Abstract
Background and Aims
Sphagnum (peatmoss) comprises a moss (Bryophyta) clade with ~300–500 species. The genus has unparalleled ecological importance because Sphagnum-dominated peatlands store almost a third of the terrestrial carbon pool and peatmosses engineer the formation and microtopography of peatlands. Genomic resources for Sphagnum are being actively expanded, but many aspects of their biology are still poorly known. Among these are the degree to which Sphagnum species reproduce asexually, and the relative frequencies of male and female gametophytes in these haploid-dominant plants. We assess clonality and gametophyte sex ratios and test hypotheses about the local-scale distribution of clones and sexes in four North American species of the S. magellanicum complex. These four species are difficult to distinguish morphologically and are very closely related. We also assess microbial communities associated with Sphagnum host plant clones and sexes at two sites.
Methods
Four hundred and five samples of the four species, representing 57 populations, were subjected to restriction site-associated DNA sequencing (RADseq). Analyses of population structure and clonality based on the molecular data utilized both phylogenetic and phenetic approaches. Multi-locus genotypes (genets) were identified using the RADseq data. Sexes of sampled ramets were determined using a molecular approach that utilized coverage of loci on the sex chromosomes after the method was validated using a sample of plants that expressed sex phenotypically. Sex ratios were estimated for each species, and populations within species. Difference in fitness between genets was estimated as the numbers of ramets each genet comprised. Degrees of clonality [numbers of genets/numbers of ramets (samples)] within species, among sites, and between gametophyte sexes were estimated. Sex ratios were estimated for each species, and populations within species. Sphagnum-associated microbial communities were assessed at two sites in relation to Sphagnum clonality and sex.
Key Results
All four species appear to engage in a mixture of sexual and asexual (clonal) reproduction. A single ramet represents most genets but two to eight ramets were dsumbers ansd text etected for some genets. Only one genet is represented by ramets in multiple populations; all other genets are restricted to a single population. Within populations ramets of individual genets are spatially clustered, suggesting limited dispersal even within peatlands. Sex ratios are male-biased in S. diabolicum but female-biased in the other three species, although significantly so only in S. divinum. Neither species nor males/females differ in levels of clonal propagation. At St Regis Lake (NY) and Franklin Bog (VT), microbial community composition is strongly differentiated between the sites, but differences between species, genets and sexes were not detected. Within S. divinum, however, female gametophytes harboured two to three times the number of microbial taxa as males.
Conclusions
These four Sphagnum species all exhibit similar reproductive patterns that result from a mixture of sexual and asexual reproduction. The spatial patterns of clonally replicated ramets of genets suggest that these species fall between the so-called phalanx patterns, where genets abut one another but do not extensively mix because of limited ramet fragmentation, and the guerrilla patterns, where extensive genet fragmentation and dispersal result in greater mixing of different genets. Although sex ratios in bryophytes are most often female-biased, both male and female biases occur in this complex of closely related species. The association of far greater microbial diversity for female gametophytes in S. divinum, which has a female-biased sex ratio, suggests additional research to determine if levels of microbial diversity are consistently correlated with differing patterns of sex ratio biases.
Keywords: Clonality, sex ratios, peatlands, peatmosses, Sphagnum, Sphagnum diabolicum, Sphagnum divinum, Sphagnum magniae, Sphagnum medium
INTRODUCTION
Asexual reproduction is widespread across the tree of life but is especially common among plants. Asexuality can bring a range of benefits, such as a hedge against local extinction by replicating individuals, spreading individuals across heterogeneous environmental conditions, and stabilizing adapted genotypes against being broken up by recombination associated with sex. In plants, the varied mechanisms of clonal spread give rise to a range of population structures that differ in how much individual clones spread and in how intermingled they grow. At two extremes of a spectrum, the so-called phalanx pattern describes a structure in which clones spread laterally and often abut but may not extensively interdigitate. The guerrilla pattern, in contrast, reflects more clonal fragmentation, and individual genotypes can be scattered and mixed. These differences can have significant impacts on patterns of sexual reproduction (Barrett, 2015).
Bryophytes (mosses, liverworts, hornworts) have an exceptionally high potential for clonal reproduction because the gametophytes of many or most species have totipotent cells that can regenerate readily when fragmented, even from a dried powder (Shaw, 1986). Plants have been shown to propagate after being frozen for hundreds and even thousands of years (La Farge et al., 2013). Moss gametophytes, the haploid perennial free-living stage of the life cycle, produce a broad diversity of deciduous vegetative propagules (Correns, 1899). In contrast, clonality in seed plants pertains almost exclusively to the sporophyte generation.
Clonal reproduction in mosses involves the haploid sexual generation and is inextricably connected with gametophyte sexuality. Gametes are produced via mitosis by haploid multicellular gametophytes so the functional egg and sperm cells are clonally replicated, and are genetically identical to the gametophytes that produce them. Thus, the asexual replication of gametophyte plants in natural moss populations can have multiplicative effects on gamete production and potential fitness. In this study, we jointly investigated clonality and gametophyte sex in four closely related species of the moss genus Sphagnum (peatmosses). Sphagnum peatmosses are dominants in relatively low-nutrient wetland communities around the Northern Hemisphere and store some 25–30 % of the entire terrestrial carbon pool in the form of partially decomposed plant material, or peat. Most (but not all) peatmosses commonly produce sporophytes and spores, and sexual reproduction appears to be common (Sundberg and Rydin, 2000; Johnson and Shaw, 2015). Nevertheless, clonal spread also occurs, at least within populations (e.g. Cronberg, 1996).
The four focal species in this study were until recently included in one widespread species, S. magellanicum. However, phylogenomic analyses have shown that there are at least seven well-differentiated clades within S. magellanicum s. lat., and four such taxa occur in eastern North America (Shaw et al., 2022). Although they are clearly distinct phylogenetically, the four species commonly grow sympatrically and are extremely difficult to distinguish morphologically (Shaw et al., 2023). The four North American species nevertheless have distinct, albeit overlapping geographic ranges and occupy different (although also overlapping) niches within peatlands (Yousefi et al., 2017; Hassel et al., 2018; Shaw et al., 2022). They are clearly important units of biodiversity and are for that reason recognized taxonomically as distinct species despite a near-absence of distinguishing morphological characters (Shaw et al., 2023). This near-absence of morphological differentiation, combined with phylogenomic patterns, suggests that taxa in the S. magellanicum complex are at an incomplete stage of speciation.
Species in the S. magellanicum complex have unisexual gametophytes and do reproduce sexually, as evidenced by sporophyte production. Gametophyte sex is determined by a U/V chromosomal system (Healey et al., 2023), from which we can expect a 1:1 ratio of male and female gametophytes from the meiotic production of spores. As Sphagnum, in general, and the S. magellanicum complex, in particular, has been developing as a model group for ecological genomics, it is important to learn more about population structure and reproduction in these species. Rapidly growing genomic resources for Sphagnum species (Weston et al., 2018; Healey et al., 2023) make this group a good focus for reproductive biology, and additional information about their local-scale genetic structure can be important for the appropriate design of field and laboratory experiments. One of two reference-quality genome sequences in Sphagnum is from S. divinum, a member of the complex. Although they are phylogenetically divergent, species in the S. magellanicum complex show evidence of interspecific introgression (Shaw et al., 2022). These evolutionary features – close phylogenetic relationships, ecological divergence and interspecific reproductive compatibility – make this group valuable for exploring ecological and reproductive divergence during the phylogenetic genesis of biodiversity.
The goals of this study were to compare the four North American species within this S. magellanicum complex in terms of gametophyte clonality and sex ratios. Specifically, we sought to answer the following questions. (1) To what extent are genets (genetically distinct haploid multi-locus genotypes) represented by multiple ramets (asexually produced gametophyte shoots)? (2) Are individual genets clonally replicated within local contiguous hummock-forming colonies within populations, between hummocks within populations, and/or between geographically distinct populations? (3) What are the relative proportions of male and female gametophytes (sex ratios) within and between populations, estimated at the ramet and genet levels? (4) Do male and female gametophytes differ in degree of clonal propagation?
In addition to addressing these questions across the four species, we had the opportunity to study plants representing two of the species at two sites, one in New York State and one in Vermont, with additional sampling that included associated prokaryotic microbes. Utilizing the plants at those two sites, we were able to address additional questions. (1) Are there differences in abundance of individual genets within sites; i.e. differences in clonal fitness (estimated as ramet frequency)? (2) Is clonal diversity spatially structured within sites? (3) Are there differences in microbial communities associated with sampling sites, species, clones or sexes?
MATERIALS AND METHODS
Focal species and sampling
Our data include 405 gametophyte stems sampled from 57 collecting sites (Supplementary Data Table S1). All voucher specimens associated with this project are preserved in the L.E. Anderson Bryophyte Herbarium at Duke University (DUKE). We hereafter refer to individual sampled stems as ‘ramets’. Some of the collecting sites were discrete, open peatlands surrounded by mesic, often wooded habitats that contained one or more of our focal Sphagnum species. Ramets were collected in an unstructured (‘haphazard’) way from throughout the open peatland as well as in the surrounding forests. Each sample consisted of a handful of ramets collected from a patch in the field. These handfuls are referred to as ‘collections’. While some sites were sampled intensively, others were represented by just one to a few collections. To estimate clonality, genetic diversity and sex ratios within sites, we grouped together collecting localities that were within 5 km of one another and defined these as ‘sites’ for downstream analyses.
Our sampling of Sphagnum ramets was hierarchical to estimate genetic diversity and clonality at the levels of within- and between-individual collections (handfuls), and within/between sites. Duplicate DNAs from 24 ramets were subjected to restriction site-associated DNA sequencing (RADseq) amplification and assembly to obtain estimates of sequencing error rates for identification of clones (‘genets’; see below).
The most intensive sampling was accomplished at two sites: St Regis Lake (Franklin County, NY) and Franklin Bog (Franklin County, VT), where our data include 54 and 30 samples, respectively. These were collected as we wandered around the sites collecting samples as they were encountered. Geocoordinates were recorded with each sample and prokaryotic microbial communities associated with the living Sphagnum samples were assessed using 16s rDNA sequencing as described in Kolton et al. (2022).
Genotyping by RADseq
RADseq libraries were prepared following the double digestion restriction site-associated DNA sequencing (ddRADseq) protocol of Parchman et al. (2012) with the modifications described in Duffy et al. (2020). Each library was cleaned and size-selected for fragments of ~350 bp using AMPur XP beads (Beckman Coulter). The cleaned libraries were inspected for quality on a BioAnalyzer (Agilent) and sequenced on a single lane of Illumina NextSeq 500 or NovaSeq 6000 with 150bp single-end reads or HiSeq 2000 with 100bp single-end reads at the Genome Sequencing Shared Resource at the Duke Center for Genomic and Computational Biology (https://biostat.duke.edu/research/duke-center-genomic-and-computational-biology). Raw Illumina reads were quality checked with FastQC (Andrews, 2010) and RADseq loci were identified with ipyrad v.0.9.50 (Eaton and Overcast, 2020) using default settings with modifications to reduce genotyping error, noted below. The in silico digested reads from 41 genomic resequencing samples were included with the Illumina reads of the RADseq library samples after the demultiplexing step as described in Shaw et al. (2022). Reads were filtered for adapter sequences or low-quality bases, trimmed to a maximum of 92 bases after removing the barcode, and filtered to remove reads shorter than 35 bp after trimming. Loci were identified using the reference assembly method against the S. divinum reference genome. Only single-nucleotide polymorphisms (SNPs) from loci on linkage groups (chromosomes) LG01–LG19 were kept; i.e. sex chromosomes or unassigned contigs were not included. To avoid miscalled genotypes resulting from ipyrad’s majority-rule haploid genotype calling method, samples were treated as diploid and all heterozygous genotype calls were converted to missing data since those calls are not statistically consistent with a single haploid allele. Homozygous genotypes were then treated as haplotypes.
