Abstract
Two plasmid-derived NcoI DNA fragments of 14 and 4.5 kb, respectively, have been isolated from the multidrug-resistant strain Enterococcus hirae S185R and analyzed. The 14-kb fragment contains two inverted (L and R) IS1216 insertion modules of the ISS1 family. These modules define a Tn5466 transposon-like structure that contains one copy of the methylase-encoding ermAM conferring erythromycin resistance and one copy of the adenylyl-transferase-encoding aadE conferring streptomycin resistance. Immediately on the left side of IS1216L there occurs a copy of pbp3r encoding the low-affinity penicillin-binding protein (PBP) PBP3r, itself preceded by a psr-like gene (psr3r) that controls the synthesis of PBP3r. ermAM, aadE, and the transposase gene (tnp) of IS1216R have the same polarities, and these are opposite those of psr3r, pbp3r, and the tnp gene of IS1216L. The 4.5-kb fragment is a copy of the 4.5-kb sequence at the 5′ end of the 14-kb fragment, although it is not a restriction product of the 14-kb fragment. It contains three genes with the same polarity: psr3r, pbp3r, and tnp in an IS1216 element. Because of the very high degree of identity (99%) with the chromosomal psrfm and pbp5fm genes of Enterococcus faecium D63R, it is proposed that both the psr3r and pbp3r genes were transferred from an E. faecium strain and inserted in a plasmid of E. hirae. E. hirae is the first known bacterial species in which a low-affinity PBP-encoding gene has been found to be plasmid borne.
The penicillin-binding proteins (PBPs) are membrane-bound serine transferases involved in wall peptidoglycan synthesis. Penicillin inactivates the PBPs in the form of stable serine ester-linked penicilloyl enzymes (14).
Resistance to penicillin in the absence of β-lactamase production can be mediated by PBPs. Enterococci gain resistance either by the overproduction of a constitutive low-affinity PBP (11, 12) or by a further reduction of the affinity of that PBP (19, 41). In some resistant enterococcal strains, two low-affinity PBPs may be present at the same time (27, 38). Resistance among staphylococci occurs by acquisition of a single PBP which has a low affinity for all the usual β-lactam antibiotics (5, 16) and has probably originated from another species within the genus Staphylococcus (39). The low-affinity PBPs confer penicillin resistance because they are able to perform the functions needed for wall peptidoglycan synthesis under conditions in which all the other PBPs are inactivated (5, 11, 16, 38).
Enterococcus hirae ATCC 9790 is moderately resistant to benzylpenicillin and produces a chromosome-encoded low-affinity PBP, PBP5 (9, 11). Overproduction of PBP5 in the highly penicillin-resistant laboratory mutant E. hirae R40 has been related to the inactivation of a negative regulatory gene psr that is located immediately upstream from pbp5 and that encodes a 33-kDa protein (20, 22).
E. hirae S185, a clinical isolate from pig intestine, produces two low-affinity PBPs, PBP5 and PBP3r (27, 28). Chemical mutagenesis of E. hirae S185 has led to the isolation of a penicillin hypersusceptible mutant, E. hirae SS22, which still produces very low levels of PBP5 but which has lost the capacity to produce PBP3r (28). Conversely, six successive passages of E. hirae S185 in broth containing benzylpenicillin (32 μg/ml) has led to the isolation of a penicillin-resistant mutant, E. hirae S185R, which selectively overproduces PBP3r (28). PBP5 and PBP3r of E. hirae S185 and S185R are structurally related to each other (78.5% amino acid sequence identity) and to the low-affinity PBP, PBP2′, of methicillin-resistant Staphylococcus aureus (33% amino acid sequence identity). In addition, PBP3r is almost identical (99.8% amino acid sequence identity) to PBP5fm found in Enterococcus faecium strains (41).
Comparison of the E. hirae wild-type strains and mutants with respect to their susceptibilities to benzylpenicillin and erythromycin suggested that pbp3r, but not pbp5, might be linked to an erythromycin resistance determinant, erm (28). The aim of the study described in this paper is to show that E. hirae S185R possesses a plasmid-borne pbp3r gene linked to two resistance determinants, those for resistance to erythromycin and streptomycin, respectively.
(The work described in this paper is part of a dissertation presented by D.R. in partial fulfilment of a Ph.D. at the University of Liège.)
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and enzymes.
E. hirae S185, SS22, and S185R were grown as described previously (27, 28). Strain S185 was shown to belong to the E. hirae species by using the API 20 Strep and API Staph test kits (BioMérieux, Marcy l’Etoile, France) and by analyzing the PBP patterns in comparison with those of the type strains of different species. In all cases, strain S185 behaved exactly like the E. hirae type strain, ATCC 9790. By a similar approach, strains D63 and D63R were shown to belong to the species E. faecium.
