ABSTRACT
Mitophagy is a selective form of autophagy that targets dysfunctional or superfluous mitochondria for degradation. During mitophagy, specific selective autophagy receptors (SARs) mark a portion of mitochondria to recruit the autophagy-related (Atg) machinery and nucleate a phagophore. The phagophore expands and surrounds the mitochondrial cargo, forming an autophagosome. Fission plays a crucial role in separating the targeted portion of mitochondria from the main body to sequester it within the autophagosome. Our recent study, utilizing fission and budding yeasts as model systems, has identified Atg44 as a mitochondrial fission factor that generates mitochondrial fragments suitable for phagophore engulfment. Atg44 resides in the mitochondrial intermembrane space (IMS) and interacts with lipid membranes, with the capacity of mediating membrane fragility and fission. Based on our findings, we propose the term mitofissin to refer to Atg44 and its homologous proteins, which might participate in diverse cellular processes requiring membrane remodeling across various species.
Abbreviations
Atg: autophagy related; IMM: inner mitochondrial membrane; IMS: intermembrane space; PAS: phagophore assembly site; SAR: selective autophagy receptor
KEYWORDS: Atg44, autophagy, mitochondria, mitochondrial fission, mitofissin, mitophagy, yeast
Mitophagy shares the core molecular machinery with bulk autophagy, wherein Atg proteins assemble at the phagophore assembly site (PAS) and generate a phagophore to engulf cytoplasmic constituents into the autophagosome vesicle. The autophagosome ultimately fuses with a lysosome/vacuole to degrade its contents. Selective autophagy targets specific cargoes, such as unnecessary organelles, protein aggregates, and invading pathogens. The selectivity is mediated by cargo-specific SARs on the surface of the targeted cargo. These SARs promote the formation and expansion of the phagophore by binding to core Atg proteins including Atg8, which is conjugated to the phagophore membrane.
In the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe, mitophagy initiation relies on a single SAR, Atg32 and Atg43, respectively, while multiple receptors have been identified in mammals. Given that mitochondria are larger than autophagosomes, mitochondrial fission is necessary to generate small fragments that can be sequestered into autophagosomes (Figure 1A). Dynamin-related proteins, such as Dnm1 in yeasts and DNM1L/DRP1 in mammals, are well-known mitochondrial fission factors. They assemble on the mitochondrial outer membrane, forming ring-like structures that constrict mitochondria for division. While Dnm1/DNM1L contributes to mitochondrial dynamics and various cellular processes, they are not essential for mitochondrial fission during mitophagy. This suggests the involvement of an alternative fission mechanism in mitophagy.
Figure 1.
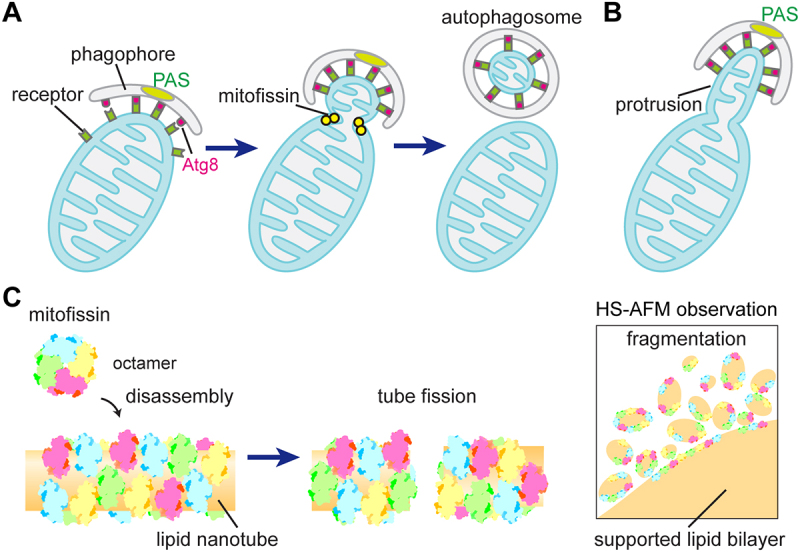
Mitofissin promotes mitochondrial fission. (A) During mitophagy, SARs such as Atg32 or Atg43 designate a region of mitochondria for degradation, and orchestrate phagophore formation and expansion at this location. Mitofissin in the IMS participates in membrane remodeling to induce mitochondrial fission, separating the targeted region of mitochondria from the rest of organelle body and generating small mitochondrial fragments that can be sequestered into autophagosomes. (B) In the absence of mitofissin, the SARs-marked region remains connected to the main body of mitochondria, resulting in the formation of mitochondrial protrusions toward the PAS. (C) Membrane fission by mitofissin in vitro. Lipid nanotubes (left) and supported lipid bilayers (right) are severed and fragmented, respectively, by the action of mitofissin, which undergoes disassembly from an octamer to smaller oligomeric form. HS-AFM stands for high-speed atomic force microscopy.
