Abstract
Background
Chronic rhinosinusitis with nasal polyposis (CRSwNP) is a multifactorial disease with no known single cause, but it is thought that bacteria play a role in the disease process.
Objective
This pilot study aims to assess the longitudinal effect of corticosteroid therapy on sinus microbiota in chronic rhinosinusitis patients with nasal polyposis (CRSwNP).
Methods
A longitudinal prospective case-control study was done on patients with CRSwNP and healthy controls. Patients with CRSwNP were randomly allocated to a corticosteroids and antibiotics treatment group (CRSwNP-SA) or a corticosteroid-only treatment group (CRSwNP-S). Data were collected at three-time points (before treatment, 1, and 3 months after treatment). Specimens were cultured and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) was used as a bacterial detection method.
Results
Data from 29 patients with CRSwNP (16 CRSwNP-SA and 13 CRSwNP-S) was compared to 15 healthy subjects. Patients reported significant symptom improvement initially (1 month), but not in the long-term (3 months). This result was found in both treatment groups, whether or not antibiotics were used. After 3 months from treatment, the prevalence of Corynebacterium genera tended to increase in the CRSwNP-SA, while Staphylococcus and Gram-negative genera (Pseudomonas) tended to increase in the CRSwNP-S. Smoking, aspirin sensitivity, and previous endoscopic sinus surgery were found to be co-factors significantly associated with the response to systemic corticosteroid therapy.
Conclusion
In this pilot study, both treatment options were effective to improve symptoms in the short-term but not in the long-term, and were not linked to any clear sinus microbiota response. As a result, this study supports the avoidance of systemic antibiotics without evidence of active infection.
Keywords: chronic rhinosinusitis, microbiota, corticosteroids, antibiotics, nasal polyps, SNOT-22
Introduction
Chronic rhinosinusitis (CRS) is a common and debilitating inflammatory disease affecting the nasal mucosa and the paranasal sinuses. 1 The estimated prevalence in the American population is up to 16% and in the European population is around 10.9%.2,3 A survey conducted on Canadian households reported the prevalence to be around 5%, with 2.89 million prescriptions dispensed in 2006 for acute rhinosinusitis (ARS) or CRS.4,5
CRS can be subdivided further phenotypically into chronic rhinosinusitis without nasal polyposis (CRSsNP), or chronic rhinosinusitis with nasal polyposis (CRSwNP), with the latter comprising 20% to 30% of CRS patients.6,7,8 CRSwNP is a multifactorial disease with no known single cause. Many theories have been proposed to better understand disease pathogenesis, which can be divided into either (a) host-related factors such as the immunity barrier hypothesis or (b) environmental factors such as the microbiome hypothesis, which has been gaining interest over the last few years. 9 However, the causative factors continue to be an area of ongoing research. Often, the most prevalent bacteria in CRS was Staphylococcus aureus. 10 However, with increased use of culture-independent bacterial identification methods, recent studies concluded that Corynebacterium and Staphylococcus were more often found in healthy sinuses, and there were changes in the abundance and richness seen in the CRSwNP patients. Pseudomonas and Moraxella were more frequently identified in CRSwNP patients as well.11,12,13
Appropriate medical therapy includes topical and oral corticosteroids, but there remains some ambiguity about the value of concurrent antibiotics. 14 Few studies have assessed the effect of these treatment options on the sinus microbiota, focusing on the immediate changes within the first month of receiving the treatment. 12 However, studying the effect of these treatments on the sinus microbiota might improve our understanding of disease pathogenesis, and lead to further refinements in management decisions.
The aim of this pilot study is to identify the longitudinal impact of corticosteroid therapy, with or without antibiotics, on the sinus microbiota in chronic rhinosinusitis patients with nasal polyposis (CRSwNP).
Methods
Study Design and Population
A longitudinal prospective case–control study with two distinct treatment arms was performed. The case group was composed of patients diagnosed with CRSwNP, and control subjects were patients undergoing either a transsphenoidal resection of pituitary tumors or a septoplasty. CRSwNP were assigned to one of the two treatment groups using simple randomization. Appropriate IRB approval from the McGill University Health Center research ethics board was obtained for this study.
