Abstract
Proteomics of Babesia microti has lagged behind other apicomplexans despite recent genome and transcriptome studies. Here, we used a combination of nanotechnology and mass spectrometry to provide a proteomic profile of B. microti acute infection. We identified ~500 parasite proteins in blood with functions such as transport, carbohydrate and energy metabolism, proteolysis, DNA and RNA metabolism, signaling, translation, lipid biosynthesis, and motility and invasion. We also identified surface antigens with roles in the immune response to the parasite. This first evaluation of the B. microti proteome in erythrocytes provides information for the study of intracellular survival and development of diagnostic tools using mass spectrometry.
Introduction
Tick-borne diseases (TBD) among humans are on the rise in North America as urbanization continues along previously unpopulated areas. Among TBDs, babesiosis has been recognized in recent years as an emerging infectious disease. Babesiosis is caused by the intraerythrocytic protozoan parasites of the genus Babesia(1). Similar to Lyme disease, transmission of babesiosis occurs primarily by Ixodes ticks, however, other routes of transmission include pregnancy and blood transfusion(2, 3). The latter is of particular concern, especially among carriers of Babesia, as to this date there is no Food and Drug Administration (FDA)-approved assay to test for the parasite in the U.S. (4).
Babesia microti is a member of the phylum Apicomplexa and the causative agent for the majority of human cases of babesiosis in the U.S., with higher prevalences occurring in the Northeastern and Upper Midwestern regions of the country (5–7). Infection is usually asymptomatic or results in mild symptoms that resolve within a few days. Severe cases feature acute anemia, thrombocytopenia, organ failure, or even death among elderly, splenectomized, and immunocompromised individuals (8, 9). Diagnosis is performed primarily by microscopy of blood smears, which requires skilled and experienced laboratory personnel and lacks sensitivity. Serological methods and molecular assays based on conventional and real-time PCR have also been developed for detection of B. microti infections but are not broadly available (10, 11).
The genome sequence of B. microti R1 strain was reported in 2012 (12). Results revealed that the genome consists of four nuclear chromosomes and a linear mitochondrial genome. The overall size of the nuclear genome is ~6.5 Mbp, encoding ~3500 polypeptides, and it is the smallest Apicomplexan genome sequenced to date. Reconstruction of metabolic pathways from the annotated genome revealed limited metabolic machinery and lack of enzymatic processes known to be important for Plasmodium (12). In addition, phylogenetic analyses revealed that B. microtibelongs to a new Apicomplexan lineage distinct from the families represented by B. bovis and Theileria species. A recent study based on genomic and transcriptomic analyses of various B. microti isolates resulted in the re-annotation of the entire B. microti R1 strain genome(13). In addition to characterizing the genomic diversity among the isolates, the authors identified the full complement of genes encoding the B. microti secretome and surface proteome (13).
The application of emerging technologies in proteomics and mass spectrometry (MS) has contributed significantly to the validation of genome annotation studies in other apicomplexan parasites such as Plasmodium, Toxoplasma and Neospora (14–17). Of note, the great majority of proteomic studies performed with these pathogens have relied on material derived from in vitro cultures. For malaria, such experiments take advantage of synchronized culture and enrichment methods for specific parasite stages, minimizing host cell contamination and reducing background noise from the host proteome during analysis of blood stage parasite proteins (27). Contrary to this, there has been little if any research on the characterization of the B. microti proteome, due in part to the absence of a stable long-term in vitro culture system for the parasite. Alternatively, proteomic studies using in vivo models of babesiosis are appealing but can be challenging, even with the support of advanced mass spectrometry instrumentation due to the masking of parasite proteins by highly abundant host proteins found in blood. A recently developed nanotechnology approach based on hydrogel affinity nanoparticles circumvents some of the analytical and physiological roadblocks mentioned above and allows protein discovery directly in complex biological fluids with increased sensitivity. The applicability of this technology has been extended to other vector-borne infections such as Lyme disease (16, 17) and Chagas disease (18, 19).
The nanoparticles used in these studies are polymeric buoyant networks of cross-linked poly(N-isopropylacrylamide) (NIPAm) functionalized with high affinity chemical baits. Once incubated with a biological fluid, the nanoparticles act as a molecular sieve and rapidly capture, concentrate and preserve low abundance biomarkers, while excluding unwanted high abundance, high molecular weight proteins such as albumin and immunoglobulins that often interfere with mass spectrometry analysis (16, 17, 20).
While size and porosity can be engineered for size-exclusion of interfering substances, different high affinity capturing baits such as Remazol Brilliant Blue (RBB), Cibacron Blue (CB), Reactive Blue 221 (RB221), Reactive Black 31 (RB31), and other synthetic organic dyes can be incorporated to entrap different classes of low-abundance proteins and peptides through a combination of ionic, hydrophobic and electrostatic interactions (21). After capture, proteins can be eluted from the nanoparticles and analyzed by mass spectrometry, Western blotting, and other immunoassays.
