Abstract
Historically, bacteria of the phylum, Actinobacteria have been a very prominent source of bioactive compounds for drug discovery. Among the actinobacterial genera, Micrococcus has not generally been prioritized in the search for novel drugs. The bacteria in this genus are known to have very small genomes (generally < 3 Mb). Actinobacteria with small genomes seldom contain the well-characterized biosynthetic gene clusters such as those encoding polyketide synthases and nonribosomal peptide synthetases that current genome mining algorithms are optimized to detect. Nevertheless, there are many reports of substantial pharmaceutically relevant bioactivity of Micrococcus extracts. On the other hand, there are remarkably few descriptions of fully characterized and structurally elucidated bioactive compounds from Micrococcus spp. This review provides a comprehensive summary of the bioactivity of Micrococcus spp. that encompasses antibacterial, antifungal, cytotoxic, antioxidant, and anti-inflammatory activities. This review uncovers the considerable biosynthetic potential of this genus and highlights the need for a re-examination of these bioactive strains, with a particular emphasis on marine isolates, because of their potent bioactivity and high potential for encoding unique molecular scaffolds.
Keywords: Micrococcus, Actinomycete natural products, Drug discovery
Graphical Abstract
Graphical Abstract.
The prevalence of documented, pharmaceutically relevant bioactivity in Micrococcus strains, contrasting with a lack of their described compounds, strongly suggests that revisitation of this genus will be productive for the discovery of novel drugs.
Introduction
Micrococcus is a genus of non-spore forming actinomycetes (family Micrococcaceae) that are ubiquitous throughout terrestrial, aquatic, and marine environments (Nuñez, 2014). This genus was first described in the late 1800s, and the type strain Micrococcus luteus was originally isolated by Alexander Fleming as Micrococcus lysodeikticus in 1922 (Cohn, 1872; Fleming & Allison, 1922; Wieser et al., 2002). Micrococcus spp. are commonly associated with the human skin microbiota as well as the microbiome of dairy products such as raw milk and cheese and have even been isolated from amber (Lakshminarasim & Iya, 1955; Bhowmik & Marth, 1990; Chiller et al., 2001; Greenblatt et al., 2004; Nuñez, 2014). In the marine environment, these bacteria have been isolated from sediments as well as marine invertebrates, including sponges and corals (Montalvo et al., 2005; Wilson et al., 2012; Wang et al., 2021). Micrococcus are generally considered nonpathogenic, although some species have been the culprit of several infections and can therefore be opportunistic pathogens (Albertson et al., 1978; Fosse et al., 1985; Nuñez, 2014). Isolates are often vividly pigmented with colonies reported to be yellow, orange, green, pink, red, and white (Kocur, 1986; Jagannadham et al., 1991). Despite considerable investigation into the pigments produced by Micrococcus spp., it was noted three quarters of a century ago, and again in the past decade, that there is a paucity of research into their biosynthetic potential (Su, 1948; Palomo et al., 2013). The first Micrococcus genome was sequenced in 2009 (Young et al., 2010) and was revealed to have a small genome of only 2.5 Mb. Extensive genomic analysis has concluded that genomes less than 3 Mb rarely contain biosynthetic gene clusters (BGCs) (Donadio et al., 2007), an observation that has likely discouraged investigations into Micrococcus spp. for novel drugs.
Current State of Knowledge
Despite the general lack of readily detectable BGCs [mainly nonribosomal peptide synthetases (NRPSs), polyketide synthases (PKSs), and ribosomally synthesized and post-translationally modified peptides (RiPPs)] in Micrococcus genomes, knowledge of the antibacterial activity of Micrococcus strains has existed since 1889, when the first photographic record of antibiosis was prepared with “Micrococcus anthracotoxicus” (Doehle, 1889; Su, 1948). Several other studies performed between 1902 and 1948 identified antibacterial activity of Micrococcus strains (Löde, 1902; Dujardin-Beaumetz, 1934; Hutchinson et al., 1943; Su, 1948). Although the original work is absent from data records, Su (1948) noted that Löde isolated the active substance from a Micrococcus strain in 1902 for animal studies that ultimately failed to demonstrate any therapeutic results. This likely makes this unidentified compound the first bioactive metabolite to ever be isolated from a Micrococcus sp.
To date, there are only 28 studies documenting pharmaceutically relevant bioactivity in terrestrial and marine Micrococcus strains, with two additional sources mentioning growth stimulation in plants (Lafi et al., 2017) and oil degradation (https://www.ncbi.nlm.nih.gov/nuccore/JN709478.1). Interestingly, 50% of these studies focus on marine isolates. Their activities range from general to selective antibacterial activity, as well as antioxidant, antifungal, anti-inflammatory, and cytotoxic activities. For a complete list of bioactivities described for published strains; see Table 1.
Table 1.
All Relevant Bioactivity of Micrococcus Strains as Described in the Literature
| Strain (Accession No.) | Origin | Activity | Source of activity | Sequence data | Reference(s) |
|---|---|---|---|---|---|
| Micrococcus luteus ATCC 53 598 | Soil (Highbridge, NJ, USA) | Antibacterial | Neoberninamycin | No | Biskupiak et al., 1988 |
| Micrococcus luteus strain R-1588–10 | Marine sponge Xestospongia sp. (Noumea, New Caledonia) |
Antibacterial | Triclosan [2,4,4′-trichloro-2′- hydroxydiphenylether] Lutoside [ acyl-1-(acyl-6′-mannobiosyl)-3-glycerol]a |
No | Bultel-Poncé et al., 1998 |
| Micrococcus luteus Otnes7 | Seawater (sea surface microlayer of Norwegian coast) |
Not specified |
Sarcinaxanthin | No | Netzer et al., 2010 |
|
Micrococcus luteus strain TUB6b (HE613658.1) |
Plectranthus tenuiflorus
(Taif, Saudi Arabia) |
Antibacterial |
Cell-free supernatant | Yes | El-Deeb et al., 2012 |
|
Micrococcus luteus strain MKVKUD 2013 (KF532949.1) |
Seawater (Marina Beach, Chennai, India) |
Antibacterial | Crude pigment | Yes | Umadevi & Krishnaveni, 2013 |
| Micrococcus luteus | Agriculture fields, gardens, and closed environments (room, kitchen, and laboratory) | Antibacterial | Culture supernatant, live colonies | No | Akbar et al., 2014 |
| Micrococcus luteus | Soil (Chennai, India) | Antibreast cancer | Crude pigment extracts | No | Ushasri & Gods Will Shalomi, 2015 |
| Micrococcus luteus strain Xp 4.2 | Marine sponge Xestospongia testudinaria (shore water of Tanjung Kasuari, Sorong, Papua) |
Antibacterial | Organic fractions, possibly due to alkaloid or steroid/triterpenoid | No | Cita et al., 2017 |
| Micrococcus luteus | Lab strain (Mustansiriyah University, Iraq) |
Antibacterial Antifungal |
Crude carotenoid pigment |
No | Majeed, 2017 |
| Micrococcus luteus isolate MRN01 | Ship hull (Arabian Sea, Cochin Port, Kerala, India) |
Antibacterial—crude exopolysaccharrides, crude pigment Antifungal, cytotoxic, antioxidant—crude pigment |
Crude pigment and exopolysaccharrides *confirmed presence of sarcinaxanthin, phytoene, and phytofluene |
No | Nisha et al., 2019 |
| Micrococcus luteus and Micrococcus roseus | Soil (Savandurga hills region, Karnataka, India) | Antibacterial, antioxidant, UV-protective | Carotenoid pigments | No | Mohana et al., 2013 |
