Abstract
Objective:
Scalable strategies to reduce the time burden and increase contact tracing efficiency are crucial during early waves and peaks of infectious transmission.
Design:
We enrolled a cohort of SARS-CoV-2-positive seed cases into a peer recruitment study testing social network methodology and a novel electronic platform to increase contact tracing efficiency.
Setting:
Index cases were recruited from an academic medical center and requested to recruit their local social contacts for enrollment and SARS-CoV-2 testing.
Participants:
A total of 509 adult participants enrolled over 19 months (384 seed cases and 125 social peers).
Intervention:
Participants completed a survey and were then eligible to recruit their social contacts with unique “coupons” for enrollment. Peer participants were eligible for SARS-CoV-2 and respiratory pathogen screening.
Main Outcome Measures:
The main outcome measures were the percentage of tests administered through the study that identified new SARS-CoV-2 cases, the feasibility of deploying the platform and the peer recruitment strategy, the perceived acceptability of the platform and the peer recruitment strategy, and the scalability of both during pandemic peaks.
Results:
After development and deployment, few human resources were needed to maintain the platform and enroll participants, regardless of peaks. Platform acceptability was high. Percent positivity tracked with other testing programs in the area.
Conclusions:
An electronic platform may be a suitable tool to augment public health contact tracing activities by allowing participants to select an online platform for contact tracing rather than sitting for an interview.
Keywords: community transmission, contact tracing, COVID-19, pandemic preparedness, snowball sampling
Highly transmissible infections, such as SARS-CoV-2,1 which can be spread via aerosols2 and while infected persons are presymptomatic or asymptomatic,3 frequently require social rather than biological interventions to slow spread.4 Effective social interventions include primary prevention (reducing contacts to prevent onward infections), secondary prevention (early diagnosis leading to isolation, reducing potential for onward infections), and tertiary prevention (isolation after symptoms, or treatment to shorten duration of infectiousness). Although tertiary efforts are generally less beneficial in reducing onward spread, isolation should be emphasized for an infection that can be transmitted through casual contact because incidental contacts are at risk, although only close contacts are generally targeted during contact tracing.
Forecasts of the size and duration of epidemic curves varied significantly early in the SARS-CoV-2 pandemic.5–8 Most, however, indicated that the public health infrastructure and human resources needed to implement testing and tracing protocols for effective social interventions would be strained. Contact tracing resources are not rapidly scalable, compounding challenges when faced with a novel infection with rapid spread.
With the goal of more efficiently identifying and contacting community members at risk of infection, we designed a protocol to identify new cases of SARS-CoV-2 in the community. We used social network analysis to streamline potential contacts of known cases based on descriptions of each case's contacts, community locations visited, and recent activities, with the aim of minimizing efforts needed to identify new cases in the community.
Methods
The Snowball Study opened to enrollment in December 2020. It included 4 elements: (1) a social network component that deployed a link tracing strategy9 to enroll participants, understand mixing patterns in the community, and identify where transmission was occurring; (2) a novel technology component consisting of a secure cloud-based platform to collect and manage study data and track relationships between participants; (3) a sampling component that developed protocols to safely sample potentially infected participants in community settings; and (4) a biometric monitoring component that enrolled a subset of participants into a sister study to use information from wearable devices to identify early signs of infection10,11 (Figure, Supplemental Digital Content 1, available at http://links.lww.com/JPHMP/B192). Using the electronic platform, which enabled self-reporting by cases, we sought to rapidly identify contacts who were recruited to the study by the case using an online enrollment pathway. To increase capacity during future epidemics, the technology developed and methods used have been made publicly available in a tool kit that is generalizable to any infection.12
Link tracing designs
Snowball sampling9,13 is a type of link tracing design in which a seed case is recruited, and that seed case collects their contacts for enrollment, who then collect their contacts for enrollment, and so on. This method not only mirrors how infections are transmitted from person-to-person but also is similar to the process used by public health contact tracing. A separate set of eligibility criteria may be applied to persons recruited into the cohort by another study member (called peer participants), depending on the goal of the cohort, which can facilitate generalizability.9,13
Snowball Platform
To support our study activities, we developed a novel online electronic data capture system, the Snowball Platform,14 which allowed us to identify potential seed cases from among new SARS-CoV-2 diagnoses made at a local academic health system (Duke University Health System [DUHS]), invite potential seed cases to enroll, validate their study eligibility, direct them to provide informed consent and complete the study survey online, and provide “coupons” to distribute to their contacts to join the study and receive SARS-CoV-2 testing. Participants could select an English- or Spanish-language track for all procedures in the platform, which was built to Fast Healthcare Interoperability Resources (FHIR) standards15 and operated in a HIPAA-compliant cloud space.16 The platform was designed so that participants could complete the entire enrollment process, from receiving an invite to recruiting their peers, without needing to interact with any member of our study team.
We invited seed cases daily from among eligible new diagnoses via informational e-mail, which included an enrollment coupon with a unique 4-word token. Invited seed cases could click a link in the coupon or a link to a study informational page and enter the token. Once the token was validated by the Snowball Platform, potential seed cases would be taken to the survey system and presented with electronic informed consent documents. Upon providing informed consent, seed cases were directed to the survey, which included modules for demographics, health history, SARS-CoV-2 and influenza testing history, respiratory symptoms, social network descriptors (cohabitants and other: demographics, relationship type, and frequency of contact), venues and activities, and health-related attitudes and behaviors.
