Abstract
Background
Epigenetic studies, particularly research on microRNA (miRNA), have flourished. The abnormal expression of miRNA contributes to the development of hematologic malignancies. miR-765 has been reported to inhibit cell proliferation by downregulating proteolipid protein 2 (PLP2), which causes apoptosis. We investigated miR-765 dysregulation in myelodysplastic syndromes (MDS).
Methods
We compared the expression profiles of miR-765 in 65 patients with MDS and 11 controls. Cell proliferation and apoptosis assays were performed to determine the in vitro effects of miR-765 on leukemia cells transfected with the miR-765 mimic. Reverse transcription quantitative PCR (RT-qPCR) and western blotting were performed to examine the targets of miR-765.
Results
We found that miR-765 levels were upregulated 10.2-fold in patients with MDS compared to controls. In refractory cytopenia with multilineage dysplasia, the percentage of patients with elevated miR-765 levels was significantly higher than in other forms of MDS. Experiments with leukemia cells revealed that transfection with a miR-765 mimic inhibited cell proliferation and induced apoptosis. RT-qPCR and western blotting demonstrated that the target of miR-765 was PLP2.
Conclusion
These findings imply that upregulation of miR-765 induces apoptosis via downregulation of PLP2 and may have a role in MDS pathogenesis.
Keywords: MDS, microRNA, Deregulation, Apoptosis, Pathogenesis
INTRODUCTION
The myelodysplastic syndromes (MDS) are clonal disorders characterized by pancytopenia of the peripheral blood, dysplasia in one or more myeloid cell lines, ineffective hematopoiesis of hematopoietic stem cells, and a high risk of progression to acute myeloid leukemia (AML) [1]. In recent decades, next-generation sequencing has enabled the identification of genetic mutations in patients with MDS. These mutations are associated with the pathophysiology of MDS and have helped us understand the biology of MDS [2].
With the recent progress in genetics, epigenetics, particularly the knowledge of microRNAs (miRNAs), has flourished. miRNAs are short, non-coding RNAs that are critical regulators of hematopoiesis, and abnormal changes in their expression levels can contribute to the development of hematologic malignancies [3]. miRNAs are located at cytogenetic locations, which are fragile sites on chromosomes, with 77% of them located at the cytogenetic locations having leukemia-associated copy-number alterations in MDS and AML cell lines [4]. The deregulation of miRNA is known to be associated with MDS pathogenesis [5].
Cytogenetic abnormalities are clonal changes that can be easily detected by routine chromosomal analysis of bone marrow samples for diagnosing MDS. Representative numerical chromosomal abnormalities in MDS include trisomy 8, deletion of 5q, and monosomy 7.
miR-765 has been reported to inhibit cell proliferation by downregulating proteolipid protein 2 (PLP2) [6]. It is located on chromosome 1, and trisomy 1q is one of the most common numerical chromosomal abnormalities in Korean patients with MDS [7, 8]. However, the association between miR-765 and MDS has not been investigated.
In the present study, we investigated the association of miR-765 with MDS by comparing the expression of miR-765 between patients with MDS and controls, and the in vitro effect of miR-765 on hematopoietic cells using an AML cell line transfected with a miR-765 mimic.
MATERIALS AND METHODS
Patients
The study population comprised 65 patients with MDS and 11 healthy controls. MDS was diagnosed based on the World Health Organization 2008 classification. The bone marrow of controls were examined to determine the presence of anemia or neutropenia. Controls showed no evidence of cancer and had normocellular bone marrow with no signs of hematologic malignancy. This study was approved by the Institutional Review Board (IRB) of Korea University Anam Hospital (Seoul, Korea) (ED12251) and IRB of Korea University Ansan Hospital (Ansan, Korea) (AS13177). Clinical and laboratory data were obtained from patients’ medical records at the time of IRB approval. Informed consent was obtained from all participants. The participants included 9 patients (13.8%) with refractory cytopenia with unilineage dysplasia, 1 patient (1.5%) with refractory anemia with ring sideroblasts, 27 patients (41.5%) with refractory cytopenia with multilineage dysplasia, 9 patients (13.8%) with refractory anemia of excess blasts type 1, 10 patients (15.4%) with refractory anemia with excess blasts type 2, 9 patients (13.8%) with unclassifiable MDS.