The definition of the American Academy of Otolaryngology–head and neck surgery clinical practice guidelines for adult sinusitis was used to select patients with CRSwNP. 6 The following inclusion criteria were used: (1) documented diagnosis of bilateral CRSwNP; (2) 18 years of age or older; and (3) no antibiotics and/or oral corticosteroids for at least one month prior to recruitment. Patients were excluded if they were found to have: (1) documented diagnosis of fungal sinusitis; (2) pregnancy, (3) diagnosed Crohn's disease or ulcerative colitis; (4) diagnosed immotile cilia syndrome; (5) diagnosed cystic fibrosis; (6) diagnosed immunodeficiency syndrome; (7) diagnosed sinonasal tumor; (8) penicillin allergy; or (9) patients on immunomodulatory medications.
As a pilot study, a sample size of 10 or more subjects per group was chosen, and considered to be satisfactory.12,15 CRSwNP patients were divided into two subgroups based on treatment. The first subgroup received corticosteroids (Prednisone tapering dose once daily for 21 days, first week: 30 mg, second week: 15 mg, and the third week: 5 mg) in addition to antibiotics (Amoxicillin Clavulanate 875 mg, by mouth twice a day for 14 days). The second subgroup received only Prednisone with the same tapering dose. Both treatment groups were concurrent to their baseline intranasal saline irrigation and topical corticosteroid.
Endoscopically guided flocked nasal swabs (Copan Italia S.p.A., Brescia, Italy) were collected from the middle meatus using a guarded technique by rotating it at least five times clockwise and counterclockwise. From each patient with CRSwNP, three different nasal swabs from the same side were collected at three different time points. The first was collected during the time of recruitment and completion the study entry questionnaire (time point 1). The second nasal swab was collected after a month, soon after finishing the course of treatment based on the assigned group (time point 2). Finally, the third nasal swab was collected three months after completing the treatment (time point 3). During each visit, every subject completed a questionnaire to track the change in disease status using SNOT-22. 16 The nasal swabs were placed on ice as soon as they were collected and transferred within four hours to the proper laboratory for processing.
Sample Processing
Each sample was cultured in a standardized fashion as described previously by striking the swab onto four different media: blood agar, chocolate agar, mannitol salt agar, and methylene blue agar. 17 Plates were then cultured in an incubator at 37°C under 5% CO2–20% O2 atmosphere for 72 h.
Bacterial colonies were differentiated by color, size, shape, consistency, and hemolysis. A 10 μl loop was used to reculture the identified colonies in the same media with the same technique described earlier and incubated for another 72 h in the same atmosphere. Furthermore, the colonies were isolated in 1 ml of 80% glycerol tube and stored in a −80°C fridge until further processing.
Microbial Extraction
In preparation for microbial identification, isolates stored in −80°C fridge were stroked on a Lysogeny broth (LB) media using the same striking technique described above and cultured in an incubator at 37°C under 5% CO2–20% O2 atmosphere for 72 h.
From the cultured isolate a scraped sample of pure cells was taken and added in a 1.5 ml Eppendorf tube filled with 300 µl of mass spectrometer water and vortexed for 10 s to suspend cells in water. Furthermore, 900 µl of 100% ethanol was added and vortexed for 10 s as well. The sample was spun for 2 min at 15 000 RPM, and then the supernatant was decanted and the sample was respun for 2 min with the same settings used before. Using a 200 µl pipette the remnant of the supernatant was removed, and the samples were left to dry for a minimum of 5 min. A Twenty-five µl of 70% formic acid was added and the sample was then vortexed for 10 s. Finally, 25 µl of acetonitrile was added by mixing the sample gently to avoid introducing bubbles, and the last spin was done at this point for 2 min at 15 000 RPM. After removing 10 µl into a 0.2 ml PCR Eppondorf tube to avoid the pellet, the sample was ready to be spotted on the microbial identification plate.
Microbial Identification
For microbial identification, 1 µl extracted bacterial isolates were placed on a 96-spot target steel plate (Bruker Daltonics, Solna, Sweden) and left to dry at room temperature. The bacterial spots were then coated with 1 µl alpha-cyano-4-hydroxycinnamic acid (HCCA) matrix solution and kept for crystallization. Samples were processed for identification in the MALDI-TOF MS, using the flexcontrol software (FlexControl 3.3, Bruker Daltonics) in a linear positive mode in a range of 2000-20 000 DA. Using MALDI Biotyper software 3.1 (Bruker Daltonics) and mass spectra were analyzed using the reference database MBT-BDAL-5627. The microbial identification criteria were used as described in the literature. 18
Statistical Analysis
Statistical analysis was done using GraphPad Prism: Version 8.1 (GraphPad, La Jolla, CA). Data obtained at the three sampled times were summarized descriptively. Continuous data were summarized using mean ± standard deviation. Then, comparisons between two-time points and the two different groups were done using t-tests. Comparisons between the three different time points were done using one-way ANOVA test.