The primary objective of this study was to perform a proteomic analysis of B. microti in whole blood using the aforementioned nanotechnology combined with MS. Using the hamster model of babesiosis, our methods resulted in the identification of ~500 B. microti proteins with functions as broad as intracellular signaling, protein translation and modification, protein transport, DNA replication, cytoskeleton support, and lipid and glucose metabolism. We also identified members of the B. microti secretome and surface proteome previously described by genomic studies (13, 22). This first evaluation of the B. microti proteome complements recent genomic and transcriptomic studies to significantly advance our knowledge of metabolism, evolution, and pathogenesis of the parasite and contribute towards the development of newer diagnostic and therapeutic tools.
Materials and Methods
Babesia isolate.
Babesia microti GI (BEI Resources NR-44070; ATCC® PRA-398™) was originally isolated from blood obtained from a human case of babesiosis in Nantucket, Massachusetts, USA, in 1983(23, 24). The isolate was propagated in Golden Syrian hamsters (Harlan Laboratories, stock: HsdHan:AURA) according to published protocols(25–27). Parasitemia was determined by microscopic examination of blood films stained with Giemsa. A minimum of 500 red blood cells (RBCs) were counted to calculate the percent parasitemia. This included all parasitized RBCs regardless of intraerythrocytic stage or number of parasites per cell. Blood samples for proteomic studies were collected after14 days of infection by peri-orbital route at 30% parasitemia (Fig. 1). All animal procedures were performedaccording to protocols approved by the ATCC® IACUC.
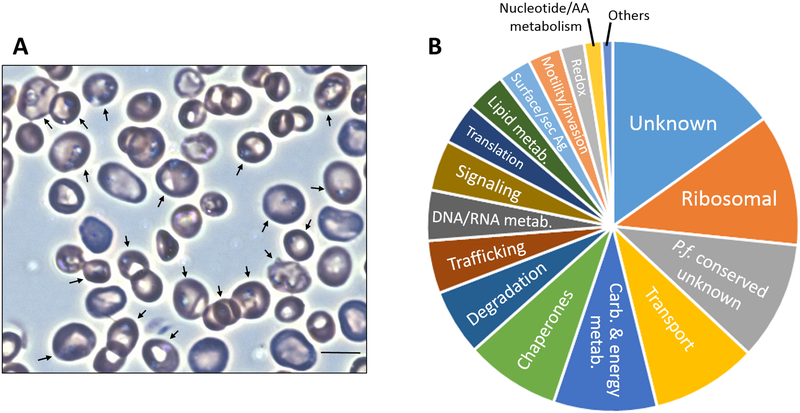
Fig. 1.
Proteomic map of Babesia microti infection in blood. (A) Light-microscopy image from Giemsa-stained hamster blood infected with the B. microti GI strain. Bar, 6 μm. Arrows indicate red blood cells harboring ring-form trophozoite stages of the parasite. (B) Classification of identified proteins based on biological function.
Nanoparticle processing of whole blood.
Detailed procedures for the synthesis of hydrogel nanoparticles, chemical functionalization, and validation of nanoparticles are described in Supplementary Materials and Methods S1. A 0.5 ml aliquot of whole blood was centrifuged at 2,000 × g for 10 min. 1 mL deionized water and 0.1 mL of nanoparticle suspension (5 mg/mL dry weight concentration) were added to the pellet and incubated at room temperature (RT) for 45 min. The RBC suspension was subjected to three freeze/thaw cycles at −80oC and centrifuged at 16,100 × g for 5 min. The resulting suspension was centrifuged at 2,000 × g for 10 min in order to remove cellular debris. The supernatant containing affinity nanoparticleswas centrifuged at 16,100 × g for 25 min and washed three times in ultrapure water (18 MΩ-cm). The nanoparticle pellet was re-suspended in 20 μl of elution buffer (Waters RapiGest™ SF surfactant with 50 mM ammonium bicarbonate and 5% tris(2-carboxyethyl)phosphine TCEP reducing agent) and incubated at RT for 20 min. Nanoparticles were centrifuged at 16,100 × g for 20 min and the supernatant was processed for MS as described in Supplementary Materials and Methods S1. Blood from an uninfected hamster was processed exactly as above for use as a negative control.
MS analysis.
