavage, which analyses the protein level changes of N-terminal gasdermin18. However, it cannot detect the aggregation of N-terminal gasdermin. Ji et al. have developed a pyroptosis reporter P30-GLuc-P20 by inserting a secreted luciferase GLuc into the GSDMD protein. Stimulated by pyroptosis inducers, cleaved P30-GLuc-P20 was released extracellularly and emitted a detectable signal53. Nevertheless, since the aggregation of GSDME-N is the downstream event of pyroptosis16, the detection of GSDME-N interaction appears more pertinent and conclusive for pyroptosis activity evaluation. Concretely, the pyroptosis reporter system in our study had several characteristics or advantages compared with the previous detection method: (1) Using GSDME instead of GSDMD as the executor of pyroptosis, the purpose was to screen anti-tumor drugs rather than anti-inflammatory drugs; (2) Using humanized GLuc as the reporter protein, its simple reaction conditions and high luminescence activity ensured rapid and smooth high-throughput analysis26,29; (3) The secretory function of GLuc was abolished, simplifying the analysis object and eliminating the need for operator to consider signals in the cell culture medium; (4) For the first time, GSDME was constructed into the protein-fragment complementation assay system, and the biological oligomerization of GSDME-N in pyroptosis were utilized to perform screening, which greatly enhanced the specificity of detection signal; (5) Simultaneous expression of two fusion proteins expressed by a dual-promoter vector, simplified the transfection procedure and reduced transfection errors; (6) Directly using PDAC cell as the tool cell, which made the screening environment consistent with the actual action environment of drugs and availed to screen out appropriate drugs for specific diseases; (7) The analysis conditions of the screening system were utterly optimized, and the stability and reliability of the screening system were verified in details. In this study, we exhibited compelling evidence that the hGLuc-hGSDME-PCA screening system was a valuable tool for monitoring the activity of GSDME and screening GSDME-mediated pyroptosis inducers. What is noteworthy is that since GSDME is specifically cleaved by pro-apoptotic caspase-3, it also appears that this screening system can identify compounds that lead to caspase-3 activation. In GSDME-sufficient cells, once caspase-3 is activated, the cleaved caspase-3 can cleave GSDME to generate GSDME-NT, the latter of which oligomerize to form pores in the plasma membrane. Consistent with the understanding, we found that basically all the GSDME activators screened out by the hGLuc-hGSDME-PCA system also activated caspase-3 protein. Thus, it can actually screen out the activator of apoptotic signaling that eventually activates caspase-3. This fact extends the research scope of the hGLuc-hGSDME-PCA system from GSDME-dependent pyroptosis to caspase-3-involved pyroptosis and apoptosis.
There are four aspects of this study that have to be addressed. Due to the rapid decay of the GLuc bioluminescence reaction, a luminometer with a built-in injector was required to shorten analysis time for high-throughput assays. Additionally, the substrate coelenterazine could permeate cell membranes and diffuse into all cellular compartments, allowing quantitative analysis in live cells in this PCA system26. We only analyzed the luminescence signal in cell lysates in the current work. In subsequent studies, we would design a new system helped with High Content Profiler to monitor pyroptotic activity in living cells dynamically and in real time. Although reliable results were obtained, our high-throughput screening assay only analyzed 106 FDA-approved anticancer drugs. Future research needs to expand the drug library to screen more effective pyroptosis inducers. Moreover, the underlying molecular mechanism involved in ponatinib-induced or perifosine-induced pyroptosis has not been completely elucidated. Studies beyond the scope of the current paper are required to understand the relationship between observed excessive ROS and the activation of caspase-3/GSDME pathway in two drugs-treated PDAC cells.