Extraction and quantification of miRNAs
We extracted miRNAs from unsorted cells (whole bone marrow cells) of archived bone marrow aspirate slides and quantified miRNAs using a previously described method [9]. The TaqMan microRNA assay (Applied Biosystems, Carlsbad, CA, USA) was used to quantify miR-765 expression. RNU48 expression was used as an endogenous control. The 2-ΔCt value of each miRNA was calculated to compare the miRNA expression between patients with MDS and controls.
Cell culture and transfection of microRNA
The AML cell line OCI-AML3 was used for the in vitro experiments. OCI-AML3 cells were cultured in α-minimum essential medium (Welgene, Gyeongsan-si, Korea) containing 10% fetal bovine serum and penicillin/streptomycin. mirVana miR-765 mimic and negative control miRNA were purchased from Applied Biosystems (Thermo Fisher Scientific, Waltham, MA, USA) and transfected (50 nM miR-765 mimic and negative control miRNA) using Lipofectamine RNAiMax (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Cell proliferation assay
Cell proliferation assays were performed as described in a previous study [10]. Briefly, OCI-AML3 cells were seeded into 96-well plates and transfected with miR-765 mimic or negative control miRNA. The CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) was used to measure cell death.
Apoptosis assay using Annexin V/propidium iodide (PI)
The FITC Annexin V Apoptosis Detection Kit (BD Biosciences, Franklin Lakes, NJ, USA) was used to quantitatively determine the percentage of apoptotic cells. OCI-AML3 cells were transfected with the miR-765 mimic or negative control miRNA for 72 hr. The cells were harvested, and apoptotic cells were measured according to the protocol provided for the FITC Annexin V apoptosis kit using FACSlyric (BD Biosciences) and FACSuite software (BD Biosciences).
Caspase activity assay
To confirm the apoptosis observed in the Annexin V/PI assay, caspase-3 activity was measured using a Caspase-3 Colorimetric Assay Kit (Abcam, Cambridge, MA, USA). Cells, transfected with miR-765 mimic and a negative control, were harvested and suspended in 150 μL of cell lysis buffer. Supernatants were transferred into new tubes, and the protein concentration was measured using the Pierce BCA Protein assay (Thermo Fisher Scientific, Rockford, IL, USA). Equal amounts of protein were transferred to a new 96-well plate and incubated with 50 and 5 μL of reaction buffer and 4 mM DEVD-p-NA substrate, respectively. After incubation for 1.5 hr at 37°C, the absorbance was measured at 405 nm.
Quantitative reverse transcription PCR (qRT-PCR)
After transfecting OCI-AML3 cells with the miR-765 mimic or negative control for 72 hr, mRNA was extracted as previously described [10]. qRT-PCR was performed as previously described [10], using the following primers: PLP2 forward, 5′-GCGCACTCGAAAGGGAATC-3′ and reverse, 5′-AGGATCATCTCAATCACCGACA-3′; GAPDH forward, 5′-AGCCTCAAGATCATCAGCAATG-3′ and reverse, 5′-CACGATACCAAAGTTGTCATGGAT-3. RT-qPCR was performed six times.
Western blot analysis
The cells were lysed in RIPA buffer (Thermo Fisher Scientific) with a protease inhibitor cocktail (Sigma-Aldrich, Saint Louis, MO, USA). Cell lysates were collected by centrifugation at 13,200 rpm for 15 min. Protein concentrations were measured using the Pierce BCA Protein Assay (Thermo Fisher Scientific). Equal amounts of protein were separated using 4–12% Bolt Bis-Tris Plus gels (Thermo Fisher Scientific), followed by electrotransfer onto polyvinylidene difluoride membranes (Invitrogen) using a Mini Blot Module (Thermo Fisher Scientific). The membranes were blocked in iBind Flex Solution (iBind Flex Buffer, iBind Flex Additive, and distilled water; Thermo Fisher Scientific) for 15 min at room temperature. Primary and secondary antibody reactions were performed for 3 hr using the iBind Flex Western System (Thermo Fisher Scientific) according to the manufacturer’s instructions. Membranes were incubated with an anti-PLP2 antibody (1:100; Abnova, Taipei, Taiwan; cat. no. H00005355) or an anti-β-actin antibody (1:100; Santa Cruz Biotechnology, Santa Cruz, CA, USA; cat. no. sc-47778), followed by a horseradish peroxidase-conjugated goat anti-mouse polyclonal secondary antibody (1:10,000; Jackson ImmunoResearch, West Grove, PA, USA; cat. no. 115-035-003). The protein-antibody complexes were visualized using Pierce ECL western blotting substrate (Thermo Fisher Scientific).
