Summary
During cytokinesis, a contractile ring consisting of unbranched filamentous actin (F-actin) and myosin II constricts at the cell equator. Unbranched F-actin is generated by formin, and without formin no cleavage furrow forms. In Caenorhabditis elegans, depletion of septin restores furrow ingression in formin mutants. How the cleavage furrow ingresses without a detectable unbranched F-actin ring is unknown. We report that, in this setting, anillin (ANI-1) forms a meshwork of circumferentially aligned linear structures decorated by non-muscle myosin II (NMY-2). Analysis of ANI-1 deletion mutants reveals that its disordered N-terminal half is required for linear structure formation and sufficient for furrow ingression. NMY-2 promotes the circumferential alignment of the linear ANI-1 structures and interacts with various lipids, suggesting that NMY-2 links the ANI-1 network with the plasma membrane. Collectively, our data reveal a compensatory mechanism, mediated by ANI-1 linear structures and membrane-bound NMY-2, that promotes furrowing when unbranched F-actin polymerization is compromised.
Keywords: cytokinesis, cell division, anillin, contractile ring, myosin, C. elegans, intrinsically disordered, liquid-liquid phase separation, formin, actin
Graphical abstract
Highlights
-
•
An anillin-dependent compensatory mechanism drives furrowing if F-actin is low
-
•
The disordered anillin N terminus forms linear structures under the plasma membrane
-
•
Non-muscle myosin II represents in independent membrane anchor during cytokinesis
-
•
The anillin ring constricts to form a cleavage furrow when septins are also reduced
During cytokinesis in animal cells, an actin-based ring constricts at the cell equator to form the cleavage furrow. Lebedev et al. discover a compensatory mechanism mediated by linear anillin structures and membrane-bound myosin II that drives cleavage furrow formation and ingression in case actin ring assembly fails.
Introduction
During cytokinesis, a contractile ring consisting of unbranched filamentous actin (F-actin) and non-muscle myosin II assembles underneath the plasma membrane between the segregating chromosomes. Constriction of the actin-myosin ring splits the mother cell into two daughter cells.1,2,3 Stimulatory and inhibitory signals emanating from the mitotic spindle activate RhoA at the cell equator, which in turn induces contractile ring assembly.4,5,6,7,8,9,10,11 Active RhoA releases the autoinhibitory conformation of formin with the help of the contractile ring component anillin and IQGAP1.12,13,14,15 Anillin is a widely conserved protein that links the actin-myosin ring with the plasma membrane.1,2,16,17 Active formin binds to the barbed end of actin filaments and induces the polymerization of long unbranched F-actin.18 F-actin circumferentially aligns around the cell equator, which is thought to facilitate furrow ingression.19,20,21,22,23 Without formin, cleavage furrow formation and cytokinesis fail.24,25,26,27
In addition to formin, RhoA also recruits and activates non-muscle myosin II (NMY-2 in Caenorhabditis elegans) via RhoA kinase. The myosin motor domain interacts with unbranched F-actin, and myosin II is required for cytokinesis in many systems.28,29,30 To be active, myosin II has to bind the phosphorylated regulatory light chain (RLC) and essential light chain (ELC).31 In addition to RhoA-dependent myosin II recruitment, a positive-feedback mechanism between ring myosin II and equatorial-directed cortical flows that brings more myosin II into the ring dictates the amount of myosin II in the contractile ring.32 Nevertheless, myosin II can enrich at the cell equator independently of its binding partner F-actin or anillin.33,34 Myosin II inhibition does not prevent the recruitment of other ring components to the equator but blocks ring constriction.28,35,36 In fission yeast, myosin II is targeted to the membrane by the anillin homolog Mid1p37,38 and, together with formin and numerous other proteins, they form defined protein assemblies called nodes.39,40,41,42 Within each node, myosin II heads point away from the membrane and grab and pull on actin filaments generated and anchored at adjacent nodes.40,41,43 This leads to the coalescence of nodes and eventually ring constriction.3
Another component of the contractile ring is the multidomain protein anillin (ANI-1 in C. elegans).1,2,16,17 Anillin is considered a scaffolding component of the contractile ring that links the actin-myosin ring with the plasma membrane. With its C terminus, anillin interacts with microtubules,44 with lipids and importin45 through the C2 domain, with membrane-associated active RhoA via the RhoA-binding domain (RBD),46,47 and with septins via the PH domain.48,49,50,51,52 With its N terminus, anillin binds to myosin II,53 F-actin,49,54,55,56,57 and formins.13,58 Anillin localization to the cell equator requires RhoA activation but not myosin II, F-actin, septins, or formins.27,33,34,53,59 Consistent with its role as a membrane-actomyosin linker, anillin depletion results in unstable furrows and in cytokinesis failure in fly and human cells.33,46,53,60 ANI-1 depletion in the one-cell C. elegans zygotes does not cause cytokinesis failure but results in symmetric furrow ingression.34,61 Anillin positively feeds back on RhoA activity46 by stabilizing active RhoA at the membrane,62 and anillin also binds and recruits septins to the contractile ring.33,34,48,50,52 Recent in vitro work suggests that anillin could contribute to force generation and ring constriction independently of myosin II.63
Besides F-actin and myosin II, septins are the third filament-forming protein of the contractile ring, and septins bind the plasma membrane.64 Similar to anillin, septins are required for cytokinesis in human cells65 but are not essential in C. elegans zygotes. Whereas humans have 13 septins, C. elegans has two septins, which are only essential for some postembryonic divisions.66 Together, septin and anillin also function during the late stages of cytokinesis.65,67,68,69
During the initial assembly phase, ring components such as F-actin, myosin II, and anillin localize in a broad equatorial band, which subsequently compacts into the contractile ring.70 Super-resolution microscopy in sea urchin embryos revealed that myosin II, anillin, and septin initially co-localize in puncta reminiscent of the nodes in yeast.71 Subsequently, a dense meshwork forms in which myosin II filaments orient parallel to actin filaments.20,72,73 This organization is consistent with myosin II mediating ring constriction by actin filament sliding in a purse-string-like fashion. Consistently myosin II motor activity is required for ring constriction.74
Although actin-based contractility appears to be the predominant mechanism for furrow ingression, alternative mechanisms were also reported sporadically. For example, the migration of the two daughter cells away from each other supports furrow ingression in the absence of a visible F-actin ring in human RPE1 cells and in Dictyostelium.75,76,77 In Chlamydomonas, furrow ingression does not require F-actin.78 In C. elegans, depletion of septin in a formin (CYK-1) temperature-sensitive (ts) mutant restored furrow ingression, although no F-actin ring was detectable.79 Here we investigate the mechanism of furrow ingression after co-depletion of CYK-1 and septins in the C. elegans embryo. We uncover a compensatory ANI-1-dependent mechanism of cleavage furrow ingression when unbranched F-actin is low.
Results
ANI-1 is required for furrow ingression after CYK-1 and septin co-depletion
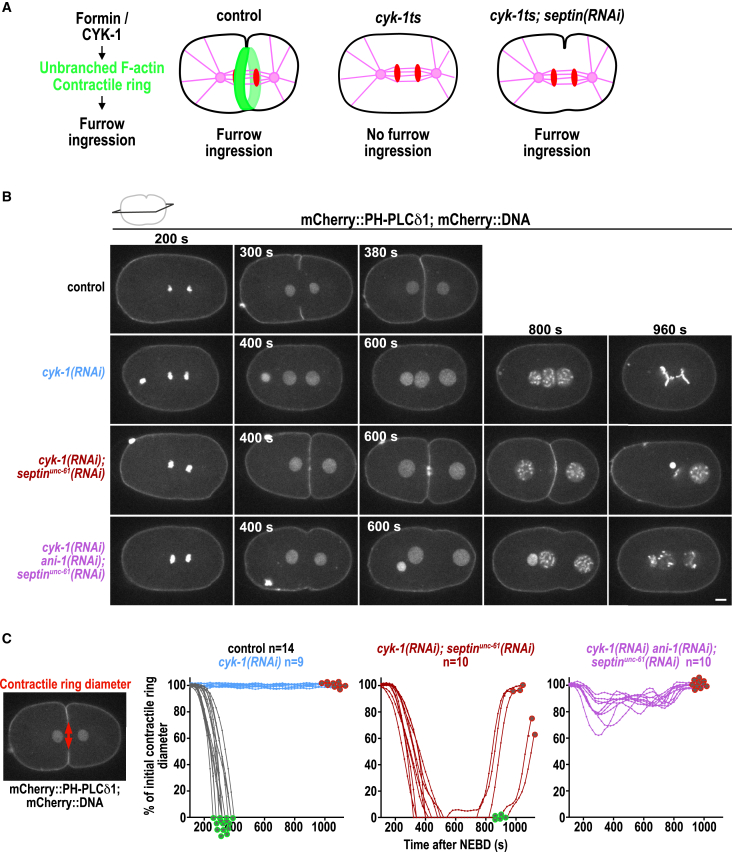
CYK-1 is the only C. elegans formin known to be essential for cytokinesis.24,25,80,81 Surprisingly, it was found that cyk-1ts mutant embryos, which have strongly reduced unbranched F-actin levels, still ingress a cleavage furrow when septins are depleted (Figure 1A).79,81 We confirmed this observation as CYK-1 depletion by RNAi prevented cleavage furrow formation but septinUNC-61 co-depletion facilitated cleavage furrow ingression in all embryos (Figures 1B and 1C; Video S1). In five out of 10 cyk-1(RNAi);septinunc-61(RNAi) embryos, the furrow remained closed until the onset of the second cell division. In the remaining embryos, the furrow eventually regressed around 800 s after nuclear envelope breakdown (NEBD) (Figures 1B and 1C). C. elegans has two septins (UNC-61 and UNC-59), which depend on each other for membrane localization66; therefore, depletion of one is expected to disrupt the function of the complex. Consistently, we observed that GFP-tagged septinUNC-59 did not localize to the cortex after septinunc-61(RNAi) or cyk-1(RNAi);septinunc-61(RNAi) (Figures S1A and S1B). Although the cortical accumulation of septinUNC-59 was strongly diminished after septinUNC-61 depletion, we tested whether the furrow still ingressed in cyk-1(RNAi) embryos after the co-depletion of both septins. In septinUNC-59 and septinUNC-61 co-depleted embryos treated with cyk-1(RNAi), the cleavage furrow fully ingressed in 88% of the cases, which demonstrates that furrowing can occur after co-depletion of both C. elegans septins (Figure S1C). Immunoblot analysis of adult worms revealed that septinunc-59(RNAi) and cyk-1(RNAi) strongly reduced endogenously tagged septinUNC-59 and CYK-1 protein levels, respectively (Figure S1D).
Figure 1.
Furrow ingression of cyk-1(RNAi);septinunc-61(RNAi) embryos requires ANI-1
(A) During cytokinesis, CYK-1 polymerizes the unbranched F-actin of the contractile ring. In cyk-1ts mutants, no contractile ring forms and cytokinesis fails, but, surprisingly, furrow ingression was restored by depleting septin.79
(B) Central plane images of embryos expressing a membrane and DNA marker, treated with the indicated RNAi conditions. Time is indicated in seconds (s) after NEBD. Scale bar, 5 μm.
(C) The contractile ring diameter is plotted over time for individual embryos for indicated RNAi conditions. Green and red encircled stars indicate whether embryos succeed or fail cytokinesis, respectively. n = number of embryos.
Images were acquired every 20 s on a Nikon Eclipse Ti spinning disk confocal controlled by NIS Elements 4.51 software equipped with a 100× 1.45-NA Plan-Apochromat oil-immersion objective and Andor DU-888 X11056 camera. Video starts 140 s after NEBD.
Septin depletion rescued furrow ingression after cyk-1(RNAi) and therefore we determined whether cortical CYK-1 levels were altered. Equatorial CYK-1 levels were slightly increased after septinUNC-61 depletion (Figure S1E), but, importantly, no equatorial enrichment of CYK-1 was observed after cyk-1(RNAi) or cyk-1(RNAi);septinunc-61(RNAi) (Figure S1F). Similarly, equatorial F-actin no longer accumulated at the cell equator after single or double cyk-1(RNAi) treatments, confirming previous observations79,81,82,83 (Figure S1F). This suggests that septinUNC-61 depletion does not rescue furrow ingression in cyk-1(RNAi) embryos by elevating either equatorial CYK-1 or F-actin levels.
In addition to formin-nucleated F-actin, the cortex contains Arp2/3 nucleated branched F-actin.83 Thus, we tested whether the remaining Arp2/3-generated F-actin contributes to furrowing in CYK-1 and septin co-depleted embryos by additional depletion of the Arp2 homolog ARX-2. All cyk-1(RNAi);septinunc-61(RNAi);arx-2(RNAi) embryos fully ingressed a cleavage furrow (Figures S2A and S2B), suggesting that Arp2/3-generated F-actin is not essential for furrowing in this condition.
To address the mechanism how CYK-1 and septin co-depleted embryos ingress a cleavage furrow, we determined whether ANI-1 is required for furrow ingression. Although ANI-1 depletion alone does not cause cytokinesis failure,34 ANI-1 depletion together with CYK-1 and septinUNC-61 prevented furrow ingression (Figures 1B and 1C). Similar to septinUNC-61 depletion, ani-1(RNAi) resulted in a increase in equatorial CYK-1 levels (Figure S1E), but, in contrast, it did not rescue furrow ingression after cyk-1(RNAi) (Figure S2C). Immunoblot analysis of ANI-1 protein levels showed that ANI-1 levels were strongly reduced (Figure S2D). Further, imaging endogenously NeonGreen-tagged ANI-1 (NG::ANI-1) confirmed that ANI-1 depletion was highly efficient in all RNAi conditions (Figure S2E). Together, ANI-1 promotes furrow ingression in case CYK-1 and septin levels are low.