To our knowledge, ponatinib is a multi-target kinase inhibitor. Its primary cellular target is the BCR-ABL tyrosine kinase protein which promotes the progression of chronic myeloid leukemia (CML). Ponatinib inhibits both native and mutant BCR-ABL, including T315I54. Additional targets of ponatinib include KIT55, RET56, and FLT357, and members of the FGFR58 VEGFR, PDGFR59, and SRC60 families of kinases. It has been reported that non-receptor tyrosine-protein kinase plays a role in many key processes linked to cell growth and survival. ABL1 not only targets mitochondria in response to oxidative stress but also phosphorylates CASP on ‘Tyr-153’ and regulates its processing in the apoptotic response to DNA damage61. By contrast, perifosine is a novel alkylphospholipid targeting PKB162, MAPK1 and PKCA63. Perifosine can alter mitogenic signal transmission, phospholipid metabolism, membrane lipid composition, and membrane permeability, resulting in cell differentiation and inhibition of cell growth64. Furthermore, this compound suppresses the anti-apoptotic mitogen-activated protein kinase (MAPK) pathway and modifies the balance between the pro-apoptotic stress-activated protein kinase (SAPK/JNK) and MAPK pathways65. In addition, as the first oral multi-kinase inhibitor that targets RAF/MEK/ERK pathway, sorafenib has been discovered and approved for advanced hepatocellular carcinoma8. A clinical trial titled ‘A Phase 1 Study of Neoadjuvant Chemotherapy, Followed by Concurrent Chemoradiation With Gemcitabine, Sorafenib, and Vorinostat in Pancreatic Cancer’ led by Virginia Commonwealth University revealed the role of sorafenib in adjuvant treatment of pancreatic cancer66. Our data concurred with the previous reports and further comprehensively demonstrated that sorafenib was a potent inducer of pyroptosis, even in PANC-1 cells expressing relatively low GSDME67,68. Hence, sorafenib might play a clinical role in pancreatic cancer by GSDME-mediated pyroptosis.
In this study, we expanded the conventional understanding to propose that GSDME-dependent pyroptosis was closely relevant to the excellent curative effect of ponatinib or perifosine in PDAC. In vitro, ponatinib and perifosine significantly suppressed PDAC cell proliferation by inducing caspase-3-activated and GSDME-mediated pyroptosis. Neither ponatinib nor perifosine had little effect on the viability and proliferation of normal pancreatic cells, indicating the specific killing of pancreatic cancer cells. In vivo, it is confirmed that perifosine and sorafenib markedly inhibited the PANC-1 xenograft tumors growth and activated the caspase-3/GSDME pathway. Furthermore, the results of knocking down GSDME in the tumor revealed that GSDME-dependent pyroptosis plays a critical role in treating PDAC by perifosine. Considering the apparent skin toxicity of ponatinib in preliminary in vivo experiments, we did not conduct further investigation. However, whether this toxicity can be ameliorated by adjusting the dosing regimen requires further experimental studies. Future work will explore the relationship between the mechanisms of ponatinib, perifosine or sorafenib-induced pyroptosis and their traditional targets. Overall, ponatinib, perifosine and sorafenib maybe three good candidates for pancreatic cancer therapy, and combination therapy with gemcitabine by targeting GSDME can reduce the resistance of pancreatic cancer to chemotherapeutics.
This study pioneered a screening method for pyroptosis inducers targeting GSDME protein for the first time, which was feasible and reliable. In addition to screening natural products and chemical compounds still in the experimental or clinical trial stage, it can also be applied to re-evaluating marketed anticancer drugs to obtain pyroptosis-inducing agents targeting GSDME efficiently. It is important to note that GSDME-mediated pyroptosis in normal cells contributes to the toxicity of chemotherapy15,18. Because normal pancreatic cells express GSDME considerably lower than pancreatic cancer cells, our work will constitute a safe strategy for treating PDAC. Together, our studies lay the foundation for developing novel therapeutic strategies for tumors with relatively higher GSDME expression. In addition, recent findings shed light on pyroptosis-induced inflammation triggers robust antitumor immunity and can synergize with checkpoint blockade21,69. Hence, the pyroptosis inducers screened for specific cancers can reduce the resistance to first-line treatment drugs and improve the efficacy of cancer immunotherapy. By establishing of GLuc-hGSDME-PCA screening system, it is promising to improve the understanding of cell pyroptosis in disease-specific contexts and clinical applications.
5. Conclusions
Overall, hGLuc-hGSDME-PCA is a powerful tool that can evaluate pyroptosis-inducing activity of chemotherapy drugs in a quantitative and high-throughput manner. Based on this system, ponatinib and perifosine were screened out from the FDA-approved anti-cancer drug library. For the first time, we found that ponatinib and perifosine could strongly trigger pyroptosis via caspase-3/GSDME signaling pathway of PDAC cells to suppress pancreatic cancer progression. In conclusion, our findings reveal the great promise of hGLuc-hGSDME-PCA in identifying compounds triggering GSDME-dependent pyroptosis and developing promising therapeutic agents for PDAC.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (No. 82174100) and the National Key R&D Program of China (No. 2022YFC3501601).