| Micrococcus roseus ATCC 516 | Not specified | Not specified | Canthaxanthin | No | Cooney et al., 1966 |
| Micrococcus roseus, psychrotrophic | Soil (Schirmacher Oasis, Antarctica) |
Membrane stabilizer | Carotenoid pigment “P3” (bisdehydro-beta-carotene-2-carboxylic acid) |
No | Jagannadham et al., 1991 |
| Micrococcus roseus (PTCC 1411) | Persian Type Culture Collection (PTCC) | Antibacterial, antifungal, anticancer, and anti-inflammatory | Crude pigment | No | Rostami et al., 2016 |
| Micrococcus roseus | Not specified | Antibacterial | Crude pigment | No | Zehra et al., 2018 |
| Micrococcus tetragenus | Human blood | Not specified | Xanthophyll, lycopene, rhodoxanthin, rubixanthin, γ-carotin, and several other unidentified pigments | No | Reimann & Eklund, 1941 |
| Micrococcus radiodurans | Not specified | Not specified | Zeaxanthin and lycophyll and derivatives | No | Lee, 1961 |
| Micrococcus yunnanensis F-256 446 | Marine sponge (Florida Keys, Florida, USA) | Anti-MRSA | Kocurin (thiazolyl peptide) | No *requested directly from authors |
Palomo et al., 2013 |
| Micrococcus yunnanensis strain rsk5 (KU991822.1) |
Root, stem, and leaf samples of Catharanthus roseus
(Rajkot, Gujarat, India) |
Antibacterial | Live colonies, extract isolated antibacterial compound |
Yes | Ranjan & Jadeja, 2017 |
| Micrococcus terreus JGI 19 (KM386643.1) | Multiple sampling sources: Lalbagh, road side, cow dung, and cow urine, diverse soil samples (in and around Bangalore, India) | Anticancer | Yellow pigment “MY3” *authors speculate bactobolin but unconfirmed |
Yes | Shukla & Nadumane, 2021 |
| Micrococcus lylae strain YH3 (MW407006.1) | Soil (El Mahmoudiyah goverrnance, Egypt) | Antibacterial, antifungal, antioxidant, and anticancer | Echinenone (β-carotene pigment) | Yes | Shahin et al., 2022 |
| Micrococcus -strain not specified | Sewage | Antibacterial/bacteriostatic | Micrococcin | No | Su, 1948 |
| Micrococcus | Marine sponge Tedania ignis | Not specified | Three diketopiperazines | No | Stierle et al., 1988 |
|
Micrococcus sp. SB58 (AF218240.1) |
Marine sponge Aplysina aerophoba or Aplysina cavernicola | Antibacterial | Live colonies | Yes | Hentschel et al., 2001 |
| Micrococcus sp. strain SCS1 | Bamboo garden waste soil (Bangladesh) | Antibacterial, cytotoxic | crude extract *authors speculate bacteriocin is responsible but unconfirmed |
No | Sharma et al., 2012 |
| Micrococcusisolate YIM 65 738 | Artemisia annua (Kunming Institute of Botany, Chinese Academy of Sciences and Xishuangbanna, Yunnan province, China) | Antifungal | Crude extract |
Yes | Li et al., 2012 |
|
Micrococcus strain
Berg02_11 Micrococcus strain Berg02_26 |
marine sponge Erylus discophorus (Berlengas Islands, Portugal) | Antibacterial—both isolates Antifungal—strain Berg02_26 |
Extracts, diffused secondary metabolites |
No | Graça et al., 2013 Santos et al., 2019 |
| Micrococcus sp. EG45 | marine sponge Spheciospongia vagabunda (Red Sea) | Antibacterial | Microluside A [4 (19-para-hydroxy benzoyloxy-O-β-d-cellobiosyl), 5 (30-para-hydroxy benzoyloxy-O-β-d-glucopyranosyl] (xanthone) | No | Eltamany et al., 2014 |
| Micrococcus sp. strain OUS9 (MN108086.1–refers to strain KLEF09) | intertidal Seawater (Nellore Krishnapatnam, India) | Wound healing | KLUF 10 (3-Hydroxy-β, ε-caroten-3′-one) KLUF 13 (1-(1-(4-methoxphyenyl)-2-(methyl amino) ethyl) cyclohexan-1-ol) |
Yes | Shanthi Kumari et al., 2020 |
|
Micrococcus sp. MP76 (KT804695.1) |
Seawater (Persian Gulf, Bushehr province, Iran) |
Antibacterial, antioxidant | Crude pigment *authors speculate an aminoglycoside antibiotic but unconfirmed |
Yes | Karbalaei-Heidari et al., 2020 |
|
Micrococcus sp. strain KRD026 (MT136106.1) Micrococcus sp. strain KRD070 (MT135986.1) Micrococcus sp. strain KRD077 (MT135795.1) Micrococcus sp. strain KRD096 (MT136510.1) Micrococcus sp. strain KRD153 (MT135519.1) |
Marine sediment (Antarctica): strain KRD026 Marine sediment (Arctic): strains KRD070, KRD077, KRD096, and KRD153 |
Antibacterial | Live colonies, crude extracts | Yes | Soldatou et al., 2021 |
| Micrococcus sp. strain RBYA34 | Marine sponge (not specified) (Rapid Bay Jetty, Australia) | Antibacterial, antifungal | Live colonies, crude extract | No | Anteneh et al., 2021 |
|
Micrococcus sp. strain XM4230A
(OP557975.1) Micrococcus sp. strain XM4230B (OP557976.1) |
marine sponge Xestospongia muta (Florida Keys, Florida, USA) | Antimycobacterial | Crude extract | Yes | Tizabi et al., 2022; Tizabi, 2022 |
| Micrococcus sp. strain R8502A1 (OP557984.1) | marine sponge Xestospongia muta (Florida Keys, Florida, USA) | Antimycobacterial | Crude extract | Yes | Tizabi, 2022 |
Note. Original studies describing isolated pigments from Micrococcus strains that were later found to be bioactive are included in the list for robustness. MRSA = methicillin-resistant Staphylococcus aureus.
There is no evidence in the literature of the antimicrobial activity of lutoside, despite several papers making this claim. This compound was isolated by Bultel-Poncé et al. in 1997; a follow-up paper in 1998 mentions that lutoside was the major component of the Micrococcus extract and that the bioactivity of this compound was under investigation at the time of publication.
Discrepancy: Micrococcus luteus strain TUB6 identified as an Acetobacter strain in El-Deeb et al. 2011.
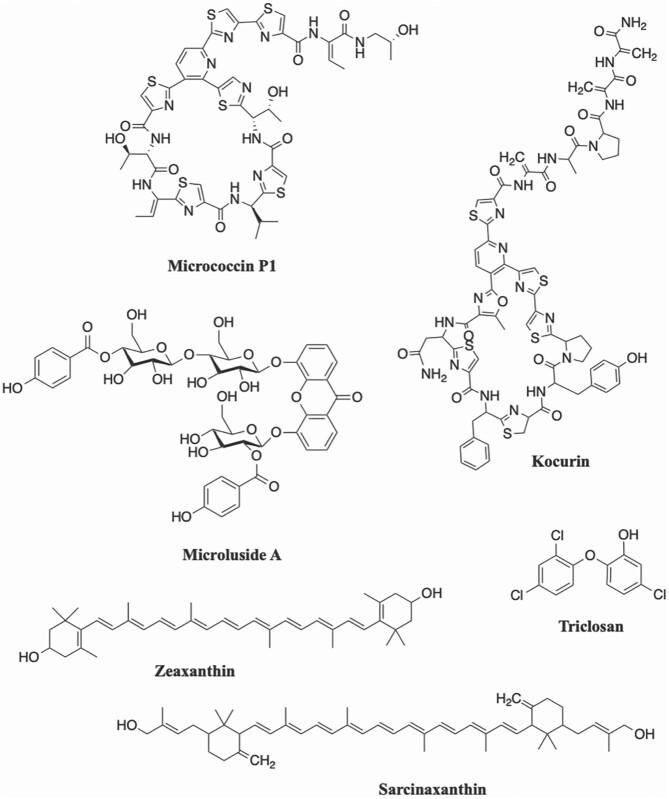
Excluding pigments, only six bioactive compounds isolated from Micrococcus spp. have been characterized (Su, 1948; Biskupiak et al., 1988; Bultel-Poncé et al., 1998; Palomo et al., 2013; Eltamany et al., 2014; Shanthi Kumari et al., 2020). The first bioactive compound to be fully characterized from a Micrococcus strain, micrococcin, was discovered in 1948 from a sewage isolate (Su, 1948). Though originally studied for its antibacterial activity against Gram-positive bacteria, including Mycobacterium tuberculosis, micrococcin has since been shown to also have antimalarial, anticancer, and gene-modulating activities (Rogers et al., 1998; Ciufolini & Lefranc, 2010). The next bioactive compound was not isolated until 40 years later, with the discovery of the antibacterial neoberninamycin from a soil isolate (Biskupiak et al., 1988). A decade later, the previously synthesized potent antimicrobial compound Triclosan (2,4,4′-trichloro-2′-hydroxydiphenylether), was isolated from a sponge-derived strain of M. luteus (Bultel-Poncé et al., 1998). A new member of the thiazolyl peptide, family of antibiotics identified as kocurin, was discovered from Micrococcus yunnanensis F-256446 as well as from two strains of Kocuria (Palomo et al., 2013). Following the discovery of kocurin in 2013 was the isolation of an antibacterial xanthone, microluside A, from another sponge-derived strain, Micrococcus sp. EG45 (Eltamany et al., 2014). Most recently, two wound-healing compounds, KLUF-10 (originally proposed to be 3-Hydroxy-β, ε-caroten-3′-one) and KLUF-13 (1-(1-(4-methoxphyenyl)-2-(methyl amino) ethyl) cyclohexan-1-ol) were isolated from the marine isolate Micrococcus sp. KLEF09 (originally published as strain OUS9) (Shanthi Kumari et al., 2020). However, KLUF-10 has since been determined by NMR analysis to in fact be zeaxanthin (Shanthi Kumari et al., 2021). It is worth noting that four of the six compounds were isolated from marine strains. The chemical structures of selected bioactive compounds isolated from Micrococcus strains are provided in Fig. 1.
Fig. 1.
Chemical structures of selected compounds synthesized by Micrococcus isolates.
