Abstract
Elements mediating VanA glycopeptide resistance in 106 diverse enterococci from humans and nonhuman sources were compared with the prototype VanA transposon, Tn1546, in Enterococcus faecium BM4147. The isolates included 64 from individual patients at 15 hospitals in the United Kingdom (isolated between 1987 and 1996) and 42 from nonhuman sources in the United Kingdom (27 from raw meat, 7 from animal feces, and 8 from sewage). VanA elements were assigned to 24 groups (designated groups A to X) with primers that amplified 10 overlapping fragments of Tn1546. Ten groups of elements were found only in human enterococci, eight groups of elements were unique to nonhuman strains, and six groups of elements were common in enterococci from all sources. Elements indistinguishable from Tn1546 (group A) were observed more frequently in enterococci from nonhuman sources (34 versus 9%) but were identified in enterococci that caused outbreaks in hospital patients between 1987 and 1995. The most common group found in human enterococci (group H; 33%) was rarely observed in enterococci from other sources (5%). Group H elements differed from Tn1546 in three regions and included a novel insertion sequence, designated IS1542, between orf2 and vanR. The VanA elements of 14 other groups had a similar insertion at this position and/or distinct insertions at other positions. We conclude that VanA elements in enterococci are heterogeneous, although all show regions of homology with Tn1546. Furthermore, the elements most common among the human and nonhuman enterococci studied were different. This approach may be useful for monitoring the evolution of VanA resistance and may also be applicable in local “snapshot” epidemiological studies. However, as transposition events involving insertion sequences accounted for the differences observed between several groups, the stability of the elements must be assessed before their true epidemiological significance can be determined.
Glycopeptide-resistant enterococci (GRE) displaying the VanA resistance phenotype have been reported widely as a cause of nosocomial infections in the United States and Europe (28). In Europe, this transferable resistance mechanism has also been identified in enterococci isolated in the community, from sewage, animal feces, and raw meat, implicating these sources as a reservoir of resistant enterococci and VanA resistance elements (1, 2, 5, 8, 9, 11, 16). The occurrence of VanA resistance in enterococci outside the hospital environment has been attributed to use of the glycopeptide avoparcin as a growth promoter for animals used for meat production, especially pigs and poultry, but this is still debated (20, 27). However, avoparcin has never been licensed for use in the United States, which, when one considers the possible absence of VanA enterococci in nonhuman sources in the United States (10, 25), demonstrates that the epidemiology of VanA resistance is complex, with multiple factors affecting its evolution and global dissemination.
The ability of VanA enterococci from nonhuman sources to colonize humans is not known, nor is their ability to transfer resistance to resident enterococci during possibly transient passage through the human gut known. Investigations of the VanA enterococci themselves by molecular biology-based methods, including pulsed-field gel electrophoresis, have failed to demonstrate significant relationships between most strains of GRE isolated from humans and those isolated from nonhuman sources (5, 15). Because the gene cluster responsible for VanA resistance is associated with transposable elements (4), comparison of resistance plasmids in diverse strains is of limited value, and furthermore, resistance may be chromosomally encoded in some strains (12). Thus, direct comparison of VanA resistance elements in enterococci from diverse sources may provide insight into the spread of these genes among enterococci.
In Enterococcus faecium BM4147, the VanA phenotype is conferred by a 10.8-kb transposon, designated Tn1546 (4). Other VanA enterococci contain elements indistinguishable from or related to this element (4, 13, 18, 26, 32). However, despite extensive similarity in their gross structures, genetic variation can be detected between elements conferring the VanA phenotype (18, 32), and the insertion of various mobile elements into intergenic regions of Tn1546-related elements has been described (3, 4, 13). The aims of the present study were to assess the distribution of elements indistinguishable from Tn1546 among enterococci from hospital patients and nonhuman sources in the United Kingdom and to explore further the possibility that VanA elements in nonhuman enterococci may pose a threat to public health.
MATERIALS AND METHODS
Bacterial isolates.
E. faecium BM4147 (4), which contains Tn1546 on plasmid pIP816, and its glycopeptide-sensitive derivative BM4147-1, which lacks this plasmid, were used in this study as positive and negative controls, respectively. Sixty-four clinical isolates of VanA enterococci, all from distinct patients at 15 hospitals in the United Kingdom, were used. These had been referred to the Laboratory of Hospital Infection during the period from 1987 to 1996 for confirmation of glycopeptide resistance, identification of the bacterial species, and, in some instances, strain characterization. Forty-two enterococci isolated from nonhuman sources between 1993 and 1995 in the United Kingdom were also studied (5, 8, 9). These included 27 isolates from raw meat (19 from chicken, 4 from pork, and 4 from beef), 7 isolates from animal feces (1 from a chicken, 3 from pigs, 1 from a turkey, 1 from a duck, and 1 from a pony), and 8 isolates from sewage.
Amplification of VanA resistance elements.
