Abstract
Background and Aims
Wind pollination has evolved repeatedly in flowering plants, yet the identification of a wind pollination syndrome as a set of integrated floral traits can be elusive. Thalictrum (Ranunculaceae) comprises temperate perennial herbs that have transitioned repeatedly from insect to wind pollination while also exhibiting mixed pollination, providing an ideal system to test for evolutionary correlation between floral morphology and pollination mode in a biotic to abiotic continuum. Moreover, the lack of floral organ fusion across this genus allows testing for specialization to pollination vectors in the absence of this feature.
Methods
We expanded phylogenetic sampling in the genus from a previous study using six chloroplast loci, which allowed us to test whether species cluster into distinct pollination syndromes based on floral morphology. We then used multivariate analyses on floral traits followed by ancestral state reconstruction of the emerging flower morphotypes and determined whether these traits are evolutionarily correlated under a Bayesian framework with Brownian motion.
Key Results
Floral traits fell into five distinct clusters, which were reduced to three after considering phylogenetic relatedness and were largely consistent with flower morphotypes and associated pollination vectors. Multivariate evolutionary analyses found a positive correlation between the lengths of floral reproductive structures (styles, stigmas, filaments and anthers). Shorter reproductive structures tracked insect-pollinated species and clades in the phylogeny, whereas longer structures tracked wind-pollinated ones, consistent with selective pressures exerted by biotic vs. abiotic pollination vectors, respectively.
Conclusions
Although detectable suites of integrated floral traits across Thalictrum were correlated with wind or insect pollination at the extremes of the morphospace distribution, a presumed intermediate, mixed pollination mode morphospace was also detected. Thus, our data broadly support the existence of detectable flower morphotypes from convergent evolution underlying the evolution of pollination mode in Thalictrum, presumably via different paths from an ancestral mixed pollination state.
Keywords: Ambophily, anemophily, entomophily, evolutionary correlation, flower morphology, integration, multivariate Brownian motion, phylogenetic comparative methods, pollination syndrome, pollination mode, Thalictrum (Ranunculaceae), wind pollination
INTRODUCTION
Pollination mode, a key life-history feature of seed plants, refers to the process by which pollen is transferred between male (anthers) and female (stigma) reproductive structures, which can occur by proximity (selfing) or via biotic or abiotic agents. Multiple aspects of floral diversity are shaped by selective pressure exerted by pollinating agents, e.g. flower shape (Smith and Kriebel, 2018), flower size and floral display (Parachnowitsch and Kessler, 2010), and nectar spur length (Whittall and Hodges, 2007). Convergent evolution on the same type of pollinator can result in an analogous suite of floral morphologies, or pollination syndrome (Fenster et al., 2004). Theoretical and empirical evidence suggests that pollinator selection can act on multiple organs within a flower (Stebbins, 1951; Fenster et al., 2015). Alternatively, selection on one floral organ can impact others owing to developmental correlation (e.g. genetic linkage, pleiotropy or structural constraint; Smith, 2016). Either of these scenarios results in evolutionary integration, whereby structures evolve in a correlated fashion within a flower under the selection pressure exerted by pollinators (Berg, 1960).
A special case of evolutionary integration within flowering plants is synorganization, whereby floral organs function as a morphological unit owing to whorled phyllotaxis and fusion, most commonly syncarpy and sympetaly (Endress, 2016). For example, petals are fused into a corolla tube (sympetaly) in many flowering plants, with variation in corolla tube size and shape emerging from specialized pollination modes. To date, evolutionary correlation between flower organs (as a proxy for integration) has been studied only in such flowers, with synorganization arising from organ fusion (Lagomarsino et al., 2017; Joly et al., 2018; Smith and Kriebel, 2018; Dellinger et al., 2019; Kriebel et al., 2020).
Here, we test whether evolutionary integration of floral organs can occur in the absence of whorled phyllotaxis and organ fusion in a non-core eudicot lineage with a variable floral ground plan (Kitazawa and Fujimoto, 2014; Kitazawa, 2021). Thalictrum (Ranunculaceae) consists of ~200 species that display variation in pollination mode, sexual system and ploidy level (Tamura, 1995). The genus lacks any form of floral organ fusion or whorled phyllotaxy (whereby organ primordia of the same kind develop synchronously), arising instead in a spiral or irregular whorl. Despite this absence of preconditions for synorganization (Endress, 2016; Phillips et al., 2020) in its floral ground plan, distinct suites of floral characters associated with different pollinating agents can be identified in Thalictrum: generalist insect pollination (entomophily); more specialized wind pollination (anemophily) that evolved at least eight times (Wang et al., 2019); and pollination by both insects and wind (ambophily), hypothesized as either an evolutionarily intermediate step or a stable state (Culley et al., 2002).