ANI-1 localizes to septin-independent linear structures, especially after the depletion of the formin CYK-1
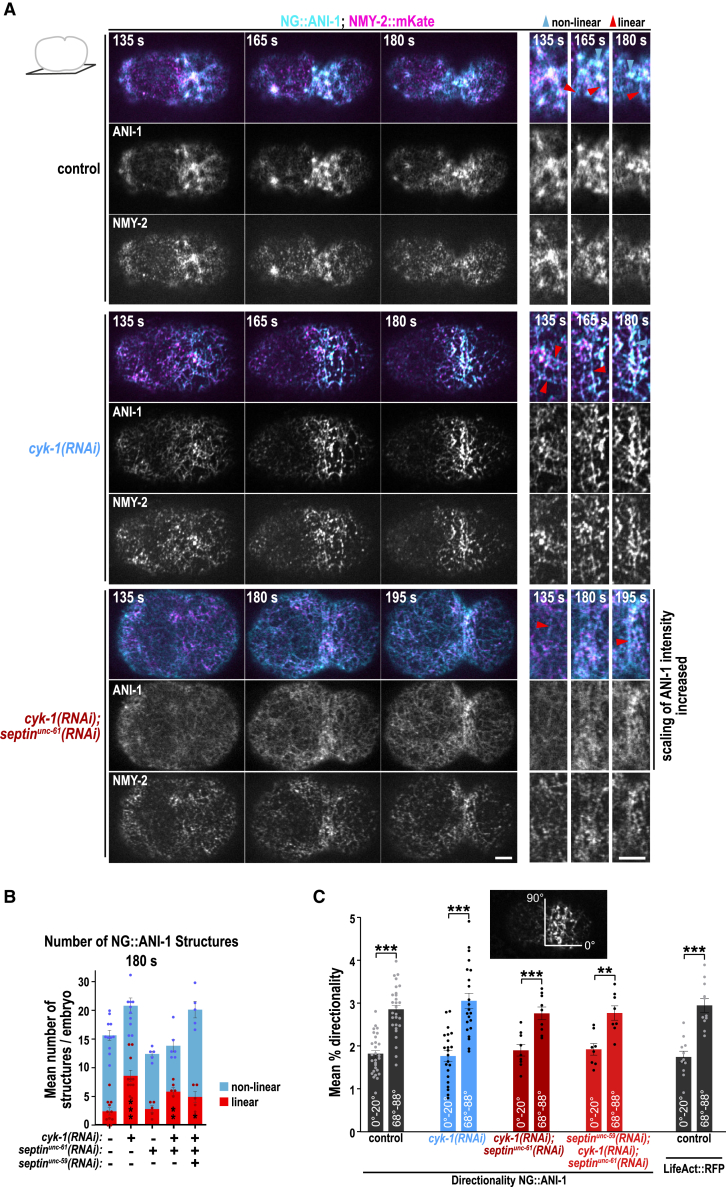
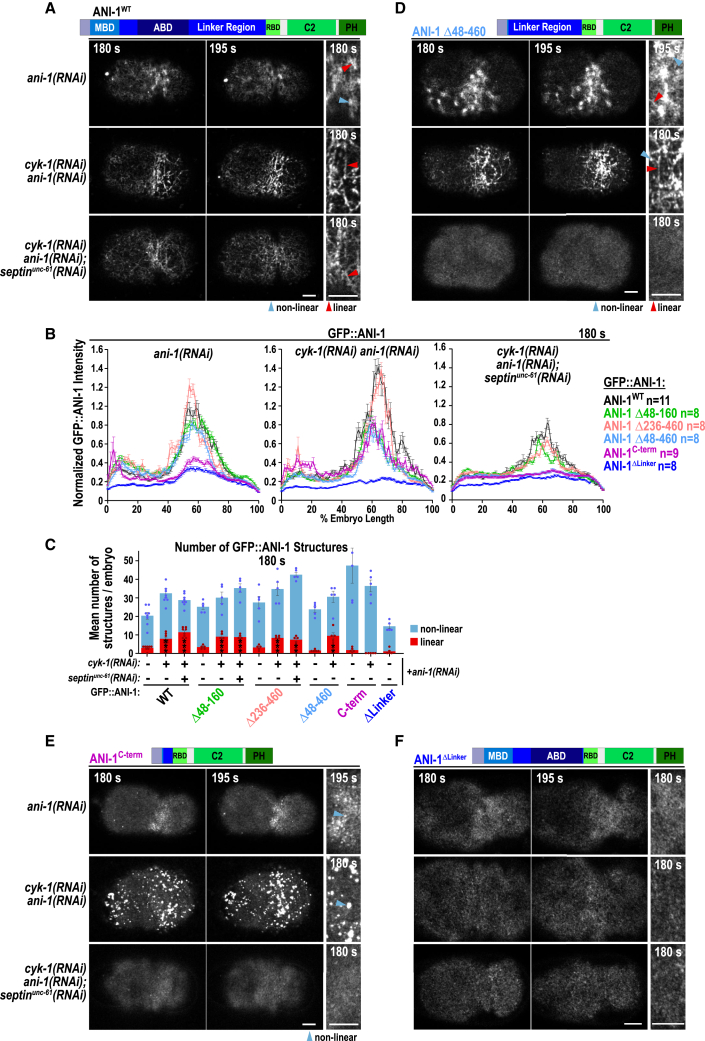
Since ANI-1 was required for furrow ingression, we analyzed ANI-1 localization on the cortex during contractile ring assembly. In control anaphase embryos, ANI-1 was enriched in cortical patches as previously published70 but also localized in linear structures, which sometimes radiate out of cortical patches (Figure 2A; Video S2). Comparison of the localization of ANI-1 with myosin II (NMY-2::mKate) revealed that NMY-2 was also present in these cortical patches84 and occasionally decorated the linear ANI-1 structures. In CYK-1-depleted embryos, cortical ANI-1 patches were absent and numerous linear structures, decorated with NMY-2, formed (Figure 2A; Video S2). Quantification revealed that linear ANI-1 structures were occasionally observed in control embryos and after cyk-1(RNAi) their number increased (Figures 2B and S3A). In cyk-1(RNAi) linear ANI-1 structures had a length of ∼2–9 μm and width of ∼0.5 μm. To determine whether the overall ANI-1 distribution was altered, ANI-1 fluorescence intensity was measured along the cell cortex. ANI-1 was still enriched in an equatorial zone after cyk-1(RNAi) and cyk-1(RNAi);septinunc-61(RNAi) and its cortical levels were decreased in the latter (Figures 2A and S3B). Reduction of cortical ANI-1 levels is consistent with the fact that septins bind and contribute to anillin recruitment in other organisms.48,49,85,86 Linear ANI-1 structures were still detected in cyk-1(RNAi) embryos co-depleted of septinUNC-61 alone or septinUNC-59 and septinUNC-61 together (Figures 2A, 2B, and S3C). Measurement of the orientation of ANI-1 revealed that ANI-1 circumferentially aligns around the cell equator similar to F-actin in control embryos (Figure 2C). Reduction of unbranched F-actin alone or together with septinUNC-61 or with septinUNC-59 and septinUNC-61 did not alter alignment of ANI-1 (Figure 2C).
Figure 2.
ANI-1 forms linear structures decorated by NMY-2 foci when CYK-1-nucleated F-actin levels are low
(A) Cortical confocal images of NG::ANI-1 (cyan) and NMY-2::mKate (magenta) for indicated RNAi conditions and time points after NEBD. Magnifications of the equatorial region are shown. Since NG::ANI-1 levels are reduced after septinUNC-61 co-depletion (Figure S3B), intensity scaling was increased. Non-linear (blue) and linear (red) structures are highlighted by arrowheads. Scale bars, 5 μm.
(B) The mean number of linear and non-linear NG::ANI-1 structures at an equatorial region (Figure S3A) for indicated RNAi conditions. The p values were calculated using Mann-Whitney U or Student’s t test: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 in comparison to control embryos treated without RNAi.
(C) Mean percentage of 0°–20° (anterior to posterior) and 68°–88° (circumferential) directionality measured for NG::ANI-1 and LifeAct::RFP at 180 s after NEBD for indicated RNAi conditions. The p values were determined with Student’s t test: ∗∗p < 0.01, ∗∗∗p < 0.001.
All error bars indicate SEM, and dots represent data points of individual embryos.
Note: scaling intensity for NG::ANI-1 after septinUNC-61 co-depletion is increased. Images were acquired every 2.5 s on a Nikon Eclipse Ti spinning disk confocal controlled by NIS Elements 4.51 software equipped with a 100× 1.45-NA Plan-Apochromat oil-immersion objective and Andor DU-888 X11056 camera. Time in seconds after NEBD is shown.
Since ANI-1 appeared to form those linear structures, we determined whether septin also localized to linear structures after cyk-1(RNAi). Indeed, we observed that, after cyk-1(RNAi), septinUNC-59 was also present in linear structures and its cortical intensity levels were increased (Figures S3D–S3F). In contrast to the linear ANI-1 structures, which could still be observed after septinunc-61(RNAi), septinUNC-59 was no longer enriched at the cell equator after ani-1(RNAi) (Figures S4D and S4E34). This shows that septinUNC-59 fully depends on the presence of ANI-1 for equatorial enrichment, but, in contrast, a pool of ANI-1 localizes independently of septins to the cortex.
Since reduction of CYK-1-nucleated unbranched F-actin altered ANI-1 localization, we analyzed whether Arp2/3-nucleated branched F-actin also has an influence on ANI-1. Consistent with previous reports, we observed a reduction of F-actin foci, a delay in furrow formation, and ANI-1 accumulation after arx-2(RNAi) (Figures S4A–S4C).36,83,87 However, ARX-2 depletion did not affect ANI-1 localization to linear structures in control or cyk-1(RNAi) embryos (Figures S4B and S4D; Video S3).
Images were acquired every 2.5 s on a Nikon Eclipse Ti spinning disk confocal controlled by NIS Elements 4.51 software equipped with a 100× 1.45-NA Plan-Apochromat oil-immersion objective and Andor DU-888 X11056 camera. Time in seconds after NEBD is shown.
In summary, CYK-1-nucleated unbranched F-actin limits the formation of linear ANI-1 structures. Septin co-depletion did not affect the formation of linear ANI-1 structures, although efficient ANI-1 targeting to the cell equator required septin function.
NMY-2 is required for the orientation of the linear ANI-1 structures and furrow ingression in embryos depleted of CYK-1 and septin
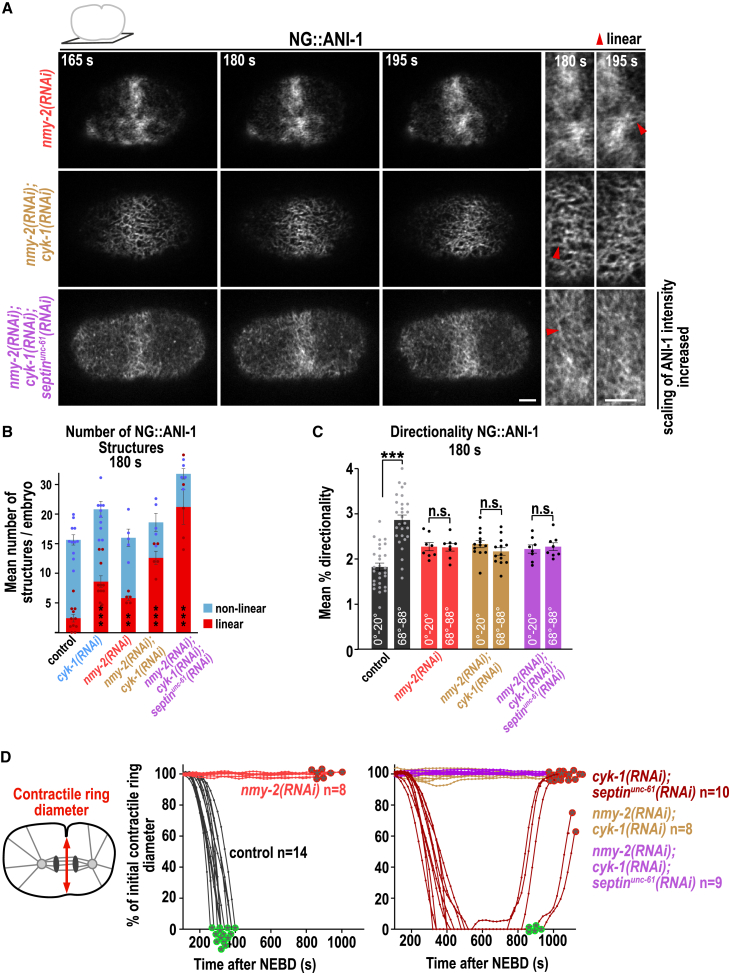
We found that ANI-1 forms linear structures decorated by NMY-2 and supports furrow ingression in cyk-1(RNAi);septinunc-61(RNAi) embryos. To test whether NMY-2 is required for the formation or alignment of the linear ANI-1 structures, we depleted NMY-2 together with CYK-1 or CYK-1 and septinUNC-61. Immunoblot analysis and quantification of cortical NMY-2 fluorescence intensity confirmed that nmy-2(RNAi) strongly reduced NMY-2 protein levels in all RNAi conditions (Figures S5A–S5C). Depletion of NMY-2 alone or in combination with CYK-1 or CYK-1 and septinUNC-61 did not prevent linear ANI-1 structure formation, although they were no longer circumferentially aligned around the cell equator (Figures 3A–3C; Video S4). This suggests that NMY-2 aligns linear ANI-1 structures, perhaps by directly binding to the ANI-1 N terminus. In normal cytokinesis, myosin II binds and aligns F-actin and mediates furrow ingression,74 so we asked whether NMY-2 still mediates furrow ingression in cyk-1(RNAi);septinunc-61(RNAi) embryos. NMY-2 co-depletion abolished furrow ingression in cyk-1(RNAi);septinunc-61(RNAi) embryos (Figures 3D and S5D), although ANI-1 was still enriched at the cell equator (Figures 3A and S5E). In conclusion, NMY-2 is absolutely required to mediate furrow ingression after the reduction of CYK-1 and septins. Further, NMY-2 is involved in aligning the linear ANI-1 structures circumferentially but is not essential for their formation.
Figure 3.
NMY-2 is not required for the formation of the linear ANI-1 structures but for their circumferential alignment
(A) Cortical confocal images of NG::ANI-1 for indicated RNAi conditions and time points after NEBD. Magnifications of the equator are shown (right). Since NG::ANI-1 levels are reduced afterseptinunc-61(RNAi), intensity scaling was increased. Linear structures are highlighted by red arrowheads. Scale bars, 5 μm.
(B) Mean number of NG::ANI-1 structures at the equator for indicated RNAi conditions. The p values were calculated using Mann-Whitney-U or Student’s t test: ∗∗p < 0.01, ∗∗∗p < 0.001 in comparison to control embryos; dots represent data points of individual embryos. Control and cyk-1(RNAi) conditions are reproduced from Figure 2B.
(C) Mean percentage of directionality for NG::ANI-1 for indicated RNAi conditions. The p values were determined with Student’s t test: ∗∗∗p < 0.001; n.s., p > 0.05. Control condition is reproduced from Figure 2C.
(D) Plotted is the contractile ring diameter over time for indicated RNAi conditions. Green and red encircled stars indicate whether embryos succeed or fail cytokinesis, respectively. Control and cyk-1(RNAi);septinunc-61(RNAi) conditions are reproduced from Figure 1C; n = number of embryos.
All error bars indicate SEM.
Intensity scaling was increased to visualize the NG::ANI-1 structures after nmy-2(RNAi);cyk-1(RNAi);septinunc-61(RNAi). Imaging conditions are the same as indicated for Video S2. Time in seconds after NEBD is shown.
ANI-1 and NMY-2 form membrane-localized structures in the absence of F-actin independently of the ANI-1 N terminus
After depolymerization of F-actin in Drosophila cells, anillin and the RLC of myosin II enrich in circular foci that evolve into linear structures, and these structures also form independently of each other.33,49,52,59 In the C. elegans zygote, NMY-2 also forms circular foci after Latrunculin A addition.88,89 However, whether ANI-1 is also present in those circular NMY-2 foci and whether linear structures still form after F-actin depolymerization have not been investigated in C. elegans. To study this, we treated embryos with Latrunculin A, which caused a rapid disappearance of F-actin from the cortex and the appearance of dynamic NMY-2 and ANI-1 structures (Figures S6A–S6E; Video S5). After loss of cortical F-actin, NMY-2 and ANI-1 co-localized in numerous non-linear structures that became linear at later time points (Figures 4A and 4B). Measurement of fluorescence intensities revealed that, upon F-actin depolymerization, NMY-2 and ANI-1 accumulated in an equatorial zone similar to control embryos, although NMY-2 fluorescence was increased at 255 s (Figure S6F). To test whether the linear structures are septin dependent, we repeated Latrunculin A treatment after septinUNC-61 depletion. In Latrunculin A-treated septinunc-61(RNAi) embryos, linear ANI-1 structures were still present; however, they were less frequent and defined than those in Latrunculin A-only-treated embryos (Figures S7A and S7B; Video S6). Finally, we tested whether septinunc-61(RNAi) restores furrow ingression in Latrunculin A-treated embryos like it does in cyk-1(RNAi) embryos. No cleavage furrow formed in Latrunculin A-treated control or septinunc-61(RNAi) embryos (Figure S7C). This suggests, although F-actin does not enrich at the cell equator in cyk-1(RNAi);septinunc-61(RNAi) embryos, some residual F-actin contributes to furrow ingression in those embryos.
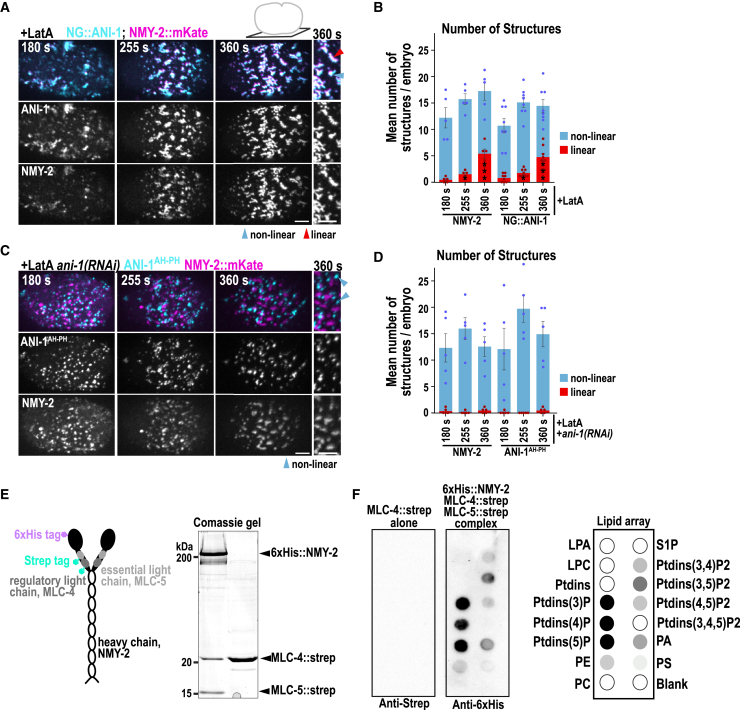
Figure 4.