Categorical data were summarized as percentages; differences between two-time points or groups were assessed using Fisher's exact test. Changes between three-time points were assessed for significance using the Pearson Chi-square test.
Multivariant regression analysis was done to assess the effect of the number of organisms isolated, gender, age of patients, asthma, aspirin sensitivity, previous endoscopic sinus surgery, smoking, and the most prevalent genera on the response to treatment in each case group. P value < 0.05 was considered statistically significant.
Results
Patient Demographics and Clinical Characteristics
A total of 29 CRSwNP patients and 15 controls were recruited. The 29 CRSwNP patients were subdivided into 16 patients treated with antibiotics (CRSwNP-SA) and 13 without antibiotics (CRSwNP-S). There was no significant difference in the mean age and gender between the control and study groups. Furthermore, several variables such as smoking, aspirin sensitivity, asthma, and previous endoscopic sinus surgery (ESS) were assessed between CRSwNP-SA and the CRSwNP-S groups and showed no statistical significance (Table 1).
Table 1.
Patient Demographics and Clinical Characteristics.
| Characteristics | Control Cohort (n = 15) |
CRSwNP-SA Cohort (n = 16) |
CRSwNP-S Cohort (n = 13) |
P value |
|---|---|---|---|---|
| Age, mean (± SD), y | 43.07 (±13.72) | 52.13 (±14.39) | 47.54 (±12.39) | 0.19 |
| Gender M:F | 11:4 | 8:7 | 6:7 | 0.33 |
| Smoking (%) | 5 (33.3) | 5 (31.25) | 4 (30) | 0.99 |
| Aspirin sensitivity (%) | 5 (31.25) | 6 (46.15) | 0.99 | |
| Asthma (%) | 8 (50) | 10 (76.9) | 0.24 | |
| Previous ESS (%) | 6 (37.5) | 9 (69.2) | 0.13 |
Abbreviation: ESS, endoscopic sinus surgery.
Microbial Profile and Comparisons
The growth rate in the control cohort was 76.2%, while the identification rate was 95.24%. Eighteen different organisms were isolated in this group, 14 Gram-positive and 4 Gram-negative. Staphylococcus was the most prevalent genera (73.3%) and Micrococcus (13.3%). On a species level, the most prevalent species in the control group was Staphylococcus aureus.
The difference in the number of isolated organisms and the difference between Gram-positive and Gram-negative isolates were not statistically significant between the control and the CRSwNP group. In CRSwNP, the most prevalent organism Staphylococcus, and there was no significant difference once compared with the controls (P = 0.46). Also, Corynebacterium was the second most prevalent organism isolated, but again it was not statistically significant (P = 0.39).
In the CRSwNP-SA group, differences in the number of organisms isolated between the three different time points were insignificant (P = 0.92). As well, there was no change in the Gram-positive microbiota or the Gram-negative microbiota. However, when comparing the three-time points using the one-way ANOVA test, the SNOT-22 score showed a significant improvement (P < 0.0001). The main improvement was between time point zero (CRSwNPSA-0) and 1 month posttreatment (CRSwNPSA-1; P = 0.019; Table 2).
Table 2.
Comparing CRSwNP Corticosteroid and Antibiotics Therapy Cohort Characteristics Over the Three Different Time Points.