Five separate injections of the B. microti-infected hamster blood sample and the uninfected control were analyzed by MS. Tandem mass spectra were searched against theoretical spectra derived from an annotated database of protein sequences from Babesia microti R1 (PiroplasmaDB.org) using the SEQUEST algorithm and tryptic cleavage constraints. High-confidence peptide identifications were obtained by applying the following filter criteria to the search results: 1) Xcorr versus charge of 1.9, 2.2, and 3.5 for 1+, 2+, 3+ ions; 2) ΔCn> 0.1; and 3) probability of randomized identification: e0.01. Acceptable false discovery rate (FDR) based on forward-reverse decoy was <1%. In addition, post-analysis filtering criteria were applied as follows: 1) peptide length >7 amino acids; 2) absence of carryover as determined by analyzing a blank sample with a 90 minute gradient; 3) manual validation of each peptide spectra, and 4) rejection of peptides showing >90% similarity with any Mesocricetus auratus (Golden hamster) protein and any other organism contained in the non-redundant database. The functional descriptions of the identified B. microti proteins were performed by searches in UniProt. In addition, searches for orthologues in other apicomplexans were performed in B. bovis, Theileria annulata and Plasmodium falciparum using PiroplasmaDB and EuPathDB.
Results
Using a combination of nanoparticle harvesting technology and mass spectrometry, we identified 522 parasite proteins from whole blood of hamsters with acute B. microti infection (Table S1). The 14 day-infected samples showed 30% parasitemia and consisted largely of trophozoite stages of the parasite (Fig. 1A). Identifications based on five or more peptides were observed in 106 proteins (20%), those based on two to four peptides were observedin 200 proteins (38%), and single peptide identifications were observed in 216 proteins (42%) (Table S1).
Categorization of proteins based on biological function determined that 134 (26%) were uncharacterized (i.e., matching to hypothetical genes in the B. microti genome). Of these, 53 were designated as conserved uncharacterized proteins with P. falciparum (Fig. 1B and Table S2). The remaining proteins were associated with common biological, cellular, and metabolicfunctions (Fig. 1B and Table 1). These functions included, among others, ribosomal (11%), transport (9%), carbohydrate and energy metabolism (9%), chaperones (8%), proteolysis (6%), trafficking (5%), DNA and RNA metabolism(4%), signal transduction (4%), protein translation and modification(4%), lipid metabolism (4%), surface/secreted antigens (3%), and motility and invasion (3%).Many of these proteins were previously found to be among the top 10% most expressed genes in B. microti by transcriptomic analysis(13).Regarding cellular location, 147 identified proteins were predicted to localize to the mitochondrion, 52 to the apicoplast, 34 to the ER/Golgi, 28 the nucleus, and 5 to secretory organelles (i.e., rhoptries and micronemes) (Table S2). Approximately 25% of the 522 B. microti proteins in our analysis were classified as ‘uncharacterized’ (i.e., matching to hypothetical genes in the B. microti genome). Of these, 53 were designated as conserved uncharacterized proteins in P. falciparum (Fig. 1B, Table 1, and Supplementary Table S2). With the purpose of supplementing previous genome annotation studies and transcriptomic analysis, we provide below an overview of components of lipid biosynthesis, carbohydrate and energy metabolism, and sero-reactive surface or secreted antigens identified in our analysis of the B. microti proteome during acute infection.
Table 1.
Classification of Identified Proteins Based on Biological Function
| Biological Function | No. of proteins | Examplesa |
|---|---|---|
| Unknown | 79 | Uncharacterized proteins matching to hypothetical genes in the genome |
| Ribosomal | 60 | Components of small subunit and large subunit ribosomal proteins |
| Plasmodium conserved, unknown | 54 | Uncharacterized Plasmodium conserved proteins |
| Transport | 48 | Ion transporters, carbohydrate transporters, protein transporters, ABC transporters |
| Carbohydrate and energy metabolism | 47 | Glycolysis proteins, TCA cycle proteins, electron transport chain proteins, mitochondrial importers, ATP synthases |
| Protein folding/chaperones | 43 | Heat shock proteins, SEC63, DnaK family proteins |
| Protein degradation | 30 | Cysteine proteases, metalloproteases, proteasome subunits, peptidases |
| Trafficking | 24 | Vesicular transport proteins, Rab proteins, SNARE-like proteins, ER lumen proteins, translocation proteins |
| DNA/RNA metabolism | 23 | DNA synthesis, replication, degradation, and repair proteins. RNA synthesis, transcription, modification, processing, and degradation proteins. Includes histones. |
| Signal transduction | 23 | Kinases, phosphatases, GTP-binding proteins |
| Protein translation and modification | 19 | Aminoacyl tRNAsynthetases, elongation factors, ubiquitin protein ligases, glycosyltransferases |
| Lipid metabolism | 18 | Proteins involved in lipid transport, biosynthesis, and degradation |
| Surface/secreted antigens | 15 | BMN family proteins, BmSA1 surface antigen |
| Motility and invasion | 15 | Actin, profilin, myosin, microneme and rhoptry proteins |
| Redox homeostasis | 11 | Thioredoxins, oxidoreductases, and other proteins that maintain the redox environment of the cell |
| Nucleotide and AA metabolism | 8 | Proteins involved in biosynthesis and degradation of nucleic acids and amino acids |
| Others | 5 | Proteins involved in porphyrin biosynthesis, chloroquine resistance, ubiquinone biosynthesis, and peripheral plastid proteins |
| Total | 522 |
ABC, ATP binding cassette; TCA, tricarboxylic acid; SNARE, soluble N-ethylmaleimide-sensitive factor activating protein receptor; ER, endoplasmic reticulum; BMN, Babesia microti sero-reactive antigen family; BmSA1, Babesia microti secreted antigen 1.
