Abstract
The high-fertility Meishan pig is currently categorized into medium sized (MMS) and small sized (SMS) based on body size. To identify causal genes responsible for the variation in body size within the two categories, we sequenced individuals representing the entire consanguinity of the existing Meishan pig. This enabled us to conduct genome selective signal analysis. Our findings revealed the genomes of MMS and SMS are stratified, with selective sweep regions formed by differential genomic intervals between the two categories enriched in multiple pig body size related quantitative trait loci (QTLs). Furthermore, the missense mutation c.575T > C of candidate causal gene NR6A1, accounting for the variation in lumbar vertebrae number in pigs, was positively selected in MMS only, leading to an increase in body length of MMS at 6 months of age. To precisely identify causal genes accounting for body size variation through multi-omics, we collected femoral cartilage and liver transcription data from MMS and SMS respectively, and re-sequencing data from pig breeds exhibiting varying body sizes. We found that two selected regions where the RSAD2-CMPK2 and COL3A1 genes are located, respectively, showed different haplotypes in pig breeds of varying body size, and was associated with body or carcass length in hybridized Suhuai pig. Additionally, the above three hub genes, were significantly greater expressed in SMS femoral cartilage and liver tissues compared to MMS. These three genes could strengthen the pathways related to bone resorption and metabolism in SMS, potentially hindering bone and skeletal development and resulting in a smaller body size in SMS. These findings provide valuable insights into the genetic mechanism of body size variation in Meishan pig population.
Keywords: body size, bone resorption and digestion, COL3A1, Meishan pig, NR6A1, RSAD2-CMPK2
This study was conducted on Meishan pigs, an internationally renowned pig breed with significant body size variation within the population, to explore the candidate genes and functional pathways that contribute to pig body size variation. The findings of this study, such as selected genes NR6A1, RSAD2-CMPK2 and COL3A1, provide insights into the genetic mechanisms underlying body size variation in pigs. Additionally, the results could serve as a reference for body size breeding not only in Meishan pigs but also in other pig breeds derived from Meishan pig lineage.
Introduction
Meishan pig is one of the famous indigenous pig breeds in Taihu Lake region of China. It is characterized by high fertility, resistant to roughage, and good meat quality (Haley and Lee, 1993; Hunter et al., 1996). The breed has been categorized into large sized, medium sized (MMS), and small sized (SMS) based on their body size. Unfortunately, the large-sized Meishan pig is now extinct, and the existing Meishan pig can be divided into MMS and SMS based on body size variation (China National Commission of Animal Genetic Resources, 2011). It is notable that MMS is relatively large (Supplementary Table S1) in all body size traits. This body size variation is the result of long-term selection by local residents. However, there is a lack of relevant reports on the identification of causal genes underlying this body size variation. Sun et al. (2018) identified some important candidate genes, such as SPDEF and PACSIN1, through Fst analysis. This provides useful information on how to identify candidate genes for body size variation in different sub-populations of Meishan pig. Additionally, body size in pigs is a highly heritable trait. In addition to many genes with small effects, there can also be genes with large effects, such as PLAG1 (Karim et al., 2011), NR6A1 (Mikawa et al., 2007), and IGF2 (Van et al., 2003), laying the foundation for understanding the inheritance pattern of animal body size traits.
Under long-term artificial selection, Meishan pigs have formed two sub-populations that differ in body size, but otherwise have highly similar appearance and performance traits (Supplementary Table S1) (China National Commission of Animal Genetic Resources, 2011). By conducting analyses such as selected signal in the above two sub-populations, it is possible to effectively identify the causal genes responsible for the variation in body size and efficiently decrease the probability of false positive signals. Using Fst analysis based on chip data (Liu et al., 2020), we have identified candidate genes such as BMP2, which has demonstrated significantly differential signals between MMS and SMS. However, the low variant density has made it challenging to identify the causal genes. To address this issue, we collected individuals from all lineages of MMS and SMS for re-sequencing, and integrated this with a re-sequencing dataset from diverse pig breeds with varying body sizes for further population and quantitative genetics analysis. Moreover, the information from genomics and transcriptomics were integrated, which would systematically facilitate elucidating the genetic mechanisms and key genes underlying the variation in body size between MMS and SMS.
In fact, the integration of multi-omics has become increasingly prevalent in animal research for identifying the causal genes that govern body size traits. For instance, Wouter et al. utilized genomic and transcriptomic methods to identify PLAG1 as a factor influencing bovine stature (Karim et al., 2011). Additionally, Wang et al. (2020) utilized genomic, transcriptomic, and epigenomic data to pinpoint multiple regulatory mutations cumulatively contributing to chicken growth traits. As cartilage and liver are among the organs most closely associated with body size (Karim et al., 2011; Zhou et al., 2018), the transcriptomes of these tissues in MMS and SMS were sequenced to aid in identifying candidate genes with a regulatory role. Finally, by integrating genomic data from different body size pig breeds, femoral cartilage and liver transcriptomic data from MMS and SMS, this study aimed to identify candidate causal genes accounting for body size variation in Meishan pigs.
Materials and Methods
Ethics Statement
All procedures employed in this study and involving animals followed the recommendations of the Regulations for the Administration of Affairs Concerning Experimental Animals of China. The ethics committee of Nanjing Agricultural University approved this study (approval number SYXK-2021-0086).
Animal Population
This study collected re-sequencing data from 139 animals, of which 28 MMS individuals and 29 SMS individuals were sequenced. In addition, a dataset of 82 genome re-sequencing data was downloaded from the public databases of the National Center for Biotechnology Information (NCBI). This dataset includes China indigenous pig breeds with varying body sizes, as well as the western pig breed Large White pig (LW) (Supplementary Tables S2 and S3). When selecting MMS and SMS individuals for sequencing, we first referred to the number of consanguinities of MMS and SMS identified by pig 50 K chip (Liu et al., 2020), and selected 1-2 individuals from each consanguinity to ensure coverage of all existing Meishan pig consanguinities. Among the other indigenous pig breeds in China, the small body size pig breeds were represented by the South China indigenous pig lineage, including Bamaxiang pig (6 individuals), Wuzhishan pig (8 individuals), and Luchuan pig (6 individuals). The large body size pig breeds were represented by the Southwest and North China indigenous pig lineage, including Neijiang pig (6 individuals), Hetaodaer pig (6 individuals), and Min pig (6 individuals). Western commercial pigs were used as an out-group large body size pig breed, specifically the Large White pig (LW) with 44 individuals. We also collected the pig 50 K chip dataset (Liu et al., 2020) of indigenous pig breeds in Taihu Lake region, which included 20 individuals each of Erhualian pig, Mi pig, Sawutou pig, Fengjing pig, Jiaxing black pig, MMS, and SMS (Supplementary Table S3).
Whole-genome Sequencing and SNP Calling
For the 28 MMS and 29 SMS sequenced individuals, DNA was extracted using the chloroform/phenol method. The DNA samples that passed the quality control were sent to Zhejiang Annoroad Biotechnology Co., Ltd. (Annoroad, Hangzhou, China) for library construction and genome re-sequencing using the Illumina NovaSeq PE150 platform (Illumina, San Diego, USA). The sequencing libraries were constructed with 350-bp paired ends. The SNP calling process was implemented in a 3-step protocol:
(1) Downloading the reference sequence
The FASTA file of the pig reference genome sequence (assembly: Sus_scrofa, version: 11.1) was downloaded from the Ensembl database. BWA v0.7.12 (BWA, RRID:SCR_010910) (Li and Durbin, 2009) was used to index reference genes.
(2) Data quality control
For the obtained raw data (including 82 pig individuals downloaded from the public database), the FastQC software (FastQC, RRID:SCR_014583) (Brandine and Smith, 2019) was used for quality control with default parameters.
(3) SNP detection
The paired-end reads were blasted against the reference genome using a MEM algorithm from BWA (Li and Durbin, 2009). The binary BAM files were obtained from SAM files using SAMtools v1.4 (SAMTOOLS, RRID:SCR_002105) (Li, 2011). Then, duplicate marking, base quality recalibration, duplicated reads removal, and mapping statistics (i.e., coverage of depth) were performed by using the Picard v1.119 (PICARD, RRID:SCR_002105), GATK v4.0 (GATK, RRID:SCR_001876) (McKenna et al., 2011), and SAMtools v1.4 (Li, 2011) softwares. The HaplotypeCaller function of GATK v4.0 (McKenna et al., 2011) was implemented for SNP detection and an SNP dataset of the 139 individuals was obtained by using the CombineGVCFs, GenotypeGVCFs, and SelectVariants module. For the original SNP dataset, VCFtools v0.1.13 (VCFtools, RRID:SCR_001235) (Danecek et al., 2011) was used for quality control with the following standards: (i) 3X < mean sequencing depth (over all included individuals), (ii) a minor allele frequency > 0.05 and a max allele frequency < 0.99, (iii) maximum missing rate < 0.1, and (iv) only two alleles.