Upon completing the survey, seed cases were given unique electronic coupons to distribute to their social contacts. Study design dictates the number of coupons given to each respondent. Here, most received 5, although up to 10 were provided if the seed cases' social network included a large number of potentially infectious people (ie, recently tested positive or symptomatic), high-risk people (congregate settings), or members of groups who are historically excluded from research (ie, Black/Latinx)17,18 or were underrepresented in the cohort compared with sociodemographic proportions of the target population (Durham County, North Carolina). Contacts who enrolled were deemed “peers” and followed the same process as seed cases; they also received sampling for SARS-CoV-2 (polymerase chain reaction [PCR] and anti-nucleocapsid immunoglobulin G [IgG] [to test for prior infection]) and a full respiratory pathogen panel (PCR) to capture causes for participants with respiratory symptoms but who had a negative SARS-CoV-2 PCR result. Peers were sampled at home during 2020 and 2021 but were given a choice of in-home or in-clinic sampling in 2022. Peers who completed the survey were eligible for their own coupons to distribute to potential next-wave peers.
Coupons provided to seed cases and peers expired if not validated within 4 days of generation; the ability to complete the survey expired 2 days after token validation. Completing the consent documents and survey online took approximately 45 minutes; participants who completed the survey before it expired were compensated. Referring seed cases and peers received additional compensation for any peer who enrolled using their coupon and completed the survey before it expired.
For most of the study, a clinical research coordinator (CRC) attempted to reach all potential seed cases to introduce the study. All participants could contact a CRC at any point during the process for assistance. This study was approved by the DUHS Institutional Review Board (#Pro00105430).
Study cohort
Seed cases were eligible if they provided informed consent, were 18 years or older, were a resident of Durham County, North Carolina, at the time of SARS-CoV-2 diagnosis (December 2020-July 2022), were diagnosed on a PCR test, had checked their test result in their electronic health record (EHR), were not admitted to inpatient care at the time of diagnosis, and had not opted out of research.
Peer participants were eligible if they were referred by another study participant, provided informed consent, were 18 years or older, resided in or sufficiently close to Durham County to be sampled, and did not have a prior SARS-CoV-2 diagnosis during an exclusionary period. This period was more than 2 weeks prior at the start of the study but amended to 2 weeks to 45 days prior between the Delta and Omicron periods, when the scientific community concluded that reinfections were common.
The positive tests performed by DUHS, which we used to identify potential cases, were submitted to the Durham County Health Department in accordance with public health reportable disease mandates. All positive results for tests performed on peers by the Snowball Study were submitted to the Durham County Health Department; testing was conducted under a Clinical Laboratory Improvement Amendments (CLIA) waiver held by our sampling team. All participants were subject to local regulations for contact tracing, isolation, quarantine, and masking, and followed guidelines for usual care.
Optional assessments
Seed cases and peers who enrolled during the Delta and Omicron waves (July 1, 2021, through study end in July 2022) and who received coupons to distribute were eligible for a follow-up survey that asked about immunizations and infections since the initial survey was completed and about perceptions of study participation. Participants who completed the follow-up survey were compensated.
Percent positivity
We used the proportion of tests administered that identified a positive contact as a metric for programmatic success, aiming for a percent positivity that matched or exceeded those of local testing programs. We aggregated all Snowball tests performed by study month and calculated the percent positivity as the number of positive PCR tests over the total number of PCR tests collected during that month (December 2020-July 2022); this proportion has a 95% confidence interval (CI). We compared this proportion to the weekly percent positivity reported by Durham County and DUHS facilities located in Durham. DUHS tests may have been administered for presurgical screening, employee surveillance, student surveillance (during some intervals), and/or testing of symptomatic patients, staff, and students as needed and according to protocols then in place.
Secondary attack rate
We calculated the secondary attack rate within households to assess risk and ability to intervene among close contacts. Using survey data from seed cases with at least one other household member, the secondary attack rate was the number of people who were documented or likely to be positive based on test results and/or the Centers for Disease Control and Prevention's SARS-CoV-2 symptom list,19 over total household members minus the respondent. For this analysis, we only included the first person enrolled by date of enrollment regardless of role (seed cases or peer), when there was a household from which multiple people were enrolled for the study.
For seed cases who believed they were infected at home, we calculated the number of other household members also diagnosed or symptomatic at the time of the seed case's survey completion, over the total number of household members minus the initially infected person, as the likely secondary infections within the household based on the cohabitants who displayed symptoms or were diagnosed after the initial household member brought the infection into the home. For seed cases who believed that they were infected outside of the home, we calculated the proportion of cohabitants who had symptoms or were diagnosed 3 to 12 days20 after the seed case's infection or until time of survey completion (if <12 days after symptom onset) as an indicator of likely onward (secondary) transmission from the seed case.
We calculated the overall household secondary attack rate and used the Kruskal-Wallis tests (at α = .05) to compare rates between the following: households with children versus not, households comprising family members or significant others versus roommates, households affiliated with Duke University or DUHS (faculty, staff, or students) versus not, by seed case's reported household social distancing, by seed case's reported mask-wearing, by seed case's number of daily contacts outside the home being above or at/below this analysis subset's median, and the predominant circulating variant when the seed case enrolled in the study. We calculated 95% CIs for the proportions for the comparisons.
Replicability
Toy data sets and generalized R code are publicly available.12
Results
Cohort
Overall, 6994 people who were eligible for the study as per their EHR received an informational e-mail to participate (Figure, Supplemental Digital Content 2, available at http://links.lww.com/JPHMP/B193). Over 19 months, 384 Durham County residents completed the survey as seed cases and successfully recruited another 125 peer participants who completed the survey (N = 509 total enrolled cohort members with completed surveys) (Table 1). These seed cases and peers described 2199 contacts in their survey (5.7 mean peers per participant; some enrolled in the study as peer participants or were duplicated across surveys). Most cohort members were female (64%); were non-Hispanic (91%) and/or were White or European American (58%) (the majority of participants identified as non-Hispanic White or European American); spoke English at home (89%); and were younger than 40 years (63%). The same was true for contacts described overall, although cohabitants were younger, more likely to be male, and more likely to be Black or African American compared with noncohabitants described (Table 1). Most (80%) cohabitants were family members, whereas 34% of noncohabitating contacts were family members.