Lipid metabolism.
The analysis of the B. microti genome shows the presence of enzymes for the de novo synthesis of phospholipids and sphingomyelin as well as glycosylphosphatidylinositol (GPI) biosynthesis (12). The majority of the 19 identified proteins involved in lipid metabolism have roles in synthesis and catabolism of sphingomyelin, phosphatidylethanolamine, phosphatidylcholine, and recycling of phospholipids (Table S2). We also detected two of the enzymes involved in the synthesis of dolichyl D-mannosyl phosphate, a substrate of the GPI anchor biosynthesis pathway derived from dolichol obtained from the host.
Carbohydrate and energy metabolism.
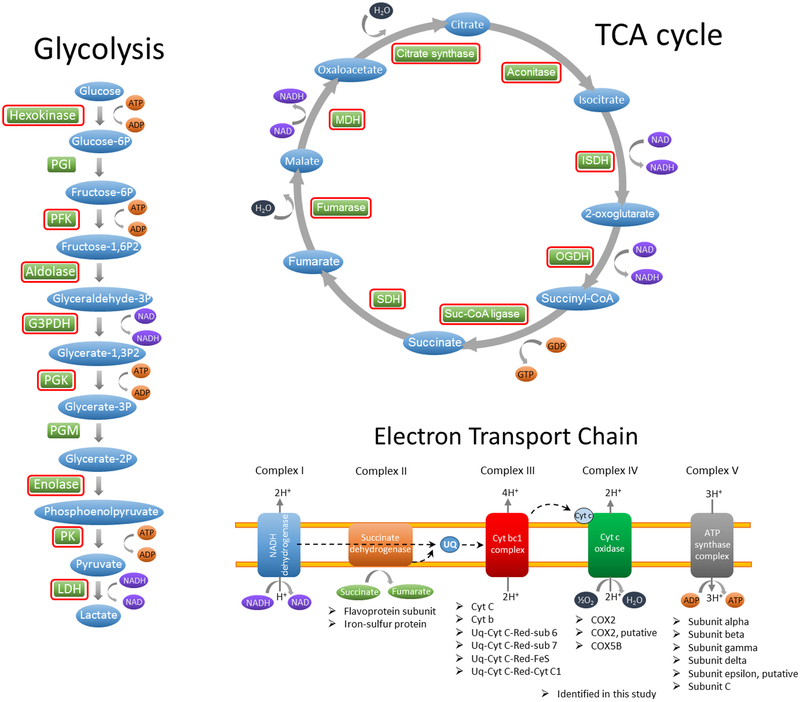
The identification of glycolytic enzymes (Supplementary Table S2, Fig. 2) is in line with the requirement of carbohydrate metabolism for energy production during the intra-erythrocytic asexual stage of the parasite. We detected the single sugar transporter found in the B. microti genome (BMR1_03g01335) (28) and eight of the ten enzymes involved in the breakdown of glucose into pyruvate(Fig. 2). These included hexokinase (BMR1_03g04600), phosphofructokinase (BMR1_02g02830), aldolase (BMR1_03g01800),glyceraldehyde 3-phosphate dehydrogenase (BMR1_01G00230), phosphoglycerate kinase (BMR1_03g00625), enolase (BmR1_04g05965), pyruvate kinase (BmR1_04g06405), and lactate dehydrogenase (BMR1_01G00020). Proteomic analysis also identified all eight enzymes of the tricarboxylic acid (TCA) cycle and several components of the four integral membrane protein complexes involved in the mitochondrial electron transport chain (Fig. 2). Subsequent MS-based studies should examine the utility of these metabolic proteins as potential diagnostic markers of active B. microti infection in the host. In addition, since glycolysis has been proposed as a potential pathway for therapeutic intervention in babesiosis (29) and other parasitic diseases (30, 31), it would be relevant to examine the proteomic profiles of components of the pathway in response to glycolytic enzyme inhibitors.
Fig. 2.
