Summary:
Convergence science teams integrating clinical, biological, engineering, and computational expertise are inventing new forecast systems to monitor and predict evolutionary changes in tumor and immune interactions during early cancer progression and therapeutic response. The resulting methods should inform a new predictive medicine paradigm to select adaptive immunotherapeutic regimens personalized to patients’ tumors at a given time during their cancer progression for durable patient response.
FRAMING THE CHALLENGE
Instructing the immune system to recognize and kill cancer has transformed the standard of care for certain malignancies. Immunotherapies have converted 20% of deadly cancers to chronic diseases and have provided patients with enhanced longevity and quality of life. Prominent among immunotherapies are immune-checkpoint inhibitors (ICI)—monoclonal antibodies that block T-cell inhibitory molecules, such as PD-1, CTLA-4, and LAG3, to unleash effector T cells to kill tumor cells. However, 80% of all cancers fail to respond because immunosuppressive cells in the tumor microenvironment (TME) impede T-cell response. Combination treatment strategies to promote T-cell trafficking and block immunosuppressive cells can enhance immunotherapy response. The cellular and molecular composition of the TME evolves over time, which can change the sensitivity of ICIs across of the same tumor subtype in different stages and limit durable response in responders with advanced cancers. Because of these dynamic changes, predictive medicine approaches that know when and how to intercept tumor progression will be critical to enhancing ICI durability, circumventing resistance, and broadening the scope of precision immunotherapy. Meeting this challenge will require new approaches to measure, in real time, patient-specific responses and temporal alterations in the tumor architecture during the evolution of advanced cancer from precancerous lesions, as well as during the development of resistance to ICIs. Convergence science teams integrating clinical, biological, technological, and mathematical expertise can design new frameworks to monitor and infer evolutionary changes in tumor and immune interactions that will empower this new predictive medicine paradigm (Fig. 1).