TABLE 1. Demographics of Cohort Members and Contactsa.
| Cohort Members With Completed Surveys (N = 509) | Seed Cases (n = 384) | Peer Cases (n = 125) | Total Contacts Described by Seed or Peer Casesb,c (N = 2199) | Cohabitants (n = 842) | Other Noncohabitating Contacts (n = 1357) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | n | % | |
| Gender | ||||||||||||
| Male | 174 | 34 | 119 | 31 | 55 | 44 | 985 | 45 | 433 | 51 | 552 | 41 |
| Female | 326 | 64 | 261 | 68 | 65 | 52 | 1 188 | 54 | 399 | 47 | 789 | 58 |
| Nonbinary/Other | 9 | 2 | 4 | 1 | 5 | 4 | 23 | 1 | 9 | 1 | 14 | 1 |
| Age, y | ||||||||||||
| 0-19 | 10 | 2 | 9 | 2 | 1 | 1 | 351 | 16 | 282 | 34 | 69 | 5 |
| 20-29 | 175 | 34 | 138 | 36 | 37 | 30 | 610 | 28 | 202 | 24 | 408 | 30 |
| 30-39 | 139 | 27 | 112 | 29 | 27 | 22 | 475 | 22 | 143 | 17 | 332 | 25 |
| 40-49 | 74 | 15 | 56 | 15 | 18 | 14 | 251 | 11 | 58 | 7 | 193 | 14 |
| 50-59 | 54 | 11 | 35 | 9 | 19 | 15 | 219 | 10 | 76 | 9 | 143 | 11 |
| 60-69 | 37 | 7 | 25 | 7 | 12 | 10 | 196 | 9 | 58 | 7 | 138 | 10 |
| 70-79 | 19 | 4 | 8 | 2 | 11 | 9 | 79 | 4 | 18 | 2 | 61 | 5 |
| 80+ | 1 | <1 | 1 | <1 | 0 | 0 | 18 | 1 | 5 | 1 | 13 | 1 |
| Ethnicity | ||||||||||||
| Hispanic or Latino | 48 | 9 | 42 | 11 | 6 | 5 | 199 | 9 | 79 | 9 | 120 | 9 |
| Not Hispanic or Latino | 461 | 91 | 342 | 89 | 119 | 95 | 1 959 | 91 | 758 | 91 | 1 201 | 91 |
| Unknownd | 0 | 0 | 0 | 41 | 5 | 36 | ||||||
| Race | ||||||||||||
| American Indian or Alaska Native | 0 | 0 | 0 | 0 | 0 | 0 | 4 | <1 | 3 | <1 | 1 | <1 |
| Asian | 59 | 12 | 46 | 12 | 13 | 10 | 210 | 10 | 90 | 11 | 120 | 9 |
| Black or African American | 105 | 21 | 76 | 20 | 29 | 23 | 408 | 19 | 213 | 25 | 195 | 15 |
| Native Hawaiian or Other Pacific Islander | 0 | 0 | 0 | 0 | 0 | 0 | 8 | <1 | 2 | <1 | 6 | <1 |
| White or European American | 294 | 58 | 220 | 57 | 74 | 59 | 1312 | 61 | 433 | 52 | 879 | 67 |
| Other/2+ races | 14 | 3 | 11 | 3 | 3 | 2 | 57 | 3 | 29 | 3 | 28 | 2 |
| None selectede | 37 | 7 | 31 | 8 | 6 | 5 | 159 | 7 | 67 | 8 | 92 | 7 |
| Unknownd | 0 | 0 | 0 | 41 | 5 | 36 | ||||||
| SARS-CoV-2 statusf | ||||||||||||
| Definitely infected | 418 | 82 | 384 | 100 | 34 | 27 | 659 | 30 | 299 | 36 | 360 | 27 |
| Probably infected | 3 | <1 | 0 | 0 | 3 | 3 | 125 | 6 | 66 | 8 | 59 | 4 |
| Not sure | 7 | 1 | 0 | 0 | 7 | 6 | 454 | 20 | 147 | 17 | 307 | 23 |
| Probably not infected | 9 | 2 | 0 | 0 | 9 | 7 | 572 | 26 | 178 | 21 | 394 | 29 |
| Definitely not infected | 72 | 14 | 0 | 0 | 72 | 58 | 389 | 18 | 152 | 18 | 237 | 18 |
aCohort members include the enrolled seed cases and enrolled peers with completed surveys. These cohort members described their social contacts in their surveys, which are shown by cohabitants versus noncohabitating other contacts.
bAttributes of contacts are as reported by the respondent describing the contact, not reported by the contact or verified.
cSome contacts may have been described as more than one respondent or may have enrolled in the study as a seed case or peer participant.
dEthnicity/race was not reported for some of the contacts described. Unknown responses are not included in the proportion.
eSome enrolled participants (seed cases and peers) or described contacts who were identified as Hispanic ethnicity declined to select a race.
fSeed cases were required to be positive on polymerase chain reaction. SARS-CoV-2 status for peers who were tested (106/125; 85%) was based on laboratory result, and the SARS-CoV-2 status for peers who were not tested (19/125; 15%) was based on that peer's survey response (Question: Do you think that you have COVID-19?). SARS-CoV-2 status for contacts described is based on the report of the seed case or peer who described the contact in the survey.