Charles Darwin (1862) famously drew on evidence of extremely long nectar spurs in the Madagascan orchid Angraecum sesquipedale to predict correctly a hawkmoth pollinator with an equally long proboscis. Yet, the predictive value of floral morphology varies widely and can be case specific, and the applicability of this concept across angiosperms is contentious at best (Ollerton et al., 2009, 2015; Rosas-Guerrero et al., 2014). Establishing the pollination mode of a species is a time-consuming task that requires the study of plants in their natural environment, and although this remains the golden standard, statistical methods can help to identify predictive morphologies by generating a training dataset from a subsample of species with known pollination mode (Lagomarsino et al., 2017; Dellinger et al., 2019; van der Niet, 2021). Such an approach could, in turn, facilitate macroevolutionary analyses requiring large datasets to investigate further the mechanisms underlying the evolution of different modes of pollination. Ecological studies of pollination mode in natural populations have been conducted for 13 Thalictrum species (Kaplan and Mulcahy, 1971; Melampy and Hayworth, 1980; Davis, 1997; Steven and Waller, 2004; Guzmán, 2005; Humphrey, 2018), and a ‘pollination index’ (PI) was devised to predict the pollination mode from morphology in the absence of field data (Kaplan and Mulcahy, 1971). This PI involves qualitatively scoring followed by averaging seven floral characters considered indicative of pollination syndrome: flower colour, flower size, anther and stigma length, filament orientation, and stamen and pistil exsertion. A PI value of one is assigned to anemophily, three to entomophily, and two to ambophily (Kaplan and Mulcahy, 1971). Here, we build upon and refine this PI using quantitative traits and phylogenetic comparative methods to increase the predictive value of flower morphology in assessing pollination mode.
Our aims were as follows: (1) to investigate the predictive power of continuous floral traits in distinguishing pollination mode within an improved phylogenetic context in Thalictrum (Ranunculaceae); (2) to reconstruct the evolution of flower morphotypes in the genus; and (3) to assess the degree of evolutionary correlation (integration) between floral traits in a phylogenetic context. To that end, we first inferred a chronogram with increased taxon and molecular sampling that constituted the framework of our phylogenetic comparative methods. We then tested the predictive value of floral morphology in assigning pollination mode by principal component analysis (PCA) and k-means clustering and reconstructed the evolution of the resulting flower morphotypes. Lastly, to identify suites of correlated floral traits that might be contributing to pollination syndromes, we characterized the degree of evolutionary integration between floral organs using multivariate Brownian motion models in a Bayesian framework.
MATERIALS AND METHODS
Taxon sampling
Ninety-nine taxa were sampled (Appendix), comprising 93 recognized species that spanned all 14 currently recognized sections of Thalictrum (Tamura, 1995) and the geographical distribution and morphological diversity of the group. Aquilegia buergeriana var. oxysepala, A. formosa, Enemion raddeanum, Isopyrum manshuricum, Leptopyrum fumarioides, Semiaquilegia adoxoides and Paraquilegia microphylla were chosen as outgroups based on previous studies (Park et al., 2015; Wang et al., 2019).
DNA extraction, amplification and sequencing
Total genomic DNA was isolated from fresh leaves or herbarium specimens as described by Park et al. (2015) and Soza et al. (2012). Six plastid regions [ndhA intron, ndhF, rbcL, trnL intron, trnL-F intergenic spacer (IGS) and rpl32-trnL IGS] were amplified using primers described by Shaw et al. (2007) or designed for this study (Supplementary data Table S1). The PCR was performed in 30 µL, including DiaStar-Taq DNA polymerase (SolGent Co., Daejeon, Korea) or GoTaq Master Mix (Promega, Madison, WI, USA) and 1 µL of genomic DNA (2–50 ng). Cycling conditions were as follows: 95 ℃ for 2–3 min; 30–40 cycles of 95 ℃ for 30–60 s, 46–60 ℃ for 40–60 s and 72 ℃ for 50–150 s; and 72 ℃ for 5 min. Sequencing of PCR products was performed at SolGent Co. or Genewiz (Seattle, WA, USA).
Phylogenetic analyses
For each plastid region, raw sequences were assembled into contigs, and consensus sequences were aligned with MUSCLE (Edgar, 2004) in Geneious R7. Phylogenetic reconstructions were performed on a concatenated alignment of six plastid regions. Maximum likelihood (ML) analyses were performed using IQ-TREE v.1.6.8 (Nguyen et al., 2015) under best-fitting partition schemes from ModelFinder (Kalyaanamoorthy et al. 2017; Supplementary data Table S2). Branch support came from 1000 ultrafast bootstrap (BS) replicates. Bayesian inference (BI) was conducted using MrBayes v.3.2 (Ronquist et al., 2012), with two runs of Markov chain Monte Carlo (MCMC) for 12 000 000 generations each and trees sampled every 100 generations. The model of molecular evolution was selected with the corrected Akaike information criterion (AICc) (Hurvich and Tsai, 1989) using jModelTest v.2.1.10 (Posada, 2008). MCMC convergence was assessed from the effective sampling size (ESS) of the combined runs. All parameter estimates had an ESS > 1000 after burn-in (first 25 % of generations discarded), indicating that the analyses had sampled the posterior distributions satisfactorily. The posterior probability (PP) of branches was estimated from the 50 % majority-rule consensus tree.
Divergence time estimation
Divergence times and topology were estimated jointly using a Bayesian MCMC method in BEAST v.2.5.2 (Drummond et al., 2012). The dataset was partitioned by each chloroplast region and its optimal model. We used a relaxed clock model (Drummond et al., 2006) and a Yule process of speciation as a tree prior. A secondary calibration point, based on a published divergence time estimate for the genus Thalictrum from a densely sampled Ranunculaceae chronogram (Fior et al., 2013), was used for the root age constraint with a normal prior distribution (mean = 26.2, s.d. = 3.6 and range = 20.3–32.3 Mya). Analyses were run for 100 000 000 generations, sampling every 1000 generations. The posterior distribution of all parameters was examined in Tracer v.1.7 (Rambaut et al., 2018). The ESS was >1000 after 10 % of samples were discarded as burn-in. A maximum clade credibility (MCC) chronogram was generated with TreeAnnotator v.1.7.1 (Drummond et al., 2012), showing mean divergence time estimates with 95 % highest posterior density intervals.
