Abstract
The selection of bacterial resistance was examined in relationship to antibiotic pharmacokinetics (PK) and organism MICs in the patients from four nosocomial lower respiratory tract infection clinical trials. The evaluable database included 107 acutely ill patients, 128 pathogens, and five antimicrobial regimens. Antimicrobial pharmacokinetics were characterized by using serum concentrations, and culture and sensitivity tests were performed daily on tracheal aspirates to examine resistance. Pharmacodynamic (PD) models were developed to identify factors associated with the probability of developing bacterial resistance. Overall, in 32 of 128 (25%) initially susceptible cases resistance developed during therapy. An initial univariate screen and a classification and regression tree analysis identified the ratio of the area under the concentration-time curve from 0 to 24 h to the MIC (AUC0–24/MIC) as a significant predictor of the development of resistance (P < 0.001). The final PK/PD model, a variant of the Hill equation, demonstrated that the probability of developing resistance during therapy increased significantly when antimicrobial exposure was at an AUC0–24/MIC ratio of less than 100. This relationship was observed across all treatments and within all organism groupings, with the exception of β-lactamase-producing gram-negative organisms (consistent with type I β-lactamase producers) treated with β-lactam monotherapy. Combination therapy resulted in much lower rates of resistance than monotherapy, probably because all of the combination regimens examined had an AUC0–24/MIC ratio in excess of 100. In summary, the selection of antimicrobial resistance appears to be strongly associated with suboptimal antimicrobial exposure, defined as an AUC0–24/MIC ratio of less than 100.
Increasing bacterial resistance, and the subsequent burden to society in terms of morbidity, mortality, and increased health care expenditures, necessitates innovative approaches to the use of antimicrobial therapy (2, 15). Considering the dearth of available innovative approaches, attention to the appropriate utilization of antimicrobials is becoming increasingly important, particularly as there are fewer new antimicrobial drugs in development (4). The elucidation of relationships between pharmacodynamic parameters and organism persistence or resistance during therapy would facilitate the design of more effective dosing regimens. Unfortunately, there have been relatively few pharmacodynamic examinations of the relationship between antibiotic dosing and resistance in patients. Most of the available data comes from animal or in vitro models integrating pharmacokinetics and pharmacodynamics, where there are actually many definitive studies and considerable amounts of data.
Reports on the relationship between antimicrobial pharmacodynamic parameters and clinical and microbiological outcomes identify the percent time above the MIC (time-dependent killing) as the parameter predictive of response to β-lactam antimicrobials (8, 17, 27) and the ratio of the peak concentration to the MIC or area under the concentration-time curve (AUC) to the MIC (concentration-dependent killing) as the predictor for response to aminoglycosides and fluoroquinolones (17, 19). Recently, the AUC-to-MIC ratio, which incorporates both concentration intensity and exposure over time, has been advocated for use as the parameter for prediction of bacteriologic response (14, 16, 18, 26).
We have previously described the pharmacodynamic parameter area under the inhibitory time curve (AUIC) and its relationship to bacteriologic eradication and clinical outcome in patients with nosocomial pneumonia (14, 16, 23, 24). Antimicrobial resistance developed to some extent in each of these trials, but in the individual trials the numbers were too small to clearly define a mathematical relationship. The purpose of this pharmacokinetic/pharmacodynamic analysis was to determine the relationship of antimicrobial exposure, expressed as the AUC/MIC ratio, antibacterial activity, and other covariates to the development of bacterial resistance in the entire patient population treated in these trials.
MATERIALS AND METHODS
All patients treated in four antimicrobial clinical trials conducted at the Millard Fillmore Hospital between 1984 and 1991 were reviewed (5, 7, 12, 22, 24). The four data sets included a total of 143 acutely ill patients, virtually all of whom were treated for lower respiratory tract infection (LRTI). The four trials included (i) an open-label study of cefmenoxime therapy, 1 to 2 g every 4 or 6 h; (ii) an open-label trial of intravenous ciprofloxacin, 200 to 300 mg every 12 h; (iii) a multicenter, double-blind, randomized trial comparing intravenous ciprofloxacin, 400 mg every 8 h, with intravenous imipenem, 1,000 mg every 8 h; and (iv) an open-label, randomized, antimicrobial exposure (target AUIC of 250 SIT−1 ×24 h [inverse serum inhibitory titer integrated from 0 to 24 h]) controlled study of intravenous ciprofloxacin (400 mg every 8 or 12 h) versus intravenous ceftazidime (1 to 2 g every 8 or 12 h). In this study, if the dosage needed to provide a target AUC/MIC ratio of 350 exceeded 1,200 mg or 6 g per day for ciprofloxacin or ceftazidime, respectively, then piperacillin was added to ciprofloxacin and tobramycin was added to ceftazidime to achieve the targeted AUIC.
All patients with LRTI from the above trials were eligible for evaluation. Exclusion criteria were the following: an infection other than an aerobic bacterial pneumonia (anaerobic infection, lung abscess, fungal infection, atypical pathogens); less than 48 h of antimicrobial therapy; inability to isolate a bacterial pathogen; lack of MIC data; and the absence of pharmacokinetic data.
Initial bacteriologic studies on tracheal aspirates were performed for the four clinical trials by the Millard Fillmore Hospital Clinical Laboratory, Department of Pathology. Standard methods for pathogen identification and susceptibility testing using microdilution techniques were employed as previously described (12, 14, 16, 22–24). Culture and sensitivity testing was performed daily, in most cases, throughout the course of therapy and during the follow-up period, for determination of microbiologic end points. Criteria for clinical and microbiologic cure were similar in all studies. The study end points were time to eradication and microbiologic cure. These study end points allowed for the determination of the extent of and the time to the development of bacterial resistance. All isolates considered as bacterial pathogens in the original studies were included in the analysis of the development of resistance. Tracheal-aspirate culture and sensitivity data were reviewed for any significant changes in MICs during therapy. The development of bacterial resistance was defined as the isolation of a bacterial strain, initially found to be susceptible to the treatment regimen, which tested resistant during therapy and/or follow-up. The National Committee for Clinical Laboratory Standards (21) document of MIC interpretive standards was utilized for defining the antimicrobial resistance breakpoints. The cefmenoxime resistance breakpoint was defined by the cefmenoxime study protocol as a MIC of ≥25 μg/ml. Time to bacterial resistance was the number of days from the isolation of an initially susceptible organism to the day when the MIC reached the resistance breakpoint.