Of 125 peers with complete surveys, 106 (85%) were sampled; 18 had positive SARS-CoV-2 PCR results (17% positivity) and 20 had positive SARS-CoV-2 antibody results (4 were positive on both). For the respiratory pathogen panel, 2 SARS-CoV-2-negative peers were positive for other respiratory infections (one for influenza A and bocavirus and the other for rhinovirus/enterovirus).
Participant engagement
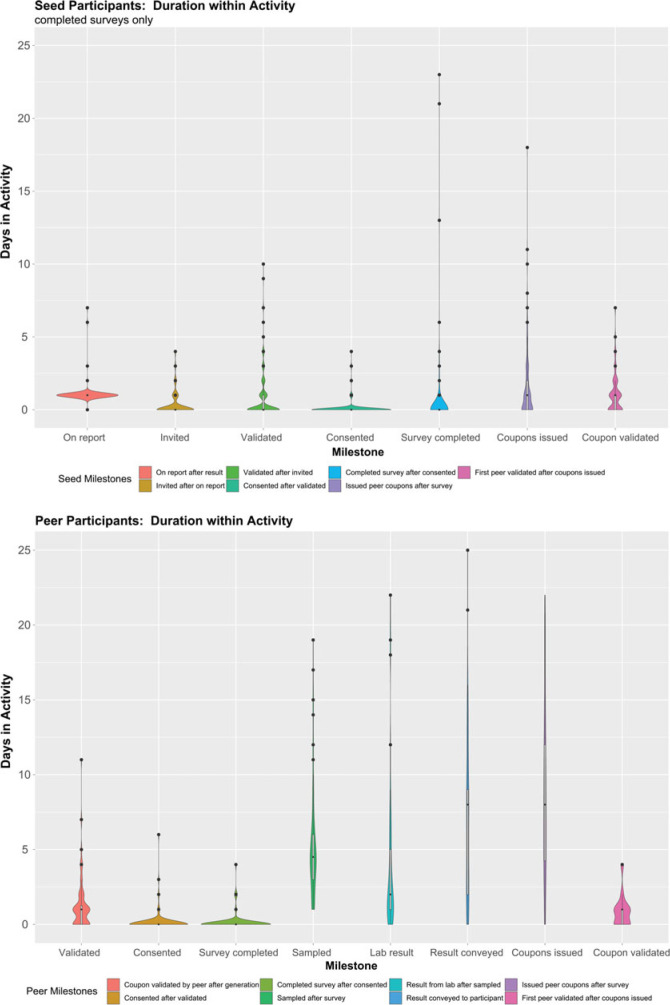
The Snowball Platform moved participants rapidly through the first few study activities, which were entirely contained within the platform (Figure 1). The number of days for each milestone increased substantially for subsequent activities (eg, time required to collect samples, process laboratory test results, and communicate results to participants) that required intervention from a clinical staff member. Among seed cases who completed the survey, two-thirds did so within 3 days of diagnosis. By the fourth day after diagnosis, 11% of seed cases who completed the survey had recruited at least one peer who completed the survey. Overall, 14% (69/509) of participants (59/384 seed cases; 10/125 peers) recruited at least one other peer who completed the survey (range, 1-10; median [IQR] = 1 [1-2] peers who completed the survey).
FIGURE 1.
Study Milestones Are Shown as Categories on the x-Axis, and the y-Axis Is Time in Days From the Prior Milestone to the Milestone Describeda
aIn these violin plots, the width is indicative of the density of the data at each y-axis value (days to complete the milestone) and the dots at higher y-values are outliers based on the interquartile range. For both seed and peer cases, we see that each of the first steps in the enrollment process was typically completed in not more than 1 day. In fact, 67% of seed cases progressed through completing the survey in 3 or fewer days after diagnosis.
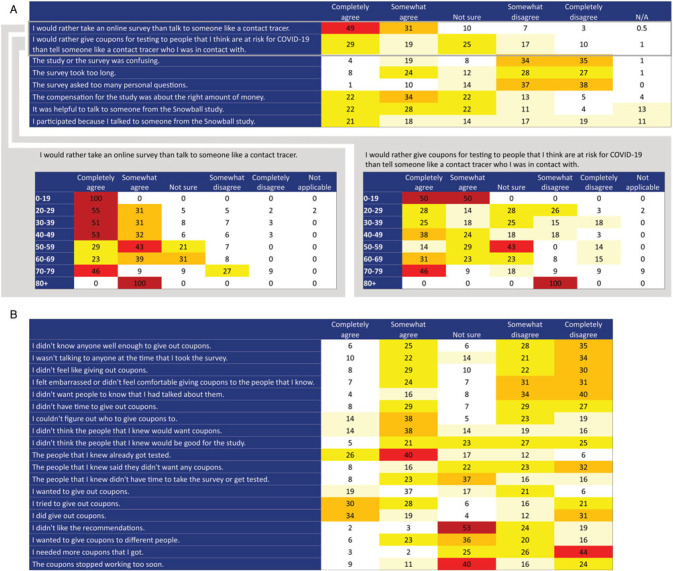
We solicited participants' attitudes toward the platform by age in the follow-up survey (n = 194 responded among 453 invited). The response was positive toward taking an online survey (Figure 2A) and in dispensing coupons (Figure 2B). Age stratification revealed a strong preference for the online survey versus talking to someone such as a contact tracer among younger age-groups (Figure 2A).
FIGURE 2.