| Characteristics | CRSwNPSA-0 | CRSwNPSA-1 | CRSwNPSA-3 | P value |
|---|---|---|---|---|
| Growth rate | 93.75% | 100% | 100% | |
| Identification rate | 100% | 93.75% | 87.5% | |
| Species no. (Mean ± SD) | 24 (1.50 ± 0.89) | 22 (1.37 ± 0.71) | 25 (1.66 ± 1.39) | 0.92 |
| Gram positive (%) | 23 (95.83) | 21 (95.45) | 24 (96) | 0.78 |
| Gram negative (%) | 2 (4.17) | 1 (4.5) | 1 | |
| SNOT-22 Mean (±SD) | 39.13 (±30.77) | 32.44 (±27.85) | 36.31 (±29.80) | <0.0001 |
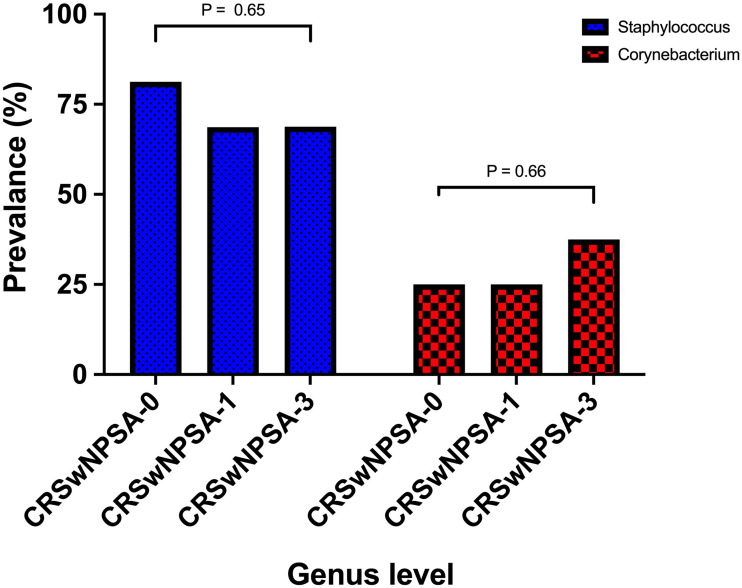
The most prevalent genera in CRSwNP-SA observed at the three different time points was Staphylococcus, with insignificant change after receiving the treatment (P = 0.65). Corynebacterium was the second most prevalent genera with an increase between 1 month posttreatment (CRSwNPSA-1) and 3 months posttreatment (CRSwNPSA-3; 17.5%). However, this slight difference was not significant (P = 0.66; Figure 1). With a deeper look at the species level Staphylococcus epidermis and Staphylococcus aureus were communally isolated at the three different time points.
Figure 1.
Comparing the most prevalent genera between the three different time points in the chronic rhinosinusitis with nasal polyposis group who received corticosteroid and antibiotics (CRSwNP-SA).
Multivariant regression analysis was done to assess the effect of several variables on the response in the CRSwNP-SA treatment group. Only smoking showed a significant impact on CRSwNPSA-1 (P = 0.04).
The number of organisms isolated in the CRSwNP-S group did not show a statistically significant change between the three different time points (P = 0.8). There was a tendency for the number of Gram-negative isolates to increase at the 3 month time point (CRSwNPS-3; ≈ 24%), however the difference between the Gram-positive and Gram-negative microbiota is insignificant. The SNOT-22 score showed a significant improvement when comparing the three measured time points (P = 0.03). The main improvement was between CRSwNPS-0 and CRSwNPS-1 (P = 0.004) (Table 3).
Table 3.
Comparing CRSwNP Corticosteroid Therapy Cohort Characteristics Over the Three Different Time Points.
| Characteristics | CRSwNPS-0 | CRSwNPS-1 | CRSwNPS-3 | P value |
|---|---|---|---|---|
| Growth rate | 86.7% | 100% | 93.4% | |
| Identification rate | 100% | 93.4% | 100% | |
| Species no. (Mean ± SD) | 21 (1.61 ± 1.12) | 21 (1.61 ± 0.86) | 21 (1.61 ± 0.86) | 0.803 |
| Gram positive (%) | 20 (95.2%) | 19 (90.4%) | 16 (76.1%) | 0.16 |
| Gram negative (%) | 1 (4.8%) | 2 (9.6%) | 5 (23.9%) | |
| SNOT-22 Mean (±SD) | 54.08 (±31.40) | 29.85 (±24.98) | 34.62 (±31.25) | 0.033 |
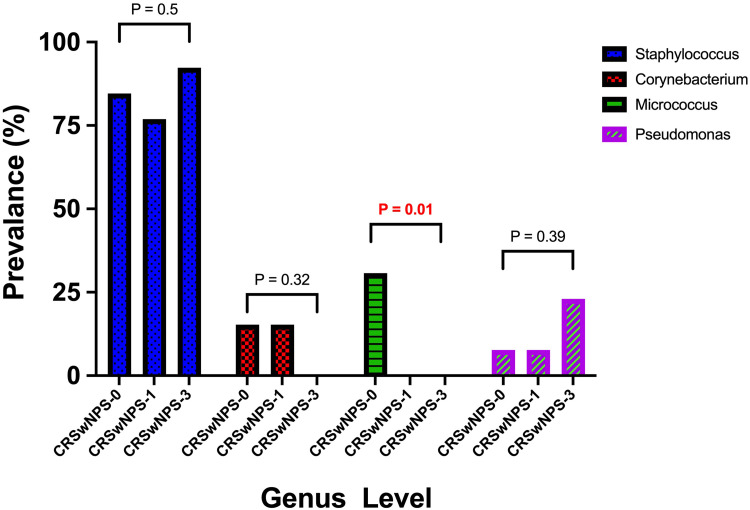
The most prevalent genera in CRSwNP-S observed at the three different time points was Staphylococcus, with a slight drop after receiving the treatment and a noticeable increase after three months of receiving the treatment. However, this pattern was not statistically significant (P = 0.5). Micrococcus was the second most prevalent organism in CRSwNPS-0, but it was not isolated in the other measurement points after treatment. This change was statistically significant (P = 0.01). Three months after corticosteroid treatment Pseudomonas identification was observed more frequently, with no statistical significance (P = 0.39) (Figure 2). At the species level, while Staphylococcus epidermis and Micrococcus luteus were the most prevalent before treatment, Staphylococcus aureus and Staphylococcus epidermis were communally isolated at the measurement points after treatment.
