Abstract
Amphibians are ideal for studying visual system evolution because their biphasic (aquatic and terrestrial) life history and ecological diversity expose them to a broad range of visual conditions. Here, we evaluate signatures of selection on visual opsin genes across Neotropical anurans and focus on three diurnal clades that are well-known for the concurrence of conspicuous colors and chemical defense (i.e., aposematism): poison frogs (Dendrobatidae), Harlequin toads (Bufonidae: Atelopus), and pumpkin toadlets (Brachycephalidae: Brachycephalus). We found evidence of positive selection on 44 amino acid sites in LWS, SWS1, SWS2, and RH1 opsin genes, of which one in LWS and two in RH1 have been previously identified as spectral tuning sites in other vertebrates. Given that anurans have mostly nocturnal habits, the patterns of selection revealed new sites that might be important in spectral tuning for frogs, potentially for adaptation to diurnal habits and for color-based intraspecific communication. Furthermore, we provide evidence that SWS2, normally expressed in rod cells in frogs and some salamanders, has likely been lost in the ancestor of Dendrobatidae, suggesting that under low-light levels, dendrobatids have inferior wavelength discrimination compared to other frogs. This loss might follow the origin of diurnal activity in dendrobatids and could have implications for their behavior. Our analyses show that assessments of opsin diversification in across taxa could expand our understanding of the role of sensory system evolution in ecological adaptation.
Keywords: gene loss, blue-sensitive opsin, Dendrobatidae, Atelopus, Brachycephalus, spectral tuning
Introduction
Natural selection has favored visual systems that maximize the detection and discrimination of light wavelengths that are relevant to organismal activity patterns and habitats (Warrant and Johnsen 2013). For instance, nocturnal or crepuscular species have visual systems that capture maximal light or permit color discrimination in dim-light conditions (Bowmaker 2008; Gutierrez et al. 2018; Mohun and Davies 2019; Guo et al. 2023). In contrast, diurnal species are active when the light spectrum is the brightest and broadest (e.g., 350–700 nm); therefore, their visual systems may be evolutionarily tuned for improved wavelength discrimination (i.e., color vision; Bowmaker 2008). In vertebrates, visual opsin genes, which encode G-protein-coupled receptors that bind to a retinal chromophore (Bowmaker 2008), have diversified and undergone selection for adaptation to various light environments. For example, diurnal primates underwent duplication and subsequent spectral tuning of the long-wavelength-sensitive opsin twice (Hunt et al. 1998), while fishes have undergone duplication and diversification of opsin genes many times (Hofmann and Carleton 2009). In addition, some groups have lost opsin genes during adaptation to new photic environments including the loss of cone opsins in coelacanths (Yokoyama et al. 1999) and the loss of a short-wavelength-sensitive cone opsin in mammals during adaptation to nocturnal lifestyles (Bowmaker 2008).
The visual opsin genes present in vertebrates include long-wavelength-sensitive (LWS or OPN1LW), middle-wavelength-sensitive (RH2, MWS, or RHB; not to be confused with an independent gene duplication that gave rise to a middle-wavelength-sensitive opsin gene in Old World primates also called MWS or OPN1MW [Nathans et al. 1986; Hunt et al. 1998]), short-wavelength-sensitive 1 (SWS1 or OPN1SW), and short-wavelength-sensitive 2 (SWS2 or OPN2SW; Bowmaker 2008; Schott et al. 2022). Other vision-related genes include the rod opsin, rhodopsin (RH1, RHO, RHA, or OPN2), which evolved after a duplication and divergence event from an ancestral opsin shared with RH2 (Okano et al. 1992). Color vision is achieved when two or more photoreceptors with opsins differing in wavelength sensitivity are simultaneously activated, and their signals are compared by the observer's central nervous system (Bowmaker 2008; Gibson 2014). For consistency, we refer to the opsin genes using the following acronyms: LWS, SWS1, SWS2, RH1, and RH2.
The repertoire of visual opsin genes and the spectral sensitivity of retinal photoreceptors have frequently undergone changes to accommodate the evolutionary transition between diurnal and nocturnal activity patterns (Surridge et al. 2003; Hauzman et al. 2017), a shift that has occurred repeatedly throughout the phylogenetic history of animals (Anderson and Wiens 2017; Akiyama et al. 2022). For instance, gene duplication has been observed independently in the common ancestor of catarrhine primates (Old World monkeys, apes, and humans) and in the New World howler monkeys as a consequence of the transition to diurnal ecologies from nocturnal ancestors (Surridge et al. 2003). The transition to diurnality has also resulted in the loss of rod photoreceptors and functional RH1 genes in some diurnal species of snakes and lizards (Hauzman et al. 2017). Similarly, spectral tuning of opsin proteins has also been involved in adaptation to bright-light environments in several diurnal species of insects (Akiyama et al. 2022), snakes (Hauzman et al. 2017), and birds (Borges et al. 2015). Vision in anuran amphibians is particularly interesting as most species are crepuscular or nocturnal, but several clades have independently evolved diurnal activity (Anderson and Wiens 2017). Some nocturnal frog species (e.g., Rana temporaria, Xenopus laevis, and Rhinella marina [Bufo marinus]) have played critical roles in discovering the biology of vision, yet much less is known regarding vision in diurnal species.
The anuran retina is thought to typically contain three types of cone cells that are active under bright-light conditions: two types of long-wavelength-sensitive cones that express LWS, and a short-wavelength-sensitive cone that expresses SWS1 (Liebman and Entine 1968; Hárosi 1982; Koskelainen et al. 1994; Mohun and Davies 2019; Donner and Yovanovich 2020). Some anuran species such as Rana pipiens (nocturnal) and Oophaga pumilio (diurnal), possess an additional fourth cone with middle-wavelength sensitivity (λmax of ∼500 nm), but the pigment contained in this type of photoreceptor cell remains to be determined (Siddiqi et al. 2004; Mohun and Davies 2019). In some vertebrates (e.g., fish and reptiles; Bowmaker 2008), the RH2 gene is responsible for green or middle-wavelength sensitivity. To date, the RH2 gene has not been found in any amphibian, and it is believed to have been lost in their last common ancestor (Mohun and Davies 2019; Schott et al. 2022). Thus, it is plausible that RH1 is the gene that is expressed in green-sensitive cones in amphibians (Schott et al. 2022), conferring them with the ability to detect middle-wavelength light spectra during the day, as has occurred in at least one species of diurnal snake, Thamnophis proximus (Schott et al. 2016). Although some vertebrates use oil droplets to further tune wavelength sensitivity, oil droplets in anurans are often absent or colorless and thus are unlikely to influence the spectral sensitivity of cone opsin proteins (Toomey and Corbo 2017). For example, oil droplets are present but colorless in O. pumilio (Siddiqi et al. 2004). In addition to these four cone cell types (single and double LWS cones, single SWS1 cones, and an MWS cone), the anuran retina has two types of rod cells that are active in dim-light conditions: one that expresses RH1, known as the “red rod,” and another that expresses SWS2, known as the “green rod” (Hunt and Collin 2014). The existence of two rod types likely allows some amphibians to have low-light (scotopic) and rod-based color discrimination (Yovanovich et al. 2017).
Despite several frog species being key to our early understanding of vision (Liebman and Entine 1968), the vast diversity of anuran visual systems is only just beginning to be unraveled (Donner and Yovanovich 2020). For example, only 108 of the >7,500 currently described anuran taxa (AmphibiaWeb 2022) have information on lens transmission properties (Yovanovich et al. 2020; Thomas et al. 2022), and the spectral sensitivity of retinal photoreceptor cells has been described for only 10 species (Donner and Yovanovich 2020). Moreover, the genetic basis of anuran vision is especially understudied (Mohun and Davies 2019), with genomic resources limited to a handful of well-assembled frog genomes until very recently (Womack et al. 2022) and one study of opsin genes in 33 frogs (Schott et al. 2022). Compared to other vertebrates, the lack of information on the molecular biology of anuran vision contrasts with the relatively large body of research into anuran visual ecology (e.g., Toledo and Haddad 2009; Bell and Zamudio 2012; Rößler et al. 2019). One example is Dendrobatidae (poison frogs), which has become a model system for understanding how natural selection by predators shapes the origin and subsequent diversification of warning signals (e.g., Clough and Summers 2000; Richards-Zawacki and Cummings 2011; Santos and Cannatella 2011; Wang 2011; Cummings and Crothers 2013; Rojas et al. 2014; Lawrence et al. 2019). Chemically defended and brightly colored (aposematic) dendrobatids are also a special focus of molecular work, including published genomes from O. pumilio and Ranitomeya imitator, with several more being assembled. Other frog families that include diurnal and aposematic species include some Neotropical genera of Bufonidae and Brachycephalidae. Atelopus (Harlequin toads, Bufonidae) and Brachycephalus (Pumpkin toadlets, Brachycephalidae) have a handful of studies on color-based intraspecific or visual signaling in brightly colored species (Rebouças et al. 2019; Rößler et al. 2019), but no genomic or transcriptomic work focusing on genes involved in vision in Atelopus and Brachycephalus have been conducted, and no publicly available genomes from either group exist.
Here, we aimed to describe patterns of selection on visual opsin genes in three diurnal, Neotropical frog clades: poison frogs (Dendrobatidae or Dendrobatoidea sensu Grant et al. [2006]), Harlequin toads (Bufonidae: Atelopus), and pumpkin toadlets (Brachycephalidae: Brachycephalus). We sequenced the four visual opsin genes known in amphibians (LWS, SWS1, SWS2, and RH1) using a target-bait capture approach including 116 species with a special emphasis on the three focal clades. We reviewed patterns of positive selection on amino acid sites relevant to the spectral sensitivity of opsin proteins, which may have been modified via substitutions to better absorb wavelengths that are most relevant to the organism's visual ecology, a process known as spectral tuning (Carvalho et al. 2007; Hunt et al. 2007; Osorio and Vorobyev 2008). Using a combination of selection analyses, we determined whether amino acid positions at or near known spectral tuning sites were associated with the transitions to diurnality in each clade and/or were under positive selection.
Results
Opsin Sequences and Phylogenies
We retrieved 28 opsin gene sequences of 13 species from GenBank and reconstructed another 49 (representing 19 species) using SRA data deposited in NCBI (see supplementary table S1, Supplementary Material online). From 484,530 Illumina reads from our bait capture experiment, we further reconstructed 289 new opsin gene sequences representing 87 anuran species. In total, we analyzed 366 sequences from 116 anuran species. The latter two-thirds of the reconstructed SWS2 gene had low coverage in many species, so sites after amino acid position 129 were excluded from downstream analyses (see Materials and Methods). Following this exclusion, all assembled sequences had an average read depth of >10×, with 82% having coverage >50 × (see supplementary table S2, Supplementary Material online). Estimated gene trees were largely concordant with recent family-level phylogenies (Feng et al. 2017; Hime et al. 2021) and hypothesized interspecific relationships (Pyron 2014; Grant et al. 2017; Jetz and Pyron 2018; see supplementary Data S3, Supplementary Material online). The phylogeny inferred using all opsin gene sequences was reciprocally monophyletic for each gene, indicating a low probability of chimeric sequences in our data set.
Analyses of Selection
To identify patterns of positive selection that might be associated with diurnality, we used a series of Contrast-FEL analyses to identify sites under differential dN/dS regimes between groups of branches (table 1). We excluded SWS2 from this analysis because no sequences from Dendrobatidae were available (see below). Contrast-FEL estimates site-specific dS values for the entire phylogeny; it then estimates and statistically compares dN values for each site between foreground and background branch sets. For sites identified by Contrast-FEL to be under different selection regimes, we used FEL to determine whether dN/dS (ω) was statistically distinguishable from 1, with values less than 1 indicating negative selection and values greater than 1 indicating positive selection. We first compared the dN value estimated for transition branches between nocturnal and diurnal lineages (the stem branches of Dendrobatidae, of Atelopus, and of Brachycephalus) to a dN value estimated for all other branches; no sites were identified as under different patterns of selection (see supplementary Data S1, Supplementary Material online). Then we directly compared dN values between diurnal and nocturnal clades (“DIURNAL” foreground, see Materials and Methods, supplementary table S1, Supplementary Material online, and the Supplementary material for how we determined activity states). This analysis identified two sites in LWS, three sites in SWS1, and four sites in RH1 to be under differential selection regimes in foreground and background lineages (table 1). As six of these sites were also identified by other methods to be under positive selection when the entire tree was included (using FEL, CODEML, FUBAR, or MEME approaches; table 2), we suspected that our large sampling of dendrobatid lineages (all diurnal) might be in part driving this pattern. We then compared dN values for Dendrobatidae branches plus its stem branch (“DENDRO” foreground, see table 1) versus other branches. This analysis identified all but one of the same sites found to be evolving under significantly different regimes in the DIURNAL foreground and background; only site 213 in RH1 was unique to the DIURNAL analysis. Interestingly site 213 in RH1 was identified to be under positive selection in the DIURNAL background (i.e., the nocturnal lineages) and neutrally evolving in the foreground, suggesting that it might be under selection in nocturnal lineages. In addition, two sites were found to be evolving under different regimes in the DENDRO foreground and background but were not identified in the DIURNAL analyses (site 316 in LWS and site 97 in RH1), suggesting that these sites are under different selective conditions in Dendrobatidae compared to other lineages.
Table 1.