Population Structure and Genetic Diversity
To ascertain whether MMS and SMS evolved from a common ancestor, we utilized the pig 50 K chip dataset (Liu et al., 2020) comprising indigenous pig breeds in Taihu Lake region for admixture analysis (Alexander et al., 2009). The K value was set to 2 to 7. Subsequently, we employed the re-sequencing dataset to more precisely evaluate the disparities in genetic structure and genetic diversity between the MMS and SMS populations. The quality-controlled SNPs were implemented to assess the population structure between MMS and SMS. To improve operational efficiency, we pruned SNPs with LD, using the pair-wise genotype correlation coefficients (r2) > 0.3 through PLINK v1.9 (Chang et al., 2015). We then transformed the PLINK format file to a MEGA format file and applied the p-distance algorithm with the neighbor-joining tree module to construct a neighbor-joining phylogenetic tree (NJ-tree) using MEGA v6 (MEGA, RRID:SCR_000667) (Tamura et al., 2013). We also repeated this algorithm to construct the NJ-tree for different pig breeds with varying body sizes. We calculated Meishan pig principal component analysis (PCA) by implementing PLINK v1.9 (Chang et al., 2015), using the command “--pca”. The first three principal components were selected.
The dataset of unpruned SNPs was used for multiple sequentially Markovian coalescent (MSMC) and genetic diversity analysis. The MSMC model was implemented in the MSMC software (Schiffels and Durbin, 2014). We set g = 1 and a rate of 1.25 × 10−8 mutations per generation to estimate the distribution of time and plotted the results with an in-house python script.
Expected heterozygosity (HET) was calculated for each individual with the command “--hardy” using PLINK v1.9 (Chang et al., 2015). We obtained the mean expected heterozygosity to represent population heterozygosity level in the overall Meishan pig population, MMS and SMS, respectively. We also applied PLINK v1.9 (Chang et al., 2015) to compute different population runs of homozygosity fragment (ROH), using the command “--homozyg”. The mean ROH of the overall Meishan pig population, MMS and SMS were calculated respectively.
To estimate the degree of linkage disequilibrium (LD) in each population, we used the command “--r2 --ld-window 9999 --ld-window-r2 0 --ldwindow-kb 1000” in PLINK v1.9 (Chang et al., 2015) to calculate pair-wise r2 values. We classified the distance between SNPs into different bins and calculated the average r2 value within each bin, starting with 25 kb and ending with 1,000 kb. Among them, 25 to 100 kb was divided into bins with a step-size of 5 kb, followed by 100 to 400 kb divided into bins with a step-size of 20 kb, 400 to 600 kb divided into bins with a step-size of 40 kb, and a step-size of 100 kb for 600 to 1,000 kb bins.
Selective Sweep Analysis
The genetic differentiation coefficients (Fst), absolute frequency differentiation (AFdiff), and Z-transformed heterozygosity (Zhet) were utilized to identify selective sweep regions. The VCFtools software (Danecek et al., 2011) was implemented to perform Fst analysis to detect differential genomic intervals between SMS and MMS, SMS and small body size pig breeds, SMS and large body size pig breeds in 50-kb windows with a 25-kb step size. For Fst analysis between SMS and MMS, we restricted our candidate selected regions descriptions to the extreme high Fst value windows with a significance level of P < 0.01 (Z test). The adjacent selected regions that were less than 100 kb in size were combined together. For AFdiff analysis, the calculation of AFdiff by following the formula: AFdiff = abs(FMMS − FSMS), where FMMS and FSMS represent the allele frequency of each SNP locus in MMS and SMS, respectively. For Zhet analysis, we first calculated the allele frequency of each SNP locus within a specific population. Then HET (expected heterozygosity) was calculated by the formula: HET = 2p*(1 − p), where p is the reference allele frequency in one specific population. The HET values were normalized into Z-scores (Zhet) by adjusting the values using the mean and standard deviation.
To calculate the region under specific selection for MMS and SMS separately, we first calculated the Zhet value for each SNP locus in a single population. We then calculated the Zhet value for each window in 50-kb windows, with a 25-kb step size. We defined the windows with Z-scores (Zhet) below −2.57 (Z test, P < 0.01) as regions under strong selection. Any adjacent regions less than 100 kb in size were combined together. For the MMS and SMS populations, we intersected the selected regions identified by Zhet with the selected regions identified by Fst analysis to determine population-specific selected regions.
To visualize the different haplotypes in selective sweep regions among diverse pig breeds with varying body size, an IBD sharing approach was utilized to construct haplotypes. Haplotypes were phased using the fastPhase function in Beagle v4.0 (BEAGLE, RRID:SCR_001789). Additionally, the IBD fragments in each individual were detected by using the fastIBD function (Browning and Browning, 2011) under the corresponding analysis population.
Annotation and Function Analysis of Identified Genomic Regions
The PigQTLdb (https://www.animalgenome.org/cgi-bin/QTLdb/SS/index) was used to annotate the potential functions of selective sweep regions. We referred to Hu et al. (2019) to identify whether these selected regions were significantly enriched in pig body size-related QTLs than expected number by Fisher’s exact test. We set the size of the intersection region between selected regions and pig body size-related QTLs to be greater than the average size of selected regions (208 kb) or at least half the size of the QTL itself for a positive annotation. Functional annotated genes involved in selective sweep regions were identified using the annotation for the Sus_scrofa assembly 11.1, accessible through the Ensembl databases (http://www.ensembl.org/index.html). The KEGG and GO databases were used for the gene functional enrichment analyses through the online website KOBAS (Bu et al., 2021). For KEGG and GO terms, multiple tests were performed using FDR method.
Association Analysis of NR6A1 Gene c.575T > C Locus
We conducted an association analysis between the c.575T > C genotype and the body length phenotypes in 165 MMS individuals at 6 months of age. We utilized the online website of SAS software (https://www.sas.com/en_us/home.html) to perform a mixed linear model analysis. The mixed linear model is represented as follows:
In the above formula, Yijklm is the phenotype of MMS 6-month-old body length trait, μ is the overall mean value, SNPi, SEXj, and Year-seaonk are the fixed effects of genotype, sex, and year-season of birth, respectively, COVl refers to age as a covariate, um is the individual random additive genetic effect, and eijklm represents the random residue.
Haplotype Association Analysis
We consulted Chen et al. (2020) to confirm the association between haplotypes in candidate selected regions and the estimated breeding value (EBV) of body length related traits in Suhuai pig. For the candidate selected regions where the RASD2-CMPK2 and COL3A1 genes are located, we randomly selected two significant differential tag SNP loci that met the conditions (AFdiffMMS-SMS > 0.55, Z-test, P < 0.01), respectively. Genotypes were determined in Suhuai pig, which is a crossbreed between Chinese indigenous Huai pig breed and western commercial pig breed LW, through PCR amplification and Sanger sequencing with primers (Tsingke, Nanjing, China) (Supplementary Table S4). Haplotypes were phased by Beagle v4.0 (Browning et al., 2011). We denoted the haplotypes combined by MMS major alleles as Q, and the other haplotypes as q. For the body length trait of 160-day-old Suhuai pigs and the carcass length of adult Suhuai pigs, respectively, the best linear unbiased prediction model of HIBLUP software (Yin et al., 2023) was applied to calculate the EBV of each trait, respectively. The specific model is:
where y is a vector of phenotypic values, β is a vector of fixed effects, such as sex, batch and age, and X is an indicator matrix of covariates. u is a vector of random additive genetic effects assumed to be normally distributed N(0, Aαu2), where A matrix is pedigree-based numerator relationship matrix, and αu2 is the additive genetic variance. Z is the random additive genetic effects indicator matrix and e is a vector of random residuals that are normally distributed N(0, Iαe2), where I is the identity matrix and αe2 is the residual variance.
Finally, the one-way ANOVA was used to test the association between haplotypes and the EBV of each trait in different Suhuai pig populations, respectively.
Transcriptome Sequencing and Analysis
We discovered that Meishan pigs exhibit a faster growth rate during the early stages of birth, based on body size characteristics of 165 MMS individuals at ages 2, 4, and 6 months (Supplementary Figure S1). To eliminate the influence of the external environment, sex, and other factors on gene expression, we selected 4 MMS and 4 SMS full-siblings, respectively, at birth (all boars). Then we collected femoral cartilage and liver tissue for RNA-seq to identify differentially expressed genes. The extraction of total RNA from the tissue was performed by the traditional trziol method. Then total RNA was sent to Beijing Nuohe Zhiyuan Bio-Information Technology Co., Ltd. (Nuohe, Beijing, China) for library construction and sequencing. In total, 16 libraries were finally produced for the RNA-seq experiment and sequenced on an Hiseq-PE150 platform (Illumina, San Diego, USA) using the 150-bp paired-end sequencing module. The average output per library was 6 Gb.
Firstly, FastQC software (Brandine and Smith, 2019) was used for quality control of the raw data. Then, the filtered and quality-controlled clean reads were aligned to Sus_scrofa 11.1 reference genome by STAR v2.7.1 (STAR, RRID:SCR_004463) (Dobin et al., 2013), and the reads were aligned to the reference genome by featureCounts v2.0.0 (featureCounts, RRID:SCR_012919) (Liao et al., 2014) and genes, exons, promoters, and other regions were counted. Finally, the R package Deseq2 (Love et al., 2014) was implemented to identify differentially expressed genes filtered by FDR < 0.05 and |log2(fold change)| ≥ 1.
PPI Analysis
First, we utilized the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database to predict protein–protein interactions (union cartilage differentially expressed genes and functional annotated genes within candidate selective sweep regions) (Franceschini et al., 2013). Additionally, we chose a combined score of ≥ 0.4 for the construction of the PPI network. Cytoscape software (Spinelli et al., 2013) was then employed to visualize the constructed network. Genes with top 10% degree centrality value (degree centrality value ≥ 16) were chosen as hub genes. Sub-module of Cytoscape software MCODE was applied to identify significantly enriched sub-networks based on the following parameters: Degree cutoff: 2, Node score cutoff: 0.2, K-core: 2, Max. depth: 100. We screened sub-networks with significant interactions involving RSAD2-CMPK2 and COL3A1 genes.