As there were a large number of different bacterial strains in these patients, the bacterial pathogens were categorized into four groups based upon similar initial-susceptibility characteristics. Group 1 contained only Pseudomonas spp. Group 2 contained gram-negative organisms, typically resistant to narrow-spectrum cephalosporins (cefazolin or cephalothin) and whose characteristics were consistent with those of type I β-lactamase producers, such as Enterobacter cloacae, Enterobacter aerogenes, Serratia marscesens, Citrobacter spp., Morganella morganii, and Proteus vulgaris. Group 3 contained other gram-negative rods typically susceptible to cephalosporins, such as Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis. Group 4 contained the remainder of the diverse organisms, i.e., Haemophilus influenzae, Staphylococcus aureus, and Streptococcus pneumoniae.
Patient-specific pharmacokinetic and pharmacodynamic parameters were determined from serial blood samples and culture MICs. Ciprofloxacin ceftazidime, cefmenoxime, and piperacillin serum drug concentrations were determined by high-performance liquid chromatography as previously described (5, 7, 22, 23, 24). Specific drug AUC values were obtained by fitting pharmacokinetic models to drug concentration data and integrating the drug concentration-versus-time curves over time. The pharmacodynamic parameter AUC0–24/MIC, where AUC0–24 is the AUC from 0 to 24 h, was calculated from each subject’s specific AUC and organism-specific MIC. In the first three studies, dosing adjustments were infrequent and the initial AUC0–24/MIC ratio was considered to be an adequate estimate of average antimicrobial exposure for the duration of therapy for these cases. In the final study, dose adjustments were made based on initial and subsequent serum concentrations by utilizing an adaptive feedback control algorithm to maintain targeted antimicrobial exposure (AUC0–24/MIC). In addition to serum drug concentrations, SITs were also obtained (5, 7). For these cases an average daily AUC0–24/MIC measure was determined by comodelling patient-specific pharmacokinetic and pharmacodynamic parameters with ADAPT II software (9, 10).
Statistical analysis.
For univariate analysis, the effect of categorical data on the likelihood of the development of resistance was evaluated by the chi-square test and Fisher’s exact test where appropriate. Continuous data, such as the patient baseline characteristics of age, weight, and time to resistance, were compared among groups by using Kruskal-Wallis analysis of variance. A P value of less than 0.05 was indicative of statistical significance. The data set was also subjected to classification and regression tree (CART) analysis with SYSTAT software (Systat, Inc., Evanston, Ill.) to identify possible significant interacting factors impacting the dependent variable, the development of resistance. Initial analysis by CART suggested that the AUC0–24/MIC ratio and several interacting factors such as prior antimicrobial therapy and the specific combinations of antimicrobial agent and organism (e.g., cefmenoxime therapy and Pseudomonas spp. and β-lactam monotherapy and group 2 organisms) were important. The final pharmacodynamic model, a variant of the Hill equation, described a relationship between antimicrobial exposure, expressed as the AUC0–24/MIC ratio, and the development of resistance for the majority of cases. Models were fit by the maximum-likelihood approach available in ADAPT II. Weighting was by the fitted-inverse observation variance, and model discrimination was accomplished by using Akaike’s information criterion (1). Kaplan-Meier survival curves were constructed to assess the probability of developing resistance beginning with the initiation of therapy.
RESULTS
A total of 143 patients were enrolled in the four clinical trials. Study inclusion criteria were met by 107 patients, with 128 organisms being evaluable. The mean age of the patients ± standard deviation was 68.6 ± 11.7 years. There were 64 males (60%) and 43 females (40%), with a mean weight of 69.4 ± 16.4 kilograms. The mean duration of therapy was 10.7 ± 3.5 days, with a range of 3 to 31 days. Patient baseline underlying disease states and case characteristics are listed in Table 1. Thirty-six patients were excluded from the evaluation; details concerning them are listed in Table 2. Serum concentrations were obtained for only ciprofloxacin in the third trial (ciprofloxacin versus imipenem); therefore, for trial 3 only the patients receiving ciprofloxacin were included in this analysis. Of the 128 organisms obtained from 107 patients, a single pathogen was evaluated for 90 patients, two pathogens were evaluated for 13 patients, and three pathogens were evaluated for 4 patients. Table 3 lists all organisms. Baseline MICs for selected pathogens are shown. Resistance developed in 32 (25%) of the evaluable cases. Three additional organisms, K. pneumonia and two strains of E. cloacae, were isolated during therapy or during the follow-up period, and were found to be resistant. These organisms were not isolated at baseline; therefore, they were not included in the analysis.
TABLE 1.