Attitudes About Snowball Study Procedures: (A) Data Collection and Contact Elicitation and (B) Recruiting Contacts for Enrollmenta
Abbreviation: N/A, not applicable.
aThe first 2 of the contact elicitation statements (panel A) are stratified by age, showing that contact elicitation via an electronic platform has higher acceptability among younger respondents. Columns show the proportion of respondents (N = 194) indicating agreement with each statement in the follow-up survey.
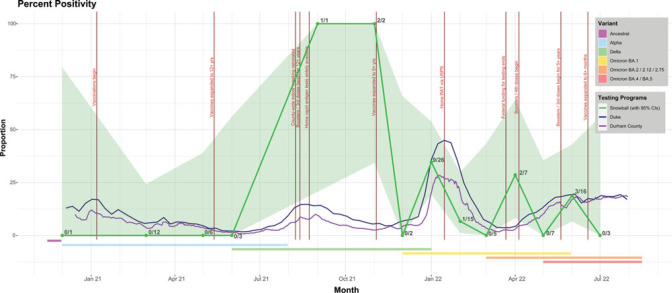
Percent positivity
For most months, there was no significant difference between the Snowball Study's performance and local testing programs (Figure 3). Although September and November 2021 reflect better performance in Snowball, the small number of tests reduces confidence in there being an actual difference.
FIGURE 3.
Percent Positivity Comparing Snowball, Local Public Health, and a Local Academic Medical Centera
Abbreviations: RAT, rapid antigen test; USPS, United States Postal Service.
aThe percent positivity was calculated as the number of positive polymerase chain reaction (PCR) tests over the number of PCR tests collected during that month (December 2020-July 2022). This proportion has a 95% confidence interval. We compared this proportion to the weekly percent positivity reported by Durham County and the Duke University Health System. The academic medical center tests may have been administered for presurgical screening, employee surveillance, possibly student surveillance during some time periods, and testing of symptomatic patients, staff, and students as needed and per the protocols at that time. Each Snowball point is labeled with the number of positive PCR tests over the total number of PCR tests. The graph also shows notable policy changes that could affect prevalence (vertical bars, labeled) and common variants circulating at the time (horizontal bars below the positivity proportions).
Secondary attack rates
Among 384 seed cases with complete surveys, 302 (79%) had at least one cohabitant who was not also a contemporaneous seed case and described the general location where they believed that they were infected. Of these, a quarter (n = 74) believed they caught SARS-CoV-2 within their own home and the other 228 believed they were infected outside the home. The overall secondary attack rate within households in this cohort was 21% (range, 0%-100%; median [IQR] = 0 [0%-33%]) (Table 2). There was no statistically significant difference in the secondary attack rate between households with Duke affiliation versus not; the seed case's number of daily contacts above or below the median; or the predominant SARS-CoV-2 variant at the time of the seed case's infection. However, secondary attack rates were significantly higher among households with children than among households without (23% vs 19%, respectively); households with any family members versus households comprising only roommates/unrelated members (23% vs 12%, respectively); households reporting “always” or “mostly” staying home in the last 2 weeks versus households reporting “sometimes,” “rarely,” or “never” staying home (28% vs 15%, respectively); and households reporting “always” or “most of the time” masking outside of the home or not leaving the home in the last 2 weeks versus households where members “sometimes,” “rarely,” or “never” wore masks outside the home (23% vs 12%, respectively).
TABLE 2. Secondary Attack Rates Within Households by Selected Characteristicsa.
| n | Secondary Attack Rate (%) (95%CI) | |
|---|---|---|
| Overall | 302 | 21 (19-23) |
| Children in householdb | ||
| Yes | 106 | 23 (20-25) |
| No | 196 | 19 (12-19) |
| Cohabitants compositionb | ||
| Some or all familial relations | 250 | 23 (20-24) |
| No familial relations | 52 | 12 (7-19) |
| Duke affiliation | ||
| Yes; affiliated | 154 | 22 (20-26) |
| No; not affiliated | 148 | 21 (17-22) |
| Household social distancing prior 2 wkb | ||
| Always, most of the time | 134 | 28 (25-31) |
| Sometimes, rarely, never | 168 | 15 (11-16) |
| Household mask wearing prior 2 wkb | ||
| Always, most of the time, did not leave house | 250 | 23 (20-24) |
| Sometimes, rarely, never | 52 | 12 (12-21) |
| Seed case's daily contacts | ||
| Above median (>15) | 144 | 20 (17-23) |
| At or below median (0-15) | 158 | 23 (19-25) |
| Predominant circulating variantc | ||
| Delta | 63 | 26 (24-33) |
| Omicron BA.1 | 138 | 22 (16-22) |
| Omicron BA.2/2.12/2.75 | 84 | 19 (12-20) |
| Omicron BA.4/BA.5 | 15 | 12 (6-20) |
Abbreviation: CI, confidence interval.
aThe secondary attack rate within the household is shown overall and by selected characteristics, which are compared using the Kruskal-Wallis test.
bDifference in groups significant at P ≤ .05.
cTwo participants recruited during the Alpha wave were dropped from this analysis due to small cell size.
Discussion
Most Snowball Study participants completed their survey within 3 days of receiving their diagnostic test result, disclosing information similar to what would have been collected during a contact tracing interview, suggesting that such surveys represent a rapid, efficient method to collect contacts during an epidemic. In addition, the scalable platform built for the Snowball Study was capable of managing all aspects of tracing contacts and identifying novel cases. Finally, the Snowball Study's percent positivity rates were similar to those reported by a local academic medical center and public health program.