The plasmids and E. coli hosts used in this study are listed in Table 1. Three different plasmids (pDML501, pDML508, and pDML510) which each contained an insert from E. hirae S185R were analyzed in the course of this study. pDML501 and pDML508 had an NcoI fragment, and pDML510 had an EcoRI fragment (28). The 4.5-kb inserts of pDML501 and pDML510 and the 14-kb insert of pDML508, obtained by restriction digestion and agarose gel electrophoresis, each hybridized with oligonucleotide O1 (see below) derived from pbp3r. In contrast, the 14-kb insert of pDML508, but not the 4.5-kb inserts of pDML501 and pDML510, hybridized with oligonucleotide O4 (erm) (see below), showing that pbp3r and erm are somehow linked to each other in the larger 14-kb DNA fragment.
TABLE 1.
Plasmids and E. coli strains used in this study
| Plasmid or strain | Genotype | Reference or source |
|---|---|---|
| Plasmids | ||
| pDML501 | pBR325 with a 4.5-kb NcoI insert from E. hirae S185R Tcr Apr Cmrpbp3r | 28 |
| pDML508 | pBR325 with a 14-kb NcoI insert from E. hirae S185R Tcr Apr Cmrerm pbp3r | 28 |
| pDML510 | pBR322 with a 4.5-kb EcoRI insert from E. hirae S185R Tcr Aprpbp3r | 28 |
| pDML540 | pBR322 with a 7-kb EcoRI insert from E. hirae R40 Tcr Aprpbp5 | 9 |
| pUCBM20 | Apr | Boehringer (Mannheim, Germany) |
| pGEM3-Zf(+) | Apr | Promega (Leiden, The Netherlands) |
| E. coli strains | ||
| JM109 | endA1 recA1 gyrA96 thi hsdR17 (rK−, mK+) relA1 supE44 λ− Δ(lac-proAB) (F′ traD36 proAB lacIqZΔM15) | Promega (Leiden, The Netherlands) |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF) U169 dcoR recA1 endA1 phoA hsdR17 (rK−, mK+) supE44 λ− thi-1 gyrA96 relA1 | Life Technologies (Gibco BRL, Merelbeke, Belgium) |
| RR1 | Δ(gpt-protA) 62 leuB6 thi-1 lacY1 hsdSB20 rpsL20 (Strr) ara-14 galK12 xyl-5, mtl-1 supE44 mcrBB | United States Biochemicals (Cleveland, Ohio) |
The restriction endonucleases (Boehringer, Mannheim, Germany) and the AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, Conn.) were used as recommended by the manufacturers.
DNA preparations.
Plasmids of Escherichia coli were prepared as described previously (28) or by using the Nucleobond kit (Macherey and Nagel, Düren, Germany). Plasmids of E. hirae S185, SS22, and S185R each were prepared by using the method of Anderson and McKay (1), which was modified as follows. Cells were grown in 100 ml of brain heart medium for 4 to 5 h (optical density at 550 nm = 2), harvested by centrifugation, and resuspended in 5 ml of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA–25% (wt/vol) sucrose–7 mg of lysozyme per ml–1 μg of mutanolysin per ml, and the suspension was incubated at 37°C for 15 min. The lysate was centrifuged for 10 min at 9,000 × g, and the pellet was resuspended in 1.5 ml of Tris-EDTA buffer. The suspension was supplemented with 1 ml of a 5% (wt/vol) sodium dodecyl sulfate (SDS) solution in Tris-EDTA buffer. After gentle mixing, the solution was incubated for 20 min at 37°C. The alkaline denaturation was performed as described by Currier and Nester (8), followed by phenol-chloroform extractions and ethanol precipitation.
DNA fragments were eluted from the agarose gels by using Sephaglas BandPrep kits (Pharmacia Biotech, Brussels, Belgium) according to the manufacturer’s recommendations.
Nucleotide sequencing.
Nucleotide sequencing was carried out as described by Sanger et al. (33) with M13 universal and reverse oligonucleotides or oligonucleotides complementary to inserts as primers. Denaturation of double-stranded DNA was performed as described by Zhang et al. (40). Sequencing reactions were carried out by using the Sequenase kit (United States Biochemicals, Cleveland, Ohio), the T7 sequencing kit (Pharmacia Biotech, Brussels, Belgium) with [35S]dATP labeling, or the Autoread sequencing kit (Pharmacia Biotech) with fluorescent primers or by the incorporation of fluorescent dUTP. For the sequencing reactions with the Autoread kit, electrophoresis was performed with an ALFexpress DNA sequencer (Pharmacia Biotech) (2).
Oligonucleotides and hybridization.
The oligonucleotides were synthesized by Eurogentec (Seraing, Belgium). Their positions are indicated in Fig. 1A and B. Oligonucleotides GCAGGAATGGCATCGAAAAAGGCAG (O1) and GCGGAAATCAAAGAAAAACAGG (O2) started 193 and 1,861 bp from the ATG of pbp3r, respectively. Oligonucleotide CCGCTAGGTTCTGTTGCAAAGTT (O3), designated an IS1216 repeat, started 80 bp upstream from the ATG of the transposase-encoding gene, tnp. The boxed sequence followed by a T is the sequence of the 18-bp IS1216 repeats. Oligonucleotides GGGCATTTAACGACGAAACTGGCTA (O4) and ACCTCTGTTTGTTAGGGAATTGAAA (O5) started 120 and 283 bp from the ATG of the ermAM gene of Enterococcus faecalis pAMβ1, respectively. Oligonucleotide O5 was based on the sequence of the complementary strand.