Figure 1.
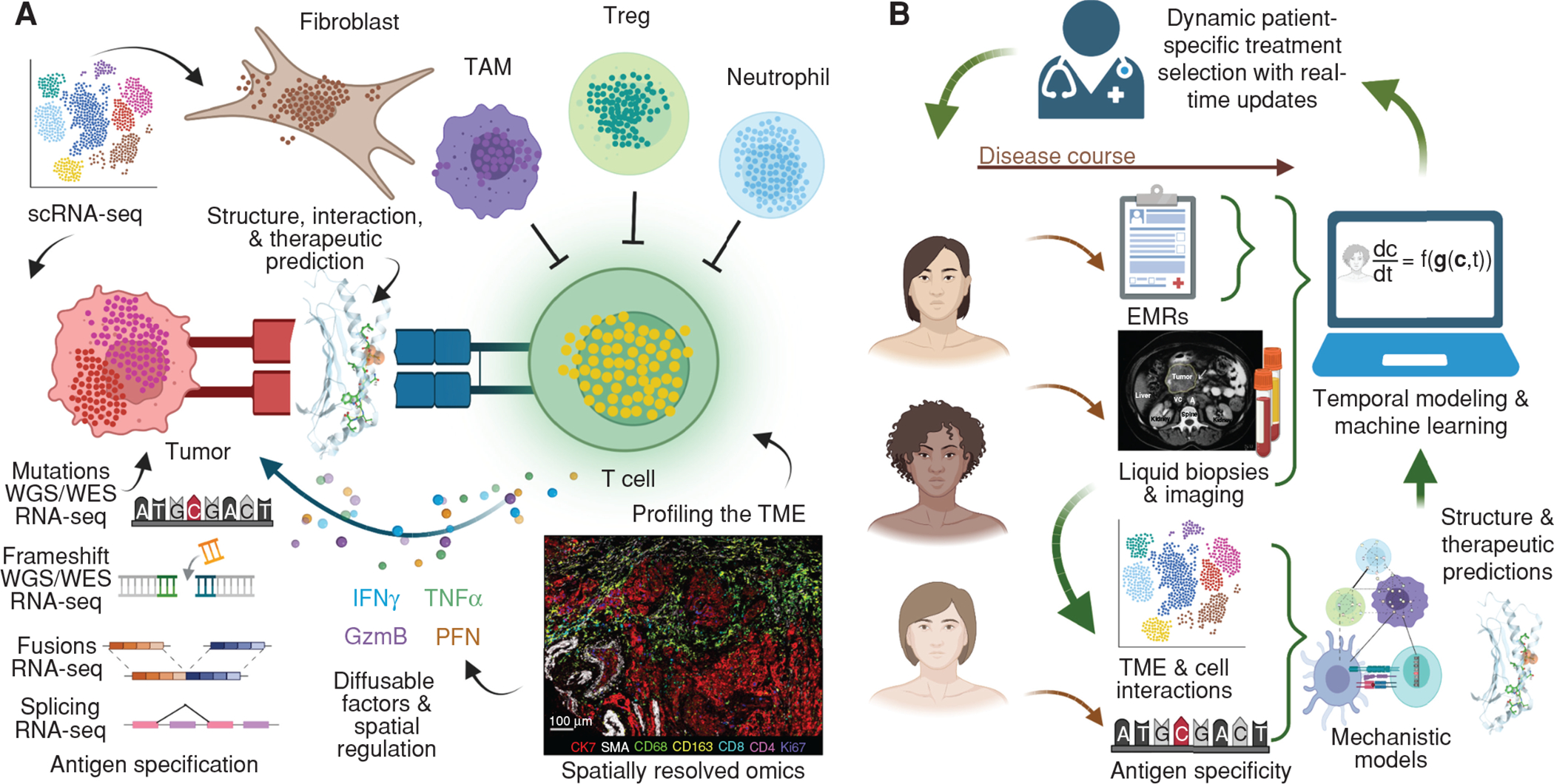
Integration of clinical, biological, technological, and mathematical expertise to design new frameworks to monitor and infer evolutionary changes in tumor and immune interactions that guide a new predictive medicine paradigm. A, Spatial and single-cell profiling of the TME over multiple time points in cancer development will allow for deeper understanding of the dynamics of immune populations and interactions with tumor cells as cancer progression occurs over time and upon intervention with therapeutics. T-cell receptor modeling with HLA–antigen complexes will also allow us to understand epitope recognition and evasion. WES/WGS/RNA-seq data will determine neoantigen evolution in tumor cells over time and upon therapeutic intervention. RNA-seq, RNA sequencing; scRNA-seq, single-cell RNA sequencing; TAM, tumor-associated macrophage; Treg, regulatory T cell; WES, whole-exome sequencing; WGS, whole-genome sequencing. B, Integration of these large datasets detailing dynamics of TME and antigenic properties of tumor cells with clinical datasets including electronic medical systems, liquid biopsies, and imaging data will allow for the development of machine learning and predictive algorithms that could guide dynamic patient-specific treatment selection in real time. EMR, electronic medical record.
WINDOW OF OPPORTUNITY FOR INTERCEPTING CANCER
At least two windows of opportunity for early interception are likely to impact mortality, morbidity, and the financial costs associated with cancers. First, there is emerging evidence for successful interception of cancer progression when ICIs are initiated during the early stages of cancers, such as in mismatch repair–deficient, localized rectal cancers. In this immunoablative strategy, patients are cured using single-agent ICI without the need for surgical intervention or radiation (1). These successes are likely due to fewer immunosuppressive signals, making early tumors more sensitive to ICI interception.