RESULTS
Differences in MMS and SMS Population Structure
The re-sequencing data of 28 MMS and 29 SMS individuals were aligned against the Sus_scrofa 11.1 reference genome. The average sequencing depth for above 57 samples was 10.5-fold, with an average genome coverage of reads at 98.35% (Supplementary Table S5). After conducting quality control, 22,281,189 SNPs with high quality were detected.
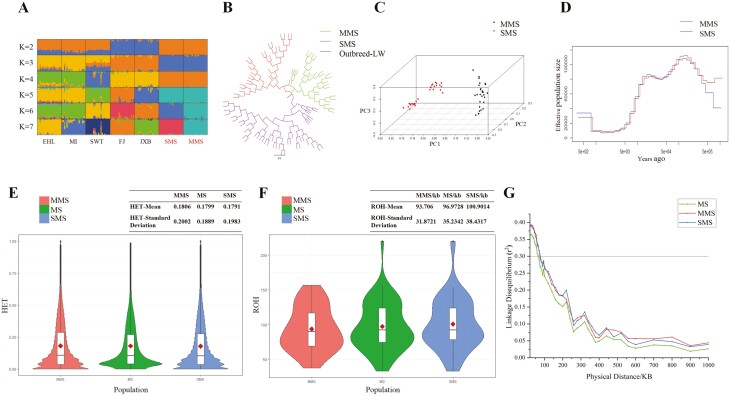
MMS and SMS are sub-populations of Meishan pigs, with differences in body size. To determine whether MMS and SMS are distinguishable populations at the genomic level, we conducted a population structure analysis. Firstly, the results of admixture analyses showed that MMS and SMS have maintained a consistent ancestral lineage from K = 2 to 5 and are different from the ancestral lineage of any other Taihu Lake region indigenous pig breeds. This suggests that MMS and SMS did indeed evolve from a common ancestor (Figure 1A). Then the neighbor-joining phylogenetic tree (NJ-tree) analysis confirmed that MMS and SMS cluster into two separate groups with no cross-clustering of individuals (Figure 1B). Additionally, PCA analysis divided MMS and SMS into two categories (Figure 1C), consistent with the clustering pattern between MMS and SMS based on a 50 K chip (Liu et al., 2020). These results suggest that although MMS and SMS belong to Meishan pig, there are genomic differences between these two sub-populations.
Figure 1.
Evolutionary structure and genetic diversity between MMS and SMS populations. (A) Admixture analysis with K values ranging from 2 to 7. EHL, MI, SWT, FJ, and JXB represent Erhualian, Mi, Shawutou, Fengjing, and Jiaxing black pig, respectively. (B) NJ-tree. (C) Principal component (PC) plot. The first (PC1), second (PC2), and third component (PC3) are displayed. (D) Demographic history of MMS and SMS. Generation time (g) = 1 year and transversion mutation rate (u) = 1.25 × 10−8 mutations per bp per generation. (E) and (F) are violin plot of MMS, SMS, and overall Meishan pig population heterozygosity (HET) and runs of homozygosity fragment (ROH) statistics, respectively, where the upper right corner shows the mean and standard deviation of the corresponding statistics for each population. (G) Linkage disequilibrium (r2 ) extents that were plotted as a function of inter-SNP distance for MMS, SMS, and overall Meishan pig population. The solid line indicates the threshold of 0.3.
To clarify the breed formation history of the two sub-populations of Meishan pigs, we conducted MSMC analysis (Schiffels and Durbin, 2014) to infer historical changes in effective population size (Ne) (Figure 1D). We found that Ne of MMS and SMS was basically the same throughout history. However, Ne of MMS began to be lower than that of SMS only 500 years ago, suggesting that MMS and SMS may have evolved from the same pig ancestor and began to differentiate into two differentiated sub-populations about 500 years ago. It can be seen that MMS and SMS both experienced severe population bottlenecks 5,000 to 10,000 years ago, when the Majiabang Culture (7 to 6 kyBP), Songze Culture (6 to 5 kyBP), and Liangzhu Culture (5 to 4 kyBP) established in Taihu Lake region. Related archaeological excavations also found that the primitive tribes already existed during this period, and boars began to be domesticated and raised. This population bottleneck may be related to the artificial domestication of the common ancestors of MMS and SMS during this period, resulting in a decrease in population diversity due to directional selection.
Genetic Diversity
To further clarify the differences in genetic diversity between MMS and SMS, we calculated the HET, ROH and LD levels of the above two populations, respectively. The results showed that the genetic diversity of MMS was slightly greater than that of SMS in terms of HET (Figure 1E; Supplementary Table S6) and ROH (Figure 1F; Supplementary Table S7). The average HET values were 0.1806 for MMS and 0.1791 for SMS, respectively. For ROH, MMS was 93.7060 kb, and SMS was 100.9014 kb. Compared with MMS, there was a greater degree of inbreeding observed for SMS. However, interestingly, the LD analysis revealed that the LD level for SMS was shorter than that of MMS. (Figure 1G; Supplementary Table S8). This could be due to the fact that SMS individuals were collected from two conservation farms, whereas MMS individuals were all collected from only one conservation farm. Even though SMS maintained a greater level of inbreeding and lower genetic diversity in each single SMS conserved population compared to MMS, there were still genomic differences between the two SMS conserved populations, which reduced the whole SMS population LD level. This can also be seen in the PCA diagram that SMS clustered into two categories, suggesting a degree of difference within two SMS conserved populations leading to the decrease in LD level and maintenance of genetic diversity. These results were also consistent with the MSMC analysis, which found that the Ne of MMS is smaller than that of SMS since modern times. In fact, the Chinese government is currently proposing a multi-site joint conservation strategy for one breed, which is an effective approach to managing African swine fever and relieve the pressure of indigenous pigs protection (Liu et al., 2020).
Identification of Differential Genomic Regions Between MMS and SMS
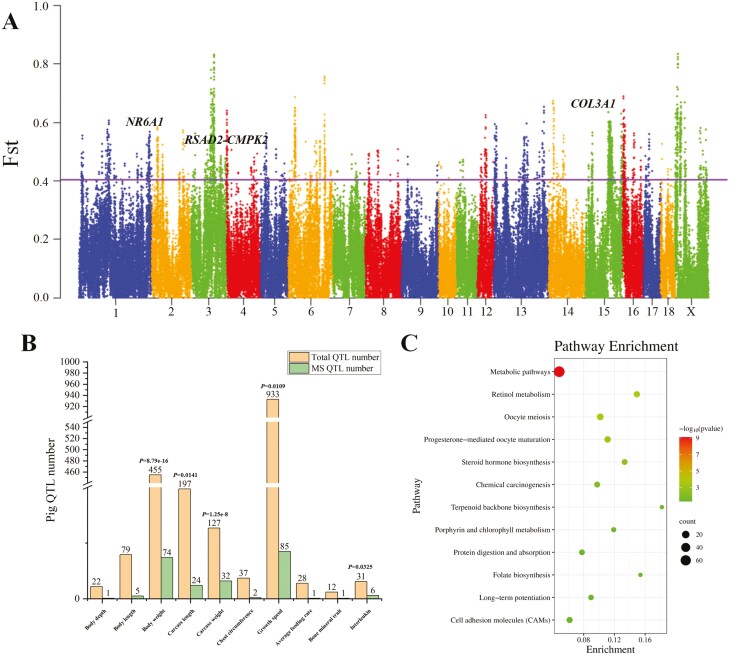
We employed MMS and SMS to conduct Fst analysis for identifying differential genomic regions between these two sub-populations, which could be responsible for body size variation. The Fst results demonstrated 272 significant selective sweep regions (Figure 2A; Supplementary Table S9), covering ~113 Mb, which accounted for ~4% of the entire genome. To preliminarily seek the physiological functions of these selective sweep regions, we annotated these selected regions with the pig QTL database. We found that these selected regions can be significantly enriched in multiple pig body size-related QTLs (Figure 2B) than expected number, such as body weight (P = 8.79e−16), carcass length (P = 0.0141), carcass weight (P = 1.25e−8), growth speed (P = 0.0109), and interleukin (P = 0.0325), suggesting that the relevant genomic regions may play important roles in pig body size formation. We also observed that in addition to traditional body size QTLs such as body length, the selective sweep regions were significantly enriched in interleukin-related QTLs, which are important for skeletal development. For example, interleukin-6 could play important roles in bone metabolism as it mediates the actions of osteoblasts and osteoclasts through sophisticated mechanisms (Wang and He, 2020). Interleukin-37 also inhibits osteoclastogenesis and alleviates inflammatory bone destruction (Tang et al., 2019). Differences in the levels of interleukins may affect bone development and cause differences in body size between MMS and SMS.
Figure 2.