Bioactive Pigments
Aside from characterization studies, the majority of the literature on Micrococcus spp. focuses on the pigments they produce, documenting their antioxidant, antibiotic, antifungal, and anticancer properties (Mohana et al., 2013; Umadevi & Krishnaveni, 2013; Ushasri & Gods Will Shalomi, 2015; Rostami et al., 2016; Majeed, 2017; Zehra et al., 2018; Nisha et al., 2019; Karbalaei-Heidari et al., 2020; Shukla & Nadumane, 2021; Shahin et al., 2022). The major carotenoid pigment of M. luteus, sarcinaxanthin, is a rare C50 carotenoid that functions as an antioxidant and has been patented for use in sunscreen, although this patent has since been abandoned (Goksøyr, 2013; Netzer et al., 2010). Several derivatives of this carotenoid also exist, including the glucosylated compounds sarcinaxanthin monoglucoside and sarcinaxanthin diglucoside (Osawa et al., 2010). Several studies have documented bioactivity in crude pigment extract prepared from M. luteus, including antibacterial activity against Staphylococcus sp., Klebsiella sp., Pseudomonas sp., and Escherichia sp. (Umadevi & Krishnaveni, 2013; Majeed, 2017), weak antifungal activity against Alternaria spp., Aspergillus niger, Cladosporium sp., and Penicillium certum (Majeed, 2017), and cytotoxicity against the breast cancer MCF-7 cell line (Ushasri & Gods Will Shalomi, 2015). However, due to the crude nature of these extracts tested, it remains unconfirmed whether sarcinaxanthin or another compound is responsible for the activity observed. In one study, carotenoids purified from M. luteus, only identified as “yellow carotenoid pigment”, were observed to have antibacterial effects against Staphylococcus aureus and Streptococcus faecalis (now classified as Enterococcus faecalis) (Mohana et al., 2013). Another study found the crude pigment extract of M. luteus to be additionally active against Salmonella typhi and several drug-resistant bacteria, including multidrug-resistant Acinetobacter sp., multidrug-resistant Pseudomonas, methicillin-resistant Staphylococcus sp., and Enterobacter carbopenum and confirmed antifungal activity (Nisha et al., 2019). Furthermore, the crude pigment displayed antitumor activity against Dalton's Lymphoma ascites cells. Although this study confirmed the presence of sarcinaxanthin in the pale orange pigment extract, phytoene derivatives and sarcinaxanthin derivatives were also found. In addition, crude exopolysaccharides from M. luteus were found to be variably active against Escherichia coli, Klebsiella sp., S. typhi, Staphylococcus sp., and Pseudomonas sp. (Nisha et al., 2019).
In mesophilic M. roseus strains, canthaxanthin is the most prominent carotenoid pigment (Cooney et al., 1966; Jagannadham et al., 1991), although in a psychrotrophic strain, the major carotenoid pigment isolated was bisdehydro-beta-carotene-2-carboxylic acid (Jagannadham et al., 1991). In a psychrotrophic M. roseus strain grown at temperatures close to freezing, production of a more polar C50 carotenoid pigment, bacterioruberin, increased (Chattopadhyay et al., 1997). Canthaxanthins have especially strong antioxidant activity and are highly valued for their applications in nutraceuticals, cosmetics, and animal feed supplements (Palozza & Krinsky, 1992; Rebelo et al., 2020). Additionally, canthaxanthin can be used to synthesize astaxanthin, a highly marketable carotenoid often employed as a nutritional supplement for its wide-ranging health benefits (Ambati et al., 2014; Rebelo et al., 2020). Aside from this major carotenoid, seven other pigments have been isolated in smaller quantities from M. roseus, including phoenicoxanthin, dihydroxy-3,4-dehydro-α-carotene, a dihydroxy-α-carotene, a diketo-α-carotene, a polyhydroxy-β-carotene, and two other uncharacterized pigments (Ungers & Cooney, 1968). Crude pigment isolated from a M. roseus strain (unspecified origin) was shown to have antibacterial activity against S. aureus (Zehra et al., 2018). The aforementioned study by Mohana et al. also described a purified red carotenoid pigment of M. roseus to be active against S. aureus and what is now classified as E. faecalis, though this pigment is never identified in the study (Mohana et al., 2013). A separate study isolating unidentified pigments from M. roseus confirmed antimicrobial activity against an extensive collection of bacteria and fungi, with stronger inhibition observed against Gram-positive pathogens than Gram-negatives, as well as antitumor, anti-inflammatory, and antioxidant activities (Rostami et al., 2016).
Less common Micrococcus strains have also been shown to produce bioactive pigments. A yellow pigment designated as MY3 was isolated from Micrococcus terreus and was found to have cytotoxic activity against cervical and liver cancer cell lines (Shukla & Nadumane, 2021). The MY3 extract was further characterized by liquid chromatography-mass spectrometry and found to putatively contain the compound bactobolin. Bactobolin isolated from a strain of Pseudomonas has previously been shown to have antitumor and antibacterial activities (Kondo et al., 1979). Echinenone, a β-carotene pigment isolated from Micrococcus lylae, demonstrates antibacterial, antifungal, cytotoxic, and antioxidant activity (Shahin et al., 2022). The crude yellow pigment extract of the marine isolate Micrococcus sp. MP76 inhibits E. coli, Pseudomonas aeruginosa, and S. aureus (Karbalaei-Heidari et al., 2020). Micrococcus tetragenus is known to produce several carotenoids including a xanthophyll, lycopene, rhodoxanthin, rubixanthin, γ-carotin, and several other unidentified pigments (Reimann & Eklund, 1941). Zeaxanthin and lycophyll and derivatives of at least one of these carotenoids were identified from Micrococcus radiodurans (Lee, 1961). Aside from the inherent antioxidant properties of these carotenoids, no additional bioactivity was documented for these pigments in the cited studies. Zeaxanthin has been found to have antiquorum sensing and antibiofilm activities (Gökalsın et al., 2017; Karpiński et al., 2022), and lycopene has been found to be useful as an adjuvant for antimicrobial treatment by way of its bactericidal activity (Lee & Lee, 2014). As previously mentioned, KLUF-10, studied for its wound-healing activity, has since been identified as zeaxanthin (Shanthi Kumari et al., 2021). Antibacterial as well as promising anticancer activity of KLUF-10 is also suggested (Shanthi Kumari et al., 2020; Shanthi Kumari et al., 2021).
Gaps in Understanding
Many studies have tested the secreted metabolites or crude Micrococcus extracts for various pharmaceutically relevant activity, but failed to isolate or fully characterize the active component (Hentschel et al., 2001; El-Deeb et al., 2012; Li et al., 2012; Sharma et al., 2012; Graça et al., 2013; Mohana et al., 2013; Umadevi & Krishnaveni, 2013; Akbar et al., 2014; Ushasri & Gods Will Shalomi, 2015; Rostami et al., 2016; Anteneh et al., 2021; Cita et al., 2017; Majeed, 2017; Ranjan & Jadeja, 2017; Zehra et al., 2018; Nisha et al., 2019; Santos et al., 2019; Karbalaei-Heidari et al., 2020; Shukla & Nadumane, 2021; Soldatou et al., 2021; Tizabi et al., 2022 ). In fact, only eight papers describe isolated bioactive compounds (including pigments examined for bioactivity) from Micrococcus spp. (Su, 1948; Biskupiak et al., 1988; Bultel-Poncé et al., 1998; Mohana et al., 2013; Palomo et al., 2013; Eltamany et al., 2014; Shanthi Kumari et al., 2020; Shahin et al., 2022). It is notable that four of these findings pertain to marine isolates, three of which are derived from sponges. Despite the fact that, historically, research has focused on terrestrially derived strains, marine strains show great promise and merit further investigation. Several studies in which pigments were isolated that were later found to be bioactive through subsequent studies are excluded from this count, as the original study does not mention any pharmaceutical relevance, but these reports are included in Table 1. Excluded from this count is also one noteworthy study concluding that a marine Micrococcus sp. is the true producer of diketopiperazines (Stierle et al., 1988) that were originally ascribed to the sponge host (Schmitz et al., 1983). Though no bioactivity is described in the original publication, subsequent research has shown diketopiperazines to have anticancer activity (van der Merwe et al., 2008; Mollica et al., 2014).