A second strategy is to intercept the evolution to early cancer from precancerous lesions. For example, with pancreatic ductal adenocarcinoma (PDAC), there is an 11-year-long window from the first genetic alteration in a normal cell to overt PDAC. To predict when and which patients are likely to respond to interception by ICIs, it is critical that we have a detailed understanding of patient-specific responses, as well as the temporal and spatial evolution of precancers at both the cellular and molecular level. Current clinical studies are testing the efficacy of vaccines to intercept cancer evolution in certain high-risk populations with known genetic predisposition as proof of concept for interception, such as for Lynch syndrome (NCT05078866). Similarly, our phase I clinical trial uses a mutant KRAS vaccine for PDAC interception in high-risk groups (NCT05013216). Vaccines targeting tumor-associated antigens such as MUC1, which is expressed in breast, ovarian, pancreatic, lung, and colorectal cancers, are currently being tested in prevention settings for colorectal adenomas (NCT02134925) and lung cancer (NCT03300817). Although the prediction of responses in high-risk individuals is likely to be challenging, mainly due to the paucity of tissue samples, development of noninvasive peripheral biomarkers and computationally defined predictive algorithms will be critical to determine and predict response and resistance in such early interception trials.
The renewed Cancer Moonshot initiative highlights immunoprevention as a key priority—it emphasizes the identification of novel targets and early interventions, such as vaccines, for high-risk individuals. Growing evidence also supports the concept of cancer immunoediting—defined by elimination, equilibrium, and escape (2)—starting in the earliest precancerous lesions. The concept is based on the idea that even early precancerous lesions can attract high-quality T cells in the “elimination phase” prior to T-cell tolerization and exhaustion, which occurs in the “equilibrium phase” and eventually leads to tumor escape. Without the appropriate immune interception agents to reeducate T cells in the elimination phase, tumor escape prevails, as an increasing number of immunosuppressive signals allow for immune evasion. We will thus need to define the precancerous immune and genomic landscape to predict when interception will need to occur. Adapting new single-cell and spatial molecular technologies to profile the TME of biospecimens of precancers and mapping these to subsequent changes in tumorigenesis in cancer are critical for predictive interception strategies (bioRxiv 2022.07.16.500312).
CIRCUMVENTING IMMUNOTHERAPY RESISTANCE
A patient’s tumor response typically falls into one of three main categories, namely, responders, nonresponders due to primary resistance, and nonresponders due to acquired resistance. Responders are patients with immunologically “hot” tumors typified by a high tumor mutation burden (TMB; ref. 3) and neoantigen loads or ones that express fewer but highly immunogenic neoantigens, such as virus-associated cancers. Neoantigens arise from nonsynonymous mutations during cancer progression and are recognized as foreign to the immune system. Nonresponders display primary resistance to single- or dual-agent ICI therapy due to a low TMB and few infiltrating T cells. These tumors include PDAC and glioblastomas, among others. A final group of tumors initially respond to ICIs but eventually progress to acquire resistance due to genetic, epigenetic, and TME changes. Several clinical biomarkers are currently being used clinically to predict responses to ICIs. Apart from TMB, these include DNA repair deficiency [e.g., microsatellite instability (MSI)–high (4), POLE mutation] and PD-L1 expression. However, the absence of these biomarkers does not always exclude response to ICIs, nor does it predict the magnitude or durability of the response. Metastasis results in a further level of complexity due to differences in the TME of different organ sites—this results in preferred ICI responses in certain metastatic sites and not in others (5).
Two immediate clinical challenges must therefore be addressed to realize the full potential of ICI therapy. The first is to identify primary and acquired resistance mechanisms and develop additional immunotherapy approaches that successfully intercept these mechanisms. Both tumor intrinsic and extrinsic escape mechanisms either exist at baseline or evolve over time to limit the breadth and durability of ICI responsiveness across cancer types. To broaden ICI responses, novel approaches must first provoke T-cell recognition of tumor antigens and then modify or bypass diverse genetic and immune-driven tumor escape mechanisms. Harnessing T-cell responses to orchestrate durable clinical responses following ICI therapy requires two steps. First, T cells need to be primed sufficiently with antigen, usually in the lymph node, to acquire the signals that program T cells to traffic into and kill tumor cells. In the second step, tumor-intrinsic and tumor-extrinsic immunosuppressive signals, immune cell subsets, and stromal cells within the TME must be reprogrammed (6). New single-cell and multiomics technologies are empowering this mechanistic interpretation, for which computational approaches are essential to determining patterns of ICI response and resistance (7). Leveraging these mechanistic insights for combination immunotherapy strategies requires further knowledge of the molecular and cellular impacts of therapeutics, enabled by enhanced structural models and multiomics treatment atlases.