NMY-2 forms membrane-localized structures independently of F-actin and the ANI-1 N terminus and interacts with lipids in vitro
(A) Maximum intensity projections of 10 cortical z planes of embryos treated with Latrunculin A (+LatA) expressing NG::ANI-1 (cyan) and NMY-2::mKate (magenta). A magnification of the equator is shown (right).
(B) Mean number of NMY-2::mKate and NG::ANI-1 structures at the cell equator after Latrunculin A treatment. The p values were calculated using Mann-Whitney-U or Student’s t test: ∗p < 0.05, ∗∗∗p < 0.001 in comparison to 180 s; dots represent data points of individual embryos.
(C) Maximum intensity projections of 10 cortical z planes of embryos expressing ANI-1AH-PH,90 (cyan) and NMY-2::mKate (magenta) treated with Latrunculin A and ani-1(RNAi). A magnification of the equator is shown.
(D) Mean number of NMY-2::mKate and ANI-1AH-PH structures at the equator after ani-1(RNAi) and Latrunculin A treatment.
(E) Coomassie-stained gel of Strep-tagged MLC-4 alone or in complex with 6xHis-tagged NMY-2 and Strep-tagged MLC-5.
(F) Purified MLC-4 alone or in complex with 6xHis-tagged NMY-2 and Strep-tagged MLC-5 were incubated with lipid arrays and blotted with anti-Strep or anti-6xHis antibodies (left). Schematics of the lipid array: LPA, lysophosphatidic acid; LPC, lysophosphatidylcholines; PtdIns, phosphatidylinositol; PE, phosphatidylethanolamine; PC, phosphatidylcholines; S1P sphingosine-1-phosphate; PA, phosphatidic acid; PS, phosphatidylserine (right).
Non-linear (blue) and linear (red) structures are highlighted by arrowheads. All error bars indicate SEM; scale bars, 5 μm; time (s) is displayed after NEBD.
Images were acquired every 5 s on inverted microscope (Ti-E, Nikon) equipped with a 60× oil-immersion Plan-Apochromat objective (NA 1.4) and an iXon Ultra 897 camera. Time in seconds after NEBD is shown.
Images were acquired every 5 s on laser scanning confocal microscope (SP8, DIVE-FALCON) equipped with a 63×/1.4 oil objective. Note: fluorescence intensity scalings are increased for septinunc-61(RNAi). Time in seconds after NEBD is shown.
Vertebrate anillin binds myosin II with its N terminus53 and the co-localization of anillin and myosin II requires the anillin N terminus in Drosophila cells.91 Since NMY-2 and ANI-1 structures co-localize after Latrunculin A treatment and NMY-2 is expected to interact with the ANI-1 N terminus, we analyzed whether ANI-1 and NMY-2 still form structures that co-localize when the N terminus of ANI-1 is absent. We used an ANI-1 fragment that comprises the RBD, C2, and PH domain (Δ1–680, ANI-1AH-PH,90) and depleted endogenous ANI-1 by RNAi to prevent NMY-2 binding via full-length endogenous ANI-1. After Latrunculin A addition, NMY-2 and ANI-1AH-PH both formed circular structures, but these did not co-localize and did not become linear over time (Figures 4C and 4D; Video S7). This shows that the N terminus of ANI-1 is required for the formation of linear but not non-linear structures and is essential for the co-localization with NMY-2 structures.
Images were acquired as described for Video 5. Time in seconds after NEBD is shown.
Since NMY-2 formed membrane-localized structures independently of its binding partner F-actin and of the ANI-1 N terminus, we hypothesized that NMY-2 could bind the membrane directly. To test this, we purified NMY-2 in a complex with the myosin ELC (MLC-4) and the myosin RLC (MLC-5). We found that the NMY-2/MLC-4/MLC-5 complex, but not MLC-4 alone, bound to several lipids known to be enriched in the plasma membrane (Figures 4E and 4F).
In summary, in the absence of F-actin, ANI-1 and NMY-2 co-localize to non-linear and linear structures and linear structure formation requires the ANI-1 N terminus and is facilitated by septinUNC-61. The co-localization but not the formation of the ANI-1 and NMY-2 non-linear structures requires the ANI-1 Nterminus, suggesting that the binding of NMY-2 to the ANI-1 N terminus pulls those structures together. The NMY-2/MLC-4/MLC-5 complex bound to several membrane lipids, suggesting that this complex likely represents an independent membrane anchor.
The linker region of ANI-1 facilitates linear structure formation
ANI-1 contains multiple distinct domains: an actin-binding domain (ABD)55 and a predicted myosin II-binding domain (MBD)34 at the N terminus, and an RBD, a C2 membrane-binding domain, and a PH domain at the C terminus47 (Figure S8A). Sequence analysis predicts that the ANI-1 N terminus contains numerous intrinsically disordered regions (IDRs) similar to human anillin (Figures S8A and S8B) and yeast Mid1p.92
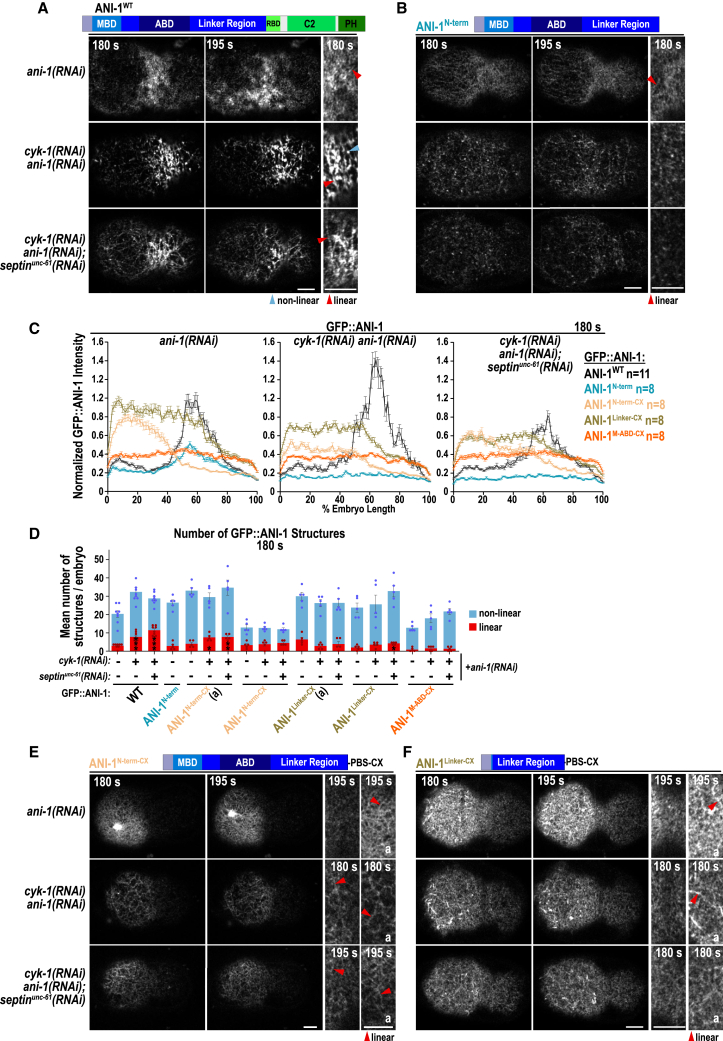
To analyze which region of ANI-1 is required for linear structure formation, we generated ani-1 RNAi-resistant transgenes in which the MBD (Δ48–160), the ABD (Δ236–460), the MBD and ABD (Δ48–460), and the entire N-terminal half (ANI-1C-term Δ48–680) were deleted (Figures 5 and S8C–S8F). As a control we generated wild-type ANI-1 transgene (ANI-1WT), which rescued embryonic lethality after ani-1(RNAi) and localized to circumferentially aligned linear structures after CYK-1 depletion, together suggesting that it is functional (Figures 5A–5C, S8F, and S9A; Video S8).
Figure 5.
The ANI-1 linker region facilitates linear structure formation
(A, D, E, and F) Representation of ANI-1 transgenes and confocal cortical images of GFP-tagged ANI-1 variants for indicated RNAi conditions and time points after NEBD. Magnifications of equatorial regions are shown (right). Non-linear (blue) and linear (red) structures are highlighted by arrowheads. To better visualize ANI-1ΔLinker after cyk-1(RNAi)ani-1(RNAi) and all transgenes after septinUNC-61 co-depletion, the signal intensity scaling was increased in those conditions. Scale bars, 5 μm.
(B) Mean normalized cortical fluorescence intensity of GFP-tagged ANI-1 variants for indicated RNAi conditions; n = number of embryos.
(C) Mean number of structures per embryo at the equator for different ANI-1 variants after RNAi depletions. The p values were calculated using Mann-Whitney U or Student’s t test: ∗∗p < 0.01 and ∗∗∗p < 0.001 in comparison to ani-1(RNAi) embryos expressing the same transgene; dots represent data points of individual embryos.
All error bars indicate SEM.
Note: fluorescence intensity scalings are increased for cyk-1(RNAi)ani-1(RNAi);septinunc-61(RNAi) conditions. Images were acquired every 5 s on laser scanning microscope (SP8) equipped with a 63×/1.3 GLYC objective and photomultiplier. Time in seconds after NEBD is shown.
All ANI-1 transgenes were found to express at similar levels as ANI-1WT (Figure S8E), and the MBD (Δ48–160), the ABD (Δ236–460), and the MBD and ABD (Δ48–460) deletion variants fully rescued embryonic lethality in the absence of endogenous ANI-1. The ANI-1C-term rescued lethality only partially after ani-1(RNAi) (Figure S8F). After ani-1(RNAi) or cyk-1(RNAi)ani-1(RNAi), the Δ48–160, Δ236–460, and Δ48–460 ANI-1 mutants enriched at the cell equator and formed linear structures that circumferentially aligned comparably to those of ANI-1WT (Figures 5A–5D and S9A–S9C; Videos S8, and S9). Furthermore, the ANI-1 Δ48–460 still formed some linear structures in the presence of Latrunculin A (Figures S9D and S9E; Video S10). After co-depletion of CYK-1 and septinUNC-61, the cortical levels of ANI-1 Δ48–460 were strongly reduced and only pulsatile accumulation was visible, but the levels of ANI-1 Δ48–160 and ANI-1 Δ236–460 were comparable to those of ANI-1WT (Figures 5D; Video S9). In those embryos, ANI-1 Δ48–160 and ANI-1 Δ236–460 still formed linear structures that aligned circumferentially around the cell equator (Figures 5C and S9A–S9C; Video S9).
Note: fluorescence intensity scalings are increased for cyk-1(RNAi)ani-1(RNAi);septinunc-61(RNAi) conditions. Images were acquired every 5 s on laser scanning microscope (SP8) equipped with a 63×/1.3 GLYC objective and photomultiplier. Time in seconds after NEBD is shown.
Images were acquired as described for Video 6. Note: fluorescence intensity scalings are increased for GFP::ANI-1N-term-CX. Time in seconds after NEBD is shown.
Since the MBD and ABD were not required for linear structure formation in cyk-1(RNAi), we analyzed the ANI-1C-term, which lacks the MBD, ABD, and the linker region. The ANI-1C-term formed very few linear but numerous non-linear structures after ani-1(RNAi). After cyk-1(RNAi)ani-1(RNAi), the few ANI-1C-term linear structures were completely absent and instead numerous non-linear structures that did not circumferentially align were present (Figures 5C, 5E, and S9A; Video S8). This demonstrates that the presence of the linker region is required for linear structure formation. Co-depletion of CYK-1 and septinUNC-61 resulted in a strong reduction of cortical ANI-1C-term levels in comparison to ANI-1WT levels, but transient pulsatile accumulation of ANI-1C-term was still observed (Figures 5B and 5E; Video S8). The absence of non-linear ANI-1C-term structures after co-depletion of CYK-1 and septinUNC-61 suggests that their formation requires septin binding.
Finally, we asked whether deletion of the linker region from the full-length ANI-1 protein also abolished linear structure formation (Figure S8C). ANI-1ΔLinker was expressed similar to ANI-1WT and partially rescued embryonic lethality after ani-1(RNAi) (Figures S8E and S8F). Cortical equatorial levels of ANI-1ΔLinker were reduced in comparison to the levels of ANI-1WT after ani-1(RNAi) and almost no linear structures formed (Figures 5B, 5C, and 5F; Video S11), consistent with the observation that the absence of the linker in ANI-1C-term abolished linear structure formation. Depletion of CYK-1 or CYK-1 and septinUNC-61 in the ANI-1ΔLinker strain caused a further reduction in cortical ANI-1ΔLinker levels, although pulsed accumulation was still visible (Figure 5F; Video S11). This contrasts with the ANI-1C-term, which formed septin-dependent non-linear structures after cyk-1(RNAi) (Figures 5C, 5E, and 5F).
Note: fluorescence intensity scalings are increased for cyk-1(RNAi)ani-1(RNAi) and cyk-1(RNAi)ani-1(RNAi);septinunc-61(RNAi) conditions. Images were acquired as described for Video 6. Time in seconds after NEBD is shown.
Together, the ANI-1 deletion analysis reveals that neither the predicted MBD nor the ABD are required for linear structure formation and that the linker region facilitates their formation.
The membrane-targeted ANI-1N-term is sufficient for linear structure formation
Since we observed that C-terminal ANI-1 half (ANI-1C-term) is not sufficient for linear structure formation, we tested whether the N-terminal half is. The ANI-1N-term, which comprises the putative MBD, the ABD, and the linker region, expressed well, partially rescued embryonic lethality, and weakly enriched at the cell equator after ani-1(RNAi) (Figures 6A–6C, S8C, S8E, and S8F). The ANI-1N-term formed some linear structures after depleting endogenous ANI-1; however, after CYK-1 or CYK-1 and septinUNC-61 depletion, the cortical levels of ANI-1N-term were strongly diminished (Figures 6B–6D; Video S12) and therefore the structure analysis was not possible. To overcome this limitation, we targeted ANI-1N-term to the plasma membrane by fusing it with a membrane-targeting region (ANI-1N-term-CX; Figure S8C). ANI-1N-term-CX was expressed similar to ANI-1WT and rescued embryonic lethality more efficiently than the non-membrane-targeted ANI-1N-term (Figures S8E and S8F). The ANI-1N-term-CX fragment was enriched at the anterior cortex (Figures 6C and 6E; Video S13) and, therefore, we analyzed ANI-1N-term-CX structures not only at the equatorial but also at the anterior cortex. The ANI-1N-term-CX formed linear structures in all RNAi conditions at the anterior cortex and some, but less, at the cell equator (Figures 6D and 6E). To determine whether full actin depolymerization prevents linear structure formation, ANI-1N-term-CX-expressing embryos were treated with Latrunculin A. Some linear ANI-1N-term-CX structures were still observed at late time points, although they were less defined than the ones formed by ANI-1WT (Figures S9D and S9E; Video S10).
Figure 6.
The membrane-targeted N-terminal half of ANI-1 is sufficient for linear structure formation
(A, B, E, and F) Representation of ANI-1 transgenes and cortical confocal images of GFP-tagged ANI-1 variants for indicated RNAi conditions and time points after NEBD. Magnifications of the cell equator and an anterior region (a) for ANI-1N-term-CX and ANI-1Linker-CX are displayed. Non-linear (blue) and linear (red) structures are highlighted by arrowheads. Scaling was increased for equator magnifications of ANI-1N-term-CX and ANI-1WT, ANI-1N-term, and ANI-1N-term-CX with septinUNC-61 co-depletion, and for the latter two also for cyk-1(RNAi)ani-1(RNAi). Scale bars, 5 μm.
(C) Mean normalized cortical fluorescence intensity of GFP-tagged ANI-1 variants for indicated RNAi treatments; n = number of embryos. ANI-1WT graphs are reproduced from Figure 5B.