Results From Contrast-FEL Analyses Comparing Selection Patterns Between Lineage Groups.
| Gene | Codon number (bovine RH1 numbering)a | Nearby spectral tuning site | Location | Foreground | Contrast-FEL | FEL | Interpretation | |||
|---|---|---|---|---|---|---|---|---|---|---|
| dS | dN | |||||||||
| Fore- ground | Back ground | Foreground (P-value) | Background (P-value) | |||||||
| LWS | 145 | None | ECD II | DIURNAL | 1.023 | 1.690 | 0.093 | Neutral (0.385) | Negative (0.007) | Similar results for DIURNAL and DENDRO |
| DENDRO | 1.019 | 2.077 | 0.086 | Neutral (0.224) | Negative (0.006) | |||||
| 176c | None | TMD IV | DIURNAL | 10.583 | 0.732 | 0.000 | Negative (<0.001) | Negative (<0.001) | Similar results for DIURNAL and DENDRO | |
| DENDRO | 10.478 | 0.901 | 0.000 | Negative (<0.001) | Negative (<0.001) | |||||
| 316 | None | C-T | DENDRO | 1.981 | 2.304 | 0.165 | Neutral (0.787) | Negative (0.001) | Under negative selection in DENDRO background | |
| 318 | None | C-T | DIURN-DEN | 1.204 | 3.000 | 0.000 | Neutral (0.323) | Negative (<0.001) | Under negative selection in DIURN-DEN background | |
| SWS1 | 218 | None | TMD V | DIURNAL | 0.797 | 1.808 | 0.096 | Neutral (0.120) | Negative (0.017) | Similar results for DIURNAL and DENDRO |
| DENDRO | 0.798 | 2.173 | 0.089 | Neutral (0.058) | Negative (0.013) | |||||
| 221 | None | TMD V | DIURNAL | 0.441 | 2.135 | 0.076 | Positive (0.024) | Neutral (0.143) | Similar results for DIURNAL and DENDRO | |
| DENDRO | 0.442 | 2.628 | 0.072 | Positive (0.010) | Neutral (0.127) | |||||
| 342 | None | C-T | DIURNAL | 0.808 | 0.838 | 0.000 | Neutral (0.0962) | Negative (0.001) | Similar results for DIURNAL and DENDRO | |
| DENDRO | 0.802 | 0.998 | 0.000 | Neutral (0.756) | Negative (0.001) | |||||
| RH1 | 22 | None | N-T | DIURN-DEN | 1.428 | 2.026 | 0.047 | Neutral (0.674) | Negative (0.001) | Under negative selection in DIURN-DEN background |
| 87c | None | TMD II | DIURNAL | 0.000 | 1.306 | 0.092 | Positive (0.002) | Neutral (0.370) | Similar results for DIURNAL and DENDRO | |
| DENDRO | 0.000 | 1.624 | 0.082 | Positive (0.001) | Neutral (0.577) | |||||
| 97b,c | 96 | TMD II | DENDRO | 0.181 | 0.000 | 1.429 | Neutral (0.251) | Positive (0.009) | Under positive selection in DENDRO background | |
| 124c | 122, 124, 125 | TMD III | DIURNAL | 1.986 | 2.204 | 0.086 | Neutral (0.817) | Negative (<0.001) | Similar results for DIURNAL and DENDRO | |
| DENDRO | 1.988 | 2.564 | 0.156 | Neutral (0.583) | Negative (<0.001) | |||||
| 169b,c | None | TMD IV | DIURNAL | 0.433 | 5.346 | 1.099 | Positive (<0.001) | Neutral (0.110) | Similar results for DIURNAL and DENDRO | |
| DENDRO | 0.422 | 6.149 | 1.197 | Positive (<0.001) | Positive (0.066) | |||||
| 213b,c | 211 | TMD V | DIURNAL | 1.031 | 0.702 | 3.565 | Neutral (0.030) | Positive (0.016) | Under positive selection in DIURNAL background | |
| 277b,c | None | TMD VI | DIURN-DEN | 0.549 | 5.530 | 0.429 | Positive (0.004) | Neutral (0.727) | Under positive selection in DIURN-DEN foreground | |
Foreground groups included “DENDRO”, that is Dendrobatidae + stem branch, “DIURNAL”, that is all diurnal branches as described in methods, or DIURN-DEN, which is all diurnal branches except for Dendrobatidae and its stem branch. A single site-specific synonymous rate (dS or α in the models) was estimated for each site using maximum likelihood. Two site-specific nonsynonymous rates (dN or β in the models) were estimated separately for the foreground and background and then compared statistically using likelihood ratio tests. We report only the sites that pass the false discovery threshold (q = 0.02). Results from FEL analyses are presented for each site identified to be under different selection regimes by Contrast-FEL. Selection patterns were estimated by FEL for specific subsets of branches using each foreground group (DENDRO, DIURNAL, or DIURN-DEN) or its background (i.e., all branches except DENDRO, DIURNAL, or DIURN-DEN) as foreground lineages. FEL results are reported as Neutral: ω not significantly different than 1; Positive: ω significantly greater than 1; Negative: ω significantly less than 1. We aligned sequences against bovine rhodopsin (NP_001014890.1) to make comparisons with known spectral tuning sites (see supplementary table S3, Supplementary Material online for references).
The numbering system used in our alignments can be determined by adding 17 to the bovine RH1 number for LWS, adding 9 for SWS2, and subtracting 5 for SWS1. However, at site 342 in SWS1, a subtraction of 6 is required.
Amino acid sites reported by Schott et al. (2022) as under positive selection.
Amino acid sites reported by FUBAR, FEL, or MEME analyses as under positive selection in this study, across the entire tree.
Table 2.
Results From Site and Branch-site Selection Analyses.
| Gene | Known spectral tuning sites in vertebrates | Codon number (bovine RH1 numbering)a |
Nearby spectral tuning site | Location | Selection test | P-value/Posterior Probability | Number of branches (MEME only) |
|---|---|---|---|---|---|---|---|
| LWS | 100, 164, 181, 214, 217, 261, 269, 292, 293 | 49b | None | TMD I | MEME | P < 0.01 | 3 |
| CODEML | PP = 1.00 | − | |||||
| 59 | None | TMD I | MEME | P = 0.02 | 1 | ||
| FEL (−) | P = 0.04 | − | |||||
| 162 | 164 | TMD IV | MEME | P < 0.01 | 7 | ||
| 166b | 164 | TMD IV | MEME | P < 0.01 | 1 | ||
| FUBAR | PP = 0.99 | − | |||||
| CODEML | PP = 0.97 | − | |||||
| 176 | None | TMD IV | MEME | P < 0.01 | 2 | ||
| FEL (−)c | P < 0.01 | − | |||||
| 205 | None | TMD V | MEME | P = 0.04 | 8 | ||
| 206 | None | TMD V | MEME | P < 0.01 | 1 | ||
| 209 | None | TMD V | MEME | P = 0.02 | 2 | ||
| 217b | 217 | TMD V | MEME | P = 0.04 | 0 | ||
| 231 | None | TMD V | MEME | P = 0.02 | 1 | ||
| FEL (−) | P < 0.01 | − | |||||
| 262 | 261 | TMD VI | MEME | P < 0.01 | 3 | ||
| SWS1 | 46, 49, 52, 81, 86, 90, 91, 93, 97, 108, 109, 113, 114, 116, 118, 207 | 50 | 49, 52 | TMD I | MEME | P = 0.04 | 0 |
| FEL (+) | P = 0.03 | − | |||||
| FUBAR | PP = 0.97 | − | |||||
| 120b | 118 | TMD III | MEME | P = 0.04 | 0 | ||
| FEL (+) | P = 0.02 | − | |||||
| 159b | None | TMD IV | MEME | P = 0.01 | 6 | ||
| 221 | None | TMD V | MEME | P < 0.01 | 5 | ||
| SWS2 (Only sites up to 129 were analyzed) | 44, 46, 49, 52, 91, 93, 94, 97, 99, 109, 116, 117, 118, 122, 164, 207, 261, 265, 269, 275, 292, 295 | −1 | None | N-T | FUBAR | PP = 0.90 | − |
| 8 | None | N-T | FUBAR | PP = 0.98 | − | ||
| 56 | None | TMD I | FUBAR | PP = 0.98 | − | ||
| 106 | None | ECD I | MEME | P = 0.02 | 1 | ||
| FEL (−) | P = 0.01 | − | |||||
| RH1 | 83, 90, 96, 102, 118, 122, 124, 125, 132, 164, 183, 189, 194, 195, 207, 208, 211, 253, 261, 265, 269, 289, 292, 295, 299, 300, 308 | 37 | None | TMD I | CODEML | PP = 1.00 | − |
| 39b | None | TMD I | MEME | P = 0.03 | 0 | ||
| FEL (+) | P = 0.02 | − | |||||
| FUBAR | PP = 0.97 | − | |||||
| 82 | 83 | TMD II | MEME | P < 0.01 | 2 | ||
| 87 | None | TMD II | MEME | P = 0.03 | 7 | ||
| FEL (+) | P = 0.03 | − | |||||
| FUBAR | PP = 0.95 | − | |||||
| 97b | 96 | TMD II | FEL (+) | P = 0.05 | − | ||
| FUBAR | PP = 0.95 | − | |||||
| 103 | 102 | ECD I | CODEML | PP = 1.00 | − | ||
| 107b | None | ECD I | MEME | P = 0.04 | 9 | ||
| 111 | None | TMD III | MEME | P < 0.01 | 2 | ||
| 124 | 122, 124, 125 | TMD III | MEME | P = 0.03 | 8 | ||
| FEL (−) | P = 0.04 | − | |||||
| 159 | None | TMD IV | MEME | P < 0.01 | 2 | ||
| 164 | 164 | TMD IV | CODEML | PP = 1.00 | − | ||
| 168 | None | TMD IV | MEME | P = 0.03 | 3 | ||
| 169b | None | TMD IV | MEME | P < 0.01 | 12 | ||
| FEL (+) | P < 0.01 | − | |||||
| FUBAR | PP = 1.00 | − | |||||
| 213b | 211 | TMD V | MEME | P < 0.01 | 9 | ||
| FUBAR | PP = 0.97 | − | |||||
| 260 | 261 | TMD VI | MEME | P < 0.01 | 1 | ||
| 262 | 261 | TMD VI | MEME | P = 0.01 | 1 | ||
| FEL (−) | P = 0.01 | − | |||||
| 277b | None | TMD VI | MEME | P = 0.01 | 3 | ||
| 281 | None | ECD III | CODEML | PP = 1.00 | − | ||
| 290 | 289, 292 | TMD VI | MEME | P = 0.02 | 2 | ||
| FUBAR | PP = 0.95 | − |
Amino acid sites in four opsin genes that were detected to be under positive selection by site (FEL, FUBAR, CODEML) or branch-site combinations (MEME). We aligned sequences against bovine rhodopsin (NP_001014890.1) to make comparisons with known spectral tuning sites (see supplementary table S3, Supplementary Material online for references). More details regarding the phylogenetic patterns of selection can be reviewed in figure 1 and supplementary S1, Supplementary Material online. We report all significant FEL results that correspond to sites identified by MEME or FUBAR and note each FEL result as indicative of positive (+) or negative (−) selection. Sites identified to be under positive selection in CODEML analyses are reported as posterior probabilities from Bayes Empirical Bayes analysis of M8 models. N-T, N-terminus; C-T, C-terminus; ECD I, extracellular domain I; TMD I–VII, transmembrane domains I–VII; −, does not apply. Reported P-values for MEME have been corrected for multiple testing using Holm–Bonferroni. Input files and results are available in supplementary Data S1, Supplementary Material online.
The numbering system used in our alignments can be determined by adding 17 to the bovine RH1 number for LWS, adding 9 for SWS2, and subtracting 5 for SWS1.
Amino acid sites reported by Schott et al. (2022) as under positive selection.
To further assess whether the amino acid sites identified to be under selection in diurnal lineages were driven by our biased sampling of Dendrobatidae and to potentially identify amino acid sites under selection in nondendrobatid diurnal lineages, we then created a third group (“DIURN-DEN”), which is the DIURNAL group excluding Dendrobatidae and its stem branch. In these analyses, three sites were identified to be under differential selective regimes (318 in LWS and 22 and 277 in RH1), and they did not overlap with any sites identified to be under differential selective regimes when using the DIURNAL or DENDRO foregrounds (table 1). Two sites were found to be neutrally evolving in the foreground but under negative selection in the background (318 in LWS and 22 in RH1) and one site was found to be under positive selection in foreground but neutrally evolving in background (277 in RH1). Thus, these sites could be related to diurnality in nondendrobatid clades. Based on our Contrast-FEL results, we note that caution should be taken when contrasting branches for selection pattern analyses based on a phylogeny that has multiple independent origins of a trait. The uneven distribution of taxa (e.g., over-representation of clades with the derived character of interest—in our case Dendrobatidae) might drive the analyses to identify sites with a signature of selection exclusive to the overrepresented group as significant in overall selection analyses.