The second challenge is to develop biomarkers that predict early response and resistance to ICI therapy. MSI-high, high TMB, and POLE mutation are tumor-intrinsic features based upon genomic sequencing data that are highly predictive of ICI response and most commonly used as biomarkers of ICI response. It is also increasingly clear that certain types of mutations may generate higher quality neoantigens and improved ICI responses. In addition to neoantigens derived from nonsynonymous mutations, durable ICI responses in MSI-high tumors are associated with indels, which result in more significant alterations in the coding peptide, as well as those from gene fusions and splice variants. However, the relative importance and immunogenicity of each type of neoantigen in driving an ICI response are unknown. Additionally, evolution in the molecular alterations and clonal selection can cause the overall neoantigen landscape to evolve over time. These factors may result in the editing of immunogenic neoantigens, which removes them from tumor, or an altered neoantigen landscape that outcompetes the development of a new T-cell response. Both mechanisms can lead to ICI resistance. Although neoantigens are detected from tumor sequencing, characterization of these alterations over time requires sequencing circulating tumor DNA for sequential clinical monitoring. The composition of additional immunosuppressive cells can also affect ICI response and may be more difficult to detect in circulating cells for clinical monitoring. The potential for large-scale databases encompassing clinical tests, imaging, disease states, and adverse reactions represents an already substantial and expanding resource for longitudinal observations across large patient cohorts. Applying machine learning approaches to large-scale databases could determine the pathways that best fit a patient’s primary or acquired resistance trajectory. Methods for analysis of time-varying data remain an open problem in computation and require the invention of new machine learning methods for inference of evolving mechanistic biomarkers from multimodal patient data. The goal here is to modify clinical paradigms from focusing solely on precision medicine at the time at which a patient presents to clinic to establishing algorithms that enable a predictive medicine framework that can accurately determine the series of combination therapies over time that would retain ICI sensitivity.
BASING THERAPEUTIC SELECTION ON OMICS CHARACTERIZATION OF TUMORS AND THE MICROENVIRONMENT
Multiomics technologies are rapidly being applied to biospecimens from malignancies and the periphery to uncover phenotypic and functional changes in the heterogeneous cell populations across stages of carcinogenesis and within ICI-treated tumors. Beyond biomarkers, prediction and characterization of patient-specific neoantigens from sequencing datasets are informing immunotherapeutic agents for antigen vaccine and adoptive T-cell therapies. Cancer displays considerable intra- and intertumoral heterogeneity. This means that the optimal treatment selection for vaccine and T-cell targeting must be unique for each patient with cancer. Current computational prediction of potentially immunogenic neoantigens uses machine learning to determine the binding affinity of specific HLA alleles to the mutated peptide. These methods compare mutational sequencing of the tumor with reference normal DNA controls in the blood or other tissues to define somatic mutations. Machine learning algorithms are then applied to compare the sequence of these alterations against databases that catalog validated MHC binding, primarily based on pathogenic antigens. Most neoantigen prediction algorithms are limited by the training data of MHC binding, which poses limitations to HLA class I and II predictions. Furthermore, MHC-binding affinity is not always an accurate predictor of neoantigen immunogenicity. Further methodologic advances are thus required to expand reference databases of antigen recognition, integrate proteomics data, and leverage 3D protein structure prediction (8). These improved algorithms should enable more accurate prediction of changes in neoantigen load or quality that may impact ICI response. Incorporating technologies for T- and B-cell receptor profiling has the potential to further formulate mechanistic biomarkers of response to these therapies.