FIG. 1.
Restriction maps of the 4.5-kb insert of pDML501 (A) and the 14-kb NcoI insert of pDML508 (B). The restriction and probe hybridization sites shown are only those necessary to understand the products generated by restriction and PCR experiments. The brackets above the maps represent the PCR products. The restriction sites are indicated by the following letters: A, AccI; C, ClaI; H, HindIII; and S, ScaI. Broken arrows identify the DNA regions whose sequences are known.
The oligonucleotide probes used for hybridization were labeled at the 3′-OH end with digoxigenin-ddUTP by using the terminal transferase from Boehringer. The hybridizations were performed under stringent conditions at temperatures 4°C below the melting temperature of the probe. Nylon filters were washed twice for 5 min each time at 64°C in 2× SSC (0.3 M NaCl plus 0.03 M sodium citrate) containing 0.1% (wt/vol) SDS and twice for 5 min at 50°C in 0.1× SSC containing 0.1% (wt/vol) SDS. Hybridizations were detected by chemiluminescence with Lumigen-PPD provided by Boehringer following the instructions of the manufacturer.
Slot blot hybridization.
Samples (62.5, 125, 250, and 500 ng) of DNA preparations (denatured at 95°C for 10 min) were deposited on a nylon membrane in a Bio-Dot SF blotting microfiltration unit (Bio-Rad, Richmond, Calif.). The DNA samples were bound to the membrane by exposure to UV light (312 nm) for 3 min. Hybridization with oligonucleotide O1 (pbp3r) was performed as described above. Quantification of the hybridization bands was done by two-dimensional densitometry of pictures taken with a video camera and processed with CAM software (Cybertech-Dalton, Berlin, Germany).
Homology searches.
Searches of the protein and DNA sequence databases were performed as described by Pearson and Lipman (26) by using the FASTA and TFASTA software packages. Alignments of nucleotide and amino acid sequences were made with BESTFIT according to the algorithm of Smith and Waterman (36) (GCG package).
Nucleotide sequence accession number.
The EMBL accession number for the sequence of the 4.5-kb NcoI insert of pDML501 is X69092, that for the sequence of IS1216 inserted in pDML501 is X81654, and that for the sequence of ermAMEH, the ermAM found in E. hirae S185, is X81655.
RESULTS
The pbp3r-containing 4.5-kb DNA insert of pDML501 possesses a psr3r gene and an insertion module IS1216.
It was previously shown that pbp3r is located in the middle of the 4.5-kb NcoI insert of pDML501 (28). Sequencing of the complete insert confirmed the restriction map shown in Fig. 1A. It also led to the identification, 417 bp upstream from pbp3r, of a 517-bp open reading frame (ORF) that was identical to the 597-bp psr gene of E. faecium D63R (psrfm) except that it lacked the first 80 bp (41). In all likelihood, the missing 80 bp at the 5′ end of psr3r of the 4.5-kb NcoI fragment of pDML501 was lost by restriction during cloning since a complete 597-bp gene, called psr3r, was subsequently identified at one end of a 4.5-kb pbp3r-containing EcoRI fragment cloned in pDML510 from a genomic library of E. hirae S185R (28). The sequence of psr3r was identical to that of psrfm. In both cases, the ATG codons each occurred 10 bp downstream from the first nucleotide of the EcoRI site and 80 bp upstream from the first nucleotide of the NcoI site (41).
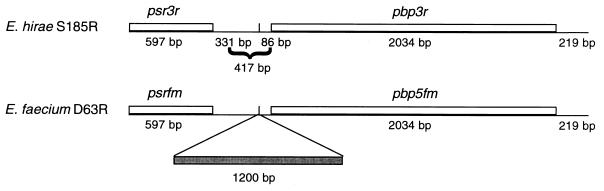
The identity also extended over the 417-bp intergenic region between psr3r and pbp3r and the 219-bp segment downstream from the pbp genes (Fig. 2). The intervening 417-bp sequence found in the NcoI insert of pDML501 consisted of two uneven parts. The sequence of a 331-bp segment was identical to that of the 331-bp segment identified immediately downstream from psrfm. The sequence of the remaining 86-bp segment was identical to that of an 86-bp segment immediately upstream from the pbp5fm. Hence, with the exception of a 1.2-kb fragment of unknown origin inserted between these two sequences in E. faecium D63R (8a, 41), the sequences of the E. hirae S185R and the E. faecium D63R DNA fragments were identical over a length of 3,267 bp (Fig. 2).
FIG. 2.
Comparison of the pbp3r and pbp5fm loci in E. hirae S185R and E. faecium D63R, respectively. The scheme highlights the genes and DNA regions that are identical in both strains. The 1.2-kb insert of E. faecium D63R is hatched.