Statistics
The relative expression of miR-765 was compared between patients and controls using the non-parametric Mann–Whitney U test. The relative expression of miR-765 was compared among patients with different MDS subtypes using the chi-squared test. The relative expression of mRNA after transfection with the miR-765 mimic was assessed using the non-parametric Mann–Whitney U test. Statistical analyses were performed using SPSS (version 23.0, SPSS, Chicago, IL, USA) and GraphPad Prism 9 (GraphPad Software, San Diego, CA, USA). The level of statistical significance was set at P<0.05.
RESULTS
Patients with MDS had upregulated miR-765, which was associated with multilineage dysplasia
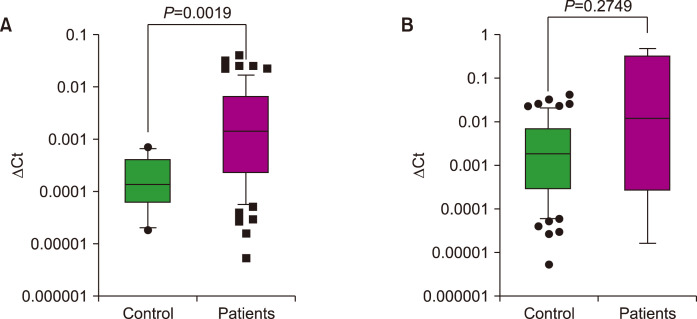
The median 2-ΔCt value of miR-765 was 0.00144 (±0.00839 standard deviation) in patients and 0.00014 (±0.00022) in controls. miR-765 expression in patients was significantly higher than that in controls (P=0.0019, Fig. 1A), indicating that the expression level was upregulated 10.2-fold in patients with MDS compared to that in controls. miR-765 expression was compared between patients with and without trisomy 1q. The number of patients with trisomy 1q was five and miR-765 expression was not significantly higher in patients with trisomy 1q than in those without it (Fig. 1B).
Fig. 1.
Relative expression of miR-765 in patients (A) with MDS and controls and (B) with and without trisomy 1q.
The median value of miR-765 in patients (0.0014) was used to define the cutoff value for the high miR-765 population. The association between high miR-765 expression and the morphological subtypes of MDS was analyzed. The distribution of MDS subclassifications based on miR-765 expression is shown in Table 1. The percentage of miR-765 was higher in refractory cytopenia with multilineage dysplasia (57.6 vs. 25%, P=0.014), implying that miR-765 upregulation may be associated with multilineage dysplasia.
Table 1.
Patients distribution according to miR-765 expression and MDS subtypes.
| MDS subtypes | N of patients (%) | |
|---|---|---|
| Low miR-765 expression | High miR-765 expression | |
| RCUD | 7 (21.9%) | 2 (6.1%) |
| RARS | 0 (0%) | 1 (3.0%) |
| RCMD | 8 (25%) | 19 (57.6%) |
| RAEB-1 | 5 (16.7%) | 4 (12.1%) |
| RAEB-2 | 4 (12.5%) | 6 (1.1%) |
| MDS U | 8 (25%) | 1 (3.0%) |
| Total | 32 (100%) | 33 (100%) |
Abbreviations: MDS, myelodysplastic syndromes; MDS U, unclassified myelodysplastic syndromes; RAEB-1, refractory anemia with excess blasts type 1; RAEB-2, refractory anemia with excess blasts type 2; RARS, refractory anemia with ring sideroblasts; RCUD, refractory cytopenia with unilineage dysplasia.
The prognostic effect of miR-765 on overall survival was also examined. The difference in overall survival between patients with high and low miR-765 expression was not significant (data not shown). The association between miR-765 expression and the International Prognostic Scoring System (IPSS-R) scores was also analyzed. IPSS-R categories distribution according to the expression of miR-765 is displayed in Table 2. The IPSS-R categories of the high miR-765 expression group were not significantly different from those of the low miR-765 expression group. These findings did not demonstrate a prognostic impact of miR-765 expression.