Flower morphology
We measured 17 floral characters from 29 species (including 10 of 13 taxa with empirical pollination mode data) from flatbed scans (Epson Perfection v.39) of fresh flowers (Supplementary data Fig. S1). Plants were grown in the University of Washington Greenhouse from wild-collected seed or purchased as adult plants from nurseries (Supplementary data Table S3). Flowers were scanned after dissecting their sepals, when anthers were beginning to dehisce in the outer stamens and before fertilization. An area of 100 cm × 50 cm was scanned at 1200–2400 dpi with a ruler for scale. The straight line or segmented line tool in ImageJ (Schneider et al., 2012) was used to measure three each of the following traits per flower (in millimetres): sepal length, sepal width, filament length, maximum filament width, minimum filament width, anther length (including mucron when present, an extension of sterile tissue at the anther apex), anther width, stigma length, stigma width at base, stigma width at apex, style length, style width at base, ovary length, ovary width, and gynophore length (when present, a stalk that elevates the gynoecium). To account for intraspecific variation, at least three organs of each type (sepals, stamens and carpels) per flower and three flowers from two to three plants per species and per sex (for dioecious and monoecious species) were measured. Missing organs were assigned a value of zero. Thalictrum revolutum female flowers and T. pubescens were measured from herbarium specimens and fixed specimens, respectively.
Pollination index
Pollination index data were compiled from the literature (Kaplan and Mulcahy, 1971; Soza et al., 2012, 2013; Wang et al., 2019) or newly calculated from images (Global Biodiversity Information Facility) for a total of 83 species.
Multivariate analysis of floral traits
Trait averages per flower (excluding count data) were log-transformed, adding one to zeros. Analyses were performed with base R functions (R Core Team, 2018), unless specified otherwise. PCA was performed on raw, replicate continuous trait values (without species averages) and on the PI dataset (on species averages). K-means clustering analysis was performed with 10 000 iterations, and the optimal value of k (the number of specified groups, or clusters) was determined with NbClust (Charrad et al., 2014) using majority-rule criteria from 30 goodness-of-fit metrics. Screen plot identified the number of principal components (PCs) including ≥80 % of the total variance (Jolliffe, 2002). Ten species with validated pollination mode were used to assign the type of pollination within their k-means cluster (Table 1). Sexual dimorphism in sepal size was tested with one-way ANOVA.
Table 1.
Flower morphotype and pollination mode assignment from multivariate analysis of floral traits in Thalictrum (Ranunculaceae)
| Species | Flower morphotype k = 5 |
Pollination mode from k-means |
Pollination index |
|---|---|---|---|
| T. thalictroides a | Petaloid sepal | Insect | 3.00 |
| T. aquilegiifolium d | Showy stamen | Insect | 2.71 |
| T. delavayi* | Petaloid sepal | Insect | 2.71 |
| T. rochebrunianum | Petaloid sepal | Insect | 2.71 |
| T. clavatum b | Showy stamen | Insect | 2.57 |
| T. ichangense | Showy stamen | Insect | 2.57 |
| T. petaloideum* | Showy stamen | Insect | 2.57 |
| T. tuberiferum | Showy stamen | Insect | 2.57 |
| T. coreanum | Showy stamen | Insect | 2.43 |
| T. lucidum | Intermediate | Wind/ambophily/insect | 2.43 |
| T. flavum d | Intermediate | Wind/ambophily/insect | 2.29 |
| T. omeiense | Intermediate | Wind/ambophily/insect | 2.29 |
| T. uchiyamae | Showy stamen | Insect | 2.29 |
| T. kiusianum | Showy stamen | Insect | 2.14 |
| T. actaeifolium | Intermediate | Wind/ambophily/insect | 2.00 |
| T. alpinum c | Intermediate | Wind/ambophily/insect | 2.00 |
| T. pubescens (S)a | Staminate | Wind | 2.00 |
| T. pubescens (H)a | Intermediate | Wind/ambophily/insect | |
| T. elegans | Intermediate | Wind/ambophily/insect | 1.86 |
| T. foetidum | Intermediate | Wind/ambophily/insect | 1.86 |
| T. isopyroides | Intermediate | Wind/ambophily/insect | 1.86 |
| T. minus d | Intermediate | Wind/ambophily/insect | 1.86 |
| T. simplex | Intermediate | Wind/ambophily/insect | 1.71 |
| T. guatemalense (S) | Staminate | Wind | 1.43 |
| T. guatemalense (H) | Intermediate | Wind/ambophily/insect | |
| T. revolutum (S)a | Staminate | Wind | 1.29 |
| T. revolutum (C)a | Carpellate | ||
| T. dasycarpum (S) | Staminate | Wind | 1.14 |
| T. dasycarpum (C) | Carpellate | ||
| T. dioicum (S)c | Staminate | Wind | 1.14 |
| T. dioicum (C)c | Carpellate | ||
| T. occidentale (S) | Staminate | Wind | 1.14 |
| T. occidentale (C) | Carpellate | ||
| T. fendleri (S)c | Staminate | Wind | 1.00 |
| T. fendleri (C)c | Carpellate | ||
| T. hernandezii (S) | Staminate | Wind | 1.00 |
| T. hernandezii (H) | Intermediate | Wind/ambophily/insect |
Field-validated pollination modes are shown in bold (aKaplan and Mulcahy, 1971; bMelampy and Hayworth, 1980; cSteven and Waller, 2004; dGuzmán, 2005).
k-means assignments (k = 5 from PCA) provide new, more defined PI threshold values (thick lines): PI = 1.00–1.29 for wind pollination and PI = 2.57–3.00 for insect pollination. Intermediate values are unresolved. Estimates of PI are from Kaplan and Mulcahy (1971), Soza et al. (2012), Wang et al. (2019) and this study.