(D) Mean number of structures at the equatorial and anterior (a) cortex for the different ANI-1 variants after RNAi depletions. The p values were calculated using Mann-Whitney U or Student’s t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 in comparison to ani-1(RNAi) embryos expressing the same transgene; dots represent data points of individual embryos. ANI-1WT graphs are reproduced from Figure 5C.
All error bars indicate SEM.
Note: fluorescence intensity scalings are increased for GFP::ANI-1WT with cyk-1(RNAi)ani-1(RNAi);septinunc-61(RNAi) and all ANI-1N-term conditions. Images were acquired as described for Video 6. Time in seconds after NEBD is shown.
Note: fluorescence intensity scalings are increased for cyk-1(RNAi)ani-1(RNAi) and cyk-1(RNAi)ani-1(RNAi); septinunc-61(RNAi). Images were acquired as described for Video 8. Time in seconds after NEBD is shown.
Since the N terminus permitted and its deletion abolished linear structure formation, we split the membrane-targeted N terminus into two fragments comprising the linker (ANI-1Linker-CX) or the MBD-ABD (ANI-1M-ABD-CX; Figure S8C). ANI-1Linker-CX and ANI-1M-ABD-CX were expressed comparable to ANI-1WT; however, embryonic lethality caused by depleting endogenous ANI-1 was only marginally rescued by ANI-1Linker-CX and not by ANI-1M-ABD-CX (Figures S8E and S8F). The ANI-1M-ABD-CX fragment formed almost no linear structures and the ANI-1Linker-CX fragment made some linear structures, although they were less frequent than observed for the ANI-1N-term-CX (Figures 6D, 6F, and S9F; Video S14).
Images were acquired as described for Video 6. Time in seconds after NEBD is shown.
Together, the membrane-targeted ANI-1 N terminus is sufficient for linear structure formation and its linker region and the M-ABD region are both necessary to form them efficiently.
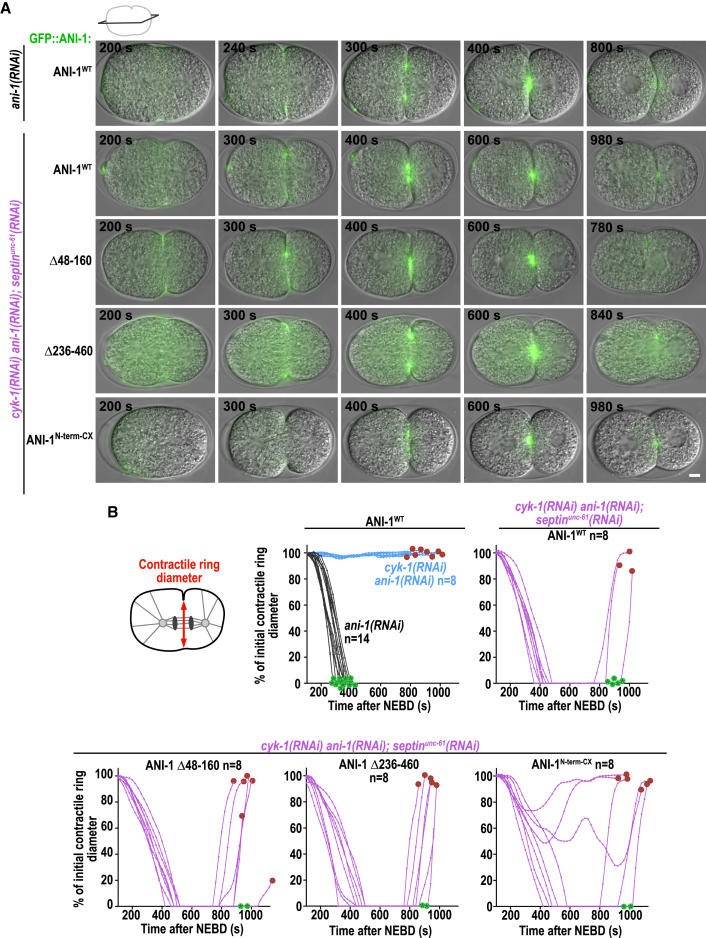
The ANI-1N-term-CX partially rescues furrow ingression in cyk-1(RNAi);septinunc-61(RNAi) embryos
To investigate whether the different ANI-1 mutants support furrow ingression, we filmed embryos in the central plane. As expected, depletion of CYK-1 in ANI-1WT-expressing embryos prevented furrow ingression and co-depletion of CYK-1 and septinUNC-61 resulted in complete furrow ingression (Figures 7 and S10A). ANI-1 Δ48–160 and ANI-1 Δ236–460 rescued cleavage furrow ingression similar to ANI-1WT (Figure 7), consistent with the fact that they formed linear structures. ANI-1 Δ48–460, ANI-1C-term, ANI-1N-term, and ANI-1ΔLinker did not rescue furrow ingression, but, as they localized only weakly to the cortex, no rescue was expected (Figure S10). Similarly, the membrane-tethered ANI-1Linker-CX or ANI-1M-ABD-CX did not rescue furrow ingression, consistent with the observation that only a few or almost no linear structures were present, respectively (Figure S10B). Importantly, five out of eight embryos expressing ANI-1N-term-CX fully ingressed a cleavage furrow and, in two of those embryos, the cleavage furrow remained closed until the onset of the second cell division (Figure 7). This demonstrates that the N terminus of ANI-1, even though not being able to fully enrich at the cell equator, is able to partially rescue the cytokinesis failure in cyk-1(RNAi)ani-1(RNAi);septinunc-61(RNAi) embryos.
Figure 7.
The membrane-targeted N-terminal ANI-1 fragment partially rescues furrow ingression in cyk-1(RNAi)ani-1(RNAi);septinunc-61(RNAi)
(A) Merged differential interference contrast (DIC) and wide-field fluorescent central plane images for indicated GFP-tagged ANI-1 variants, RNAi conditions, and time points after NEBD. Scale bar, 5 μm.
(B) Plotted is the contractile ring diameter for indicated RNAi conditions and ANI-1 variants; n = number of embryos. Green and red encircled stars indicate whether embryos succeed or fail cytokinesis, respectively.
Discussion
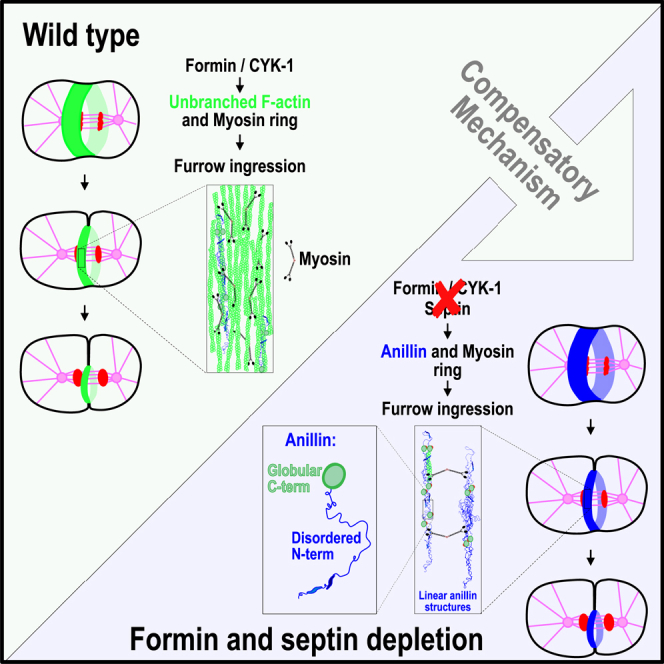
In animal cells, the contractile ring consists of formin-nucleated unbranched F-actin, which circumferentially aligns around the cell equator. Myosin-dependent constriction of the contractile ring mediates cleavage furrow ingression.93 Here we discover an ANI-1-dependent mechanism that promotes furrow ingression when the levels of formin-nucleated F-actin and septins are strongly reduced (Figure S11). We propose that, in this situation, active RhoA recruits ANI-1 and NMY-2 to the cell equator during anaphase. ANI-1 is tethered to the equatorial membrane by its C2 and RBD domain and NMY-2 directly binds lipids of the plasma membrane. The disordered ANI-1 N terminus extends into the cytoplasm, interacts with neighboring ANI-1 molecules, and forms linear structures that circumferentially align around the cell equator in an NMY-2-dependent manner. The linear ANI-1 structures together with the residual F-actin form a contractile ring that is decorated by NMY-2 and is able to constrict. The generated force is conveyed to the plasma membrane by membrane-bound ANI-1 and NMY-2 to form the cleavage furrow. Some linear ANI-1 structures are present in wild-type embryos and could also assist with furrow ingression by a similar mechanism in the presence of normal levels of formin-nucleated F-actin.
NMY-2 is an independent membrane anchor that links the actin cortex with the membrane
NMY-2 enriches in foci on the equatorial plasma membrane after F-actin depolymerization even in the absence of the ANI-1 N terminus. Furthermore, in vitro binding assays show that NMY-2, together with the RLC and ELC, interacts with multiple plasma membrane lipids. Together, our data suggest that NMY-2, after activation by RhoA signaling, binds the membrane directly. Membrane binding of myosin II seems conserved in other organisms as, in Drosophila cells, myosin also localizes to the membrane independently of anillin and F-actin.33 In fission yeast, Mid1p targets Myo2 to the cytokinesis nodes during ring assembly37,38 and, with the onset of ring constriction, Mid1p but not Myo2 leaves the division sites,94 suggesting that Myo2 could bind the membrane independently of Mid1p. A previous study found that myosin II interacts with lipids through its RLC-binding site and lipid binding resulted in displacement of the RLC.95 This implies that membrane-bound myosin II is in the inactive state, since the RLC is required for myosin II activity. In contrast, we observed that NMY-2 interacts with membrane lipids in the presence of the RLC and ELC, suggesting that NMY-2 can bind the membrane in its active state. Thus, in C. elegans, membrane-bound active NMY-2 might represent an independent membrane anchor that links the plasma membrane with the actin cortex and thereby could transmit forces generated in the contractile ring to the plasma membrane. NMY-2 membrane tethering might function redundantly with other membrane-binding components such as ECT2,96 anillin,48 and RacGAP1.97
Formation of linear ANI-1 structures is modulated by the amount of F-actin
We discover that ANI-1 localizes not only to cortical patches34,70 but also to some linear structures during cytokinesis in the one-cell C. elegans embryo. After the reduction of formin-nucleated F-actin, numerous linear ANI-1 structures form and circumferentially align around the cell equator. How could F-actin limit the formation of linear ANI-1 structures? We speculate that, when unbranched F-actin levels are high, ANI-1 binds F-actin and distributes along the filaments, which limits contacts between ANI-1 molecules or restrains extension or flexibility of the disordered ANI-1 N terminus. When unbranched F-actin levels are low, such as in the case of CYK-1 depletion, ANI-1 N termini are free, accessible to self-interact and form linear structures. In the complete absence of F-actin, ANI-1WT, and to some extent even ANI-1 Δ48–460 and ANI-1N-term-CX, first forms circular structures that become linear at late anaphase time points. The delayed formation of linear ANI-1 structures in the absence of F-actin strongly suggests that the residual short actin filaments might bridge adjacent linear ANI-1 structures to accelerate the process. We did not observe a change in ANI-1 structure formation after the reduction of Arp2/3-generated branched F-actin, suggesting that ANI-1 preferentially binds CYK-1-nucleated unbranched F-actin.
Anillin binds septins, which can oligomerize into higher-order structures such as filaments.64,98 Septins are required for the formation of filamentous anillin structures in Latrunculin A-treated Drosophila S2 cells.33,52 Similarly, our data suggest that, in the absence of F-actin, septins facilitate linear structure formation of full-length ANI-1. However, a C-terminal ANI-1 fragment (ANI-1AH-PH), which still harbors the septin binding PH domain, was insufficient for linear structure formation in the absence of F-actin, suggesting that linear structures form via the N terminus. Linear ANI-1 structures induced by cyk-1(RNAi) were not affected by septinUNC-61 or septinUNC-61 and septinUNC-59 co-depletion, although cortical ANI-1 levels were clearly reduced. This shows that linear ANI-1 structures form efficiently after a strong reduction of both C. elegans septins as long as residual F-actin is present. Only in cases when F-actin is completely absent do septins facilitate linear structure formation. This suggests that the residual F-actin and septin support linear structure formation, probably stabilizing and/or bundling ANI-1. Although the septin depletion was highly efficient, we cannot entirely exclude a contribution of some remaining septin in linear ANI-1 structure formation, and future work will have to generate null mutations in both septin genes to test this.
Filamentous-type anillin was not only observed in Latrunculin A-treated Drosophila cells33,52 but also along F-actin bundles in unperturbed vertebrate cells49 after myosin II inhibition.53 Thus, the competence of anillin to form linear structures may be a conserved feature from worms to humans.
The C-terminal half of ANI-1 concentrates at the cell equator in septin-dependent circular structures
Anillin’s C-terminal half harbors three membrane interaction elements: the RBD, the C2, and the PH domain, and each of them contributes to membrane localization of anillin in human cells.46,47,48 Importantly, the ANI-1 C-terminal half forms circular and not linear structures in cyk-1(RNAi) and Latrunculin A-treated embryos. The localization of the ANI-1C-term to the circular structures strongly depends on septins, thus C. elegans ANI-1, similar to anillin in other organisms, should bind to septins via the PH domain.46,48,49,86
Human anillin binds septins not only directly with the PH domain but also indirectly via CIN85 with amino acids 1–150 at its very N terminus.85 Thus it is possible that ANI-1 has a second septin-binding site at its very N-terminal end. We found that removal of amino acids 48–160 from ANI-1 did not prevent linear structure formation or furrow ingression after CYK-1 and septin co-depletion. Further, the presence of amino acids 1–48 in the ANI-1C-term fragment and amino acids 1–459 in the ANI-1M-ABD-CX fragment was not sufficient for linear structure formation. Together, indirect septin binding of ANI-1 via its very N terminus is either not conserved in C. elegans or is not essential for the process studied here.
Since ANI-1 is reduced after septin depletion and septin recruitment to the contractile ring entirely depends on ANI-1,34,48 ANI-1 and septin stabilize each other on the plasma membrane.
After septin and CYK-1 co-depletion, a residual pulsatile membrane localization of ANI-1C-term, ANI-1 Δ48–460, and ANI-1ΔLinker persisted. This could reflect direct binding to the plasma membrane and/or binding to active RhoA. Given that the ANI-1C-term membrane localization strongly depends on septins, changes in ANI-1C-term dynamics cannot be exclusively attributed to active RhoA dynamics. Therefore, employing the ANI-1C-term as a biosensor for active RhoA22,35,89 should be done with caution.
The disordered ANI-1 N terminus is sufficient for linear structure formation
The membrane-targeted N-terminal ANI-1 half localized in linear structures. Further, our mutant analysis of full-length ANI-1 revealed that neither the MBD nor the ABD nor both domains together are required for ANI-1 linear structure formation in control and cyk-1(RNAi) embryos. Only further shortening of ANI-1 by removal of the linker region prevented the formation of linear structures, and circular septin-dependent foci formed instead. Splitting the N-terminal ANI-1 half revealed that the linker region formed a few and the M-ABD no linear structures, indicating that both regions contribute to efficient linear structure formation when tethered to the membrane. Since the linker region of ANI-1 was sufficient for linear structures formation when bound to the membrane by the ANI-1 C terminus (ANI-1Δ48-460), it suggests that the employed membrane anchor might not mimic all necessary functional features such as enrichment in certain lipids domains48 and/or the correct distance from the membrane. Nevertheless, the membrane-targeted N-terminal fragment rescued embryonic lethality more efficiently than the non-targeted version, demonstrating that membrane binding of the Nterminus improves its functionality. Deletion of the linker region from full-length ANI-1 reduced linear structures after ani-1(RNAi), supporting our conclusion that the linker is a key region to form them. Unfortunately, deletion of the linker appears to compromise the function of both the M-ABD and the Cterminus, since the ANI-1ΔLinker mutant is more lethal than the ANI-1C-term mutant, which lacks the entire N terminus, including the linker region. In addition, the ANI-1ΔLinker forms no circular septin-dependent structures after cyk-1(RNAi), although it contains the PH domain. We therefore speculate that the M-ABD and C terminus bind each other in the ANI-1ΔLinker mutant and thereby block each other’s function.