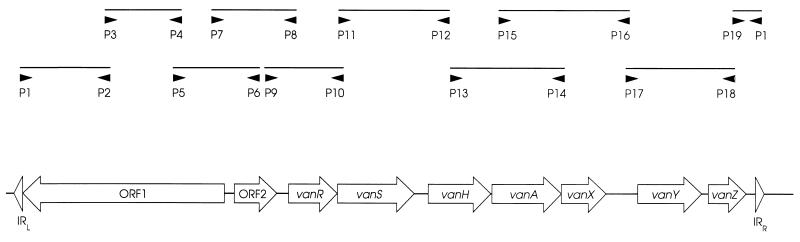
Crude preparations of template DNA were made by suspending two colonies of the enterococcus in 100 μl of distilled water (Tissue Culture Grade; Sigma Chemical Co.), vortexing briefly, and then pulsing on a microcentrifuge for 10 s. Two microliters of each of these suspensions was used in 25-μl PCR mixtures. Nineteen primers (p1 to p19) were used as described previously (4, 31) to amplify 10 overlapping fragments of the VanA elements (see Fig. 1). All amplifications were performed on Touchdown (Hybaid, Taddington, United Kingdom) or Genius (Techne, Cambridge, United Kingdom) thermal cyclers under previously published cycling conditions (29). The primers were derived from the sequence of Tn1546 (GenBank accession no. m97297). Each pair of primers except primers p1, p2, and p19 was used in a separate assay (with 50 ng of each primer used per reaction mixture); primers p1, p2, and p19 were tested together. The p1p2 and p19p1 products of Tn1546 have predicted sizes of 1,309 and 428 bp, respectively, and so they can easily be distinguished on gels. Control strains BM4147(Tn1546) and BM4147-1 were used throughout. PCR products were analyzed by electrophoresis through 2% agarose gels. The VanA element in each isolate was scored for the presence or absence of each of the 10 amplicons, and the size of each amplicon was compared with that obtained from Tn1546.
FIG. 1.
Ten overlapping fragments of prototype VanA element Tn1546 amplified with primers p1 to p19 (4, 31). IR, inverted repeat.
Sequencing of orf2-vanR intergenic regions.
The p9p10 amplicons from control strain BM4147(Tn1546) and from an enterococcus that had a product larger than the prototype VanA element were purified with a Hybaid Recovery DNA Purification Kit II (Hybaid) and were resuspended in sterile distilled water to a concentration of 30 ng/μl. Cycle sequencing was performed with an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer, Warrington, United Kingdom) by using 60 ng of template DNA and 5 pmol of either forward or reverse primer in a final reaction volume of 20 μl, as recommended by the supplier of the kit. Primers p9 (forward) or p8 (reverse) (4, 31) were used to begin sequencing, but extension of the strands was continued with primers designed from the obtained sequences. Amplification was carried out on a Touchdown (Hybaid) thermal cycler with an initial denaturation step of 95°C for 60 s, followed by 25 cycles of 96°C for 30 s, 50°C for 15 s, and 60°C for 4 min. The ramp rate was set at 1°C per s throughout this protocol. After amplification, each sample was carefully transferred to a fresh 500-μl microcentrifuge tube containing 50 μl of 95% ethanol and 2 μl of 3 M sodium acetate (pH 5.6), the contents were mixed briefly, and the tube was placed on ice for 10 min. The tubes were centrifuged for 30 min, after which time the supernatant was aspirated so as not to disturb any pellet. The pellets were washed once with 250 μl of 70% ethanol, and after spinning for 5 min and aspiration of the alcohol, they were dried at 90°C for 2 min. Samples were analyzed on an ABI PRISM 310 Genetic Analyzer (PE Applied Biosystems, Warrington, United Kingdom).
Analysis of DNA sequences.
Traces from the automated sequencer were visualized and edited by using Chromas 1.2 (the program is available on the World Wide Web at http://trishul.sci.gu.edu.au/∼conor/chromas.html) on an IBM-compatible personal computer. Comparison of the sequence with known DNA sequences, manipulation, and design of further forward and reverse sequencing primers were achieved with the GCG Wisconsin Package (version 8.1; UNIX), access to which was provided by the Human Genome Mapping Project Resource Centre of the Medical Research Council of the United Kingdom.
Generation and use of an IS1542 probe.
Two primers internal to the deduced partial sequence of IS1542 (see Results) were used to construct a probe to investigate its distribution among the enterococci studied. These were 5′-GAA TCG CTT TTA CTG CTT CTC (forward) and 5′-TTC TAA AGC TGC CAT ATT GC (reverse). The product of ca. 250 bp was amplified as described previously (29), labelled with digoxigenin-11-dUTP by PCR as described previously (30), and used in hybridization studies with DNA bound to nylon membranes. The target DNA consisted either of intact PCR amplicons or genomic DNA extracted with guanidium thiocyanate (21) and then digested with EcoRI (Life Technologies, Paisley, United Kingdom).