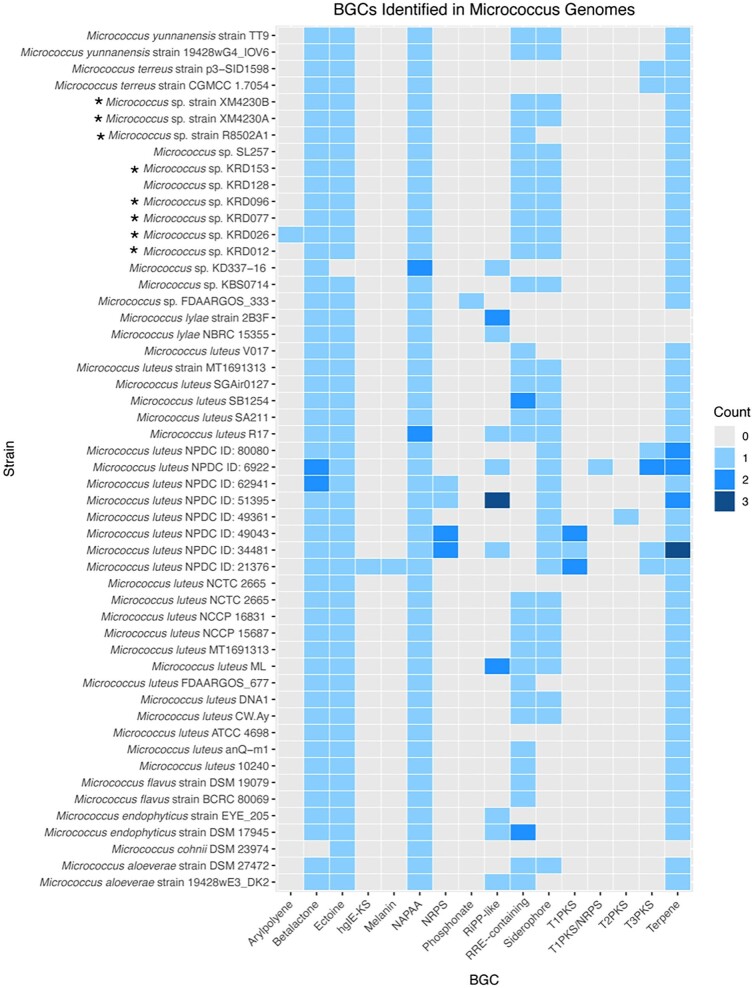
Historically, bioprospecting efforts have focused on actinomycetes with large genomes, typically larger than 6 Mb and ranging up to 13 Mb, as these strains tend to encode more of the common BGCs, such as PKSs and NRPSs (Baltz, 2014, 2017, 2019; Katz & Baltz, 2016). Investigations into these BGCs take advantage of the fact that the genomic sequence of these multimodular enzymes often provide a clear link between the biochemical processes required for synthesis and the final structure of the compound (Donadio et al., 2007; Katz & Baltz, 2016). To better understand the overall biosynthetic potential of Micrococcus spp., the genomes of Micrococcus strains with high quality assemblies available from GenBank and the Natural Products Discovery Center (NPDC) Portal were analyzed for secondary metabolite clusters using anti-SMASH version 7 beta (Fig. 2). In total, 52 Micrococcus spp. genomes ranging in size from 2.3 to 4.4 Mb were mined for BGCs, with the total number of putative clusters detected per genome ranging between 2 and 12 (Supplementary Fig. 1). Generally, the overall pattern described in the literature that genomes less than 3 Mb in size rarely contain BGCs was observed. However, while the majority of strains were detected to encode the same five or six putative clusters (betalactone, ectoine, NAPAA, terpene, siderophore, and RRE-containing), several isolates contain significantly more BGCs belonging to a diverse array of cluster types. Interestingly, Micrococcus sp. SL257 was isolated from the microbiome of the freshwater sponge Spongilla lacustris and assessed for putative BGCs along with 32 other representative isolates. Despite maintaining the smallest genome size out of all the bacteria studied (only 2.5 Mb), this genome was found to contain six putative BGCs, more than the number of BGCs detected in 13 of the other genomes analyzed (Graffius et al., 2023).
Fig. 2.
Heatmap showing abundance of various BGC types (as identified by anti-SMASH version 7 beta) in Micrococcus genomes. At least two isolates belonging to the same species were included when possible. Strains with confirmed bioactivity are denoted by “*”. Several BGC categories as identified by anti-SMASH were combined here to improve visualization. The BGC category “RiPP-like” includes any clusters identified as “RiPP-like”, “thiopeptide”, “lanthipeptide”, “linaridin”, or “bacteriocin”. The category ‘NRPS” includes any clusters identified as “NRPS” or “NRPS-like”. “T1/T2/T3PKS” refer to type I, type II, and type III PKSs, respectively. “HgIE-KS” refers to “heterocyst glycolipid synthase-like PKS”. The number of putative BGCs of any particular category detected in each strain is represented by a color code between gray and dark blue.
It is important to note that certain BGC types are associated much more frequently with drug-like activity than others (Baltz, 2014, 2017, 2019). The most common BGCs detected in the Micrococcus genomes analyzed are not commonly associated with drug-like activity. Unfortunately, only eight of the strains profiled for BGCs have been investigated for bioactivity. It should be noted that Micrococcus sp. strain R8502A1 has been observed to have very potent antimycobacterial activity, including against Mycobacterium tuberculosis (Tizabi, 2022). This strain is closely related to Micrococcus sp. strains XM4230A and XM4230B, both of which also have been shown to have potent inhibitory activity against several Mycobacterium spp. including M. tuberculosis (Tizabi et al., 2022). Nevertheless, all three strains have identical, uninformative BGC profiles when analyzed with anti-SMASH. The five additional strains were tested for bioactivity in a single study (Soldatou et al., 2021). The paucity of BGCs detected by bioinformatic analysis supports the notion that many Micrococcus-derived secondary metabolites effecting various inhibitory activities detected by bioassays are likely not synthesized by typical BGCs. Even though BGC profiling has revealed few BGCs in these strains, their diverse and potent bioactivity profiles indicate biosynthetic potential, perhaps of even more interest than activities from BGC-rich strains because these activities are not encoded by the genes routinely found by BGC analysis.
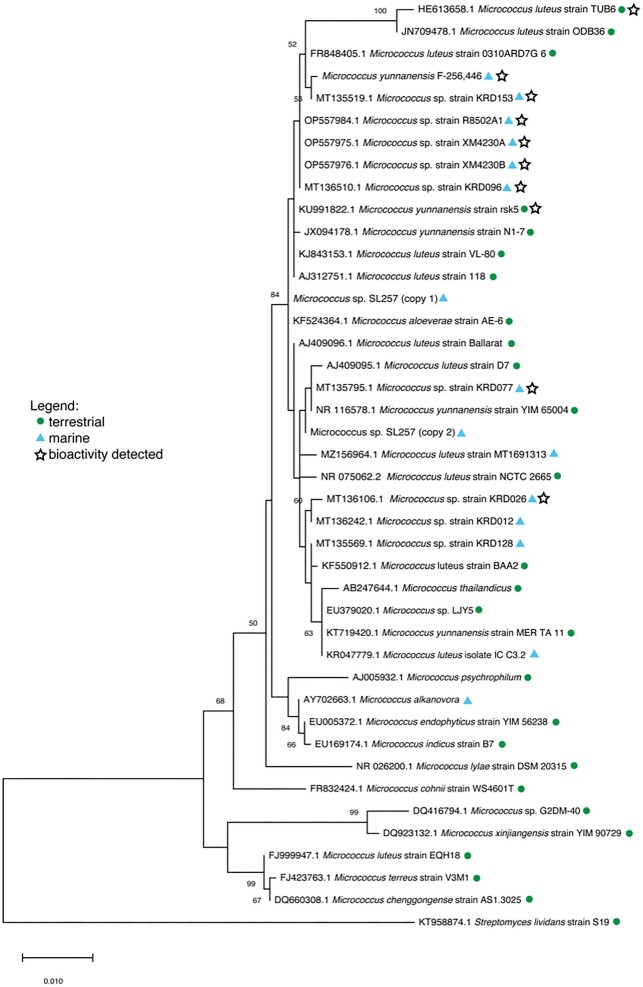
In an attempt to establish a relationship between different bioactive Micrococcus strains, a phylogenetic tree based on 16S rRNA gene sequence analysis was constructed from an exhaustive literature search of bioactive terrestrial and marine-derived strains (Fig. 3). Unfortunately, the majority of Micrococcus strains discovered to produce bioactive compounds, including antibacterial pigments [Micrococcus luteus strain MKVKUD 2013 (AN: KF532949.1) and Micrococcus sp. MP76 (AN: KT804695.1)], anticancer compounds [Micrococcus sp. OUS9 (AN: MN108086.1)] and antibacterial metabolites [Micrococcus sp. strain SB58 (AN: AF218240.1)] had to be excluded from this phylogenetic analysis either due to insufficient 16S rRNA gene sequence length for analysis or poor sequence quality. In fact, of the 28 studies that identify bioactivity in Micrococcus strains, only 11 provide 16S rRNA gene sequence data, from which only eight strains were included in the phylogenetic analysis (excluded from this tally is Micrococcus yunnanensis F-256446, as in this case sequence data were obtained directly from the authors) (Palomo et al., 2013). Exclusion of these strains from phylogenetic analysis precludes a comprehensive understanding of the chemotaxonomic relationship among Micrococcus isolates. As a result, additional strains lacking bioactivity were included in the analysis to provide phylogenetic robustness. M. yunnanensis F-256446 is the only strain included in the phylogenetic tree from which a bioactive compound was actually isolated, rather than bioactivity being reported from an extract. This sponge-derived isolate produces kocurin, a thiopeptide antibiotic (RiPP) with anti-MRSA activity (Palomo et al., 2013).
Fig. 3.
Maximum-likelihood phylogenetic tree based on partial 16S rRNA gene sequences of Micrococcus strains from literature. Adapted from MegaX: The evolutionary history was inferred by using the Maximum Likelihood method and Tamura–Nei model (Tamura & Nei, 1993). The tree with the highest log likelihood (−3425.14) is shown. Bootstrap values are calculated from 1 000 sampling replicates. The percentage of trees in which the associated taxa clustered together is shown next to the branches (only values > 50% shown). Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura–Nei model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 42 nucleotide sequences. Micrococcus sp. SL257 16S rRNA sequence was extracted from WGS data and found to contain two distinct 16S rRNA genes (“copy 1” and “copy 2”), both of which are included in the tree. Codon positions included were 1st + 2nd + 3rd + Noncoding. There were a total of 1 325 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018; Stecher et al., 2020). Streptomyces lividans strain S19 (AN: KT958874.1) was used as the outgroup.