Only SNPs present in a minimum of 80 % of samples after filtering were included in the final datasets. To avoid systematic error rate differences that affected the ability to distinguish clones from closely related genotypes, loci and SNPs in the Franklin Bog and St Regis Lake samples (HiSeq 2000 reads) were identified and analysed separately from the rest of the RADseq library samples. Datasets with SNPs present in 80 % of specific sets of samples were generated for all samples excluding the Franklin Bog and St Regis Lake samples (the clonality dataset), for all samples from sites with at least four samples (the four-deep dataset), and for the Franklin Bog and St Regis Lake samples (the FBSRL dataset).
Identification of clones
Because of the potential for error in RADseq genotype calls and missing data, it is possible that genetically identical individuals (clones) may not have identical multi-locus genotypes and that very similar genotypes could appear identical. Pairwise genetic distances for all samples within species in the clonality and FBSRL datasets were calculated in R v.4.1.1 (R Core Team, 2021) with the adegenet package. Genetic distances between pairs from duplicated DNAs were compared to identify a maximum observed distance between samples known to be genetically identical. Twenty-one of 24 duplicated DNA pairs had genetic distances of zero and the remaining three had low distances relative to most sample pairwise distances. Clusters of samples where any of the pairwise distances between them were less than twice the maximum observed clonal distance were grouped and evaluated as possible clones. For many of these clusters, all pairwise distances between samples in the cluster were equal to zero and the cluster was inferred to be a clone. When pairwise distances were not all equal to zero, each sample with non-zero distances was evaluated and kept in the cluster if all or most distances were less than the maximum observed clonal distance, or removed from the cluster if all or most distances were greater than the maximum observed clonal distance.
Maximum likelihood and neighbour-joining methods were used to reconstruct phylogeny using the clonality and FBSRL datasets. Potential clones inferred through clustering were further evaluated for consistency with the phylogeny. For the maximum likelihood analysis in IQ-TREE2 v.2.1.2 (Minh et al., 2020), we used ModelFinder (Kalyaanamoorthy et al., 2017) to perform automatic model selection and kept the most likely tree from ten independent runs. Model selection incorporated corrections for ascertainment bias (Lewis, 2001) and bipartition support was assessed using the ultrafast bootstrap method with 1000 pseudoreplicates (Hoang et al., 2018). Neighbour-joining was performed with the R ape package (Paradis and Schliep, 2019).
The four-deep dataset was filtered to replace the SNPs contributing to non-zero distances between ramets of an inferred clone with missing data so that all ramets of a genet had identical genotypes in analyses of population diversity.
Distinguishing male and female gametophytes
Using the pre-filtered loci from ipyrad, male and female samples were distinguished based on their locus coverage on the female sex chromosome (linkage group LG20) relative to their locus coverage on autosomal chromosomes (LG01–LG19). Because of similarity between the male and female sex chromosomes, even males have RADseq reads that map to the female sex chromosome, but in a male sample the proportion of LG20 loci that are identified is substantially lower than the proportion of autosomal loci. In a female sample, the proportions of LG20 loci identified were similar to the proportions of autosomal loci. Samples differ in their locus coverage, but by calculating the mean percent autosomal locus coverage for each sample and plotting the LG20 coverage as a percentage of that mean in R, two clear groups of samples with higher and lower LG20 coverage emerge (Supplementary Data Fig. S1).
To validate sample sex inferences using this informatic method, a subset of samples was evaluated using primers previously demonstrated to distinguish sexually expressing male and female samples (Healey et al., 2023). Multiplexed PCR reactions contained 0.25 μm each of male and female forward and reverse primers, 0.3 mm dNTPs, 2.5 mm MgCl2, 0.5 μg μL−1 BSA, 3 % DMSO, 1.0–2.8 ng μL−1 template DNA, 1× buffer, and 4 units of Choice Blue DNA Taq Polymerase (Denville Scientific) in 20-μL reactions. Thermocycler conditions were 3 minutes at 95°C, followed by 35 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s, with a final elongation at 72°C for 5 minutes. Bands were visualized by running 1 h at 95 V on a 3 % agarose SB gel with 4 μL SYBR Safe DNA Gel Stain (Invitrogen) in 80 mL agarose solution. Sex was inferred based on band migration distance relative to a size-standard ladder (female expected band size = 394 bp and male expected band size = 444 bp). As a final check, inferences of sample sex were compared with inferences of sample clone membership to confirm that all members of a clone were also the same sex.
Genetic differentiation and diversity
We estimated relative genetic differentiation between collection sites for each species in the four-deep dataset with pairwise FST and Nei’s unbiased genetic distance using the hierfstat R package (Goudet, 2005). Significance of pairwise FST values after Benjamini and Hochberg FDR correction was tested with 1E4 permutations in R. Population diversity statistics by site, sex and species were calculated for the four-deep and FBSRL datasets using the poppr R package (Kamvar et al., 2014). Distributions of pairwise genetic distance within and between sites were compared for the clonality dataset (after removing clones) by testing for differences in mean, mode or skewness with 1E5 permutations in R. Differences in number of ramets per genotype by sex and species for the clonality dataset were tested using 1E5 permutations in R. Differences in sex ratio by species in the clonality dataset and by species and site in the four-deep dataset were also tested using 1E5 permutations.
Identification of microbial associates based on 16S rDNA sequencing
Identification of prokaryotic microbes associated with Sphagnum samples from St Regis Lake (NY) and Franklin Bog (VT) were included in the larger study by Kolton et al. (2022). We utilized data for the two sites in our current analyses. See that publication for collection, sampling and sequencing protocols. Datasets were normalized by cumulative sum scaling (CSS) and variance components of β diversity were determined using non-metric multidimensional scaling (NMDS) of Bray–Curtis and weighted UniFrac dissimilarities with the phyloseq package in R (McMurdie and Holmes, 2013). Effects of site, species and sex on microbial β diversity were tested by PERMANOVA with 1000 permutations using the vegan package in R (Oksanen et al. 2022) to evaluate marginal and term effects.
Each Sphagnum collection was georeferenced so we could assess physical clustering of ramets and genets by sex. The St Regis Lake site had only S. divinum, but both this species and S. diabolicum were detected at Franklin Bog. It was only at the latter site, therefore, that we could assess species-specificity of microbial communities. Spatial clustering of S. divinum ramets and genets (using the centroid geographic location of all ramets of a genet) by sex was evaluated using the mark connection function of physical locations to inspect the pattern of dependence between samples of the same or different sexes with the spatstat package in R (Baddeley et al., 2015), and by testing for significance of the difference in mean geographic distances between samples of the same or different sexes or genotypes using 1E4 permutations in R. Isolation by distance of ramets and genets were evaluated for S. divinum samples within each site using Mantel tests in R with 1E5 permutations.
RESULTS
Between-population and geographic patterns in genetic structure and clonality
Our sampling included all four eastern North American species of the ‘S. magellanicum complex’ and phylogenetic relationships among them (Supplementary Data Figs S2 and S3) were consistent with previous reconstructions based on whole-genome resequencing and RADseq (Shaw et al., 2022). Among the North American species, S. medium is sister to a clade including S. divinum, S. magniae and S. diabolicum, and within that clade S. divinum is sister to S. magniae + S. diabolicum. The last two species are especially closely related and have not reached reciprocal monophyly across their genomes (Shaw et al., 2022).
Samples from many localities comprise site-specific clades relative to those from other sites (sometimes related to clonality within sites), but intraspecific samples from some sites are scattered within their respective species (with varying levels of support; Supplementary Data Figs S2 and S3). For example, all 12 samples of S. diabolicum from Shark Cove Bog (ME) form a strongly supported clade (Supplementary Data Fig. S2). Similarly, all ten plants of S. diabolicum from Jam Pond Bog (NY) comprise a single clade and appear to represent just two replicated clones. In contrast, ten plants from Birch Harbor (ME) comprise a single clade within S. diabolicum while four other plants from the site form a separate clade, and a 15th sample falls into a third phylogenetic location within the species. Within-site sample sizes for S. magniae tend to be smaller because the populations are themselves smaller. Eleven samples from the Green Swamp (NC) fall into a clade, but that clade also includes samples from other sites in MS and elsewhere in NC (Supplementary Data Fig. S2). Eight other samples of S. magniae from the Green Swamp are scattered across at least two other divergent clades. Within S. divinum, 12 samples from Jam Pond Bog (NY, where it is sympatric with S. diabolicum) are scattered across at least four intraspecific clades (Supplementary Data Fig. S2). In contrast, 11 samples of S. divinum from Dolly Sods (WV), at or near the southern edge of its range, all form a single clade. Similar patterns seem to hold in the relatively uncommon species S. medium (Supplementary Data Fig. S2). Samples from most sites form separate clades, though at the 1000 Acre Bog (ME), seven samples of S. medium fall into three different clades.
Genetic differentiation among sites within species, estimated by FST and Nei’s unbiased genetic distances, was generally low, although some pairwise comparisons were significant (Supplementary Data Table S2). Most FST values were below 0.2 and a few higher values reflect limited sampling within some sites rather than strong differentiation. The distributions of between-site genetic distances relative to those within sites differed in each of S. diabolicum, S. divinum and S. medium (Fig. 1) with statistical significance depending on which measure of distribution shape/position was compared (Supplementary Data Table S3). Between- and within-site genetic distances do not differ in S. magniae – plants are on average as genetically divergent within sites as between them (Fig. 1; Supplementary Data Table S3). Moreover, in all species other than S. magniae, the distributions of inter-individual genetic distances within populations are skewed left, indicating some proportion of especially closely related (but not clonal) individuals, presumably a reflection of local mating and segregation of offspring.
Fig. 1.
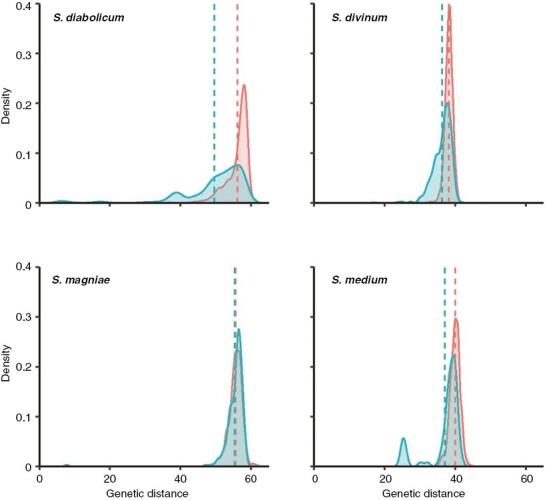
Distribution of pairwise genetic distances in species of the S. magellanicum complex after removing inferred clones. Red, pairwise comparisons of samples from different sites; blue, pairwise comparisons of samples from the same site. Dashed lines indicate the mean of each distribution.