An insertion module designated IS1216 was also identified in the 4.5-kb NcoI insert of pDML501. IS1216 started 637 bp downstream from the 3′ end of pbp3r and shared 65% nucleotide sequence identities with the methicillin-resistant strain S. aureus IS257 sequence (also called IS431mec [4]) and 75 to 76% sequence identities with the Lactococcus lactis IS946 (31) and ISS1 sequences (29), and its transposase-encoding gene tnp had the same orientation as pbp3r.
Finally, one may mention that the EcoRI site at one end of the EcoRI insert of pDML510 was only 82 bp away from the inverted repeat (IR) sequence of one IS1216 element. This latter element was partially sequenced. A 411-bp sequence was found to be identical to the sequence occurring immediately upstream from the EcoRI site at the 3′ end of the 4.5-kb NcoI insert in pDML501 (as depicted in Fig. 1A). Hence, in all likelihood, the NcoI and EcoRI inserts of pDML501 and pDML510, respectively, were identical.
The 14-kb DNA insert of pDML508 has a pbp3r gene adjacent to a transposon-like structure that carries erythromycin and streptomycin resistance markers.
The 14-kb insert of pDML508, the restriction map of which is also shown in Fig. 1B, comprises three regions, as described below.
(i) The 4.5-kb terminal portion of the 6.5-kb NcoI-EcoRI fragment at the 5′ end of the insert (as defined in Fig. 1B) had the same restriction map as the 4.5-kb NcoI insert of pDML501 except that one ClaI site was not detected by restriction analysis. Upon PCR amplification with oligonucleotides O2 (pbp3r) and O3 (IS1216 repeats), both the NcoI-EcoRI fragment of pDML508 and the NcoI insert of pDML501 generated several 0.8-kb IS1216 copies (because O3 annealed on both inverted repeats) and one 1.6-kb DNA fragment that encompasses the 637 bp of intervening pbp3r-IS1216 sequence. The restriction fragments of the PCR products of IS1216 and the 1.6-kb fragment were those expected from the restriction map (Fig. 1B). As a consequence, the 4.5-kb terminal portion of the 5′ region of the pDML508 14-kb insert was a duplicate of the 4.5-kb NcoI fragment of pDML501 except that it lacked one ClaI site and the EcoRI and NcoI sites which, in the 4.5-kb insert, were immediately downstream from IS1216. However (as also shown in Fig. 1B), the 14-kb insert of pDML508 possessed an EcoRI site 6.5 kb downstream from the 5′ end. Partial sequencing of that 6.5-kb fragment showed that the NcoI-EcoRI fragments of the pDML508 and pDML501 inserts had identical 809-bp IS1216 elements and psr3r sequences (300 bp) but different 3′ end sequences.
(ii) The 2.6-kb EcoRI-HindIII central fragment of the 14-kb insert of pDML508 possessed the unique ClaI, SphI, PvuII, and AccI sites. It did not hybridize with any of the pbp3r, IS1216, and erm probes.
(iii) The 4.6-kb HindIII-NcoI fragment of the 3′ region of the 14-kb insert of pDML508, shown in Fig. 1B (cloned in pUCBM20; plasmid pDML528), did not hybridize with oligonucleotide O1 (pbp3r) but it hybridized with oligonucleotides O3 (IS1216 repeats) and O4 (erm). The HindIII-NcoI fragment was restricted with ScaI and with ScaI-PvuII, respectively, and the products were subcloned into pUCBM20 or pGEM3-Zf(+). Double-stranded DNA sequencing of the subclones led to the identification of the insertion module IS1216R, the erythromycin resistance determinant, erm (consisting of the methylase-encoding gene and the flanking ORFs, ORFs 1 and 3), and the streptomycin resistance determinant, aad. These ORFs each had the same orientation opposite that of tnp in IS1216L of the 6.5-kb NcoI-EcoRI fragment.
IS1216R was identical to IS1216 (of the pDML501 insert) and to IS1216L (of the NcoI-EcoRI fragment of the pDML508 insert). The result of the analysis of the 80-bp flanking sequences was that the regions upstream from both IS1216 and IS1216L (oriented as shown in Fig. 1) were identical. In contrast, the downstream sequences diverged immediately after the right IR sequence, with IS1216R being inserted in a totally different DNA sequence (as shown in Fig. 3). More importantly, short direct repeats (3 to 14 bp) were not identified in the sequences on both sides of IS1216, IS1216L, and IS1216R.
FIG. 3.
Comparison of the 30-bp sequences identified immediately upstream and downstream from each of the IS1216 elements.
The entire sequence of the erm determinant was almost identical (98.5% identities) to those of the ermAM determinants of pAMβ1 (21), pAM77 (18), and Tn917 (35). Accordingly, it was designated ermAMEH because it was found in E. hirae S185. The ATG codon of ermAMEH ORF1 and that of the methylase-encoding gene started 73 and 276 bp downstream from the left inverted repeat of IS1216R, respectively. The 73-bp sequence was identical to those of pAM77 and Tn917. It contained a −10 motif that might be involved in the formation of a hybrid promoter with a putative −35 hexamer found in the left IR of IS1216R (13). Note that the sequences of the ermAM in Tn917 and pAM77 diverged from that of ermAMEH 2 and 71 bp downstream from the stop codon of ORF3, respectively.