*Species with flowers split into more than one cluster were classified by the majority cluster (six of nine for T. delavayi and five of six for T. petaloideum).
For dioecious and andromonoecious species: C, carpellate; H, hermaphroditic; and S, staminate.
Phylogenetic comparative methods
Phylogenetic PCA.
The estimated chronogram from above was trimmed to the 29 taxa with floral trait data using phytools (Revell, 2012). All species were monophyletic on the trimmed phylogeny except T. aquilegiifolium; we chose accession Kawahara & al. 666 [TI] to represent this species. Phylogenetic PCA (pPCA) was performed under a model of Brownian motion and lambda with the phyl.pca function (Revell, 2009) in phytools (Revell, 2012).
Ancestral state reconstruction of flower morphology.
The ancestral flower morphotype was inferred by fitting a continuous-time Markov model of discrete character evolution in corHMM (Boyko and Beaulieu, 2021). ‘Equal rates’, ‘symmetrical rates’ and ‘all rates different’ models under ‘equal’, ‘empirical’ and ‘stationary’ root priors (nine models total) were fitted using the rayDISC function. The AICc was used to select the best-fitting model. To estimate the number of transitions between character states, we implemented stochastic character mapping with make.simmap in phtyools to simulate 1000 character histories under the best-fitting model (Revell, 2012). Two cases where different flowers from the same species did not fall within the same cluster (T. delavayi and T. petaloideum) were scored based on the cluster with majority representation.
Evolutionary correlation of floral traits.
We aimed to identify statistical correlations among floral traits, accounting for within-species variation and phylogenetic relationships while accommodating data missing because of sexual system variation. To that end, we inferred the evolutionary rate matrix under a model of multivariate Brownian motion (Revell and Harmon, 2008). This matrix describes both the rates of evolution of individual traits and the evolutionary covariance (correlation coefficient) between pairs of traits, allowing us to test whether pairs of traits are evolutionarily correlated or independent of each other. We fitted a multivariate Brownian motion model of correlated evolution for all pairwise combinations of traits using the R package MCMCglmm (Hadfield, 2010), with parameter-expanded uninformative priors. In addition to direct measurements, we included three composite traits: stamen length (filament length + anther length), carpel length (ovary length + style length) and anther size (anther length × anther width). Given that stigmatic papillae mostly run along the style in Thalictrum and the two measurements often coincide, we chose to use style length (together with ovary length) for the carpel length estimate. Gaussian distributions (Brownian motion) and Poisson distributions were used to model the evolution of continuous traits and flower organ counts, respectively. Three MCMC chains were run for 100 000 iterations until convergence (ESS > 200 for all parameters), as calculated by CODA (Plummer et al., 2006). Correlation coefficients that did not overlap with zero were considered significant (Harmon, 2018). The 95 % confidence interval (CI) for posterior distributions was calculated with LaplacesDemon (Statisticat, 2021). Maximum a posteriori estimates from RevGadgets were used to summarize posterior distributions (Tribble et al., 2022). Plots were generated with ggplot2, Cowplot and ggtree (Wickham, 2011; Yu et al., 2017; Wilke, 2020).
We tested whether floral traits showed evolutionary correlation for all pairwise combinations (except gynophore length, present in 9 of 29 species, hence not amenable to this type of analysis). Dioecious taxa have sepal data from staminate and carpellate flowers, whereas andromonoecious taxa (staminate and hermaphroditic flowers on the same plant) and cryptically dioecious T. pubescens (male-sterile hermaphroditic flowers and staminate flowers on separate plants) have sepal and stamen data from staminate and hermaphroditic flowers. To account for this intraspecific flower dimorphism, two types of analyses were performed: on reproductive organs (stamens and carpels) and on all organs (including sepals). For reproductive organs, we combined stamen and carpel measurements from different flowers (dioecious taxa) or averaged stamen measurements between male and hermaphroditic flowers (andromonoecious taxa). This enabled the computation of phylogenetic correlations between stamen and carpel traits found on separate flowers within a species. For all floral organs (perianth included), we divided the data into a ‘carpellate dataset’ (carpellate + hermaphroditic flowers) and a ‘staminate dataset’ (staminate + hermaphroditic flowers), with stamen and sepal averages across flower types for andromonoecious species. This analysis allowed us to infer evolutionary correlations between the perianth and reproductive organs, while also accounting for sexual dimorphism of sepals.
RESULTS
An expanded phylogeny for Thalictrum
The current phylogeny has the most comprehensive sampling to date for the genus, 93 of 196 species (47 % taxon coverage), using six concatenated chloroplast regions to improve resolution and provide support along the backbone, with all but one node being strongly supported (i.e. BS > 75 % and PP ≥ 0.95) (Supplementary data Figs S2, S3). Consistent with prior analyses (Soza et al., 2012, 2013; Wang et al., 2019), the present phylogeny identified two major clades (I and II) in the genus and three strongly supported subclades in clade II with divergent sexual systems: one consisting mostly of andromonoecious species (except for hermaphroditic T. decipiens, renamed subclade A) and two consisting of dioecious species (renamed subclades B and C).