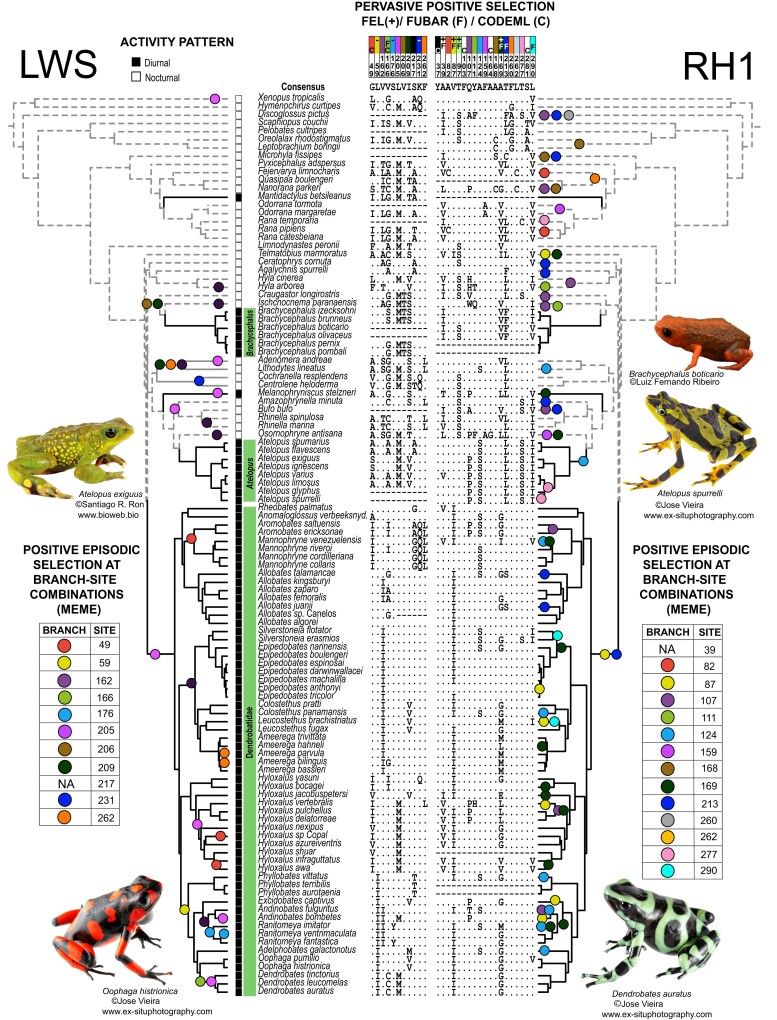
Next, we aimed to determine whether other amino acid sites experienced positive selection along the entire phylogeny, with the goal of identifying sites of functional importance for anurans and potentially for diurnal vision. We conducted four types of selection tests that detect positive selection at specific sites (FEL, FUBAR, CODEML) or branch-site combinations (MEME). In CODEML analyses, the M8 model including positive selection was a better fit than M7 and M8a models (which exclude positive selection) for LWS and RH1 but not for SWS1 or SWS2 (see supplementary table S4, Supplementary Material online). Thus, we only report CODEML results for RH1 and LWS. In combination with Contrast-FEL results, site and branch-site selection tests identified a total of 44 sites across the four opsin genes as having experienced positive selection across all branches (FEL, FUBAR, CODEML) or in a subset of branches (MEME, Contrast-FEL) (tables 1 and 2). Of the identified sites, 14 were in LWS, 6 in SWS1, 4 in SWS2, and 20 in RH1. Based on published data, 3 of the 44 sites are known spectral tuning sites including site 217 in LWS and sites 124 and 164 in RH1; 12 other sites are within three amino acids from a known spectral tuning site (table 2; supplementary Data S1, Supplementary Material online; supplementary table S3, Supplementary Material online). While results from FEL, FUBAR, and CODEML reflect signatures of positive selection that are strong enough to provide enough signal across the entire phylogeny, MEME detects signals of selection in subsets of branches and thus provides more precision about where in the phylogeny sites are under positive selection. Most sites identified by MEME were found to be under positive selection in only one or two branches, but 13 (sites 49, 162, 205, and 262 in LWS, sites 159 and 221 in SWS1, and sites 87, 107, 124, 168, 169, 213, and 277 in RH1) were found to be under positive selection in three or more branches, suggesting that these sites may be of particular functional importance in frogs (fig. 1 and supplementary S1, Supplementary Material online).
Fig. 1.
A summary of activity trait data (see supplementary table S1, Supplementary Material online for references) and results from site and branch-site selection analyses of two opsin genes, LWS and RH1 (see supplementary fig. S1, Supplementary Material online for SWS1 and SWS2 and table 2 for details) using bovine RH1 numbering. Branch-site combinations that were detected by Mixed Effect Models of Evolution (MEME) as evolving under positive selection are mapped onto the phylogenetic tree using filled circles where circle colors correspond to specific sites in the alignment. One site in LWS and one in RH1 were significant in MEME analyses but no specific branch-site combination was reported; these are marked by “NA” in the MEME tables. Amino acid sites marked with “F” or “C” above the alignment were reported to be under positive selection by FUBAR or CODEML, respectively. FEL results are indicated using + to indicate positive selection and − to indicate negative selection (negative selection results are only shown for sites found to be under positive selection by other analyses; see supplementary Data S1, Supplementary Material online for full FEL results). We abbreviated Anomaloglossus verbeeksnyderorum as Anomaloglossus verbeeksnyd. for brevity. Black branches indicate branches categorized as diurnal.
Among the list of sites in table 2, a few present complex characterizations. For example, site 277 in RH1 was reported to be under positive selection by MEME, but we observed that there were no nonsynonymous changes on the branches identified by MEME (Atelopus spurrelli and Node 33, the ancestral branch leading to A. spurrelli, Atelopus glyphus, Atelopus limosus, and Atelopus varius; fig. 1) Instead, these branches have two mutations that do not result in an amino acid substitution (Ser-AGC → Ser-TCC), but likely require a nonsynonymous intermediate that is not recovered in our analysis. Another six sites were found by MEME to be under positive selection in a subset of branches and by FEL to be under negative selection across the entire phylogeny (table 2, denoted with “FEL (−)”). Five of these sites are characterized by high conservation, with an amino acid change present in only one or two branches (fig. 1 and supplementary S1, Supplementary Material online). In contrast, the sixth site, site 124 in RH1, presents an alanine-to-serine replacement in eight branches. Finally, one site in LWS, two sites in SWS1, and one site in RH1 (table 2) were found to be under positive selection by MEME but without any branch identified, suggesting that the signal for selection is diffuse, which occurs when there is enough signal to report positive selection, but no individual branch rises to the level of significance (Spielman et al 2019).
Verifying Absence of SWS2 in Poison Frogs
During our exome capture reconstructions, we failed to recover any exon or fragment of the SWS2 gene in dendrobatids. We considered at first that our baiting design might have been unsuccessful. While it is possible that our SWS2 sequence baits (designed using a consensus sequence matrix) were not similar enough to capture this gene in Dendrobatidae, especially given that part of SWS2 was not captured efficiently in other frogs, we had no issues obtaining partial sequences from all other clades including the other aposematic species of Atelopus and Brachycephalus (see supplementary fig. S1, Supplementary Material online). Therefore, we needed to confirm that this gene was not present with additional transcriptomic and genomic data. For the transcriptome approach, we extracted mRNA from the O. pumilio eye, and we were unable to amplify SWS2 cDNA. This suggests that at least in this species, SWS2 is not expressed.
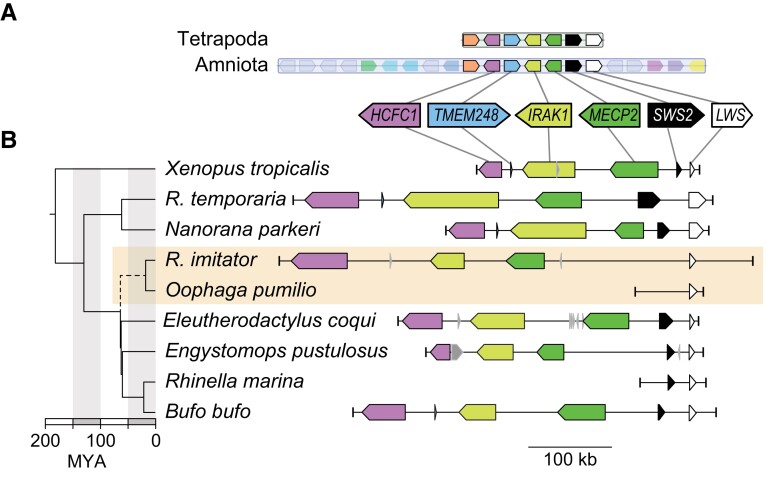
For the genomic approach, we used synteny analyses with BLAST, and we found that SWS2 is located in a syntenic block with LWS and MECP2 in the ancestor of all tetrapods, in the ancestor of all amniotes (fig. 2A), and in five frog families with genomic data that is publicly accessible (fig. 2B: Bufonidae, Leptodactylidae, Eleutherodactylidae, Pipidae, and Ranidae). For dendrobatids, we explored the reassembled O. pumilio scaffolds and found that scaffold70671 contained LWS and was syntenic with other amphibians (two short and highly conserved regions between SWS2 and LWS could be aligned between Nanorana and O. pumilio [supplementary Data S2, Supplementary Material online]), but we could not identify any coding region of SWS2. Then, we explored the more complete scaffold of this region from R. imitator (scaffold934), which included LWS and MECP2, the latter of which is expected to be upstream of SWS2, yet we were unable to find SWS2 between these two genes on this scaffold (fig. 2B). In fact, we could not detect an SWS2 gene or pseudogene on any scaffold in O. pumilio or R. imitator genomes (see supplementary table S5, Supplementary Material online). In contrast, using the same methods, we were able to detect SWS2 upstream of LWS in other hyloid frogs including Bufo, Eleutherodactylus, Engystomops, and Rhinella (fig. 2B, supplementary Data S4, Supplementary Material online). The hyloid clade originated ∼70 Ma and the age of the last common ancestor of Dendrobatidae has been estimated to be ∼40 My (Feng et al. 2017). Thus, the most parsimonious explanation is that a functional SWS2 gene is not present in living dendrobatids, including O. pumilio or R. imitator, and that the SWS2 gene may have been pseudogenized (to a point beyond recognition) or lost in the ancestor of dendrobatids between 70 and 40 Ma.
Fig. 2.
(A) SWS2 is found in a syntenic block with LWS in the ancestors of all tetrapods (block 355) and in the ancestor of all amniotes (block 65). (B) The region in Xenopus tropicalis (chromosome 8, NC_030684:32108538-32360324) shows synteny with Rana (R.) temporaria (chromosome 9, NC_053497.1:22641469-22170306; Ranidae), Nanorana parkeri (unplaced scaffold, NW_017306743:511492-806940; Ranidae), Rhinella marina (contig ctg22529_RHIMB, ONZH01019223.1:1-80000; Bufonidae), Bufo bufo (chromosome 8, NC_053396.1:25948637-26357822; Bufonidae), Eleutherodactylus coqui (chromosome 9, CM034094.1:11013102-11351932; Eleutherodactylidae), Engystomops pustulosus (chromosome 10, CM033650.1:68367373-68196643; Leptodactylidae), which also all contain SWS2, MECP2, IRAK1, TMEM187, and HCFC1 upstream of LWS. However, SWS2 is absent in the scaffold containing LWS in two dendrobatid genomes (Oophaga pumilio [scaffold70671:1-49101] and Ranitomeya [R.] imitator [CAJOBX010072427.1, scaffold934:531527-1]). A dated chronogram (Feng et al. 2017) indicates that SWS2 may have been lost in the ancestral lineage (dashed line) leading to Dendrobatidae (orange box), between ∼70 and 40 Ma. Where coding regions were identified in genome assemblies but not annotated, we added annotations using blastp and blastn. Coding regions in gray had ambiguous blast results and thus were left without annotations.
Discussion
In this study, we reviewed the evolution of the visual opsin gene repertoire (LWS, SWS1, SWS2, and RH1) in anurans by analyzing a dataset of 116 frog species from 20 families, including 78 diurnal and 38 nocturnal species. Our analyses report 44 amino acid sites as under positive selection in our sampling of Anura, including sites in or near locations that have been implicated in visual tuning. Many of these sites are undergoing positive selection in branches within diurnal and aposematic Neotropical frog clades. Moreover, our results provide evidence for the loss of the SWS2 gene in the ancestor of Dendrobatidae between 70 and 40 Ma, an event that coincides with the evolution of diurnal activity patterns in this clade. Diurnality in poison frogs has been hypothesized as a prerequisite for the origin of many of their unique adaptations including aposematism, audiovisual communication, mating choice, and parental care (Santos et al. 2003; Santos and Grant 2011; Yang et al. 2019; Carvajal-Castro et al. 2021).