Author contributions
Jun Chen and Ping Li conceived and designed the research. Yang Liu performed most of the experiments, analyzed data, and wrote the manuscript. Xiaowei Zhang, Ping Zhang, Tingting He and Weitao Zhang performed partial experiments and acquired the data. Weitao Zhang wrote a part of the manuscript and improved the manuscript. Dingyuan Ma provided reagents, mice, and conceptual advice. Jun Chen directed and supervised the research. All authors reviewed the results and approved the final version of the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.07.018.
Contributor Information
Ping Li, Email: liping2004@126.com.
Jun Chen, Email: chenj2002cpu@126.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Chen X., Zeh H.J., Kang R., Kroemer G., Tang D. Cell death in pancreatic cancer: from pathogenesis to therapy. Nat Rev Gastroenterol Hepatol. 2021;18:804–823. doi: 10.1038/s41575-021-00486-6. [DOI] [PubMed] [Google Scholar]
- 2.Huang L., Holtzinger A., Jagan I., BeGora M., Lohse I., Ngai N., et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med. 2015;21:1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:804–823. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 4.Collisson E.A., Bailey P., Chang D.K., Biankin A.V. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2019;16:207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 5.Neoptolemos J.P., Kleeff J., Michl P., Costello E., Greenhalf W., Palmer D.H. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15:333–348. doi: 10.1038/s41575-018-0005-x. [DOI] [PubMed] [Google Scholar]
- 6.Chen X., Kang R., Kroemer G., Tang D. Targeting ferroptosis in pancreatic cancer: a double-edged sword. Trends Cancer. 2021;7:891–901. doi: 10.1016/j.trecan.2021.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Quiñonero F., Mesas C., Doello K., Cabeza L., Perazzoli G., Jimenez-Luna C., et al. The challenge of drug resistance in pancreatic ductal adenocarcinoma: a current overview. Cancer Biol Med. 2019;16:688–699. doi: 10.20892/j.issn.2095-3941.2019.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Kang R., Kroemer G., Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18:280–296. doi: 10.1038/s41571-020-00462-0. [DOI] [PubMed] [Google Scholar]
- 9.Wang M., Wu M., Liu X., Shao S., Huang J., Liu B., et al. Pyroptosis remodeling tumor microenvironment to enhance pancreatic cancer immunotherapy driven by membrane anchoring photosensitizer. Adv Sci (Weinh) 2022;9 doi: 10.1002/advs.202202914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu S.K., Chu Y.H., Syue W.J., Lin H.Y.H., Chang W.T., Chen J.Y.F., et al. The role of nonapoptotic programmed cell death‒ferroptosis, necroptosis, and pyroptosis‒in pancreatic ductal adenocarcinoma treatment. Front Oncol. 2022;12:872–883. doi: 10.3389/fonc.2022.872883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers C., Fernandes-Alnemri T., Mayes L., Alnemri D., Cingolani G., Alnemri E.S. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8 doi: 10.1038/ncomms14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu W., Che Y., Zhang Q., Huang H., Ding C., Wang Y., et al. Apaf-1 pyroptosome senses mitochondrial permeability transition. Cell Metabol. 2021;33:424–436. doi: 10.1016/j.cmet.2020.11.018. [DOI] [PubMed] [Google Scholar]
- 13.Shi J., Gao W., Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci. 2017;42:245–254. doi: 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen X., Wang H., Weng C., Jiang H., Chen J. Caspase 3/GSDME-dependent pyroptosis contributes to chemotherapy drug-induced nephrotoxicity. Cell Death Dis. 2021;12:186–201. doi: 10.1038/s41419-021-03458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broz P., Pelegrín P., Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol. 2020;20:143–157. doi: 10.1038/s41577-019-0228-2. [DOI] [PubMed] [Google Scholar]