The plastid dataset was subsequently used to reconstruct a chronogram with Bayesian divergence estimation and one calibration point (Fig. 1), which was implemented in subsequent analyses. Resulting divergence estimates that coincided with previously supported clades were within the range of previous estimates (Soza et al., 2013): a crown age of 8.9–21.8 Mya for Thalictrum, 4.8–13.4 Mya for clade I, 6.6–16.9 Mya for clade II, 1.6–5.3 Mya for clade A, 0.9–2.7 Mya for clade B, and 0.9–3.4 Mya for clade C.
Fig. 1.
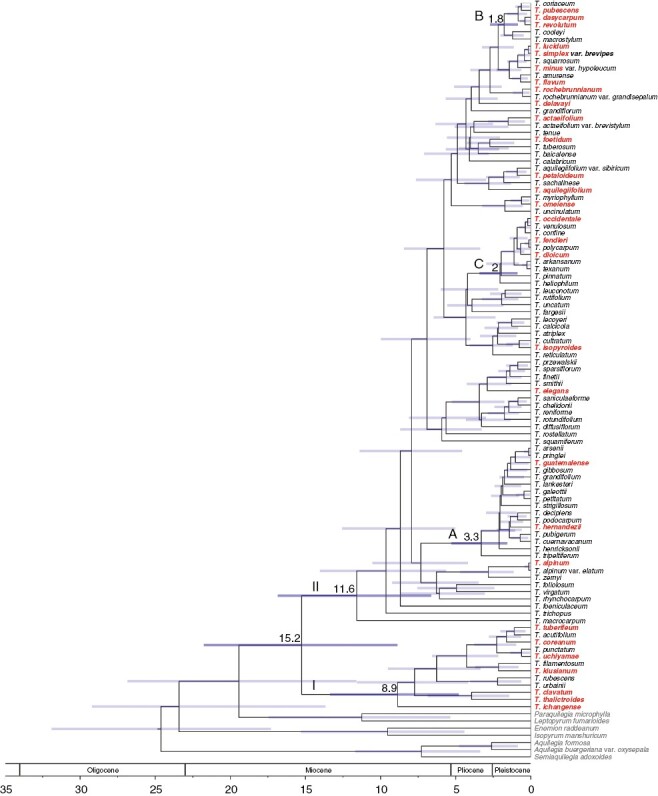
Thalictrum (Ranunculaceae) chronogram, showing Bayesian phylogeny and divergence time estimates for 99 Thalictrum taxa and seven outgroups based on the analysis of six combined plastid regions in BEAST. Estimated mean ages (for main clades) and 95 % highest posterior density intervals are shown at nodes. Two major clades, I and II, and three subclades with andromonoecious (A) and dioecious (B and C) members are indicated. Taxa used for flower trait analyses are shown in red. Geological epochs are after Walker et al. (2018).
Distinct flower morphotypes are associated with different pollination modes
To address whether suites of integrated floral traits segregate species by pollination mode, we sampled continuous floral traits across the genus, representing all major clades and floral morphotypes. Thalictrum flowers exhibited a wide range of variation in flower morphology that included a >3-fold difference in the sum of all trait values, a potential proxy for floral size (Supplementary data Fig. S4A). Within-species variation was also present in dioecious and andromonoecious taxa as sexual dimorphism of sepals, which were larger (longer and/or wider) in staminate flowers compared with carpellate or hermaphroditic flowers (Supplementary data Fig. S4B), a relationship previously reported for other unisexual wind-pollinated species, including T. dioicum (Delph et al., 1996).
The first four PCs in multivariate analyses explained the majority of the total variance (83.23 % combined; Supplementary data Fig. S5A), resulting in five k-means clusters (Supplementary data Fig. S5B) best visualized in PC1 vs. PC3 (Fig. 2A; Supplementary data Fig. S5C, D). PC1 segregated the data into distinct carpellate (green, left), hermaphroditic (grey + blue + pink, centre) and staminate (yellow, right) clusters (Fig. 2A; Supplementary data Video S1), with predominant contributions from carpel and stamen traits (left- and right-pointing biplot vectors, respectively; Fig. 2B) and was therefore interpreted as a sexual system axis. PC2 segregated flowers that had a higher sum of all trait values from those with a lower sum of all trait values, a potential proxy for flower size (Supplementary data Fig. S5C, E, biplot vectors pointing down). PC3 further separated the three hermaphroditic clusters, distinguishing flowers with larger petaloid sepals and narrower (filiform to weakly dilated, i.e. wider at the top) stamen filaments (e.g. T. thalictroides; Fig. 2A, pink cluster) from those with smaller, early deciduous sepals and wider (strongly dilated or clavate) stamen filaments, resulting in showier stamens, and carpels elevated on gynophores (e.g. T. aquilegiifolium; Fig. 2A, grey cluster; and Fig. 2B, biplot vectors for sepal width and length, filament width and gynophore length). Finally, PC4 separated species with shorter reproductive organs and more sepals, such as T. thalictroides, from those with longer reproductive organs and fewer sepals, such as T. hernandezii (Supplementary data Fig. S5D, F, biplot vector for style, stigma, anther and filament length). In summary, hermaphroditic flowers fell into three morphotypes: ‘petaloid sepals’, ‘showy stamens’ and ‘intermediate’ with small white sepals, white filiform or weakly dilated stamen filaments and yellow anthers (e.g. T. lucidum; Fig. 2A, blue cluster). We called this morph ‘intermediate’ because, although none of these flowers has large petaloid sepals or strongly dilated filaments, their white and weakly dilated filaments and non-green sepals make them potentially attractive to insects. Hermaphroditic flowers of sexually dimorphic taxa, including cryptically dioecious T. pubescens (Kaplan and Mulcahy, 1971; Davis, 1997) and andromonoecious T. hernandezii and T. guatemalense, were found at the boundary of the intermediate cluster (Fig. 2A, encircled blue symbols). Likewise, staminate flowers of T. pubescens were found on the edge of the staminate cluster (Fig. 2A, encircled yellow symbols).