Well-known genetic alterations in tumor cells can regulate oncogenic pathways that can, in turn, alter the cellular populations within the TME. Furthermore, immune cells, each with a wide array of subtypes and transition states, can change their function as cancer develops over time. For example, a tumor with high T-cell infiltration may become nonresponsive to immunotherapy if the quality of infiltrating T cells becomes poor. The immune cell subtypes in the TME also interact to further alter T-cell function in the TME. For example, regulatory T cells, myeloid-derived suppressor cells, and tumor-associated M2 macrophages, as well as cytokines and immune metabolites, may together render the TME immunosuppressive (6). Furthermore, in some tumors such as PDAC, a dense stromal compartment consisting of cancer-associated fibroblasts can create a formidable physical barrier to T-cell infiltration.
Single-cell and spatial multiomics technologies can now characterize the states of the cells in the TME and determine the more complex relationships between altered tumor cells and signaling within the TME. These technologies can characterize the cellular composition of the microenvironment and select therapeutics to target predominant immunosuppressive cells. However, most therapeutics that target immunosuppressive cells have diverse effects on the molecular and phenotypic states of additional cells in the TME. Single-cell and spatial transcriptomics data provide higher dimensional data to infer cellular phenotypes and cell–cell interaction networks that affect carcinogenesis and therapeutic response. Complementary computational advances for inference of cellular networks, ligand–receptor networks, and cellular interactions based upon colocalization are emerging to empower these inferences (ref. 7; bioRxiv 2022.06.02.490672). We have shown that applying these methods to single-cell reference atlases can lead to novel hypotheses of how specific agents would alter the cellular and molecular landscape of the TME (9). Mechanistic preclinical studies, cross-species analysis between human tumor atlases and multiomics treatment atlases in mouse models, and further in silico validation in independent human datasets can provide further evidence to translate single-cell predictions of TME response into new human clinical trials.
APPROACHES FOR MEASURING AND MODELING PREDICTIVE IMMUNOTHERAPY
Large-scale tumor atlas projects through national and international consortia are providing comprehensive resources to uncover the molecular and cellular landscape of tumors. However, these atlases provide static, population-level snapshots rather than patient-specific dynamics of the immune landscape of tumors. Furthermore, many such efforts are focused on primary tumors, whereas metastatic disease introduces a complex set of challenges, as each site may have meaningful differences in antigen presentation and TME composition that render them harder to treat (5). Additional characterization of changes at the earliest stages of premalignancy are required for interception. Thus, characterizing the dynamics of alterations across all stages of oncogenesis should provide important tools for stage-specific therapeutic design and selection.
Tracking dynamic changes in tumors and the TME is critical for three key reasons: to discern the TME’s evolution as oncogenesis progresses from precancerous lesions and early cancer to advanced disease; to determine changes in response to immunotherapy; and to study cellular heterogeneity as cancer metastasizes to distant sites. This should allow appropriate timing for interception and therapy with personalized ICIs. Characterizing molecular alterations in tumors and changes to the cellular pathways in the TME throughout a patient’s disease progression can enable new predictive medicine strategies to allow clinicians to adapt therapy over time. Realizing the potential of this paradigm requires new advances in computational methods, clinical study designs, and patient monitoring tailored to the dynamic characterization of tumor–immune evolution in patients. Clinical trials should therefore incorporate serial tumor biopsies (before, during, and at time of progression) and blood draws to measure the evolution of tumor antigens and oncogenic pathways that influence the TME’s immune and nonimmune cell populations. However, this level of longitudinal profiling presents a major challenge clinically, because, in addition to patient consent and compliance, acquiring the samples is often expensive and invasive. Furthermore, multiple longitudinal tumor biopsies may be difficult to acquire, may be insufficient in quality or quantity for single-cell profiling, and may lack full representation of the entire tumor’s heterogeneity. Even when samples can be acquired or states can be predicted using machine learning, it is not guaranteed that they are obtained from specimens that are representative of disease, particularly with multiple disease sites. Therefore, noninvasive and systemic measurements are essential to enable comprehensive longitudinal profiling of patients’ tumors and their response to ICIs. For example, circulating tumor DNA from liquid biopsies is an emerging blood-based biomarker to guide treatment and monitor disease progression (10). Likewise, mechanism-driven computational methods for ICI monitoring will also benefit from expanding features of genomic and proteomic assays, imaging technologies, and radiomics.
