Abstract
We studied the DNA sequence variation of van genes of 34 isolates of Enterococcus spp. The isolates containing the vanB gene exhibited between 0 and 41 base pair changes per 801 bp studied when the vanB sequences were compared to that of the reference strain Enterococcus faecalis V583. The isolates carrying the vanC-2 gene exhibited between 0 and 23 base pair changes per 346 bp studied when the vanC-2 sequences were compared to that of the reference strain E. casseliflavus ATCC 25788. Little variation was noted in the vanA and vanC-1 genes.
We have previously described a multiplex PCR-restriction fragment length polymorphism (RFLP) assay which detects and discriminates vanA, vanB, vanC-1, and vanC-2/3 genes in Enterococcus spp. (5). In our original study we noted that in 4 of 63 isolates of vancomycin-resistant enterococci (VRE), a PCR product was produced but that it was produced with an amplicon which had RFLPs which differed from those found with the reference vanA, vanB, vanC-1, and vanC-2 strains. We detected sequence variability to account for the unique MspI restriction enzyme patterns observed. Since our assay detected sequence variation only in resistance genes that had variations located at their restriction enzyme sites, we hypothesized that there would be further sequence variation present in the van genes of enterococci. The objective of the present study was to determine the sequence variation of the vanA, vanB, vanC-1, and vanC-2/3 genes in VRE.
Thirty-four clinical isolates of Enterococcus spp. were studied (5). The 34 isolates included 10 isolates which we previously identified as carrying the vanA gene (E. faecium [n = 9] and E. gallinarum [n = 1] [vancomycin MIC, ≥256 μg/ml; teicoplanin MIC, >16 μg/ml]), 8 isolates previously identified as carrying the vanB gene (E. faecium [n = 5] and E. faecalis [n = 3] [vancomycin MIC, 128 to >256 μg/ml; teicoplanin MIC, ≤8 μg/ml]), 9 isolates previously identified as carrying the vanC-1 gene (E. gallinarum), and 7 isolates previously identified as carrying the vanC-2 gene (E. casseliflavus [n = 6] and E. flavescens [n = 1] [vancomycin MIC, 4 to 8 μg/ml; teicoplanin MIC, ≤8 μg/ml]) (5). The following reference strains were used: E. faecium B7641 (vanA), E. faecalis V583 (vanB), E. gallinarum GS (vanC-1), and E. casseliflavus ATCC 25788 (vanC-2) (all of which were kindly provided by Daniel F. Sahm) (4–6).
Identification and antimicrobial susceptibility testing, PCR, and amplicon sequencing of isolates of Enterococcus spp. were performed as previously described (5). For pulsed-field gel electrophoresis (PFGE), a 7-ml sample of log-phase bacterial cells in brain heart infusion broth was centrifuged, the supernatant was discarded, and the cell pellet was added to 500 μl of 0.5 M EDTA–0.1 M EGTA–1 M Tris (pH 8) in distilled water. One-half milliliter of 1.6% SeaPlaque GTG agarose in the same solution was added. An insert was prepared, removed, and placed in a solution of 6 mM Tris-hydrochloride (pH 7.6), 1 M NaCl, 0.1 M EDTA, 0.5% Brij 58 (Sigma, St. Louis, Mo.), 0.2% deoxycholate (Sigma), 0.5% N-lauroylsarcosine (Sigma), 20 μg of RNase per ml, and 1 mg of lysozyme (Sigma) per ml in distilled water. Following overnight incubation, the lysis buffer was discarded and 1 ml of 0.5 M EDTA–0.1 M EGTA–1 M Tris (pH 8)–1% sodium dodecyl sulfate and 1 mg of proteinase K (Boehringer Mannheim, Indianapolis, Ind.) per ml in distilled water were added. This mixture was incubated at 50°C in a water bath, and then the insert was washed four times with Tris-EDTA wash buffer. For restriction endonuclease digestion, the inserts were removed from the buffer and equilibrated for 10 min in a solution containing 450 μl of distilled water and 50 μl of restriction buffer. SmaI (3 μl) was added, and the mixture was incubated at 37°C overnight. The inserts were then placed in 0.5× TAE. Thin slices of the inserts were loaded into the gel, and PFGE was performed. Previously published guidelines for interpreting chromosomal DNA restriction patterns produced by PFGE were used for the interpretation of PFGE findings (7).
We sequenced 825-bp DNA sequences for the vanA isolates, 801-bp DNA sequences for the vanB isolates, 360-bp DNA sequences for the vanC-1 isolates, and 346-bp DNA sequences for the vanC-2 isolates.