This Web-based system is rapidly deployable and easily scalable to manage the number of cases requiring contact tracing. This was demonstrated during surges in COVID-19 infection, when rapid increases in cases and enrollment activity did not require additional personnel. This suggests an obvious benefit to public health, given that surges could be met without the need for hiring and training new contact tracers.21–24 In addition, the Snowball Platform enabled the collection of information similar to that gathered by contact tracing, including demographics, social contacts, household composition, workplace information, venues frequented, and health beliefs and behaviors, without time spent by contact tracers to collect the information.
Most infections have longer serial intervals to onward infection. Applying the Snowball Platform14 and social analysis methods to other settings may yield an advantage that could help reduce community transmission rates overall. For example, because persons with sexually transmitted infections tend to be younger or middle-aged, the Snowball Platform might be a suitable tool in this population, given the high acceptability seen in our study among these age-groups. Cases or contacts could be offered the platform as an alternative to traditional contact tracing, with a provisionally scheduled interview that would be canceled if the person completed the survey within a specified time window. Several contact tracing programs developed during the SARS-CoV-2 pandemic reported multiple days or multiple attempts to engage cases and complete contact tracing interviews.25,26 The platform could thus augment public health activities and reduce time and effort needed to elicit risk behaviors and contacts for tracing.
Household transmission was high among the Snowball Study cohort, with 1 in 5 household members infected on average along with the seed case. However, only 11% of seed cases with completed surveys had at least one peer enroll by day 4 postdiagnosis. This represents the outer margin of the intervention window for slowing onward transmission for SARS-CoV-2,27 particularly given that most seed cases were diagnosed a few days into their infection cycles, similar to other programs,28 and that the serial interval time for SARS-CoV-2 for the majority of seed cases (enrolled during Delta and Omicron waves) may have been as short as 3 to 4 days.29 Disrupting transmission in this setting would require a combination of the Snowball Platform to distribute or make available highly accurate rapid home tests with a way to report results, combined with biometric monitoring as a second layer of targeting resources to highly infectious or even preinfectious persons. Providing bar-coded home tests and developing a way to securely upload results might extend the efficiency of the platform further into the disease transmission process.
We expected to see a lower secondary attack rate in households affiliated with Duke, as these households are likely better resourced than other Durham County households and had excellent access to testing, presumably enabling earlier detection, and subsequently less onward transmission. The absence of significant difference may be due to the fact that seed cases in the cohort were drawn from Durham County residents who had access to care (as evidenced by having been tested within this single health system). We did not expect that households reporting higher levels of mitigating social behaviors (masking, social distancing) at the time of household infection would have significantly higher secondary attack rates than households that were less likely to report such behaviors; these results require further investigation. We expected the higher secondary attack rates among households with children and households where family members resided together that we observed. The Delta variant appeared to have a higher secondary attack rate than other variants among this cohort. This differs from estimated effective reproduction numbers for the variants, where Omicron and its sublineages appear more transmissible.30,31 Higher vaccination rates and less severe symptoms may have led to fewer Omicron secondary attacks being detected within households and could indicate that a lower proportion of cases were or are being counted in the communities.
In April 2022, Snowball performed slightly better than local testing programs due to a multiwave peer chain among a group that was not tested regularly and also had several members who tested positive for SARS-CoV-2. The major limitation of the percent positivity analysis is the small sample sizes, which limits interpretation. Some months had large CIs and the collection of more tests would have changed the proportion and interval. Second, we compared a study population comprising contacts identified by an infected person with a hospital-based and a county testing program, which included screening and surveillance testing. However, one of the goals of the study was to test the efficiency of the platform and the methods by investigating whether we could successfully identify positive cases and match percent positivity with less effort and fewer human resources.
Multiple biases affected selection into this cohort, which limits inference. First, we invited seed cases from an academic medical center that does not serve a population that is representative of this geographic area. This is especially problematic, given that this cohort failed to engage representative proportions of groups that have historically been excluded from research. Because most participant recruitment chains terminated at the seed case or first-wave peer, we were not able to reach into the community and mitigate bias from seed case selection.32–34 Second, employees of the health system/university and students are overrepresented in the cohort. Third, invitation to the study required an e-mail address, access to the Internet, and the ability to access one's EHR. The first 3 biases described parallel marginalization of some groups that occurred during the pandemic. Fourth, study activities were only available in English and Spanish. Fifth and finally, cohort members who elected to participate in the study were offered a small monetary incentive and received coupons to distribute for free testing. However, free testing was widely available by the time the study began enrollment and thus this strategy may not have constituted a suitable incentive.35 Selection bias may have been compounded for the follow-up survey, as people who were amenable to online data collection in the first place or who had positive feelings toward the study may have been more likely to agree to participate, which would bias toward more acceptable attitudes toward the platform and study process. However, the concentrated geographic area from which the cohort was drawn lends strength to the analysis.
Employing a link tracing design that permitted study participants to determine and recruit their own contacts at risk yielded similar percent positivity in this setting as more time- and labor-intensive local sampling programs. The electronic platform required resources to develop and deploy but was scalable to the infection and is now publicly available,14 obviating the need for intensive development to address future outbreaks of COVID-19 or other novel or known infectious agents. The technology and methods used here have the potential to increase capacity as they are acceptable, adaptable, and scalable, reducing the need for additional human resources during typically labor-intensive contact tracing steps.
Implications for Policy & Practice
An electronic platform was developed to collect information analogous to what is typically collected during a contact tracing interview and was made freely available.
The platform captures case-contact relationships and permits cases to refer their contacts for testing.
The platform has the ability to expand public health capacity: in the setting described here, the percentage of positive COVID-19 tests mirrored that of local testing programs, but with far fewer human resources needed.
The electronic platform is scalable to any epidemic size, customizable to any infection, and can be used by public health systems across administrative levels.
The platform had high acceptability among respondents who chose to use it.