Despite the currently limited literature describing known Micrococcus strains and the compounds they produce, the phylogenetic analysis is interesting in regard to the relationship (or lack thereof) between bioactivity and taxonomy. The three novel strains previously isolated from Xestospongia muta by Montalvo et al. (2005) (strains R8502A1, XM4230A, and XM4230B) cluster together, which is not surprising since they were isolated from a single sponge species collected in a single region. Micrococcus yunnanensis F-256446 was also isolated from a marine sponge in the Florida Keys and clusters with the aforementioned novel isolates, along with terrestrial strains isolated from a medicinal plant from Saudi Arabia (either Coleus forskohlii or Plectranthus tenuiflorus) (M. luteus strain TUB6), Indian soil (M. luteus strain ODB36), the air in a subterranean Spanish show cave (M. luteus strain 0310ARD7G_6), and several strains isolated from Arctic marine sediment (Micrococcus spp. strain KRD153 and KRD096) (Fernandez-Cortes et al., 2011; El-Deeb et al., 2012; Soldatou et al., 2021). Micrococcus sp. strain KRD153 yielded an extract with antibacterial activity against S. aureus, E. coli and P. aeruginosa. Micrococcus sp. strain KRD093 is active against E. coli (Soldatou et al., 2021). M. luteus strain TUB6 produces a crude extract with antimicrobial activity against the human pathogen Proteus mirabilis (El-Deeb et al., 2012). Within a larger branch is M. yunnanensis strain rsk5, isolated from a plant from India (Catharanthus roseus) and shown to produce a crude extract with broad-spectrum antibacterial activity, as well as three more marine strains isolated from Polar regions (Ranjan & Jadeja, 2017; Soldatou et al., 2021). An extract of Micrococcus sp. strain KRD026 inhibited S. aureus and E. coli, while no antibacterial activity was observed from the extracts of strains KRD128 or KRD012 (Soldatou et al., 2021).
The study analyzing M. yunnanensis F-256446, along with two Kocuria strains for their production of the anti-MRSA thiopeptide, kocurin, noted that although all three microbes were isolated from the same region and found to produce the same compound, they all exhibited different metabolic gene amplification patterns (Palomo et al., 2013). Coupled with the fact that actinomycetes of distantly related genera isolated from Antarctica have also been found to produce kocurin, this observation suggests that geographic proximity does not necessarily correlate with chemosimilarity (Palomo et al., 2013). Furthermore, studies show that geographic location can have a dramatic effect on the specialized metabolism of Micrococcus isolates of the same genus sampled from distinct regions (Parra & Duncan, 2019). All six strains included in the phylogenetic analysis (terrestrial and marine isolates) identified as having antibacterial properties cluster together. No definitive conclusions can be made from this relationship until activity is confirmed or excluded from all other strains included in the analysis. This highlights the lack of data regarding the biosynthetic potential of Micrococcus sp. and emphasizes the need for continued research in this area. One final and critical note on the phylogenetic analysis is that the results are entirely dependent on the accuracy of the sequence data provided. Several sequences were removed from this analysis based on highly unlikely base diversions; we cannot be certain that the 16S rRNA gene sequences of the included strains are all 100% accurate. Nevertheless, the analysis performed here provides insight into the taxonomic relationship of Micrococcus strains isolated from widely disparate environments and emphasizes the need for additional study of the biosynthetic potential of this genus.
Taken together, the results from the genome mining analysis and phylogenetic analysis reveal a major disconnect in the current state of Micrococcus research. The most significant obstacle to truly understanding the potential of Micrococcus spp. as a source of novel drugs is the lack of overlapping bioactivity screening data and genomic data available for any given strain. Strains for only 8 of the 52 Micrococcus genomes mined for BGCs have been assayed for various bioactivities (Micrococcus sp. strains XM4230A, XM4230B, R8502A1, KRD153, KRD128, KRD 096, KRD077, KRD026, and KRD012). Similarly, only 13 of the 41 Micrococcus strains included in the phylogenetic analysis (7 of which were detected to have pharmaceutically relevant bioactivity) provide corresponding whole genome sequencing data, hindering assessment of the biosynthetic potential of these remaining strains. That there are only eight Micrococcus strains in the literature for which both genome assemblies and bioactivity screening data are provided starkly contrasts with the fact that there are nearly 30 separate studies documenting antibacterial, antifungal, antioxidant, anti-inflammatory, and anticancer activities in Micrococcus isolates. As Baltz recently noted, the availability of complete and high-quality genomes encoding known secondary metabolites is critical for subsequent research in order to facilitate bioinformatics dereplication and to avoid rediscovery (2019, 2021). It is imperative that future investigations into bioactivity of Micrococcus spp. provide corresponding genomic data for BGC analysis. However, because it is unlikely that PKs or NRPs are responsible for the bioactivity observed in these bacteria, in-depth chemical analysis is still essential to characterizing the novel compounds. Elucidation of the novel chemical structures will better inform subsequent genomic analyses and help facilitate the discovery of novel biosynthetic pathways.
Concluding Remarks
A recent global assessment of antimicrobial resistance (AMR) estimated that bacterial AMR was responsible for approximately 1.27 million deaths and associated with an additional 4.95 million deaths in 2019 alone. The infamous ESKAPE pathogens (Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter spp.), which are the main culprit of nosocomial infections worldwide, and often drug-resistant, accounted for 1 million of these deaths and were associated with an additional 3.57 million deaths (Murray et al., 2022). The rising threat of antibiotic resistance demands therapeutics with novel mechanisms of action to treat these infectious diseases. Research has repeatedly shown that rare actinomycetes are highly diverse and maintain a high degree of novelty, as well as the ability to synthesize complex metabolites, often with low toxicity (Bérdy, 2005; Kurtböke, 2012; Schorn et al., 2016). In fact, only approximately 10% of biosynthetic compounds produced by rare actinomycetes are also found in Streptomyces (Bérdy, 2005). Much of the literature reviewed here describes bioactivity of Micrococcus-derived compounds that targets the infamous ESKAPE pathogens. Yet, as recently as 2012, literary reviews documenting the biosynthetic potential of rare actinomycete species exclude Micrococcus spp., indicating that this genus has historically been overlooked in drug discovery efforts (Bérdy, 2005; Kurtböke, 2012).
Investigations into the bioactivity of Micrococcus strains are few and far between. To effectively identify novel compounds from this genus with pharmaceutical relevance, analyses must prioritize strains in which bioactivity has already been observed or for which sequence data already exists. The Shen lab at the University of Florida Scripps Biomedical Research has recently launched the (NPDC Portal (https://npdc.rc.ufl.edu/home), which offers an extensive actinobacterial genome database and provides evidence that Micrococcus genomes contain more BGCs than expected. There are currently 50 M. luteus genomes in the NPDC Portal, most of which contain BGCs belonging to the same five clusters (betalactone, siderophore, terpene, ectoine, and NRPS-like cluster). Interestingly, four M. luteus genomes (all > 3 Mb) encode at least 10 BGCs, with varying similarities to known antitumor, antibiotic, antiparasitic, and antifungal compounds. This genomic data are insufficient as a standalone tool for compound discovery, but provides valuable insight into the theoretical chemical arsenal of these Micrococcus isolates, which can be corroborated with bioassays. Resources such as NPDC, which offer access to strains for experimental analysis, as well as free access to their genome assemblies, will facilitate efficient prioritization of Micrococcus strains for drug discovery.
The paucity of information regarding Micrococcus-derived compounds in the literature is at once both frustrating and intriguing. The scarcity of sequence data, combined with the limited success of genome mining strategies, severely limits the capacity of genomic-based analyses and makes it difficult to speculate on the bioactivity of novel strains. Genome mining tools are only as strong as their databases, as they rely on the availability of known biosynthetic pathways to make conjectures (Bachmann et al., 2014). Due to the absence or scarcity of BGCs in most Micrococcus genomes, it is more probable that these smaller genomes encode more elusive compounds of a group with more scaffold versatility such as alkaloids, quinones, or xanthones, as opposed to an NRPS, PKS, or RiPP, the latter three of which are more readily detectable by typical genome mining algorithms (Schorn et al., 2016). Extensive chemical approaches are thus necessary to characterize these bioactive compounds responsible for the various antibacterial, anticancer, and antifungal activities observed from bioassays. Further research should revisit the bioactive strains discussed in this review and use bioassay-guided fractionation to further isolate the compound(s) of interest in these promising strains. Additionally, emphasis should be placed on elucidating the bioactivity of marine Micrococcus isolates, as these have been shown to consistently retain pharmaceutical relevance. With at least 71% of known marine scaffolds being used exclusively by marine organisms, and 53% of marine scaffolds detected from only one source thus far (Kong et al., 2010), the probability of discovering novel chemistry from the marine environment is all but guaranteed.
Supplementary Material
Acknowledgements
We would like to thank Dr. Olga Genilloud for providing sequence data for Micrococcus yunnanensis F-256446. Sabeena Nazar in the BioAnalytical Services Laboratory at the Institute of Marine and Environmental Technology is thanked for performing sequencing on several Micrococcus strains discussed in this review.