The streptomycin resistance gene started 169 bp downstream from the ermAMEH ORF3 stop codon. Its established 864-bp sequence had 89.6% identity with the 906-nucleotide ant(6)Ia (or aadE) of E. faecalis that encodes the 302-amino-acid residue adenylyl-transferase (25). It was called aadEEH.
The sequence of the 8.8-kb structure comprising ermAMEH, aadEEH, IS1216L and IS1216R described above resembled those of composite transposons (13). It was designated Tn5466.
Amplification of the sequence pbp3r-IS1216 of E. hirae S185R.
As described below, restriction digestions of a plasmid-enriched preparation of E. hirae S185R led to the identification of two EcoRI fragments of 4.5 and 6.6 kb, respectively, and two NcoI fragments of 4.5 and 14 kb, respectively. The four fragments each hybridized with oligonucleotide O1 (pbp3r), indicating that E. hirae S185R contained at least two copies of pbp3r. However, the intensities of the 4.5-kb bands were higher than those of the 6.6- and 14-kb bands, respectively, suggesting that the 4.5-kb fragments occurred at a higher copy number (data not shown).
At variance with the plasmid-enriched preparations of E. hirae S185R, only a 6.6-kb EcoRI fragment and a 14-kb NcoI fragment were identified by hybridization in restricted total DNA and plasmid-enriched preparations of E. hirae S185 (from which mutant S185R was isolated). These two fragments each hybridized with oligonucleotides O1 (pbp3r) and O3 (IS1216 repeats), and the 14-kb NcoI fragment also hybridized with oligonucleotide O4 (erm). In all likelihood, these fragments were similar to those present in the E. hirae S185R digestion products, and they almost completely overlapped (Fig. 1B). Arising from this view, the 4.5-kb EcoRI and NcoI fragments derived from the E. hirae S185R plasmid preparations might be the result of rearrangements at the 5′ end of the 14-kb NcoI fragment. The 20-fold increase in the pbp3r content of resistant strain S185R in comparison with that of the wild-type strain S185 (shown by the slot blot technique) supported this hypothesis. One may note that in these experiments, E. hirae SS22 DNA and pDML501 were used as negative and positive controls, respectively (see below).
The antibiotic resistance determinants are plasmid borne.
E. hirae S185, SS22, and S185R (see the Introduction) were examined with respect to their susceptibilities to antibiotics and their plasmid patterns. In comparison with the wild-type strain S185, which was moderately susceptible to gentamicin, resistant to benzylpenicillin, and highly resistant to erythromycin and streptomycin, strain SS22, which was derived from strain S185 after chemical mutagenesis (28), had greatly increased susceptibilities to benzylpenicillin, erythromycin, and streptomycin (MICs, 0.1, 1, and 50 μg/ml, respectively), while strain S185R was sixfold more resistant to benzylpenicillin (MIC, 100 μg/ml) and had the same high levels of resistance to erythromycin (MIC, >360 μg/ml) and streptomycin (MIC, >2,000 μg/ml). Expression of erythromycin resistance in E. hirae S185 was inducible by a 1-h pretreatment with a low concentration of the antibiotic (0.5 μg/ml). Pretreated and control cells were grown in the presence of a high concentration of erythromycin (500 μg/ml). The antibiotic did not inhibit the pretreated cells, which grew as well as untreated control cells, but it retarded the growth of control cells.
E. hirae S185, SS22, and S185R were analyzed by procedures which allowed small and large plasmids (up to 130 kb) to be prepared. Agarose gel electrophoresis and detection with ethidium bromide (Fig. 4A) showed that (i) each of the preparations obtained from the three strains each contained an 80-kb plasmid; (ii) the preparations obtained from strains S185 and S185R, but not that from strain SS22, contained a 40-kb plasmid; and (iii) the preparations obtained from strain S185R contained additional plasmid bands, the origins of which remain unknown. The smallest plasmid was 14 kb. The bands between 40 and 80 kb were not visible in 0.7 or 0.8% agarose gels, but they were detected in small quantities just below the thick 80-kb band in more concentrated agarose gels (Fig. 4C). A supercoiled DNA ladder (not shown in Fig. 4) and plasmids whose sizes were known (e.g., pIP964) were used to determine the sizes of the different bands. Oligonucleotide O1 (pbp3r) hybridized with the 40-kb plasmid of strains S185 and S185R and with one of the upper bands present in strain S185R. It did not hybridize with the NcoI- or EcoRI-digested total DNA of SS22 (data not shown) or with the 80-kb plasmid-enriched preparation of the same strain (Fig. 4B), showing that pbp3r is exclusively plasmid borne in E. hirae S185 and S185R and is borne mainly on the 40-kb plasmid. One may note that the 164-bp erm PCR product (Fig. 1B) corresponding to the segment of ermAM from nucleotides 120 to 283 (oligonucleotides O4 and O5) also hybridized with the 40-kb plasmids of strains S185 and S185R and the 14-kb plasmid and probably with one of the upper bands of strain S185R. Finally, oligonucleotide O3 (IS1216 repeats) hybridized with the genomic DNA and all the plasmids of the three strains (data not shown).