Identification and functional enrichment of differential genome fragments between MMS and SMS. (A) Manhattan plot of Fst values between MMS and SMS by using 50-kb windows with a step size of 25-kb. The solid line indicates the significance threshold (Fst = 0.4029, Z test, P < 0.05). (B) Histogram plot of pig QTL enrichment analysis for selective sweep regions. The x axis represents the QTL categories. The y-axis represents the number of corresponding QTLs. Orange represents the total number of related category QTLs in the pig QTL database, and green represents the number of related category QTLs annotated by selective sweep regions. The P values above the histogram indicate significant enrichment of selective sweeps in the corresponding trait QTLs. (C) Significantly enriched KEGG pathways of protein-encoding genes located in selective sweep regions
We annotated the protein-coding genes within the selective sweep regions and identified 858 genes (Supplementary Table S10). Through enrichment for these genes with KEGG functions, we found these genes were enriched in pathways related to growth and metabolism (Figure 2C; Supplementary Table S11), including metabolic pathways, steroid hormone synthesis and protein metabolism and absorption. These results indicated that the models of nutrient digestion and absorption may differ between MMS and SMS. Additionally, it has been reported that steroid hormones are important for bone development and growth (Bland, 2000), suggesting that body size variation between MMS and SMS may also be caused by factors such as differences in metabolic patterns and hormone release.
To further identify population-specific selected regions for MMS and SMS, respectively, we utilized Zhet analyses to detect regions with low genomic polymorphisms, possibly due to strong natural or artificial selection (Supplementary Figures S2A and S2C). We then intersected these regions with the ones identified through Fst analyses to determine candidate regions. The candidate selected regions were annotated for protein-codable genes. In the MMS population, genes annotated to 43 intersections were considered (Supplementary Table S12), while in the SMS population, genes annotated to 36 intersections were considered (Supplementary Table S13). Subsequently, we conducted functional enrichment analyses of protein-coding genes within these candidate regions using KEGG and GO. There were a total of 10 GO pathways significantly enriched in the selected region of the MMS population (Supplementary Figure S2B and Table S14). Of these pathways, the response to growth hormone pathway showed significant enrichment (FDR test, P = 0.0147). Because growth hormone plays a crucial role in pig growth and development (Etherton et al., 1987), this finding suggests that MMS experiences specific selection in terms of its response to growth hormone. For the SMS population, there were a total of 2 KEGG pathways and 8 GO pathways significantly enriched in the selected regions (Supplementary Figure S2D and Table S15). These pathways encompassed growth and metabolism-related processes such as metabolic pathways (FDR test, P = 0.0478) and fibroblast growth factor receptor signaling pathway (FDR test, P = 0.0475). Additionally, a disease resistance-related pathway, Influenza A (FDR test, P = 0.0475), was also found to be significantly enriched. This suggests that SMS is specifically selected for traits related to growth metabolism and disease resistance.
Candidate Gene NR6A1 Accounting for a Portion of Body Length Variation between MMS and SMS
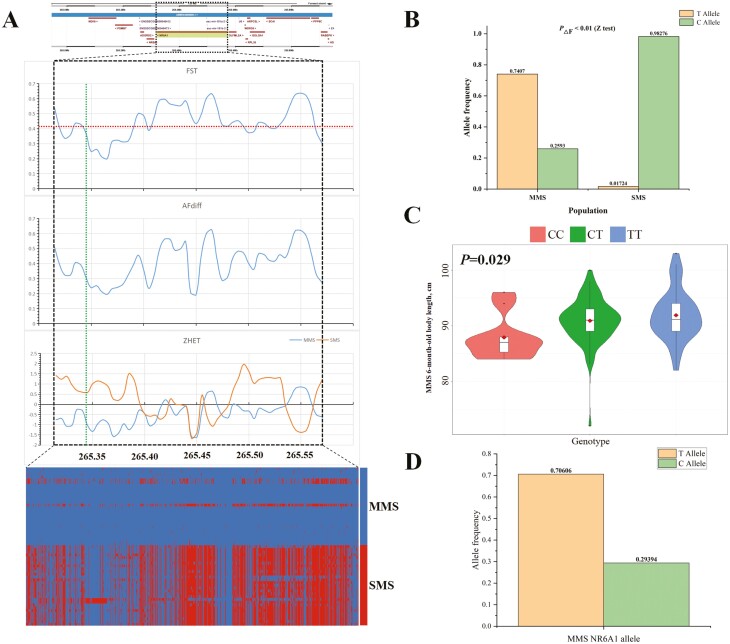
One annotated protein-coding gene, NR6A1, is located within one of the selective sweep regions. The NR6A1 gene is a causal gene, influencing variations in the number of lumbar vertebrae in pigs. The missense mutation at the c.575T > C locus in NR6A1 alters the binding activity of NR6A1 to its corepressors, changing its protein function, and ultimately resulting in the generation of lumbar vertebrae changing (Mikawa et al., 2007). NR6A1 has also been reported to impact changes in body size such as pig body length (Li et al., 2021). The analysis of Fst, AFdiff, and haplotype between MMS and SMS showed that this selected region, where the NR6A1 gene located was significantly differentiated. Zhet analysis showed that MMS exhibited lower polymorphism in this gene region, particularly the region around candidate causal mutation c.575T > C (Figure 3A), indicating that MMS is under greater selection pressure. Additionally, the causal mutation c.575T > C allele frequency indicated that the advantageous T allele is the major allele in MMS, but has not completely fixed. The C allele in SMS is basically fixed (Figure 3B). To further clarify the effect of the c.575T > C locus on body size in Meishan pigs, an association analysis was conducted between 6-month-old body length and this locus in MMS (n = 165). The results showed a significant association (P = 0.029) for this locus with an additive effect (Figure 3C; Supplementary Table S16). It has been reported that the T allele is fixed in large body size pigs, while the C allele dominates in small body size pigs (Ijiri et al., 2021; Yang et al., 2009). Additionally, the allele frequency of c.575T > C in the 165 MMS individuals in the above association analysis showed that the T allele still dominates the population and the frequency is greater than 0.7 (Figure 3D). These results suggest that the NR6A1 gene may be one of the candidate causal genes responsible for the body size variation between MMS and SMS. Due to selection pressure in MMS, the frequency of the advantageous T allele has increased, resulting in an increase in the number of lumbar vertebrae and body length in MMS.
Figure 3.
The NR6A1 gene is one of the candidate causal genes accounting for body size variation between MMS and SMS. (A) Genome scans along the NR6A1 region. At the top panel is the genomic location for NR6A1. Fst, AFdiff, and Zhet statistics are shown in the middle panels. The plots for above three selection signals were all based on 10- kb windows with a step size of 5-kb. The significance threshold of Fst is shown with dashed line. The vertical dashed line is the location of the candidate causal mutation c.575T > C of NR6A1 gene. The bottom is a heat map of MMS and SMS haplotypes within NR6A1 region. Blue is MMS major allele and red is SMS major allele. (B) Histogram plot of allele frequency of c.575T > C of NR6A1 gene between sequenced 28 MMS and 29 SMS individuals. Z test found that the absolute frequency differentiation (AF) of c.575T > C between MMS and SMS showed significant difference. (C) Violin plot of association analysis between NR6A1 c.575T > C locus and the 6-month-old body length phenotype with 165 MMS individuals. (D) Histogram plot of allele frequencies of NR6A1 c.575T > C for 165 MMS individuals for above association analysis.
Differential Analysis of Gene Expression Patterns Between MMS and SMS in Cartilage and Liver Tissues
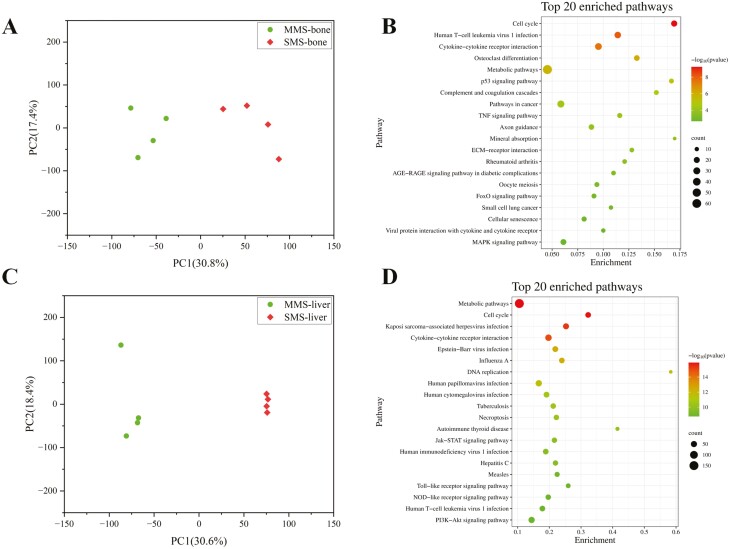
To precisely identify the candidate causal gene, RNA-seq was performed on the cartilage and liver tissues of MMS and SMS to identify differentially expressed genes, respectively. The RNA-seq results of the cartilage revealed significant differences in gene expression patterns between MMS and SMS (Figure 4A). A total of 923 differentially expressed genes were identified, of which 388 were upregulated and 535 were downregulated compared to MMS (Supplementary Figure S3A). These differentially expressed genes could be significantly enriched in multiple pathways related to skeletal development (Figure 4B; Supplementary Table S17), especially osteoclast differentiation (FDR test, P = 7.94E−07), which directly regulates the differentiation and activation of osteoclasts and the process of bone resorption and digestion (Asagiri and Takayanagi, 2007). Additionally, cytokine–cytokine receptor interaction (Cai et al., 2020), mineral absorption (Whisner and Castillo, 2018), and MAPK signaling pathway have been reported to be involved in bone development, particularly in bone resorption and digestion.
Figure 4.