Patterns of diversity and clonality within and among species
Some degree of clonality [estimated by its reciprocal, actually an estimate of sexuality: (G − 1)/(R − 1), where G is number of genets and R is number of ramets] was detected within each of the four species (Table 1). Levels of clonality did not differ significantly among species (Supplementary Data Table S4). On average, fewer than two ramets represented each genet across the four species (Table 2). In all four species, most genets are represented by a single ramet (Fig. 2), but a few genets are represented by a greater number of ramets, up to eight. Given limited sample sizes, these are likely underestimates, but it is clear that none of the species is strictly or even predominantly clonal.
Table 1.
Clonality statistics for species in the S. magellanicum complex
| S. diabolicum | S. divinum | S. magniae | S. medium | |
|---|---|---|---|---|
| Total ramets | 101 | 128 | 52 | 42 |
| Collections | 95 | 105 | 45 | 36 |
| Collection sites | 18 | 25 | 16 | 12 |
| Unique genets | 61 | 92 | 41 | 29 |
| Genets/ramets (G − 1)/(R − 1) | 0.60 | 0.72 | 0.78 | 0.68 |
| Mean ramets/genet | 1.66 | 1.39 | 1.27 | 1.44 |
| Genets with >1 ramet | 18 | 28 | 7 | 7 |
| Multi-site genets | 0 | 0 | 1 | 0 |
| Multi-collection genets | 14 | 13 | 5 | 4 |
| Collections with >1 stem sampled | 6 | 16 | 5 | 5 |
| Collections with >1 genet | 1 | 3 | 1 | 1 |
| Collection sites with >1 stem sampled | 11 | 21 | 8 | 8 |
| Collection sites with >1 genet | 8 | 19 | 6 | 7 |
Sample (ramet) counts do not include technical replicates.
Multi-site genets: number of genets represented by ramets at more than one site.
Multi-collection genets: number of genets represented by ramets in more than one collection.
Collections with >1 stem sampled: numbers of individual collections (handfuls) with more than one ramet sampled.
Collections with >1 genet: numbers of collections where more than one genet was found.
Collection sites with >1 stem sampled: numbers of sites where more than one ramet was sampled.
Collection sites with >1 genet: numbers of sites where more than one genet was found.
Table 2.
Population summary statistics for species in the S. magellanicum complex by site and sex
| Site or sex | Ramets | Genets | R/G | Hexp | PPL | PPr | (G − 1)/(R − 1) |
|---|---|---|---|---|---|---|---|
| Sphagnum diabolicum | |||||||
| ME-BirchHarbor | 15 | 11 | 1.36 | 0.109 | 40.8 | 13.7 | 0.71 |
| ME-GreatWassIsland | 12 | 10 | 1.2 | 0.29 | 41.8 | 14.4 | 0.82 |
| ME-HadleyLakeRd | 8 | 1 | 8 | 0 | 0 | 1.5 | 0.00 |
| ME-IndianRiverRdFen | 19 | 12 | 1.58 | 0.199 | 33.4 | 9.3 | 0.61 |
| ME-SharkCoveBog | 12 | 8 | 1.5 | 0.232 | 31.2 | 9.5 | 0.64 |
| ME-SteubenBog | 4 | 3 | 1.33 | 0.052 | 17 | 4.7 | 0.67 |
| ME-WhalesbackRidge | 7 | 2 | 3.5 | 0.057 | 0.5 | 1.8 | 0.17 |
| NY-JamPondBog | 10 | 2 | 5 | 0 | 6.5 | 2.9 | 0.11 |
| Female | 27 | 18 | 1.5 | 0.337 | 59.1 | 21.1 | 0.65 |
| Male | 60 | 31 | 1.94 | 0.345 | 78.6 | 40.7 | 0.51 |
| Total | 87 | 49 | 1.78 | 0.404 | 100 | NA | 0.56 |
| Sphagnum divinum | |||||||
| BC-CampbellRiver | 4 | 2 | 2 | 0.199 | 2.5 | 0.4 | 0.33 |
| LB-Schefferville11 | 4 | 1 | 4 | 0 | 0 | 0.3 | 0.00 |
| ME-1000AcreBog | 6 | 3 | 2 | 0.041 | 23.1 | 1.9 | 0.40 |
| ME-JemtlandBog | 8 | 6 | 1.33 | 0.166 | 36.2 | 4.3 | 0.71 |
| ME-MillinocketSawdustBog | 6 | 5 | 1.2 | 0.01 | 33.6 | 3.3 | 0.80 |
| MN-Spruce | 14 | 13 | 1.08 | 0.311 | 49.9 | 5.4 | 0.92 |
| NH-CoosCoWoodedPeatlands | 5 | 4 | 1.25 | 0.076 | 24.5 | 1.6 | 0.75 |
| NH-HurlbertSwamp | 5 | 4 | 1.25 | 0.179 | 21.3 | 1.7 | 0.75 |
| NH-PhilbrickCricentiBog | 6 | 4 | 1.5 | 0.024 | 25.4 | 2.1 | 0.60 |
| NY-JamPondBog | 12 | 6 | 2 | 0.003 | 36.8 | 3.2 | 0.45 |
| NY-McLeanBog | 4 | 3 | 1.33 | 0.006 | 24.6 | 1.3 | 0.67 |
| QC-Schefferville13 | 4 | 3 | 1.33 | 0.005 | 24.3 | 1.6 | 0.67 |
| QC-SeptIles | 5 | 4 | 1.25 | 0.307 | 20.7 | 2.3 | 0.75 |
| VT-MooseBog | 7 | 5 | 1.4 | 0.005 | 30.6 | 2.3 | 0.67 |
| WI-BeulahBog | 11 | 8 | 1.38 | 0.144 | 34.5 | 2.5 | 0.70 |
| WV-DollySods | 12 | 8 | 1.5 | 0.012 | 34.9 | 3.4 | 0.64 |
| Female | 70 | 50 | 1.4 | 0.353 | 85.8 | 27.2 | 0.71 |
| Male | 42 | 28 | 1.5 | 0.28 | 72.1 | 13.7 | 0.66 |
| Total | 113 | 79 | 1.43 | 0.382 | 100 | NA | 0.70 |
| Sphagnum magniae | |||||||
| FL-Apalachicola | 4 | 4 | 1 | 0.259 | 20.6 | 3.2 | 1.00 |
| GA-OkefenokeeCI | 4 | 3 | 1.33 | 0.057 | 25.8 | 3.5 | 0.67 |
| NC-AlligatorRiver | 5 | 4 | 1.25 | 0.136 | 33.8 | 7.4 | 0.75 |
| NC-GreenSwamp | 19 | 15 | 1.27 | 0.401 | 74.8 | 30.7 | 0.78 |
| NC-LakeWaccamaw | 5 | 4 | 1.25 | 0.202 | 33.2 | 7 | 0.75 |
| Female | 20 | 18 | 1.11 | 0.429 | 79.6 | 35.8 | 0.89 |
| Male | 17 | 12 | 1.42 | 0.364 | 64.2 | 20.3 | 0.69 |
| Total | 37 | 30 | 1.23 | 0.444 | 100 | NA | 0.81 |
| Sphagnum medium | |||||||
| ME-1000AcreBog | 7 | 5 | 1.4 | 0.263 | 44.8 | 14.6 | 0.67 |
| ME-BirchHarbor | 6 | 6 | 1 | 0.366 | 47.3 | 14 | 1.00 |
| ME-SharkCoveBog | 4 | 4 | 1 | 0.056 | 39.1 | 11.3 | 1.00 |
| NH-PhilbrickCricentiBog | 9 | 2 | 4.5 | 0.08 | 3.7 | 3.8 | 0.13 |
| QC-SeptIles | 4 | 3 | 1.33 | 0.026 | 36.9 | 10.3 | 0.67 |
| Female | 22 | 13 | 1.69 | 0.388 | 79.3 | 39.9 | 0.57 |
| Male | 8 | 7 | 1.14 | 0.377 | 59.9 | 20.5 | 0.86 |
| Total | 30 | 20 | 1.5 | 0.445 | 100 | NA | 0.66 |
Ramets, stems sampled; Genets, unique multi-locus genotypes; R/G, ramets/genet; Hexp, expected heterozygosity; PPL, percent polymorphic loci; PPR, percent private alleles.
Fig. 2.
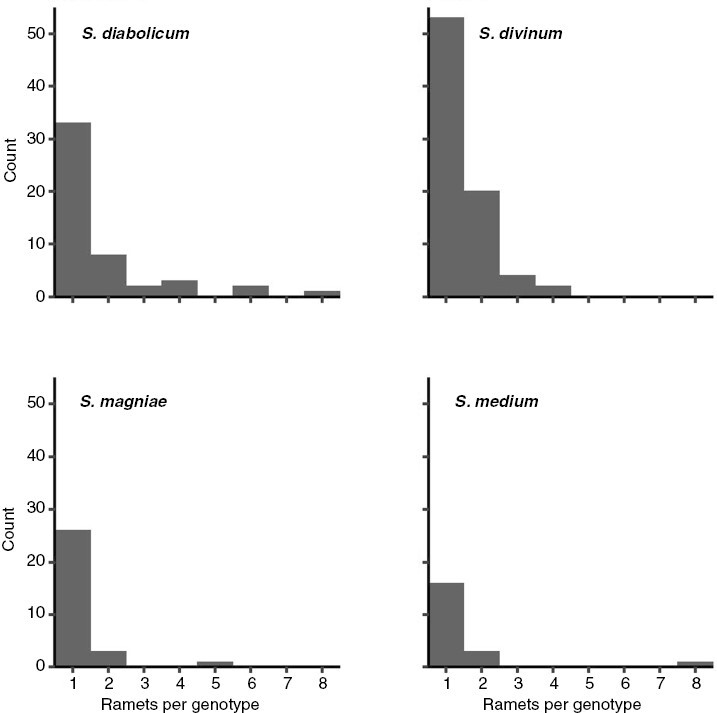
Histograms showing numbers of ramets per genet in species of the S. magellanicum complex.
Only two of 34 species populations appeared to be monoclonal. The Hadley Lake (ME) site for S. diabolicum (with eight samples) and the Schefferville (Quebec) site for S. divinum (with four samples) yielded a single genet from each (Table 2). At the other end of the spectrum, the Spruce site (MN) for S. divinum revealed 13 distinguishable genets among 14 sampled shoots. Data from the Appalachicola site (FL) for S. magniae indicate that each of four samples represents a unique genet, and sampling at other sites for this species suggest that most shoots belong to different genets (Table 2). When multiple ramets from a single genet were detected, they were, with one exception, restricted to a single site. One clonally replicated genet, in S. magniae, was detected at two different collecting sites, but all other clones were limited to a single collection (handful of plants collected together), or less commonly from different collections made at a single site (Table 1).