Figure 2.
Comparing the most prevalent genera between the three different time points in the chronic rhinosinusitis with nasal polyposis group who received corticosteroid therapy (CRSwNP-S).
A multivariable regression analysis was done to assess the effect of several variables on the response in the CRSwNP-S treatment group. It was found that smoking, previous endoscopic sinus surgery, and aspirin sensitivity had a significant impact (P = 0.04, P = 0.04, P = 0.02, respectively) on clinical improvement in CRSwNPS-3.
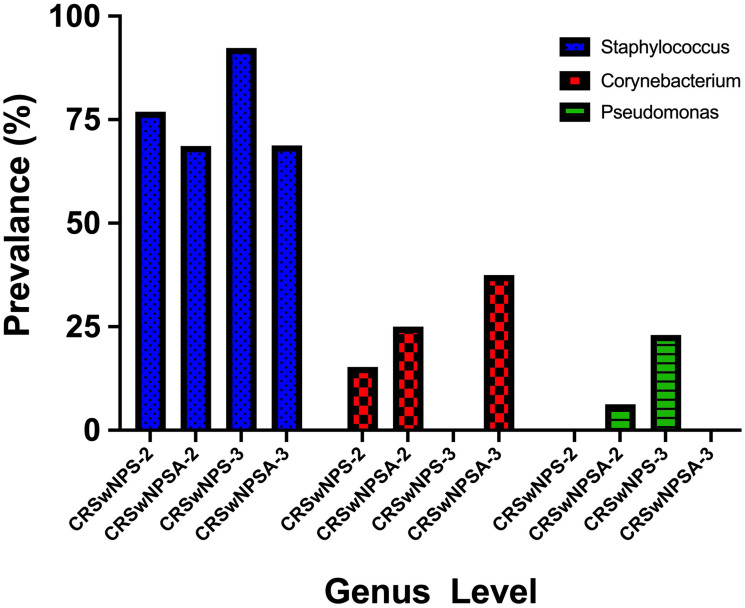
The difference between CRSwNPSA-1 and CRSwNPS-1 in the number of species isolated was not statistically significant (P = 0.61). Neither was the difference between Gram-positive and Gram-negative isolates (P = 0.6). Also, the response to treatment that was measured by the SNOT-22 scoring system was not significantly different between both groups (P = 0.79). In the two cohorts, the most prevalent microbiota isolates were Staphylococcus and Corynebacterium, with no significant difference (Figure 3).
Figure 3.
Comparing the most prevalent genera between the two different treatment groups; chronic rhinosinusitis with nasal polyposis who received corticosteroid and antibiotics therapy (CRSwNP-SA) and chronic rhinosinusitis with nasal polyposis who received corticosteroid therapy after (CRSwNP-S) at the one month and three months’ time points.
On the other hand, when comparing CRSwNSA-3 and CRSwNPS-3 patients. The prevalence of Gram-negative was higher in CRSwNPS-3 (38.4%) while the percentage in CRSwNPSA-3 was (6.25%). However, the difference was insignificant (P = 0.06). At 3 months posttreatment, Staphylococcus was the most prevalent bacteria in both groups, with 24% increase in CRSwNPS-3 compared to CRSwNPSA-3(P = 0.18). With the increase of Staphylococcus prevalence in CRSwNPS-3, there was a decrease in the prevalence of Corynebacterium, which led to a significant statistical difference when compared to CRSwNPSA-3 (P = 0.02). Moreover, in CRSwNPS-3, the prevalence of Gram-negative isolates was more frequent and Pseudomonas organisms were isolated in 23% of the participants (P = 0.07) (Figure 3).