Since the N terminus of Mid1p forms high-molecular-weight oligomers,99 one possibility is that the ANI-1 N terminus assembles into linear structures by self-interaction-mediated oligomerization. Furthermore, liquid-liquid phase separation mechanisms might drive the formation of linear ANI-1 structures. Our structure prediction reveals that the N terminus of ANI-1 is highly disordered but the Cterminus is not. This feature of ANI-1 is conserved from fission yeast92 to worms and humans. Disordered regions frequently mediate the formation of biomolecular condensates by liquid-liquid phase separation,100 and indeed the disordered N terminus of Mid1p forms condensates in vitro.92 Liquid droplets are typically isotropic but their shape and material properties are tunable in multiple ways.101 For example, isotropic liquid condensates of fused in sarcoma (FUS) transform into spindle-shaped condensates when F-actin is incorporated in the droplets.102 We speculate that linear ANI-1 structures could form by similar principles: the isotropic spherical shape of the biomolecular ANI-1 condensates is biased to an extended anisotropic form by additional factors such as remaining F-actin polymers or posttranslational modifications of ANI-1.
The ANI-1 N terminus promotes furrow ingression after co-depletion of formin and septins
After CYK-1 depletion, ANI-1 forms linear structures and, at the same time, cortical septinUNC-59 levels increase. The reduction in F-actin levels after cyk-1(RNAi) might increase the free pool of ANI-1, which then binds and stabilizes septin and thereby causes an elevation in cortical septin levels. Septins or the excess thereof could then tether ANI-1 tightly to the membrane and thereby hinder cleavage furrow ingression in CYK-1-depleted embryos. A reduction in septin levels might weaken the interaction between ANI-1 and the plasma membrane and consequently lead to increased mobility of ANI-1 on the membrane, which could facilitate the interaction between ANI-1 molecules and the binding of ANI-1 to NMY-2 and the residual F-actin. All cyk-1(RNAi);septinunc-61(RNAi) embryos fully ingress a cleavage furrow, but half of them fail in abscission and the furrow regresses. Septins are essential for abscission in Drosophila and human cells65,68 but not in C. elegans embryos.67 Thus, our data indicate a functional redundancy between unbranched F-actin and septins during abscission in C. elegans.
Our data suggest that the N terminus of ANI-1 interacts with NMY-2, since co-localization of NMY-2 and ANI-1 after Latrunculin A treatment requires its presence. Consistent with that, NMY-2 was required for the circumferential alignment of the linear ANI-1 structures. Surprisingly, deletion of the putative MBD did not abrogate linear ANI-1 structure alignment. Thus, either the NMY-2 binding region is not properly predicted or NMY-2 does not align ANI-1 via direct binding. Since CYK-1-depleted embryos only exhibit short-range cortical flows,103 the alignment of the linear ANI-1 structures around the cell equator is unlikely to be flow dependent, as suggested for unbranched F-actin.22 Therefore, we favor alternative models where localized motor activity of NMY-2 and crosslinkers promotes equatorial circumferential alignment.23
Furrow ingression in CYK-1 and septin-depleted embryos required residual F-actin, NMY-2, and ANI-1. Furthermore, furrow ingression was only supported by ANI-1 fragments that formed linear structures and that circumferentially aligned around the cell equator, similar to unbranched F-actin in control embryos. Therefore, it is tempting to speculate that those ANI-1 structures have a prominent role in furrowing in septin and CYK-1 co-depleted embryos. Although CYK-1 depletion was highly efficient, residual CYK-1 could generate some actin filaments at the cell equator. Additionally, actin filaments could be polymerized at the polar cortex and transported to the equator by equatorial-directed cortical flows.104 However, we consider flow-mediated actin transport unlikely since cortical flows are compromised in cyk-1(RNAi) embryos103 and actin filament turnover is typically too high for efficient long-range transport.105 Even though those mechanisms might result in some F-actin at the cell equator, we and others could not detect equatorial F-actin enrichment.79,81,82,83
It is known that several anillin homologs, including C. elegans ANI-1, bind and bundle F-actin.50,55,56 Human anillin even mediates the formation of actin rings from actin filaments in vitro.63 Thus, the circumferentially aligned linear ANI-1 structures could organize the few remaining actin filaments into a stable equatorial ring by providing a scaffold and by bridging the residual actin filaments. Although it has been reported that ANI-1-induced actin rings can contract in the absence of myosin in vitro,63 ANI-1 rings formed in vivo after co-depletion of CYK-1 and septins do require NMY-2 for constriction. Anillin organizes NMY-2 into cortical patches in the C. elegans zygote,34 stabilizes myosin II in the contractile ring, and interacts with NMY-2.33,46,53 Therefore, ANI-1 could mediate the higher-order organization of NMY-2, stabilize NMY-2 in the furrow region, and bring NMY-2 in close proximity to residual F-actin so that it can generate force as a motor or tension as an F-actin crosslinker, similar to what happens in normal contractile rings.93
However, since detectable F-actin did not accumulate at the equator after CYK-1 and septin co-depletion and the ABD of ANI-1 was not required for furrow ingression, we favor an alternative explanation. We hypothesize that linear ANI-1 structures represent a novel type of network that contracts by biomolecular condensation. ANI-1 is predicted to have numerous IDRs at the N terminus, and we showed that the Nterminus is sufficient for linear structure formation and furrow ingression. IDRs are frequently found in proteins that form biomolecular condensates by liquid-liquid phase separation. Recent reports demonstrate that biomolecular condensation can also exert forces on cellular structures and alter the shape of membranes.106,107,108 For example, the formation of biomolecular condensates of FUS on the surface of membrane vesicles generates compressive stress and causes inward membrane bending.109 We speculate that condensation of linear ANI-1 structures results in the contraction of the ANI-1 network and ultimately in the inward bending of the plasma membrane (Figure S11). The contractile force generated by ANI-1 condensation is then propagated around the ANI-1 network by NMY-2 filaments, which bind and crosslink the ANI-1 linear structures. As a first step, future work will need to show that C. elegans ANI-1, like Mid1p from fission yeast,92 also undergoes liquid-liquid phase separation. Linear ANI-1 structures are also observed in wild-type embryos that have normal F-actin levels. So, it is possible that ANI-1 linear structures may assist with furrow ingression in a normal situation and take over the process when unbranched F-actin levels are reduced.
In summary, we find that ANI-1 forms linear structures and supports furrow ingression when unbranched F-actin levels are low. Although actomyosin-dependent constriction is the most actively investigated mechanism for cytokinetic furrowing, alternative ways have been reported to exist. For example, furrow ingression is mediated in Dictyostelium and human RPE1 by the opposite migration direction of the two daughter cells75,76,77 and in archaea by supercoiling of the elastic ESCRT-III filament.110,111 Our findings highlight the existence of a compensatory mechanism of cleavage furrow ingression that is likely to contribute to the enormous robustness of cytokinesis.
Limitations of the study
The study employs RNAi-mediated protein depletions to analyze the function of various cytokinesis regulators. Although the study shows that the RNAi depletions are highly efficient, it cannot be excluded that residual amounts of proteins contribute to the processes. In addition, the condensation-based model of ring constriction is based on the prediction that ANI-1 undergoes liquid-liquid phase separation, which still must be demonstrated for C. elegans ANI-1.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-GFP (clones 7.1 and 13.1) | Roche | Cat#11814460001; RRID: AB_390913 |
| Mouse monoclonal anti-Actin (clone AC-15) | Sigma-Aldrich | Cat#A1978; RRID: AB_476692 |
| Mouse monoclonal anti-FLAG (clone M2) | Sigma-Aldrich | Cat#F1804; RRID: AB_262044 |
| Mouse monoclonal anti-Strep-tag™ II | IBA Lifesciences | Cat#2-1507-001; RRID: AB_513133 |
| Mouse monoclonal anti-His-tag (clone HIS.H8) | Millipore | Cat#05-949; RRID: AB_492660 |
| Goat anti-Mouse IgG (H/L)-HRP Conjugate antibody (polyclonal) | Bio-Rad | Cat#1706516; RRID: AB_11125547 |
| Bacterial and virus strains | ||
| Escherichia coli OP50 | Caenorhabditis Genetics Center (CGC) | N/A |
| Escherichia coli DH5a | Thermo Fisher Scientific | Cat#18265017 |
| Escherichia coli HT115 (DE3) | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| ECL Prime Western Blotting Detection Reagent | Cytiva Amersham™ | Cat#RPN2236 |
| Pierce™ ECL Western Blotting Substrate | Thermo Fisher Scientific | Cat#32109 |
| Latrunculin A | Enzo Life Sciences; Sigma-Aldrich | Cat#BML-T119; Cat#L5163 |
| FM™ 4–64 Dye | Thermo Fisher Scientific | Cat#T13320 |
| Critical commercial assays | ||
| PIP Strips – Lipid-Protein Interaction Assay | Echelon Bioscience | Cat#P-6001 |
| MEGAscript™ T7 Transcription Kit | Thermo Fisher Scientific | Cat#AM1334 |
| Experimental models: organisms/strains | ||
| Wild type (Caenorhabditis elegans) | Caenorhabditis Genetics Center (CGC) | N2 |
| xsSi5 [pie-1p::GFP::ani-1AH-PH::pie-1 3′UTR + Cbr-unc-119(+)]II | (Tse et al.)90 | MG617 |
| ani-1(mon7[mNeonGreen^3xFlag::ani-1]) III | (Rehain-Bell et al.)112 | MDX29 |
| zbls2[pie-1p::LifeAct::RFP + unc-119(+)] | Zhirong Bao | BV70 |
| nmy-2(cp52[nmy-2::mkate2 + LoxP unc-119(+) LoxP]) I; unc-119(ed3) III | (Dickinson et al.)113 | LP229 |
| cyk-1::gfp | (Reymann et al.)22 | SWG004 |
| oxTi179 II; unc-119(ed3) III | (Frøkjær-Jensen et al.)114 | EG8079 |
| unc-59(qy50[unc-59::GFP::3xflag::AID]) I | (Chen et al.)115 | NK2225 |
| ItIs157 [pAC16; ppie-1::LifeAct::GFP; unc-119 (+)]; unc- 119(ed3) III; prtSi2[pAC71; Pnmy-2:: re-encoded nmy- 2::mCherry::StrepTagII::3′UTR nmy-2; cb-unc-119(+)] II | (Osório et al.)74 | GCP22 |
| nmy-2(cp13[nmy-2::gfp + LoxP]) I; OD56 (mCherry::histone H2B; ltIs44[pie-1p::mCherry::PH(PLC1delta1)+unc-119(+)] | This study | ZAN23 |
| ani-1(mon7[mNeonGreen^3xFlag::ani-1]) III; zbls[pie-1:: LifeAct::RFP + unc-119(+)] | This study | ZAN348 |
| nmy-2(cp52[nmy-2::mkate2 + LoxP unc-119(+) LoxP]) I; unc-119(ed3) III; ani-1(mon7[mNeonGreen^3xFlag::ani-1]) III | This study | ZAN351 |
| nmy-2(cp52[nmy-2::mkate2 + LoxP unc-119(+) LoxP]) I; unc-119(ed3) III; xsSi5 [pie-1p::GFP::ani-1AH-PH::pie-1 3′UTR + Cbr-unc-119(+)] II | This study | ZAN368 |
| cyk-1::gfp, zbls2[pie-1p::LifeAct::RFP + unc-119(+)] | This study | ZAN371 |
| Si192[pEZ379; pani-1::GFP::ANI-1RE-WT; cb-unc-119(+)]II; unc-119(ed3) III | This study | ZAN373 |
| Si193[pEZ388; pani-1::GFP::ANI-1RE-Δ236-460AA; cb-unc-119(+)]II; unc-119(ed3) III | This study | ZAN375 |
| Si194[pEZ389; pani-1::GFP::ANI-1RE-Δ48-460AA; cb-unc-119(+)]II; unc-119(ed3) III | This study | ZAN376 |
| Si197[pEZ390; pani-1::GFP::ANI-1N−term-CX(1-763AA-PBS-CX); cb-unc-119(+)]II; unc-119(ed3) III | This study | ZAN379 |
| Si200[pEZ391; pani-1::GFP::ANI-1C−term(Δ48-680AA); cb-unc-119(+)]II; unc-119(ed3) III | This study | ZAN382 |
| Si201[pEZ387; pani-1::GFP::ANI-1RE-Δ48–160; cb-unc-119(+)]II; unc-119(ed3) III | This study | ZAN383 |
| Si203[pEZ419; pani-1::GFP::ANI-1N−term(1-763AA); cb-unc-119(+)]II; unc-119(ed3) III | This study | ZAN386 |
| Si204[pEZ422; pani-1::GFP::ANI-1Linker-CX(1-47AA, 441–763 AA-PBS-CX); cb-unc-119(+)]II; unc-119(ed3) III | This study | ZAN387 |
| Si205[pEZ421; pani-1::GFP::ANI-1M-ABD-CX(1-459AA-PBS-CX); cb-unc-119(+)]II; unc-119(ed3) III | This study | ZAN390 |
| Si206[pEZ420; pani-1::GFP::ANI-1ΔLinker(Δ460-763AA); cb-unc-119(+)]II; unc-119(ed3) III | This study | ZAN391 |
| unc-59(qy50[unc-59::GFP::AID])I; ltIs37 [pAA64; pie-1/mCherry::his-58; unc-119 (+)] | This study | GCP1126 |
| Recombinant DNA | ||
| pACEbac1-6xHis:nmy-2 | This study | N/A |
| pACEbac1-StrepTagII:mlc-4 | (Osório et al.)74 | pAC437 |
| pACEbac1-StrepTagII:mlc-5 | (Osório et al.)74 | pAC438 |
| pCFJ350: pani-1::gfp::ani-1RE-WT::ani-1 | This study | pEZ379 |
| pCFJ350: pani-1::gfp::ani-1RE-Δ48-160AA::ani-1 | This study | pEZ387 |
| pCFJ350: pani-1::gfp::ani-1RE-Δ236-460AA::ani-1 | This study | pEZ388 |
| pCFJ350: pani-1::gfp::ani-1RE-Δ48-460AA::ani-1 | This study | pEZ389 |
| pCFJ350: pani-1::gfp::ani-1N−term(1-763AA-PBS-CX)::ani-1 | This study | pEZ390 |
| pCFJ350: pani-1::gfp::ani-1C−term (Δ48-680AA)::ani-1 | This study | pEZ391 |
| pCFJ350: pani-1::gfp::ani-1N−term (1-763AA)::ani-1 | This study | pEZ419 |
| pCFJ350: pani-1::gfp::ani-1ΔLinker (Δ460-763AA)::ani-1 | This study | pEZ420 |
| pCFJ350: pani-1::gfp::ani-1Linker−CX (1-47AA, 441-763AA-PBS-CX)::ani-1 | This study | pEZ422 |
| pCFJ350: pani-1::gfp::ani-1M−ABD-CX (1-459AA-PBS-CX)::ani-1 | This study | pEZ421 |
| Software and algorithms | ||
| NIS Elements 5.21.03 | Nikon | https://www.microscope.healthcare.nikon.com/en_EU/products/software/nis-elements |
| LAS X 3.5.7.23225 | Leica | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Andor iQ3 | Oxford Instruments | https://andor.oxinst.com/ |
| Fiji (ImageJ) 1.54 | Schindelin et al.116 | https://imagej.nih.gov/ij/ |
| Prism 9 | GraphPad | https://www.graphpad.com/ |
| Image Lab v. 5.2.1 | Bio-Rad | http://www.bio-rad.com/ |
| MS Excel | Microsoft | https://www.microsoft.com/ |
| KNIME Analytics Platform 4.6.3 | KNIME | https://www.knime.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Esther Zanin (esther.zanin@fau.de).
Materials availability
C. elegans strains and plasmids generated in this study will be made available upon request via the lead contact.
Experimental model and subject details
C. elegans strains were maintained at 20°C on NGM plates seeded with Escherichia coli (OP50).117 Details of the used worm strains112,113,115 are provided in key resources table.