Fig. 2.
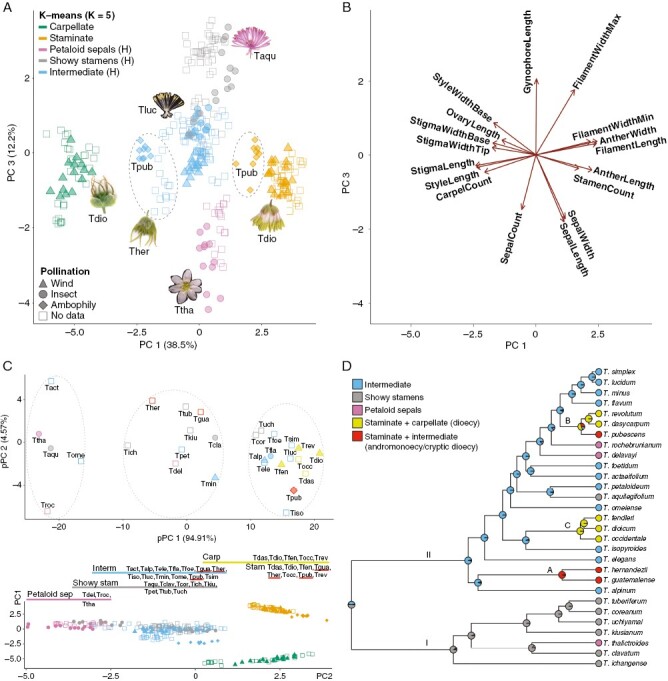
Multivariate cluster analysis of floral traits. (A) Principal component analysis (PCA) of 17 floral traits across 29 species. Data points represent single flowers (N = 309) coloured by k-means cluster with ad hoc assignment of flower morphology (top left), with symbols representing the pollination mode (bottom left); representative flowers are shown for each of five k-means clusters. Principal component (PC)1 (as a percentage of total variance explained) discriminates flowers mostly by sexual system, segregating dioecious, wind-pollinated taxa (at both extremes of the axis) from hermaphrodites (H) comprising all three pollination modes at the centre of the axis. PC3 separates the central hermaphroditic flower cluster further into three morphotypes, interpreted as petaloid sepals, showy stamens and an intermediate morph, separating insect-pollinated species at its extremes from mixed-pollinated taxa towards the centre. Filled symbols identify species with field-validated pollination mode data. (B) Biplot corresponding to the PCA shown in panel A, with loadings for the different floral traits represented as arrows towards larger values; the direction of the arrows indicates the contribution of each trait to the respective PC. (C) Top: phylogenetic PCA (pPCA) of flower morphology in Thalictrum (Ranunculaceae). The dataset (from panel A) uses species averages, with species names abbreviated to the first three letters. K-means analysis resulted in three clusters (k = 3), enclosed in dashed lines, and the available pollination mode information is shown with filled shapes as in panel A; colour-coding is as in panel D. Bottom: PC combination from panel A that best matches the pPCA outcome (PC1 vs. PC2), for a direct comparison, showing a continuum of flower morphotypes and pollination modes along PC2. (D) Ancestral state reconstruction of flower morphotypes for 29 Thalictrum species from multivariate analysis shown in panel A. The first three categories correspond to hermaphroditic sexual systems, while the last two encompass dioecy, andromonoecy and cryptic dioecy (T. pubescens), respectively. Clades A–C are as in Fig. 1. Pie charts represent the marginal probability of observing a certain flower morphotype (character state) at any given node.
To prevent circularity in assigning the pollination mode from morphology based on k-means, we validated the pollination mode for each cluster based on membership by species whose pollination mode had been investigated in the field (Table 1; Fig. 2A, filled symbols). The petaloid sepal cluster was designated as insect-pollinated based on T. thalictroides membership, and the showy stamen cluster was designated as insect-pollinated based on T. clavatum and T. aquilegiifolium. Staminate and carpellate clusters were classified as wind-pollinated based on T. fendleri, T. dioicum and T. revolutum. The intermediate cluster was unresolved with respect to pollination mode, containing ambophilous T. pubescens (hermaphroditic flowers), wind-pollinated T. alpinum and T. minus, and insect-pollinated T. flavum. In summary, although the edges of the PCA could be assigned more readily to insect or wind pollination, complexity in the data resulted in representatives of each of the three potential syndromes (wind, insect or ambophily) at the centre of the morphospace, representing an intermediate flower morphotype unresolved with respect to pollination mode.