Discussion
Although the effect of systemic corticosteroids and antibiotics on the sinus microbiota is poorly understood, they are still the main treatment for CRS. This is a pilot longitudinal study, aimed to investigate the effect of systemic corticosteroid therapy with and without antibiotics on the sinus microbiota in CRSwNP patients and compare it with controls. This is the first study to assess the longitudinal effect of corticosteroid therapy on sinus microbiota in CRSwNP patients. We were unable to detect a significant change in sinus microbiota, but in the CRSwNP-S treatment group, a slight decrease in Staphylococcus genera was noted immediately posttreatment, with an increase in both Staphylococcus and Gram-negative genera at the long-term posttreatment period associated with worsening of clinical symptoms.
In this study, all patients received prednisone, and a short-term Amoxicillin Clavulanate was added to one of the treatment groups to assess their effect on the sinus microbiota. Amoxicillin Clavulanate was added for a broad spectrum coverage and patients with a history of penicillin allergy were excluded. The findings of this study suggest that antibiotics do little to alter the microbial environment in patients with CRSwNP in the setting of empirical use without infection. For multiple reasons, including side effects and the emergence of bacterial resistance, the trend has been to move away from antibiotics in general in the absence of clear infection. Steroid irrigations have been an essential part of the appropriate medical therapy for CRS, and the use in this study was in keeping with multiple clinical practice guidelines recommendations.6,14
This study was able to identify Staphylococcus and Corynebacterium as the most prevalent microbiota in patients with CRSwNP. This finding was compatible with Paramasivan et al, the largest study to date describing the sinus microbiota. 19 In addition, there was no significant difference in the sinus microbiota prevalence between CRSwNP patients and normal subjects, although recent reports discussed a significant drop in the prevalence of Corynebacterium genera and an increase in Streptococcus genera associated with CRSwNP. 19
Moreover, both treatment options used in this study showed significant short-term clinical improvement when assessed using SNOT-22 at the second time point. In both groups, there was a slight decrease in Staphylococcus prevalence but no difference in Corynebacterium genera. This might explain this improvement with no notable change in the number of the bacteria isolated. An additional change was seen in the CRSwNP-S group: Micrococcus genera was isolated before the treatment but was not isolated posttreatment. However, Micrococcus is not a common organism isolated reported in the CRS literature. Thus, further investigation is required to clarify this finding. Jain et al recently compared the effect of antibiotics and corticosteroids on sinus microbiota. They found a decrease in relative abundance in Corynebacterium genera after corticosteroid treatment and an increase of relative abundance in the Propionibacterium genera after antibiotics treatment. However, this change was not assessed in this study due to the difference in the methodologies used. 12
Three months after completing the medical treatment, there was a worsening of clinical symptoms in both groups. In CRSwNPSA-3 (corticosteroids and antibiotics group at 3 months posttreament), Corynebacterium genera showed a tendency to increase in prevalence, which could be linked to the deterioration in clinical symptoms seen in this group. A meta-analysis published recently found Corynebacterium genus was linked to CRS and worsening of the disease as well. 20
In CRSwNPS-3, there was an increase in the Gram-negative-genera prevalence with around 24% mostly Pseudomonas and an increase as well in Staphylococcus genera. In the past, Staphylococcus and Pseudomonas have been the most commonly cultured microbiota in CRS. Several reports linked disease recalcitrance postoperatively to positive cultures for Staphylococcus and Pseudomonas.21,22,23,24 Understanding this pattern further might improve the medical treatment of CRSwNP in general.
A Multivariant analysis was done to assess the different cofactors that might impact the response to corticosteroid therapy in CRSwNP patients 3 months after receiving the treatment. It was found that smoking, previous endoscopic sinus surgery, and aspirin sensitivity were all significantly associated with the responses. In this study, there is an imbalance in the number of patients who had a previous ESS with a 37.5% (6/16) in the CRSwNP-SA cohort and 69.2% (9/13) in the CRSwNP-S cohort. This is a limitation of this study, future studies are recommended to assess the effect of corticosteroids on CRSwNP patients who have not undergone a previous ESS. Jain et al, have assessed the changes in bacterial communities in CRS patients before and after surgeries and showed an increase in the Staphylococcus genus levels after surgery with a reduction in most other genera. Microbiome changes in this study did not show an effect on the outcome of ESS. 25
In 2013, a study discussing the factors affecting the sinus microbiota found distribution of species in smokers differed qualitatively with a significant increase in the species Staphylococcus aureus, where Propionibacterium acnes and Corynebacteria tended to decrease. 26
Microbiotyping using MALDI-TOF-MS provides an effective method of microbial identification in the sinuses of patients with CRSwNP and in normals; in the current study, the most prevalent bacteria were readily identified. In addition, it was possible to better understand the effect of corticosteroid therapy on sinus microbiota longitudinally and to compare it with the additional effect of antibiotics. Although, there was not a significant difference in clinical response between the two treatment groups. This agrees with the results from other studies and stresses the benefits of decreasing antibiotics use such as decrease in cost, medication side effects, drug-resistance, and morbidity. An advantage in the current study (in addition to the longitudinal design) is the standardized treatment between participants, which limits the intervention bias.