Contributor Information
Daniela Tizabi, Institute of Marine and Environmental Technology, University of Maryland Center for Environmental Science, Baltimore, MD 21202, USA.
Russell T Hill, Institute of Marine and Environmental Technology, University of Maryland Center for Environmental Science, Baltimore, MD 21202, USA.
Funding
This work was supported by the funding provided by the NIST-IMET Graduate Fellowship in Environmental Biotechnology, the University of Maryland College Park College of Mathematical and Natural Sciences Dean's Fellowship, Ratcliffe Environmental Entrepreneur Fellowship, the Chateaubriand Fellowship STEM, and the American Association of University Women's American Dissertation Fellowship.
Conflict of Interest
The authors declare no conflict of interest.
References
- Akbar A., Sitara U., Ali I., Muhammad N., Khan S. A. (2014); Isolation and characterization of biotechnologically potent micrococcus luteus strain from environment. Pakistan Journal of Zoology, 46(4), 967–973. [Google Scholar]
- Albertson D., Natsios G. A., Gleckman R. (1978); Septic shock with micrococcus luteus. Archives of Internal Medicine, 138(3), 487–488. 10.1001/ARCHINTE.1978.03630270093032 [DOI] [PubMed] [Google Scholar]
- Ambati R. R., Moi P. S., Ravi S., Aswathanarayana R. G. (2014); Astaxanthin: Sources, extraction, stability, biological activities and its commercial applications—A review. Marine Drugs, 12(1), 128. 10.3390/MD12010128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anteneh Y. S., Yang Q., Brown M. H., Franco C. M. M. (2021); Antimicrobial activities of marine sponge-associated bacteria. Microorganisms, 9(1), 1–19. 10.3390/MICROORGANISMS9010171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. O., Van Lanen S. G., Baltz R. H. (2014); Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making? Journal of Industrial Microbiology & Biotechnology, 41(2):175–84. 10.1007/s10295-013-1389-9. Epub 2013 Dec 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R. H. (2014); MbtH homology codes to identify gifted microbes for genome mining. Journal of Industrial Microbiology & Biotechnology, 41(2), 357–369. 10.1007/s10295-013-1360-9 [DOI] [PubMed] [Google Scholar]
- Baltz R. H. (2017); Gifted microbes for genome mining and natural product discovery. Journal of Industrial Microbiology & Biotechnology, 44(4–5), 573–588. 10.1007/s10295-016-1815-x [DOI] [PubMed] [Google Scholar]
- Baltz R. H. (2019); Natural product drug discovery in the genomic era: Realities, conjectures, misconceptions, and opportunities. Journal of Industrial Microbiology and Biotechnology, 46(3–4), 281–299. 10.1007/s10295-018-2115-4 [DOI] [PubMed] [Google Scholar]
- Baltz R. H. (2021); Genome mining for drug discovery: Progress at the front end. Journal of Industrial Microbiology and Biotechnology, 48(9–10), kuab044. 10.1093/jimb/kuab044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérdy J. (2005); Bioactive microbial metabolites. The Journal of Antibiotics, 58(1), 1–26. 10.1038/ja.2005.1 [DOI] [PubMed] [Google Scholar]
- Bhowmik T., Marth E. H. (1990); Rote of Micrococcus and Pediococcus species in cheese ripening: A review. Journal of Dairy Science, 73(4), 859–866. 10.3168/JDS.S0022-0302(90)78740-1 [DOI] [Google Scholar]
- Biskupiak J. E., Meyers E., Gillum A. M., Dean L., Trejo W. H., Kirsch D. R. (1988); Neoberninamycin, a new antibiotic produced by Micrococcus luteus. The Journal of Antibiotics, 41(5), 684–687. 10.7164/antibiotics.41.684. [DOI] [PubMed] [Google Scholar]
- Bultel-Poncé V., Debitus C., Berge J. P., Cerceau C., Guyot M. (1998); Metabolites from the sponge-associated bacterium micrococcus luteus. Journal of Marine Biotechnology, 6(4), 233–236. [PubMed] [Google Scholar]
- Bultel-Poncé V., Debitus C., Blond A., Cerceau C., Guyot M. (1997); Lutoside: An acyl-1-(acyl-6′-mannobiosyl)-3-glycerol isolated from the sponge-associated bacterium Micrococcus luteus. Tetrahedron Letters, 38(33), 5805–5808. [Google Scholar]
- Chattopadhyay M. K., Jagannadham M. V., Vairamani M., Shivaji S. (1997); Carotenoid pigments of an antarctic psychrotrophic bacterium Micrococcus roseus: Temperature dependent biosynthesis, structure, and interaction with synthetic membranes. Biochemical and Biophysical Research Communications, 239(1), 85–90. 10.1006/BBRC.1997.7433 [DOI] [PubMed] [Google Scholar]
- Chiller K., Selkin B. A., Murakawa G. J. (2001); Skin microflora and bacterial infections of the skin. The Journal of Investigative Dermatology. Symposium Proceedings, 6(3), 170–174. 10.1046/J.0022-202X.2001.00043.X [DOI] [PubMed] [Google Scholar]
- Cita Y. P., Suhermanto A., Radjasa O. K., Sudharmono P. (2017); Antibacterial activity of marine bacteria isolated from sponge Xestospongia testudinaria from Sorong, Papua. Asian Pacific Journal of Tropical Biomedicine, 7(5), 450–454. 10.1016/J.APJTB.2017.01.024 [DOI] [Google Scholar]
- Ciufolini M. A., Lefranc D. (2010); Micrococcin P1: Structure, biology and synthesis. Natural Product Reports, 27(3), 330–342. 10.1039/B919071F [DOI] [PubMed] [Google Scholar]
- Cohn F. (1872); Untersuchungen über Bakterien. Beiträge zur Biologie der Pflanzen, 1, 127–244. [Google Scholar]
- Cooney J. J., Marks H. W., Smith A. M. (1966); Isolation and identification of canthaxanthin from Micrococcus roseus. Journal of Bacteriology, 92(2), 342–345. 10.1128/JB.92.2.342-345.1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehle P. (1889); Beobachtunger über Einen Antagonisten des Milzbrandes. [Thesis]. Schmidt & Klaunig. [Google Scholar]
- Donadio S., Monciardini P., Sosio M. (2007); Polyketide synthases and nonribosomal peptide synthetases: The emerging view from bacterial genomics. Natural Product Reports, 24(5), 1073–1079. 10.1039/b514050c [DOI] [PubMed] [Google Scholar]
- Dujardin-Beaumetz E. (1934); Comptes rendus des seances de la Societe de Biologie et de ses filiales. (Vol. 117), Société de biologie (Paris, France). . [Google Scholar]
- El-Deeb B., Bazaid S., Gherbawy Y., Elhariry H. (2011); Characterization of endophytic bacteria associated with rose plant (Rosa damascena trigintipeta) during flowering stage and their plant growth promoting traits. Journal of Plant Interactions, 7(3), 248–253. 10.1080/17429145.2011.637161 [DOI] [Google Scholar]
- El-Deeb B., Fayez K., Gherbawy Y. (2012Isolation and characterization of endophytic bacteria from Plectranthus tenuiflorus medicinal plant in Saudi Arabia desert and their antimicrobial activities. Journal of Plant Interactions, 8(1), 56–64. 10.1080/17429145.2012.680077 [DOI] [Google Scholar]
- Eltamany E. E., Abdelmohsen U. R., Ibrahim A. K., Hassanean H. A., Hentschel U., Ahmed S. A. (2014); New antibacterial xanthone from the marine sponge-derived Micrococcus sp. EG45. Bioorganic & Medicinal Chemistry Letters, 24(21), 4939–4942. 10.1016/J.BMCL.2014.09.040 [DOI] [PubMed] [Google Scholar]
- Fernandez-Cortes A., Cuezva S., Sanchez-Moral S., Cañaveras J. C., Porca E., Jurado V., Martin-Sanchez P. M., Saiz-Jimenez C. (2011); Detection of human-induced environmental disturbances in a show cave. Environmental Science and Pollution Research, 18(6), 1037–1045. 10.1007/S11356-011-0513-5/TABLES/2 [DOI] [PubMed] [Google Scholar]
- Fleming A., Allison V. D. (1922); Observations on a bacteriolytic substance (“lysozyme”) found in secretions and tissues. British Journal of Experimental Pathology, 3(5), 252. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2047739/ [Google Scholar]
- Fosse T., Toga B., Peloux Y., Granthil C., Bertrando J., Sethian M. (1985). Meningitis due to Micrococcus luteus. Infection, 13(6), 280–281. 10.1007/BF01645439 [DOI] [PubMed] [Google Scholar]
- Gökalsın B., Aksoydan B., Erman B., Sesal N. C. (2017); Reducing virulence and biofilm of Pseudomonas aeruginosa by potential quorum sensing inhibitor carotenoid: Zeaxanthin. Microbial Ecology, 74(2), 466–473. 10.1007/S00248-017-0949-3 [DOI] [PubMed] [Google Scholar]
- Goksøyr A. (2013); Carotenoid sunscreen (U.S. Patent No. US20130078203A1). U.S. Patent and Trademark Office. https://patents.google.com/patent/US20130078203A1/en