Use of proteomic analysis to support models for carbohydrate and energy metabolism of Babesia microti. Similar to other apicomplexans, Babesia possess all 10 enzymes of glycolysis in the genome (12). Enzymes circled in red were detected in the present study and support the presence of a functional glycolytic pathway and tricarboxylic acid cycle (TCA) cycle in B. microti. The flow charts for glycolysis, TCA cycle, and electron transport chain were adapted from Babesia bovis as described in the Liverpool Library of Apicomplexan Metabolic Pathways (LAMP) (www.llamp.net/) (35). PGI, phosphoglucose isomerase; PFK, phosphofructose kinase; G3PDH, glyceraldehyde-3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; PGM, phosphoglycerate mutase; PK, pyruvate kinase; LDH, lactate dehydrogenase; ISDH, isocitrate dehydrogenase; OGDH, oxoglutarate dehydrogenase; Suc-CoA, succinate-CoA; SDH, succinate dehydrogenase; MDH, malate dehydrogenase; UQ, ubiquinone; Cyt, cytochrome; COX, cytochrome c oxidase; IMS, intermembrane space; IMM, inner mitochondrial membrane; MCM, mitochondrial matrix.
Sero-reactive surface or secreted antigens.
Nanoparticles were also useful in trapping surface or secreted Babesiaantigensknown toelicit host immune responses in vivo (13). These included GPI-anchored, secreted, and transmembrane proteins, some of which share homologies to P. falciparum proteins (Table 2). Seven of the eleven identified seroreactive antigens in Table 2 were among the top 10% most expressed genes in B. microti (13).Four of these proteins were correspondingly highly abundant in the proteome based on identifications by seven or more peptides. These included proteins reported to elicit strong IgM/IgG responses such as BmSA1 (BMR1_03g00785), S1/P1 nuclease (BMR1_02g03140), and the conserved protein BMR1_03g00947, as well as the strong IgG inducer BmGPI10 (BMR1_02g04275) (Table 2). Other proteins known to trigger significant IgG responses were the N1–15 maltese-cross seroactive antigen (BmR1_04g07535) and MDN1 (BmR1_04g05531). The remaining groupof identified proteins includedtwo members of the BMN2 family (BMR1_02g04280 and BMR1_03g00020) and uncharacterized protein BmR1_04g07556 which werepreviously reported to elicit strong IgM responses in vivo (13).
Table 2.
Identification of B. microti proteins with reported immune responses in vivo1
| Gene ID | Description | Class2 | Ig Response1 | AA length | MW (kDa) | Orthologues in other piroplasmids and P. falciparum3 |
|---|---|---|---|---|---|---|
| BMR1_03g00785 | BmGPI12, BMN1 family, BMN1–9, BmSA1 orthologue | GPI | IgM/IgG | 328 | 35.4 | |
| BMR1_02g04275 | BmGPI10, BMN1 family, N1–21a orthologue | GPI | IgG | 304 | 34.4 | |
| BMR1_02g03140 | S1/P1 Nuclease | SEC | IgM/IgG | 373 | 42.8 | PF3D7_1411900 |
| BMR1_03g00947 | Conserved Plasmodium protein, unknown function | SEC | IgM/IgG | 438 | 48.7 | PF3D7_1324300 |
| BMR1_01G00985 | Conserved Plasmodium protein, unknown function | SEC | IgM/IgG | 1025 | 114.4 | PF3D7_0822900 |
| BMR1_02g04280 | BMN2 family, possible orthologue of N1-10-2 | TM | IgM | 519 | 60.9 | |
| BMR1_04g07535 | N1–15 protein, maltese-cross seroactive antigen | SEC | IgG | 2396 | 265.7 | |
| BMR1_03g04695 | Rhoptry neck protein 2 | IgM/IgG | 1483 | 165.7 | BBOV_I001630 TA19390 PF3D7_1452000 | |
| BMR1_03g00020 | BMN2 family | SEC | IgM | 474 | 55.4 | |
| BMR1_04g07556 | Uncharacterized protein | TM | IgM | 218 | 24.4 | |
| BMR1_04g05531 | MDN1, REA1, midasin | IgG | 4337 | 493.1 | BBOV_III002250 TA18495 PF3D7_1434500 |
As reported in Silva, J.C., et al. 2016. Sci. Rep. 6: 35284.
GPI, Glycosylphosphatidylinositol; SEC, secreted; TM, transmembrane.
BBOV, Babesia bovis T2Bo; TA, Theileria annulata Muguga; PF3D7, Plasmodium falciparum 3D7. B. bovis and T. annulata orthologues were found in PiroplasmaDB.org. P. falciparum orthologues were previously reported by Silva et al. (13).