Table 2.
Patients distribution according to miR-765 expression and IPSS-R subgroups.
| Prognostic subgroups | N of patients (%) | |
|---|---|---|
| Low miR-765 expression | High miR-765 expression | |
| Very low | 2 (6.7%) | 2 (6.7%) |
| Low | 4 (13.3%) | 6 (16.7%) |
| Intermediate | 12 (40.0%) | 10 (33.3%) |
| High | 5 (16.7%) | 4 (13.3%) |
| Very high | 7 (23.3%) | 5 (16.7%) |
| Total | 30 (100%) | 30 (100%) |
Abbreviation: IPSS-R, International Prognostic Scoring System.
Overexpression of miR-765 induces apoptosis in leukemic cells
miR-765 is upregulated in patients with MDS and is associated with dysplasia; therefore, we assumed that the upregulation of miR-765 may be associated with cell death. In vitro experiments were performed using human AML cells (OCI-AML3) to investigate the effects of miR-765. OCI-AML3 cells were transfected with a miR-765 mimic or negative control miRNA, and a cell proliferation assay was performed. The results of the assay revealed that cell growth was significantly inhibited following transfection with miR-765 compared to the negative control (P<0.0001) (Fig. 2).
Fig. 2.
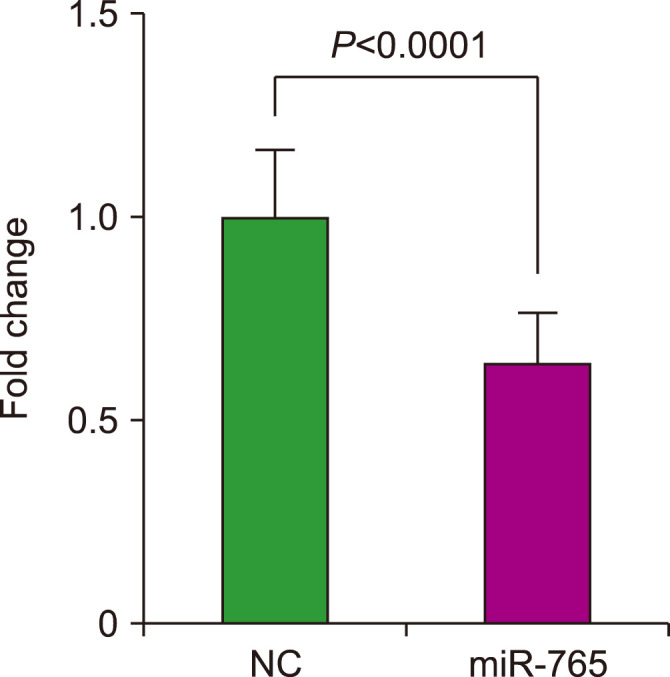
Cell proliferation after transfection with a miR-765 mimic (miR-765) or a negative control miRNA.
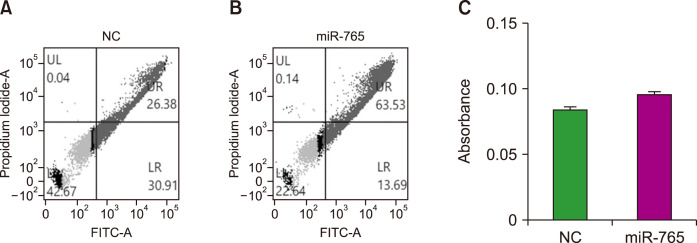
To further explore the inhibition of cell growth induced by miR-765 transfection, Annexin V/PI and caspase activity assays were performed. The Annexin V/PI assay demonstrated a marked increase in the number of apoptotic cells upon transfection with the miR-765 mimic compared to the negative control (Fig. 3A, B). Caspase assays revealed that caspase-3 activity was increased in cells transfected with the miR-765 mimic compared to cells transfected with the negative control miRNA (Fig. 3C). These findings indicate that the upregulation of miR-765 expression leads to cell death through apoptosis.
Fig. 3.