Individual populations (sites) within the four species did not typically display high levels of genetic diversity (Table 2). Expected heterozygosity among (unsampled) diploid sporophytes produced by random mating within sites is low, mostly <0.4. Nevertheless, every site for each species is characterized by at least one private allele, and some sites by as many as 14 such private alleles (Table 2). Despite low levels of overall population differentiation within species, each intraspecific population has a unique genomic signature. Interestingly, male plants of S. diabolicum have approximately twice as many private alleles within populations as female plants. This species also has a male-biased sex ratio (see below). The other three species, S. magniae, S. divinum and S. medium, which have female-biased sex ratios, have roughly twice the numbers of private alleles in female gametophytes compared with males (Table 2).
Gametophyte sexuality
Based on known genomic differentiation between the U sex chromosome in Sphagnum female gametophytes and the V chromosome in males, we predicted that RADseq coverage of U chromosome loci should approximately equal coverage of autosomal loci in females, whereas coverage of U chromosome loci in males should be considerably lower than the average autosomal locus coverage (see Materials and methods section). A plot of these values supports this prediction as most of our samples clearly belong to one of these two groups based on coverage (Supplementary Data Fig. S1). To confirm that this method of inferring gametophyte sex from RADseq data is accurate, we compared inferences about gametophyte sexuality from our RADseq data with PCR amplification of U and V loci in 20 gametophyte plants using primers previously demonstrated to differentiate plants of known sex based on production of male or female gametangia (Healey et al., 2023). The correspondence of inferred sex was 100 % (Fig. 3).
Fig. 3.
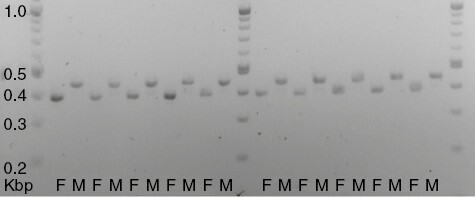
Gel image demonstrating gametophyte sex identification by PCR. Expected band size for female samples: 394 bp; expected band size for male samples: 444 bp. F (female) and M (male) indicate the sex of each sample as inferred from RADseq LG20 locus coverage.
When ramets of each species are treated as independent samples, S. diabolicum displays a significant male gametophyte bias overall and S. divinum has a significant female bias (Table 3). The other two species, S. magniae and S. medium, both display female biases but are not significant. When sex ratios were evaluated among unique genets, only the female bias in S. divinum is significant. These differences in significance appear to mainly reflect sample sizes (i.e. degrees of freedom) rather than differences in levels of clonality between male and female gametophytes, as we otherwise detected no differences in clonality related to sex (Supplementary Data Table S4).
Table 3.
Sex ratio statistics for species in the S. magellanicum complex
| By ramet | ||||
|---|---|---|---|---|
| Species | Samples | Counts (F, M, ?) |
Sex ratio | P |
| S. diabolicum | 101 | 38, 63, 0 | 0.62 | 0.0219 |
| S. divinum | 128 | 82, 45, 1 | 0.35 | 0.0032 |
| S. magniae | 52 | 29, 22, 1 | 0.43 | 0.2290 |
| S. medium | 42 | 26, 16, 0 | 0.38 | 0.1646 |
| By genet | ||||
|---|---|---|---|---|
| Species | Genotypes | Counts (F, M, ?) |
Sex ratio | P |
| S. diabolicum | 61 | 27, 34, 0 | 0.56 | 0.2300 |
| S. divinum | 92 | 61, 30, 1 | 0.33 | 0.0000 |
| S. magniae | 41 | 24, 16, 1 | 0.40 | 0.2100 |
| S. medium | 29 | 17, 12, 0 | 0.41 | 0.2300 |
Sample counts do not include technical replicates.
Sex ratio is males/(males + females).
P-values testing whether the sex ratio differs from 0.5 were calculated using permutation tests with 100 000 permutations. Statistically significant values (P < 0.05 after Benjamini and Hochberg FDR correction) are indicated in bold.
‘?’ indicates samples for which sex could not be reliably determined.
Despite sample size limitations on statistical significance, differences in sex ratios among species appear to be real. In S. diabolicum, four populations all exhibited a male bias; in three, only male plants were sampled (Table 4). In S. divinum, three populations had female biases, with two containing no males (albeit with very limited sampling). When unisexual, individual populations of S. magniae and S. medium contained only female gametophytes (Table 4).
Table 4.
Sex ratio statistics by collection site for species in the S. magellanicum complex, by sample (ramets) and genotype (genet)
| Site | By ramet | By genet | ||||||
|---|---|---|---|---|---|---|---|---|
| N | F, M | Sex ratio | P | N | F, M | Sex ratio | P | |
| S. diabolicum | ||||||||
| ME-BirchHarbor | 15 | 10, 5 | 0.33 | 0.1761 | 11 | 7, 4 | 0.36 | 0.3843 |
| ME-GreatWassIsland | 12 | 3, 9 | 0.75 | 0.1016 | 10 | 3, 7 | 0.70 | 0.3843 |
| ME-HadleyLakeRd | 8 | 0, 8 | 1.00 | 0.0137 | 1 | 0, 1 | 1.00 | 0.5833 |
| ME-IndianRiverRdFen | 19 | 4, 15 | 0.79 | 0.0168 | 12 | 2, 10 | 0.83 | 0.1344 |
| ME-SharkCoveBog | 12 | 8, 4 | 0.33 | 0.1940 | 8 | 4, 4 | 0.50 | 0.6366 |
| ME-WhalesbackRidge | 7 | 0, 7 | 1.00 | 0.0168 | 2 | 0, 2 | 1.00 | 0.3843 |
| NY-JamPondBog | 10 | 0, 10 | 1.00 | 0.0070 | 2 | 0, 2 | 1.00 | 0.3843 |
| S. divinum | ||||||||
| ME-1000AcreBog | 6 | 4, 2 | 0.33 | 0.5000 | 3 | 2, 1 | 0.33 | 0.6874 |
| ME-JemtlandBog | 8 | 8, 0 | 0.00 | 0.0390 | 6 | 6, 0 | 0.00 | 0.0930 |
| ME-MillinocketSawdustBog | 6 | 5, 1 | 0.17 | 0.2623 | 5 | 4, 1 | 0.20 | 0.5436 |
| MN-Spruce | 14 | 12, 2 | 0.14 | 0.0390 | 13 | 11, 2 | 0.18 | 0.0930 |
| NH-CoosCoWoodedPeat-lands | 5 | 3, 2 | 0.40 | 0.5000 | 4 | 2, 2 | 0.50 | 0.6874 |
| NH-HurlbertSwamp | 5 | 3, 2 | 0.40 | 0.5000 | 4 | 2, 2 | 0.50 | 0.6874 |
| NH-PhilbrickCricentiBog | 6 | 1, 5 | 0.83 | 0.2623 | 4 | 1, 3 | 0.75 | 0.6270 |
| NY-JamPondBog | 12 | 5, 7 | 0.58 | 0.5000 | 6 | 3, 3 | 0.50 | 0.6874 |
| QC-SeptIles | 5 | 5, 0 | 0.00 | 0.1244 | 4 | 4, 0 | 0.00 | 0.2492 |
| VT-MooseBog | 7 | 4, 3 | 0.43 | 0.5000 | 5 | 3, 2 | 0.40 | 0.6874 |
| WI-BeulahBog | 10 | 7, 3 | 0.30 | 0.3326 | 7 | 5, 2 | 0.29 | 0.5436 |
| WV-DollySods | 12 | 8, 4 | 0.33 | 0.3326 | 8 | 4, 4 | 0.50 | 0.6874 |
| S. magniae | ||||||||
| NC-AlligatorRiver | 5 | 5, 0 | 0.00 | 0.0933 | 4 | 4, 0 | 0.00 | 0.1869 |
| NC-GreenSwamp | 19 | 8, 11 | 0.58 | 0.4860 | 15 | 8, 7 | 0.47 | 0.5000 |
| NC-LakeWaccamaw | 5 | 3, 2 | 0.40 | 0.5000 | 4 | 3, 1 | 0.25 | 0.4703 |
| S. medium | ||||||||
| ME-1000AcreBog | 7 | 4, 3 | 0.43 | 0.5000 | 5 | 3, 2 | 0.40 | 0.5000 |
| ME-BirchHarbor | 6 | 4, 2 | 0.33 | 0.5000 | 6 | 4, 2 | 0.33 | 0.5000 |
| NH-PhilbrickCricentiBog | 9 | 9, 0 | 0.00 | 0.0060 | 2 | 2, 0 | 0.00 | 0.5000 |
Sample counts do not include technical replicates and only collection sites with at least five samples were included.
N, number of samples or genotypes; F, M, counts of females and males.
Sex ratio is males/(males + females).
P-values indicate whether the sex ratio differs from 0.5 and statistically significant values (P < 0.05 after Benjamini and Hochberg FDR correction) are indicated in bold.
St Regis Lake (NY) and Franklin Bog (VT): clonality and gametophyte sex ratios
Larger sample sizes for these two sites, with spatially explicit coordinates for all samples, combined with the 16S rRNA gene microbial data associated with each Sphagnum sample, permitted more detailed analyses.
Two species were detected among our samples: S. divinum was abundant at both sites whereas S. diabolicum was found only at Franklin Bog and was less common. We collected only five samples of S. diabolicum at Franklin Bog and they were all ramets of a single female genet (Supplementary Data Figs S4 and S5). We therefore focused our analyses of genetic structure and sex on S. divinum (but see below for microbial associations). Plants of S. divinum were sampled as they were encountered as we walked through the peatlands at each site. The species was sporadic near the margins of both sites but was substantially more common well into the peatlands (Fig. 4).
Fig. 4.
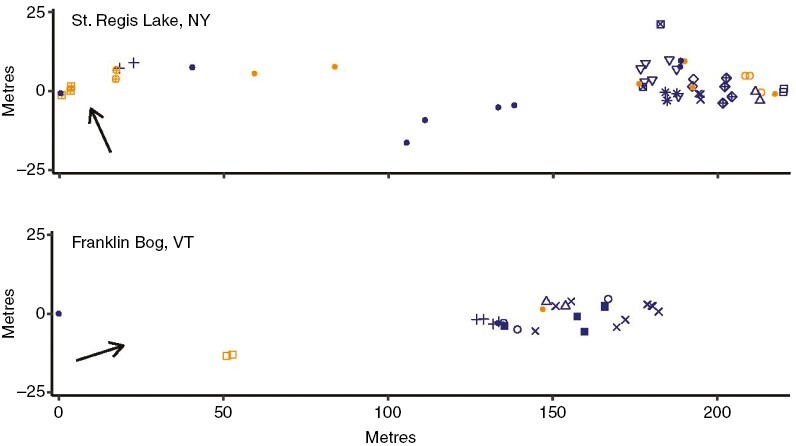
Plotted positions of S. divinum collections at St Regis Lake (NY) and of S. divinum and S. diabolicum collections at Franklin Bog (VT). Arrows indicate north. Male ramets are in orange and female ramets in blue. St Regis Lake: plot origin (0,0) = 44.426237N 74.244877W; filled circles, S. divinum ramets with unique genotypes; other shapes, S. divinum ramets of the same genet (clones). Franklin Bog: plot origin (0,0) = 44.988260N 72.899248W; filled squares, S. diabolicum ramets (all share the same genotype); filled circles, S. divinum ramets with unique genotypes; other shapes, S. divinum ramets sharing genotypes (clones).