Discussion
As mass spectrometry instrumentation has advanced sufficiently to allow protein discovery directly in complex biological fluids, the present study examined the B. microti proteome in whole blood in vivo by performing a prior nanoparticle-based approach to sequester proteins undergoing analysis. From the technical point of view, there are advantages to directly analyzing proteins of interest trapped in specialized buoyant porous hydrogel nanoparticles added to Babesia-infected blood. This approach avoids the cumbersome separation of infected RBCs and/or free parasites, which, to our knowledge, has been rarely documented with success in Babesia (32, 33). The method also circumvents pre-fractionation strategies and removal of detergents and other reagents used in conventional protein extractions, is simple to perform, and prevents proteolysis (20). The specificity of the approach for identification of parasite proteins is not derived from the bait itself but from the downstream MS analytical system.
Despite the ability of our nanoparticles to efficiently capture Babesia proteins, certain targets may exist in exceedingly low concentrations below the limit of detection of the nanoparticle/MS workflow (1 pg/ml) (34) or may avoid detection by the mass analyzer due to poor ionization efficiency. Preliminary enrichment steps may still be required for subsets of proteins such as parasite cytoskeletal proteins absent from our analysis (i.e., tubulin, dynein, and inner membrane complex proteins). Importantly, the resulting MS data set presented herein is a snapshot of the B. microti proteome during acute infection rather than a representation of protein expression over time. We found overall agreement between previous transcriptomic analysis and our study, particularly among the top 10 most highly expressed genes, which included seroreactive antigens, proteins involved in carbohydrate and energy metabolism, chaperones, transporters, and proteins with functions in translation, redox homeostasis, and degradation. Combined with other components of lipid metabolism, nucleotide and amino acid synthesis, and certain proteins associated with apicoplast metabolism, the catalog of proteins presented in this study largely mirrors metabolically active B. microti trophozoite populations present in acutely infected animals.
In summary, ours is the first known report to present a preliminary proteome map of B. microti in vivo. In the short term, this information can serve as a blueprint for studies of the proteomic response of the parasite at different times and under different conditions, as proteomes are not stable, but rather highly dynamic entities, reflecting the biological environment of the cell at the time of analysis. Importantly, studies of protein expression in the context of persistent infection could be facilitated with our nanoparticle-MS approach and the animal model presented herein as hamsters infected with the GI strain of B. microti can survive for months despite parasitemias of ~30%. Lastly, as recent advances in MS technologies are currently being used for the rapid and reproducible identification of pathogens in clinical microbiological laboratories, we foresee adapting our methods to an MS-based diagnostic test for the multiplex identification of B. microti and other tick-borne pathogens in human samples.
Supplementary Material
Acknowledgements
We thank Debra Adams-Fish from the ATCC® Specialty Laboratory Facilities, USA, for her assistance in the animal experiments. R.E. Molestina was supported by the ATCC® Internal Research and Development Program. R. Magni, A. Luchini, and L. Liotta and were supported by grants W81XWH-17-1-0175 from the U.S. Army Medical Research Acquisition Activity, and R21AI138135, 1R33CA206937, and 1R01AR068436 from the U.S. National Institutes of Health. ATCC® is a registered trademark of the American Type Culture Collection.
Footnotes
Note added in proof
The U.S. FDA approved the first tests to screen for Babesia microti in March 2018 (www.fda.gov/NewsEvents).
References
- 1.Hunfeld KP, Hildebrandt A, Gray JS. 2008. Babesiosis: recent insights into an ancient disease. Int J Parasitol 38: 1219–37 [DOI] [PubMed] [Google Scholar]