RNAi experiments were performed by injection or feeding and details of the RNAi experiments are listed in Table S1. For the generation of dsRNA for injection the target sequence was amplified by PCR from cDNA or genomic DNA with sequence specific primers containing T7 overhangs on both sides (Table S1). The purified PCR product was used as a template for the in vitro transcription reaction using the MEGAscript T7 kit (AM1334 Invitrogen, Thermo Fischer). dsRNA was injected into young hermaphrodites mounted on 2% agarose pads. After injections worms were rescued on OP50 seeded NGM plates and incubated at 20°C for 15–19 h (perm-1), 24–28 h (nmy-2), or 39–49 h (cyk-1, ani-1, arx-2, septinunc−59, septinunc−61). Since septinunc−61(RNAi) and septin co-depletion showed a similar phenotype, we pursued most experiments with septinunc−61(RNAi). For perm-1 and ani-1 double RNAi (Figure S9D), worms were first injected with ani-1 dsRNA and after 30 h incubation time at 20°C injected again with perm-1 dsRNA. For cyk-1 or cyk-1; septinunc−61 and nmy-2 double/triple RNAi (Figure 3), worms were first injected with cyk-1 or cyk-1; septinunc−61 dsRNA and after 22 h incubation time at 20°C injected again with nmy-2 dsRNA.
For feeding RNAi, L4440 vectors carrying part of the sequence of ani-1 or perm-1 were obtained from the Ahringer library (Source Bioscience), sequenced to confirm the gene target, and transformed into HT115 (DE3) bacteria. Briefly, the RNAi bacterial clones were grown until an OD of 1.6 in 50 mL Lysogeny broth (LB) medium containing 12.5 μg/mL tetracycline and 50 μg/mL ampicillin overnight at 37°C. The overnight culture was centrifuged at 4000 g for 10 min, the supernatant was removed, and the cell pellet was resuspended in 2.5 mL LB medium containing 12.5 μg/mL tetracycline, 50 μg/mL ampicillin, and 0.23 mg/mL IPTG (isopropyl β-D-1-thiogalactopyranoside). In parallel, unseeded NGM plates were dried for 1 h in a 37°C incubator. Then, 100 μL of a 1:1:1 mix of 50 μg/mL ampicillin, 12.5 μg/mL tetracycline, and 0.1 g/mL IPTG was added. These treated plates were inoculated with 100 μL of bacterial culture. To obtain permeabilized embryos in Figures 4 and S6, the inoculated bacterial culture consisted of 20 μL of bacteria expressing perm-1 dsRNA118 and 80 μL of LB (perm-1(RNAi) alone), or 20 μL of bacteria expressing perm-1 dsRNA and 80 μL of bacteria expressing ani-1 (perm-1(RNAi)). The expression of dsRNA was induced for 5 h at 37°C, in the dark. 25–30 L4 stage hermaphrodites were added to the treated plates and incubated at 20°C for 45–48 h before dissection for imaging. For Latrunculin A treatment in Figure S9D hermaphrodites were injected first injected with ani-1 and subsequently with perm-1 day RNA as described above.
Method details
Embryonic lethality counts
To determine embryonic lethality after ani-1(RNAi), young adult worms were injected with ani-1 dsRNA and incubated at 20°C for 48 h. Afterward, injected worms were singled on NGM plates, incubated 20 h and sacrificed after 24 h. The number of larvae and dead embryos was counted 24 h later.
Immunoblotting
For immunoblotting, worms were picked in MPEG (M9 plus 0.05% polyethylene glycol 8000 (PEG)) buffer and washed three times. The volume of the MPEG buffer was reduced and sample buffer was added to reach approximately 1 worm/μL. Worms were incubated at 95°C for 5 min, followed by centrifugation at room temperature for 10 min at 20000 g to spin down debris and subsequently 20 μL of supernatant were loaded in each lane.119 Membranes were incubated with anti-Actin (A1978 Sigma, 1:6000), anti-FLAG (F1804 Sigma, 1:2500) or anti-GFP (11814460001 Roche, 1:600) primary antibodies. HRP-conjugated mouse (1706516 Bio-Rad, 1:7500) were used as secondary antibodies.
Mos1-mediated single copy insertion (MosSCI) of ANI-1 variants
For the gfp::ani-1 transgenes the genomic ani-1 locus (6507 bp) with its promotor (565 bp) and 3′UTR (319 bp) were cloned together with GFP into pCFJ350 by Gibson cloning. ANI-1 transgenes were ani-1 RNAi resistant so that we could specifically deplete endogenous ANI-1 by RNAi and test the function of the ANI-1 variants. For this, exon 5 was rendered RNAi resistant by amino acid codon shuffling (Figure S8D). For the ANI-1N−term-CX, ANI-1Linker−CX, ANI-1M−ABD-CX variants the PBS and the CX motif of RHO-1 (C. elegans RhoA, AA181-192) were fused to the C-terminal end. We chose this region as a membrane anchor, since it was sufficient for membrane localization and did not exhibit any specific cortical enrichment during anaphase. All transgenes are listed in the key resources table.
Ani-1 transgenes were integrated on chromosome II (EG8079) using the MosSCI method.114 Young hermaphrodites were mounted on 2% agarose pads and injected with a mix containing the vector carrying the ani-1 transgene, the transposase (pCFJ601) and co-injection markers (pCFJ90 and pCFJ104). Worms were singled on NGM plates seeded with OP50 and incubated at 25°C for 7–10 days until starved. Wild type movers, negative for the mCherry-tagged array markers, were checked for homozygous GFP expression and correct integration was verified using PCR.
Fluorescence microscopy and image analysis
C. elegans gravid hermaphrodites were dissected in 4 μL M9 buffer on a 18 × 18 mm coverslip. The embryos were mounted on a 2% agarose pad by inverting the coverslip onto the pad and then sealed with vaseline. Confocal images (Figures 1, 2, 3, S1A, S1F, S2, S3A, S4, and S5) were acquired using a Nikon inverted microscope (Eclipse Ti) equipped with a confocal spinning disk unit, a 100×1.45-NA Plan-Apochromat oil immersion objective and an Andor DU-888 X-11056 camera (1024x1024 pixels). The system was controlled by NIS Elements software. GFP and red-fluorescent probes were imaged using 488 nm and 561 nm lasers, respectively, and for some experiments transmission images were acquired in the central plane. Confocal images of Figures 5A, 5D, 5E, 6E, S9B, and S9C were acquired on the Leica SP8 confocal laser scanning microscope equipped with a white light laser, 63x/1.3 GLYC objective and a photomultiplier and of Figures 5F, 6A, 6B, 6F, S1E, S3C, S7, and S9D, on a Leica TCS SP8 DIVE-FALCON equipped with 488 nm argon and 561 nm lasers and APO CS2 63x/1.4 oil objective. To display images acquired on the Leica SP8, a Gaussian Blur filter with a radius of 0.8 was applied to reduce noise. Cortical images in Figures 4 and S6 were acquired on a spinning disk confocal system (Andor Revolution XD Confocal System; Andor Technology) with a confocal scanner unit (CSU-X1; Yokogawa Electric Corporation) mounted on an inverted microscope (Ti-E, Nikon) equipped with a 60×1.4 NA Plan-Apochromat oil objective and solid-state lasers of 488 nm (50 mW) and 561 nm (50 mW). For image acquisition, 12 × 0.5 μm z-stacks were collected every 5 s by using an electron multiplication back-thinned charge coupled device camera (iXon Ultra 897; Andor Technology). Acquisition parameters, shutters and focus were controlled by Andor iQ3 software. Wide-field fluorescent and DIC images in Figures 7A and S10A were acquired on either a Nikon Ti2-Eclipse microscope equipped with a CFI Apochromat 100×1.49 NA oil objective and a Prime 95B, A18E203001 camera or on a Zeiss AxioImager.Z1 ApoTome microscope equipped with a 63×1.4-NA Plan-Apochromat oil immersion objective (Zeiss) and AxioCamMR3 camera to measure contractile ring diameter.
Image analysis and quantification was performed in Fiji.116 All data analysis was performed in Excel, Prism (GraphPad) or KNIME analytics (http://www.knime.org) and figures were assembled with the Affinity Designer software.
For high-time resolution data, imaging was started during pronuclear migration with central planes images acquired every 16 s (Figures 2, 3, 5, 6, S1, S2E, S3, S4, S5A, S7, and S9). At the time of NEBD (defined as the time point when the border of the nucleus was no longer visible in the DIC channel) imaging was stopped and resumed at the cell cortex before anaphase onset with a time interval of 2.5 s (Figures 2, 3, S1A, S1F, S2E, S4, and S5A) or 5 s (Figures 5, 6, S1E, S3C, S7, and S9).
Fluorescence intensity along cortical plane images (Figures 5B, 6C, S1B, S1E, S1F, S2E, S3B, S3E, S4C, S5B, S5E, and S6F) was measured by drawing a wide line from the anterior to the posterior cortex of the embryo 180 s after NEBD, which is around the time of contractile ring assembly (Figure S1B). Cortical fluorescence intensity was normalized by dividing by the mean cytoplasmic intensity measured in a rectangular box in the posterior region of central plane images at the time of NEBD. The mean normalized cortical intensity was calculated for 101 intervals from the anterior (0%) to the posterior (100%) for each embryo. To correct the fluorescent signal for bleaching, the mean cortical intensity was measured over time in a rectangular box in the posterior region for each embryo. The mean fluorescence intensity relative to the initial intensity was calculated for control embryos (n ≥ 11) for each time point and used as a ‘correction coefficient’. To correct for bleaching, the fluorescent intensity measured at a specific time point was multiplied with the ‘correction coefficient’ of this time point.
Directionality of ANI-1 and LifeAct was determined on cortical images that were processed by subtracting the mean fluorescence intensity measured outside the embryos. Subsequently a Gaussian Blur filter with a radius of 0.5 was applied followed by an Unsharp Mask filter with a radius of 2 pixels and a Mask Weight of 0.6. Images were also rotated (anterior – left, posterior – right). To measure directionality a rectangular box was placed at the cell equator at selected time points after NEBD and the ImageJ plugin ‘Directionality’ with the method local gradient orientation120 was used. The orientation of ANI-1 and LifeAct was classified into 45 bins from 0° to 90° orientation. The mean percentage of ANI-1 and LifeAct structures with 0°–20°, reflecting anterior-posterior orientation, and 68°–88°, reflecting circumferentially orientation, was calculated for each embryo.
The number of linear and non-linear structures was analyzed on cortical images that were processed as described for the directionality analysis. Additionally, the mean fluorescence intensity measured at the anterior cortex was subtracted from the cortical image at the end. A rectangular box was positioned at the cell equator (and anterior for ANI-1N−term-CX and ANI-1Linker−CX embryos) and the length and width of all structures was determined manually. For each structure the length/width ratio was calculated and structures with the length/width ratio <4 were classified as non-linear and structures with a length/width ratio ≥4 were classified as linear (Figure S3A).
The contractile ring diameter was measured manually on central plane images acquired every 20 s starting at NEBD. The initial ring diameter NEBD was set to 100% diameter width. Embryos were filmed from NEBD of the first cell division until to onset of the second cell division.
Latrunculin A treatment
Adult hermaphrodites that had been treated with perm-1 dsRNA (see above) were dissected, and permeabilized embryos were filmed in meiosis medium (25 mM HEPES, pH 7.4, 0.5 mg/mL inulin, 20% heat-inactivated fetal bovine serum, and 60% Leibowitz-15 medium) without compression. Embryos were either imaged in an custom build microdevice containing microwells118 (Figures 4 and S6) or in a self-made imaging chamber (Figures S7A and S9D). For the self-made chamber double sided tape was folded into two layers, a 5 mm well was introduced with a puncher and the tape was glued onto a freshly 0.1% poly-L-lysine coated 24 × 60 mm glass coverslip. Using a few drops of water, the coverslip was tapped onto a metal slide containing a central whole surrounding the well and 10 μL meiosis medium were added to the well in which the worms were dissected. 10 μM Latrunculin A (L5163 Sigma, BML-T119 Enzo Life Sciences) was added between metaphase and anaphase onset. At the end of each video, medium containing 33 μM FM 4–64 (T13320 Invitrogen/Thermo Scientific) was added to the imaging chamber to confirm that the imaged embryo was permeable.
6xHIS::NMY-2 cloning and NMY-2/MLC-4/MLC-5 protein expression
NMY-2 full length was amplified from C. elegans cDNA and cloned into the pACEbac1 expression vector using Gibson assembly with an N-terminal 6×Histidine tag followed by a flexible linker (GlySerGlySerGly). The primers used were oAC1233 - 5′-ACCATGGCTCTGGTAGCGGCACATCATCTCGACAAAAAGATGATGAG-3’ (forward) and oAC1234 - 5′-TAGTACTTCTCGACAAGCTTTTAGTTGCGAACTGAGTCGCG GTCT-3’ (reverse). The backbone was amplified from pACEbac1 vector using the following primers oAC1235 – 5′-AAGCTTGTCGAGAAGTACTAGAGGATCATAATC-3’ (forward) and oAC1236 5′-GCCGCTACCAGAGCCATGG-3’ (Reverse). Baculovirus expression of the 6xHIS::NMY-2 full-length/StreptagII::MLC-4/StreptagII:MLC-5 complex, cell lysates and tandem streptactin/Ni-NTA affinity chromatography were performed as described previously for NMY-2 HMM complexes.74 The concentrated eluate of the Ni-NTA purification was dialyzed overnight in high salt buffer (10 mM MOPS, 500 mM NaCl, 3 mM NaN3, 1 mM EDTA, 1 mM DTT (pH 7.3)). Glycerol and DTT were added to a final concentration of 10% (v/v) and 1 mM, respectively. Aliquots were flash-frozen in liquid nitrogen and stored at −80°C.
PIP strips and immunoblotting
To analyze lipid binding, PIP strips (P-6001 Echelon Bioscience) were used according to the manufacturer’s instructions. Each membrane consists of 16 spots: 15 contain several lipids and 1 empty (blank). Additionally, on the empty space on the top of the membranes, was spotted with two controls for proper detection consisting of 1 μL of each protein sample (0.5–1 μg) or 1 μL of the secondary antibody pre-diluted 1:100 (secondary antibody control). The two control spots were allowed to dry before proceeding with the protocol. Membranes were then incubated with blocking solution (3% BSA fatty-acid free (Sigma-Aldrich) in PBS (Gibco, Thermo Fisher) with 0.1% (v/v) Tween 20) (PBS-T) for one 1 h at RT. Membranes were then incubated with 4 μg/mL of either StreptagII::MLC-4 or 6xHIS::NMY-2/Strep-tag II::MLC-4/Strep-tag II:MLC-5 complex in blocking buffer for 1 h at RT, followed by three washes in PBS-T with gentle agitation for 5 min. To detect bound proteins, membranes were incubated for 1 h at room temperature in blocking solution with mouse anti-Strep-tagII antibody (2-1507-001 IBA Lifesciences, 1:750) for Strep-tagII::MLC-4 detection or mouse anti-6xHistidine tag antibody (05–949 Millipore, 1:2,500) for 6xHIS::NMY-2 detection. After washing, membranes were incubated with HRP-conjugated anti-mouse antibody at 1:5,000 in blocking solution and incubated for 1 h at RT. This was followed by another round of three washes and PBS was used for the final wash. All steps were performed under gentle agitation. Immunoblots were visualized by chemiluminescence using Pierce ECL Western Blotting Substrate (Thermo Fisher) and a ChemiDoc XRS+ System with Image Lab Software (Bio-Rad).
Quantification and statistical analysis
Statistical analysis was performed in Prism (GraphPad). Mean values are presented with error bars representing standard error of the mean (SEM) as indicated in the figure legends. For data with normal distribution a parametric (two sided Student’s t-test) and with nonnormal distribution a Mann-Whitney-U test for was performed.
Acknowledgments
We are grateful to Christin Nöcker, Maysoon Noureddine, Suraj Shaji, and Isabel Schultz-Pernice for helping with experiments. Microscopy was performed either at the Center for Advanced Light Microscopy (CALM, Ludwig-Maximilians-Universität München) or at the Optical Imaging Center Erlangen (OICE, Friedrich-Alexander-Universität Erlangen-Nürnberg). We are grateful to Christof Osman for access to the Nikon Ti2-Eclipse microscope. For critical comments on the manuscript, we thank Friederike Wolff and Sabine Müller. The Deutsche Forschungsgemeinschaft supported E. Zanin (ZA619/3, 265449562), T. Mikeladze-Dvali (MI1867/1-3), and the Leica SP8 DIVE-FALCON (INST90/1074-1 FUGG for Benedikt Kost). A.-X.C. was supported by the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme (640553–ACTOMYO) and by a Principal Investigator position from the Portuguese Foundation for Science and Technology (FCT) (CEECIND/01967/2017). F-Y.C. has an FCT junior researcher position (DL 57/2016/CP1355/CT0013). J.B. and E.R. were members of the Life Science Munich graduate program. For C. elegans strains, we thank Amy Maddox, Stephan Grill, and Zhirong Bao and the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Author contributions
Conceptualization, E.Z., T.M.-D., and A.X.C.; methodology, E.Z., A.X.C., M.L., and F.Y.C.; investigation, M.L., F.Y.C., A.L., J.B., D.S.O., and E.R.; formal analysis, M.L.; writing – original draft, E.Z.; writing – review & editing, E.Z., T.M.-D., and A.X.C.; visualization, E.Z., funding acquisition, E.Z., T.M.-D., F.Y.C., and A.X.C.; supervision, E.Z., T.M.-D., and A.X.C.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: September 3, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113076.