Patterns of variation and clonality within S. divinum at St Regis Lake and Franklin Bog were similar to the patterns in the species overall. Expected heterozygosity was low, the degree of clonality suggested a mix of sexual and asexual reproduction, and plants from each site harboured differentially fixed SNPs (Table 5). Also, as with the broader regional sampling, female gametophytes of S. divinum at these two sites were characterized by higher numbers of fixed alleles. In fact, females had two to three times the numbers of fixed differences than males (Table 5).
Table 5.
Population summary statistics for Sphagnum divinum from St Regis Lake, NY, and Franklin Bog, VT
| Site/sex | Ramets | Genets | R/G | Hexp | PPL | PPr | (G − 1)/(R − 1) |
|---|---|---|---|---|---|---|---|
| NY-StRegis Lake | 54 | 29 | 1.86 | 0.226 | 16.2 | 31.9 | 0.53 |
| Female | 37 | 18 | 2.06 | 0.137 | 13.8 | 19.4 | 0.47 |
| Male | 17 | 11 | 1.55 | 0.188 | 10.6 | 8.2 | 0.63 |
| VT-FranklinBog | 23 | 8 | 2.88 | 0.108 | 8.6 | 5.5 | 0.32 |
| Female | 20 | 6 | 3.33 | 0.068 | 7.0 | 21.6 | 0.26 |
| Male | 3 | 2 | 1.50 | 0.018 | 2.7 | 5.8 | 0.50 |
| Both sites | 77 | 37 | 2.08 | 0.236 | 100 | NA | 0.47 |
| Female | 57 | 24 | 2.38 | 0.171 | 15.3 | 21.9 | 0.41 |
| Male | 20 | 13 | 1.54 | 0.198 | 11.6 | 8.7 | 0.63 |
Ramets, stems sampled; Genets, unique multi-locus genotypes; R/G, ramets/genets; Hexp, expected heterozygosity; PPL, percent polymorphic loci; PPR, percent private alleles.
Both male and female gametophytes were sampled at both localities but sex ratios were biased toward females in S. divinum (Table 5), as observed for this species overall. Both sexes within S. divinum were represented by clonal ramets but the numbers of ramets per genet were mostly limited to four or fewer. Two female genets were, however, represented by seven and nine ramets each (Fig. 5). These occurred towards the centre of the site where S. divinum was most abundant (Fig. 4). It is clear from the spatial plot (Fig. 4) that ramets of each genet were spatially clustered. The average geographic distance between ramets of a genet is significantly smaller than the average distance between ramets of different genets at both sites (Supplementary Data Fig. S6, Supplementary Data Table S5). There was a significant correlation between spatial separation of plants and genetic distance [i.e. isolation by distance, (IBD); Supplementary Data Fig. S7]. That the correlation is significant when all ramets were included but not when analyses were based on unique genotypes again reflects reduced sample sizes in the latter (Supplementary Data Table S6). However, in this case the pattern also corroborates that ramets of a given genet tend to be clustered.
Fig. 5.
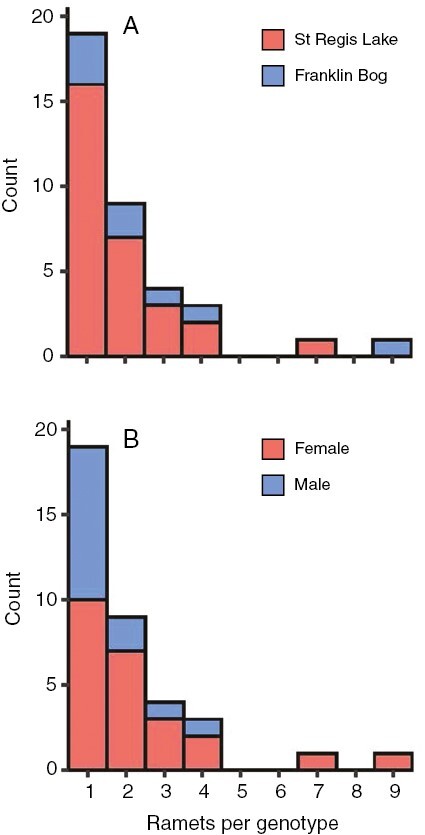
Histogram of ramets per genotype for samples of S. divinum from St Regis Lake (NY) and Franklin Bog (VT) by (A) site and (B) sex.
The physical locations of female gametophytes at both sites are positively associated with other female gametophytes and negatively associated with male gametophytes at the ramet but not genet level (Fig. 6). Distances between pairs of females at the ramet level are significantly smaller than distances between male–female pairs (Supplementary Data Fig. S8), suggesting that female ramets are clustered separately from male samples. Sample size for male gametophytes was smaller because of the female-biased sex ratio and clustering of males was not significant (Supplementary Data Table S7).
Fig. 6.
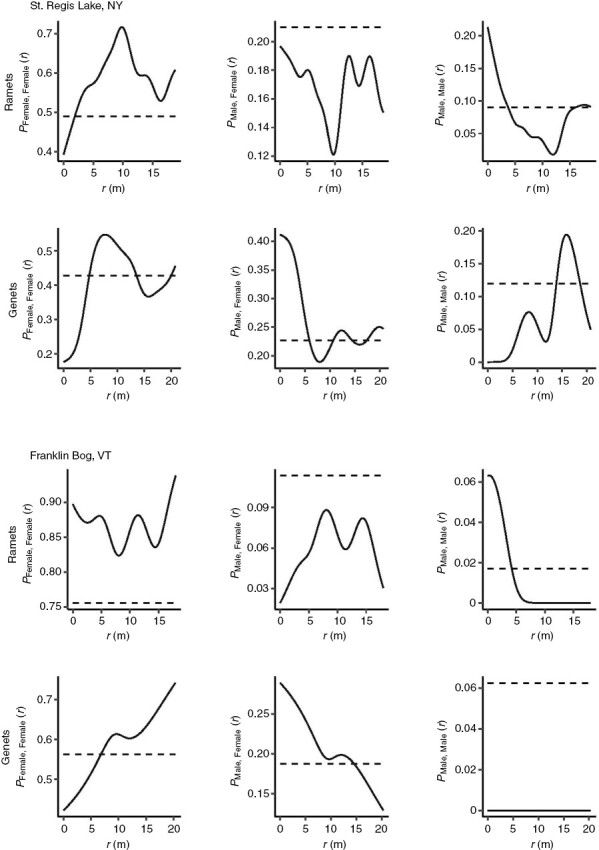
Association or clustering by sex of Sphagnum ramets or genets from St Regis Lake (NY) and Franklin Bog (VT). Solid lines represent the observed mark connect function of the probability of finding female:female pairs, male:female pairs or male:male pairs of ramets or genets at r distance. Dotted lines indicate the expected probability of finding the specified pair type. Observed values above the expected value suggest a positive association of the pair type at that distance. Observed values below the expected value suggest a negative association of the pair type. Distances between genets were based on the geographic centroid location of all ramets of a genotype.
St Regis Lake (NY) and Franklin Bog (VT): microbial communities relative to Sphagnum genetic structure
The two sites had distinct microbial communities (Fig. 7, Table 6). The site effect on microbial communities was significant for both the ordination on Bray–Curtis dissimilarity, which takes into account relative frequencies of microbe amplified sequence variants (ASVs), and for the ordination on weighted UniFrac dissimilarity, which also takes into account the phylogenetic relationships among the ASVs. There was no significant effect of species, genet or gametophyte sex on microbial community makeup when the site differences were taken into account using marginal tests (Table 6). There was also no significant site × sex interaction effect in tests of term effect models (F = 0.8828, P = 0.675).
Fig. 7.
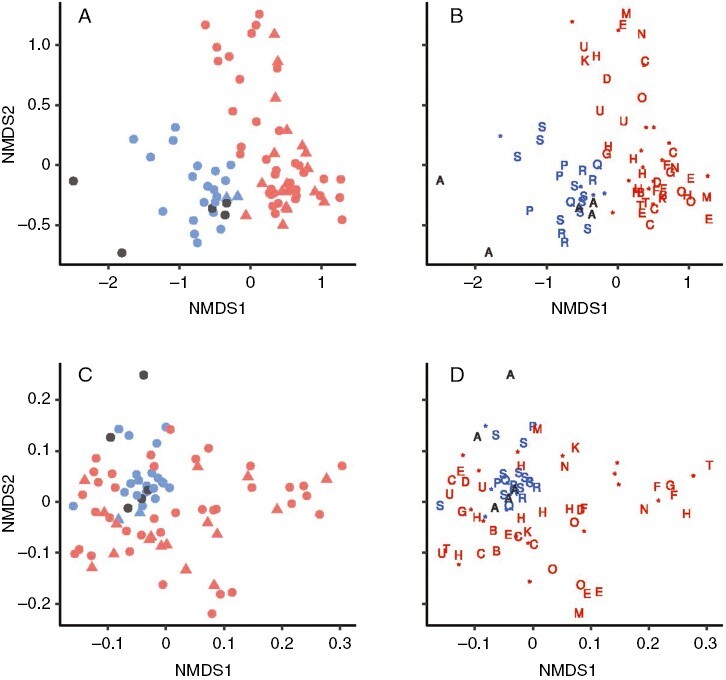
Similarity between Sphagnum-associated microbial communities from Sphagnum plants at St Regis Lake (NY) and Franklin Bog (VT). Community similarity is visualized by site and sex (A, C) and by site and genotype (B, D) with non-metric multidimensional scaling (NMDS) of β diversity indices estimated by (A, B) Bray–Curtis dissimilarity and (C, D) weighted UniFrac dissimilarity. Red, St Regis Lake; blue, Franklin Bog; circles, female Sphagnum plants; triangles, male Sphagnum plants; letters, samples sharing a genotype; asterisks, samples with unique genotypes.
Table 6.
ANOVA for microbial communities associated with S. divinum and S. diabolicum at St Regis Lake, NY, and Franklin Bog, VT
| Bray–Curtis dissimilarity | Weighted UniFrac dissimilarity | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Degrees of freedom | SS | R 2 | F | P | SS | R 2 | F | P | |
| Site | 1 | 2.4069 | 0.0925 | 8.2739 | 0.001 | 0.2844 | 0.0829 | 7.3625 | 0.001 |
| Sex | 1 | 0.3389 | 0.0130 | 1.1650 | 0.186 | 0.0550 | 0.0160 | 1.4227 | 0.200 |
| Species | 1 | 0.3825 | 0.0147 | 1.3148 | 0.090 | 0.0256 | 0.0075 | 0.6639 | 0.726 |
| Residual | 77 | 22.399 | 0.8606 | 2.9747 | 0.8665 | ||||
SS, sums of squares. PERMANOVA significance tests of marginal effects were conducted using 1000 permutations.
In addition to community structure (see above), microbial taxon richness (the numbers of different ASVs per sample) differed significantly between the two sites, and also between male and female plants of S. divinum (Fig. 8). Sphagnum samples from St Regis Lake had, on average, 282 associated microbial taxa whereas Franklin Bog had 382 (P < 0.0003). Females of S. divinum across the two sites had, on average, 336 associated microbial ASVs whereas males had 248 (t = 3.823, P < 0.0004). The same pattern of sexual dimorphism in microbial taxon richness for S. divinum was evident at St Regis Lake (Fig. 8) alone (female mean = 303 microbial ASVs, male mean = 237 ASVs, t = 2.267, P < 0.03).