(A, B) Annexin V (FITC)/PI assay and (C) caspase assay after transfection with a negative control miRNA (NC) and miR-765 mimic.
miR-765 induces apoptosis through PLP2 inhibition
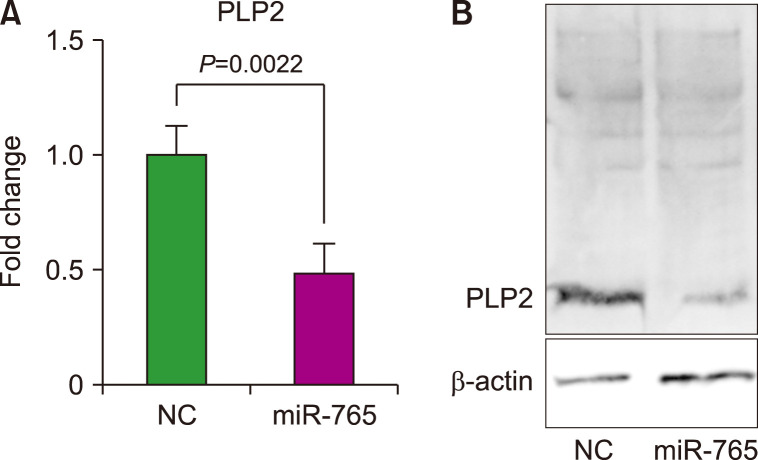
To investigate the mechanisms underlying miR-765-mediated apoptosis in AML cells, we examined the targets of miR-765. miR-765 has been reported to inhibit cell proliferation by inhibiting PLP2 [6]. The relative mRNA levels of PLP2 were measured after transfection of OCI-AML3 cells with the miR-765 mimic or negative control miRNA. The mRNA level of PLP2 was significantly downregulated (P=0.0022; Fig. 4A). Western blot analysis revealed decreased PLP2 protein levels in miR-765-transfected cells compared to those in control cells (Fig. 4B). Therefore, it can be inferred that the upregulation of miR-765 leads to apoptosis through PLP2 inactivation.
Fig. 4.
(A) mRNA analysis of PLP2 and (B) western blot analysis after transfection with a miR-765 mimic (miR-765) or negative control miRNA (NC).
DISCUSSION
In the present study, in patients with MDS, miR-765 was upregulated, which induced apoptosis and was associated with multilineage dysplasia. These findings indicate that the upregulation of miR-765 can lead to apoptosis in hematopoietic cells and may manifest as dysplasia, which is a phenotype of apoptosis in hematopoietic cells.
The role of miR-765 in various cancers has been investigated. It is upregulated in clear-cell renal carcinoma, and its overexpression suppresses cell proliferation [6]. A study reported that miR-765 expression was significantly increased in renal cell carcinoma and that high expression of miR-765 was associated with a poor prognosis [11]. In gastric cancer, miR-765 is upregulated in cancer cell lines and specimens obtained from patients with gastric cancer who do not respond to chemotherapy [12]. Similarly, miR-765 was overexpressed in breast carcinoma tissue and cancer cells [13]. These studies reveal that miR-765 can act as either a tumor suppressor or an oncogene, implying that the function of miR-765 in cancer depends on its target and tissue expression.
This study showed that miR-765 upregulation induced apoptosis and decreased the expression of PLP2. PLP2 is a small integral membrane protein located in the endoplasmic reticulum (ER). PLP2-knockout mice display increased susceptibility to ER stress, which leads to apoptosis [14]. Similarly, downregulation of PLP2 induces ER stress, promotes apoptosis, and reduces glioma cell survival [15]. These results imply that the upregulation of miR-765 induces apoptosis via the downregulation of PLP2. Apoptosis is a characteristic feature of MDS. Therefore, it can be inferred that miR-765 upregulation contributes to the pathogenesis of MDS by inducing apoptosis.
PLP2 has been investigated in multiple myeloma and other hematological malignancies. PLP2 expression increases with the progression and prognosis in multiple myeloma [16]; however, this has not been studied in MDS. Further studies on PLP2 in MDS are required to elucidate its pathophysiological role.
Overall, the present study indicates that miR-765 upregulation induces apoptosis via PLP2 downregulation and is associated with multilineage dysplasia, suggesting a possible role for miR-765 in the pathogenesis of MDS.
Funding Statement
*This study was supported by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (NRF-2021R1F1A1045955).
Footnotes
Authors’ Disclosures of Potential Conflicts of Interest
No potential conflicts of interest relevant to this article were reported.