Differentially expressed genes in cartilage and liver tissues between MMS and SMS. (A) Principal component (PC) plot based on all expressed genes in MMS and SMS cartilage tissue. The first (PC1) and second (PC2) are displayed. (B) Top 20 significantly enriched KEGG pathways of differentially expressed genes in cartilage tissue between MMS and SMS. (C) Principal component (PC) plot based on all expressed genes in MMS and SMS liver tissue. The first (PC1) and second (PC2) are displayed. (D) Top 20 significantly enriched KEGG pathways of differentially expressed genes in liver tissue between MMS and SMS.
The RNA-seq results of the liver showed that the liver gene expression patterns of MMS and SMS were also different (Figure 4C). A total of 2,159 differentially expressed genes were identified, of which 855 were upregulated and 1,304 were downregulated compared with MMS (Supplementary Figure S3B). The top significant pathways that the differentially expressed genes enriched in were mainly related to the function of the liver (Figure 4D; Supplementary Table S18), such as metabolic pathways (FDR test, P = 1.22E−16) and cell cycle (FDR test, P = 1.35E−16). It has been reported that metabolic patterns are important for reprogramming in osteoclasts (Park-Min, 2019), and the liver cell cycle is also crucial for maintaining liver function (Mao et al., 2014).
Body Size Variation Candidate Causal Genes RAD2, CMPK2, and COL3A1
By intersecting the genes located in selective sweep regions, cartilage, and liver differential genes, we have identified 11 protein-coding genes with significant signals in all three analyses. These genes are MAPK4, PCLAF, FAM81A, PCYOX1, CENPA, RSAD2, CMPK2, RACGAP1, SGO1, COL3A1, ZNF622, and KLHL13 genes (Supplementary Table S19). Many of these genes have been reported to possess functions related to animal body size. For instance, in a GWAS analysis of body size traits in the Yorkshire population, the MAPK4 gene exhibited a significant signal (Liu et al., 2021). These 11 genes are considered important candidate causal genes affecting body size variation between MMS and SMS.
To determine if these 11 candidate genes exhibit different haplotypes among varying body size pig breeds, we conducted a population comparison of these genes. Preliminary studies have indicated that Chinese indigenous pig breeds can be categorized into three pure original regions: Taihu Lake region, South China, and Southwest China, while North China pigs mixed with Western pig consanguinity (Liu et al., 2020). Hence, we selected representative small body size pig breeds Bamaxiang, Wuzhishan, and Luchuan from South China, as well as representative Southwest China indigenous pig breeds Neijaing and representative North China indigenous pigs Hetaodaer and Min for their larger body size (Supplementary Table S20). Based on the constructed NJ-tree, Bamaxiang, Wuzhishan, and Luchuan clustered together, while Neijiang, Hetaodaer, and Min formed another cluster. MMS and SMS, with independent origins, formed separate clusters from the aforementioned pig breeds. However, MMS and large body size pig breeds exhibited a closer clustering relationship (Supplementary Figure S4), indicating that we have chosen small and large body size pig breeds reasonably.
Then, above diverse pig breeds were used for comparison in population genetics. By integrating the results of three Fst analyses (SMS compared with MMS, SMS compared with large body size pig breeds, SMS compared with small body size pig breeds), it was discovered that there are common significant signals for the RSDA2, CMPK2, and COL3A1 genes in the above analysis.
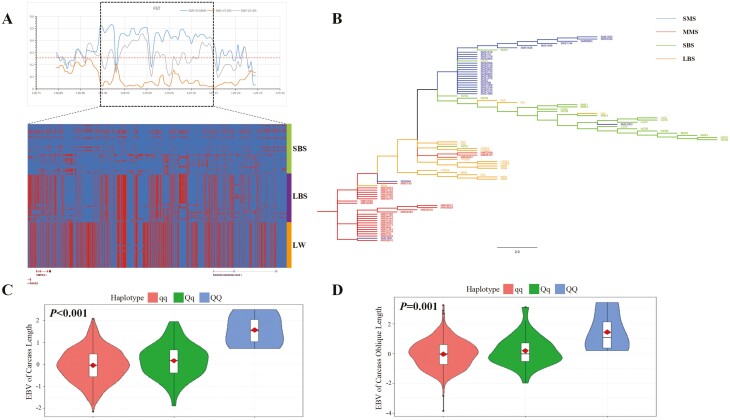
Specifically, the selected region where the RSDA2-CMPK2 genes are located was on SSC3: 128,875,001 bp to 129,150,000 bp. For SMS, there was obvious genetic differentiation compared with MMS (Z test, P < 0.05) and large body size pig breeds (Z test, P < 0.05), and demonstrated considerably lower genetic differentiation compared with small body size pig breeds (Figure 5A). To further substantiate this trend, an NJ-tree for this genomic interval was constructed among all pig breeds mentioned above. Clustering patterns showed that MMS and large body size pig breeds clustered together while SMS and small body size pig breeds clustered together, except one Wuzhishan and two SMS individuals (Figure 5B). The haplotype heat map showed the same trend (Figure 5A), where SMS and small body size pig breeds displayed similar haplotypes, but exhibited much different haplotypes from MMS, large body size pig breeds, and LW. Based on the above evidence, it appears that the genomic interval nearby RSDA2-CMPK2 genes may play a crucial role in body size variation between MMS and SMS.
Figure 5.
RSAD2-CMPK2 region is a candidate causal genomic region affecting body size between MMS and SMS. (A) RSAD2-CMPK2 region demonstrated significant selection signals between MMS and SMS. At the top panel is three Fst statistics: SMS compared with MMS (SMS VS MMS), SMS compared with large body size pig breeds (SMS VS LBS), SMS compared with small body size pig breeds (SMS VS SBS). The dashed line is the Fst significance threshold. At the bottom panel is heat map of MMS and SMS haplotypes within RSAD2-CMPK2 region. Blue is MMS major allele and red is SMS major allele. SBS represents small body size pig breeds including SMS, LBS represents large body size pig breeds including MMS and LW represents Large White pig. (B) NJ-tree of different body size pig breeds for RSAD2-CMPK2 region. Blue lines represent SMS, green lines represent SBS except SMS, red lines represent MMS, and orange lines represent LBS except MMS. SBS represents small body size pig breeds, LBS represents large body size pig breeds. (C) and (D) are violin plot for haplotype association analysis of Suhuai pig carcass length trait and carcass oblique length trait, respectively. Among which, Q is MMS major haplotype and the others are q haplotype.
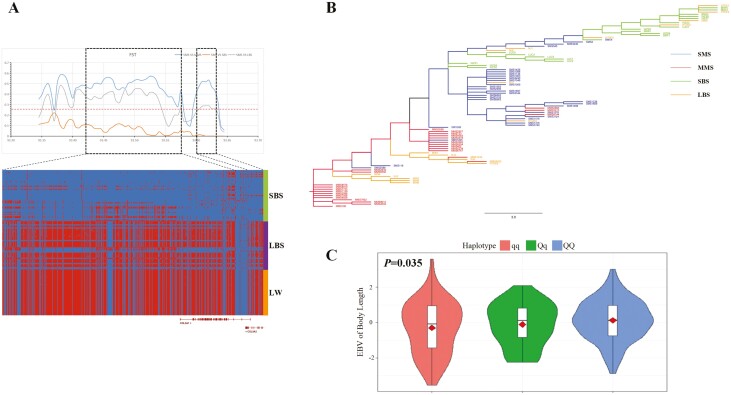
Furthermore, we focused on the genomic region SSC15: 84,925,001 bp to 93,925,000 bp where another important selected gene, COL3A1 is located. Using the same analysis method as previously mentioned, we found that the trends for this region were similar to those of the RSDA2 and CMPK2 genes. There was significant genetic differentiation between SMS and MMS (Z test, P < 0.05) and large body size pig breeds (Z test, P < 0.05), but less differentiation compared to small body size pig breeds (Figure 6A). The NJ-tree analysis showed that SMS and small body size pig breeds clustered together, while MMS and large body size pig breeds clustered together. However, some large body size pig breeds individuals clustered into small body size pig breeds cluster, while all small body size pig breeds individuals basically did not cluster with the other category (Figure 6B). This could be because the so-called large body size pig breeds are only relatively larger than the small breeds in China, but they are still relatively smaller than western commercial pigs, and there is still a degree of variation in body size within breeds (Li et al., 2021). The haplotype heat map for this genomic region showed consistent haplotypes between SMS and small body size pig breeds, and different haplotypes compared with MMS, large body size pig breeds and LW. Especially around the region where the COL3A1 gene is located, the differentiation was evident (Figure 6A). These results suggest that the COL3A1 gene and its adjacent genomic region may also play an important role in explaining the body size variation between MMS and SMS
Figure 6.
COL3A1 region is a candidate causal genomic region affecting body size differentiation between MMS and SMS. (A) COL3A1 region demonstrated significant selection signals between MMS and SMS. At the top panel is three Fst statistics: SMS compared with MMS (SMS vs. MMS), SMS compared with large body size pig breeds (SMS vs. LBS), SMS compared with small body size pig breeds (SMS vs. SBS). The dashed line is the Fst significance threshold. At the bottom panel is heat map of MMS and SMS haplotypes within COL3A1 region. Blue is MMS major allele and red is SMS major allele. SBS represents small body size pig breeds including SMS, LBS represents large body size pig breeds including MMS and LW represents Large White pig. (B) NJ-tree of different body size pig breeds for COL3A1 region. Blue lines represent SMS, green lines represent SBS except SMS, red lines represent MMS, and orange lines represent LBS except MMS. SBS represents small body size pig breeds, LBS represents large body size pig breeds. (C) is violin plot for haplotype association analysis of Suhuai body length trait. Among which, Q is MMS major haplotype and the others are q haplotype.