- 2.Joseph JT, Purtill K, Wong SJ, Munoz J, Teal A, Madison-Antenucci S, Horowitz HW, Aguero-Rosenfeld ME, Moore JM, Abramowsky C, Wormser GP. 2012. Vertical transmission of Babesia microti, United States. Emerg Infect Dis 18: 1318–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lobo CA, Cursino-Santos JR, Alhassan A, Rodrigues M. 2013. Babesia: an emerging infectious threat in transfusion medicine. PLoS Pathog 9: e1003387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gubernot DM, Nakhasi HL, Mied PA, Asher DM, Epstein JS, Kumar S. 2009. Transfusion-transmitted babesiosis in the United States: summary of a workshop. Transfusion 49: 2759–71 [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease C, Prevention. 2012. Babesiosis surveillance - 18 States, 2011. MMWR Morb Mortal Wkly Rep 61: 505–9 [PubMed] [Google Scholar]
- 6.Johnson ST, Cable RG, Tonnetti L, Spencer B, Rios J, Leiby DA. 2009. Seroprevalence of Babesia microti in blood donors from Babesia-endemic areas of the northeastern United States: 2000 through 2007. Transfusion 49: 2574–82 [DOI] [PubMed] [Google Scholar]
- 7.Kogut SJ, Thill CD, Prusinski MA, Lee JH, Backerson PB, Coleman JL, Anand M, White DJ. 2005. Babesia microti, upstate New York. Emerg Infect Dis 11: 476–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krause PJ, Gewurz BE, Hill D, Marty FM, Vannier E, Foppa IM, Furman RR, Neuhaus E, Skowron G, Gupta S, McCalla C, Pesanti EL, Young M, Heiman D, Hsue G, Gelfand JA, Wormser GP, Dickason J, Bia FJ, Hartman B, Telford SR, 3rd, Christianson D, Dardick K, Coleman M, Girotto JE, Spielman A. 2008. Persistent and relapsing babesiosis in immunocompromised patients. Clin Infect Dis 46: 370–6 [DOI] [PubMed] [Google Scholar]
- 9.Vannier EG, Diuk-Wasser MA, Ben Mamoun C, Krause PJ. 2015. Babesiosis. Infect Dis Clin North Am 29: 357–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosqueda J, Olvera-Ramirez A, Aguilar-Tipacamu G, Canto GJ. 2012. Current advances in detection and treatment of babesiosis. Curr Med Chem 19: 1504–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson M, Glaser KC, Adams-Fish D, Boley M, Mayda M, Molestina RE. 2015. Development of droplet digital PCR for the detection of Babesia microti and Babesia duncani. Exp Parasitol 149: 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornillot E, Hadj-Kaddour K, Dassouli A, Noel B, Ranwez V, Vacherie B, Augagneur Y, Bres V, Duclos A, Randazzo S, Carcy B, Debierre-Grockiego F, Delbecq S, Moubri-Menage K, Shams-Eldin H, Usmani-Brown S, Bringaud F, Wincker P, Vivares CP, Schwarz RT, Schetters TP, Krause PJ, Gorenflot A, Berry V, Barbe V, Ben Mamoun C. 2012. Sequencing of the smallest Apicomplexan genome from the human pathogen Babesia microti. Nucleic Acids Res 40: 9102–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva JC, Cornillot E, McCracken C, Usmani-Brown S, Dwivedi A, Ifeonu OO, Crabtree J, Gotia HT, Virji AZ, Reynes C, Colinge J, Kumar V, Lawres L, Pazzi JE, Pablo JV, Hung C, Brancato J, Kumari P, Orvis J, Tretina K, Chibucos M, Ott S, Sadzewicz L, Sengamalay N, Shetty AC, Su Q, Tallon L, Fraser CM, Frutos R, Molina DM, Krause PJ, Ben Mamoun C. 2016. Genome-wide diversity and gene expression profiling of Babesia microti isolates identify polymorphic genes that mediate host-pathogen interactions. Sci Rep 6: 35284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro-Sesquen YE, Gilman RH, Galdos-Cardenas G, Ferrufino L, Sanchez G, Valencia Ayala E, Liotta L, Bern C, Luchini A, Working Group on Chagas Disease in B, Peru. 2014. Use of a novel chagas urine nanoparticle test (chunap) for diagnosis of congenital chagas disease. PLoS Negl Trop Dis 8: e3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castro-Sesquen YE, Gilman RH, Mejia C, Clark DE, Choi J, Reimer-McAtee MJ, Castro R, Valencia-Ayala E, Flores J, Bowman N, Castillo-Neyra R, Torrico F, Liotta L, Bern C, Luchini A, Chagas HIVWGiB, Peru. 2016. Use of a Chagas Urine Nanoparticle Test (Chunap) to Correlate with Parasitemia Levels in T. cruzi/HIV Co-infected Patients. PLoS Negl Trop Dis 10: e0004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglas TA, Tamburro D, Fredolini C, Espina BH, Lepene BS, Ilag L, Espina V, Petricoin EF, Liotta LA, Luchini A. 2011. The use of hydrogel microparticles to sequester and concentrate bacterial antigens in a urine test for Lyme disease. Biomaterials 32: 1157–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magni R, Espina BH, Shah K, Lepene B, Mayuga C, Douglas TA, Espina V, Rucker S, Dunlap R, Petricoin EF, Kilavos MF, Poretz DM, Irwin GR, Shor SM, Liotta LA, Luchini A. 2015. Application of Nanotrap technology for high sensitivity measurement of urinary outer surface protein A carboxyl-terminus domain in early stage Lyme borreliosis. J Transl Med 13: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castro-Sesquen YE, Gilman RH, Galdos-Cardenas G, Ferrufino L, Sánchez G, Valencia Ayala E, Liotta L, Bern C, Luchini A, Peru WGoCDiBa. 