Selection Patterns Relating to the Transition to Diurnality in Neotropical Anurans
Initially, we aimed to identify sites and patterns of selection that were associated with the transition to diurnality in the three focal anuran clades sampled for this study (Dendrobatidae, Atelopus and Brachycephalus). However, we found no evidence for unique patterns of selection on the three branches where transitions to diurnality occurred in our dataset compared to other branches in our tree. Nevertheless, we were able to parse apart some of the patterns in Dendrobatidae and other diurnal lineages and propose the following. Seven sites (176 in LWS, 218 and 221 in SWS1, and 87, 124, 169, and 277 in RH1) were found to be under positive selection in diurnal lineages including Dendrobatidae, and two sites (318 in LWS and 22 in RH1) were found to be under positive selection in diurnal lineages excluding Dendrobatidae; these nine sites may be associated with adaptation to diurnality. For example, site 124 in RH1 (a known spectral tuning site) was found to be under positive selection in diurnal lineages and in dendrobatid lineages but under negative selection in other branches. This site was also found to be under positive selection by MEME in eight branches (R. imitator, Adelphobates galactonotus, Andinobates fulguritus, Phyllobates vittatus, Colostethus panamansis, Lithodytes lineatus, Mannophryne venezuelensis, and the branch leading to the Atelopus clade). The amino acid type at site 124 of those species is serine instead of alanine as seen in other anuran species. Mutagenesis experiments in bovine RH1 indicate that alanine-to-threonine and alanine-to-serine substitutions at this position provide a slight blue shift (Lin et al. 1998; Castiglione and Chang 2018). In addition, A124S in combination with L119F significantly increases the stability of the active conformation of bovine RH1 (Castiglione and Chang 2018). Therefore, it is possible that an anuran RH1 with the A124S substitution has a blue-shifted λmax, and that such changes may be associated with adaptation to diurnality as has been suggested in species of diurnal snakes (Schott et al. 2016; Hauzman et al. 2017). The loss of the cone opsin RH2 in the ancestor of all amphibians resulted in a decreased sensitivity to a significant portion of the visual spectrum, mainly due to the absence of overlap between LWS and SWS1. However, a blue shift in the absorption spectrum of RH1 would coincide with the wavelength detection range previously covered by RH2. This shift would enhance chromatic discrimination in diurnal clades by providing more uniform coverage across the color spectrum (Schott et al. 2016). Previous studies have suggested the expression of RH1 in the green-sensitive cones of amphibians (Schott et al. 2022), supporting a potential role for such a blue shift.
Our failure to identify sites under differential selection in branches where the transition to diurnality occurred suggests either that we may have limited power to detect such sites and/or that relatively stronger selection occurred following the transition rather than during it. We identify many sites under selection within the three diurnal clades on which we focused (i.e., poison frogs, Atelopus and Brachycephalus), even if these were not found to be specifically associated with diurnality in Contrast-FEL assessments. As many diurnal animals rely on color-based signals (i.e., detecting ripe fruits; [Melin et al. 2009]) or perceiving sexual signals (Carleton et al. 2005), they are expected to have a suite of opsins that are tuned to perceive the relevant signal under the corresponding light environment while balancing other aspects of protein function (e.g., kinetics; Hauser et al. 2017). Thus, our selection results suggest that the transition to diurnality is complex and idiosyncratic such that each lineage differently accumulated changes to accommodate the diel habit. In other words, the opsin genes in each clade experienced the process of visual tuning independently and may have been influenced by other confounding factors (e.g., mating visual signal recognition); not surprisingly, this resulted in some parallel and some different substitution patterns.
Identification of Putative Functional Sites in Anuran Opsin Genes
Tables 1 and 2 provide an extensive list of opsin amino acid sites putatively involved in frog vision. Changes at these sites may work alone or together to shift the wavelength sensitivity of anuran opsin proteins as they adapt to diverse light environments, including diurnality. The sites inferred to be under positive selection occur predominantly in the transmembrane domains. Such domains impact the tertiary structure, thermal stability, and properties of the retinal-binding pocket (Andrés et al. 2001; Yokoyama et al. 2006). Spectral sensitivity is related directly to interactions between the amino acid residues in the transmembrane domains and the chromophore (Yokoyama et al. 2006). Although it is possible that replacements at these sites might influence spectral tuning in frogs, without more experimental data, their implications are difficult to interpret. For this reason, we conservatively discuss their possible role in anuran vision below.
Of the 44 sites we found to be under positive selection, only three are previously known spectral tuning sites in vertebrates. Site 124 in RH1 is discussed above. Site 217 in LWS (233 in the human LWS) has been previously shown to play a role in the differentiation of the spectral sensitivities between the long-wave-sensitive and the middle-wave-sensitive pigments in humans and other mammals (Yokoyama and Yokoyama 1990; Asenjo et al. 1994; Fasick and Robinson 1998; Yokoyama and Radlwimmer 2001, but see Hiramatsu et al. 2004). In our study, MEME detected positive selection on this site without indicating any specific branches, suggesting a diffuse signal of selection at this site across the tree (fig. 1; table 2). Moreover, this site was also reported by Schott et al. (2022) to be under positive selection, confirming that this is a key site that could be responsible for shifting the sensitivity of LWS in frogs, including the diurnal taxa explored in this study. In our alignment, amino acid substitutions at site 217 are present in several species including several diurnal clades such as the dendrobatid genera Mannophryne (whose females have a colorful yellow collar), Rheobates, Aromobates, and toxic Phyllobates, as well as two species of Atelopus. However, this site was not identified in Contrast-FEL analyses as under positive selection specifically in diurnal lineages.
The third known spectral tuning site, 164 in RH1, was found by CODEML to be under positive selection; other analyses did not identify this site to be under selection. In our data set, 102 species at this position had an alanine, while just Osornophryne antisana expressed a glycine. Substitutions at this site are known to contribute to a red shift in the absorption maxima of bovine RH1 (Chan et al. 1992). Although an A164S substitution seems to result in a rather small (2 nm) red shift in absorption, it has been shown that an additive effect is achieved when it is in combination with other substitutions including F261S and A269T (Chan et al. 1992). Thus, it is difficult to predict how changes at this site might impact the spectral sensitivity of O. antisana or other frog species, and our analyses do not offer strong support for the involvement of site 164 in RH1 in adaptation to diurnality.
Considering the general lack of data on spectral tuning sites in frog opsins, it is possible that other sites we identified to be under positive selection (tables 1 and 2) could be directly or indirectly involved in spectral tuning in anurans. Eleven sites found to be under positive selection are not known spectral tuning sites but are located within three amino acids of a known spectral tuning site: 162, 166, and 262 in LWS; 50 and 120 in SWS1; 82, 97, 103, 213, 260, 262, and 290 in RH1. Additionally, 13 sites, including three at or near spectral tuning sites, were found to be under selection in three or more branches: 49, 162, 205, and 262 in LWS; 159 and 221 in SWS1; 87, 107, 124, 168, 169, 213, and 277 in RH1 (table 2, fig. 1). We speculate that some of these sites may be of functional importance in the vision of frogs. For example, site 169 in RH1 was found to be under positive selection in 12 lineages. Experiments using site-directed mutagenesis could help verify which sites are important for spectral tuning in amphibians.
A prior study (Schott et al. 2022) identified 16 sites in opsin genes of anurans to be under positive selection; we found 11 of these to be under positive selection in our study. We failed to identify five sites (65, 212, and 270 for RH1; 154 for LWS; −2 for SWS2), which might reflect differences in the number of taxa in our studies (i.e., 116 in our study vs. 33 in theirs) or the specific focus on diurnal species in this article. Schott et al. (2022) also discussed additional variation in other known spectral tuning sites. We exclude them from our discussion because we could not find evidence of positive selection on these sites. Our comparison depicts the conserved nature of opsins but also reports new amino acid changes that contribute to the repertoire of variants that might contribute to opsin spectral sensitivity.
Visual Signals in Focal Frog Clades
Due to the selective pressure of diurnal and visually guided predators, many diurnal frogs are adorned with colorful visual signals that evolved as part of a defensive strategy known as aposematism, in which conspicuous signals warn predators of prey defenses (Bell and Zamudio 2012). Each of our three focal clades (i.e., poison frogs, Atelopus and Brachycephalus) contain species that possess aposematic signals. Although aposematic signals initially evolve under selective pressure from predators and might intensify over evolutionary time (Mappes et al. 2005; Sherratt 2008; Loeffler-Henry et al. 2023), warning signals can also become entangled with mating behaviors and further diversify under sexual selection pressures (Cummings and Crothers 2013; Rojas et al. 2018), if the mate recognition and visual sensory system is appropriately tuned to the visual cues. We hypothesize that some of the sites identified to be under selection are related to intraspecific signaling within these aposematic clades. For instance, color perception seems to be key for mate recognition and territorial display in dendrobatids. In the aposematic species O. pumilio, tadpoles imprint on their mother's color (Yang et al. 2019), females show assortative mate preferences (Summers et al. 1999; Reynolds and Fitzpatrick 2007; Maan and Cummings 2008; Yang et al. 2016), male–male competition relies on color-mediated aggressive behaviors towards phenotypically similar rivals (Yang et al. 2018; Yang and Richards-Zawacki 2020), and there may be directional sexual selection on male coloration brightness (Maan and Cummings 2009). Assortative mating by coloration and pattern is present but less pronounced in other dendrobatid species including additional Oophaga species (Oophaga histrionica and Oophaga lehmanni; Medina et al. 2013), species of Mannophryne with sexual dichromatism where throat coloration in males (gray) and females (bright yellow) plays a role in territoriality and mate choice (Greener et al. 2020), and in many species of Allobates that are sexually dimorphic for throat color that is visible in vocalizing males.
In our other focal clades, visual signaling has been investigated in only a few species. For example, Atelopus zeteki appears to use limb motions as visual signals to conspecifics and this might be related to their lack of tympanic middle ear (Lindquist and Hetherington 1998); yet, it is unknown if other Atelopus use their conspicuously colored soles for intraspecific communication or aposematic signals (Rößler et al. 2019). Likewise, at least one species of Brachycephalus is inferred to be aposematic based on experimental evidence (Goutte et al. 2019; Rebouças et al. 2019); Brachycephalus ephippium and Brachycephalus pitanga also have fluorescent dermal bones visible through their skins with a potential function as a signal (Goutte et al. 2019). While territorial males of some Brachycephalus species display a foot-waving behavior to warn off encroaching males similar to some species of Atelopus (Pombal et al. 1994), it is unclear how important a role color and/or contrast play in the display. Future studies could investigate species- or population-level variation in opsin sequences to assess whether the sites listed in tables 1 and 2 have any effect on the recognition or evaluation of intraspecific signals.
Loss of SWS2 in Dendrobatidae and the Implications for Their Visual Ecology
Based on our inability to sequence SWS2 from an O. pumilio eye transcriptome, the complete absence of SWS2 from bait-capture data from all sequenced dendrobatids (60 species representing all major lineages), and our failure to identify any trace of an SWS2 gene or pseudogene in the O. pumilio and R. imitator genomes, we hypothesize that dendrobatids lost this gene early in their history. Such gene-loss events are not surprising as other comparative genomic studies have shown (Borges et al. 2015; Xu et al. 2021). The loss of short-wavelength photopigment genes has occurred multiple times in the evolutionary history of vertebrates and coincides with shifts in activity patterns, habitat occupancy, and the evolution of other aspects of the sensory capacity of animals (Bowmaker 2008; Jacobs 2013). For example, the loss of SWS2 in therian mammals and coelacanths is considered an adaptation to nocturnality and deep-sea environments, respectively. The loss of a functional SWS1 pigment in several bat species coincides with the origin of a specialized form of echolocation (i.e., high-duty-cycle echolocation; Jacobs 2013).
In dendrobatids, it remains unclear how the loss of SWS2 may have impacted their vision and visual ecology. To answer such a general question, we can explore some specific ones. First, how has SWS2 loss potentially impacted the physiology of dendrobatid visual systems? The limited empirical data from anurans suggest that SWS2 expression is restricted to green rods (Yovanovich et al. 2017; Mohun and Davies 2019). However, green rods are absent from the O. pumilio retina (Siddiqi et al. 2004). Thus, the loss of SWS2 and of the green rod cell type suggests that under low-light levels, dendrobatids might be less capable of discriminating color (scotopic vision) than other diurnal frogs.
Second, when and how did SWS2 disappear from the genome of ancestral dendrobatids? Based on our results, we could not find a SWS2 pseudogene (i.e., nonfunctional segments of DNA that resemble functional SWS2) in the available genomic data for the poison frog species R. imitator and O. pumilio. In contrast, close relatives of dendrobatids do have a functional SWS2 (fig. 2). Thus, our data suggest that only dendrobatids lost SWS2. Dendrobatidae is nested within the superfamily Hyloidea, whose last ancestor lived about ∼70 Ma (Feng et al. 2017; Hime et al. 2021). As the approximate age of the crown clade of Dendrobatidae is ∼40 Ma (Feng et al. 2017), SWS2 must have been lost between 40 and 70 Ma. Two alternative explanations exist for the loss of SWS2 during this 30-My timeframe: (1) The ancestor of dendrobatids had a functional gene which pseudogenized and changed beyond recognition during that period or (2) SWS2 was lost without pseudogenization. Given the lack of evidence of SWS2 pseudogene relics, the most parsimonious explanation is a complete deletion of the gene. Additional data may show that other hyloid groups have lost SWS2, but until then, we consider the loss of SWS2 as a synapomorphy of Dendrobatidae (i.e., Aromobatidae + Dendrobatidae sensu Grant et al. [2006]).