- 17.Rogers C., Erkes D.A., Nardone A., Aplin A.E., Fernandes-Alnemri T., Alnemri E.S. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat Commun. 2019;10:1689–1705. doi: 10.1038/s41467-019-09397-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Gao W., Shi X., Ding J., Liu W., He H., et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 19.Cai J., Yi M., Tan Y., Li X., Li G., Zeng Z., et al. Natural product triptolide induces GSDME-mediated pyroptosis in head and neck cancer through suppressing mitochondrial hexokinase-ΙΙ. J Exp Clin Cancer Res. 2021;40:190–207. doi: 10.1186/s13046-021-01995-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang R., Xu J., Zhang B., Liu J., Liang C., Hua J., et al. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol. 2020;13:110–128. doi: 10.1186/s13045-020-00946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z., Zhang Y., Xia S., Kong Q., Li S., Liu X., et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579:415–420. doi: 10.1038/s41586-020-2071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X., Chen G., Yu K.N., Yang M., Peng S., Ma J., et al. Cold atmospheric plasma induces GSDME-dependent pyroptotic signaling pathway via ROS generation in tumor cells. Cell Death Dis. 2020;11:295–306. doi: 10.1038/s41419-020-2459-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv J., Liu Y., Mo S., Zhou Y., Chen F., Cheng F., et al. Gasdermin E mediates resistance of pancreatic adenocarcinoma to enzymatic digestion through a YBX1-mucin pathway. Nat Cell Biol. 2022;24:364–372. doi: 10.1038/s41556-022-00857-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P., Wang L., Di L.J. Applications of protein fragment complementation assays for analyzing biomolecular interactions and biochemical networks in living cells. J Proteome Res. 2019;18:2987–2998. doi: 10.1021/acs.jproteome.9b00154. [DOI] [PubMed] [Google Scholar]
- 25.Remy I., Campbell-Valois F.X., Michnick S.W. Detection of protein‒protein interactions using a simple survival protein-fragment complementation assay based on the enzyme dihydrofolate reductase. Nat Protoc. 2007;2:2120–2125. doi: 10.1038/nprot.2007.266. [DOI] [PubMed] [Google Scholar]
- 26.Remy I., Michnick S.W. A highly sensitive protein‒protein interaction assay based on Gaussia luciferase. Nat Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 27.Larionova M.D., Markova S.V., Vysotski E.S. Bioluminescent and structural features of native folded Gaussia luciferase. J Photochem Photobiol, B. 2018;183:309–317. doi: 10.1016/j.jphotobiol.2018.04.050. [DOI] [PubMed] [Google Scholar]
- 28.Tannous B.A., Kim D.-E., Fernandez J.L., Weissleder R., Breakefield X.O. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Maguire C.A., Deliolanis N.C., Pike L., Niers J.M., Tjon-Kon-Fat L.A., Sena-Esteves M., et al. Gaussia luciferase variant for high-throughput functional screening applications. Anal Chem. 2009;81:7102–7106. doi: 10.1021/ac901234r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luker K.E., Mihalko L.A., Schmidt B.T., Lewin S.A., Ray P., Shcherbo D., et al. In vivo imaging of ligand receptor binding with Gaussia luciferase complementation. Nat Med. 2011;18:172–177. doi: 10.1038/nm.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X., Tian X., Wang F., Ma Y., Kornmann M., Yang Y. BRG1 promotes chemoresistance of pancreatic cancer cells through crosstalking with Akt signalling. Eur J Cancer. 2014;50:2251–2262. doi: 10.1016/j.ejca.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 32.Bernier M., Catazaro J., Singh N.S., Wnorowski A., Boguszewska-Czubara A., Jozwiak K., et al. GPR55 receptor antagonist decreases glycolytic activity in PANC-1 pancreatic cancer cell line and tumor xenografts. Int J Cancer. 2017;141:2131–2142. doi: 10.1002/ijc.30904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voskoglou-Nomikos T., Pater J.L., Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res. 2003;9:4227–4239. [PubMed] [Google Scholar]
- 34.Hollingshead M.G. Antitumor efficacy testing in rodents. J Natl Cancer Inst. 2008;100:1500–1510. doi: 10.1093/jnci/djn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nan Y., Luo Q., Wu X., Chang W., Zhao P., Liu S., et al. HCP5 prevents ubiquitination-mediated UTP3 degradation to inhibit apoptosis by activating c-Myc transcriptional activity. Mol Ther. 2023;31:552–568. doi: 10.1016/j.ymthe.2022.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long Y., Chen R., Yu X., Tong Y., Peng X., Li F., et al. Suppression of tumor or host intrinsic CMTM6 drives antitumor cytotoxicity in a PD-L1-independent manner. Cancer Immunol Res. 2023;11:241–260. doi: 10.1158/2326-6066.CIR-22-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou T., Sang Y.H., Cai S., Xu C., Shi M.H. The requirement of mitochondrial RNA polymerase for non-small cell lung cancer cell growth. Cell Death Dis. 2021;12:751–763. doi: 10.1038/s41419-021-04039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopra K., van Adrichem A.J., Salo-Ahen O.M.H., Peltonen J., Wennerberg K., Härmä H. High-throughput dual screening method for Ras activities and inhibitors. Anal Chem. 2017;89:4508–4516. doi: 10.1021/acs.analchem.6b04904. [DOI] [PubMed] [Google Scholar]