RESULTS
Groups of VanA elements found in enterococci.
By using 10 pairs of primers derived from the sequence of the prototype VanA element, Tn1546, the elements responsible for VanA glycopeptide resistance in 107 enterococci of diverse origins were placed into 24 groups (Table 1). Twenty (19%) isolates (including control strain BM4147) contained elements indistinguishable from Tn1546 by this method. These were designated group A, and the VanA elements of isolates of other groups were designated B to X, in accordance with their assortment by the Microsoft Access (version 2.0) database package. The only amplicons shared by elements of all 24 groups were those amplified with primer pairs p11p12 and p13p14, which correspond to the vanS, vanH, and vanA genes on Tn1546 (Fig. 1). Twenty-one groups (84 isolates; 79%) lacked one or more of the five amplicons obtained with primer pairs p1p2 to p9p10 inclusive (Table 1), suggesting alterations in the region of Tn1546 associated with transposition functions. One of these, designated group D (seven isolates), had nine amplicons in common with Tn1546 but did not give a product with primer pair p1p2. Nineteen groups (76 isolates; 71%) showed alterations in one or more of the amplicons obtained with the three primer pairs p15p16, p17p18, and p19p1 (Table 1), indicating changes downstream of vanA. Amplicons larger than those of Tn1546 were observed frequently with primers p7p8 (7 groups), p9p10 (7 groups), and p15p16 (10 groups), while amplicons were obtained with primers p17p18 that were both larger (2 groups) and smaller (3 groups) than predicted. Negative control strain BM4147-1 did not give amplicons with any of the 10 primer pairs.
TABLE 1.
Groups of VanA elements recognized among 107 enterococci of diverse origins using 10 primer pairs derived from the sequence of the prototype element, Tn1546
| Group | PCR product with the following primer
paira:
|
No. of isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| p1p2 | p3p4 | p5p6 | p7p8 | p9p10 | p11p12 | p13p14 | p15p16 | p17p18 | p19p1 | ||
| A | + | + | + | + | + | + | + | + | + | + | 20 |
| B | + | + | + | + | + | + | + | ++ | + | + | 2 |
| C | + | + | + | + | + | + | + | ++ | ++ | + | 1 |
| D | − | + | + | + | + | + | + | + | + | + | 7 |
| E | − | + | + | + | + | + | + | ++ | + | + | 3 |
| F | − | + | + | + | + | + | + | − | +/− | + | 4 |
| G | − | + | + | + | + | + | + | − | − | + | 7 |
| H | − | + | + | ++ | ++ | + | + | ++ | + | + | 23 |
| I | − | + | + | ++ | ++ | + | + | − | + | + | 1 |
| J | − | + | + | ++ | ++ | + | + | − | − | + | 1 |
| K | − | + | + | ++ | ++ | + | + | − | − | − | 2 |
| L | − | + | − | ++ | + | + | + | − | +/− | + | 1 |
| M | − | − | + | + | + | + | + | + | + | + | 2 |
| N | − | − | + | + | + | + | + | − | − | + | 2 |
| O | − | − | + | ++ | ++ | + | + | + | + | + | 1 |
| P | − | − | − | + | + | + | + | + | + | + | 1 |
| Q | − | − | − | + | + | + | + | ++ | + | + | 1 |
| R | − | − | − | ++ | ++ | + | + | ++ | + | + | 1 |
| S | − | − | − | − | + | + | + | ++ | + | + | 1 |
| T | − | − | − | − | ++ | + | + | ++ | + | + | 8 |
| U | − | − | − | − | − | + | + | ++ | + | + | 9 |
| V | − | − | − | − | − | + | + | ++ | ++ | + | 1 |
| W | − | − | − | − | − | + | + | − | +/− | + | 7 |
| X | − | − | − | − | − | + | + | − | − | + | 1 |
+, ++, +/−, amplicons the same size as, larger than, and smaller than those obtained for E. faecium BM4147 (containing Tn1546), respectively; −, absence of an amplicon with the particular pair of primers.
VanA elements in enterococci from humans and nonhuman sources.