Third, was the loss of SWS2 consequential in that it affected other aspects of dendrobatid vision? Other diurnal hyloid clades in our dataset do not appear to have lost SWS2: all Atelopus and Brachycephalus frogs maintain this gene. Some uncertainty remains regarding SWS2 functionality in nondendrobatids because we were unable to recover the entire gene from many species using target-bait capture, yet complete SWS2 sequences are available in assembled genomes (e.g., fig. 2, Eleutherodactylus). Whether diurnal hyloid species have the green rod cell type (which expresses SWS2) remains unknown. Nevertheless, dendrobatids are known to differ in at least two visual properties from other hyloids. First, the lenses of some dendrobatid species (O. pumilio, Epipedobates tricolor, Dendrobates auratus, Dendrobates leucomelas, Allobates femoralis, and Adelphobates castaneoticus) transmit less short-wavelength light (lens λt50 413–425 nm; except for D. leucomelas lens λt50 326 nm) than those of bufonids (R. marina, R. icterica, R. ornata, Bufo bufo, Rhaebo guttatus, Sclerophis maculata, and Atelopus varius lens λt50 331–365 nm) and brachycephalids (Brachycephalus rotenbergae, Ischnocnema parva, Ischnocnema henseli lens λt50 314–356 nm) (Donner and Yovanovich 2020; Yovanovich et al. 2020; Thomas et al. 2022). As the typical peak wavelength sensitivity of SWS2 rods (∼430 nm; Yovanovich et al. 2017) is much closer to the limit of lens transmission in dendrobatids than other frogs, it is plausible that lens transmission properties were adjusted in some species following the loss of SWS2, or vice versa. Additionally, in O. pumilio, the short-wavelength-sensitive cone (likely SWS1), which absorbs light wavelengths ∼430 nm in other anurans (Yovanovich et al. 2017), was found to absorb light at 466 nm (Siddiqi et al. 2004), suggesting that it has undergone a shift in spectral sensitivity. Further investigation of these patterns will clarify whether loss of SWS2 in dendrobatids in combination with a diurnal lifestyle led to altered lens transmission properties and a change in SWS1 wavelength sensitivity, or vice versa.
Despite the apparent loss of SWS2 in dendrobatid frogs and the lack of RH2 pigment in amphibians (Mohun and Davies 2019), microspectrophotometry data revealed an MWS photoreceptor in the O. pumilio retina (Siddiqi et al. 2004). In accordance with prior literature (Schott et al. 2022), we speculate that these frogs are instead using RH1 in those photoreceptors as the microspectrophotometry absorbance graphs for rods and MWS cones appear nearly identical (Siddiqi et al. 2004). Whether this is the case and if there are any modifications to the RH1 pigment or to any proteins in the O. pumilio MWS cone phototransduction pathway required to make the rod protein function in cone cells requires further investigation. We did find that a large number of amino acid sites (i.e., 20) are under selection in RH1 within frogs, which could be a result of this dual function, though only a few of these sites were noted to be specifically under selection in Dendrobatidae or experiencing dendrobatid-specific patterns of selection. Further, our genome mining, transcriptome, and synteny analyses do not provide any evidence suggesting that RH1 has been duplicated in frogs, but at least one species (Pyxicephalus adspersus) has a duplication of LWS (Schott et al. 2022). Empirical data of spectral sensitivity and opsin protein function in frogs are sparse, and further studies using microspectrophotometry of isolated frog rods and cones will be necessary to recapitulate the evolution of vision in frogs (Donner and Yovanovich 2020).
Conclusion
Our results fill gaps in our knowledge by illustrating how diversification in ecology and life history may have affected opsin evolution in amphibians. We uncover evidence of new putative tuning sites in frogs and show strong evidence suggesting that poison frogs have lost the SWS2 gene. Further work is needed to elucidate the functional consequences of its loss, and of the potential role of RH1 in facilitating color vision in some frogs. Like Donner and Yovanovich (2020), we expect that additional studies of opsin evolution in amphibians will reveal noncanonical visual adaptations and broaden our understanding of the many ways in which animals adapt to diverse light environments.
Materials and Methods
Bait Capture Design, Library Preparation, and Sequencing
Using publicly available sequences (see supplementary table S1, Supplementary Material online) and previously generated transcriptomes (Santos et al. 2018), we designed a custom bait-capture array with myBaits (Arbor Biosciences). The 120-bp baits were synthesized at >10 × tiling across all exons of the genes of interest. Tissues from representative species of Dendrobatidae, Atelopus, and Brachycephalus and their relatives were obtained from the field or from museum collections (see supplementary table S1, Supplementary Material online for source information), under approved protocols (UT Austin AUP-2012-00032 and AUP-0709210, STRI 200715122207, 2015-00205 Tulane #0453) and collection permits (001-13 IC-FAU-DNB/MA and 001-11 IC-FAU-DNB/MA [Ecuador], IBD0359 Res 1177-2014 and Isla Gorgona PIDB DTPA 020 - 16 Res 061-16 [Colombia], and SE/A-47-07 and CITES export permit SE/A-47-07 [Panama]). Tissue samples from Ecuador and Peru were loaned by Luis A. Coloma (Fundación Otonga and Centro Jambatu de Investigación y Conservación de Anfibios; Ecuador) and César Aguilar (Universidad Nacional Mayor de San Marcos; Lima, Peru). We note there is some controversy over the elevation of subgenera such as Oophaga to genera (Santos et al. 2009; Grant et al. 2017), but we have followed taxonomy following (AmphibiaWeb 2022).
DNA was extracted using Qiagen DNeasy kits (Germantown, MD). DNA quality was reviewed with a 0.8% electrophoresis gel and highly degraded samples were excluded. RNA was removed from extractions with RNAse A (E1008, Zymo Research, Irvine, CA), and extractions were further purified and concentrated with Genomic DNA Clean and Concentrate (D4011, Zymo Research) and then quantified using a Qubit 3.0 fluorometer (Thermo Fisher Scientific, Waltham, MA) following manufacturer protocols. Given the large size of amphibian genomes, we used 400–1600 ng of starting DNA for each sample for library preparation. DNA was sheared to ∼300 bp using a Covaris S2 Focused-ultrasonicator (Covaris, Inc, Woburn, MA; settings as Intensity: 5; Duty Cycle: 10%; Cycles per Burst: 200; Time: 50 s; Temp: 7C; Water Level: 12; Sample Volume: 50 µl). Whole-genome libraries were prepared from sheared samples using the KAPA Hyper Plus library preparation kit (KK8514, Roche Diagnostics, Santa Clara, CA), NEBNext Multiplex Oligos for Illumina (E7600, New England BioLabs, Ipswich, MA), home-made SPRI beads (Rohland and Reich 2012) (Lydia Smith, personal communication), and manufacturer protocols. Uniquely barcoded libraries from 4–10 closely related species were pooled to a total of 1.6–5 µg DNA and then size-selected for an insert size of 250 ± 25 bp (total length with 120-bp adaptors: 345 ± 25 bp) using a Blue Pippin (Sage Science, Beverly, MA) and 2% gel cassette. For samples with low concentrations, a second batch of libraries were constructed with the same protocol, except that these were size-selected to 250 ± 50 bp with a 1.5% gel cassette (total length with 120-bp adaptors: 370 ± 50 bp). All size-selected libraries were cleaned with Qiagen MinElute PCR purification kits, eluted in 10 µl, and quantified with Qubit 3.0.
Seven microliter of each pooled library set (50–275 ng/pool) was then hybridized with custom myBaits biotinylated RNA, C0t-1 DNA, and xGen Universal Blockers–TS Mix (1075474, Integrated DNA Technologies, Redwood City, CA) according to myBaits v4.01 recommended protocols. As amphibian genomes are large and contain many repetitive sequences, we used a large amount of starting DNA (1.6–5 µg at size selection) and a larger quantity of blocking oligos than described in the manufacturer's protocol (8 µg human C0t-1 and 8 µg salmon C0t-1 per hybridization reaction). We also used the xGen blockers rather than those provided in the myBaits kit. Following hybridization with C0t-1, universal blockers, and baits for 36 h, pooled libraries were washed and cleaned with DynaBeads MyOne Streptavidin C1 beads (65002, Thermo Fisher Scientific). 12–17 µl of resulting libraries were amplified using the NEBNext Illumina primers (that came with the multiplex oligos) and the KAPA Library Amplification Kit (KK2611, Roche Diagnostics) in two separate PCR reactions with 15–17 cycles. These were purified and eluted in 10 µl with the Qiagen MinElute purification kit. Duplicate PCR reactions and libraries from all 97 samples were normalized by concentration and according to the number of samples per pool and then sequenced across two lanes of the Illumina HiSeq4000 at the Genome Sequencing and Analysis Facility (GSAF) at University of Texas at Austin, yielding approximately 4 M reads per sample.
Assembly of Opsin Genes
Read quality was checked with FastQC (Andrews 2010), barcodes were excluded with Trimmomatic (Bolger et al. 2014) and, because combining multiple assembly methods better characterize multigene families (Holding et al. 2018), the reads were assembled with default parameters for de novo with MEGAHIT (Li et al. 2015), Trinity (Grabherr et al. 2011), and SPAdes (Bankevich et al. 2012). Final assemblies were reduced using CD-HIT set to >98% similarity. Reduced assemblies were annotated using a custom library generated with the opsin sequences used to design the baits and BLASTX. Reference sequences for BLASTX were derived from amphibian opsin genes available in GenBank: X. laevis (Pipidae), Nanorana parkeri (Dicroglossidae) and other species with opsin sequences (e.g., Rana catesbeianus [Ranidae]). Any sequence matching one of the reference opsin genes with an e-value <10−6 was pulled out for downstream analyses.
Given that we de novo assembled genomic DNA that was captured using baits designed from mRNA, the sequences were often only partially assembled, and some shorter sequences were identified erroneously by BLASTX. We used the program BLAT v.36 × 2 (Kent 2002) with reference sequences from the Nanorana parkeri genome (LWS, XM_018560714.1; SWS1, NW_017306744.1; SWS2, NW_017307939.1; RH1, NW_017306456.1) to verify sequences from our dataset that were putatively identified as opsins using BLASTX as described above. A BLAT server was prepared using N. parkeri reference sequences with the “-trans” option to translate the database into protein for all six reading frames. Then BLAT was run with options “-t = dnax” and “-q = dnax” to specify that the format of the database and query were both DNA sequences translated in six frames to protein. Sequences that matched N. parkeri references were pulled out from the BLAT results file generated using “-o = pslx”. These query sequences were then aligned to the reference genome using MAFFT v47.19 (Katoh and Standley 2013) with the options “−auto” and “−adjustdirection”. The smaller fragments whose sequences did not align to the exons were manually removed, and exons from the same individual of each species were merged.
For most of these genes, some edges of exons and several short exons were still missing from the assemblies. To recover the sequences or fill these gaps, we implemented an in silico target capture using MITObim (Hahn et al. 2013). For this procedure, we used the recovered exon sequences as bait sequences and the raw exome capture as target data. We included the following parameters “−quick” (starts process with initial baiting using provided fasta reference), “−mismatch” (% number of allowed mismatches in mapping), and “−kbait” (set kmer for baiting stringency). For the last two parameters we used low stringency (12–17 and 10–15, respectively) to bait more raw reads beyond those obtained from standard assemblers. This approach allowed MITObim to progressively expand the extremes of the bait sequences beyond the original gene reconstructions. The resulting sequences were then aligned with the original sequence matrices for each gene and full sequences from NCBI references (e.g., Xenopus, Nanorana, and Rana). In most cases, these extended sequences overlapped and were merged with the original ones to recover the missing regions. Lastly, we generated a clean alignment containing only coding regions that we used for the subsequent analyses. Raw sequencing coverage for SWS2 past position 389 was low (perhaps because probes designed for that region did not effectively pull-down exons compared to other probes), so we excluded the data past position 389 to avoid any potential errors in its reconstruction.
Gene Tree Construction
To check for any sequence contaminants, misidentifications, or assembly errors, we estimated a gene tree for each opsin, partitioned by codon position, using raxml-ng v.0.9.0 (Kozlov et al. 2019) implemented with the GTR + G model and 200 bootstrap replicates to assess support. We found several sequences of different species to be identical for each gene, although in each case the identical sequences were from closely related species. For LWS, there were two sets of identical sequences: Atelopus glyphus and A. spurrelli; Brachycephalus boticario and B. olivaceus. For SWS1 and SWS2, there was one set of identical sequences for each gene: Atelopus glyphus, A. limosus, and A. varius for SWS1; Brachycephalus pernix and B. pombali for SWS2. For RH1, we found two sets of identical species: B. pernix and B. pombali; Phyllobates aurotaenia and P. terribilis. Because there are no genomes for these species, it was not possible to verify the sequences, so we conservatively excluded all identical sequence sets from the following analyses. Finally, to check for chimeras, we aligned all four gene sequence sets to each other using MAFFT (Katoh and Standley 2013) and estimated a gene tree using raxml-ng v.0.9.0 (Kozlov et al. 2019), the GTR + G model, and 1000 bootstrap replicates to assess support (with bootstrapping enabled).
Determination of Diel Habits
Data on time of activity were compiled from primary and secondary literature, including species descriptions, taxonomic revisions, and books (see supplementary table S1, Supplementary Material online for details and Supplementary Information for references). Activity patterns were categorized as diurnal or nocturnal, with the latter category conservatively including species with crepuscular or mixed activity patterns. When not clearly stated, the time of activity was inferred from reports on time of calling, breeding activity, and behavior (asleep or active) at the time of collection. Diurnal groups include all of Dendrobatidae, Atelopus, and Brachycephalus (and the stem branches leading to each of these clades), as well as Mantella baroni, Mantidactylus betsileanus, and Melanophryniscus stelzneri. Species of the following genera were classified as nocturnal: Adenomera, Agalychnis, Amazophrynella, Amietia, Bufotes, Centrolene, Ceratophrys, Cochranella, Craugastor, Discoglossus, Fejervarya, Hyla, Hymenochirus, Ischnocnema, Leptobrachium, Limnodynastes, Lithodytes, Microhyla, Nanorana, Odorrana, Oreolalax, Osornophryne, Pelobates, Pyxicephalus, Quasipaa, Rana, Rhinella, Scaphiopus, Telmatobius, and Xenopus.