The integration of phylogenetic relationships via pPCA and species averages reduced the number of distinct morphological clusters from five to three (Fig. 2C, K = 3, colour-coding as in Fig. 2D), underscoring the importance of shared evolutionary history and species-level variation. Phylogenetic principal component (pPC)1 explained the majority of the variance (94.91 %), largely discriminating taxa with petaloid sepals validated as insect pollinated at the far left (e.g. T. thalictroides, pink) from unisexual flowers validated as wind pollinated at the far right (e.g. T. dioicum, citrine). This pattern along pPC1 largely matches the distribution of species along PC2 (Fig. 2C, bottom inset; Supplementary data Fig. S5C), suggesting that the associated floral trait combinations are mostly independent of shared ancestry (i.e. certain trait clusters remain distinct despite being weighted by the phylogenetic variance–covariance matrix), hence convergent evolution to pollination vector is one likely explanation. Exceptions to that match include a member of the petaloid sepal group, T. delavayi, that fell into the middle cluster in pPCA (Fig. 2C), suggesting less distinction (less data granularity) when using species averages and phylogeny. Flowers with showy stamens and intermediate flower morphotypes were distributed across all three pPCA clusters, implying that the trait contribution to those morphotypes was decreased when accounting for phylogeny. For the showy stamen cluster, gynophore length and the maximum width of the stamen filaments were the largest discriminating trait contributions (Fig. 2B, upward arrows) that were, presumably, attenuated in the pPCA. Taken together, phylogenetically informed multivariate analysis in Thalictrum still broadly discriminated between insect-pollinated flower types with petaloid sepals and wind-pollinated small and mostly unisexual flower types.
An intermediate, transitional flower type is inferred as the ancestral state for clade II
Given that the intermediate cluster from PCA contains taxa with all three pollination modes that are morphologically intermediate between those in the wind and insect clusters (Fig. 2A), we asked whether it represents the ancestral condition for clade II (Fig. 1), where all major transitions occurred (to polyploidy, wind pollination and unisexual flowers; Soza et al., 2012, 2013). To test this hypothesis, we inferred discrete ancestral states using the k-means cluster scheme on a trimmed phylogeny of the 29 species with flower trait data, representing all major clades (Fig. 2D). To capture properly all the flower morphotypes emerging from the k-means analysis, sexually dimorphic species were assigned the combined score [staminate + intermediate] for andromonoecy (T. guatemalense and T. hernandezii) and cryptic dioecy (staminate and male-sterile hermaphroditic flowers, T. pubescens) or [staminate + carpellate] for dioecious species (Table 1). Root prior assumptions did not have a significant effect on the model for ancestral state inference for the three transition rate model classes (e.g. equal rates + stationary root prior and equal rates + empirical root prior, ΔAICc < 2). The equal rates model with an empirical root prior best fitted the data (ΔAICc to next non-equivalent model = 23.41), inferring the intermediate flower type as the most likely ancestral state for clade II. In clade II, small flowers [staminate + carpellate] evolved on average 2.2 times with dioecy (95 % CI [1.7–2.8]), [staminate + intermediate] evolved on average 2.2 times with andromonoecy/cryptic dioecy (95 % CI [2.1–2.4]), petaloid sepal flower types 2.2 times (95 % CI [1.7–2.7]) and showy stamen flower types 1.4 times (95 % CI [1–1.7]) (Fig. 2D). The ancestral state for clade I was inferred as most likely consisting of flowers with showy stamens, from which the petaloid sepal morphotype evolved once on average (95 % CI [0.8–1.3]). The genus-level ancestral flower type could not be resolved confidently owing to the inability to include outgroups using the flower morphotypes arising from our analyses within Thalictrum. Nevertheless, we were able to restrict the marginal probability (MP) for the ancestral flower type for the genus to two of the floral morphotypes: flowers with showy stamens (MP = 51.6 %) or the intermediate morphotype (MP = 41.3 %), with a much lower probability for flowers with petaloid sepals or unisexual flowers in the two dimorphic states (all three latter character states had MP < 3 %).
Refining pollination index boundaries in Thalictrum
We used the PI (Kaplan and Mulcahy, 1971) as a summary indicator of pollination mode and as a separate method of assigning pollinator that can be scored from flower photographs or herbarium specimens, enabling wider taxonomic sampling (83 species, compared with 29 in our morphology dataset). Initially, we calculated PI ranges for the five K clusters from PCA representing the three pollination modes, identifying the highest PI value for wind pollination and the lowest for insect pollination based on validated species. The more refined PI ranges were 1.00–1.29 for wind pollination and 2.57–3.00 for insect pollination, while intermediate values (1.3–2.56) remained ambiguous (wind/ambophily/insect) (Table 1). Thalictrum coreanum had the lowest PI for an insect-pollinated group (2.43), but because it matched that of T. lucidum in the intermediate group, we conservatively set the next available value of 2.57 (T. petaloideum, excluding one outlier) as the lower bound for the insect-pollinated group. The intermediate cluster had a PI ranging from 1.43 (T. guatemalense) to 2.43 (T. lucidum).
To address whether multivariate analysis of PI would mirror the results from our continuous floral trait analyses, we conducted PCA of PI values. PC1 and PC2 accounted for 49.57 % and 19 % of the total variance, respectively (Supplementary data Fig. S6A), and k-means cluster analysis grouped species into ‘wind’ (cluster 1), ‘wind/ambophilous’ (cluster 2) and ‘insect’ clusters (cluster 3) (k = 3; Supplementary data Fig. S6B). These results mostly coincide with the outcome of the pPCA, supporting the usefulness of the PI in capturing the most informative morphological parameters in the absence of more comprehensive measurements from field-validated observations.