Baseline characteristics by clinical trial
| Characteristic or disease condition | % of cases in indicated antibiotic treatment group
|
||||
|---|---|---|---|---|---|
| All studies (n = 107) | Cefmenoxime (n = 26) | Ciprofloxacin (n = 39) | Imipenem vs cipro- floxacin (n = 8) | Ceftazidime vs cipro- floxacin (n = 34) | |
| Ventilator dependent | 72.9 | 76.9 | 59.0 | 75.0 | 85.3 |
| COPD | 32.7 | 42.3 | 23.1 | 25.0 | 38.2 |
| Major surgery | 36.4 | 23.1 | 35.9 | 37.5 | 44.1 |
| Diabetes mellitus | 25.3 | 11.5 | 25.6 | 0 | 41.2 |
| Steroids | 25.2 | 26.9 | 23.1 | 12.5 | 29.4 |
| Malignancy or neoplasm | 14.0 | 23.1 | 12.8 | 0 | 8.8 |
| Chemotherapy or radiation therapy | 3.7 | 11.5 | 2.6 | 0 | 0 |
| Microbiologic cure | 67.6 | 53.8 | 64.1 | 62.5 | 84.8 |
| Clinical cure | 68.3 | 65.4 | 61.5 | 100 | 72.7 |
| Bacterial eradication | 67.0 | 46.1 | 64.1 | 87.5 | 82.3 |
| Prior antibiotics | 63.2 | 84.6 | 79.5 | 0 | 44.1 |
| Serum albumin | |||||
| ≥3.9 g/dl | 1.9 | 0 | 0 | 28.6 | 0 |
| 3.0–3.8 g/dl | 22.4 | 19.2 | 23.1 | 14.3 | 29.0 |
| 1.9–2.9 g/dl | 72.0 | 76.9 | 76.9 | 42.9 | 64.5 |
| <1.9 g/dl | 3.7 | 3.8 | 0 | 14.3 | 6.5 |
TABLE 2.
Patients excluded from the analysis
| Study | No. of patients
|
Reason for patient exclusion
|
|||||
|---|---|---|---|---|---|---|---|
| In study | Ex- cluded | ≤48 H of therapy | No organism | NO PK or PDa | Resistant at baseline | Missing case report | |
| Cefmenoxime | 30 | 4 | 1 | 3 | |||
| Ciprofloxacin | 50 | 11 | 2 | 3 | 5 | 1 | |
| Imipenem vs ciprofloxacin | 14 | 6 | 3 | 3 | |||
| Ceftazidime vs ciprofloxacin | 49 | 15 | 5 | 5 | 2 | 3 | |
PK, pharmacokinetic data; PD, pharmacodynamic data.
TABLE 3.
Median MICs for study organisms at baseline and at the end point (resistance)
| Organism (n)a | Median MIC (μg/ml) ofb:
|
|||||
|---|---|---|---|---|---|---|
| Ceftazidime
|
Cefmenoxime
|
Ciprofloxacin
|
||||
| Pre TX | Resistant | Pre TX | Resistant | Pre TX | Resistant | |
| E. aerogenes (8) | 8.1 (2) | >32 (1) | 1.6 (2) | 37.5 (2) | 0.02 (4) | 4 (1) |
| E. cloacae (11) | 8.2 (2) | NR | 1.6 (3) | 50 (3) | 0.02 (6) | NR |
| E. coli (14) | 0.2 (4) | NR | 0.5 (4) | NR | 0.01 (6) | NR |
| H. influenzae (6) | 0.4 (2) | NR | ND | NR | 0.01 (4) | NR |
| K. pneumoniae (17) | 2.0 (3) | 64 (1) | 1.0 (3) | NR | 0.1 (11) | 8 (3) |
| P. aeruginosa (37) | 6 (4) | >32 (1) | 12.5 (9)c | 50 (5) | 0.5 (24) | 5 (12) |
| P. mirabilis (6) | 0.1 (1) | NR | 3.1 (2) | NR | 0.06 (4) | NR |
| S. aureus (5) | 8 (1) | NR | ND | NR | 0.4 (4) | NR |
| S. marcescens (11) | 0.7 (3) | 32 (1) | 0.4 (3) | 25 (1) | 0.1 (5) | NR |
Other organisms (number of isolates): Acinetobacter calcoaceticus (1); Citrobacter freundii (1); Citrobacter sp. (1); Klebsiella oxytoca (3); M. morganii (1); Pseudomonas fluorescens (2), P. vulgaris (2); one was resistant [MIC = 50]); S. pneumoniae (1); Serratia rubrifaciens (1).
Pre TX, baseline value; Resistant, end point value; NR, no resistance; ND, not done. Values in parentheses are numbers of isolates.
Range, 3.1 to 12.5 μg/ml.
The rates of resistance development as determined for organism groupings are listed in Table 4. The greatest frequency of selected resistance was observed for Pseudomonas (group 1 organisms) (46.1%), followed by group 2 organisms (27%) and group 3 organisms (10%). Resistance was not observed in the diverse group of remaining organisms (group 4). When resistance was evaluated by treatment, the greatest rate of resistance was observed for cefmenoxime (42.9%), followed by ciprofloxacin (27.6%), ceftazidime (20%), and the ceftazidime-tobramycin combination (9.1%). Resistance was not observed in the ciprofloxacin-piperacillin combination treatment arm. For monotherapy the rate of selected resistance development was 30.7% (31 of 101 cases), while resistance developed in only 3.7% (1 of 27) of the cases of combination therapy. The rates of resistance development by treatment groups, monotherapy and combination therapy, and by organisms are listed in Table 4.
TABLE 4.
Selected resistance rates by organism grouping and treatment
| Groupa | No. of isolates | % Rb (n) | Rate of resistance developmentc (%) for indicated treatment
|
||||
|---|---|---|---|---|---|---|---|
| CIP | CEFMNX | CAZ | CIP/PIP | CAZ/TOB | |||
| 1 | 39 | 46.1 (18) | 66.7 (12/18) | 55.5 (5/9) | 0 (0/3) | 0 (0/7) | 50 (1/2) |
| 2 | 37 | 27 (10) | 6.7 (1/15) | 70 (7/10) | 50 (2/4) | 0 (0/3) | 0 (0/5) |
| 3 | 40 | 10 (4) | 17.6 (3/17) | 0 (0/9) | 20 (1/5) | 0 (0/5) | 0 (0/4) |
| 4 | 12 | 0 (0) | 0 (0/8) | ND | 0 (0/3) | 0 (0/1) | ND |
| All organisms | 131 | 25 (32) | 27.6 (16/58) | 42.9 (12/28) | 20 (3/15) | 0 (0/16) | 9.1 (1/11) |
For a description of organisms in each group, see Materials and Methods.