Analyses of Selection
An updated phylogeny of the focal taxa was derived from alignments provided in the two largest phylogenetic reconstructions of amphibians (Pyron 2014; Jetz and Pyron 2018). Both alignments were appended, taxa not included in our analysis were removed, and duplicate taxa were removed by choosing the one with more sequence data. Taxonomic nomenclature was updated following AmphibiaWeb (2022). The final alignment was realigned with MAFFT (Katoh and Standley 2013), then reviewed and adjusted manually, particularly the mitochondrial ribosomal gene sequences. Multiple alignment programs provide a good starting point, but they usually need to be examined and adjusted by eye (Baum and Smith 2013). The optimized alignment was then used for phylogenetic estimation. A maximum likelihood tree was estimated using IQ-TREE2 v2.1.3 (Minh et al. 2020) with five replicate runs. The tree was constrained so that the topology among the families of Hyloidea matched that found by Feng et al. (2017) and Hime et al. (2021). This method was preferred because the trees presented in Pyron (2014) and Jetz and Pyron (2018) are largely dictated by the abundance of data from mitochondrial genes. In contrast, the trees found by Feng et al. (2017) and Hime et al. (2021) are based on 100–1000 s of nuclear genes. The sequences were partitioned by gene and codon and the best-partitioned model was determined using -TESTMERGE option (Chernomor et al. 2016; Kalyaanamoorthy et al. 2017) within IQTREE2. Ultrafast bootstrap values (Hoang et al. 2018) were calculated using 50,000 replicates and were plotted on the best likelihood tree. The data matrix, IQTREE2 scripts, constraint tree, and analysis log files are included in supplementary Data S3, Supplementary Material online in the folder IQTree.
For each selection analysis, we pruned the tree using the “ape” R package (Paradis and Schliep 2019) to contain the subset of species available for each opsin gene. In some cases, we replaced a tip in the tree with a closely related species that was used in our study (see supplementary table S1, Supplementary Material online for details). We then used the resulting tree as the backbone for three types of site-based selection analyses (with P-value set to 0.05) in HyPhy v2.5.14 (Kosakovsky Pond et al. 2005). The type of selection is determined by the value of omega (ω), which is calculated by rate of nonsynonymous substitutions (dN) divided by rate of synonymous substitutions (dS). ω values significantly less than 1 indicate negative selection, while neutral selection is indicated by ω values not significantly different from 1, and positive selection is detected by ω values significantly greater than 1. To conduct hypothesis testing, we first used Contrast-FEL, which statistically compares dN substitution rates for each site between two sets of branches (foreground and background) to determine whether different selection regimes occur in each set of branches (Kosakovsky Pond et al. 2021). Because there were no SWS2 sequences for Dendrobatidae, we did not include this gene in Contrast-FEL analyses. We created five sets of foreground lineages to compare with background lineages in Contrast-FEL: “DENDRO”, that is all Dendrobatidae plus its stem branch, “DIURNAL”, that is all diurnal branches as described above, and “DIURN-DEN”, which is all diurnal branches except for the Dendrobatidae lineages and its stem branch. We also compared substitution rates between transition branches (the stem branches of Dendrobatidae, of Atelopus, and of Brachycephalus) and all other branches, but no sites were identified to be under different selection regimes in this foreground (see supplementary Data S1, Supplementary Material online). Once sites under different selection regimes were identified with Contrast-FEL, we used FEL (see below) with the same sets of foreground and background lineages to determine whether ω values at sites of interest were significantly different from 1. For these analyses, we designated foreground groupings according to the list above (DENDRO, DIURNAL, and DIURN-DEN). We repeated FEL analyses using the inverse of each grouping (e.g., all lineages except DENDRO) to estimate background ω values (reported in table 1).
We then conducted a series of complementary approaches to detect site-specific and branch-site combinations of selection patterns across the entire phylogeny using methods in HyPhy and PAML. Fixed effects likelihood (FEL) estimates the rate of synonymous (dS) and nonsynonymous (dN) substitutions per site with maximum likelihood and compares them using likelihood ratio tests to determine if ω is greater or less than 1 (Kosakovsky Pond and Frost 2005); this test provides an estimate of ω for the entire phylogeny (or only for foreground branches when specified) for each site, which can be used to show pervasive positive or negative selection at specific sites. Fast, Unconstrained Bayesian AppRoximation (FUBAR) is similar to FEL in that it compares single estimates of dN and dS values for each site but uses a hierarchical Bayesian method rather than maximum likelihood (Murrell et al. 2013); this test is more sensitive than FEL to weaker signatures of positive selection (i.e., when ω is close to 1). Mixed effects model of evolution (MEME) is a branch-site model that uses maximum likelihood to test for positive selection at each site (Murrell et al. 2012); this method does not assume a single ω value across the tree and thus can be used to detect selection at a specific site in a subset of branches on a tree. To compare our results to those of Schott et al. (2022), we also conducted CODEML analyses comparing the M7, M8, and M8a models in the PAML software suite (Yang 1997, 2007). The fit of M7 and M8 models for each gene were compared using a likelihood ratio test with two degrees of freedom; the fit of M8 and M8a models were compared using a likelihood ratio test with one degree of freedom.
Following convention, the amino acid sites are numbered based on the bovine rhodopsin sequence (NP_001014890.1); sequences from each opsin gene were aligned with bovine rhodopsin using MAFFT v7.419, and the numbering of the amino acid site was determined by referring to the numbering of bovine rhodopsin starting with the start codon as 1 (see supplementary Data S1, Supplementary Material online). Table 1 indicates how bovine RH1 positions compare to positions in our alignments. We report our selection results referring to the location of each amino acid according to the 3D structure of opsins, which encompasses seven transmembrane domains (TMD I–VII), three extracellular domains (ECD I–III), and the amino- and carboxyl- termini (N and C) (Palczewski et al. 2000; tables 1 and 2, fig. 1 and supplementary S1, Supplementary Material online).
Attempts to find SWS2 in O. pumilio and R. imitator. As we did not recover SWS2 sequences from any dendrobatid species in our bait-capture data set, we attempted to verify whether the SWS2 gene had been lost in this clade using transcriptomics and genome skimming.
First, we attempted to determine which opsin genes are expressed in eye tissue of O. pumilio. As part of another project, one eye from each of 11 O. pumilio populations (Aguacate, Almirante, Bastimentos “Cemetery”, Bastimentos “Green”, Bastimentos “Orange”, Cayo Agua, Colón, Pastores, Popa, San Cristóbal, Solarte; see Maan and Cummings (2012) for details) was taken out of RNAlater and placed immediately in Trizol (Life Technologies, Grand Island, NY). RNA was extracted according to manufacturer instructions. Equal concentrations of total RNA from each of the 11 samples were pooled into one sample from which poly-adenylated RNA was isolated using the Poly(A) purist kit (Life Technologies) and manufacturer instructions. Lack of contaminating rRNA was confirmed using an Agilent 2100 Bioanalyzer. Strand-specific libraries for 100-bp paired-end sequencing were prepared and sequenced on the Illumina HiSeq 2000 according to manufacturer library kit instructions. A total of 83,168,029 reads were obtained. We preprocessed the reads using Trimmomatic (Bolger et al. 2014) by removing adapter sequence and sequence artifacts as well as trimming low-quality nucleotides based on the Phred score (Ewing et al. 1998) greater than 20, which corresponds to a 1% sequencing error rate.
De novo sequence assembly was then completed using Trinity (Grabherr et al. 2011) on the Odyssey cluster supported by the FAS Science Division Research Computing Group at Harvard University. We remapped reads to the raw assembly using BWA (Li and Durbin 2009) and then used eXpress (http://bio.math.berkeley.edu/eXpress/) to generate FPKM (fragments per kilobase per million mapped) scores for each contig. Low-confidence contigs that had an FPKM value of less than one were removed from the draft assembly. After removing contigs with low confidence, we used CD-HIT-EST (Li and Godzik 2006) to remove contig redundancy. Given that redundant contigs can represent alternative splice variants, polymorphisms among the pooled individuals, or sequencing errors, we used a conservative threshold of 98% sequence similarity. The final assembly was annotated using Trinotate (Bryant et al. 2017); it was found to contain only three opsin genes: RH1, LWS, and SWS1. The SWS2 gene was absent. Thus, we hypothesized that SWS2 might have been lost in the ancestor of dendrobatids.
Genomics-based approaches included a synteny analysis with LWS and mining of publicly available dendrobatid genomes. For the first approach, we predicted that remnants of SWS2 might be detectable in genomic data as a pseudogene either near its expected location or translocated elsewhere in the genome. We thus reviewed the genomes of O. pumilio (Rogers et al. 2018; Rodríguez et al. 2020) and R. imitator (Stuckert et al. 2021) for evidence of SWS2 using synteny and genome mining. We searched for the syntenic block containing LWS using Genomicus v.100.01 (Nguyen et al. 2018) with search term opn1lw and Xenopus tropicalis as the focal species. To visualize gene order in the syntenic block, we used BLAST to locate SWS2, LWS, and the gene predicted to be directly upstream of LWS, MECP2 (methyl-CpG binding transcription factor) in N. parkeri, and X. tropicalis genomes. We hypothesized that a degraded ancestral SWS2 sequence might be maintained in O. pumilio and R. imitator upstream of LWS. We mined the published genome assembly of O. pumilio (Rogers et al. 2018) and a rescaffolded version of this genome (Rodríguez et al. 2020), as well as the recently published R. imitator genome for LWS using BLAST v2.10.0 (Altschul et al. 1990). We then attempted to align the scaffolds containing LWS against SWS2 sequences from other amphibians using MAFFT v7.453 (Katoh and Standley 2013). We note that the O. pumilio scaffold contains two sections of Ns upstream of LWS (2093 and 2391 nucleotides long), which may inflate our perceived length of coverage of this chromosome. Likewise, it is possible that this scaffold might be misassembled and require further refinement and deeper coverage. The rescaffolded version (Rodríguez et al. 2020) of the O. pumilio scaffold containing LWS is identical to the original and thus did not alter our conclusions. As a positive control for this analysis, we also ran our pipeline on the draft genome of R. marina, which is known to contain SWS2 based on our data. This analysis is implemented in the Dryad folder named “synteny” (see supplementary Data S4, Supplementary Material online).
Lastly, we also mined two dendrobatid genomes for SWS2. Because we were unable to detect SWS2 upstream of the O. pumilio and R. imitator LWS scaffolds, we then hypothesized that the syntenic relationship between LWS and SWS2 might have been broken in O. pumilio and R. imitator. Therefore, we used tblastn v2.10.0 to screen all scaffolds from the rescaffolded genome assembly of O. pumilio and the draft assembly of R. imitator for any potential SWS2 orthologs. This approach uncovered several candidate sequences with low E-values (<1e−10). We screened the top candidates using tblastn against Nanorana parkeri, a well-annotated frog genome more closely related to dendrobatids than Xenopus. None of the candidate sequences returned SWS2 as the most likely ortholog; the highest scoring sequences were annotated as related proteins (e.g., pinopsin, rhodopsin; supplementary table S5, Supplementary Material online). As a positive control for this analysis, we also ran tblastn on 43 known SWS2 sequences from other frog species against Nanorana parkeri. As SWS2 is known to be present in Nanorana, we would expect our pipeline to correctly detect and annotate these sequences. We found that the top hit from N. parkeri correctly identified SWS2 as the most likely ortholog for all 43 known SWS2 sequences (see supplementary table S5, Supplementary Material online). As another positive control, we ran an identical search for LWS, which identified LWS as the top hit for 101 other frog sequences and for one of the O. pumilio scaffolds (see supplementary table S5, Supplementary Material online). Lastly, we ran the reciprocal best-hit pipeline for SWS2 using the R. marina genome as the query. Since R. marina is known to contain SWS2, we would expect that our pipeline should identify SWS2 in the R. marina genome. We found that the pipeline identifies the same contig in R. marina that is identified in our synteny-based analysis (ONZH01019223.1) as a likely SWS2 ortholog, while other sequences with low E-values were annotated with other related opsin genes (see supplementary table S5, Supplementary Material online). This analysis is implemented in the Dryad folders named “SWS2_search” and “LWS_search” (see supplementary Data S4, Supplementary Material online).
Supplementary Material
Acknowledgments
This research was funded by NSF DEB-1556967 to D.C.C.; NSF GROW, NSF DDIG (DEB-1404409), National Geographic Society 2014 Young Explorer Grant, and UC Berkeley start-up to RDT; NIH 5P50GM068763, NSF IOS-1822025, and William F. Milton Fund from Harvard Medical School to L.A.O.; L.A.O. is a New York Stem Cell Foundation—Robertson Investigator. J.C.S. was supported by SJU start-up funds and NSF DEB-2016372. M.J.N. was supported by the American Association of University Women. We thank Roberto Ibañez and Brian Gratwicke (Amphibian Rescue and Conservation Project), Vicky Poole, and Kevin Barrett (The Maryland Zoo), Luis A. Coloma (Fundación Otonga and Centro Jambatu de Investigación y Conservación de Anfibios; Quito, Ecuador), César Aguilar (Universidad Nacional Mayor de San Marcos; Lima, Peru), and Travis LaDuc (Biodiversity Center, UT Austin) for facilitating access to tissues and specimens. We thank two anonymous reviewers for critiques that improved the quality of this article. We also acknowledge and express gratitude for the lives of the animals involved in this study. The publication of this article was made possible in part by support from the Berkeley Research Impact Initiative (BRII) sponsored by the UC Berkeley Library.