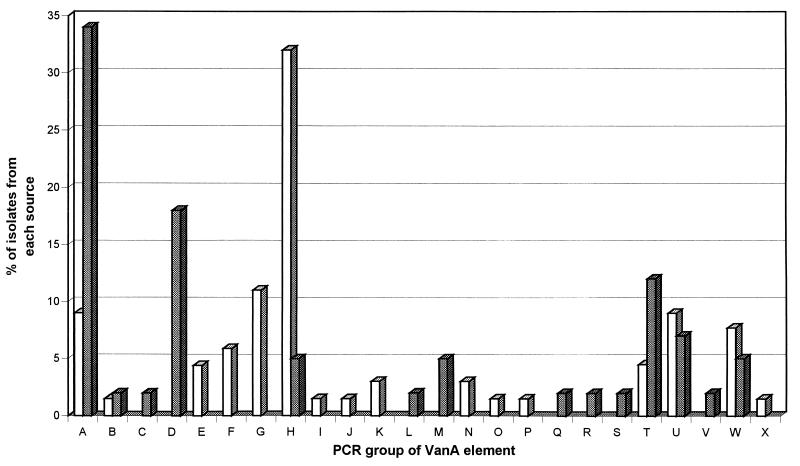
Ten of the 24 recognized groups of VanA elements were found only in enterococci isolated from hospital patients, eight groups of elements were found only in enterococci from nonhuman sources, but six groups of elements were common to isolates from all sources (Fig. 2). VanA elements belonging to the shared groups of elements from isolates from all sources (groups A, B, H, T, U, and W) were present in 42 (65%) GRE from humans and 27 (64%) GRE from nonhuman sources. However, the distribution of these groups among enterococci from humans versus those from nonhuman sources differed (Fig. 2). Fourteen of 42 (34%) GRE from nonhuman sources contained VanA elements of group A (Table 2), whereas only 6 of 65 (9%) GRE from hospital patients contained VanA elements of group A (chi square = 8.2; P < 0.005). The six human enterococci that contained VanA elements of group A (including BM4147) were isolated from patients during hospital outbreaks or from patients with sporadic cases of infection throughout the period from 1987 to 1995. The commonest group of VanA elements among GRE from humans was group H (21 isolates; 33%), but this group of elements was observed in only 2 isolates (5%) from other sources (chi square = 9.9; P < 0.005). Differences in the numbers of GRE containing elements of groups B, T, U, and W were not statistically significant.
FIG. 2.
Distribution of 24 different groups of VanA elements among 107 enterococci isolated from hospital patients (□) and nonhuman sources (▩).
TABLE 2.
Diversity of VanA elements in enterococci isolated from nonhuman sources
| Group | No. (%) of isolates from the following
sources:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Raw
meat
|
Feces
|
Sewage | Total | |||||
| Chicken | Pork | Beef | Chicken | Pig | Others | |||
| A | 7 | 1 | 1 | 3 | 2 | 14 (34) | ||
| B | 1 | 1 (2) | ||||||
| C | 1 | 1 (2) | ||||||
| D | 2 | 1 | 3 | 1 | 7 (18) | |||
| H | 2 | 2 (5) | ||||||
| L | 1 | 1 (2) | ||||||
| M | 2 | 2 (5) | ||||||
| Q | 1 | 1 (2) | ||||||
| R | 1 | 1 (2) | ||||||
| S | 1 | 1 (2) | ||||||
| T | 4 | 1 | 5 (12) | |||||
| U | 1 | 1 | 1 | 3 (7) | ||||
| V | 1 | 1 (2) | ||||||
| W | 2 | 2 (5) | ||||||
| Total | 19 | 4 | 4 | 1 | 3 | 3 | 8 | 42 (100) |
The groups of VanA elements identified in GRE from nonhuman sources are listed in Table 2. Although the number of GRE isolated from each source was small, the VanA elements in GRE recovered from raw meat appeared to be more diverse than those in GRE from animal feces; groups A and D were found in 12 of 27 (44%) and 6 of 7 (86%) GRE, respectively. The VanA elements in GRE from sewage were also diverse.
VanA elements in isolates of a single strain.
In order to examine the stability of VanA elements within a disseminated strain, 12 isolates (from 10 London hospitals) of an epidemic strain of a glycopeptide-resistant enterococcus designated EVREM-3 (epidemic vancomycin-resistant E. faecium) (19) were studied. The VanA elements in these isolates belonged to six groups: groups H (five isolates), I (one isolate), J (one isolate), N (two isolates), U (one isolate), and W (two isolates). Elements of groups H, U, and W were also found in non-EVREM-3 isolates of GRE from both hospital patients and nonhuman sources, but elements of groups I, J, and N were observed only in this epidemic strain. Three isolates of EVREM-3 from various stages of a prolonged outbreak on a single hospital unit contained elements of groups H and I and differed only in their reactions with primer pair p15p16, suggesting that the VanA element may have undergone some alteration during the outbreak.
Preliminary characterization of IS1542.
VanA elements belonging to seven groups (groups H, I, J, K, O, R, and T) gave identically sized amplicons with primer pairs p7p8 and/or p9p10 that were ca. 1,300 bp larger (as deduced from migration through agarose gels) than those derived from Tn1546 (Table 1) and which were consistent with an insertion(s) in this region. Because 28 of the 37 isolates in these groups had normally sized p5p6 amplicons (excluding those of groups R and T), it seemed likely that if a single insertion was present, it would be located within the p9p8 region of Tn1546 and most probably would be present in the intergenic region between orf2 and vanR (Fig. 1). The sequence of the orf2-vanR intergenic region of Tn1546 obtained from control strain BM4147 was identical to that described previously (4). One isolate of group H (the most frequently encountered group in GRE isolated from hospital patients, including EVREM-3) was chosen to represent isolates carrying the ca. 1,300-bp insertion. Partial sequencing of the intergenic region in this isolate revealed an 8-bp direct repeat of CTATAATC, corresponding to nucleotides 3925 to 3932 of the published Tn1546 sequence, on both sides of a proposed insertion sequence. This element has been designated IS1542, and partial forward and reverse sequencing indicated homology (detected with the FASTA program) with the staphylococcal element IS256 (data not shown). Figure 3 shows a comparison of the 26-bp imperfect, inverted repeats flanking IS1542 with those of IS256.