FIG. 4.
Plasmid patterns. Agarose gel electrophoresis of plasmid preparations of E. hirae S185R, S185, and SS22. (A and C) Detection with ethidium bromide on 0.8 and 1% (wt/vol) agarose gels, respectively. (B) Southern blot of the gel described in panel A with oligonucleotide O1 (pbp3r). Controls were pDML501 with a 4.5-kb NcoI insert containing pbp3r and pDML540 with a 7-kb EcoRI insert containing pbp5. The upper bands observed in the pDML501 preparation are probably another form and dimers of pDML501, because restrictions yielded only the 4.5-kb insert and the pBR325 vector. Standards were linear bacteriophage λ HindIII fragments and supercoiled pIP964 (a generous gift from T. Horaud, Pasteur Institute, Paris, France). Ch, linearized chromosomal DNA contaminant.
DISCUSSION
E. hirae S185R carries several copies of the insertion module IS1216 and penicillin resistance marker pbp3r, as well as the streptomycin resistance marker aadEEH and the erythromycin resistance marker ermAMEH. These elements are plasmid borne, although some IS1216 modules also seem to be present on the chromosome. Most likely, aadEEH, ermAMEH, and the two IS1216 modules form a transposon-like structure: IS1216L-aadEEH-ermAMEH-IS1216R. Since the unknown portion of the plasmid replicon may contain additional insertion sequence modules and markers, this putative transposon only has been numbered Tn5466.
The enterococcal IS1216 and the S. aureus IS257 are highly similar. They belong to the ISS1 family (32). Members of this family are known to be involved in cointegration and recombination processes in L. lactis (29).
Recently, IS1216-like modules were found to be associated with antibiotic resistance determinants in other enterococcal strains. IS1216V was proposed to mediate the horizontal spread of the vancomycin resistance transposon Tn5506 in E. faecium (17). IS1216V modules were also described in another vancomycin resistance transposon, Tn5482 (15). A similar module was found on the chromosome of E. faecalis CX19 downstream from the β-lactamase gene (30).
IS257 is probably involved in the integration of the low-affinity PBP2′-encoding mecA in the chromosomes of methicillin-resistant S. aureus strains, in the amplification phenomena observed in highly resistant mutants, and in the evolution of the mec locus (23). In the multidrug-resistant methicillin-resistant S. aureus strains, mecA is surrounded by Tn554 or ψTn544, by the heavy metal resistance determinant mer, and/or by an integrated plasmid, pUB110 or pT181. All of these elements except the transposons are flanked by IS257 copies, and the transposons and plasmids each carry additional antibiotic resistance genes (3, 37), suggesting that IS257 is involved in the clustering of the resistance genes. IS257 also facilitates the cointegration of plasmids in staphylococci (24).
The IS1216 modules of E. hirae lack adjacent direct target sequences, a situation which is reminiscent of IS257 in the mec region of the chromosome of S. aureus (37). Yet, by analogy with IS257 and ISS1, the E. hirae IS1216 modules might be responsible for the transposition of pbp3r in E. hirae S185R and for the emergence of other plasmids. Further studies are needed to understand how IS1216 elements mediate DNA rearrangements and exchanges leading to an increase in the pbp3r copy number. However, because of the almost complete overlap of the 6.6-kb EcoRI fragment and the 14-kb NcoI fragment in strain S185 and probably in strain S185R, it is highly probable that both the EcoRI and the NcoI sites at the 5′ ends of these fragments are separated only by a 90-bp sequence identical to that found in pDML510. Assuming that transposition occurred during selection and led to the formation of a repetitive DNA structure on one plasmid, restriction with NcoI or EcoRI could produce only the 4.5-kb fragments, in addition to either the 14-kb fragment or the 6.6-kb fragment. Thus, one of the plasmids, at least in strain S185R, might have a repetitive DNA organization.
In addition to DNA rearrangements, an increase in the copy number of the pbp3r bearing plasmids might also have occurred due to a mutation that modified the regulatory mechanism of the copy number. Copy numbers were not determined during this work mainly because the yields of the plasmid preparations of E. hirae S185 and S185R were not highly reproducible.
By analogy with mecA in methicillin-resistant S. aureus strains, pbp3r in E. hirae S185R (and probably S185) occurs next to Tn5466. An answer to the question of whether pbp3r could be transferred with the help of Tn5466 or a larger Tn5466-containing composite transposon requires further investigation. At variance with the location of integration of mecA in the naturally occurring MRSA strains, pbp3r is integrated into a large plasmid, not in the chromosome. E. hirae is the first known bacterial species in which a low-affinity PBP-encoding gene that confers penicillin resistance is plasmid borne.
The origins of pbp3r and psr3r are still unknown. Because both genes are inserted in a 3.2-kb DNA portion whose sequence is 99% homologous to the sequence of a fragment cloned from the E. faecium D63R chromosome (41), one may hypothesize that a DNA fragment of at least 3.2 kb was derived from an E. faecium strain and inserted directly in a plasmid of E. hirae or in a plasmid of E. faecium which was then transferred to E. hirae.