R, overall rate of resistance development.
CIP, ciprofloxacin; CEFMNX, cefmenoxime; CAZ, ceftazidime; CIP/PIP, ciprofloxacin plus piperacillin; CAZ/TOB, ceftazidime plus tobramycin; ND, not done. The numbers in parentheses are numbers of resistant isolates/total numbers of isolates.
An initial univariate screen of patient, organism, and antimicrobial factors and their relationship to the development of resistance revealed that age, sex, weight, ventilator status, surgery, chronic obstructive pulmonary disease (COPD), diabetes mellitus, steroid use, malignancy, chemotherapy and/or radiation therapy, ciprofloxacin therapy, ceftazidime therapy, and the presence of group 2 gram-negative rods were not significant as predictors of resistance. However, several factors, including the AUC0–24/MIC ratio, the presence of group 1 or group 3 gram-negative rods, cefmenoxime therapy, group 2 organisms treated with β-lactam monotherapy, and previous antimicrobial therapy, were determined to be significant by univariate analysis (Table 5).
TABLE 5.
Univariate analysis results of resistance development by patient risk factors and characteristics
| Risk factor or characteristica | Pb |
|---|---|
| Age | 0.16c |
| Weight | 0.17c |
| AUC0–24/MIC ratio | <<0.001c |
| Gender | 0.77 |
| Infection with: | |
| Pseudomonas spp. | 0.0002 |
| Type I β-lac+ organisms | 0.74 |
| Other gram-negative rods | 0.008 |
| Miscellaneous organisms | 0.04 |
| Treatment with: | |
| CIP + PIP | 0.11 |
| CAZ | 0.76 |
| Cefmenoxime | 0.01 |
| CIP | 0.54 |
| CAZ + TOB | 0.29 |
| Ventilator dependent | 0.91 |
| Major surgery | 0.24 |
| COPD | 0.67 |
| Steroids | 0.82 |
| Diabetes mellitus | 0.82 |
| Malignancy | 0.22 |
| Chemotherapy or radiation therapy | 0.57 |
| Prior antibiotics | 0.02 |
| Type I β-lac+ organisms; β-lactam monotherapy | 0.001 |
β-lac+, β-lactamase producing; CIP, ciprofloxacin; PIP, piperacillin; CAZ, ceftazidime; TOB, tobramycin.
As evaluated by the chi-square test.
Determined by Kruskal-Wallis one-way analysis of variance.
The median AUC0–24/MIC values by organism and treatment groupings are presented in Table 6. The designation of susceptible or resistant indicates the antimicrobial exposure and applies to those organisms that either remained susceptible or became resistant, respectively, during treatment. Although the parameter AUC0–24/MIC has great variability, there is an apparent trend of emergent resistance at lower levels of antimicrobial exposure.
TABLE 6.
Summary statistics for AUC0–24/MIC ratios by organism groups and by treatment groups
| Organism or treatment groupa | AUC0–24/MIC
|
||
|---|---|---|---|
| Median | Mean (SD) | Range | |
| Organism group | |||
| 1 | |||
| Susceptible | 274 | 735 (1,501) | 73–6,967 |
| Resistant | 74 | 145 (190) | 15–733 |
| 2 | |||
| Susceptible | 667 | 1,604 (1,720) | 33–5,541 |
| Resistant | 693 | 3,517 (5,042) | 217–14,190 |
| 3 | |||
| Susceptible | 2,396 | 4,849 (8,510) | 169–45,263 |
| Resistant | 290 | 379 (399) | 48–887 |
| 4 | |||
| Susceptible | 843 | 5,937 (15,970) | 130–56,423 |
| Therapy group | |||
| CIP/PIP | |||
| Susceptible | 302 | 1,130 (1,693) | 118–6,103 |
| CAZ | |||
| Susceptible | 5,303 | 13,346 (19,174) | 218–56,423 |
| Resistant | 887 | 5,246 (7,746) | 661–14,190 |
| CEFMNX | |||
| Susceptible | 668 | 1,999 (2,305) | 73–6,852 |
| Resistant | 278 | 1,655 (3,259) | 18–9,822 |
| CIP | |||
| Susceptible | 582 | 1,516 (1,904) | 33–8,158 |
| Resistant | 72 | 185 (275) | 15–1,024 |
| CAZ/TOB | |||
| Susceptible | 2,499 | 3,069 (3,075) | 199–10,950 |
| Resistant (n = 1) | 733 | ||
Organism groups are as defined in Materials and Methods. CIP/PIP, ciprofloxacin plus piperacillin; CAZ, ceftazidime; CEFMNX, cefmenoxime; CIP, ciprofloxacin; CAZ/TOB, ceftazidime plus tobramycin.
CART analysis identified four factors as significant: AUC0–24/MIC ratio, cefmenoxime treatment, and organisms of group 1 and group 2. Treatment and organism interactions, such as Pseudomonas treated with cefmenoxime and β-lactamase-producing organisms treated with cefmenoxime and/or ceftazidime, were scrutinized for possible significance. Further inspection of these findings and analysis with pharmacodynamic models revealed that Pseudomonas spp. and all other organisms, with the exception of β-lactamase-producing organisms (group 2 organisms) treated with β-lactam monotherapy (cefmenoxime or ceftazidime), exhibited an inverse relationship between the probability of developing resistance and the AUC0–24/MIC ratio.