Nine of the 10 vanA-containing isolates had identical vanA amplicon sequences, and these sequences differed from vanA of reference strain B7641 by one base pair and one amino acid change. One isolate had a vanA amplicon sequence identical to that of the reference strain. PFGE on the 10 vanA isolates revealed that two isolates were closely related (one fragment in each isolate differed from the other) and that the remainder were different (7).
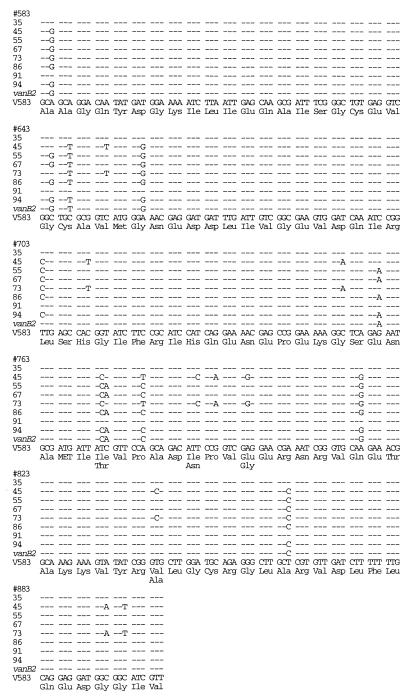
Marked sequence variation was found among the vanB-containing isolates (Fig. 1). One E. faecalis and one E. faecium isolate had sequences identical to the vanB sequence of the reference strain V583, which was used to determine the published sequence of the vanB gene (1, 2). The other six isolates, however, exhibited 41 base pair changes (5%) and 13 amino acid changes (two E. faecalis isolates, i.e., isolates 45 and 73), 31 base pair changes (4%) and 9 amino acid changes (three E. faecium isolates), and 30 base pair changes (4%) and 8 amino acid changes (one E. faecium isolate) compared to the vanB sequence of the reference strain V583. We found 22 base pair changes in each of these six isolates that were identical among these isolates but that differed from the vanB sequence of the reference strain V583. PFGE on the eight vanB isolates revealed that isolates 45 and 73 were indistinguishable, isolates 67 and 86 were closely related (one fragment in each isolate differed from the other), and the remaining four isolates were different (7).
FIG. 1.
Comparison of DNA sequences of vanB genes from VRE isolates to the vanB sequence from the reference strain V583 and to vanB2 (3).
The 584-bp sequence common to vanB2 (3) was compared to those of our vanB study isolates with marked sequence differences from the reference vanB strain V583 (Fig. 1). We found 16 base pair changes in each of our six vanB study isolates with marked sequence differences from the reference vanB strain that were identical among these isolates and that were also found in the vanB2 sequence. Isolates 45 and 73 had 16 base pair changes in common and 21 base pair changes (3.6%) that were different (resulting in five amino acid differences) from the published sequence of vanB2. The sequences of the remaining four isolates (isolates 55, 67, 86, and 94) shared 21 base pair changes with vanB2 and differed from vanB2 by only 3 to 4 (0.5 to 0.7%) base pair changes.
Our vanB isolates 45 and 73, which were identical by van gene sequencing and PFGE, showed marked sequence diversion from both the vanB gene and the vanB2 gene. We have designated the genes found in isolates 45 and 73 vanB3. Gold and associates proposed the vanB2 genotype based on a 3.6% base pair difference from vanB (3). Our isolates 45 and 73 had a 5% base pair difference from the vanB sequence from the reference strain V583 and a 3.6% base pair difference from the published sequence of vanB2.
Sequence variation was found in the vanC-1 amplicon in four of nine vanC-1 isolates. These four isolates exhibited between one and six base pair changes (up to 2% variation) resulting in no amino acid changes when they were compared to the vanC-1 sequence of the reference strain E. gallinarum GS. PFGE on the vanC-1 isolates revealed that two isolates were closely related (at most, one fragment was different from the corresponding fragment in the other isolate), one isolate had an uninterpretable PFGE pattern, and the remaining six isolates had different RFLPs (7).