REFERENCES
- 1.Raza A, Galili N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat Rev Cancer. 2012;12:849–59. doi: 10.1038/nrc3321. [DOI] [PubMed] [Google Scholar]
- 2.Cazzola M. Myelodysplastic syndromes. N Engl J Med. 2020;383:1358–74. doi: 10.1056/NEJMra1904794. [DOI] [PubMed] [Google Scholar]
- 3.Fang J, Varney M, Starczynowski DT. Implication of microRNAs in the pathogenesis of MDS. Curr Pharm Des. 2012;18:3170–9. doi: 10.2174/1381612811209023170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Starczynowski DT, Morin R, McPherson A, et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood. 2011;117:595–607. doi: 10.1182/blood-2010-03-277012. [DOI] [PubMed] [Google Scholar]
- 5.Rhyasen GW, Starczynowski DT. Deregulation of microRNAs in myelodysplastic syndrome. Leukemia. 2012;26:13–22. doi: 10.1038/leu.2011.221. [DOI] [PubMed] [Google Scholar]
- 6.Xiao W, Wang C, Chen K, et al. miR-765 functions as a tumour suppressor and eliminates lipids in clear cell renal cell carcinoma by downregulating PLP2. EBioMedicine. 2020;51:102622. doi: 10.1016/j.ebiom.2019.102622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee DS, Kim SH, Seo EJ, et al. Predominance of trisomy 1q in myelodysplastic syndromes in Korea: is there an ethnic difference? A 3-year multi-center study. Cancer Genet Cytogenet. 2002;132:97–101. doi: 10.1016/S0165-4608(01)00533-7. [DOI] [PubMed] [Google Scholar]
- 8.Jung SW, Lee SY, Jekarl DW, et al. Cytogenetic characteristics and prognosis analysis in 231 myelodysplastic syndrome patients from a single institution. Leuk Res. 2011;35:735–40. doi: 10.1016/j.leukres.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Choi JS, Nam MH, Yoon SY, Kang SH. MicroRNA-194-5p could serve as a diagnostic and prognostic biomarker in myelodysplastic syndromes. Leuk Res. 2015;39:763–8. doi: 10.1016/j.leukres.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Kang SH, Kim HB, Choi JS. Upregulation of microRNA-597 in myelodysplastic syndromes induces apoptosis through FOSL2 inhibition. Eur J Haematol. 2022;109:680–5. doi: 10.1111/ejh.13852. [DOI] [PubMed] [Google Scholar]
- 11.Meng K, Li Z, Cui X. Three LHPP gene-targeting co-expressed microRNAs (microRNA-765, microRNA-21, and microRNA-144) promote proliferation, epithelial-mesenchymal transition, invasion, and are independent prognostic biomarkers in renal cell carcinomas patients. J Clin Lab Anal. 2021;35:e24077. doi: 10.1002/jcla.24077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin W, Miao Y, Meng X, Huang Y, Zhao W, Ruan J. miRNA-765 mediates multidrug resistance via targeting BATF2 in gastric cancer cells. FEBS Open Bio. 2020;10:1021–30. doi: 10.1002/2211-5463.12838.2f225db1118043f3845c488ad3b680f7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao Y, Yuan C, Wu H, Li X, Yu J. Oncogenic microRNA-765 promotes the growth and metastasis of breast carcinoma by directly targeting ING4. J Cell Biochem. 2020;121:3887–900. doi: 10.1002/jcb.29545. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Wang T, Valle D. Reduced PLP2 expression increases ER-stress-induced neuronal apoptosis and risk for adverse neurological outcomes after hypoxia ischemia injury. Hum Mol Genet. 2015;24:7221–6. doi: 10.1093/hmg/ddv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Z, Zhou W, Wang J, et al. Reduced expression of proteolipid protein 2 increases ER stress-induced apoptosis and autophagy in glioblastoma. J Cell Mol Med. 2020;24:2847–56. doi: 10.1111/jcmm.14840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai H, Zhu Y, Xu P, Chen B. PLP2 expression as a prognostic and therapeutic indicator in high-risk multiple myeloma. Biomed Res Int. 2020;2020:4286101. doi: 10.1155/2020/4286101. [DOI] [PMC free article] [PubMed] [Google Scholar]