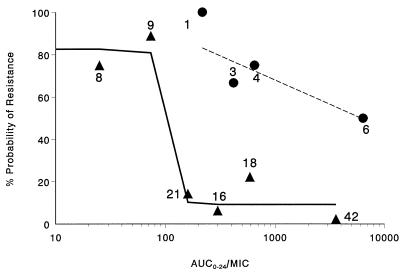
The final pharmacodynamic model describing this relationship is represented by the equation %P = [P0 − (P0 − P∞) · AUICH/(AUICmH + AUICH)] · (1 − R2) + P2 · R2, where P0 is the asymptotic maximum percent probability of resistance as the AUC0–24/MIC ratio goes to 0; P∞ is the asymptotic minimum percent probability of resistance as the AUC0–24/MIC ratio goes to infinity; R2 is an indicator, either 0 or 1, of group 2 organisms (β-lactamase-producing organisms treated with β-lactam monotherapy); P2 is the percent resistance for cases when R2 is 1; AUICm is the AUC0–24/MIC ratio at which %P = 0.5 · (P0 + P∞); and H is Hill’s constant, which reflects the degree of sigmoidicity. Parameters fitted by this model were %P0 = 82.6%, %Pmin = 9.2%, %P2 = 64.3%, AUC0–24/MIC (AUICm) = 100, and H = 40 (fixed). A log-linear regression approach with weighting was utilized to determine the line of best fit for the group 2 organisms treated with β-lactam therapy. The equation describing the line is y = 1.409 − 0.2548 · log10 AUC0–24/MIC. The final model fitted the data extremely well. The results of the model goodness-of-fit analysis are presented in Table 7. As described by this model, an inverse-effect relationship, which applies to all organisms and treatments with the exception of β-lactam monotherapy for group 2 organisms (consistent with type I β-lactamase-producing gram-negative organisms), exists. The observed and modelled data are graphically presented in Fig. 1. As shown in the figure the solid line represents the modelled response surface for all cases, classified as either susceptible or resistant, within the data set for which the AUC0–24/MIC inverse-effect relationship applied. The dashed line represents the line of best fit for the group 2 organisms treated with β-lactam monotherapy. Observed cases identified by symbols and numbers are plotted as single points within an AUC0–24/MIC ratio category at the median value for that category.
TABLE 7.
Pharmacodynamic model goodness of fit
| Antimicrobial exposure (AUC0–24/MIC)a | n | % Observed resistance | % Modelled fits |
|---|---|---|---|
| <100 | 17 | 82.4 | 82.6 |
| ≥100 | 97 | 9.3 | 9.2 |
| 0–50 | 8 | 75.0 | 82.6 |
| 50–100 | 9 | 88.9 | 80.9 |
| 100–250 | 21 | 14.3 | 10.2 |
| 250–500 | 16 | 6.3 | 9.2 |
| 500–1,000 | 18 | 22.2 | 9.2 |
| >1,000 | 42 | 2.4 | 9.2 |
| BLAC, all | 14 | 64.3 | 64.3 |
| BLAC, 100–250 | 1 | 100 | 83.2 |
| BLAC, 250–500 | 3 | 66.7 | 76.6 |
| BLAC, 500–1,000 | 4 | 75.0 | 72.3 |
| BLAC, >1,000 | 6 | 50.0 | 49.6 |
BLAC, type I β-lactamase-producing organisms treated with β-lactam monotherapy. The numbers following BLAC indicate the AUC0–24/MIC ranges; all, all AUC0–24/MIC values.
FIG. 1.
Relationship between the percent probability of resistance and the antimicrobial exposure as expressed by the average daily AUC0–24/MIC ratio. •, observed data for type I β-lactamase-producing gram-negative rods treated with β-lactam monotherapy; ▴, observed data for all other cases. The solid line represents the modelled response surface for all cases, classified as either susceptible or resistant, within the data set for which the AUC0–24/MIC inverse-effect relationship applied. The dashed line represents the line of best fit for the group 2 organisms treated with β-lactam monotherapy. Observed cases identified by symbols and numbers are plotted as single points within an AUC0–24/MIC ratio category at the median value for that category.
For those cases in which the model fits applied, Pseudomonas aeruginosa treated with ciprofloxacin represented a large number of the fitted cases. The observed percent resistance for ciprofloxacin monotherapy for pseudomonas organisms was 66.7%. This extremely high incidence of resistance was associated with the level of antimicrobial exposure. For example, the observed percent resistance for pseudomonas organisms treated with ciprofloxacin monotherapy, determined by utilizing the fitted AUC0–24/MIC breakpoint, was 100% (10 of 10 cases) when the AUC0–24/MIC ratio was <100 and 25% (2 of 8 cases when it was ≥100). In the case of group 3 organisms (other gram-negative rods), including all treatments, the observed percent resistance was 100% (2 of 2 cases) below the breakpoint, and 5.3% (2 of 38 cases) above the breakpoint. The observed data points of β-lactam monotherapy for group 2 organisms are clearly not reflective of the modelled response surface. The high percentage of resistance (>60%) within this group could not be explained by the pharmacodynamic measure of antimicrobial exposure, AUC0–24/MIC, as resistance occurred throughout a large range of AUC0–24/MIC ratios (217 to 14,190).