Contributor Information
Yin Chen Wan, Museum of Vertebrate Zoology and Department of Integrative Biology, University of California Berkeley, Berkeley, CA, USA; Department of Molecular Genetics, University of Toronto, Toronto, Ontario, Canada; School of Biomolecular and Biomedical Science, University College Dublin, Belfield, Dublin, Ireland.
María José Navarrete Méndez, Museum of Vertebrate Zoology and Department of Integrative Biology, University of California Berkeley, Berkeley, CA, USA; Museo de Zoología, Facultad de Ciencias Exactas y Naturales, Pontificia Universidad Católica del Ecuador, Quito, Ecuador.
Lauren A O'Connell, Department of Biology, Stanford University, Palo Alto, CA, USA.
Lawrence H Uricchio, Museum of Vertebrate Zoology and Department of Integrative Biology, University of California Berkeley, Berkeley, CA, USA; Department of Biology, Tufts University, Medford, MA, USA.
Alexandre-Benoit Roland, FAS Center for Systems Biology, Harvard University, Cambridge, MA, USA; Research Centre on Animal Cognition (CRCA), Centre for Integrative Biology (CBI), UMR5169 CNRS, Toulouse University, Toulouse, France.
Martine E Maan, Groningen Institute for Evolutionary Life Sciences, University of Groningen, Groningen, The Netherlands.
Santiago R Ron, Museo de Zoología, Facultad de Ciencias Exactas y Naturales, Pontificia Universidad Católica del Ecuador, Quito, Ecuador.
Mileidy Betancourth-Cundar, Departamento de Ciencias Biológicas, Universidad de los Andes, Bogotá, Colombia.
Marcio R Pie, Department of Zoology, Universidade Federal do Paraná, Curitiba, Brazil; Biology Department, Edge Hill University, Ormskirk, United Kingdom.
Kimberly A Howell, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA, USA.
Corinne L Richards-Zawacki, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA, USA.
Molly E Cummings, Department of Integrative Biology, University of Texas at Austin, Austin, TX, USA.
David C Cannatella, Department of Integrative Biology, University of Texas at Austin, Austin, TX, USA; Biodiversity Center, University of Texas at Austin, Austin, TX, USA.
Juan C Santos, Department of Biological Sciences, St. John's University, New York City, NY, USA.
Rebecca D Tarvin, Museum of Vertebrate Zoology and Department of Integrative Biology, University of California Berkeley, Berkeley, CA, USA.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
R.D.T. and J.C.S. conceptualized the study; Y.C.W., R.D.T., J.C.S., L.H.U., L.A.O., M.J.N., A.R., and M.E.C. collected data; Y.C.W., J.C.S., L.H.U., and R.D.T. performed analyses; D.C.C., L.A.O., R.D.T., M.E.C., and J.C.S. acquired funding; R.D.T., J.C.S., M.B.C., and M.E.M. conducted fieldwork; M.R.P., M.E.C., M.E.M., C.L.R.-Z., S.R.R., M.B.C., R.D.T., and J.C.S. contributed samples and helped with permits; Y.C.W., M.J.N., L.H.U., J.C.S., L.H., and R.D.T. prepared the original draft; R.D.T. coordinated the project; R.D.T. and J.C.S. supervised the study; all authors reviewed and revised the manuscript.
Data Availability
Raw Illumina sequencing data are deposited in the Sequence Read Archive (BioProject PRJNA856227). Specimens are available in QCAZ at Pontificia Universidad Católica del Ecuador (Quito, Ecuador), Centro Jambatu de Investigación y Conservación de Anfibios (Quito, Ecuador), Museo de Historia Natural de la Universidad Nacional Mayor de San Marcos (MUSM, Lima, Peru), Coleção de Herpetologia, Departamento de Zoologia, Universidade Federal do Paraná, (Paraná, Brazil), or Museo de Historia Natural ANDES at the Universidad de los Andes. All scripts and other data are available in the supporting material on DataDryad (DOI: 10.5061/dryad.zw3r2289j).
References
- Akiyama T, Uchiyama H, Yajima S, Arikawa K, Terai Y. 2022. Parallel evolution of opsin visual pigments in hawkmoths by tuning of spectral sensitivities during transition from a nocturnal to a diurnal ecology. J Exp Biol. 225:jeb244541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- AmphibiaWeb . 2022. <https://amphibiaweb.org> University of California, Berkeley, CA, USA. Accessed 17 May 2022.
- Anderson SR, Wiens JJ. 2017. Out of the dark: 350 million years of conservatism and evolution in diel activity patterns in vertebrates. Evolution. 71:1944–1959. [DOI] [PubMed] [Google Scholar]
- Andrés A, Kosoy A, Garriga P, Manyosa J. 2001. Mutations at position 125 in transmembrane helix III of rhodopsin affect the structure and signalling of the receptor. Eur J Biochem. 268:5696–5704. [DOI] [PubMed] [Google Scholar]
- Andrews SM. 2010. FastQC: a quality control tool for high throughput sequence data. Available from:http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Asenjo AB, Rim J, Oprian DD. 1994. Molecular determinants of human red/green color discrimination. Neuron. 12:1131–1138. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum DA, Smith SD. 2013. Tree thinking: an introduction to phylogenetic biology. Greenwood village. CO: Roberts. [Google Scholar]
- Bell RC, Zamudio KR. 2012. Sexual dichromatism in frogs: natural selection, sexual selection and unexpected diversity. Proc R Soc B: Biol Sci. 279:4687–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges R, Khan I, Johnson WE, Gilbert MTP, Zhang G, Jarvis ED, O’Brien SJ, Antunes A. 2015. Gene loss, adaptive evolution and the co-evolution of plumage coloration genes with opsins in birds. BMC Genomics. 16:751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowmaker JK. 2008. Evolution of vertebrate visual pigments. Vision Res. 48:2022–2041. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, Lee TJ, Leigh ND, Kuo T-H, Davis FG, et al. 2017. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 18:762–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton KL, Parry JWL, Bowmaker JK, Hunt DM, Seehausen O. 2005. Colour vision and speciation in Lake Victoria cichlids of the genus Pundamilia. Mol Ecol. 14:4341–4353. [DOI] [PubMed] [Google Scholar]
- Carvajal-Castro JD, Vargas-Salinas F, Casas-Cardona S, Rojas B, Santos JC. 2021. Aposematism facilitates the diversification of parental care strategies in poison frogs. Sci Rep. 11:19047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho LS, Cowing JA, Wilkie SE, Bowmaker JK, Hunt DM. 2007. The molecular evolution of avian ultraviolet- and violet-sensitive visual pigments. Mol Biol Evol. 24:1843–1852. [DOI] [PubMed] [Google Scholar]
- Castiglione GM, Chang BS. 2018. Functional trade-offs and environmental variation shaped ancient trajectories in the evolution of dim-light vision. Elife. 7:e35957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T, Lee M, Sakmar TP. 1992. Introduction of hydroxyl-bearing amino acids causes bathochromic spectral shifts in rhodopsin. Amino acid substitutions responsible for red-green color pigment spectral tuning. J Biol Chem. 267:9478–9480. [PubMed] [Google Scholar]
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65:997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough M, Summers K. 2000. Phylogenetic systematics and biogeography of the poison frogs: evidence from mitochondrial DNA sequences. Biol J Linn Soc. 70:515–540. [Google Scholar]
- Cummings ME, Crothers LR. 2013. Interacting selection diversifies warning signals in a polytypic frog: an examination with the strawberry poison frog. Evol Ecol. 27:693–710. [Google Scholar]
- Donner K, Yovanovich CAM. 2020. A frog's eye view: foundational revelations and future promises. Semin Cell Dev Biol. 106:72–85. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier LD, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185. [DOI] [PubMed] [Google Scholar]
- Fasick JI, Robinson PR. 1998. Mechanism of spectral tuning in the dolphin visual pigments. Biochemistry. 37:433–438. [DOI] [PubMed] [Google Scholar]
- Feng Y-J, Blackburn DC, Liang D, Hillis DM, Wake DB, Cannatella DC, Zhang P. 2017. Phylogenomics reveals rapid, simultaneous diversification of three major clades of gondwanan frogs at the Cretaceous–Paleogene boundary. Proc Natl Acad Sci U S A. 114:E5864–E5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JJ. 2014. The ecological approach to visual perception: classic edition. New York: Psychology Press. [Google Scholar]
- Goutte S, Mason MJ, Antoniazzi MM, Jared C, Merle D, Cazes L, Toledo LF, El-Hafci H, Pallu S, Portier H, et al. 2019. Intense bone fluorescence reveals hidden patterns in pumpkin toadlets. Sci Rep. 9:5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Frost DR, Caldwell JP, Gagliardo R, Haddad CFB, Kok PJR, Means DB, Noonan BP, Schargel WE, Wheeler WC. 2006. Phylogenetic systematics of dart-poison frogs and their relatives (Amphibia: Athesphatanura: Dendrobatidae). Bull Am Mus Nat Hist. 299:1–262. [Google Scholar]
- Grant T, Rada M, Anganoy-Criollo M, Batista A, Dias PH, Jeckel AM, Machado DJ, Rueda-Almonacid JV. 2017. Phylogenetic systematics of dart-poison frogs and their relatives revisited (Anura: Dendrobatoidea). South Am J Herpetol. 12:S1–S90. [Google Scholar]
- Greener MS, Hutton E, Pollock CJ, Wilson A, Lam CY, Nokhbatolfoghahai M, Jowers MJ, Downie JR. 2020. Sexual dichromatism in the neotropical genus Mannophryne (Anura: Aromobatidae). PLoS One. 15:e0223080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Chi H, Zhang L, Song S, Rossiter SJ, Liu Y. 2023. Convergent evolutionary shifts in rhodopsin retinal release explain shared opsin repertoires in monotremes and crocodilians. Proc R Soc B. 290:20230530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez EA, Castiglione GM, Morrow JM, Schott RK, Loureiro LO, Lim BK, Chang BSW. 2018. Functional shifts in bat dim-light visual pigment are associated with differing echolocation abilities and reveal molecular adaptation to photic-limited environments. Mol Biol Evol. 35:2422–2434. [DOI] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hárosi FI. 1982. Recent results from single-cell microspectrophotometry: cone pigments in frog, fish, and monkey. Color Res Appl. 7:135–141. [Google Scholar]
- Hauser FE, Ilves KL, Schott RK, Castiglione GM, López-Fernández H, Chang BSW. 2017. Accelerated evolution and functional divergence of the dim light visual pigment accompanies cichlid colonization of Central America. Mol Biol Evol. 34:2650–2664. [DOI] [PubMed] [Google Scholar]
- Hauzman E, Bonci DMO, Suárez-Villota EY, Neitz M, Ventura DF. 2017. Daily activity patterns influence retinal morphology, signatures of selection, and spectral tuning of opsin genes in colubrid snakes. BMC Evol Biol. 17:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hime PM, Lemmon AR, Lemmon ECM, Prendini E, Brown JM, Thomson RC, Kratovil JD, Noonan BP, Pyron RA, Peloso PLV, et al. 2021. Phylogenomics reveals ancient gene tree discordance in the amphibian tree of life. Syst Biol. 70:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu C, Radlwimmer FB, Yokoyama S, Kawamura S. 2004. Mutagenesis and reconstitution of middle-to-long-wave-sensitive visual pigments of new world monkeys for testing the tuning effect of residues at sites 229 and 233. Vision Res. 44:2225–2231. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann CM, Carleton KL. 2009. Gene duplication and differential gene expression play an important role in the diversification of visual pigments in fish. Integr Comp Biol. 6:630–643. [DOI] [PubMed] [Google Scholar]
- Holding ML, Margres MJ, Mason AJ, Parkinson CL, Rokyta DR. 2018. Evaluating the performance of de novo assembly methods for venom-gland transcriptomics. Toxins (Basel). 10:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DM, Carvalho LS, Cowing JA, Parry JWL, Wilkie SE, Davies WL, Bowmaker JK. 2007. Spectral tuning of shortwave-sensitive visual pigments in vertebrates. Photochem Photobiol. 83:303–310. [DOI] [PubMed] [Google Scholar]
- Hunt DM, Collin SP. 2014. The evolution of photoreceptors and visual photopigments in vertebrates. In: Hunt DMHankins MWCollin SP and Marshall NJ, editors. Evolution of visual and non-visual pigments. Boston (MA): Springer. p. 163–217. [Google Scholar]
- Hunt DM, Dulai KS, Cowing JA, Julliot C, Mollon JD, Bowmaker JK, Li WH, Hewett-Emmett D. 1998. Molecular evolution of trichromacy in primates. Vision Res. 38:3299–3306. [DOI] [PubMed] [Google Scholar]
- Jacobs GH. 2013. Losses of functional opsin genes, short-wavelength cone photopigments, and color vision—a significant trend in the evolution of mammalian vision. Vis Neurosci. 30:39–53. [DOI] [PubMed] [Google Scholar]
- Jetz W, Pyron RA. 2018. The interplay of past diversification and evolutionary isolation with present imperilment across the amphibian tree of life. Nat Ecol Evol. 2:850–858. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 22:1208–1222. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. 2005. Hyphy: hypothesis testing using phylogenies. Bioinformatics. 21:676–679. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Wisotsky SR, Escalante A, Magalis BR, Weaver S. 2021. Contrast-FEL—a test for differences in selective pressures at individual sites among clades and sets of branches. Mol Biol Evol. 38:1184–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskelainen A, Hemilä S, Donner K. 1994. Spectral sensitivities of short- and long-wavelength sensitive cone mechanisms in the frog retina. Acta Physiol Scand. 152:115–124. [DOI] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35:4453–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JP, Rojas B, Fouquet A, Mappes J, Blanchette A, Saporito RA, Bosque RJ, Courtois EA, Noonan BP. 2019. Weak warning signals can persist in the absence of gene flow. Proc Natl Acad Sci U S A. 116:19037–19045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 31:1674–1676. [DOI] [PubMed] [Google Scholar]