Supplemental information
Data and code availability
This paper does not report original code. Primary data associated with the paper is available upon reasonable request.
References
- 1.Mishima M. Centralspindlin in Rappaport’s cleavage signaling. Semin. Cell Dev. Biol. 2016;53:45–56. doi: 10.1016/j.semcdb.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 2.D’Avino P.P., Giansanti M.G., Petronczki M. Cytokinesis in animal cells. Cold Spring Harb. Perspect. Biol. 2015;7:a015834. doi: 10.1101/cshperspect.a015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollard T.D., O’Shaughnessy B. Molecular Mechanism of Cytokinesis. Annu. Rev. Biochem. 2019;88:661–689. doi: 10.1146/annurev-biochem-062917-012530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangal S., Sacher J., Kim T., Osório D.S., Motegi F., Carvalho A.X., Oegema K., Zanin E. TPXL-1 activates Aurora A to clear contractile ring components from the polar cortex during cytokinesis. J. Cell Biol. 2018;217:837–848. doi: 10.1083/jcb.201706021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zanin E., Desai A., Poser I., Toyoda Y., Andree C., Moebius C., Bickle M., Conradt B., Piekny A., Oegema K. A conserved RhoGAP limits M phase contractility and coordinates with microtubule asters to confine RhoA during cytokinesis. Dev. Cell. 2013;26:496–510. doi: 10.1016/j.devcel.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneid S., Wolff F., Buchner K., Bertram N., Baygün S., Barbosa P., Mangal S., Zanin E. The BRCT domains of ECT2 have distinct functions during cytokinesis. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108805. [DOI] [PubMed] [Google Scholar]
- 7.Dechant R., Glotzer M. Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev. Cell. 2003;4:333–344. doi: 10.1016/s1534-5807(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 8.Yüce O., Piekny A., Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 2005;170:571–582. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basant A., Glotzer M. Spatiotemporal Regulation of RhoA during Cytokinesis. Curr. Biol. 2018;28:R570–R580. doi: 10.1016/j.cub.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-Cavazos J.S., Lee K.-Y., Lara-Gonzalez P., Li Y., Desai A., Shiau A.K., Oegema K. A Non-canonical BRCT-Phosphopeptide Recognition Mechanism Underlies RhoA Activation in Cytokinesis. Curr. Biol. 2020:1–27. doi: 10.1016/j.cub.2020.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prokopenko S.N., Brumby A., O'Keefe L., Prior L., He Y., Saint R., Bellen H.J. A putative exchange factor for Rho1 GTPase is required for initiation of cytokinesis in Drosophila. Genes Dev. 1999;13:2301–2314. doi: 10.1101/gad.13.17.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen A., Arora P.D., Lai C.C., Copeland J.W., Moraes T.F., McCulloch C.A., Lavoie B.D., Wilde A. The scaffold-protein IQGAP1 enhances and spatially restricts the actin-nucleating activity of Diaphanous-related formin 1 (DIAPH1) J. Biol. Chem. 2020;295:3134–3147. doi: 10.1074/jbc.RA119.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen A., Arora P.D., McCulloch C.A., Wilde A. Cytokinesis requires localized β-actin filament production by an actin isoform specific nucleator. Nat. Commun. 2017;8:1530. doi: 10.1038/s41467-017-01231-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otomo T., Otomo C., Tomchick D.R., Machius M., Rosen M.K. Structural Basis of Rho GTPase-Mediated Activation of the Formin mDia1. Mol. Cell. 2005;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Li F., Higgs H.N. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- 16.Piekny A.J., Maddox A.S. The myriad roles of Anillin during cytokinesis. Semin. Cell Dev. Biol. 2010;21:881–891. doi: 10.1016/j.semcdb.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Naydenov N.G., Koblinski J.E., Ivanov A.I. Anillin is an emerging regulator of tumorigenesis, acting as a cortical cytoskeletal scaffold and a nuclear modulator of cancer cell differentiation. Cell. Mol. Life Sci. 2021;78:621–633. doi: 10.1007/s00018-020-03605-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kühn S., Geyer M. Formins as effector proteins of Rho GTPases. Small GTPases. 2014;5:e29513. doi: 10.4161/sgtp.29513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeder T.E. Cytokinesis: filaments in the cleavage furrow. Exp. Cell Res. 1968;53:272–276. doi: 10.1016/0014-4827(68)90373-x. [DOI] [PubMed] [Google Scholar]
- 20.Henson J.H., Ditzler C.E., Germain A., Irwin P.M., Vogt E.T., Yang S., Wu X., Shuster C.B. The organization of actin and myosin II filaments in the contractile ring: new support for an old model of cytokinesis. Mol. Biol. Cell. 2017;28:613–623. doi: 10.1091/mbc.E16-06-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spira F., Cuylen-Haering S., Mehta S., Samwer M., Reversat A., Verma A., Oldenbourg R., Sixt M., Gerlich D.W. Cytokinesis in vertebrate cells initiates by contraction of an equatorial actomyosin network composed of randomly oriented filaments. Elife. 2017;6:e30867. doi: 10.7554/eLife.30867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reymann A.-C., Staniscia F., Erzberger A., Salbreux G., Grill S.W. Cortical flow aligns actin filaments to form a furrow. Elife. 2016;5:e17807. doi: 10.7554/eLife.17807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leite J., Chan F.Y., Osório D.S., Saramago J., Sobral A.F., Silva A.M., Gassmann R., Carvalho A.X. Equatorial Non-muscle Myosin II and Plastin Cooperate to Align and Compact F-actin Bundles in the Cytokinetic Ring. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.573393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swan K.A., Severson A.F., Carter J.C., Martin P.R., Schnabel H., Schnabel R., Bowerman B. cyk-1: a C. elegans FH gene required for a late step in embryonic cytokinesis. J. Cell Sci. 1998;111:2017–2027. doi: 10.1242/jcs.111.14.2017. [DOI] [PubMed] [Google Scholar]
- 25.Severson A.F., Baillie D.L., Bowerman B. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr. Biol. 2002;12:2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- 26.Pelham R.J., Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe S., Ando Y., Yasuda S., Hosoya H., Watanabe N., Ishizaki T., Narumiya S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol. Biol. Cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straight A.F., Cheung A., Limouze J., Chen I., Westwood N.J., Sellers J.R., Mitchison T.J. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 29.De Lozanne A., Spudich J.A. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 30.Cuenca A.A., Schetter A., Aceto D., Kemphues K., Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard T.D. Myosins in Cytokinesis. Adv. Exp. Med. Biol. 2020;1239:233–244. doi: 10.1007/978-3-030-38062-5_11. [DOI] [PubMed] [Google Scholar]
- 32.Khaliullin R.N., Green R.A., Shi L.Z., Gomez-Cavazos J.S., Berns M.W., Desai A., Oegema K. A positive-feedback-based mechanism for constriction rate acceleration during cytokinesis in Caenorhabditis elegans. Elife. 2018;7:e36073. doi: 10.7554/eLife.36073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hickson G.R.X., O’Farrell P.H. Rho-dependent control of anillin behavior during cytokinesis. J. Cell Biol. 2008;180:285–294. doi: 10.1083/jcb.200709005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maddox A.S., Habermann B., Desai A., Oegema K. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development. 2005;132:2837–2848. doi: 10.1242/dev.01828. [DOI] [PubMed] [Google Scholar]
- 35.Tse Y.C., Piekny A., Glotzer M. Anillin promotes astral microtubule-directed cortical myosin polarization. Mol. Biol. Cell. 2011;22:3165–3175. doi: 10.1091/mbc.E11-05-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies T., Jordan S.N., Chand V., Sees J.A., Laband K., Carvalho A.X., Shirasu-Hiza M., Kovar D.R., Dumont J., Canman J.C. High-Resolution Temporal Analysis Reveals a Functional Timeline for the Molecular Regulation of Cytokinesis. Dev. Cell. 2014;30:209–223. doi: 10.1016/j.devcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motegi F., Mishra M., Balasubramanian M.K., Mabuchi I. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J. Cell Biol. 2004;165:685–695. doi: 10.1083/jcb.200402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saha S., Pollard T.D. Anillin-related protein Mid1p coordinates the assembly of the cytokinetic contractile ring in fission yeast. Mol. Biol. Cell. 2012;23:3982–3992. doi: 10.1091/mbc.E12-07-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willet A.H., McDonald N.A., Bohnert K.A., Baird M.A., Allen J.R., Davidson M.W., Gould K.L. The F-BAR Cdc15 promotes contractile ring formation through the direct recruitment of the formin Cdc12. J. Cell Biol. 2015;208:391–399. doi: 10.1083/jcb.201411097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laplante C., Huang F., Tebbs I.R., Bewersdorf J., Pollard T.D. Molecular organization of cytokinesis nodes and contractile rings by super-resolution fluorescence microscopy of live fission yeast. Proc. Natl. Acad. Sci. USA. 2016;113:E5876–E5885. doi: 10.1073/pnas.1608252113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laporte D., Coffman V.C., Lee I.J., Wu J.Q. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald N.A., Lind A.L., Smith S.E., Li R., Gould K.L. Nanoscale architecture of the Schizosaccharomyces pombe contractile ring. Elife. 2017;6:e28865. doi: 10.7554/eLife.28865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vavylonis D., Wu J.-Q., Hao S., O'Shaughnessy B., Pollard T.D. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 44.van Oostende Triplet C., Jaramillo Garcia M., Haji Bik H., Beaudet D., Piekny A. Anillin interacts with microtubules and is part of the astral pathway that defines cortical domains. J. Cell Sci. 2014;127:3699–3710. doi: 10.1242/jcs.147504. [DOI] [PubMed] [Google Scholar]
- 45.Beaudet D., Pham N., Skaik N., Piekny A. Importin binding mediates the intramolecular regulation of anillin during cytokinesis. Mol. Biol. Cell. 2020;31:1124–1139. doi: 10.1091/mbc.E20-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piekny A.J., Glotzer M. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr. Biol. 2008;18:30–36. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- 47.Sun L., Guan R., Lee I.-J., Liu Y., Chen M., Wang J., Wu J.-Q., Chen Z. Mechanistic Insights into the Anchorage of the Contractile Ring by Anillin and Mid1. Dev. Cell. 2015;33:413–426. doi: 10.1016/j.devcel.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J., Fairn G.D., Ceccarelli D.F., Sicheri F., Wilde A. Cleavage Furrow Organization Requires PIP2-Mediated Recruitment of Anillin. Curr. Biol. 2012;22:64–69. doi: 10.1016/j.cub.2011.11.040. [DOI] [PubMed] [Google Scholar]
- 49.Oegema K., Savoian M.S., Mitchison T.J., Field C.M. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J. Cell Biol. 2000;150:539–552. doi: 10.1083/jcb.150.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Field C.M., Coughlin M., Doberstein S., Marty T., Sullivan W. Characterization of anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development. 2005;132:2849–2860. doi: 10.1242/dev.01843. [DOI] [PubMed] [Google Scholar]
- 51.El Amine N., Kechad A., Jananji S., Hickson G.R.X. Opposing actions of septins and Sticky on Anillin promote the transition from contractile to midbody ring. J. Cell Biol. 2013;203:487–504. doi: 10.1083/jcb.201305053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carim S.C., Hickson G.R.X. The Rho1 GTPase controls anillo-septin assembly to facilitate contractile ring closure during cytokinesis. iScience. 2023;26 doi: 10.1016/j.isci.2023.106903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Straight A.F., Field C.M., Mitchison T.J. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol. Biol. Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Field C.M., Alberts B.M. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian D., Diao M., Jiang Y., Sun L., Zhang Y., Chen Z., Huang S., Ou G. Anillin Regulates Neuronal Migration and Neurite Growth by Linking RhoG to the Actin Cytoskeleton. Curr. Biol. 2015;25:1135–1145. doi: 10.1016/j.cub.2015.02.072. [DOI] [PubMed] [Google Scholar]
- 56.Jananji S., Risi C., Lindamulage I.K.S., Picard L.-P., Van Sciver R., Laflamme G., Albaghjati A., Hickson G.R.X., Kwok B.H., Galkin V.E. Multimodal and Polymorphic Interactions between Anillin and Actin: Their Implications for Cytokinesis. J. Mol. Biol. 2017;429:715–731. doi: 10.1016/j.jmb.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 57.Matsuda K., Sugawa M., Yamagishi M., Kodera N., Yajima J. Visualizing dynamic actin cross-linking processes driven by the actin-binding protein anillin. FEBS Lett. 2020;594:1237–1247. doi: 10.1002/1873-3468.13720. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe S., Okawa K., Miki T., Sakamoto S., Morinaga T., Segawa K., Arakawa T., Kinoshita M., Ishizaki T., Narumiya S. Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol. Biol. Cell. 2010;21:3193–3204. doi: 10.1091/mbc.E10-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D’Avino P.P., Takeda T., Capalbo L., Zhang W., Lilley K.S., Laue E.D., Glover D.M. Interaction between Anillin and RacGAP50C connects the actomyosin contractile ring with spindle microtubules at the cell division site. J. Cell Sci. 2008;121:1151–1158. doi: 10.1242/jcs.026716. [DOI] [PubMed] [Google Scholar]
- 60.Goldbach P., Wong R., Beise N., Sarpal R., Trimble W.S., Brill J.A. Stabilization of the actomyosin ring enables spermatocyte cytokinesis in Drosophila. Mol. Biol. Cell. 2010;21:1482–1493. doi: 10.1091/mbc.e09-08-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maddox A.S., Lewellyn L., Desai A., Oegema K. Anillin and the Septins Promote Asymmetric Ingression of the Cytokinetic Furrow. Dev. Cell. 2007;12:827–835. doi: 10.1016/j.devcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 62.Budnar S., Husain K.B., Gomez G.A., Naghibosadat M., Varma A., Verma S., Hamilton N.A., Morris R.G., Yap A.S. Anillin Promotes Cell Contractility by Cyclic Resetting of RhoA Residence Kinetics. Dev. Cell. 2019;49:894–906.e12. doi: 10.1016/j.devcel.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 63.Kučera O., Siahaan V., Janda D., Dijkstra S.H., Pilátová E., Zatecka E., Diez S., Braun M., Lansky Z. Anillin propels myosin-independent constriction of actin rings. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-24474-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mostowy S., Cossart P. Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2012;13:183–194. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- 65.Estey M.P., Di Ciano-Oliveira C., Froese C.D., Bejide M.T., Trimble W.S. Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J. Cell Biol. 2010;191:741–749. doi: 10.1083/jcb.201006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nguyen T.Q., Sawa H., Okano H., White J.G. The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J. Cell Sci. 2000;113:3825–3837. doi: 10.1101/gad.831500. [DOI] [PubMed] [Google Scholar]