Fig. 8.
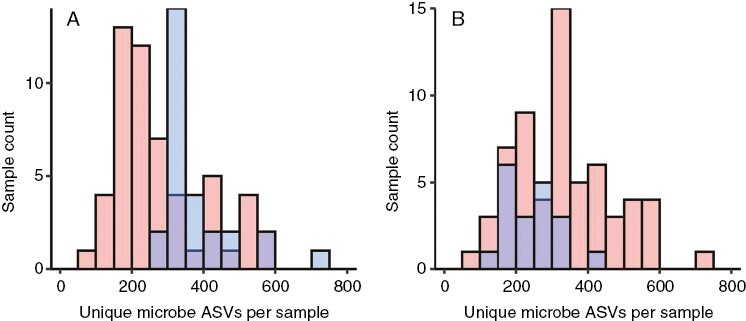
Microbial taxon richness associated with male and female gametophytes of S. divinum at St Regis Lake (NY) and Franklin Bog (VT). Histograms of (A) number of unique microbes (ASVs) per sample by site, and (B) number of unique microbe ASVs per sample by sex. In (A) red, St Regis Lake; blue, Franklin Bog. In (B) red, female; blue, male; purple, both males and females.
DISCUSSION
All four North American species in the S. magellanicum complex appear to reproduce via a mixture of sexual and asexual means. All the species produce sporophytes that result from sexual fusion, but our informal observations suggest that there can be much year-to-year variation in sporophyte/spore production. Summer of 2022 was a strong year for sporophyte production in New England, possibly because of a wet previous spring and summer. Precise details about sexual reproductive phenology for these species have not been described but, extrapolating from other Sphagnum species in northern latitude regions, we expect that male gametes are formed and dispersed in spring, summer, and/or fall (autumn), fertilization occurs at this time, and sporophytes mature the following summer. The majority of multi-locus genets in our study are represented by a single ramet, suggesting that sexual reproduction is the rule rather than the exception. Our observation that genetic similarities among individuals within sites are skewed towards highly similar individuals, more so than conspecific plants from different sites, suggests that closely related, sexually produced relatives at least sometimes establish within populations. Nevertheless, we cannot rule out somatic mutation as a significant factor in these haploid, potentially fragmenting gametophytes. Both male and female gametophytes occur in most populations. Sexual reproduction in Sphagnum was often considered relatively uncommon, partly because sporelings are rarely observed in natural populations and conditions for spore germination were thought to be restrictive. But in recent years the importance of sexuality and spore reproduction in Sphagnum has been documented (Sundberg, 2002; Sundberg and Rydin, 2002).
The degree of clonality across Sphagnum is variable both within and between species. A summary of published data (Table 7) from microsatellite variation shows that clonality versus sexual reproduction are in some cases a feature of populations and/or regional metapopulations rather than species. In S. subnitens, for example, plants in western North America are entirely clonal whereas plants in Europe are much more diverse. In S. cribrosum, some eastern North American populations are nearly or completely clonal whereas others are very diverse (Table 7). In S. angermannicum, eastern North American plants are highly diverse, although the species is uncommon, and in Europe it is restricted to Scandinavia and much more clonal. In the hornwort, Nothoceros aenigmaticus, all eastern North American populations appear to be strictly clonal, and both male-only and female-only populations occur (Alonso-García et al., 2020). Underlying reasons for intraspecific variation in clonality clearly include a historical component. Estimates of clonality in the four focal species of this study fall in the middle range, indicating a mix of clonal propagation and sexuality. We did not observe high levels of variation among populations or regions in these species.
Table 7.
Clonality in Sphagnum
| Species | Ramets | Genets | (G − 1)/(R − 1) | Reference |
|---|---|---|---|---|
| S. angermannicum (eastern North America) | 37 | 31 | 0.83 | Stenøien et al., 2011 |
| S. angermannicum (Scandinavia) | 118 | 19 | 0.15 | Stenøien et al., 2011 |
| S. carolinianum (eastern North America) | 61 | 37 | 0.60 | Duffy et al., 2022 |
| S. comosum (New Zealand) | 21 | 6 | 0.25 | Karlin et al., 2008 |
| S. cribrosum (FL8 population FL) | 7 | 5 | 0.67 | Johnson et al., 2012 |
| S. cribrosum (GA25 population GA) | 8 | 8 | 1.00 | Johnson et al., 2012 |
| S. cribrosum (GA30 population GA) | 11 | 11 | 1.00 | Johnson et al., 2012 |
| S. cribrosum (GA32 population GA) | 75 | 29 | 0.38 | Johnson et al., 2012 |
| S. cribrosum (Jones Lake ‘waveform’, NC) | 4 | 1 | 0.00 | Johnson et al., 2012 |
| S. cribrosum (Singletary Lake ‘normal’, NC) | 20 | 3 | 0.11 | Johnson et al. 2012 |
| S. cribrosum (Singletary Lake ‘waveform’, NC) | 25 | 2 | 0.04 | Johnson et al., 2012 |
| S. diabolicum | 101 | 61 | 0.60 | This study |
| S. divinum | 128 | 92 | 0.72 | This study |
| S. macrophyllum (eastern North America) | 149 | 65 | 0.43 | Johnson and Shaw, 2016 |
| S. macrophyllum (eastern North America) | 233 | 131 | 0.56 | Duffy et al., 2022 |
| S. magniae | 52 | 41 | 0.78 | This study |
| S. medium | 42 | 29 | 0.68 | This study |
| S. myabeanum (Asia, western North America) | 158 | 121 | 0.76 | Shaw et al., 2015 |
| S. novo-zelandicum (New Zealand) | 18 | 8 | 0.41 | Karlin et al., 2008 |
| S. pylaesii (eastern North America) | 110 | 67 | 0.61 | Duffy et al., 2022 |
| S. subnitens (Europe) | 18 | 15 | 0.82 | Karlin et al., 2010 |
| S. subnitens (New Zealand) | 31 | 2 | 0.03 | Karlin et al., 2010 |
| S. subnitens (western North America) | 22 | 1 | 0.00 | Karlin et al., 2010 |
| S. tumidulum (Reunion) | 48 | 31 | 0.64 | Liu et al., 2014 |
Dispersal of clones across long or even moderate distances appears to be limited. We detected no clones from multiple populations in our four focal species with the exception of one clone in S. magniae. For most genets where we detected multiple ramets they were limited to a single collection (‘handful’). At the Franklin Bog (VT) and St Regis Lake (NY) sites, where sufficient georeferenced samples were available, the physical occurrence of clonal ramets was significantly clumped. With this limited separation and dispersal of clones, the four species we studied fall between strictly ‘phalanx’ and ‘guerrilla’ patterns of clonality, but perhaps closer to the phalanx end of the spectrum.
The distribution of numbers of ramets per genet is an estimate of clonal fitness. Large genets that produce multiple ramets occupy more space and, in the case of female ramets, they can carry large numbers of spore-producing sporophytes. Large male genets potentially release more sperm. The distributions of ramet numbers per genet appeared similar across the four species – most genets were represented by a single ramet and a limited number of other genets were represented by 2–6 (–9) ramets within sites. The distributions of ramets per genet for S. divinum at the St Regis Lake (NY) site, where sampling was especially intensive, is very similar to the overall pattern for this species, so that seems to be general, both within sites and across the species overall. The variation in ramet numbers across genets can reflect differences in genet ages, growth rates or some combination.
Sex ratios in all four species appear to be biased, although the skew was not always significant. Nevertheless, the pattern seems to be consistent and biologically real. A significant male-biased sex ratio characterizes S. diabolicum, whereas the other three species have female-biased ratios, significantly so in S. divinum. Sphagnum diabolicum and S. magniae are closely related sister species and the two are in fact not reciprocally monophyletic across their genomes (Shaw et al., 2023), yet the direction of their sex ratio biases are reversed.
Most studies that have documented sex ratios in bryophytes have depended on sexually expressive gametophytes; i.e. those bearing male or female gametangia. Only a few have utilized molecular markers to determine the sexes of plants without gametangia, which allows determination of genetic sex ratios independent of sexual expression (e.g. Korpelainen et al., 2008; Bisang et al., 2020). Hedenäs and Bisang (2015) combined information about clonality and sexuality to derive estimates of genet sex ratios and found greater haplotypic diversity among males than females in the Swedish Archipelago. Many species of Sphagnum produce sporophytes (direct evidence of sexuality) regularly. Nevertheless, in their taxonomic treatment of Sphagnum for the Flora of North America project, McQueen and Andrus (2007) recognized 89 species for the continent and stated that sporophytes were ‘unknown’ or ‘not seen’ in 21 species, or almost 25 % of the flora. For at least 14 of these species, even gametophyte sexuality (unisexual versus hermaphroditic) was ‘unknown’. Male and female gametophytes are only distinguishable by gametangium production for limited times of the growing season and we did not gather phenotypic sexual data for taxa in the S. magellanicum complex.
We detected no distributional differences between male and female gametophytes in any species, or overall, in the complex. Bisang et al. (2020) found that sex ratios are less strongly female-biased within Drepanocladus lycopodioides in the Stockholm Archipelago than in other regions. In our study, there appears to be no correlation between sex ratio and climate region. A (statistically non-significant) female bias characterizes the warm-temperate/subtropical species, S. magniae, as in two of the northern species, S. divinum and S. medium. The three northern species (including S. diabolicum, with its male-biased sex ratios) commonly occur sympatrically within peatlands so any environmental determinants of sex ratio would have to be at a microenvironmental scale.
In contrast to seed plants, where male-biased sex ratios are common, most mosses that have unbalanced sex ratios are female-biased (Longton and Greene, 1969; Bisang and Hedenäs, 2005; Bisang et al., 2014). This is surprising because female gametophytes physically support, and to some extent nourish, attached sporophytic offspring (Proctor, 1977) and bear that energy cost. In a field study, female sporophyte-bearing stems of the moss Hylocomium splendens subsequently produced less growth and fewer branches than females without sporophytes (Rydgren and Økland, 2002). Rydgren et al. (2010) modelled population growth rates relative to sex ratios and the frequencies at which female gametophytes successfully bore sporophytic offspring. They found that, despite the cost of bearing sporophytes, if female genets without sporophytes produced more ramets than males, even though females with sporophytes produced the least numbers of ramets, a female-biased sex ratio could develop. We found stronger evidence of sex ratio biases at the ramet than genet level but attributed that mainly to smaller sample sizes; an explicit test failed to show any difference in ramet numbers between male and female genets within species or across the complex. Nevertheless, sex ratios are more biased at the ramet than at the genet levels.
Ecological differences between male and female gametophytes have been documented in both mosses and liverworts (Benassi et al., 2011; Blackstock, 2015; Pereira et al., 2016). Female plants of some moss and liverwort species appear to be more tolerant of desiccation than male plants, and this has sometimes been supported by experimental manipulations (Marks et al., 2016). In southern Sweden, female gametophytes of the moss Drepanocladus lycopodioides were more frequent in the wetter microsites within populations (Bisang et al., 2020). In the Mojave Desert, the moss Syntrichia caninervis rarely produces sporophytes and more than 50 % of ramets do not form gametangia, at least not regularly. Bowker et al. (2000) found that male gametophytes of S. caninervis were restricted to moister, shadier microsites whereas females occurred at both shadier and sunnier sites.