The platform can be deployed rapidly and customized to a new infection in the event of a future outbreak, epidemic, or pandemic.
The platform can be integrated if there are existing disease surveillance systems by uploading data from the platform through the back end of any existing system.
For standardizing data collection for diseases that are reportable, the public health agencies mandating disease reporting can customize the survey module before disseminating the platform to local agencies, ensuring the data are collected in a standard manner.
Supplementary Material
Footnotes
The authors acknowledge the contributions of Victoria Christian, Jordan Hairston, Micah McClain, Yuhan McGee, Giovanna Merli, Dianne Oliver-Clapsaddle, Margaret Pendzich, and the CovIdentify Study to the work described here. The authors thank Duke AI Health for support provided. The authors thank the clinical research coordinators in the Duke University Center for Infectious Disease Diagnostics & Innovation and the coordinators at the Pickett Road Clinic for their assistance in data collection. The authors acknowledge the BIG IDEAS laboratory members at Duke University and HumanFirst team for their help with the project.
The project described was supported by Grant/Cooperative Agreement Number 75D30120C09551 made to Duke University from the Centers of Disease Control and Prevention (CDC), US Department of Health and Human Services (HHS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of CDC or US HHS. This work was also supported in part by Duke OIT, Duke Bass Connections Fellowship, Duke Margolis Center for Health Policy, Duke MEDx, Microsoft AI in Health, Duke CTSI (UL1TR002553), and NC Biotech (2020-FLG-3884). D.K.P. and J.M. were also supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institutes of Health (grant awards 1R25HD079352 and 1R21HD101268) and National Science Foundation (grant award SES-2029790). M.Y. was also supported by the Duke Population Research Center (DPRC) and its NICHD Center Grant (P2C HD0065563) and the Duke Center for Population Health and Aging (CPHA) and its NIA Center Grant (P30 AG034424).
J.McCall is an employee of Duke University and serves as a contractor for the US Food and Drug Administration. J.P.D. is a Scientific Advisor at Veri, Inc. C.W.W. is employed by Duke University, Durham Veterans Affairs Healthcare System, and Biomeme; C.W.W. is founder and holds equity in Predigen, Inc; C.W.W. has grants from National Institutes of Health (ARLG, VTEU, NIMHO, NIGMS), DARPA, CDC, PCORI, USAMRAA, DOD, Abbott, Najit, Pfizer, and Sanofi; C.W.W. is a consultant for Arena, Biofire, FHI Clinical, and Karius; C.W.W. is on the advisory board for FHI Clinical and Regeneron, and he is a member of the Board of Directors for Global Health Innovation Alliance Accelerator; C.W.W. serves on a DSMB for Janssen. E.S.H. is an employee of and has equity in Verily Life Sciences, is founder of and has equity in KēlaHealth, and is founder of and has equity in Clinetic. For the remaining authors, no conflicts of interest were declared.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (http://www.JPHMP.com).
Contributor Information
Dana K. Pasquale, Email: dana.pasquale@duke.edu.
Whitney Welsh, Email: wew2@duke.edu.
Andrew Olson, Email: andrew.olson@duke.edu.
Mark Yacoub, Email: mark.yacoub@duke.edu.
James Moody, Email: jmoody77@duke.edu.
Brisa A. Barajas Gomez, Email: Brisa.barajas.gomez@duke.edu.
Keisha L. Bentley-Edwards, Email: Keisha.bentley.edwards@duke.edu.
Jonathan McCall, Email: jonathan.mccall@duke.edu.
Maria Luisa Solis-Guzman, Email: maria.solis-guzman@duke.edu.
Jessilyn P. Dunn, Email: jessilyn.dunn@duke.edu.
Christopher W. Woods, Email: chris.woods@duke.edu.
Elizabeth A. Petzold, Email: Elizabeth.petzold@duke.edu.
Aleah C. Bowie, Email: bowiealeah@gmail.com.
Karnika Singh, Email: karnika.singh@duke.edu.
Erich S. Huang, Email: erichdata@verily.com.
References
- 1.Killingley B, Mann AJ, Kalinova M, et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat Med. 2022;28(5):1031–1041. [DOI] [PubMed] [Google Scholar]
- 2.Azimi P, Keshavarz Z, Cedeno Laurent JG, Stephens B, Allen JG. Mechanistic transmission modeling of COVID-19 on the Diamond Princess cruise ship demonstrates the importance of aerosol transmission. Proc Natl Acad Sci U S A. 2021;118(8):e2015482118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–675. [DOI] [PubMed] [Google Scholar]
- 4.Cevik M, Marcus JL, Buckee C, Smith TC. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission dynamics should inform policy. Clin Infect Dis. 2021;73(suppl 2):S170–S176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.James LP, Salomon JA, Buckee CO, Menzies NA. The use and misuse of mathematical modeling for infectious disease policymaking: lessons for the COVID-19 pandemic. Med Decis Making. 2021;41(4):379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cramer EY, Ray EL, Lopez VK, et al. Evaluation of individual and ensemble probabilistic forecasts of COVID-19 mortality in the United States. Proc Natl Acad Sci U S A. 2022;119(15):e2113561119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cramer EY, Huang Y, Wang Y, et al. The United States COVID-19 Forecast Hub dataset. Sci Data. 2022;9(1):462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nixon K, Jindal S, Parker F, et al. Real-time COVID-19 forecasting: challenges and opportunities of model performance and translation. Lancet Digit Health. 2022;4(10):e699–e701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heckathorn DD. Snowball versus respondent-driven sampling. Sociol Methodol. 2011;41(1):355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shandhi MMH, Cho PJ, Roghanizad AR, et al. A method for intelligent allocation of diagnostic testing by leveraging data from commercial wearable devices: a case study on COVID-19. NPJ Digit Med. 2022;5(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn J, Shaw RJ, Ginsburg GS, et al. CovIdentify: a Duke University study. https://covidentify.covid19.duke.edu/. Published 2020. Accessed Febuary 17, 2023.