FIG. 3.
Comparison of the imperfect, inverted repeats (IRs) of IS1542 with those of IS256 (mismatches are indicated in large boldface characters).
Distribution of IS1542 in GRE.
Based upon the partial sequence available for IS1542, a ca. 250-bp fragment internal to this element was amplified, labelled with digoxigenin, and used to investigate the distribution of IS1542 in the GRE studied. This probe hybridized with the p9p10 amplicons of all 37 isolates that had the ca. 1,300-bp insertion at this position but not with the amplicons of BM4147(Tn1546) or other isolates lacking the insertion. In addition, the probe failed to hybridize with the p15p16 amplicons of 10 isolates chosen to represent VanA elements of groups B, C, E, H (the isolate of group H had the orf2-vanR copy used for sequencing), Q, R, S, T, U, and V, all of which gave amplicons with this primer pair larger than that of Tn1546, which had indicated a probable insertion at this position (data not shown). The probe hybridized with the p17p18 product of the single element of group V but not with that of group C.
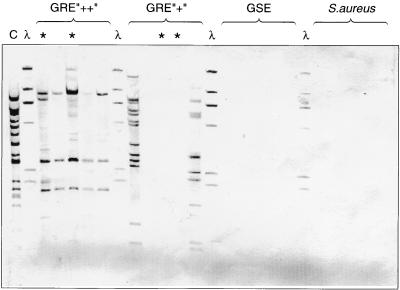
The probe was also used in hybridization studies with EcoRI-digested genomic DNA to gain a preliminary indication of the occurrence of IS1542 elements in enterococci. The probe hybridized with multiple EcoRI fragments from the group H control strain (from which the IS1542 partial sequence was derived), which was consistent with the presence of IS1542 elements at multiple locations in the genome of this strain (Fig. 4). Five other GRE with an orf2-vanR copy of IS1542 (including two EVREM-3 isolates) also gave multiple hybridization signals with the probe, although the banding patterns varied. Two of five GRE lacking the orf2-vanR copy of IS1542 also hybridized at multiple positions, indicating that the element was present at other positions in the genome. However, the probe did not hybridize with three GRE lacking the orf2-vanR copy (including two other EVREM-3), with any of five glycopeptide-sensitive enterococci (GSE) or with any of five clinical isolates of Staphylococcus aureus (Fig. 4). Because IS256 occurs frequently in clinical isolates of S. aureus and because the GSE tested included Enterococcus faecalis HH22, which is known to contain copies of IS256 (14), these data suggest that the probe used was specific for IS1542 and that the signals generated did not represent cross hybridization with copies of IS256.
FIG. 4.
Hybridization of EcoRI-digested genomic DNA with an IS1542-specific probe. HindIII digests of phage λ DNA are shown as size markers. Lane C, DNA of the strain from which the partial sequence of IS1542 was determined; lanes GRE “++”, GRE “+”, and GSE, DNA from enterococci with and without the orf2-vanR copy of IS1542 and from glycopeptide-sensitive enterococci, respectively; lanes ∗, DNA from four isolates of the epidemic strain of GRE, EVREM-3.
DISCUSSION
We have investigated the genetic elements responsible for VanA glycopeptide resistance in enterococci isolated from hospital patients and from nonhuman sources (animal feces, raw meat, and sewage) in the United Kingdom and have compared them with the prototype VanA transposon, Tn1546, which was characterized from a strain of E. faecium isolated from a French hospital patient in the late 1980s (4, 17). In Europe, the food chain has been implicated as a possible route by which enterococci with VanA glycopeptide resistance may be transmitted to humans and pose a threat to public health (1, 2, 5, 9, 11, 16). The isolation of VanA enterococci from the gastrointestinal tracts of people who eat meat but not from the gastrointestinal tracts of vegetarians lends support to this hypothesis (24). However, the actual strains of GRE isolated from humans and nonhuman sources rarely show similarity (5, 15), which suggests that the transmission of the elements that mediate resistance has contributed significantly to the dissemination of VanA resistance. Consistent with this, in the present study we identified VanA elements that were indistinguishable from Tn1546 (herein called group A elements) in 34% of enterococci from nonhuman sources, while in a further 18% of nonhuman isolates we found Tn1546-related elements that lacked a PCR product only with primer pair p1p2 (group D). A variant VanA element analogous to group D was recognized in one strain during the original characterization of these elements (4). Thus, our data indicate that VanA elements indistinguishable from those originally described in enterococci from hospital patients were also present in the majority of GRE studied from nonhuman sources and isolated in the United Kingdom during the period from 1993 to 1995. However, despite the high incidence of VanA elements of groups A and D in the GRE from nonhuman sources, elements belonging to group A were found in only 9% of GRE from hospital patients and group D elements were not observed among GRE from hospital patients. This finding is in contrast to those in previous reports that the VanA elements in most enterococci isolated from humans were indistinguishable or highly related to Tn1546 (6, 26).