Several limitations have been identified in this study. First, this study examined the effect of systemic corticosteroid therapy with and without antibiotics on a relatively small number of patients. As a result, some of the notable trends in the bacterial prevalence between treatment groups and the clinical scores did not reach statistical significance. A further study with a larger sample size should better clarify changes in the sinus microbiota in response to medical treatment. A second limitation of this study was that the microbiota identification method used focused on aerobic bacteria. Thus, corticosteroid effects on the anaerobic bacteria, and the associated changes, were not investigated.
The microbiota identification method used in this study had multiple advantages. It was cost-effective, fast, accurate, and gave sufficient microbiota information at the strain level. However, a DNA sequencing identification method can provide more information to better understand the change in the sinus microbiota by investigating the microbiome abundance and diversity. Some limitation of the DNA sequencing identification method includes the inability to differentiate between live and dead organisms, commensal and inhaled organisms will still amplify, the need for extensive training, and the high cost. The current literature on microbiota in CRS remains limited due to the lack of standardization of sampling, variation in detection methods protocols, and the presence of confounding factors in many studies such as different antibiotic regimens and ESS.
Finally, while this study focused on examining the effect of corticosteroid therapy on sinus microbiota in CRSwNP, other subtypes of CRS such as CRSsNP (chronic rhinosinusitis without nasal polyposis) and cystic fibrosis still requires further study to clarify the relation between these varies disease subtypes, response to treatment and the microbiota.
Conclusion
In this pilot study, both treatment options were effective to improve symptoms in the short-term but not in the long-term, and were not linked to any clear sinus microbiota response. As a result, this study supports the avoidance of systemic antibiotics without evidence of active infection. It also highlights a potential role of Staphylococcus and Pseudomonas in the disease response to medical treatment. Finally, as a future direction, a continuation project using one of the nucleic acid bacterial identification methods to compare with a larger study group will add a better understanding of long-term medical treatment effects on sinus bacterial communities.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Ethical Approval and Informed Consent: The McGill University Research Ethics Committee approved this research with the number (MP-37-2017-3073) and consent was obtained from all participating subjects.
ORCID iD: Yousif Alammar MDhttps://orcid.org/0000-0003-2791-4550
References
- 1.Desrosiers M, Evans GA, Keith PK, et al. Canadian Clinical practice guidelines for acute and chronic rhinosinusitis. Allergy Asthma Clin Immunol. 2011;7(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackwell DL, Collins JG, Coles R. Summary health statistics for U.S. adults: National health interview survey, 1997. Vital Health Stat. 2002;10(205):1–109. [PubMed] [Google Scholar]
- 3.Hastan D. Chronic rhinosinusitis in Europe – an underestimated disease. A GA(2)LEN study. Allergy. 2011;66(9):1216–1223. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y. The epidemiology of chronic rhinosinusitis in Canadians. Laryngoscope. 2003;113(7):1199–1205. [DOI] [PubMed] [Google Scholar]
- 5.IMS Health: Canadian Disease and Therapeutic Index (CDTI) database.2006.