- 12.Pasquale DK, Olson A, Welsh WE, et al. Snowball Study Tool Kit. Durham, NC: Duke Network Analysis Center; 2022. https://sites.duke.edu/dnac/resources/snowballtoolkit. Accessed February 17, 2023. [Google Scholar]
- 13.Heckathorn DD, Cameron CJ. Network sampling: from Snowball and multiplicity to respondent-driven sampling. Ann Rev Sociol. 2017;43(1):101–119. [Google Scholar]
- 14.Duke Crucible. Snowball: An Integrated Cloud Platform for Respondent-driven Sampling. Durham, NC: Duke University; 2021. [Google Scholar]
- 15.Mandl KD, Kohane IS. Time for a patient-driven health information economy? N Engl J Med. 2016;374(3):205–208. [DOI] [PubMed] [Google Scholar]
- 16.McGee Y, Olson A, Burdick D, et al. Design and implementation of a cloud-based platform supporting the trial of a respondent-driven sampling method for identifying and contacting individuals at risk of COVID-19 infection. Podium presentation at: the American Medical Informatics Association (AMIA) Informatics Summit; 2023; Seattle, WA. [Google Scholar]
- 17.Kaiksow FA, Carter J. A comprehensive literature review addressing the range of factors that prevent inclusion in clinical trials and research. https://nap.nationalacademies.org/resource/26479/Kaiksow_Lit_Review-Factors_that_Prevent_Inclusion_Clinical_Trials_and_Research.pdf. Accessed January 18, 2023.
- 18.Bibbins-Domingo K, Helman A, eds; National Academies of Sciences, Engineering, and Medicine. Improving Representation in Clinical Trials and Research: Building Research Equity for Women and Underrepresented Groups. 1st ed. Washington, DC: The National Academies Press; 2022. [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention. Symptoms of COVID-19. https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html. Published 2020. Accessed February 17, 2023.
- 20.Kremer C, Braeye T, Proesmans K, André E, Torneri A, Hens N. Serial intervals for SARS-CoV-2 Omicron and Delta variants, Belgium, November 19-December 31, 2021. Emerg Infect Dis. 2022;28(8):1699–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lash RR, Moonan PK, Byers BL, et al. COVID-19 case investigation and contact tracing in the US, 2020. JAMA Netw Open. 2021;4(6):e2115850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shelby T, Schenck C, Weeks B, et al. Lessons learned from COVID-19 contact tracing during a public health emergency: a prospective implementation study. Front Public Health. 2021;9:721952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harper-Hardy P, Ruebush E, Allen M, Carlin M, Plescia M, Blumenstock JS. COVID-19 case investigation and contact tracing programs and practice: snapshots from the field. J Public Health Manag Pract. 2022;28(4):353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macomber K, Hill A, Coyle J, Davidson P, Kuo J, Lyon-Callo S. Centralized COVID-19 contact tracing in a home-rule state. Public Health Rep. 2022;137(2)(suppl):35S–39S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lash RR, Donovan CV, Fleischauer AT, et al. COVID-19 contact tracing in two counties—North Carolina, June-July 2020. MMWR Morb Mortal Wkly Rep. 2020;69(38):1360–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelton M, Medina D, Sood N, et al. Efficacy of a student-led community contact tracing program partnered with an academic medical center during the coronavirus disease 2019 pandemic. Ann Epidemiol. 2021;56:26–33.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kretzschmar ME, Rozhnova G, Bootsma MCJ, van Boven M, van de Wijgert JHHM, Bonten MJM. Impact of delays on effectiveness of contact tracing strategies for COVID-19: a modelling study. Lancet Public Health. 2020;5(8):e452–e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blaney K, Foerster S, Baumgartner J, et al. COVID-19 case investigation and contact tracing in New York City, June 1, 2020, to October 31, 2021. JAMA Netw Open. 2022;5(11):e2239661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng K, Santhya S, Soong A, et al. Serial intervals and incubation periods of SARS-CoV-2 omicron and delta variants, Singapore. Emerg Infect Dis. 2023;29(4):814–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madewell ZJ, Yang Y, Longini IM, Jr, Halloran ME, Dean NE. Household secondary attack rates of SARS-CoV-2 by variant and vaccination status: an updated systematic review and meta-analysis. JAMA Netw Open. 2022;5(4):e229317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jørgensen SB, Nygård K, Kacelnik O, Telle K. Secondary attack rates for Omicron and Delta variants of SARS-CoV-2 in Norwegian households. JAMA. 2022;327(16):1610–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salganik MJ, Heckathorn DD. Sampling and estimation in hidden populations using respondent driven sampling. Sociol Methodlogy. 2004;34:193–240. [Google Scholar]
- 33.Johnston LG, Hakim AJ, Dittrich S, Burnett J, Kim E, White RG. A systematic review of published respondent-driven sampling surveys collecting behavioral and biologic data. AIDS Behav. 2016;20(8):1754–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gyarmathy VA, Johnston LG, Caplinskiene I, et al. A simulative comparison of respondent driven sampling with incentivized snowball sampling–the “Strudel effect”. Drug Alcohol Depend. 2014;135:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helms YB, Hamdiui N, Kretzschmar MEE, et al. Applications and recruitment performance of Web-based respondent-driven sampling: scoping review. J Med Internet Res. 2021;23(1):e17564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.