A screening of a fission yeast knockout collection identified mutants defective in mitophagy. Along with genes coding known Atg proteins and the SAR Atg43, the atg44+ gene (SPAC26A3.14c) was discovered to be essential for mitophagy [1]. Furthermore, a gene coding an Atg44 ortholog (YIL156W-B) is present in budding yeast. In both yeasts, Atg44 is required for mitophagy while it is dispensable for selective autophagy of other organelles or bulk autophagy. Mitophagy deficiency in fission yeast atg44∆ cells can be rescued by expressing budding yeast Atg44, and vice versa, indicating functional conservation of these proteins. Notably, Atg44 was not initially identified in comprehensive screens conducted in budding yeast that identified Atg32, because the atg44∆ mutant was not included in the knockout library.
In both yeasts, Atg44 localizes to the IMS. Cells lacking Atg44 exhibit abnormal mitochondrial morphology, similar to dnm1∆ cells that are defective in mitochondrial fission, regardless of mitophagy induction. Additionally, overexpression of Atg44 induces fragmentation of mitochondria independently of Dnm1, indicating that Atg44 functions as a mitochondrial fission factor. Therefore, we propose the term mitofissin (mitochondrial fission protein) to describe Atg44 and its homologous proteins.
Functional analyses of Atg44 in budding yeast support the notion that Atg44 facilitates mitophagy by generating mitochondrial fragments that are compatible with the size of autophagosomes. In atg44∆ cells, Atg32 is normally localized to the mitochondrial surface and recruits Atg11, an autophagy scaffold protein that connects Atg32 to the Atg machinery. Despite the normal function of Atg32, atg44∆ cells fail to form autophagosomes containing mitochondria as cargo. Additionally, atg44∆ cells exhibit mitochondrial protrusions that extend toward the vacuolar region where the PAS is formed. These protrusions are associated with the SAR Atg32, as well as the phagophore marked by Atg8 and the core Atg protein Atg11, all of which are necessary for protrusion formation. Hence, the mitochondrial region marked by Atg32, where the Atg machinery assembles and the phagophore is formed, fails to be separated from the main body of mitochondria in the absence of Atg44, hindering the phagophore engulfment into an autophagosome (Figure 1B). Consistently, artificial fragmentation of mitochondria by inhibiting mitochondrial fusion restores mitophagy in both budding and fission yeast cells lacking Atg44.
Structural analyses revealed that Atg44 is an amphiphilic protein that forms an octamer in both crystal and solution, with its hydrophobic residues buried inside. In vitro studies using recombinant Atg44 demonstrate its preferential binding to lipid membranes with high positive curvature through an electrostatic interaction. This interaction depends on cardiolipin, a phospholipid specific to the inner mitochondrial membrane (IMM), suggesting that Atg44 functions on the IMM. Remarkably, Atg44 possesses the ability to induce membrane fission without the requirement of cofactors (Figure 1C). Both simulations and in vitro observations suggest that Atg44 interacts with lipid membranes as a monomer, dimer, or tetramer, rather than as an octamer. Moreover, the interaction of Atg44 with lipid membranes causes membrane fragility, leading to the fragmentation of supported lipid bilayers as observed by high-speed atomic force microscopy. Based on these findings, we infer that Atg44 binds to membranes as a tetramer or a smaller oligomer, with its hydrophobic residues inserted into the lipid membrane. This hydrophobic binding generates membrane-area differences, imposes steric pressure on the membrane through protein crowding, or both, resulting in membrane remodeling that drives mitochondrial fission.
Collectively, we have identified and characterized a novel mitochondrial fission protein, Atg44, that we termed mitofissin. Atg44 is able to generate mitochondrial fragments for engulfment within the autophagosome. Notably, mitofissin is also involved in mitochondrial fission during vegetative growth in which mitophagy is suppressed. The function and mechanism underlying mitofissin-promoted mitophagy-independent fission are of great interest for investigating the relationship between mitofissin and Dnm1/DNM1L. Moreover, mitofissin-like proteins are not limited to fungi but also present in other eukaryotic organisms, including Dictyostelium species, algae, and bryophytes. Exploring their functions in mitochondria and other organelles would be intriguing. It is possible that metazoans and higher plants harbor proteins functionally related to mitofissin or have developed processes analogous to mitofissin-driven mitochondrial fission to facilitate mitophagy.
Funding Statement
This work was supported by grants from the Japan Society for the Promotion of Science (23K05679 to TF and 19H05712 to KT), the Institute for Fermentation, Osaka (IFO) (TF), and the Takeda Science Foundation (TF).
Disclosure statement
No potential conflict of interest was reported by the authors.
Reference
- [1].Fukuda T, Furukawa K, Maruyama T, et al. The mitochondrial intermembrane space protein mitofissin drives mitochondrial fission required for mitophagy. Mol Cell. 2023;83(12):2045–2058.e9. doi: 10.1016/j.molcel.2023.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]