- 67.Green R.A., Mayers J.R., Wang S., Lewellyn L., Desai A., Audhya A., Oegema K. The midbody ring scaffolds the abscission machinery in the absence of midbody microtubules. J. Cell Biol. 2013;203:505–520. doi: 10.1083/jcb.201306036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kechad A., Jananji S., Ruella Y., Hickson G.R.X. Anillin Acts as a Bifunctional Linker Coordinating Midbody Ring Biogenesis during Cytokinesis. Curr. Biol. 2012;22:197–203. doi: 10.1016/j.cub.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Renshaw M.J., Liu J., Lavoie B.D., Wilde A. Anillin-dependent organization of septin filaments promotes intercellular bridge elongation and Chmp4B targeting to the abscission site. Open Biol. 2014;4:130190. doi: 10.1098/rsob.130190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lewellyn L., Dumont J., Desai A., Oegema K. Analyzing the effects of delaying aster separation on furrow formation during cytokinesis in the Caenorhabditis elegans embryo. Mol. Biol. Cell. 2010;21:50–62. doi: 10.1091/mbc.E09-01-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garno C., Irons Z.H., Gamache C.M., McKim Q., Reyes G., Wu X., Shuster C.B., Henson J.H. Building the cytokinetic contractile ring in an early embryo: Initiation as clusters of myosin ii, anillin and septin, and visualization of a septin filament network. PLoS One. 2021:1–27. doi: 10.1371/journal.pone.0252845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fenix A.M., Taneja N., Buttler C.A., Lewis J., Van Engelenburg S.B., Ohi R., Burnette D.T. Expansion and concatenation of non-muscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol. Biol. Cell. 2016;27:1465–1478. doi: 10.1091/mbc.E15-10-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beach J.R., Shao L., Remmert K., Li D., Betzig E., Hammer J.A., III Nonmuscle Myosin II Isoforms Coassemble in Living Cells. Curr. Biol. 2014;24:1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Osório D.S., Chan F.Y., Saramago J., Leite J., Silva A.M., Sobral A.F., Gassmann R., Carvalho A.X. Crosslinking activity of non-muscle myosin II is not sufficient for embryonic cytokinesis in C. elegans. Development. 2019;146 doi: 10.1242/dev.179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neujahr R., Heizer C., Gerisch G. Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow. J. Cell Sci. 1997;110:123–137. doi: 10.1242/jcs.110.2.123. [DOI] [PubMed] [Google Scholar]
- 76.Nagasaki A., Kanada M., Uyeda T.Q. Cell adhesion molecules regulate contractile ring-independent cytokinesis in Dictyostelium discoideum. Cell Res. 2009;19:236–246. doi: 10.1038/cr.2008.318. [DOI] [PubMed] [Google Scholar]
- 77.Dix C.L., Matthews H.K., Uroz M., McLaren S., Wolf L., Heatley N., Win Z., Almada P., Henriques R., Boutros M., et al. The Role of Mitotic Cell-Substrate Adhesion Re- modeling in Animal Cell Division. Dev. Cell. 2018;45:132–145.e3. doi: 10.1016/j.devcel.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 78.Onishi M., Umen J.G., Cross F.R., Pringle J.R. Cleavage-furrow formation without F-actin in Chlamydomonas. Proc. Natl. Acad. Sci. USA. 2020;117:18511–18520. doi: 10.1073/pnas.1920337117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jordan S.N., Davies T., Zhuravlev Y., Dumont J., Shirasu-Hiza M., Canman J.C. Cortical PAR polarity proteins promote robust cytokinesis during asymmetric cell division. J. Cell Biol. 2016;212:39–49. doi: 10.1083/jcb.201510063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pruyne D. Revisiting the Phylogeny of the Animal Formins: Two New Subtypes, Relationships with Multiple Wing Hairs Proteins, and a Lost Human Formin. PLoS One. 2016;11 doi: 10.1371/journal.pone.0164067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davies T., Kim H.X., Romano Spica N., Lesea-Pringle B.J., Dumont J., Shirasu-Hiza M., Canman J.C. Cell-intrinsic and -extrinsic mechanisms promote cell-type-specific cytokinetic diversity. Elife. 2018;7:e36204. doi: 10.7554/eLife.36204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ding W.Y., Ong H.T., Hara Y., Wongsantichon J., Toyama Y., Robinson R.C., Nédélec F., Zaidel-Bar R. Plastin increases cortical connectivity to facilitate robust polarization and timely cytokinesis. J. Cell Biol. 2017;216:1371–1386. doi: 10.1083/jcb.201603070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chan F.Y., Silva A.M., Saramago J., Pereira-Sousa J., Brighton H.E., Pereira M., Oegema K., Gassmann R., Carvalho A.X. The ARP2/3 complex prevents excessive formin activity during cytokinesis. Mol. Biol. Cell. 2019;30:96–107. doi: 10.1091/mbc.E18-07-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Werner M., Munro E., Glotzer M. Astral Signals Spatially Bias Cortical Myosin Recruitment to Break Symmetry and Promote Cytokinesis. Curr. Biol. 2007;17:1286–1297. doi: 10.1016/j.cub.2007.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Panagiotou T.C., Chen A., Wilde A. An anillin-CIN85-SEPT9 complex promotes intercellular bridge maturation required for successful cytokinesis. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111274. [DOI] [PubMed] [Google Scholar]
- 86.Kinoshita M., Field C.M., Coughlin M.L., Straight A.F., Mitchison T.J. Self- and Actin-Templated Assembly of Mammalian Septins. Dev. Cell. 2002;3:791–802. doi: 10.1016/S1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- 87.Shivas J.M., Skop A.R. Arp2/3 mediates early endosome dynamics necessary for the maintenance of PAR asymmetry in Caenorhabditis elegans. Mol. Biol. Cell. 2012;23:1917–1927. doi: 10.1091/mbc.E12-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sobral A.F., Chan F.Y., Norman M.J., Osório D.S., Dias A.B., Ferreira V., Barbosa D.J., Cheerambathur D., Gassmann R., Belmonte J.M., Carvalho A.X. Plastin and spectrin cooperate to stabilize the actomyosin cortex during cytokinesis. Curr. Biol. 2021;31:5415–5428.e10. doi: 10.1016/j.cub.2021.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Michaux J.B., Robin F.B., McFadden W.M., Munro E.M. Excitable RhoA dynamics drive pulsed contractions in the early C. elegans embryo. J. Cell Biol. 2018;217:4230–4252. doi: 10.1083/jcb.201806161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tse Y.C., Werner M., Longhini K.M., Labbé J.-C., Goldstein B., Glotzer M. RhoA activation during polarization and cytokinesis of the early Caenorhabditis elegans embryo is differentially dependent on NOP-1 and CYK-4. Mol. Biol. Cell. 2012;23:4020–4031. doi: 10.1091/mbc.E12-04-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carim S.C., Kechad A., Hickson G.R.X. Animal Cell Cytokinesis: The Rho-Dependent Actomyosin-Anilloseptin Contractile Ring as a Membrane Microdomain Gathering, Compressing, and Sorting Machine. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.575226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chatterjee M., Pollard T.D. The Functionally Important N-Terminal Half of Fission Yeast Mid1p Anillin Is Intrinsically Disordered and Undergoes Phase Separation. Biochemistry. 2019;58:3031–3041. doi: 10.1021/acs.biochem.9b00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Leite J., Osorio D.S., Sobral A.F., Silva A.M., Carvalho A.X. Network Contractility During Cytokinesis—from Molecular to Global Views. Biomolecules. 2019;9 doi: 10.3390/biom9050194. 194–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Willet A.H., DeWitt A.K., Beckley J.R., Clifford D.M., Gould K.L. NDR Kinase Sid2 Drives Anillin-like Mid1 from the Membrane to Promote Cytokinesis and Medial Division Site Placement. Curr. Biol. 2019;29:1055–1063.e2. doi: 10.1016/j.cub.2019.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu X., Shu S., Billington N., Williamson C.D., Yu S., Brzeska H., Donaldson J.G., Sellers J.R., Korn E.D. Mammalian Nonmuscle Myosin II Binds to Anionic Phospholipids with Concomitant Dissociation of the Regulatory Light Chain. J. Biol. Chem. 2016;291:24828–24837. doi: 10.1074/jbc.M116.739185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Su K.-C., Takaki T., Petronczki M. Targeting of the RhoGEF Ect2 to the equatorial membrane controls cleavage furrow formation during cytokinesis. Dev. Cell. 2011;21:1104–1115. doi: 10.1016/j.devcel.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 97.Lekomtsev S., Su K.-C., Pye V.E., Blight K., Sundaramoorthy S., Takaki T., Collinson L.M., Cherepanov P., Divecha N., Petronczki M. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature. 2012;492:276–279. doi: 10.1038/nature11773. [DOI] [PubMed] [Google Scholar]
- 98.Cavini I.A., Leonardo D.A., Rosa H.V.D., Castro D.K.S.V., D'Muniz Pereira H., Valadares N.F., Araujo A.P.U., Garratt R.C. The Structural Biology of Septins and Their Filaments: An Update. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.765085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Celton-Morizur S., Bordes N., Fraisier V., Tran P.T., Paoletti A. C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast. Mol. Cell Biol. 2004;24:10621–10635. doi: 10.1128/MCB.24.24.10621-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ong J.Y., Torres J.Z. Phase Separation in Cell Division. Mol. Cell. 2020;80:9–20. doi: 10.1016/j.molcel.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weirich K.L., Banerjee S., Dasbiswas K., Witten T.A., Vaikuntanathan S., Gardel M.L. Liquid behavior of cross-linked actin bundles. Proc. Natl. Acad. Sci. USA. 2017;114:2131–2136. doi: 10.1073/pnas.1616133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scheff D.R., Weirich K.L., Dasbiswas K., Patel A., Vaikuntanathan S., Gardel M.L. Tuning Shape and Internal Structure of Protein Droplets via Biopolymer Filaments. Soft Matter. 2020:1–10. doi: 10.1039/C9SM02462J. [DOI] [PubMed] [Google Scholar]
- 103.Naganathan S.R., Fürthauer S., Rodriguez J., Fievet B.T., Jülicher F., Ahringer J., Cannistraci C.V., Grill S.W. Morphogenetic degeneracies in the actomyosin cortex. Elife. 2018;7:354. doi: 10.7554/eLife.37677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hird S.N., White J.G. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J. Cell Biol. 1993;121:1343–1355. doi: 10.1083/jcb.121.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Y., Munro E. Filament-guided filament assembly provides structural memory of filament alignment during cytokinesis. Dev. Cell. 2021;56:2486–2500.e6. doi: 10.1016/j.devcel.2021.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ganar K.A. Shaping Synthetic Cells through Cytoskeleton-Condensate-Membrane Interactions. Curr. Opin. Colloid Interface Sci. 2021:1–12. doi: 10.1016/j.cocis.2021.101459. [DOI] [Google Scholar]
- 107.Quail T., Golfier S., Elsner M., Ishihara K., Murugesan V., Renger R., Jülicher F., Brugués J. Force generation by protein–DNA co-condensation. Nat. Phys. 2021;17:1007–1012. doi: 10.1038/s41567-021-01285-1. [DOI] [Google Scholar]
- 108.Bergeron-Sandoval L.-P., Kumar S., Heris H.K., Chang C.L.A., Cornell C.E., Keller S.L., François P., Hendricks A.G., Ehrlicher A.J., Pappu R.V., Michnick S.W. Endocytic proteins with prion-like domains form viscoelastic condensates that enable membrane remodeling. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2113789118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yuan F., Alimohamadi H., Bakka B., Trementozzi A.N., Day K.J., Fawzi N.L., Rangamani P., Stachowiak J.C. Membrane bending by protein phase separation. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2017435118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harker-Kirschneck L., Hafner A.E., Yao T., Vanhille-Campos C., Jiang X., Pulschen A., Hurtig F., Hryniuk D., Culley S., Henriques R., et al. Physical mechanisms of ESCRT-III-driven cell division. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2107763119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tarrason Risa G., Hurtig F., Bray S., Hafner A.E., Harker-Kirschneck L., Faull P., Davis C., Papatziamou D., Mutavchiev D.R., Fan C., et al. The proteasome controls ESCRT-III-mediated cell division in an archaeon. Science. 2020;369 doi: 10.1126/science.aaz2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bell K.R., Werner M.E., Doshi A., Cortes D.B., Sattler A., Vuong-Brender T., Labouesse M., Maddox A.S. Novel cytokinetic ring components drive negative feedback in cortical contractility. Mol. Biol. Cell. 2020;31:1623–1636. doi: 10.1091/mbc.E20-05-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dickinson D.J., Schwager F., Pintard L., Gotta M., Goldstein B. A Single-Cell Biochemistry Approach Reveals PAR Complex Dynamics during Cell Polarization. Dev. Cell. 2017;42:416–434.e11. doi: 10.1016/j.devcel.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.-P., Grunnet M., Jorgensen E.M. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen D., Hastie E., Sherwood D. Endogenous Expression of UNC-59/Septin in C. elegans. MicroPubl. Biol. 2019 doi: 10.17912/micropub.biology.000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Carvalho A., Olson S.K., Gutierrez E., Zhang K., Noble L.B., Zanin E., Desai A., Groisman A., Oegema K. Acute Drug Treatment in the Early C. elegans Embryo. PLoS One. 2011;6:e24656–e24658. doi: 10.1371/journal.pone.0024656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zanin E., Dumont J., Gassmann R., Cheeseman I., Maddox P., Bahmanyar S., Carvalho A., Niessen S., YatesIII J.R., Oegema K., Desai A. Second edition. Elsevier Inc.; 2011. Affinity Purification of Protein Complexes in C. elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu Z.Q. Scale space approach to directional analysis of images. Appl. Opt. 1991;30:1369–1373. doi: 10.1364/AO.30.001369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Images were acquired every 20 s on a Nikon Eclipse Ti spinning disk confocal controlled by NIS Elements 4.51 software equipped with a 100× 1.45-NA Plan-Apochromat oil-immersion objective and Andor DU-888 X11056 camera. Video starts 140 s after NEBD.
Note: scaling intensity for NG::ANI-1 after septinUNC-61 co-depletion is increased. Images were acquired every 2.5 s on a Nikon Eclipse Ti spinning disk confocal controlled by NIS Elements 4.51 software equipped with a 100× 1.45-NA Plan-Apochromat oil-immersion objective and Andor DU-888 X11056 camera. Time in seconds after NEBD is shown.
Images were acquired every 2.5 s on a Nikon Eclipse Ti spinning disk confocal controlled by NIS Elements 4.51 software equipped with a 100× 1.45-NA Plan-Apochromat oil-immersion objective and Andor DU-888 X11056 camera. Time in seconds after NEBD is shown.
Intensity scaling was increased to visualize the NG::ANI-1 structures after nmy-2(RNAi);cyk-1(RNAi);septinunc-61(RNAi). Imaging conditions are the same as indicated for Video S2. Time in seconds after NEBD is shown.
Images were acquired every 5 s on inverted microscope (Ti-E, Nikon) equipped with a 60× oil-immersion Plan-Apochromat objective (NA 1.4) and an iXon Ultra 897 camera. Time in seconds after NEBD is shown.
Images were acquired every 5 s on laser scanning confocal microscope (SP8, DIVE-FALCON) equipped with a 63×/1.4 oil objective. Note: fluorescence intensity scalings are increased for septinunc-61(RNAi). Time in seconds after NEBD is shown.
Images were acquired as described for Video 5. Time in seconds after NEBD is shown.
Note: fluorescence intensity scalings are increased for cyk-1(RNAi)ani-1(RNAi);septinunc-61(RNAi) conditions. Images were acquired every 5 s on laser scanning microscope (SP8) equipped with a 63×/1.3 GLYC objective and photomultiplier. Time in seconds after NEBD is shown.
Note: fluorescence intensity scalings are increased for cyk-1(RNAi)ani-1(RNAi);septinunc-61(RNAi) conditions. Images were acquired every 5 s on laser scanning microscope (SP8) equipped with a 63×/1.3 GLYC objective and photomultiplier. Time in seconds after NEBD is shown.
Images were acquired as described for Video 6. Note: fluorescence intensity scalings are increased for GFP::ANI-1N-term-CX. Time in seconds after NEBD is shown.
Note: fluorescence intensity scalings are increased for cyk-1(RNAi)ani-1(RNAi) and cyk-1(RNAi)ani-1(RNAi);septinunc-61(RNAi) conditions. Images were acquired as described for Video 6. Time in seconds after NEBD is shown.
Note: fluorescence intensity scalings are increased for GFP::ANI-1WT with cyk-1(RNAi)ani-1(RNAi);septinunc-61(RNAi) and all ANI-1N-term conditions. Images were acquired as described for Video 6. Time in seconds after NEBD is shown.
Note: fluorescence intensity scalings are increased for cyk-1(RNAi)ani-1(RNAi) and cyk-1(RNAi)ani-1(RNAi); septinunc-61(RNAi). Images were acquired as described for Video 8. Time in seconds after NEBD is shown.
Images were acquired as described for Video 6. Time in seconds after NEBD is shown.
Data Availability Statement
This paper does not report original code. Primary data associated with the paper is available upon reasonable request.