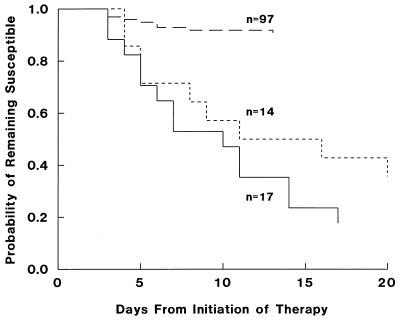
The median time to the observation of selected bacterial resistance was 6 days. The median time to resistance for all cases below the AUC0–24/MIC ratio breakpoint was 7 days versus 6 days for those cases above the breakpoint. This difference was not significant, irrespective of treatment or organism. Figure 2 is a Kaplan-Meier plot of the probability of remaining susceptible over time, from the initiation of therapy. The three curves represent three distinct groups: (i) the cases fit by the model below the AUC0–24/MIC breakpoint of 100 (n = 17); (ii) the cases consistent with type I β-lactamase-producing organisms treated with β-lactam monotherapy, which did not exhibit an AUC0–24/MIC relationship (n = 14); and (iii) the cases fit by the model above the AUC0–24/MIC breakpoint of 100 (n = 97). For the organisms represented by curve i (n = 17), the times to selection of 25, 50, and 75% resistance occurred by days 5, 10, and 14, respectively. For the gram-negative rods represented by curve ii (n = 14), the time to selection of 25% resistance occurred by day 5, and the time to selection of 50% or greater resistance occurred by day 16. For all other organisms, the cumulative rate of resistance development was approximately 9% and remained relatively consistent over time, with all cases of resistance occurring by day 13. A statistically significant difference was noted between groups 1 and 3 (P < 0.001) and between groups 1 and 2 (P < 0.001). Groups 2 and 3 did not differ (P = 0.322). However, the numbers of cases in these two groups are small and a type II error may exist. For these two groups, which exhibit different relationships of antimicrobial exposure to response, the rates of selection of bacterial resistance are similar.
FIG. 2.
Relationship between the probability of the development of resistance and treatment duration (days). Solid line, data for cases in which the AUC0–24/MIC ratio was <100; small dashed line, data for type I β-lactamase-producing gram-negative rods treated with β-lactam monotherapy; large dashed line, data for cases in which the AUC0–24/MIC ratio was ≥100.
DISCUSSION
In this population of acutely ill patients with LRTIs, there was an inverse-effect relationship between the probability of the development of bacterial resistance and the AUC0–24/MIC ratio. This relationship was strongest for Pseudomonas aeruginosa treated with ciprofloxacin, but was also found within other organism groups and antibiotic treatments. These findings support previous reports of the selection of resistance within Pseudomonas aeruginosa strains from studies utilizing in vitro pharmacodynamic models with various ciprofloxacin dosing regimens (18, 19). In the in vitro model studies, bacterial resistance did not occur when a dose of 1,200 mg of ciprofloxacin was administered once daily, but it was associated with a Cmax/MIC (Cmax, maximum concentration of the drug in serum) ratio of <10 (19). In a similar in vitro model, the investigators were able to select subvariant bacterial populations with increasing MICs related to a Cmax/MIC50 (MIC50, MIC at which 50% of isolates are inhibited) ratio of 7.3 and a breakpoint AUC/MIC50 ratio of 95 SIT−1 h (18), essentially identical to our AUC0–24/MIC breakpoint of 100. These results suggest that the AUC0–24/MIC ratio may be a useful parameter to guide therapy with the goal of preventing the selection of resistance (18).
In a neutropenic rat model of Pseudomonas sepsis, Drusano and colleagues studied lomefloxacin and found that either the Cmax/MIC ratio or the AUC/MIC ratio was a significant predictor of survivorship (11). However, when the AUC was held constant but the dose was changed to produce very high Cmax/MIC ratios (≥20:1), outcomes were significantly improved. Success in this model may reflect successful eradication of bacterial subpopulations that remain viable and that are selected for resistance by lower exposures. Unfortunately, dosing regimens of the fluoroquinolones which could be tolerated in human patients would not achieve these very high Cmax/MIC ratios for Pseudomonas. Finally, our data provide support for the observation that a Cmax/MIC ratio of approximately 5:1 may correspond to an AUC0–24/MIC ratio of 100 for ciprofloxacin.
We agree with Madaras-Kelly (18), that the AUC0–24/MIC ratio is the most reasonable parameter, as it combines both concentration intensity and exposure over time. In addition, we do not advocate Cmax/MIC targets over AUC/MIC targets because the dosing regimens suggested by Marchbanks and Drusano to achieve their effective Cmax/MIC ratios are not achievable in human pseudomonas infections, without unacceptable side effects. The other problem is that unlike AUC0–24/MIC ratios, Cmax/MIC ratios are not additive. Thus, it is not possible to optimize two antibiotics based on the Cmax/MIC ratio.
A recent review investigating the frequency of bacterial resistance included 173 clinical trials, incorporating 14,000 patients and seven antibiotic classes (13). One study included within our data set was among the 173 trials reviewed (22).
The authors reported an overall rate of bacterial resistance development of 4%, and in LRTI patients the incidence was 8.9%. The rates of resistance development reported for the intensive-care unit (ICU) population (7%) and the mechanically ventilated population (9%) were less than the 25% overall rate of resistance development reported in our study. However, our patients were primarily ICU patients and 72% were mechanically ventilated. Factors within the ICU setting such as mechanical ventilation and multiple underlying diseases appeared to contribute to bacterial resistance. Our observation of a lower frequency of resistance with combination therapy was also consistent with the finding of the review. Combination regimens had median AUC0–24/MIC ratios similar to those of the monotherapy regimens (Table 6), but the minimum values more frequently exceeded the AUC0–24/MIC ratio of 100. When two antibiotics are used, there is always a greater chance that one of the two may have an AUC0–24/MIC ratio above 100, thus explaining the better overall protection from resistance of combination therapy in pharmacodynamic terms.