Sphagnum has a U/V sex chromosome system (Healey et al., 2023), which is expected to yield a 1:1 ratio of male and female spores. Nevertheless, differential mortality among spores can give rise to biased ratios at germination. Shaw and Gaughan (1993) and Norrell et al. (2014) grew single spore isolates of the moss Ceratodon purpureus. Both groups found that many individual sporophytes yielded unbalanced sex ratios of opposite direction among spore progeny isolated at the time of germination, although Shaw and Gaughan based that inference on sexually expressive plants whereas Norrell et al. used molecular markers to estimate sex ratios. Similar inconsistent sex ratio biases occur in natural populations (Shaw, 1993; Shaw and Gaughan, 1993). Slate et al. (2017) grew clonally propagated plants from three Ceratodon populations and found that males and females differ in growth, cell traits and photosynthesis, with females outperforming males. They nevertheless also found that natural populations were sometimes female-biased and in other cases male-biased.
Although the sex chromosomes in our four Sphagnum focal species are both very small (~25 % of a typical autosome) they have functional genes and an epistatic interaction between loci on the sex chromosome and autosomal loci impacts the growth response of another Sphagnum species, S. angustifolium, to microenvironmental pH (Healey et al., 2023). Similar ecologically important interactions involving sex chromosomes in the current species could underlie sex ratio variation in natural populations.
Our study of microbes associated with Sphagnum showed that two sites at St Regis Lake (Adirondack Mountains, NY) and Franklin Bog (VT) have strongly differentiated microbial communities, consistent with results from a broader survey of Sphagnum-associated prokaryotic microbes across North America (Kolton et al., 2022). In that study, site specificity was strong but (Sphagnum) species specificity was weak. We did not detect differences in microbial community makeup between genets or sexes within S. divinum at St Regis Lake, although the microbial communities differed in terms of both presence/absence of taxa and their relative frequencies between the two sites. We did, however, find that female gametophytes of S. divinum harboured significantly higher microbe taxon (ASV) richness than males, sometimes with up to three times the number of distinct microbial taxa. It is possible that differential taxon richness in associated microbes is related to the female sex bias in this species. If sex ratios are related to taxon richness of associated microbes, we can predict that females of S. medium and S. magniae could also have greater microbe diversity whereas S. diabolicum, with its male-biased sex ratio, might have greater microbe diversity associated with male plants. This prediction could be tested.
Bragina et al. (2012) detected species specificity in the microbial communities associated with S. magellanicum s. lat. versus S. fallax (subg. Cuspidata). It is not clear which of the current species their ‘S. magellanicum’ plants represent as the taxonomy of the complex had not been investigated at that time and all European plants were referred to S. magellanicum. It would have to be S. divinum or S. medium, as S. diabolicum and S. magniae are restricted to North America. Most components of the Sphagnum microbiome are not species-specific (Weston et al., 2018; Kolton et al., 2022). Nitrogen (N)-fixing bacteria provide N to their Sphagnum host plants (Lindo et al., 2013) and are important sources of N to peatland ecosystems (Kostka et al., 2016; Kolton et al., 2022). Warm-adapted microbial associates can facilitate positive responses to high temperatures in their Sphagnum hosts (Carrell et al., 2022). Sphagnum–microbe interactions comprise a ripe area for ecological research in peatlands.
Male and female conspecifics in sporophyte-dominant vascular plants can differ in many parameters of clonality, including ramet production, mortality patterns and micro-niche (McLetchie and Puterbaugh, 2000; Dering et al., 2015). Male and female plants may exhibit different patterns of local-scale genetic structure that is impacted by sex-specific modes of clonality. There is evidence that clonality can increase within-population spatial genetic structure across diverse tree species (Dering et al., 2015). In Populus alba, male and female trees differ in how clones spread within populations and males exhibit stronger spatial structure than females (Dering et al., 2015), but the two sexes do not differ in overall levels of genetic diversity. Comparable life history traits in Sphagnum that include clonality, sex ratio variation and sexual reproduction both reflect and determine evolutionary processes in peatlands.
Our study has both practical implications and is relevant to the ecology of Sphagnum and Sphagnum-dominated peatlands. These plants continue to be a focus for ecological and physiological experiments because of their global important in the carbon cycle. When designing experiments, it can often be advantageous to know something about clonal structure. Are plants that form a discrete hummock of limited size likely to be ramets of a single clone? Do plants sampled from scattered locations around a peatland reliably represent distinct genets? These questions are important if ‘replicate’ sampling is designed to minimize or maximize within-site variation.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following.
Figure S1: plot to infer sample sex from RADseq data or RADseq-like data generated from in silico digested genome sequences. Figure S2: maximum likelihood tree. Figure S3: neighbour-joining tree. Figure S4: maximum likelihood tree of samples from St Regis Lake, NY, and Franklin Bog, VT, with additional samples included to help identify the species clades. Figure S5: neighbour-joining tree of samples from St Regis Lake, NY, and Franklin Bog, VT, with additional samples included to help identify the species clades. Figure S6: distribution of pairwise geographic distances between Sphagnum ramets of the same genet and ramets of different genets from St Regis Lake, NY, and Franklin Bog, VT. Figure S7: genetic distance versus geographic distance for pairs of S. divinum ramets and genets at St Regis Lake, NY, and Franklin Bog, VT. Figure S8: distribution of pairwise geographic distances between Sphagnum ramets or genets from St Regis Lake, NY, and Franklin Bog, VT. Table S1: voucher table with collection information for Sphagnum samples included in RADseq analyses. Table S2: pairwise FST and genetic distance between collection sites of each species. Table S3: comparisons of distributions of pairwise genetic distance of Sphagnum samples within and between collection sites after removing inferred clones. Table S4: permutation tests of differences in ramets per genotype between sexes, sexes within species and between species. Table S5: tests of differences between mean pairwise geographic distances of ramets of the same genet and ramets of different genets from St Regis Lake, NY, and Franklin Bog, VT. Table S6: Mantel tests of isolation by distance for S. divinum ramets and genets at St Regis Lake, NY, and Franklin Bog, VT, based on 100 000 replicates. Table S7: tests of differences between mean pairwise geographic distances of female:female, male:female and male:male Sphagnum ramet and genet pairs from St Regis Lake, NY, and Franklin Bog, VT.
Contributor Information
A Jonathan Shaw, Department of Biology & L. E. Anderson Bryophyte Herbarium, Duke University, Durham, NC, 27708, USA.
Aaron M Duffy, Department of Biology & L. E. Anderson Bryophyte Herbarium, Duke University, Durham, NC, 27708, USA.
Marta Nieto-Lugilde, Department of Biology & L. E. Anderson Bryophyte Herbarium, Duke University, Durham, NC, 27708, USA.
Blanka Aguero, Department of Biology & L. E. Anderson Bryophyte Herbarium, Duke University, Durham, NC, 27708, USA.
Scott Schuette, Pennsylvania Natural Heritage Program, Western Pennsylvania Conservancy, Pittsburgh, PA, 15222, USA.
Sean Robinson, Department of Biology, SUNY Oneonta, Oneonta, NY, 13820, USA.
James Loveland, Department of Biology, Kenyon College, Gambier, OH 43022, USA.
Karen A Hicks, Department of Biology, Kenyon College, Gambier, OH 43022, USA.
David Weston, Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831, USA.
Bryan Piatkowski, Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831, USA.
Max Kolton, The Jacob Blaustein Institutes for Desert Research, Ben-Gurion University of the Negev, Midreshet Ben-Gurion, 8499000, Israel.
Joel E Koska, Center for Microbial Dynamics and Infection, Georgia Institute of Technology, Atlanta, GA 30332, USA.
Adam L Healey, HudsonAlpha Institute for Biotechnology, Huntsville, AL 35806, USA.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
FUNDING
This research was supported by U.S. National Science Foundation grants DEB-1737899 and DEB-1928514 (to A.J.S.).
DATA AVAILABILITY
The data underlying this article are available in Dryad at https://doi.org/10.5061/dryad.2ngf1vhv5.
Literature Cited
- Alonso-García M, Villarreal JC, McFarland K, Goffinet B.. 2020. Population genomics and phylogeography of a clonal bryophyte with spatially separated sexes and extreme sex ratios. Frontiers in Plant Science 11: 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/(25 August 2022, date last accessed).