- 39.Hou W.R., Xie S.N., Wang H.J., Su Y.Y., Lu J.L., Li L.L., et al. Intramuscular delivery of a naked DNA plasmid encoding proinsulin and pancreatic regenerating III protein ameliorates type 1 diabetes mellitus. Pharmacol Res. 2011;63:320–327. doi: 10.1016/j.phrs.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Luthra P., Anantpadma M., De S., Sourimant J., Davey R.A., Plemper R.K., et al. High-throughput screening assay to identify small molecule inhibitors of Marburg virus VP40 protein. ACS Infect Dis. 2020;6:2783–2799. doi: 10.1021/acsinfecdis.0c00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitman N.A., Lin Z.-W., DiProspero T.J., McIntosh J.C., Lockett M.R. Screening estrogen receptor modulators in a paper-based breast cancer model. Anal Chem. 2018;90:11981–11988. doi: 10.1021/acs.analchem.8b02486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 43.Zhou B., Zhang J.Y., Liu X.S., Chen H.Z., Ai Y.L., Cheng K., et al. Tom20 senses iron-activated ROS signaling to promote melanoma cell pyroptosis. Cell Res. 2018;28:1171–1185. doi: 10.1038/s41422-018-0090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han X., Li Y., Xu Y., Zhao X., Zhang Y., Yang X., et al. Reversal of pancreatic desmoplasia by re-educating stellate cells with a tumour microenvironment-activated nanosystem. Nat Commun. 2018;9:3390–3408. doi: 10.1038/s41467-018-05906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X., Zhang Z., Ruan J., Pan Y., Magupalli V.G., Wu H., et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Z., He H., Wang K., Shi X., Wang Y., Su Y., et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020;368:eaaz7548. doi: 10.1126/science.aaz7548. [DOI] [PubMed] [Google Scholar]
- 47.Deng W., Bai Y., Deng F., Pan Y., Mei S., Zheng Z., et al. Streptococcal pyrogenic exotoxin B cleaves GSDMA and triggers pyroptosis. Nature. 2022;602:496–502. doi: 10.1038/s41586-021-04384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z., Zhang Y., Lieberman J. Lighting a fire: can we harness pyroptosis to ignite antitumor immunity? Cancer Immunol Res. 2021;9:2–7. doi: 10.1158/2326-6066.CIR-20-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong Y., Feng Z., Chen A., Qi Q., Han M., Wang S., et al. The natural flavonoid galangin elicits apoptosis, pyroptosis, and autophagy in glioblastoma. Front Oncol. 2019;9:942–955. doi: 10.3389/fonc.2019.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson D.C., Taabazuing C.Y., Okondo M.C., Chui A.J., Rao S.D., Brown F.C., et al. DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nat Med. 2018;24:1151–1156. doi: 10.1038/s41591-018-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.An H., Heo J.S., Kim P., Lian Z., Lee S., Park J., et al. Tetraarsenic hexoxide enhances generation of mitochondrial ROS to promote pyroptosis by inducing the activation of caspase-3/GSDME in triple-negative breast cancer cells. Cell Death Dis. 2021;12:159–173. doi: 10.1038/s41419-021-03454-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui J., Zhou Z., Yang H., Jiao F., Li N., Gao Y., et al. MST1 suppresses pancreatic cancer progression via ROS-induced pyroptosis. Mol Cancer Res. 2019;17:1316–1325. doi: 10.1158/1541-7786.MCR-18-0910. [DOI] [PubMed] [Google Scholar]
- 53.Ji M., Wang X., Zheng H., Mao W., Shi X., Chen S., et al. A secreted reporter for blood monitoring of pyroptotic cell death. Anal Chem. 2020;92:15565–15572. doi: 10.1021/acs.analchem.0c03629. [DOI] [PubMed] [Google Scholar]