- Liebman PA, Entine G. 1968. Visual pigments of frog and tadpole (Rana pipiens). Vision Res. 8:761–775. [DOI] [PubMed] [Google Scholar]
- Lin SW, Kochendoerfer GG, Carroll KS, Wang D, Mathies RA, Sakmar TP. 1998. Mechanisms of spectral tuning in blue cone visual pigments: visible and Raman spectroscopy of blue-shifted rhodopsin mutants. J Biol Chem. 273:24583–24591. [DOI] [PubMed] [Google Scholar]
- Lindquist ED, Hetherington TE. 1998. Semaphoring in an earless frog: the origin of a novel visual signal. Anim Cogn. 1:83–87. [DOI] [PubMed] [Google Scholar]
- Loeffler-Henry K, Kang C, Sherratt TN. 2023. Evolutionary transitions from camouflage to aposematism: hidden signals play a pivotal role. Science. 379:1136–1140. [DOI] [PubMed] [Google Scholar]
- Maan ME, Cummings ME. 2008. Female preferences for aposematic signal components in a polymorphic poison frog. Evolution. 62:2334–2345. [DOI] [PubMed] [Google Scholar]
- Maan ME, Cummings ME. 2009. Sexual dimorphism and directional sexual selection on aposematic signals in a poison frog. Proc Natl Acad Sci U S A. 106:19072–19077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maan ME, Cummings ME. 2012. Poison frog colors are honest signals of toxicity, particularly for bird predators. Am Nat. 179:E1–E14. [DOI] [PubMed] [Google Scholar]
- Mappes J, Marples NM, Endler JA. 2005. The complex business of survival by aposematism. Trends Ecol Evol. 20:598–603. [DOI] [PubMed] [Google Scholar]
- Medina I, Wang IJ, Salazar C, Amézquita A. 2013. Hybridization promotes color polymorphism in the aposematic harlequin poison frog, Oophaga histrionica. Ecol Evol. 3:4388–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin AD, Fedigan LM, Hiramatsu C, Hiwatashi T, Parr N, Kawamura S. 2009. Fig foraging by dichromatic and trichromatic Cebus capucinus in a tropical dry forest. Int J Primatol. 30:753–775. [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37:1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohun SM, Davies WIL. 2019. The evolution of amphibian photoreception. Front Ecol Evol. 7:321. [Google Scholar]
- Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: a fast, unconstrained Bayesian AppRoximation for inferring selection. Mol Biol Evol. 30:1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8:e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness DS. 1986. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 232:196–202. [DOI] [PubMed] [Google Scholar]
- Nguyen NTT, Vincens P, Roest Crollius H, Louis A. 2018. Genomicus 2018: karyotype evolutionary trees and on-the-fly synteny computing. Nucleic Acids Res. 46:D816–D822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano T, Kojima D, Fukada Y, Shichida Y, Yoshizawa T. 1992. Primary structures of chicken cone visual pigments: vertebrate rhodopsins have evolved out of cone visual pigments. Proc Natl Acad Sci U S A. 89:5932–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio D, Vorobyev M. 2008. A review of the evolution of animal colour vision and visual communication signals. Vision Res. 48:2042–2051. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. 2000. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 289:739–745. [DOI] [PubMed] [Google Scholar]
- Paradis E, Schliep K. 2019. . Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 35:526–528. [DOI] [PubMed] [Google Scholar]
- Pombal JP, Sazima I, Haddad CF. 1994. Breeding behavior of the pumpkin toadlet, Brachycephalus ephippium (Brachycephalidae). J Herpetol. 28:516–519. [Google Scholar]
- Pyron RA. 2014. Biogeographic analysis reveals ancient continental vicariance and recent oceanic dispersal in amphibians. Syst Biol. 63:779–797. [DOI] [PubMed] [Google Scholar]
- Rebouças R, Carollo AB, Freitas MDO, Lambertini C, Dos Santos RMN, Toledo LF. 2019. Is the conspicuous dorsal coloration of the Atlantic forest pumpkin toadlets aposematic? Salamandra. 55:39–47. [Google Scholar]
- Reynolds RG, Fitzpatrick BM. 2007. Assortative mating in poison-dart frogs based on an ecologically important trait. Evolution. 61:2253–2259. [DOI] [PubMed] [Google Scholar]
- Richards-Zawacki CL, Cummings ME. 2011. Intraspecific reproductive character displacement in a polymorphic poison dart frog, Dendrobates pumilio. Evolution. 65:259–267. [DOI] [PubMed] [Google Scholar]
- Rodríguez A, Mundy NI, Ibáñez R, Pröhl H. 2020. Being red, blue and green: the genetic basis of coloration differences in the strawberry poison frog (Oophaga pumilio). BMC Genomics. 21:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RL, Zhou L, Chu C, Márquez R, Corl A, Linderoth T, Freeborn L, MacManes MD, Xiong Z, Zheng J, et al. 2018. Genomic takeover by transposable elements in the strawberry poison frog. Mol Biol Evol. 35:2913–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Reich D. 2012. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22:939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas B, Burdfield-Steel E, De Pasqual C, Gordon S, Hernández L, Mappes J, Nokelainen O, Rönkä K, Lindstedt C. 2018. Multimodal aposematic signals and their emerging role in mate attraction. Front Ecol Evol. 6:93. [Google Scholar]
- Rojas B, Devillechabrolle J, Endler JA. 2014. Paradox lost: variable colour-pattern geometry is associated with differences in movement in aposematic frogs. Biol Lett. 10:20140193. [Google Scholar]
- Rößler DC, Lötters S, Mappes J, Valkonen JK, Menin M, Lima AP, Pröhl H. 2019. Sole coloration as an unusual aposematic signal in a Neotropical toad. Sci Rep. 9:1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JC, Cannatella DC. 2011. Phenotypic integration emerges from aposematism and scale in poison frogs. Proc Natl Acad Sci U S A. 108:6175–6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JC, Coloma LA, Canatella DC. 2003. Multiple, recurring origins of aposematism and diet specialization in poison frogs. Proc Natl Acad Sci U S A. 100:12792–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JC, Coloma LA, Summers K, Caldwell JP, Ree R, Cannatella DC. 2009. Amazonian amphibian diversity is primarily derived from late Miocene Andean lineages. PLoS Biol. 7:0448–0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RR, Grant T. 2011. Diel pattern of migration in a poisonous toad from Brazil and the evolution of chemical defenses in diurnal amphibians. Evol Ecol. 25:249–258. [Google Scholar]
- Santos JC, Tarvin RD, O’Connell LA, Blackburn DC, Coloma LA. 2018. Diversity within diversity: parasite species richness in poison frogs assessed by transcriptomics. Mol Phylogenet Evol. 125:40–50. [DOI] [PubMed] [Google Scholar]
- Schott RK, Müller J, Yang CGY, Bhattacharyya N, Chan N, Xu M, Morrow JM, Ghenu AH, Loew ER, Tropepe V, et al. 2016. Evolutionary transformation of rod photoreceptors in the all-cone retina of a diurnal garter snake. Proc Natl Acad Sci U S A. 113:356–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott RK, Perez L, Kwiatkowski MA, Imhoff V, Gumm JM. 2022. Evolutionary analyses of visual opsin genes in frogs and toads: diversity, duplication, and positive selection. Ecol Evol. 12:e8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt TN. 2008. The evolution of Müllerian mimicry. Naturwissenschaften. 95:681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi A, Cronin TW, Loew ER, Vorobyev M, Summers K. 2004. Interspecific and intraspecific views of color signals in the strawberry poison frog Dendrobates pumilio. J Exp Biol. 207:2471–2485. [DOI] [PubMed] [Google Scholar]
- Spielman SJ, Weaver S, Shank SD, Magalis BR, Li M, Kosakovsky Pond SL. 2019. Evolution of viral genomes: interplay between selection, recombination, and other forces. Evol Genomics Stat Comput Methods. 1910:427–468. [DOI] [PubMed] [Google Scholar]
- Stuckert AMM, Chouteau M, McClure M, LaPolice TM, Linderoth T, Nielsen R, Summers K, MacManes MD. 2021. The genomics of mimicry: gene expression throughout development provides insights into convergent and divergent phenotypes in a Müllerian mimicry system. Mol Ecol. 30:4039–4061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Summers K, Symula R, Clough M, Cronin T. 1999. Visual mate choice in poison frogs. Proc Biol Sci. 266:2141–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surridge AK, Osorio D, Mundy NI. 2003. Evolution and selection of trichromatic vision in primates. Trends Ecol Evol. 18:198–205. [Google Scholar]
- Thomas KN, Gower DJ, Streicher JW, Bell RC, Fujita MK, Schott RK, Liedtke HC, Haddad CFB, Becker CG, Cox CL, et al. 2022. Ecology drives patterns of spectral transmission in the ocular lenses of frogs and salamanders. Funct Ecol. 36:850–864. [Google Scholar]
- Toledo LF, Haddad CFB. 2009. Colors and some morphological traits as defensive mechanisms in anurans. Int J Zool. 2009:910892. [Google Scholar]
- Toomey MB, Corbo JC. 2017. Evolution, development and function of vertebrate cone oil droplets. Front Neural Circuits. 11:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IJ. 2011. Inversely related aposematic traits: reduced conspicuousness evolves with increased toxicity in a polymorphic poison-dart frog. Evolution. 65:1637–1649. [DOI] [PubMed] [Google Scholar]
- Warrant EJ, Johnsen S. 2013. Vision and the light environment. Curr Biol. 23:R990–R994. [DOI] [PubMed] [Google Scholar]
- Womack MC, Steigerwald E, Blackburn D, Cannatella DC, Catenazzi A, Che J, Koo MS, McGuire JA, Ron SR, Spencer C, Vredenburg VT, Tarvin RD. 2022. State of the Amphibia 2020: A review of five years of amphibian research and existing resources. Ichthyol Herpetol. 110:638–661. [Google Scholar]
- Xu Y, Lei Y, Su Z, Zhao M, Zhang J, Shen G, Wang L, Li J, Qi J, Wu J. 2021. A chromosome-scale Gastrodia elata genome and large-scale comparative genomic analysis indicate convergent evolution by gene loss in mycoheterotrophic and parasitic plants. Plant J. 108:1609–1623. [DOI] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl BioSci. 13:555–556. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol and Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Y, Dugas MB, Sudekum HJ, Murphy SN, Richards-Zawacki CL. 2018. Male-male aggression is unlikely to stabilize a poison frog polymorphism. J Evol Biol. 31:457–468. [DOI] [PubMed] [Google Scholar]
- Yang Y, Richards-Zawacki CL. 2020. Male–male contest limits the expression of assortative mate preferences in a polymorphic poison frog. Behav Ecol. 32:151–158. [Google Scholar]
- Yang Y, Richards-Zawacki CL, Devar A, Dugas MB. 2016. Poison frog color morphs express assortative mate preferences in allopatry but not sympatry. Evolution. 70:2778–2788. [DOI] [PubMed] [Google Scholar]
- Yang Y, Servedio MR, Richards-Zawacki CL. 2019. Imprinting sets the stage for speciation. Nature. 574:99–102. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB. 2001. The molecular genetics and evolution of red and green color vision in vertebrates. Genetics. 158:1697–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Starmer WT, Takahashi Y, Tada T. 2006. Tertiary structure and spectral tuning of UV and violet pigments in vertebrates. Gene. 365:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Yokoyama S. 1990. Convergent evolution of the red- and green-like visual pigment genes in fish, Astyanax fasciatus, and human. Proc Natl Acad Sci U S A. 87:9315–9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Zhang H, Radlwimmer FB, Blow NS. 1999. Adaptive evolution of color vision of the Comoran coelacanth (Latimeria chalumnae). Proc Natl Acad Sci U S A. 96:6279–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yovanovich CAM, Koskela SM, Nevala N, Kondrashev SL, Kelber A, Donner K. 2017. The dual rod system of amphibians supports colour discrimination at the absolute visual threshold. Philos Trans R Soc Lond B Biol Sci. 372:20160066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yovanovich CAM, Pierotti MER, Kelber A, Jorgewich-Cohen G, Ibáñez R, Grant T. 2020. Lens transmittance shapes ultraviolet sensitivity in the eyes of frogs from diverse ecological and phylogenetic backgrounds. Proc Biol Sci. 287:20192253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw Illumina sequencing data are deposited in the Sequence Read Archive (BioProject PRJNA856227). Specimens are available in QCAZ at Pontificia Universidad Católica del Ecuador (Quito, Ecuador), Centro Jambatu de Investigación y Conservación de Anfibios (Quito, Ecuador), Museo de Historia Natural de la Universidad Nacional Mayor de San Marcos (MUSM, Lima, Peru), Coleção de Herpetologia, Departamento de Zoologia, Universidade Federal do Paraná, (Paraná, Brazil), or Museo de Historia Natural ANDES at the Universidad de los Andes. All scripts and other data are available in the supporting material on DataDryad (DOI: 10.5061/dryad.zw3r2289j).