Furthermore, to complement the static data of atlases, system biology approaches can use mathematical and physical modeling derived from prior knowledge of molecular and cellular interactions—this will convert snapshots into mechanistic movies (11). Whereas data-driven analyses of high-throughput data are limited to the time points of measurements, these systems biology approaches can leverage the biological mechanisms to predict future, unseen states of tumors during cancer progression and therapeutic response. The accuracy of predictions from computational models depends on the prior knowledge of the mechanistic rules of biological systems and their abstraction into equations. Pairing these computational approaches with experimental systems can better define the components of these models and provide the range of parameters for the variables in these models and test their predictions of therapeutic response. In the same way that data assimilation approaches that embed high-throughput data into mathematical models have improved forecasting in the weather, personalizing mechanistic models with high-throughput data should provide significant insight into the underlying response dynamics that result in primary or acquired resistance (12). Further computational advances that incorporate both systems-level effects of therapeutics with cellular behavior of tumor–immune interactions can model the systemic effects of therapeutic delivery to enable in silico clinical trials (13). Paired with the single-cell multiomics profiling data, systems biology models could also potentially predict the timing of when a patient may become resistant, allowing clinicians to best sequence therapies or, in other words, administer the right ICI at the right time.
FUTURE PLATFORM FOR PREDICTIVE IMMUNOTHERAPY
We posit that the availability of tools to understand tumor biology at a single-cell resolution and the application of mathematical models will yield an integrated unified platform for the early interception of cancer progression and ICI resistance for each individual patient. Accomplishing this goal requires first developing the theory to characterize the evolution of the TME and then the infrastructure to trace it. Methodologic advances in computational biology and machine learning are needed to invent algorithms that can predict the course of disease in a given patient. These methodologic advances require new technological advances that can trace the molecular and cellular states of tumors as they change systemically throughout a patient’s body and over time. Together, these advances form a new area of basic science in engineering and data science that are essential to answer fundamental questions about the duration of therapeutic response, mechanisms of tumor and immune evolution, and predictability of disease.
Infrastructure advances can serve current efforts to translate precision immunotherapy and provide a foundation for future implementation of predictive medicine as the basic science matures. First, we must create unique pan-tumor atlases that incorporate not only static but also dynamic immune, stromal, and cancer cell changes over time using tumor profiling datasets from patients treated with ICIs across the globe. Second, we must integrate these atlases with clinical data from expansive clinical databases. Third, and given the challenges in obtaining multiple biopsies, there is also an urgent need to develop and utilize noninvasive biomarkers, such as blood-based cell-free tumor DNA and molecular imaging data, to monitor disease development and progression. Finally, methodologic advances in computational biology and machine learning can leverage these high-throughput datasets to personalized therapeutic regimens to target a patient’s disease. As computational approaches develop to forecast the cellular and molecular pathways that will ultimately cause disease to progress or therapeutic resistance to occur, these predictions can be used as the basis of therapeutic selection to empower interception and prevention of resistance in the new predictive medicine paradigm. There is an urgent need for an international effort to move this forward and to provide relevance to all patients with cancer regardless of their nationality, culture, and race.