In this study we observed a high degree of heterogeneity among VanA elements and categorized these elements into 24 groups using 10 pairs of PCR primers. These groups had variable degrees of homology with Tn1546, although all had amplicons after PCR with p11p12 and p13p14, consistent with the presence of the vanS, vanH, and vanA genes (4). The vanX gene is also essential for the expression of VanA resistance (23), so it was somewhat surprising that some isolates lacked an amplicon after PCR with p15p16. However, it is likely that use of a vanX-specific probe would confirm the presence of this gene in all of the GRE studied because difficulties with amplifying this region of some VanA elements have been reported previously (4, 18).
VanA elements belonging to 15 of the 24 groups gave amplicons larger than those of Tn1546 with one or more of the 10 primer pairs used, suggesting the presence of DNA insertions. Previous studies have identified variants of Tn1546 that carry novel insertion sequence elements in the vanS-vanH (13) and vanX-vanY (3) intergenic regions or have reported variable restriction fragment length polymorphisms corresponding to alterations in these regions (18). None of the isolates in this study appeared to carry insertions in the vanS-vanH region because the amplicon size after PCR with p11p12 for all isolates was identical to that of Tn1546. However, larger amplicons were observed frequently with primer pair p15p16. Because the amplicons of these isolates obtained with p13p14 were the same size as that of Tn1546, the insertion(s) was located between p14 and p17 (i.e., between vanA and vanY), but because vanX is essential (23), the insertions are most likely to be in the vanX-vanY region. A novel DNA insertion, designated IS1542, was identified in the orf2-vanR intergenic region of seven groups of VanA elements (35% of the isolates studied) from GRE from both humans and nonhuman sources. Insertions at this position have not been reported previously. Partial sequencing of this element indicated that it was flanked by 26-bp, imperfect, inverted repeats that showed homology with those of IS256 (7) and also by an 8-bp direct repeat of the Tn1546 sequence, consistent with a target site duplication following a transposition event. Comparison of the orientation and partial sequence of IS1542 with the sequence of IS256 indicated that the former may be transcribed in the direction opposite that in which orf2 and vanR are transcribed (4, 7). Despite the availability of only a partial sequence, single-passage sequencing was sufficient to allow the generation of a probe for IS1542. Multiple copies of this element were present in the genomes of GRE with the orf2-vanR copy and also in some GRE that lacked this copy. However, it was not detected in any of five GSE or S. aureus studied. Further studies with larger numbers of isolates are required to determine the distribution of this element in enterococci. The insertions at other positions of VanA elements noted in this study were distinct from IS1542 except for those for one isolate (of group V) that carried a copy of this element on the amplicon obtained with p17p18.
Many of the VanA elements studied (groups D to X) lacked one or more amplicons in the p1 to p10 region of Tn1546, corresponding to genes orf1 and orf2 associated with transposition functions (4). Further work is needed to determine how many groups do indeed contain these genes and for how many the lack of amplicons reflects a total or partial absence of these sequences. Transposition is believed to play a role in the dissemination of both VanA and VanB glycopeptide resistance (4, 22, 28). Thus, the absence of orf1 and/or orf2 in some VanA elements may affect their ability to spread, although the resistance genes may be part of larger composite transposons, as documented for isolates with chromosomally located VanA (12) or VanB (22) resistance. In this study, we did not determine whether the various VanA elements were located on plasmids or the chromosome, but such considerations would also influence their ability to be transmitted and require further investigation.