The elements so far identified highlight the strong potential for the spread of resistance to penicillin (mediated by PBP3r), erythromycin, and streptomycin among enterococci and other bacteria by transposition and/or cointegration.
Finally, one may note that as observed in most of the enterococcal species except E. faecalis, E. hirae has an l-Lys-d-Asp type of peptidoglycan (6, 7, 10, 34). Given that PBP3r must function as a transpeptidase, it is highly probable that the spread of PBP3r-mediated penicillin resistance remains limited to bacterial species whose peptidoglycan is structurally related to that of E. hirae.
ACKNOWLEDGMENTS
This work was supported in part by the Belgian Programme on Interuniversity Poles of Attraction initiated by the Belgian Prime Minister’s Office, Services Fédéraux des Affaires Scientifiques, Techniques et Culturelles (PAI no. 19 and P4/03).
REFERENCES
- 1.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansorge W, Sproat B, Stegemann J, Schwager C, Zenke M. Automated DNA sequencing: ultrasensitive detection of fluorescent bands during electrophoresis. Nucleic Acids Res. 1987;15:4593–4602. doi: 10.1093/nar/15.11.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer G L, Niemeyer D M. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 1994;2:343–347. doi: 10.1016/0966-842x(94)90608-4. [DOI] [PubMed] [Google Scholar]
- 4.Barberis-Maino L, Berger-Bächi B, Weber H, Beck W D, Kayser F H. IS431, a staphylococcal insertion sequence-like element related to IS26 from Proteus vulgaris. Gene. 1987;59:107–113. doi: 10.1016/0378-1119(87)90271-x. [DOI] [PubMed] [Google Scholar]
- 5.Brown F J, Reynolds P E. Intrinsic resistance to β-lactam antibiotics in Staphylococcus aureus. FEBS Lett. 1980;122:275–278. doi: 10.1016/0014-5793(80)80455-8. [DOI] [PubMed] [Google Scholar]
- 6.Collins M D, Farrow J A E, Jones D. Enterococcus mundtii sp. nov. Int J Syst Bacteriol. 1986;36:8–12. [Google Scholar]
- 7.Collins M D, Jones D, Farrow J A E, Kilpper-Bälz R, Schleifer K H. Enterococcus avium nom. rev., comb. nov.; E. casseliflavus nom. rev., comb. nov.; E. durans nom. rev., comb. nov.; E. gallinarum comb. nov.; and E. malodoratus sp. nov. Int J Syst Bacteriol. 1984;34:220–223. [Google Scholar]
- 8.Currier T C, Nester E W. Isolation of covalently closed circular DNA of high molecular weight from bacteria. Anal Biochem. 1976;76:431–441. doi: 10.1016/0003-2697(76)90338-9. [DOI] [PubMed] [Google Scholar]
- 8a.Dardenne, O. Unpublished results.
- 9.El Kharroubi A, Jacques P, Piras G, Van Beeumen J, Coyette J, Ghuysen J M. The Enterococcus hirae R40 penicillin-binding protein 5 and the methicillin-resistant Staphylococcus aureus penicillin-binding protein 2′ are similar. Biochem J. 1991;280:463–469. [PMC free article] [PubMed] [Google Scholar]
- 10.Farrow J A E, Collins M D. Enterococcus hirae, a new species that includes amino acid assay strains NCDO 1258 and strains causing growth depression in young chickens. Int J Syst Bacteriol. 1985;35:73–75. [Google Scholar]
- 11.Fontana R, Cerini R, Longoni P, Grossato A, Canepari P. Identification of a streptococcal penicillin-binding protein that reacts very slowly with penicillin. J Bacteriol. 1983;155:1343–1350. doi: 10.1128/jb.155.3.1343-1350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontana R, Grossato A, Rossi L, Cheng Y R, Satta G. Transition from resistance to hypersusceptibility to β-lactam antibiotics associated with loss of low affinity PBP in a Streptococcus faecium mutant highly resistant to penicillin. Antimicrob Agents Chemother. 1985;26:678–683. doi: 10.1128/aac.28.5.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 14.Ghuysen J M. Molecular structures of penicillin-binding proteins and β-lactamases. Trends Microbiol. 1994;2:372–380. doi: 10.1016/0966-842x(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 15.Handwerger S, Skoble J. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1995;37:2446–2453. doi: 10.1128/aac.39.11.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartman B J, Tomasz A. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heaton M P, Discotto L F, Pucci M J, Handwerger S. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faecium sex pheromone-response plasmid. Gene. 1996;171:9–17. doi: 10.1016/0378-1119(96)00022-4. [DOI] [PubMed] [Google Scholar]
- 18.Horinouchi S, Byeon W H, Weisblum B. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J Bacteriol. 1983;154:1252–1262. doi: 10.1128/jb.154.3.1252-1262.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klare I, Rodloff A C, Wagner J, Witte W, Hakenbeck R. Overproduction of a penicillin-binding protein is not the only mechanism of penicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1992;36:783–787. doi: 10.1128/aac.36.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ligozzi M, Pittaluga F, Fontana R. Identification of a genetic element (psr) which negatively controls expression of Enterococcus hirae penicillin-binding protein 5. J Bacteriol. 1993;175:2046–2051. doi: 10.1128/jb.175.7.2046-2051.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin B, Alloing G, Mejean V, Claverys J P. Constitutive expression of erythromycin resistance mediated by the ermAM determinant of plasmid pAMβ1 results from deletion of 5′ leader peptide sequences. Plasmid. 1987;18:250–253. doi: 10.1016/0147-619x(87)90068-0. [DOI] [PubMed] [Google Scholar]