To further validate the effects of these two selected regions on body size variation, we performed SNP typing for the relevant interval in the hybridized pig breed Suhuai pig with abundant heterozygotes. We then identified haplotypes and conducted association analysis between body length traits EBV and haplotypes. As a Chinese-Western hybrid pig breed, there was a degree of variation in body length-related traits within 160-day-old and adult Suhuai pig populations (Supplementary Tables S21/S22). Through association analysis, we found that the MMS major haplotype (QQ), where the RSDA2-CMPK2 genes are located, extremely significantly greater than other haplotypes (Qq or qq) for carcass length and carcass oblique length EBV (Figure 5C-D; Supplementary Tables S23 and S24). For selected region where the COL3A1 gene is located, it was found that for body length EBV, the MMS major haplotype (QQ) was significantly greater than the other haplotypes (Figure 6C; Supplementary Table S25). These results again confirm that the RSAD2-CMPK2 and COL3A1 genes and their adjacent genomic regions may be important candidate genomic intervals explaining body size variation between MMS and SMS.
The Effects of RSAD2, CMPK2, and COL3A1 Genes on Bone Development
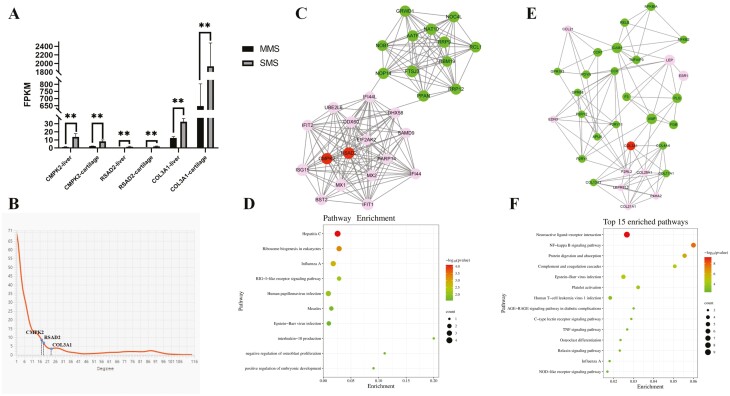
The RNA-seq data revealed significant differential expression of the RSAD2, CMPK2, and COL3A1 genes in cartilage and liver between MMS and SMS (FDR test, P < 0.001). The expression levels of all three genes in SMS were significantly greater (Figure 7A) with highly consistent expression within the population (Supplementary Figure S5A-F), particularly CMPK2 and COL3A1 genes. Both were highly expressed in cartilage and liver (Figure 7A; Supplementary Figure S5C-F). To preliminarily verify the physiological functions of these three highly significant differentially expressed genes, we integrated the genes annotated in selective sweep regions with differentially expressed genes of cartilage and identified a total of 1,731 protein-coding genes. PPI interaction analysis revealed that the RSAD2, CMPK2, and COL3A1 genes were hub genes due to their strong ability to interact with other genes (Figure 7B). Overall, hub genes often serve important biological functions, implying that there can be profound impact on physiological processes for alterations in the expression levels of RSAD2, CMPK2, and COL3A1 genes.
Figure 7.
RSAD2-CMPK2 and COL3A1 genes effects on bone or skeletal development of MMS and SMS. (A) Cartilage and liver FPKM values of RSAD2 CMPK2 and COL3A1 in four MMS individuals and four SMS individuals. ** represents extremly significant difference (P < 0.01). (B) The degree of interaction between genes. The x-axis represents the degree of gene interaction and the y-axis represents the number of genes. blue dots represent RSAD2 CMPK2 and COL3A1 gene. (C) and (E) are sub-networks constructed by the MCODE module of cytoscape software, which are significant interacted with RSAD2-CMPK2 or COL3A1, respectively. (D) Significantly enriched KEGG and GO pathways of the genes interacting with RSAD2-CMPK2. (F) Top 15 significantly enriched KEGG pathways of the genes interacting with COL3A1.
Based on the results of sub-networks found by the MCODE module of the Cytoscape software, the RSAD2-CMPK2 genes were significantly enriched in one sub-network (Figure 7C), and the COL3A1 gene was significantly enriched in another sub-network (Figure 7E). The enrichment of the sub-network by the MCODE module reaffirmed that the above three genes, as hub genes, could interact with other genes to exert biological function. The genes that significantly interacted with RSAD2-CMPK2 were mostly enriched in immune-related pathways (Figure 7D; Supplementary Table S26). Additionally, GO functional enrichment found that some significant GO pathways were related to skeletal development, such as interleukin-10 production (FDR test, P = 0.01), negative regulation of osteoblast proliferation (FDR test, P = 0.02), and positive regulation of embryonic development (FDR test, P = 0.03).
The genes that significantly interacted with COL3A1 were mainly enriched in pathways associated with skeletal body development (Figure 7F; Supplementary Table S27), particularly osteoclast differentiation and cytokine–cytokine receptor interaction pathways. These pathways are also found in enrichment analyses of differentially expressed genes between MMS and SMS cartilage, which have been proven to play an important role in bone digestion and resorption. Additionally, the IL-17 signaling pathway (Li et al., 2019), PI3K-Akt signaling pathway (Sun et al., 2020), and other bone metabolism-related pathways were significantly enriched. These results confirmed that the genes RSAD2, CMPK2, and COL3A1, particularly CMPK2 and COL3A1 genes, were highly expressed in the cartilage and liver tissues of MMS and SMS, and their expression levels were significantly greater in SMS than in MMS (FDR test, P < 0.001). Through PPI analysis, we found that these three genes, as important hub genes, may be involved in multiple pathways of bone development, particularly those pathways related to bone digestion and resorption, which played an important role in inhibition of bone growth and development.
Discussion
Body size traits have been widely used as one of the main breeding selection criteria to monitor pig growth and evaluate the selection response. Identifying genes that contribute to pig body size variation would be valuable, because body size traits are complex quantitative traits that are affected by polygenes and the environment. Many effective genes only contribute minor effects on body size. For example, Yengo et al (2018) implemented meta-analysis of genome-wide association studies on height and body mass index (BMI), identifying 3,290 and 941 near-independent SNPs that were associated with height and BMI, respectively. The near-independent genome-wide significant SNPs explain only 24.6% of the variance of height and only 6.0% of the variance of BMI. This shows the complexity of body size traits.
Body size traits can vary significantly among different breeds of the same species and even among different sub-populations within the same breed. It is possible to screen out breeds of different body sizes for genome comparison, with combination of transcriptomics to mapping causal genes. Using the methods of comparative genomics, researchers have identified candidate causal genes or genomic regions that affect pig body size variation, such as genomic intervals located on the X chromosome (Ai et al., 2015; Reimer et al., 2018). Although a large number of candidate selective sweep regions underlying for body size variation could be identified, it may be difficult to mine key genes.
In this study, Meishan pig was selected to explore the variation in body size within the breed. We initially identified genomic regions with significant differentiation by conducting Fst analysis between MMS and SMS. Within these selected regions, we focused on the protein-coding genes, which emerged as important candidate genes. To understand their significance further, we compared these candidate genes with the already reported major effector genes that influence animal body size traits. Moreover, we performed multiple selection signal analyses and association analyses between genotypes and body length traits in Meishan pigs. These analyses enabled us to preliminarily ascertain the roles played by the reported major effector genes in body size variation within the Meishan pig population. To further explore potential candidate genes influencing the variation in body size within Meishan pigs, we conducted an integration of transcriptome data from cartilage and liver in MMS and SMS populations. This analysis aimed to identify common differential genes at three levels: the genome and the transcriptomes of the two tissues. Based on these findings, we subsequently screened for candidate selected genes that could be linked to body size variation in Meishan pigs, considering the discrepancies in gene expression observed in both cartilage and liver. We additionally screened and validated the effects of the candidate genes using various approaches, including comparative genomics of multiple populations, haplotype association analysis, and PPI analysis. Through Fst and other selection signal related methods, we observed significant differentiation in the NR6A1 gene region between MMS and SMS. However, the NR6A1 gene showed no differential expression trends in neither cartilage nor liver between MMS and SMS. The NR6A1 gene is one of the most important causal genes for variations in the number of lumbar vertebrae in pigs. NR6A1 gene changes its protein function through missense mutation c.575T > C, resulting in the function change for generating lumbar vertebrae (Mikawa et al., 2007). NR6A1 has been associated with the number of lumbar vertebrae and body size traits in various pig breeds, particularly Chinese indigenous pig breeds (Yang et al., 2009; Ijiri et al., 2021; Li et al., 2021). Therefore, NR6A1 regulates phenotype by altering gene function instead of gene expression.
In our study, NR6A1 gene causal missense mutation c.575T > C exhibited a significant allele frequency difference between this two populations (Z test, P < 0.01). Most MMS individuals carried the T allele, which can increase the number of lumbar vertebrae, while SMS individuals carried the wild-type allele C and was nearly fixed. Our association analysis found that the MMS individuals with the advantageous TT genotype was indeed significantly longer than those with the wild-type CC genotype in the 6-month-old body length trait. This finding indicates that the NR6A1 gene may be an important candidate causal gene accounting for the variation in body length between MMS and SMS. MMS might increase lumbar vertebrae number and body length directly by selecting for the advantageous genotype TT. Moreover, our study found that the wild-type C allele at the c.575T > C locus in SMS was nearly fixed, and there was no room for breeding subsequently. However, the T allele in MMS was not entirely fixed, and there is still some space for breeding.