Acknowledgments
We are grateful to Rebecca Riggins for feedback. We acknowledge funding from the Skip Viragh Center for Pancreatic Cancer at Johns Hopkins; The Bloomberg-Kimmel Institute for Cancer Immunotherapy at Johns Hopkins; the NCI (P01CA247886–01 to E.M. Jaffee, E.J. Fertig, D.T. Le, and N. Zaidi; K08CA248624–02 to N. Zaidi; U01 CA253403, U54CA274371, and U01CA271273 to E.J. Fertig); Stand Up To Cancer; the Lustgarten Foundation; and Break Through Cancer. N. Zaidi is a recipient of the 2021 American Society of Clinical Oncology Career Development Award. G.L. Stein-O’Brien is supported by the Kavli NDI Distinguished Postdoctoral Fellowship, the Johns Hopkins Provost Postdoctoral Fellowship, and the BRAIN Initiative in partnership with the National Institute of Neurologic Disorders (K99NS122085).
Authors’ Disclosures
D.T. Le reports grants and other support from Merck, Bristol Myers Squibb, and Nouscom, and grants from G1 Therapeutics, Janssen, Merus, Curegenix, and AbbVie outside the submitted work. E.M. Jaffee reports other support from Abmeta and Adventris, personal fees from Achilles, DragonFly, Parker Institute, Surge, Mestag, and Medical Home Group, and grants from Lustgarten, Genentech, Bristol Myers Squibb, and Break Through Cancer outside the submitted work. E.J. Fertig reports personal fees from Resistance Bio during the conduct of the study, as well as personal fees from Mestag Therapeutics and Merck outside the submitted work. N. Zaidi reports nonfinancial support from Bristol Myers Squibb, personal fees from Genentech, and other support from Adventris Pharmaceuticals outside the submitted work. No disclosures were reported by the other author.
REFERENCES
- 1.Cercek A, Lumish M, Sinopoli J, Weiss J, Shia J, Lamendola-Essel M, et al. PD-1 blockade in mismatch repair–deficient, locally advanced rectal cancer. N Engl J Med 2022;386:2363–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immuno-surveillance to tumor escape. Nat Immunol 2002;3:991–8. [DOI] [PubMed] [Google Scholar]
- 3.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377:2500–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho WJ, Erbe R, Danilova L, Phyo Z, Bigelow E, Stein-O’Brien G, et al. Multi-omic profiling of lung and liver tumor microenvironments of metastatic pancreatic cancer reveals site-specific immune regulatory pathways. Genome Biol 2021;22;154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popovic A, Jaffee EM, Zaidi N. Emerging strategies for combination checkpoint modulators in cancer immunotherapy. J Clin Invest 2018;1283209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis-Marcisak EF, Deshpande A, Stein-O’Brien GL, Ho WJ, Laheru D, Jaffee EM, et al. From bench to bedside: single-cell analysis for cancer immunotherapy. Cancer Cell 2021;391062–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaidi N, Soban M, Chen F, Kinkead H, Mathew J, Yarchoan M, et al. Role of in silico structural modeling in predicting immunogenic neoepitopes for cancer vaccine development. JCI Insight 2020;5:e136991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li K, Tandurella JA, Gai J, Zhu Q, Lim SJ, Thomas DL, et al. Multi-omic analyses of changes in the tumor microenvironment of pancreatic adenocarcinoma following neoadjuvant treatment with anti–PD-1 therapy. Cancer Cell 2022;401374–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivapalan L, Murray JC, Canzoniero JVL, Landon B, Jackson J, Scott S, et al. Liquid biopsy approaches to capture tumor evolution and clinical outcomes during cancer immunotherapy. J Immunother Cancer 2023;11e005924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szeto GL, Finley SD. Integrative approaches to cancer immunotherapy. Trends Cancer 2019;5400–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fertig EJ, Jaffee EM, Macklin P, Stearns V, Wang C. Forecasting cancer: from precision to predictive medicine. Cell Med 2021;2;1004–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milberg O, Gong C, Jafarnejad M, Bartelink IH, Wang B, Vicini P, et al. A QSP model for predicting clinical responses to monotherapy, combination and sequential therapy following CTLA-4, PD-1, and PD-L1 checkpoint blockade. Sci Rep 2019;911286. [DOI] [PMC free article] [PubMed] [Google Scholar]