- Graça A. P., Bondoso J., Gaspar H., Xavier J. R., Monteiro M. C., de la Cruz M., Oves-Costales D., Vicente F., Lage O. M. (2013); Antimicrobial activity of heterotrophic bacterial communities from the marine sponge Erylus discophorus (Astrophorida, Geodiidae). PLoS ONE, 8(11), e78992. 10.1371/JOURNAL.PONE.0078992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graffius S., Garzón J. F. G., Zehl M., Pjevac P., Kirkegaard R., Flieder M., Loy A., Rattei T., Ostrovsky A., Zotchev S. B. (2023); Secondary metabolite production potential in a microbiome of the Freshwater sponge spongilla lacustris. Microbiology Spectrum, 11(2):e0435322. 10.1128/spectrum.04353-22. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt C. L., Baum J., Klein B. Y., Nachshon S., Koltunov V., Cano R. J. (2004); Micrococcus luteus—survival in amber. Microbial Ecology, 48(1), 120–127. 10.1007/S00248-003-2016-5/FIGURES/6 [DOI] [PubMed] [Google Scholar]
- Hentschel U., Schmid M., Wagner M., Fieseler L., Gernert C., Hacker J. (2001); Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiology Ecology, 35(3), 305–312. 10.1111/J.1574-6941.2001.TB00816.X [DOI] [PubMed] [Google Scholar]
- Hutchinson D., Weaver R. H., Scherage M. (1943); The incidence and significance of microorganisms antagonistic to E. coli in water. Journal of Bacteriology, 45, 29–34. [Google Scholar]
- Jagannadham M. V., Rao V. J., Shivaji S. (1991). The major carotenoid pigment of a psychrotrophic Micrococcus roseus strain: Purification, structure, and interaction with synthetic membranes. Journal of Bacteriology, 173(24), 7911. 10.1128/JB.173.24.7911-7917.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbalaei-Heidari H. R., Partovifar M., Memarpoor-Yazdi M. (2020); Evaluation of the bioactive potential of secondary metabolites produced by a new marine Micrococcus species isolated from the Persian Gulf. Avicenna Journal of Medical Biotechnology, 12(1), 61–65. /pmc/articles/PMC7035459/ [PMC free article] [PubMed] [Google Scholar]
- Karpiński T. M., Ożarowski M., Alam R., Łochyńska M., Stasiewicz M. (2022); What do we know about antimicrobial activity of astaxanthin and fucoxanthin? Marine Drugs, 20(1), 36. 10.3390/MD20010036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Baltz R. H. (2016); Natural product discovery: Past, present, and future. Journal of Industrial Microbiology and Biotechnology, 43(2–3), 155–176. 10.1007/s10295-015-1723-5 [DOI] [PubMed] [Google Scholar]
- Kocur M. (1986); Genus I. Micrococcus Cohn 1872, 151AL. In Krieg N. R., Holt J. G. (Eds.), Bergey's Manual of Systematic Bacteriology (Vol. 2, pp. 1004–1008). The Williams & Wilkins Co. [Google Scholar]
- Kondo S., Horiuchi Y., Hamada M., Takeuchi T., Umezawa H. (1979); A new antitumor antibiotic, bactobolin produced by Pseudomonas. The Journal of Antibiotics, 32(10), 1069–1071. 10.7164/antibiotics.32.1069 [DOI] [PubMed] [Google Scholar]
- Kong D. X., Jiang Y. Y., Zhang H. Y. (2010); Marine natural products as sources of novel scaffolds: Achievement and concern. Drug Discovery Today, 15(21–22), 884–886. 10.1016/j.drudis.2010.09.002 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018); MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549. 10.1093/MOLBEV/MSY096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtböke D. I. (2012); Biodiscovery from rare actinomycetes: An eco-taxonomical perspective. Applied Microbiology and Biotechnology, 93, 1843–1852. 10.1007/s00253-012-3898-2 [DOI] [PubMed] [Google Scholar]
- Lafi F. F., Ramirez-Prado J. S., Alam I., Bajic V. B., Hirt H., Saad M. M. (2017); Draft genome sequence of plant growth-promoting Micrococcus luteus strain K39 isolated from Cyperus conglomeratus in Saudi Arabia. Genome Announcements, 5(4), e01520–16. 10.1128/GENOMEA.01520-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarasim V., Iya K. K. (1955); Studies on the micrococci in milk. Part I. Incidence and distribution. Indian Journal of Dairy Science, 8, 67. [Google Scholar]
- Lee J. S. (1961); Spectrophotometric Characterization of the Carotenoid Pigments Isolated from Micrococcus Radiodurans. [Thesis, Oregon State University; ]. https://ir.library.oregonstate.edu/concern/graduate_thesis_or_dissertations/6m311r734 [Google Scholar]
- Lee W., Lee D. G. (2014); Lycopene-induced hydroxyl radical causes oxidative DNA damage in Escherichia coli. Journal of Microbiology and Biotechnology, 24(9), 1232–1237. 10.4014/JMB.1406.06009 [DOI] [PubMed] [Google Scholar]
- Li J., Zhao G. Z., Huang H. Y., Qin S., Zhu W. Y., Zhao L. X., Xu L. H., Zhang S., Li W. J., Strobel G. (2012); Isolation and characterization of culturable endophytic actinobacteria associated with Artemisia annua L. Antonie van Leeuwenhoek. International Journal of General and Molecular Microbiology, 101(3), 515–527. 10.1007/S10482-011-9661-3/TABLES/4 [DOI] [PubMed] [Google Scholar]
- Löde A. (1902); Zentralblatt für bakteriologie. Mikrobiologie und Hygiene (1 Orig), 33, 196. [Google Scholar]
- Majeed H. Z. (2017); Antimicrobial activity of Micrococcus luteus cartenoid pigment. Al-Mustansiriyah Journal of Science, 28(1), 64. 10.23851/MJS.V28I1.314 [DOI] [Google Scholar]
- Mohana D. C., Thippeswamy S., Abhishek R. U. (2013); Antioxidant, antibacterial, and ultraviolet-protective properties of carotenoids isolated from Micrococcus spp. Radiation Protection and Environment, 36(4), 168. 10.4103/0972-0464.142394 [DOI] [Google Scholar]
- Mollica A., Costante R., Fiorito S., Genovese S., Stefanucci A., Mathieu V., Kiss R., Epifano F. (2014); Synthesis and anti-cancer activity of naturally occurring 2,5-diketopiperazines. Fitoterapia, 98, 91–97. 10.1016/J.FITOTE.2014.07.010 [DOI] [PubMed] [Google Scholar]
- Montalvo N. F., Mohamed N. M., Enticknap J. J., Hill R. T. (2005); Novel actinobacteria from marine sponges. Antonie van Leeuwenhoek, International Journal of General and Molecular Microbiology, 87(1), 29–36. 10.1007/s10482-004-6536-x [DOI] [PubMed] [Google Scholar]
- Murray C. J., Ikuta K. S., Sharara F., Swetschinski L., Robles Aguilar G., Gray A., Han C., Bisignano C., Rao P., Wool E., Johnson S. C., Browne A. J., Chipeta M. G., Fell F., Hackett S., Haines-Woodhouse G., Kashef Hamadani B. H., Kumaran E. A. P., McManigal B., Naghavi M. (2022); Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. The Lancet, 399(10325), 629–655. 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer R., Stafsnes M. H., Andreassen T., Goksøyr A., Bruheim P., Brautaset T. (2010); Biosynthetic pathway for γ-cyclic sarcinaxanthin in Micrococcus luteus: Heterologous expression and evidence for diverse and multiple catalytic functions of C50 carotenoid cyclases. Journal of Bacteriology, 192(21), 5688–5699. 10.1128/JB.00724-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisha P., John N., Mamatha C., Thomas M. (2019); Characterization of bioactive compound produced by microfouling actinobacteria (Micrococcus luteus) isolated from the ship hull in Arabian Sea, Cochin. Kerala. Materials Today: Proceedings, 25, 257–264. 10.1016/j.matpr.2020.01.362 [DOI] [Google Scholar]
- Nuñez M. (2014); Encyclopedia of Food Microbiology (2nd ed., pp. 627–633). Elsevier. [Google Scholar]
- Osawa A., Ishii Y., Sasamura N., Morita M., Kasai H., Maoka T., Shindo K. (2010); Characterization and antioxidative activities of rare C 50 carotenoids-Sarcinaxanthin, sarcinaxanthin monoglucoside, and sarcinaxanthin diglucoside-obtained from Micrococcus yunnanensis. Journal of Oleo Science, 59(12), 653–659. http://www.jstage.jst.go.jp/browse/jos/ [DOI] [PubMed] [Google Scholar]
- Palomo S., González I., De La Cruz M., Martín J., Tormo J. R., Anderson M., Hill R. T., Vicente F., Reyes F., Genilloud O. (2013); Sponge-derived Kocuria and Micrococcus spp. as sources of the new thiazolyl peptide antibiotic kocurin. Marine Drugs, 11(4), 1071. 10.3390/MD11041071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palozza P., Krinsky N. I. (1992); Astaxanthin and canthaxanthin are potent antioxidants in a membrane model. Archives of Biochemistry and Biophysics, 297(2), 291–295. 10.1016/0003-9861(92)90675-M [DOI] [PubMed] [Google Scholar]
- Parra J., Duncan K. R. (2019); Assessing metabolite biogeography of Micrococcus spp. and Pseudonocardia spp. isolated from marine environments. Access Microbiology, 1(1A), 587. 10.1099/ACMI.AC2019.PO0359 [DOI] [Google Scholar]