In contrast to the NR6A1 gene, we identified RSAD2-CMPK2 and COL3A1 genes that might regulate bone development by regulating gene expression. At the level of multi-population genomic comparisons and association analysis in Suhuai pigs, the selected regions harboring RSAD2-CMPK2 and COL3A1 genes showed significant signals. These three genes exhibited significant differences in gene expression in cartilage and liver between MMS and SMS, especially CMPK2 and COL3A1 genes were highly expressed in Meishan pig cartilage. Skeletal development, including the development of the length of various bones, especially the long bones, is crucial for body size development. Bone development occurs through a series of synchronous events that result in the formation of the body scaffold, leading to body size variation. The balance of activity between bone-forming osteoblasts and bone-resorbing osteoclasts is accountable for bone repair capacity.
There are numerous pathways that play a key role in governing bone and skeletal development. In terms of osteogenesis, WNT signaling pathway, BMP signaling pathway, FGF signaling, notch signaling are among the most important pathways in bone formation (Little et al., 2002; Bandyopadhyay et al., 2006). In terms of bone resorption and digestion, osteoclast pathway (Asagiri and Takayanagi, 2007), cytokine–cytokine receptor (Cai et al., 2020), among others, play important roles.
In our study, we conducted enrichment analysis for genes that were differentially expressed in the cartilage between MMS and SMS. All of these differentially expressed genes were associated with multiple bone resorption and digestion-related functions, including osteoclast pathway, cytokine-cytokine receptor. This suggests that variation in body size between MMS and SMS could be mainly reflected in differences in bone resorption. In addition, the genes strongly interacting with RASD2 and CMPK2 were significantly enriched in interleukin-10 production, negative regulation of osteoblast proliferation and other GO pathways related to skeletal development. The genes strongly interacting with COL3A1 gene were significantly enriched in pathways, such as osteoclast differentiation, cytokine–cytokine receptor interaction, IL-17 signaling pathway, PI3K-Akt signaling pathway. These above pathways were found to be related to bone development, especially bone resorption and digestion-related pathways. Moreover, RASD2, CMPK2, and COL3A1 genes have also been reported to be involved in multiple bone metabolism and bone resorption related diseases, such as osteoarthritis and osteoporosis (Lodewyckx et al., 2012; Xiao et al., 2012; Dong et al., 2017). These results suggest that body size variation between MMS and SMS could likely be due to differentiation in skeletal development, including multiple pathways involved in bone resorption and digestion. Interestingly, genes involved in bone resorption and digestion-related pathways, such as RASD2-CMPK2 and COL3A1, were highly expressed and significantly enriched in SMS. The possible reason was that the bone resorption and bone digestion process of the SMS are greater than that of MMS, which leads to the retardation of skeletal development and growth rate in SMS. As a result, the overall body size of SMS is smaller than that of MMS.
In conclusion, we screened individuals covering the overall Meishan pig consanguinity for population structure and genetic diversity analysis between MMS and SMS. Through comparative genomics, cartilage and liver transcriptome analysis, association analysis of hybridized pig breed and bioinformatics verification, we identified NR6A1, RSAD2, CMPK2 and COL3A1 genes were candidate causal genes that caused body size variation between MMS and SMS. The causal missense mutation c.575T > C in the NR6A1 gene was positively selected in MMS, leading to a greater frequency of the advantageous allele T, and an increase in body length. The remaining candidate genes were mainly highly expressed in SMS and were significantly enriched in bone resorption and digestion related pathways, indicating that SMS could have enhanced bone resorption and bone digestion processes, resulting in a smaller body size compared to MMS.
Supplementary Material
Acknowledgments
We express gratitude toward Kunshan Meishan Pig Breeding Co., Ltd. in Suzhou, Jiangsu Province, China for providing us with Meishan pig individuals for analysis.
Glossary
Abbreviations
- AFdiff
absolute frequency differentiation
- BMI
body mass index
- EBV
estimated breeding value
- Fst
genetic differentiation coefficients
- HET
expected heterozygosity
- LD
linkage disequilibrium;
- LSB
large body size pig breeds
- LW
Large White pig
- MMS
medium-sized Meishan pig
- MSMC
multiple sequentially Markovian coalescent
- NCBI
National Center for Biotechnology Information
- Ne
effective population size
- NJ-tree
neighbor-joining phylogenetic tree
- PCA
principal component
- QTL
quantitative trait locus
- r2
genotype correlation coefficients;
- ROH
runs of homozygosity fragment
- SBS
small body size pig breeds
- SMS
small-sized Meishan pig
- Zhet
Z-transformed heterozygosity
Contributor Information
Chenxi Liu, Institute of Swine Science (Key Laboratory of Pig Genetic Resources Evaluation and Utilization, Ministry of Agriculture and Rural Affairs (Nanjing)), Nanjing Agricultural University, Nanjing 210095, China.
Liming Hou, Institute of Swine Science (Key Laboratory of Pig Genetic Resources Evaluation and Utilization, Ministry of Agriculture and Rural Affairs (Nanjing)), Nanjing Agricultural University, Nanjing 210095, China.
Qingbo Zhao, Institute of Swine Science (Key Laboratory of Pig Genetic Resources Evaluation and Utilization, Ministry of Agriculture and Rural Affairs (Nanjing)), Nanjing Agricultural University, Nanjing 210095, China.
Wuduo Zhou, Institute of Swine Science (Key Laboratory of Pig Genetic Resources Evaluation and Utilization, Ministry of Agriculture and Rural Affairs (Nanjing)), Nanjing Agricultural University, Nanjing 210095, China.
Kaiyue Liu, Institute of Swine Science (Key Laboratory of Pig Genetic Resources Evaluation and Utilization, Ministry of Agriculture and Rural Affairs (Nanjing)), Nanjing Agricultural University, Nanjing 210095, China.
Qian Liu, Institute of Swine Science (Key Laboratory of Pig Genetic Resources Evaluation and Utilization, Ministry of Agriculture and Rural Affairs (Nanjing)), Nanjing Agricultural University, Nanjing 210095, China.
Tengbin Zhou, Kunshan Animal Disease Prevention and Control Center, Suzhou 215000, China.
Binbin Xu, Kunshan Meishan Pig Breeding Co., Ltd., Suzhou 215000, China.
Pinghua Li, Institute of Swine Science (Key Laboratory of Pig Genetic Resources Evaluation and Utilization, Ministry of Agriculture and Rural Affairs (Nanjing)), Nanjing Agricultural University, Nanjing 210095, China; Huaian Academy, Nanjing Agricultural University, Huaian 223001, China.
Ruihua Huang, Institute of Swine Science (Key Laboratory of Pig Genetic Resources Evaluation and Utilization, Ministry of Agriculture and Rural Affairs (Nanjing)), Nanjing Agricultural University, Nanjing 210095, China.
Funding
This work was supported by Jiangsu Seed Industry Revitalization Project (JBGS (2021) 024).
Conflict of Interest Statement
The authors declare that they have no competing interests.
Data Availability
The WGS sequence reads of 57 Meishan pig individuals are publicly available at the NCBI Sequence Read Archive under accession PRJNA954987. Also, the RNA-seq sequence reads of 8 liver samples and 8 cartilage samples from Meishan pig at birth are publicly available at the NCBI Sequence Read Archive under accession PRJNA956716.