- 54.O'Hare T., Shakespeare W.C., Zhu X., Eide C.A., Rivera V.M., Wang F., et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garner A.P., Gozgit J.M., Anjum R., Vodala S., Schrock A., Zhou T., et al. Ponatinib inhibits polyclonal drug-resistant KIT oncoproteins and shows therapeutic potential in heavily pretreated gastrointestinal stromal tumor (GIST) patients. Clin Cancer Res. 2014;20:5745–5755. doi: 10.1158/1078-0432.CCR-14-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Falco V., Buonocore P., Muthu M., Torregrossa L., Basolo F., Billaud M., et al. Ponatinib (AP24534) is a novel potent inhibitor of oncogenic RET mutants associated with thyroid cancer. J Clin Endocrinol Metab. 2013;98:E811–E819. doi: 10.1210/jc.2012-2672. [DOI] [PubMed] [Google Scholar]
- 57.Smith C.C., Lasater E.A., Zhu X., Lin K.C., Stewart W.K., Damon L.E., et al. Activity of ponatinib against clinically-relevant AC220-resistant kinase domain mutants of FLT3-ITD. Blood. 2013;121:3165–3171. doi: 10.1182/blood-2012-07-442871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren M., Hong M., Liu G., Wang H., Patel V., Biddinger P., et al. Novel FGFR inhibitor ponatinib suppresses the growth of non-small cell lung cancer cells overexpressing FGFR1. Oncol Rep. 2013;29:2181–2190. doi: 10.3892/or.2013.2386. [DOI] [PubMed] [Google Scholar]
- 59.Guo M., Duan Y., Dai S., Li J., Chen X., Qu L., et al. Structural study of ponatinib in inhibiting SRC kinase. Biochem Biophys Res Commun. 2022;598:15–19. doi: 10.1016/j.bbrc.2022.02.001. [DOI] [PubMed] [Google Scholar]
- 60.Sourbier C., Ricketts C.J., Matsumoto S., Crooks D.R., Liao P.J., Mannes P.Z., et al. Targeting ABL1-mediated oxidative stress adaptation in fumarate hydratase-deficient cancer. Cancer Cell. 2014;26:840–850. doi: 10.1016/j.ccell.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hossain M.B., Shifat R., Johnson D.G., Bedford M.T., Gabrusiewicz K.R., Cortes-Santiago N., et al. TIE2-mediated tyrosine phosphorylation of H4 regulates DNA damage response by recruiting ABL1. Sci Adv. 2016;2 doi: 10.1126/sciadv.1501290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shome D., Trent J., Espandar L., Hatef E., Araujo D.M., Song C.D., et al. Ulcerative keratitis in gastrointestinal stromal tumor patients treated with perifosine. Ophthalmology. 2008;115:483–487. doi: 10.1016/j.ophtha.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 63.Knowling M., Blackstein M., Tozer R., Bramwell V., Dancey J., Dore N., et al. A phase II study of perifosine (D-21226) in patients with previously untreated metastatic or locally advanced soft tissue sarcoma: a National Cancer Institute of Canada Clinical Trials Group trial. Invest N Drugs. 2006;24:435–439. doi: 10.1007/s10637-006-6406-7. [DOI] [PubMed] [Google Scholar]
- 64.Burgeiro A., Pereira C.V., Carvalho F.S., Pereira G.C., Mollinedo F., Oliveira P.J. Edelfosine and perifosine disrupt hepatic mitochondrial oxidative phosphorylation and induce the permeability transition. Mitochondrion. 2013;13:25–35. doi: 10.1016/j.mito.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 65.Li X., Luwor R., Lu Y., Liang K., Fan Z. Enhancement of antitumor activity of the anti-EGF receptor monoclonal antibody cetuximab/C225 by perifosine in PTEN-deficient cancer cells. Oncogene. 2006;25:525–535. doi: 10.1038/sj.onc.1209075. [DOI] [PubMed] [Google Scholar]
- 66.Booth L., Poklepovic A., Dent P. Neratinib decreases pro-survival responses of [sorafenib + vorinostat] in pancreatic cancer. Biochem Pharmacol. 2020;178:114067–114077. doi: 10.1016/j.bcp.2020.114067. [DOI] [PubMed] [Google Scholar]
- 67.Hage C., Hoves S., Strauss L., Bissinger S., Prinz Y., Pöschinger T., et al. Sorafenib induces pyroptosis in macrophages and triggers natural killer cell-mediated cytotoxicity against hepatocellular carcinoma. Hepatology. 2019;70:1280–1297. doi: 10.1002/hep.30666. [DOI] [PubMed] [Google Scholar]
- 68.Zhang X., Zhang P., An L., Sun N., Peng L., Tang W., et al. Miltirone induces cell death in hepatocellular carcinoma cell through GSDME-dependent pyroptosis. Acta Pharm Sin B. 2020;10:1397–1413. doi: 10.1016/j.apsb.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Q., Wang Y., Ding J., Wang C., Zhou X., Gao W., et al. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature. 2020;579:421–426. doi: 10.1038/s41586-020-2079-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.