The organisms which failed to demonstrate a statistically significant relationship between the development of resistance and the AUC0–24/MIC ratio were all β-lactamase-producing organisms treated with β-lactam monotherapy. Type I β-lactamase production is readily selected by the broad-spectrum cephalosporins in Enterobacter spp. and is associated with the development of resistance by the simple eradication of the susceptible subpopulations (3, 6, 20). The selection of resistant mutants during therapy results in a greater proportion of organisms producing adequate quantities of enzyme, which readily hydrolyzes most β-lactams (20), and the net effect is a rise in MIC. We suggest that in this situation there are bacterial subpopulations with baseline enzyme production present in the culture which will not be eradicated even with exposure of the entire population above the AUC0–24/MIC ratio of 100. Although the initial MICs seem to be low, they quickly increase as the susceptible organisms are eradicated leaving the resistant mutants behind. These findings provide further argument to avoid the use of monotherapy with broad-spectrum cephalosporins in these organisms, since our data show that variable rates of selection of bacterial subpopulations cannot be predicted by baseline MIC testing. Combination therapy with an antibiotic not susceptible to this mechanism is the necessary procedure. We surmise that an AUC0–24/MIC ratio above 100 is required for the second (non-β-lactam) drug, or the probability for failure will also be increased for the combination regimen. This may explain why aminoglycosides added to broad-spectrum cephalosporins did not always prevent the selection of resistance in E. cloacae (3), since aminoglycosides alone do not often produce AUC0–24/MIC ratios above 100.
A mechanistic discussion of how AUC0–24/MIC relationships might explain the selection of each type of resistance during therapy is clearly beyond the scope of this paper. However, the mechanism of resistance in many of our patients is most likely the selection of preexisting subpopulations of resistant mutants. In gram-negative pathogens the mutant subpopulations may have alterations in porin channels, plasmids containing resistant genes, active efflux pumps, or mutations in regulatory genes controlling β-lactamase production (6, 15, 20). In each of these cases, the action of the antibiotic is to eliminate the susceptible majority, leaving the selected remainder intact. In the absence of host defense or in the presence of foreign surfaces, the selected remainder quickly becomes the dominant population.
This study has several limitations. First, the retrospective nature of the review and the potential for bias in selection of resistant cases must be acknowledged, as ICU patients with nosocomial pneumonia are frequently colonized with gram-negative organisms and many have received previous courses of antimicrobial therapy. These data are not epidemiologic study data useful for determining the comparative frequency of resistance across patient populations, among ICUs, or among different antibiotics. Second, controversy will continue over the relative value of the different methods of lower respiratory tract specimen sampling and culture. The patients were clinically similar in all four trials (Table 1), and all the patients were treated in one institution under observation by the same investigators. There were common inclusion criteria for all of our nosocomial pneumonia trials, and most cases reported here were eventually used as part of new drug application submissions. Third, this study assumed that the resistant organisms isolated during therapy and follow-up were the same organisms as those isolated in the original cultures. Molecular typing (DNA) methods would be required to compare strain homologies, and these tests were not performed. Thus, the impact of AUC0–24/MIC ratios below 100 on selection versus the impact of new mutations or the introduction of new organisms cannot be definitively resolved. We can state with statistical certainty that when the AUC0–24/MIC ratio is below 100, 82.4% of the organisms developed resistance via some mechanism, the most likely being the selection of a subpopulation. When the AUC0–24/MIC ratio was above 100, only 9% of similar patients with similar organisms developed resistance (Table 7 and Fig. 2).
These results provide clinical data to support previously reported findings from in vitro models and from animal studies. They demonstrate a relationship between the antimicrobial pharmacodynamic parameter AUC0–24/MIC and the development of bacterial resistance. The association between resistance and bacterial MIC “coverage” by drug concentration is clearly stronger than any of the previously elucidated clinical risk factors. This finding is intuitively logical, since exactly the same conclusion arises from studies conducted in vitro and with animal models in the absence of patient factors and since the nosocomial pneumonia patient represents one of the situations where there is relatively little contribution of host defense to outcome.
Our results also suggest resistance can also be avoided with attention to dosing, since dosing regimens which provide an AUC0–24/MIC ratio of at least 100 appear to reduce the rate of the development of bacterial resistance in acutely ill patients with nosocomial pneumonia. Additional molecular studies of subpopulations will be needed to identify the relationship between β-lactam monotherapy and the selection of resistant organisms consistent with type I β-lactamase producers. Finally, clinical trials evaluating new antimicrobial agents should measure antimicrobial exposure parameters, such as the AUC0–24/MIC ratio, in order to relate these findings to both microbiologic and clinical outcomes and to determine the relationship between these indices and the development of antibacterial resistance.
REFERENCES
- 1.Akaike H. A bayesian extension of the minimum AIC procedure of autoregressive model fitting. Biometrika. 1979;66:237–242. [Google Scholar]
- 2.ASM Task Force on Antibiotic Resistance. Report of the ASM Task Force on Antibiotic Resistance. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 3.Ballow C H, Schentag J J. Trends in antibiotic utilization and bacterial resistance. Report of the national nosocomial resistance surveillance group. Diagn Microbiol Infect Dis. 1992;15:37S–42S. [PubMed] [Google Scholar]
- 4.Bax, R. P. 1997. Antibiotic resistance: a view from the pharmaceutical industry. Clin. Infect. Dis. 24(Suppl. 1):S151–S153. [DOI] [PubMed]
- 5.Bhavnani S M, Cheng A, Ballow C H, Forrest A. Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1996. Modelling the PDs of SITs for ceftazidime (Ctz), Ctz/tobramycin (C+T), ciprofloxacin (Cip), and Cip/piperacillin (C+P), in acutely ill patients, abstr. A99; p. 20. [Google Scholar]
- 6.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, A., A. Forrest, C. H. Ballow, and J. J. Schentag. Pharmacodynamics of intravenous ciprofloxacin with and without piperacillin in seriously ill patients. Submitted for publication.