Although we have identified 24 groups of VanA elements in the GRE studied here, elements that represent groups not identified in our sample undoubtedly exist, such as those that lack only an amplicon obtained with p15p16 (4) and those with an IS1251 element in the vanS-vanH region (13). Increasing evidence suggests that many VanA elements differ from Tn1546 as the result of the insertion, by transposition, of various insertion sequence elements in intergenic regions (3, 13; this study). The genetic stability of the groups must therefore be examined. As an example, the commonest group of elements in human isolates, those of group H, differed from group D elements only by the insertion of IS1542 between orf2 and vanR and a second distinct insertion, probably between vanX and vanY. Because we could demonstrate the presence of IS1542 in some isolates that did not have it within their VanA elements, a transposition event from elsewhere in the genome would alter the group to which the element is allocated for these isolates. We are investigating the various elements further by several methods, including long PCR (31, 32). Detailed characterization will permit the evolution of VanA glycopeptide resistance to be monitored and may also be applicable in “snapshot” epidemiological surveys of GRE on hospital units. However, the issue of the stability of the elements must be resolved before the true epidemiological significance of their heterogeneity can be determined. This was highlighted by the six different groups of VanA elements observed in 12 isolates of the designated epidemic strain EVREM-3 in the United Kingdom and, in one instance, by the possible change in the VanA element during an extended hospital outbreak caused by this strain. However, because IS1542 was present at multiple locations in two isolates of this strain but was not present in two further isolates, it is possible that isolates of EVREM-3 may not all represent a single strain.
In conclusion, we have shown that the elements mediating VanA glycopeptide resistance in enterococci are heterogeneous. Six groups of VanA elements were shared by enterococci from human and nonhuman sources, but elements indistinguishable from Tn1546 were more common in enterococci isolated from nonhuman sources than in those from patients in hospitals in the United Kingdom. The greater diversity of elements in enterococci from humans may reflect the greater selection pressures exerted in the hospital environment, which may lead to a more rapid alteration of the elements. However, in Europe at least, nonhuman sources cannot be excluded as the reservoir of VanA resistance elements found in enterococci currently affecting public health.
ACKNOWLEDGMENTS
We thank the numerous colleagues in microbiology laboratories in the United Kingdom who have referred GRE to us in the last 10 years. We are particularly grateful to Paul Chadwick, Zoe Jordens, and Mehrnaz Seyed-Akhavani who referred enterococci of nonhuman origin that were included in this study, to Patrice Courvalin for kindly supplying control strains BM4147 and BM4147-1, to Michel Arthur for advice on primer sequences, and Patricia Woodford and the other staff of the Department of Medical Microbiology, Imperial College of Science, Technology & Medicine at St. Mary’s, London, United Kingdom, for analyzing samples on the automated DNA sequencer.
REFERENCES
- 1.Aarestrup F M. Occurrence of glycopeptide resistance among Enterococcus faeciumisolates from conventional and ecological poultry farms. Microb Drug Resist. 1995;1:255–257. doi: 10.1089/mdr.1995.1.255. [DOI] [PubMed] [Google Scholar]
- 2.Aarestrup F M, Ahrens P, Madsen M, Pallesen L V, Poulsen R L, Westh H. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalisisolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrob Agents Chemother. 1996;40:1938–1940. doi: 10.1128/aac.40.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes in Tn1546and related elements in the absence of induction. J Bacteriol. 1997;179:97–106. doi: 10.1128/jb.179.1.97-106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faeciumBM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates J, Jordens J Z, Griffiths D T. Farm animals as a putative reservoir for vancomycin-resistant enterococcal infection in man. J Antimicrob Chemother. 1994;34:507–514. doi: 10.1093/jac/34.4.507. [DOI] [PubMed] [Google Scholar]
- 6.Biavasco F, Miele A, Vignaroli C, Manso E, Lupidi R, Varaldo P E. Genotypic characterization of a nosocomial outbreak of VanA Enterococcus faecalis. Microb Drug Resist. 1996;2:231–237. doi: 10.1089/mdr.1996.2.231. [DOI] [PubMed] [Google Scholar]
- 7.Byrne M E, Rouch D A, Skurray R A. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin resistance transposon Tn4001. Gene. 1989;81:361–367. doi: 10.1016/0378-1119(89)90197-2. [DOI] [PubMed] [Google Scholar]
- 8.Casewell M, Seyed-Akhavani M, Hill R L R, Morrison D, Woodford N, Beighton D. A molecular comparison of vancomycin-resistant Enterococcus faeciumisolates from patients and from chickens. J Med Microbiol. 1996;44:v. [Google Scholar]
- 9.Chadwick P R, Woodford N, Kaczmarski E B, Gray S, Barrell R A, Oppenheim B A. Glycopeptide-resistant enterococci from uncooked meat. J Antimicrob Chemother. 1996;38:908–909. doi: 10.1093/jac/38.5.908. [DOI] [PubMed] [Google Scholar]