2014. Use of a novel chagas urine nanoparticle test (chunap) for diagnosis of congenital chagas disease. PLoS Negl Trop Dis 8: e3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castro-Sesquen YE, Gilman RH, Mejia C, Clark DE, Choi J, Reimer-McAtee MJ, Castro R, Valencia-Ayala E, Flores J, Bowman N, Castillo-Neyra R, Torrico F, Liotta L, Bern C, Luchini A, Peru CHWGiBa. 2016. Use of a Chagas Urine Nanoparticle Test (Chunap) to Correlate with Parasitemia Levels in T. cruzi/HIV Co-infected Patients. PLoS Negl Trop Dis 10: e0004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longo C, Patanarut A, George T, Bishop B, Zhou W, Fredolini C, Ross MM, Espina V, Pellacani G, Petricoin EF, 3rd, Liotta LA, Luchini A 2009. Core-shell hydrogel particles harvest, concentrate and preserve labile low abundance biomarkers. PLoS One 4: e4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shafagati N, Patanarut A, Luchini A, Lundberg L, Bailey C, Petricoin E, III, Liotta L, Narayanan A, Lepene B, Kehn-Hall K 2014. The use of Nanotrap particles for biodefense and emerging infectious disease diagnostics. Pathogens and Disease 71: 162–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornillot E, Dassouli A, Pachikara N, Lawres L, Renard I, Francois C, Randazzo S, Bres V, Garg A, Brancato J, Pazzi JE, Pablo J, Hung C, Teng A, Shandling AD, Huynh VT, Krause PJ, Lepore T, Delbecq S, Hermanson G, Liang X, Williams S, Molina DM, Ben Mamoun C. 2016. A targeted immunomic approach identifies diagnostic antigens in the human pathogen Babesia microti. Transfusion 56: 2085–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray J, von Stedingk LV, Gurtelschmid M, Granstrom M. 2002. Transmission studies of Babesia microti in Ixodes ricinus ticks and gerbils. J Clin Microbiol 40: 1259–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piesman J, Karakashian SJ, Lewengrub S, Rudzinska MA, Spielmank A. 1986. Development of Babesia microti sporozoites in adult Ixodes dammini. Int J Parasitol 16: 381–5 [DOI] [PubMed] [Google Scholar]
- 25.Cullen JM, Levine JF. 1987. Pathology of experimental Babesia microti infection in the Syrian hamster. Lab Anim Sci 37: 640–3 [PubMed] [Google Scholar]
- 26.Oz HS, Hughes WT. 1996. Acute fulminating babesiosis in hamsters infected with Babesia microti. Int J Parasitol 26: 667–70 [DOI] [PubMed] [Google Scholar]
- 27.Wozniak EJ, Lowenstine LJ, Hemmer R, Robinson T, Conrad PA. 1996. Comparative pathogenesis of human WA1 and Babesia microti isolates in a Syrian hamster model. Lab Anim Sci 46: 507–15 [PubMed] [Google Scholar]
- 28.Mahmud O, Kissinger JC. 2017. Evolution of the Apicomplexan Sugar Transporter Gene Family Repertoire. Int J Genomics 2017: 1707231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bork S, Okamura M, Boonchit S, Hirata H, Yokoyama N, Igarashi I. 2004. Identification of Babesia bovis L-lactate dehydrogenase as a potential chemotherapeutical target against bovine babesiosis. Mol Biochem Parasitol 136: 165–72 [DOI] [PubMed] [Google Scholar]
- 30.Bruno S, Pinto A, Paredi G, Tamborini L, De Micheli C, La Pietra V, Marinelli L, Novellino E, Conti P, Mozzarelli A. 2014. Discovery of covalent inhibitors of glyceraldehyde-3-phosphate dehydrogenase, a target for the treatment of malaria. J Med Chem 57: 7465–71 [DOI] [PubMed] [Google Scholar]
- 31.Haanstra JR, Gerding A, Dolga AM, Sorgdrager FJH, Buist-Homan M, du Toit F, Faber KN, Holzhütter HG, Szöör B, Matthews KR, Snoep JL, Westerhoff HV, Bakker BM. 2017. Targeting pathogen metabolism without collateral damage to the host. Sci Rep 7: 40406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alkhalil A, Hill DA, Desai SA. 2007. Babesia and plasmodia increase host erythrocyte permeability through distinct mechanisms. Cell Microbiol 9: 851–60 [DOI] [PubMed] [Google Scholar]
- 33.Rossouw I, Maritz-Olivier C, Niemand J, van Biljon R, Smit A, Olivier NA, Birkholtz LM. 2015. Morphological and Molecular Descriptors of the Developmental Cycle of Babesia divergens Parasites in Human Erythrocytes. PLoS Negl Trop Dis 9: e0003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim B, Araujo R, Howard M, Magni R, Liotta LA, Luchini A. 2018. Affinity enrichment for mass spectrometry: improving the yield of low abundance biomarkers. Expert Rev Proteomics 15: 353–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanmugasundram A, Gonzalez-Galarza FF, Wastling JM, Vasieva O, Jones AR. 2013. Library of Apicomplexan Metabolic Pathways: a manually curated database for metabolic pathways of apicomplexan parasites. Nucleic Acids Res 41: D706–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.