- 22.Massidda, O., O. Dardenne, M. B. Whalen, W. Zorzi, J. Coyette, G. D. Shockman, and L. Daneo-Moore. The psr gene of Enterococcus hirae ATCC9790 is longer than previously reported. Submitted for publication. [DOI] [PubMed]
- 23.Matthews P R, Stewart P R. Amplification of a section of chromosomal DNA in methicillin-resistant Staphylococcus aureus following growth in high concentration of methicillin. J Gen Microbiol. 1988;134:1455–1464. doi: 10.1099/00221287-134-6-1455. [DOI] [PubMed] [Google Scholar]
- 24.Needham C, Noble W C, Dyke K G. The staphylococcal insertion sequence IS257 is active. Plasmid. 1995;34:198–205. doi: 10.1006/plas.1995.0005. [DOI] [PubMed] [Google Scholar]
- 25.Ounissi H, Courvalin P. Nucleotide sequences of streptococcal genes. In: Ferretti J J, Curtiss III R, editors. Streptococcal genetics. Washington, D.C: American Society for Microbiology; 1987. p. 275. [Google Scholar]
- 26.Pearson W R, Lipman D J. Improved tools for biological sequences comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piras G, El Kharroubi A, Van Beeumen J, Coeme E, Coyette J, Ghuysen J M. Characterization of an Enterococcus hirae penicillin-binding protein 3 with low penicillin affinity. J Bacteriol. 1990;172:6856–6862. doi: 10.1128/jb.172.12.6856-6862.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piras G, Raze D, El Kharroubi A, Hastir D, Englebert S, Coyette J, Ghuysen J M. Cloning and sequencing of the low-affinity penicillin-binding protein 3r-encoding gene of Enterococcus hirae S185. Modular design and structural organization of the protein. J Bacteriol. 1993;175:2844–2852. doi: 10.1128/jb.175.10.2844-2852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polzin K M, Shimizu-Kadota M. Identification of a new insertion element, similar to gram-negative IS26, on the lactose plasmid of Streptococcus lactis ML3. J Bacteriol. 1987;169:5481–5488. doi: 10.1128/jb.169.12.5481-5488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice L B, Marshall S H. Insertions of IS256-like element flanking the chromosomal β-lactamase gene of Enterococcus faecalis CX19. Antimicrob Agents Chemother. 1994;38:693–701. doi: 10.1128/aac.38.4.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero D A, Klaenhammer T D. Characterization of insertion sequence IS946, an iso-ISS1 element isolated from the conjugative lactococcal plasmid pTR2030. J Bacteriol. 1990;172:4151–4160. doi: 10.1128/jb.172.8.4151-4160.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouch D A, Skurray R A. IS257 from Staphylococcus aureus: member of an insertion sequence superfamily prevalent among gram-positive and gram-negative bacteria. Gene. 1989;76:195–205. doi: 10.1016/0378-1119(89)90160-1. [DOI] [PubMed] [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schleifer K H, Klipper-Bälz R. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int J Syst Bacteriol. 1984;34:31–34. [Google Scholar]
- 35.Shaw J H, Clewell D B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985;164:782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith J F, Waterman M S. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 37.Stewart P R, Dubin D T, Chikramane S G, Inglis B, Matthews P R, Poston S M. IS257 and small plasmid insertions in the mec region of the chromosome of Staphylococcus aureus. Plasmid. 1994;31:12–20. doi: 10.1006/plas.1994.1002. [DOI] [PubMed] [Google Scholar]
- 38.Williamson R, Le Bouguénec C, Gutmann L, Horaud T. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J Gen Microbiol. 1985;131:1933–1940. doi: 10.1099/00221287-131-8-1933. [DOI] [PubMed] [Google Scholar]
- 39.Wu S, Piscitelli C, de Lencastre H, Tomasz A. Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri Microb. Drug Resist. 1996;2:435–441. doi: 10.1089/mdr.1996.2.435. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Scholl R, Browse J, Sommerville C. Double strand DNA sequencing as a choice for DNA sequencing. Nucleic Acids Res. 1988;16:1220. doi: 10.1093/nar/16.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zorzi W, Zhou X Y, Dardenne O, Lamotte J, Raze D, Pierre J, Gutmann L, Coyette J. Structure of the low-affinity penicillin-binding protein 5, PBP5fm, in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J Bacteriol. 1996;178:4948–4957. doi: 10.1128/jb.178.16.4948-4957.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]