- 6.Rosenfeld RM. Clinical practice guideline (update): adult sinusitis executive summary. Otolaryngol Head Neck Surg. 2015;152(4):598–609. [DOI] [PubMed] [Google Scholar]
- 7.Crombruggen K. Pathogenesis of chronic rhinosinusitis: inflammation. J Allergy Clin Immunol. 2011;128(4):728–732. [DOI] [PubMed] [Google Scholar]
- 8.Hamilos DL. Chronic rhinosinusitis patterns of illness. Clin Allergy Immunol. 2007;20:1–13. [PubMed] [Google Scholar]
- 9.Lam K, Schleimer R, Kern RC. The etiology and pathogenesis of chronic rhinosinusitis: a review of current hypotheses. Curr Allergy Asthma Rep. 2015;15(7):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feazel LM, Robertson CE, Ramakrishnan VR, Frank DN. Microbiome complexity and Staphylococcus aureus in chronic rhinosinusitis. Laryngoscope. 2012;122(2):467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoggard M, Biswas K, Zoing M, Wagner Mackenzie B, Taylor MW, Douglas RG. Evidence of microbiota dysbiosis in chronic rhinosinusitis. Int Forum Allergy Rhinol. 2017;7(3):230–239. [DOI] [PubMed] [Google Scholar]
- 12.Jain R, Hoggard M, Zoing M, et al. The effect of medical treatments on the bacterial microbiome in patients with chronic rhinosinusitis: a pilot study: the CRS microbiome after medical therapy. Int Forum Allergy Rhinol. 2018;8(8):890–899. [DOI] [PubMed] [Google Scholar]
- 13.Biswas K, Hoggard M, Jain R, Taylor MW, Douglas RG. The nasal microbiota in health and disease: variation within and between subjects. Front Microbiol. 2015;9:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fokkens WJ, Lund VJ, Hopkins C, et al. European position paper on rhinosinusitis and nasal polyps 2020. Rhinology. 2020;58(Suppl S29):1–464. [DOI] [PubMed] [Google Scholar]
- 15.Liu CM, Soldanova K, Nordstrom L, et al. Medical therapy reduces microbiota diversity and evenness in surgically recalcitrant chronic rhinosinusitis: treatment decreases microbiota diversity in CRS. Int Forum Allergy Rhinol. 2013;3(10):775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopkins C, Gillett S, Slack R, Lund VJ, Browne JP. Psychometric validity of the 22-item sinonasal outcome test. Clin Otolaryngol. 2009;34(5):447–454. [DOI] [PubMed] [Google Scholar]
- 17.Koeller K, Herlemann DPR, Schuldt T, et al. Microbiome and culture based analysis of chronic rhinosinusitis compared to healthy sinus mucosa. Front Microbiol. 2018:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mårtensson A, Abolhalaj M, Lindstedt M, et al. Clinical efficacy of a topical lactic acid bacterial microbiome in chronic rhinosinusitis: a randomized controlled trial. Laryngoscope Investig Otolaryngol. 2017;2(6):410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paramasivan S, Bassiouni A, Shiffer A, et al. The international sinonasal microbiome study: a multicentre, multinational characterization of sinonasal bacterial ecology. Allergy. 2020;75(8):2037–2049. [DOI] [PubMed] [Google Scholar]
- 20.Wagner Mackenzie B, Waite DW, Hoggard M, Douglas RG, Taylor MW, Biswas K. Bacterial community collapse: a meta-analysis of the sinonasal microbiota in chronic rhinosinusitis. Environ Microbiol. 2017;19(1):381–392. [DOI] [PubMed] [Google Scholar]
- 21.Bendouah Z, Barbeau J, Hamad WA, Desrosiers M. Biofilm formation by Staphylococcus aureus and Pseudomonas aeruginosa is associated with an unfavorable evolution after surgery for chronic sinusitis and nasal polyposis. Otolaryngol Head Neck Surg. 2006;134(6):991–996. [DOI] [PubMed] [Google Scholar]
- 22.Coffey CS. Endoscopically guided aerobic cultures in postsurgical patients with chronic rhinosinusitis. Am J Rhinol. 2006;20(1):72–76. [PubMed] [Google Scholar]
- 23.Jervis-Bardy J, Foreman A, Field J, Wormald PJ. Impaired mucosal healing and infection associated with Staphylococcus aureus after endoscopic sinus surgery. Am J Rhinol Allergy. 2009;23(5):549–552. [DOI] [PubMed] [Google Scholar]
- 24.Singhal D, Foreman A, Jervis-Bardy J, Wormald P-J. Staphylococcus aureus biofilms: nemesis of endoscopic sinus surgery. Laryngoscope. 2011;121(7):1578–1583. [DOI] [PubMed] [Google Scholar]
- 25.Ramakrishnan VR, Feazel LM, Gitomer SA, Ir D, Robertson CE, Frank DN. The microbiome of the middle meatus in healthy adults. PLoS One. 2013;8(12):e85507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain R, Hoggard M, Biswas K, Zoing M, Jiang Y, Douglas RG. Changes in the bacterial microbiome of patients with chronic rhinosinusitis after endoscopic sinus surgery. Int Forum Allergy Rhinol. 2017;7(1):7–15. [DOI] [PubMed] [Google Scholar]