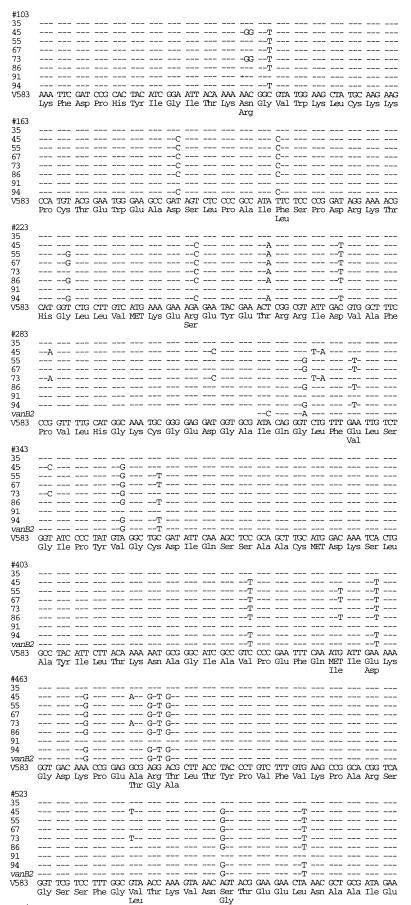
Sequence variation was found in the vanC-2 amplicons of all of the vanC-2 isolates (Fig. 2). The sequence of the reference vanC-2 strain ATCC 25788 (GenBank accession no. L29639) was confirmed. Our seven isolates exhibited between 0 and 23 base pair changes (up to 7% variation), resulting in zero to six amino acid changes from the consensus vanC-2 sequence of these isolates and of the reference strain ATCC 25788 (Fig. 2). PFGE on the vanC-2 isolates revealed that all had different RFLPs (7).
FIG. 2.
Comparison of DNA sequences of vanC-2 genes from VRE isolates, the reference strain ATCC 25788, and the consensus sequence (shown at the bottom) of these isolates.
The finding of sequence variation among the vanC-2 genes is important. Current breakpoint guidelines of the National Committee for Clinical Laboratory Standards do not accurately detect vanC-containing organisms. Therefore, in order to reliably detect vanC-containing organisms, molecular techniques must be used. The sequence variation which we have noted, however, indicates that caution is needed when ostensibly stringent molecular approaches for the detection of vancomycin resistance genes are applied to this group of microorganisms. That is, probes and PCR primers which anneal to areas of van genes prone to sequence variation may fail to detect these genes.
The sequence variation noted raises the question of whether sequencing of genes associated with vancomycin resistance may be used to determine relatedness of genes of VRE carrying vanB and vanC-2 genes for epidemiologic purposes. The greater the variation in a genetic sequence, the greater the opportunity to see relatedness and differences.
In conclusion, we have found relatively large sequence variation in the vanB and vanC-2 genes in enterococci but not, to any great extent, in the vanA and vanC-1 genes by a PCR sequencing assay. We have designated the gene found in two of our isolates vanB3. Knowledge of the sequence variation in van genes which we have identified will assist in the accurate design of PCR primers to amplify these genes.
Nucleotide sequence accession numbers.
The nucleotide sequences of isolates 45, 55, 94, 12, 21, 52, 77, 82, 44, and 32 have been submitted to GenBank and have been given accession no. U72704, U94528, U94526, U94521, U94522, U94523, U94524, U94525, U72706, and U72705, respectively.
REFERENCES
- 1.Evers S, Reynolds P E, Courvalin P. Sequence of the vanB and ddl genes encoding d-alanine:d-lactate and d-alanine:d-alanine ligases in vancomycin-resistant Enterococcus faecalis V583. Gene. 1994;140:97–102. doi: 10.1016/0378-1119(94)90737-4. [DOI] [PubMed] [Google Scholar]
- 2.Evers S, Sahm D F, Courvalin P. The vanB gene of vancomycin-resistant Enterococcus faecalis V583 is structurally related to genes encoding d-Ala:d-Ala ligases and glycopeptide-resistance proteins VanA and VanC. Gene. 1993;124:143–144. doi: 10.1016/0378-1119(93)90779-3. [DOI] [PubMed] [Google Scholar]
- 3.Gold H S, Unal S, Cercenado E, Thauvin-Eliopoulos C, Eliopoulos G M, Wennersten C B, Moellering R C. A gene conferring resistance to vancomycin but not to teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes in enterococci. Antimicrob Agents Chemother. 1993;37:1604–1609. doi: 10.1128/aac.37.8.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayden M K, Trenholme G M, Schultz J E, Sahm D F. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J Infect Dis. 1993;167:1224–1227. doi: 10.1093/infdis/167.5.1224. [DOI] [PubMed] [Google Scholar]
- 5.Patel R, Uhl J R, Kohner P, Hopkins M K, Cockerill F R., III Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J Clin Microbiol. 1997;35:703–707. doi: 10.1128/jcm.35.3.703-707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahm D F, Free L, Handwerger S. Inducible and constitutive expression of vanC-1-encoded resistance to vancomycin in Enterococcus gallinarum. Antimicrob Agents Chemother. 1995;39:1480–1484. doi: 10.1128/aac.39.7.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]