References
- Ai, H., Fang X., Yang B., Huang Z., Chen H., Mao L., Zhang F., Zhang L., Cui L., He W.,. et al. 2015. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat. Genet. 47:217–225. doi: 10.1038/ng.3199. [DOI] [PubMed] [Google Scholar]
- Alexander, D. H., Novembre J., and Lange K... 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asagiri, M., and Takayanagi H... 2007. The molecular understanding of osteoclast differentiation. Bone 40:251–264. doi: 10.1016/j.bone.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay, A., Tsuji K., Cox K., Harfe B. D., Rosen V., and Tabin C. J... 2006. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland, R. 2000. Steroid hormone receptor expression and action in bone. Clin. Sci 98:217–240. 10.1042/cs0980217. [DOI] [PubMed] [Google Scholar]
- Brandine, G., and Smith A. D... 2019. Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res 8:1874. doi: 10.12688/f1000research.21142.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning, B. L., and Browning S. R... 2011. A fast, powerful method for detecting identity by descent. Am. J. Hum. Genet. 88:173–182. doi: 10.1016/j.ajhg.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning, B. L., Tian X., Zhou Y., and Browning S. R.. 2021. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 108(10):1880–1890. doi: 10.1016/j.ajhg.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu, D., Luo H., Huo P., Wang Z., Zhang S., He Z., Wu Y., Zhao L., Liu J., Guo J.,. et al. 2021. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49:W317–W325. doi: 10.1093/nar/gkab447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, W., Li H., Zhang Y., and Han G... 2020. Identification of key biomarkers and immune infiltration in the synovial tissue of osteoarthritis by bioinformatics analysis. PeerJ 8:e8390. doi: 10.7717/peerj.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., and Lee J. J... 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Huang M., Yang B., Wu Z., Deng Z., Hou Y., Ren J., and Huang L... 2020. Introgression of Eastern Chinese and Southern Chinese haplotypes contributes to the improvement of fertility and immunity in European modern pigs. GigaScience 9:giaa014. doi: 10.1093/gigascience/giaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- China National Commission of Animal Genetic Resources. 2011. Animal Genetic Resources in China Pigs. In: Guo, Y. Y., Zhang L. L., and Zhang Y. J., editors, Meishan pig. Beijing, China: Agriculture Press; p. 68–73. [Google Scholar]
- Danecek, P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T.,. et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davi C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R... 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y., Song C., Wang Y., Lei Z., Xu F., Guan H., Chen A., and Li F... 2017. Inhibition of PRMT5 suppresses osteoclast differentiation and partially protects against ovariectomy-induced bone loss through downregulation of CXCL10 and RSAD2. Cell. Signal. 34:55–65. doi: 10.1016/j.cellsig.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Etherton, T. D., Wiggin J. P., Evock C. M., Chung C. S., Rebhun J. F., Walton P. E., and Steele N. C... 1987. Stimulation of pig growth performance by porcine growth hormone: determination of the dose-response relationship. J. Anim. Sci. 64:433–443. doi: 10.2527/jas1987.642433x. [DOI] [PubMed] [Google Scholar]
- Franceschini, A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., Mering C. V.,. et al. 2013. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley, C. S., and Lee G. J... 1993. Genetic basis of prolificacy in Meishan pigs. J. Reprod. Fertil. Suppl. 48:247–259. https://pubmed.ncbi.nlm.nih.gov/8145208/. [PubMed] [Google Scholar]
- Hu, Z. L., Park C. A., and Reecy J. M... 2019. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 47:D701–D710. doi: 10.1093/nar/gky1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, M. G., Picton H. M., Biggs C., Mann G. E., McNeilly A. S., and Foxcroft G. R... 1996. Periovulatory endocrinology in high ovulating Meishan sows. J. Endocrinol. 150:141–147. doi: 10.1677/joe.0.1500141. [DOI] [PubMed] [Google Scholar]
- Ijiri, M., Lai Y. C., Kawaguchi H., Fujimoto Y., Miura N., Matsuo T., and Tanimoto A... 2021. NR6A1 allelic frequencies as an index for both miniaturizing and increasing pig body size. In Vivo 35:163–167. doi: 10.21873/invivo.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim, L., Takeda H., Lin L., Druet T., Arias J. A., Baurain D., Cambisano N., Davis S. R., Farnir F., Grisart B.,. et al. 2011. Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nat. Genet. 43:405–413. doi: 10.1038/ng.814. [DOI] [PubMed] [Google Scholar]
- Li, H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and Durbin R... 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. Y., Yu M., Tyagi A. M., Vaccaro C., Hsu E., Adams J., Bellido T., Weitzmann M. N., and Pacifici R... 2019. IL-17 receptor signaling in osteoblasts/osteocytes mediates PTH-induced bone loss and enhances osteocytic RANKL production. J. Bone Miner. Res. 34:349–360. doi: 10.1002/jbmr.3600. [DOI] [PubMed] [Google Scholar]
- Li, L., Xiao S., Tu J., Zhang Z., Zheng H., Huang L., Huang Z., Yan M., Liu X., and Guo Y... 2021. A further survey of the quantitative trait loci affecting swine body size and carcass traits in five related pig populations. Anim. Genet. 52:621–632. doi: 10.1111/age.13112. [DOI] [PubMed] [Google Scholar]
- Liao, Y., Smyth G. K., and Shi W... 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Little, R. D., Recker R. R., and Johnson M. L... 2002. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 347:943–4; author reply 943. doi: 10.1056/NEJM200209193471216. [DOI] [PubMed] [Google Scholar]
- Liu, C., Li P., Zhou W., Ma X., Wang X., Xu Y., Jiang N., Zhao M., Zhou T., Yin Y.,. et al. 2020. Genome data uncover conservation status, historical relatedness and candidate genes under selection in Chinese Indigenous Pigs in the Taihu Lake Region. Front. Genet. 11:591. doi: 10.3389/fgene.2020.00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Song H., Jiang Y., Jiang Y., Zhang F., Liu Y., Shi Y., Ding X., and Wang C... 2021. A single-step genome wide association study on body size traits using imputation-based whole-genome sequence data in Yorkshire pigs. Front. Genet. 12:629049. doi: 10.3389/fgene.2021.629049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodewyckx, L., Cailotto F., Thysen S., Luyten F. P., and Lories R. J... 2012. Tight regulation of wingless-type signaling in the articular cartilage - subchondral bone biomechanical unit: transcriptomics in Frzb-knockout mice. Arthritis Res. Ther. 14:R16. doi: 10.1186/ar3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. I., Huber W., and Anders S... 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, S. A., Glorioso J. M., and Nyberg S. L... 2014. Liver regeneration. Transl. Res. 163:352–362. doi: 10.1016/j.trsl.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., and Daly M., et al. 2010. The genome analysis toolkit: a map reduce framework for analyzing next-generation DNA sequencing data. Genome Res.20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa, S., Morozumi T., Shimanuki S., Hayashi T., Uenishi H., Domukai M., Okumura N., and Awata T... 2007. Fine mapping of a swine quantitative trait locus for number of vertebrae and analysis of an orphan nuclear receptor, germ cell nuclear factor (NR6A1). Genome Res. 17:586–593. doi: 10.1101/gr.6085507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park-Min, K. H. 2019. Metabolic reprogramming in osteoclasts. Semin. Immunopathol. 41:565–572. doi: 10.1007/s00281-019-00757-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer, C., Rubin C. J., Sharifi A. R., Ha N. T., Weigend S., Waldmann K. H., Distl O., Pant S. D., Fredholm M., Schlather M.,. et al. 2018. Analysis of porcine body size variation using re-sequencing data of miniature and large pigs. BMC Genomics 19:687. doi: 10.1186/s12864-018-5009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels, S., and Durbin R... 2014. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli, L., Gambette P., Chapple C. E., Robisson B., Baudot A., Garreta H., Tichit L., Guénoche A., and Brun C... 2013. Clust&See: a Cytoscape plugin for the identification, visualization and manipulation of network clusters. Biosystems 113:91–95. doi: 10.1016/j.biosystems.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Sun, H., Wang Z., Zhang Z., Xiao Q., Mawed S., Xu Z., Zhang X., Yang H., Zhu M., Xue M.,. et al. 2018. Genomic signatures reveal selection of characteristics within and between Meishan pig populations. Anim. Genet. 49:119–126. doi: 10.1111/age.12642. [DOI] [PubMed] [Google Scholar]
- Sun, K., Luo J., Guo J., Yao X., Jing X., and Guo F... 2020. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: a narrative review. Osteoarthritis Cartilage 28:400–409. doi: 10.1016/j.joca.2020.02.027. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Stecher G., Peterson D., Filipski A., and Kumar S... 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, R., Yi J., Yang J., Chen Y., Luo W., Dong S., and Fei J... 2019. Interleukin-37 inhibits osteoclastogenesis and alleviates inflammatory bone destruction. J. Cell. Physiol. 234:7645–7658. doi: 10.1002/jcp.27526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laere, A. S., Nguyen M., Braunschweig M., Nezer C., Collette C., Moreau L., Archibald A. L., Haley C. S., Buys N., Tally M.,. et al. 2003. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425:832–836. doi: 10.1038/nature02064. [DOI] [PubMed] [Google Scholar]
- Wang, T., and He C... 2020. TNF-α and IL-6: the link between immune and bone system. Curr. Drug Targets 21:213–227. doi: 10.2174/1389450120666190821161259. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Cao X., Luo C., Sheng Z., Zhang C., Bian C., Feng C., Li J., Gao F., Zhao Y.,. et al. 2020. Multiple ancestral haplotypes harboring regulatory mutations cumulatively contribute to a QTL affecting chicken growth traits. Commun. Biol. 3:472. doi: 10.1038/s42003-020-01199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisner, C. M., and Castillo L. F... 2018. Prebiotics, bone and mineral metabolism. Calcif. Tissue Int. 102:443–479. doi: 10.1007/s00223-017-0339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, H., Shan L., Zhu H., and Xue F... 2012. Detection of significant pathways in osteoporosis based on graph clustering. Mol. Med. Rep. 6:1325–1332. doi: 10.3892/mmr.2012.1082. [DOI] [PubMed] [Google Scholar]
- Yang, G., Ren J., Zhang Z., Huang L... 2009. Genetic evidence for the introgression of Western NR6A1 haplotype into Chinese Licha breed associated with increased vertebral number. Anim. Genet. 40:247–250. doi: 10.1111/j.1365-2052.2008.01820.x. [DOI] [PubMed] [Google Scholar]
- Yengo, L., Sidorenko J., Kemper K. E., Zheng Z., Wood A. R., Weedon M. N., Frayling T. M., Hirschhorn J., Yang J., and Visscher P. M... 2018. Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry. Hum. Mol. Genet. 27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, L., Zhang H., Tang Z., Yin D., Fu Y., Yuan X., Li X., Liu X., and Zhao S... 2023. HIBLUP: an integration of statistical models on the BLUP framework for efficient genetic evaluation using big genomic data. Nucleic Acids Res. 51:3501–3512. doi: 10.1093/nar/gkad074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Li M., Cheng H., Fan W., Yuan Z., Gao Q., Xu Y., Guo Z., Zhang Y., Hu J.,. et al. 2018. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat. Commun. 9:2648. doi: 10.1038/s41467-018-04868-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The WGS sequence reads of 57 Meishan pig individuals are publicly available at the NCBI Sequence Read Archive under accession PRJNA954987. Also, the RNA-seq sequence reads of 8 liver samples and 8 cartilage samples from Meishan pig at birth are publicly available at the NCBI Sequence Read Archive under accession PRJNA956716.