- 10.Coque T M, Tomayko J F, Ricke S C, Okhyusen P C, Murray B E. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob Agents Chemother. 1996;40:2605–2609. doi: 10.1128/aac.40.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devriese L A, Ieven M, Goossens H, Vandamme P, Pot B, Hommez J, Haesebrouck F. Presence of vancomycin-resistant enterococci in farm and pet animals. Antimicrob Agents Chemother. 1996;40:2285–2287. doi: 10.1128/aac.40.10.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handwerger S, Skoble J. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1995;39:2446–2453. doi: 10.1128/aac.39.11.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handwerger S, Skoble J, Discotto L F, Pucci M J. Heterogeneity of the vanAgene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob Agents Chemother. 1995;39:362–368. doi: 10.1128/aac.39.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodel-Christian S L, Murray B E. Characterization of the gentamicin resistance transposon Tn5281 from Enterococcus faecalis and comparison to staphylococcal transposons Tn4001 and Tn4031. Antimicrob Agents Chemother. 1997;35:1147–1152. doi: 10.1128/aac.35.6.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klare I, Heier H, Claus H, Bohme G, Marin S, Seltmann G, Hakenbeck R, Antanassova V, Witte W. Enterococcus faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community. Microb Drug Resist. 1995;1:265–272. doi: 10.1089/mdr.1995.1.265. [DOI] [PubMed] [Google Scholar]
- 16.Klare I, Heier H, Claus H, Reissbrodt R, Witte W. vanA-mediated high-level glycopeptide resistance in Enterococcus faeciumfrom animal husbandry. FEMS Microbiol Lett. 1995;125:165–172. doi: 10.1111/j.1574-6968.1995.tb07353.x. [DOI] [PubMed] [Google Scholar]
- 17.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 18.Miele A, Bandera M, Goldstein B. Use of primers selective for vancomycin resistance genes to determine vangenotype in enterococci and to study gene organization in VanA isolates. Antimicrob Agents Chemother. 1995;39:1772–1778. doi: 10.1128/aac.39.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison D, Woodford N, Cookson B D. Epidemic vancomycin-resistant Enterococcus faeciumin the UK. Clin Microbiol Infect. 1996;1:146–147. doi: 10.1111/j.1469-0691.1995.tb00460.x. [DOI] [PubMed] [Google Scholar]
- 20.Mudd A. Vancomycin resistance and avoparcin. Lancet. 1996;347:1412. doi: 10.1016/s0140-6736(96)91055-7. [DOI] [PubMed] [Google Scholar]
- 21.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 22.Quintiliani R J, Courvalin P. Characterization of Tn1547, a composite transposon flanked by IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalisBM4281. Gene. 1996;172:1–8. doi: 10.1016/0378-1119(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds P E, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of d-alanyl-d-alanine. Mol Microbiol. 1994;13:1065–1070. doi: 10.1111/j.1365-2958.1994.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 24.Schouten M A, Voss A, Hoogkamp-Korstanje J A A. VRE and meat. Lancet. 1997;349:1258. doi: 10.1016/s0140-6736(05)62461-0. [DOI] [PubMed] [Google Scholar]
- 25.Thal L A, Chow J W, Mahayni R, Bonilla H, Perri M B, Donabedian S A, Silverman J, Taber S, Zervos M J. Characterization of antimicrobial resistance in enterococci of animal origin. Antimicrob Agents Chemother. 1996;39:2112–2115. doi: 10.1128/aac.39.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van de Auwera P, Pensart N, Korten V, Murray B E, Leclercq R. Influence of oral glycopeptides on the fecal flora of human volunteers: selection of highly glycopeptide-resistant enterococci. J Infect Dis. 1996;173:1129–1136. doi: 10.1093/infdis/173.5.1129. [DOI] [PubMed] [Google Scholar]
- 27.Wise R. Avoparcin and animal feedstuff. Lancet. 1996;347:1835. doi: 10.1016/s0140-6736(96)91654-2. [DOI] [PubMed] [Google Scholar]
- 28.Woodford N, Johnson A P, Morrison D, Speller D C E. Current perspectives on glycopeptide resistance. Clin Microbiol Rev. 1995;8:585–615. doi: 10.1128/cmr.8.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodford N, Jones B L, Baccus Z, Ludlam H A, Brown D F J. Linkage of vancomycin and high-level gentamicin resistance genes on the same plasmid in a clinical isolate of Enterococcus faecalis. J Antimicrob Chemother. 1995;35:179–184. doi: 10.1093/jac/35.1.179. [DOI] [PubMed] [Google Scholar]
- 30.Woodford N, Morrison D, Johnson A P, Briant V, George R C, Cookson B. Application of DNA probes for rRNA and vanAgenes to investigation of a nosocomial cluster of vancomycin-resistant enterococci. J Clin Microbiol. 1993;31:653–658. doi: 10.1128/jcm.31.3.653-658.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodford, N., and J. M. Stigter. Molecular investigation of glycopeptide resistance in gram-positive bacteria. In N. Woodford and A. P. Johnson. (ed.), Molecular bacteriology: protocols and clinical applications, in press. Humana Press Inc., Totowa, N.J.
- 32.Woodford N, Watson A P, Chadwick P R. Investigation by long PCR of the genetic elements mediating VanA glycopeptide resistance in enterococci from uncooked meat in South Manchester. In: Horaud T, Bouvet A, Leclercq R, de Montclos H, editors. Streptococci and the host. New York, N.Y: Plenum Publishing Corporation; 1997. pp. 409–412. [DOI] [PubMed] [Google Scholar]