- Baddeley A, Rubak E, Turner R.. 2015. Spatial point patterns: methodology and applications with R. London: Chapman and Hall/CRC Press, 2015. https://www.routledge.com/Spatial-Point-Patterns-Methodology-and-Applications-with-R/Baddeley-Rubak-Turner/p/book/9781482210200/. [Google Scholar]
- Barrett SCH. 2015. Influences of clonality on plant sexual reproduction. Proceedings of the National Academy of Sciences USA; 112: 8859–8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benassi M, Stark LR, Brinda JC, McLetchie DN, Bonine M, Mishler BD.. 2011. Plant size, sex expression and sexual reproduction along an elevation gradient in a desert moss. Bryologist 114: 277–288. doi: 10.1639/0007-2745-114.2.277. [DOI] [Google Scholar]
- Bisang I, Hedenäs L. 2005. Sex ratio patterns in dioicous bryophytes re-visited. Journal of Bryology 27: 207–219. [Google Scholar]
- Bisang I, Ehrlén J, Persson C, Hedenäs L. 2014. Family affiliation, sex ratio and sporophyte frequency in unisexual mosses. Botanical Journal of the Linnean Society 174: 163–172. [Google Scholar]
- Bisang I, Ehrlen J, Hedenas L.. 2020. Sex expression and genotypic sex ratio vary with region and environment in the wetland moss Drepanocladus lycopodioides. Botanical Journal of the Linnean Society 192: 421–434. [Google Scholar]
- Blackstock TH. 2015. Sex expression and sporophyte frequency in Frullania tamarisci (L.) Dumort. Journal of Bryology 37: 202–208. doi: 10.1179/1743282015y.0000000018. [DOI] [Google Scholar]
- Bowker MA, Stark LR, McLetchie DN, Mishler BD.. 2000. Sex expression, skewed sex ratios, and microhabitat distribution in the dioecious desert moss Syntrichia caninervis (Pottiaceae). American Journal of Botany 87: 517–526. [PubMed] [Google Scholar]
- Bragina A, Berg C, Cardinale M, Shcherbakov A, Chebotar V, Berg G.. 2012. Sphagnum mosses harbour highly specific bacterial diversity during their whole lifecycle. ISME Journal 6: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell AA, Lawrence TJ, Cabugao KGM, et al. 2022. Habitat-adapted microbial communities mediate Sphagnum peatmoss resilience to warming. New Phytologist 234: 2111–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correns C. 1899. Untersuchungen über die Vermehrung der Laubmoose durch Brutorgane und Stecklinge. Jena: Fischer. [Google Scholar]
- Cronberg N. 1996. Clonal structure and fertility in a sympatric population of the peat mosses Sphagnum rubellum and Sphagnum capillifolium. Canadian Journal of Botany 74: 1375–1385. doi: 10.1139/b96-167. [DOI] [Google Scholar]
- Dering M, Chybicki IJ, Rączka G.. 2015. Clonality as a driver of spatial genetic structure in populations of clonal tree species. Plant Research 128: 731–745. [DOI] [PubMed] [Google Scholar]
- Duffy A, Aguero B, Stenøien H, et al. 2020. Phylogenetic structure in the Sphagnum recurvum complex (Bryophyta: Sphagnaceae) relative to taxonomy and geography. American Journal of Botany 107: 1283–1295. [DOI] [PubMed] [Google Scholar]
- Duffy AD, Ricca M, Robinson S, et al. 2022. Heterogeneous genetic structure in eastern North American peat mosses (Sphagnum). Biological Journal of the Linnean Society 135: 692–707. [Google Scholar]
- Eaton DAR, Overcast I.. 2020. ipyrad: interactive assembly and analysis of RADseq datasets. Bioinformatics 36: 2592–2594. doi: 10.1093/bioinformatics/btz966. [DOI] [PubMed] [Google Scholar]
- La Farge C, Williams KH, England JH.. 2013. Regeneration of Little Ice Age bryophytes emerging from a polar glacier with implications of totipotency in extreme environments. Proceedings of the National Academy of Sciences of the USA 110: 9839–9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. 2005. hierfstat, a package for R to compute and test hierarchical F-statistics. Molecular Ecology Notes 5: 184–186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- Hassel K, Kyrkjeeide MO, Yousefi N, et al. 2018. Sphagnum divinum (sp. nov.) and S. medium Limpr. and their relationship to S. magellanicum Brid. Journal of Bryology 40: 197–222. [Google Scholar]
- Healey AL, Piatkowski BT, Lovell JT, et al. 2023. Newly identified sex chromosomes in the Sphagnum (peat moss) genome alter carbon sequestration and ecosystem dynamics. Nature Plants 9: 238–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenäs L, Bisang I.. 2015. Infraspecific diversity in a spore-dispersed species with limited distribution range. Systematics and Biodiversity 13: 17–27. [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MG, Shaw AJ.. 2015. Genetic diversity, sexual condition, and microhabitat preference determine mating patterns in Sphagnum (Sphagnaceae) peat-mosses. Biological Journal of the Linnean Society 115: 96–113. doi: 10.1111/bij.12497. [DOI] [Google Scholar]
- Johnson MG, Shaw AJ.. 2016. The effects of quantitative fecundity in the haploid stage on reproductive success and diploid fitness in the aquatic peat moss Sphagnum macrophyllum. Heredity 116: 523–530. doi: 10.1038/hdy.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MG, Shaw B, Zhou P, Shaw AJ.. 2012. Genetic analysis of the peatmoss Sphagnum cribrosum (Sphagnaceae) indicates independent origins of an extreme infraspecific morphology shift. Biological Journal of the Linnean Society 106: 137–153. doi: 10.1111/j.1095-8312.2012.01842.x. [DOI] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamvar ZN, Tabima JF, Grünwald NJ.. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2: e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin EF, Boles SB, Shaw AJ.. 2008. Systematics of Sphagnum section Sphagnum in New Zealand: a microsatellite-based analysis. New Zealand Journal of Botany 46: 105–118. doi: 10.1080/00288250809509758. [DOI] [Google Scholar]
- Karlin EF, Andrus RE, Boles SB, Shaw AJ.. 2010. One haploid parent contributes 100% of the gene pool for a widespread species in northwest North America. Molecular Ecology 20: 753–767. doi: 10.1111/j.1365-294x.2010.04982.x. [DOI] [PubMed] [Google Scholar]
- Kolton M, Weston DJ, Mayali X, et al. 2022. Defining the Sphagnum core microbiome across the North American continent reveals a central role for diazotrophic methanotrophs in the nitrogen and carbon cycles of boreal peatland ecosystems. Microbial Ecology 13: 1–17. [Google Scholar]
- Korpelainen H, Bisang I, Hedenäs L, Kolehmainen J.. 2008. The first sex-specific molecular marker discovered in the moss Pseudocalliergon trifarium. Journal of Heredity 99: 581–587. doi: 10.1093/jhered/esn036. [DOI] [PubMed] [Google Scholar]
- Kostka JE, Weston DJ, Glass JB, Lilleskov EA, Shaw AJ, Turetsky MR.. 2016. The Sphagnum microbiome: new insights from an ancient plant lineage. New Phytologist 211: 57–64. doi: 10.1111/nph.13993. [DOI] [PubMed] [Google Scholar]
- Lewis PO. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Systematic Biology 50: 913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- Lindo Z, Nilsson MC, Gundale MJ.. 2013. Bryophyte–cyanobacteria associations as regulators of the northern latitude carbon balance in response to global change. Global Change Biology 19: 2022–2035. doi: 10.1111/gcb.12175. [DOI] [PubMed] [Google Scholar]
- Liu YL, Ah-Peng C, Wilding N, et al. 2014. Population structure in the tropical peatmoss, Sphagnum tumidulum Besch. (Sphagnaceae). Bryologist 117: 329–335. [Google Scholar]
- Longton RE, Green SW.. 1969. The growth and reproductive cycle of Pleurozium schreberi (Brid.) Mitt. Annals of Botany 33: 83–105. [Google Scholar]
- Marks RA, Burton JF, McLetchie DN.. 2016. Sex differences and plasticity in dehydration tolerance: insight from a tropical liverwort. Annals of Botany 118: 347–356. doi: 10.1093/aob/mcw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLetchie DN, Puterbaugh MN.. 2000. Population sex ratios, sex-specific clonal traits and tradeoffs among these traits in the liverwort Marchantia inflexa. Oikos 90: 227–237. doi: 10.1034/j.1600-0706.2000.900203.x. [DOI] [Google Scholar]
- McMurdie PJ, Holmes S.. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen CB, Andrus RE.. 2007. Sphagnaceae. In: Flora of North America Editorial Committee, ed. Bryophytes: mosses, part 1. Flora of North America, Vol. 27. New York: Oxford University Press, 45–101. [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, et al. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrell TE, Jones KS, Payton AC, McDaniel SF.. 2014. Meiotic sex ratio variation in natural populations of Ceratodon purpureus (Ditrichaceae). American Journal of Botany 101: 1572–1576. doi: 10.3732/ajb.1400156. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Simpson G, Blanchet F, et al. 2022. vegan: community ecology package. R package version 2.6-4. https://CRAN.R-project.org/package=vegan. [Google Scholar]
- Paradis E, Schliep K.. 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35: 526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Parchman TL, Gompert Z, Mudge J, Schilkey FD, Benkman CW, Buerkle CA.. 2012. Genome-wide association genetics of an adaptive trait in lodgepole pine. Molecular Ecology 21: 2991–3005. doi: 10.1111/j.1365-294X.2012.05513.x. [DOI] [PubMed] [Google Scholar]
- Pereira MR, Dambros CS, Zartman CE.. 2016. Prezygotic resource-allocation dynamics and reproductive trade-offs in Calymperaceae (Bryophyta). American Journal of Botany 103: 1838–1846. doi: 10.3732/ajb.1600240. [DOI] [PubMed] [Google Scholar]
- Proctor MCF. 1977. Evidence on the carbon nutrition of moss sporophytes from 14CO2 uptake and the subsequent movement of labelled assimilate. Journal of Bryology 9: 375–386. doi: 10.1179/jbr.1977.9.3.375. [DOI] [Google Scholar]
- R Core Team. 2021. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. https://www.R-project.org/. [Google Scholar]
- Rydgren K, Økland RH.. 2002. Ultimate costs of sexual reproduction in the clonal moss Hylocomium splendens. Ecology 83: 1573–1579. doi: 10.1890/0012-9658(2002)083[1573:ucospi]2.0.co;2. [DOI] [Google Scholar]
- Rydgren K, Halvorsen R, Cronberg N. 2010. Infrequent sporophyte production maintains a female-biased sex ratio in the unisexual clonal moss Hylocomium splendens. Journal of Ecology 98: 1224–1231. [Google Scholar]
- Shaw J. 1986. A new approach to the experimental propagation of bryophytes. Taxon 35: 671–675. doi: 10.2307/1221609. [DOI] [Google Scholar]
- Shaw AJ. 1993. Control of sex ratios in haploid populations of the moss, Ceratodon purpureus. American Journal of Botany; 80: 584–591. [DOI] [PubMed] [Google Scholar]
- Shaw AJ, Gaughan JF.. 1993. Control of sex ratios in haploid populations of the moss, Ceratodon purpureus. American Journal of Botany 80: 584–591. doi: 10.1002/j.1537-2197.1993.tb13844.x. [DOI] [PubMed] [Google Scholar]
- Shaw AJ, Piatkowski BT, Duffy A, et al. 2022. Phylogenomic structure and speciation in an emerging model: the Sphagnum magellanicum complex (Bryophyta). New Phytologist 136: 1497–1512. doi: 10.1111/nph.18429. [DOI] [PubMed] [Google Scholar]
- Shaw AJ, Nieto-Lugilde M, Aguero B, et al. 2023. Sphagnum diabolicum sp. nov. and S. magniae sp. nov.; morphological variation and taxonomy of the ‘S. magellanicum complex’. Bryologist 126: 69–89. [Google Scholar]
- Slate ML, Rosenstiel TN, Eppley SM.. 2017. Sex-specific morphological and physiological differences in the moss Ceratodon purpureus (Dicranales). Annals of Botany 120: 845–854. doi: 10.1093/aob/mcx071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenøien HK, Shaw AJ, Shaw B, Hassel K, Gunnarsson U.. 2011. North American origin and recent European establishment of the amphi-Atlantic peat moss: Sphagnum angermannicum. Evolution 65: 1181–1194. doi: 10.1111/j.1558-5646.2010.01191.x. [DOI] [PubMed] [Google Scholar]
- Sundberg S. 2002. Sporophyte production and spore dispersal phenology in Sphagnum: the importance of summer moisture and patch characteristics. Canadian Journal of Botany 80: 543–556. doi: 10.1139/b02-060. [DOI] [Google Scholar]
- Sundberg S, Rydin H. 2000. Experimental evidence for a persistent spore bank in Sphagnum. New Phytologist 148: 105–116. [DOI] [PubMed] [Google Scholar]
- Sundberg S, Rydin H.. 2002. Habitat requirements for establishment of Sphagnum from spores. Journal of Ecology 90: 268–278. doi: 10.1046/j.1365-2745.2001.00653.x. [DOI] [Google Scholar]
- Weston DJ, Turetsky MR, Johnson MG, et al. 2018. The Sphagnome Project: enabling ecological and evolutionary insights through a genus-level sequencing project. New Phytologist 217: 16–25. [DOI] [PubMed] [Google Scholar]
- Yousefi N, Hassel K, Flatberg KI, et al. 2017. Divergent evolution and niche differentiation within the common peatmoss Sphagnum magellanicum. American Journal of Botany 104: 1060–1072. doi: 10.3732/ajb.1700163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in Dryad at https://doi.org/10.5061/dryad.2ngf1vhv5.