- Ranjan R., Jadeja V. (2017); Isolation, characterization and chromatography based purification of antibacterial compound isolated from rare endophytic actinomycetes Micrococcus yunnanensis. Journal of Pharmaceutical Analysis, 7(5), 343–347. 10.1016/J.JPHA.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo B. A., Farrona S., Rita Ventura M., Abranches R. (2020); Canthaxanthin, a red-hot carotenoid: Applications, synthesis, and biosynthetic evolution. Plants (Basel), 9(8), 1–18. 10.3390/PLANTS9081039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann H. A., Eklund C. M. (1941). The pigments of Micrococcus tetragenus: VI. Micrococcus tetragenus infection. Journal of Bacteriology, 42(5), 605. 10.1128/JB.42.5.605-614.1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. J., Cundliffe E., McCutchan T. F. (1998); The antibiotic micrococcin is a potent inhibitor of growth and protein synthesis in the malaria parasite. Antimicrobial Agents and Chemotherapy, 42(3), 715. 10.1128/AAC.42.3.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostami H., Hamedi H., Yolmeh M. (2016); Some biological activities of pigments extracted from Micrococcus roseus (PTCC 1411) and Rhodotorula glutinis (PTCC 5257). International Journal of Immunopathology and Pharmacology, 29(4), 684–695. 10.1177/0394632016673846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J. D., Vitorino I., De la Cruz M., Díaz C., Cautain B., Annang F., Pérez-Moreno G., Martinez I. G., Tormo J. R., Martín J. M., Urbatzka R., Vicente F. M., Lage O. M. (2019); Bioactivities and extract dereplication of Actinomycetales isolated from marine sponges. Frontiers in Microbiology 10, 727. 10.3389/FMICB.2019.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz F. J., Vanderah D. J., Hollenbeak K. H., Enwall C. E. L., Gopichand Y., SenGupta P. K., Hossain M. B., van der Helm D. (1983); Metabolites from the marine sponge tedania ignis. A new atisanediol and several known diketopiperazines. Journal of Organic Chemistry, 48(22), 3941–3945. 10.1021/jo00170a011 [DOI] [Google Scholar]
- Schorn M. A., Alanjary M. M., Aguinaldo K., Korobeynikov A., Podell S., Patin N., Lincecum T., Jensen P. R., Ziemert N., Moore B. S. (2016); Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters. Microbiology (Reading, England), 162(12), 2075. 10.1099/MIC.0.000386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin Y. H., Elwakil B. H., Ghareeb D. A., Olama Z. A. (2022); Micrococcus lylae MW407006 pigment: Production, optimization, nano-pigment synthesis, and biological activities. Biology, 11(8), 1171. 10.3390/BIOLOGY11081171/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanthi Kumari K., Shivakrishna P., Ganduri V. S. R. (2020); Wound healing activities of the bioactive compounds from Micrococcus sp. OUS9 isolated from marine water. Saudi Journal of Biological Sciences, 27(9), 2398–2402. 10.1016/J.SJBS.2020.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanthi Kumari K., Shivakrishna P., Sreenivasulu K. (2021); Molecular docking analysis of two bioactive molecules KLUF10 and KLUF13 isolated from the marine bacteria Micrococcus sp. OUS9 with TNF alpha. Bioinformation, 17(5), 530–535. 10.6026/97320630017530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. C. D., Shovon M. S., Jahan M. G. S., Asaduzzaman A. K. M., Khatun B., Yeasmin T., Roy N. (2012); Antibiotic sensitivity and antibacterial activity of Micrococcus spp. SCS1. Research & Reviews in BioSciences, 6(10), 304–310. [Google Scholar]
- Shukla M., Nadumane V. K. (2021); Yellow pigment from a novel bacteria, Micrococcus terreus, activates caspases and leads to apoptosis of cervical and liver cancer cell lines. Journal of Applied Pharmaceutical Science, 11(08), 077–084. [Google Scholar]
- Soldatou S., Eldjárn G. H., Ramsay A., van der Hooft J. J. J., Hughes A. H., Rogers S., Duncan K. R. (2021); Comparative metabologenomics analysis of polar actinomycetes. Marine Drugs, 19(2), 103. 10.3390/MD19020103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher G., Tamura K., Kumar S. (2020); Molecular evolutionary genetics analysis (MEGA) for macOS. Molecular Biology and Evolution, 37(4), 1237–1239. 10.1093/MOLBEV/MSZ312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierle A. C., Cardellina J. H., Singleton F. L. (1988); A marine Micrococcus produces metabolites ascribed to the sponge Tedania ignis. Experientia, 44(11–12), 1021. 10.1007/BF01939910 [DOI] [PubMed] [Google Scholar]
- Su T. L. (1948); Micrococcin, an antibacterial substance formed by a strain of Micrococcus. British Journal of Experimental Pathology, 29(5), 473–481. [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Nei M. (1993); Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution, 10(3), 512–526. 10.1093/OXFORDJOURNALS.MOLBEV.A040023 [DOI] [PubMed] [Google Scholar]
- Tizabi D. (2022); Bioprospecting Marine Actinomycetes for Novel Anti-tuberculosis Drugs [Doctoral dissertation, University of Maryland]. Digital Repository at the University of Maryland. [Google Scholar]
- Tizabi D., Bachvaroff T., Hill R. T. (2022); Comparative analysis of assembly algorithms to optimize biosynthetic gene cluster identification in novel marine actinomycete genomes. Frontiers in Marine Science, 9, 914197. 10.3389/FMARS.2022.914197/BIBTEX [DOI] [Google Scholar]
- Umadevi K., Krishnaveni M. (2013); Antibacterial activity of pigment produced from Micrococcus luteus KF532949. International Journal of Chemical and Analytical Science, 4(3), 149–152. 10.1016/J.IJCAS.2013.08.008 [DOI] [Google Scholar]
- Ungers G. E., Cooney J. J. (1968); Isolation and characterization of carotenoid pigments of Micrococcus roseus. Journal of Bacteriology, 96(1), 234–241. 10.1128/JB.96.1.234-241.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushasri R., Gods Will Shalomi C. (2015); A study on in vitro anti breastcancer activity of crude ethanol and acetone pigment extracts of Micrococcus luteus by MTT assay and analysis of pigment by thin layer chromatography. International Journal of Pharmacy and Biological Science, 5(1), 59–65. www.ijpbs.comorwww.ijpbsonline.com [Google Scholar]
- van der Merwe E., Huang D., Peterson D., Kilian G., Milne P. J., Van de Venter M., Frost C. (2008); The synthesis and anticancer activity of selected diketopiperazines. Peptides, 29(8), 1305–1311. 10.1016/J.PEPTIDES.2008.03.010 [DOI] [PubMed] [Google Scholar]
- Wang Z., Feng J., Li X., Zhang J. (2021); Whole-genome sequencing of Micrococcus luteus MT1691313, isolated from the Mariana Trench. Microbiology Resource Announcements, 10(23), e0036921. 10.1128/MRA.00369-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser M., Denner E. B. M., Kämpfer P., Schumann P., Tindall B., Steiner U., Vybiral D., Lubitz W., Maszenan A. M., Patel B. K. C., Seviour R. J., Radax C., Busse H. J. (2002); Emended descriptions of the genus Micrococcus, Micrococcus luteus (Cohn 1872) and Micrococcus lylae (Kloos et al. 1974). International Journal of Systematic and Evolutionary Microbiology, 52(2), 629–637. 10.1099/00207713-52-2-629/CITE/REFWORKS [DOI] [PubMed] [Google Scholar]
- Wilson B., Aeby G. S., Work T. M., Bourne D. G. (2012); Bacterial communities associated with healthy and Acropora white syndrome-affected corals from American Samoa. FEMS Microbiology Ecology, 80(2), 509–520. 10.1111/J.1574-6941.2012.01319.X [DOI] [PubMed] [Google Scholar]
- Young M., Artsatbanov V., Beller H. R., Chandra G., Chater K. F., Dover L. G., Goh E.-B., Kahan T., Kaprelyants A. S., Kyrpides N., Lapidus A., Lowry S. R., Lykidis A., Mahillon J., Markowitz V., Mavromatis K., Mukamolova G. V., Oren A., Rokem J. S., Greenblatt C. L. (2010); Genome sequence of the fleming strain of Micrococcus luteus, a simple free-living Actinobacterium. Journal of Bacteriology, 192(3), 841. 10.1128/JB.01254-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehra A., Begum M. A., Achiffa A. K., Rahiman A., Ushasri R. (2018); Micrococcus roseus pigment—Biocolour as novel antibacterial agent against Staphylococcus aureus isolate from currency notes. European Journal of Pharmaceutical Sciences, 5(5), 528–532. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.