- 8.Craig W A, Andes D. Pharmacokinetics and pharmacodynamics of antibiotics in otitis media. Pediatr Infect Dis J. 1996;15:255–259. doi: 10.1097/00006454-199603000-00015. [DOI] [PubMed] [Google Scholar]
- 9.D’Argenio D Z, Schumitzky A. A program package for simulation and parameter estimation in pharmacokinetic systems. Comput Programs Biomed. 1979;9:115–134. doi: 10.1016/0010-468x(79)90025-4. [DOI] [PubMed] [Google Scholar]
- 10.D’Argenio D Z, Schumitzky A. ADAPT II users manual. Biomedical simulations resource. Los Angeles, Calif: University of Southern California; 1992. [Google Scholar]
- 11.Drusano G L, Johnson D E, Rone M, Standiford H C. Pharmacodynamics of a fluoroquinolone antimicrobial agent in a neutropenic rat model of Pseudomonas sepsis. Antimicrob Agents Chemother. 1993;37:483–490. doi: 10.1128/aac.37.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fink M P, Snydman D R, Niederman M S, Leeper K V, Johnson R H, Heard S O, Wunderink G, Caldwell J W, Schentag J J, Siami G A, Zameck R L, Haverstock D C, Reinhart H H, Echols R M. Treatment of severe pneumonia in hospitalized patients: results of a multicenter, randomized, double-blind trial comparing intravenous ciprofloxacin with imipenem-cilastatin. Antimicrob Agents Chemother. 1994;38:547–557. doi: 10.1128/aac.38.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fish D N, Piscitelli S C, Danziger L H. Development of resistance during antimicrobial therapy: a review of antibiotic class and patient characteristics in 173 studies. Pharmacotherapy. 1995;15:279–291. [PubMed] [Google Scholar]
- 14.Forrest A, Nix D E, Ballow C H, Goss T F, Birmingham M C, Schentag J J. Pharmacodynamics of intravenous ciprofloxacin in seriously ill patients. Antimicrob Agents Chemother. 1993;37:1073–1081. doi: 10.1128/aac.37.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gold H S, Moellering R C. Antimicrobial-drug resistance. N Engl J Med. 1996;335:1445–1453. doi: 10.1056/NEJM199611073351907. [DOI] [PubMed] [Google Scholar]
- 16.Goss T F, Forrest A, Nix D E, Ballow C H, Birmingham M C, Cumbo T J, Schentag J J. Mathematical examination of dual individual principles. II. The rate of bacterial eradication at the same area under the inhibitory curve is more rapid for ciprofloxacin than for cefmenoxime. Ann Pharmacother. 1994;28:863–868. doi: 10.1177/106002809402800707. [DOI] [PubMed] [Google Scholar]
- 17.Hyatt J M, McKinnon P S, Zimmer G S, Schentag J J. The importance of pharmacokinetic/pharmacodynamic surrogate markers to outcome. Clin Pharmacokinet. 1995;28:143–160. doi: 10.2165/00003088-199528020-00005. [DOI] [PubMed] [Google Scholar]
- 18.Madaras-Kelly K J, Ostergaard B E, Hovde L B, Rotschafer J C. Twenty-four-hour area under the concentration-time curve/MIC ratio as a generic predictor of fluoroquinolone antimicrobial effect by using three strains of Pseudomonas aeruginosa and an in vitro pharmacodynamic model. Antimicrob Agents Chemother. 1996;40:627–632. doi: 10.1128/aac.40.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchbanks C R, McKiel J R, Gilbert D H, Robillard N J, Painter B, Zinner S H, Dudley M N. Dose ranging and fractionation of intravenous ciprofloxacin against Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro model of infection. Antimicrob Agents Chemother. 1993;37:1756–1763. doi: 10.1128/aac.37.9.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medeiros, A. A. 1997. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin. Infect. Dis. 24:(Suppl.):S19–S45. [DOI] [PubMed]
- 21.National Committee for Clinical Laboratory Standards. Minimum inhibitory concentration interpretive standards for organisms other than Haemophilus spp., Neisseria gonorrheae, and Streptococcus, table 2. M7-A3 (M100-S6). Wayne, Pa: National Committee for Clinical Laboratory Standards; 1995. [Google Scholar]
- 22.Peloquin C A, Cumbo T J, Nix D E, Sands M F, Schentag J J. Evaluation of intravenous ciprofloxacin in patients with nosocomial lower respiratory tract infections. Arch Intern Med. 1989;149:2269–2273. [PubMed] [Google Scholar]
- 23.Schentag, J. J., D. P. Reitberg, and T. J. Cumbo. 1984. Cefmenoxime efficacy, safety, and pharmacokinetics in critical care patients with nosocomial pneumonia. Am. J. Med. 77:(Suppl.6A):34–42. [DOI] [PubMed]
- 24.Schentag, J. J., I. L. Smith, D. J. Swanson, C. DeAngelis, J. E. Fracasso, A. Vari, and J. W. Vance. 1984. Role for dual individualization with cefmenoxime. Am. J. Med. 77:(Suppl.):43–50. [DOI] [PubMed]
- 25.Schentag J J, Nix D E, Adelman M H. Mathematical examination of dual individualization principles. I. Relationships between AUC and MIC and the area under the inhibitory curve for cefmenoxime, ciprofloxacin, and tobramycin. Drug Intell Clin Pharm. 1991;25:1050–1057. doi: 10.1177/106002809102501003. [DOI] [PubMed] [Google Scholar]
- 26.Schentag J J, Nix D E, Forrest A, Adelman M H. AUIC—the universal parameter within the constraint of a reasonable dosing interval. Ann Pharmacother. 1996;30:1029–1031. doi: 10.1177/106002809603000920. [DOI] [PubMed] [Google Scholar]
- 27.Vogelman B, Gudmundsson S, Leggett J, Turnidge J, Ebert S, Craig W A. Correlation of antimicrobial pharmacokinetic parameters with therapeutic efficacy in an animal model. J Infect Dis. 1988;158:831–847. doi: 10.1093/infdis/158.4.831. [DOI] [PubMed] [Google Scholar]