Reproductive organs show significant positive evolutionary correlation
To identify potential suites of co-evolving floral characters, we tested for evolutionary correlation while accounting for species variation, by fitting pairwise multivariate Brownian motion models in a Bayesian framework (Fig. 3). No significant negative correlations were found between floral traits, nor did most traits exhibit a significant evolutionary correlation (the posterior estimate of the correlation coefficient overlapped with zero for most traits tested; Fig. 3A–C, grey cells). However, 27 trait pairs exhibited strong positive evolutionary correlation (Fig. 3A–C, orange cells): 17 between reproductive traits (Fig. 3A), five between stamen and sepal traits (Fig. 3B), and five between carpel and sepal traits (Fig. 3C). Between reproductive traits, stamen and carpel lengths were positively correlated (Fig. 3A). The correlations between these two organs were probably driven by the positive correlation between their component parts: anther length with style length and anther length with stigma length (Fig. 3A). However, not all flower organ component parts exhibited correlation. Importantly, filament length was not positively correlated with any carpellate feature, and ovary length did not show a positive correlation with staminate features, highlighting the modularity of stamens and carpels. Anther size, a composite trait between length and width, exhibited a positive correlation with style length (Fig. 3C) that was probably driven, in part, by the positive correlation between anther length and style length. There was also a positive evolutionary correlation between sepal number and size (width and length) in the carpellate and staminate datasets (Fig. 3B, C), and between stamen number and sepal size in the staminate dataset (Fig. 3C).
Fig. 3.
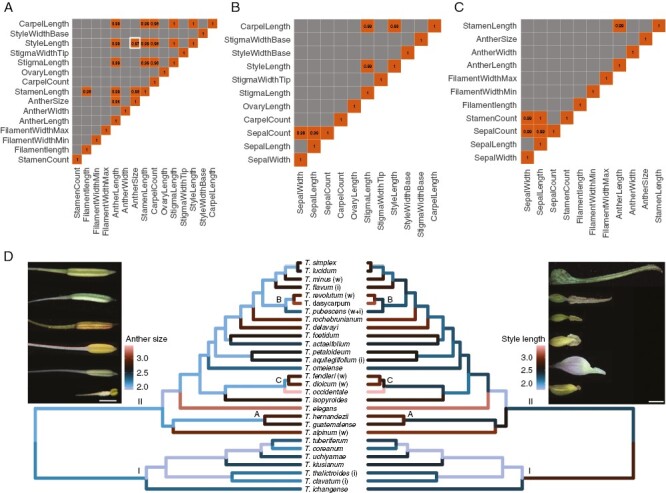
Evolutionary correlation among floral traits in Thalictrum by multivariate Brownian motion. Correlation matrices are shown for: (A) reproductive floral organs (stamens and carpels); (B) the carpellate dataset (from carpellate and hermaphroditic flowers); and (C) the staminate dataset (from staminate and hermaphroditic flowers). The heat map shows the maximum a posteriori estimates of the correlation coefficient for significant correlations; grey denotes no significant correlation, while orange indicates a positive correlation (except on the diagonal, where it represents each trait against itself). No negative correlations were found. (D) Mirror trees exemplifying ancestral state reconstructions for two floral traits: anther size (left, with images to exemplify range) and style length (right, with images to exemplify range), highlighted in white in panel A. Inferred maximum a posteriori values (inverse log + 1) for ancestral nodes and tips are depicted with a colour gradient on the phylogeny. The known pollination mode for species with field studies is shown for reference: i, insect pollination; w, wind pollination; and w + i, ambophily (both). Scale bar within images = 1 mm.
Traits with significant positive evolutionary correlation mirrored each other when mapped onto the phylogeny, as exemplified by anther size and style length (Fig. 3D; Fig. 3A, highlighted cell). The observed evolutionary correlations are consistent with pollination syndromes in Thalictrum, whereby insect-pollinated taxa tend to have smaller (shorter) anthers and short capitate stigmas, whereas wind-pollinated taxa tend to have larger (longer) anthers and longer styles and stigmas (Fig. 3D). Hence, clade I contains experimentally validated insect-pollinated species and exhibits cooler colours (i.e. shorter styles and smaller anthers), whereas clade II, where wind pollination has evolved repeatedly, contains a mix of dark and warmer colours. Subclade C provides further validation, with its confirmed wind-pollinated taxa having longer reproductive structures (warmer branch colours denoting longer styles and larger anthers).
DISCUSSION
We set out to detect different pollination syndromes in flowers of the ranunculid genus Thalictrum that lack synorganization (floral organ fusion), petals and nectar, while exhibiting at least two distinct pollination modes (wind and insect). A substantially stronger reconstruction of relationships at the genus level, with increased taxon sampling and better resolved and supported subclades, was implemented to guide an unbiased exploration of floral morphospace via comparative analyses in the context of the evolution of wind pollination from insect-pollinated flowers. Flower morphology can predict the pollination mode in a subset of the species surveyed at the extremes of the distribution, possibly as a consequence of morphological convergence to pollination vectors combined with the evolutionary integration of floral traits. We propose that this evolutionary integration of traits is likely to compensate for the lack of floral synorganization, allowing for a certain degree of specialization to different pollination vectors. An intermediate floral morphotype identified via multivariate analysis was reconstructed as most likely ancestral for the clade where wind pollination evolved repeatedly, suggesting that this ancestral state might have provided an evolutionary testing ground that potentially led to adaptation to multiple pollination vectors.
Floral morphology as a proxy for pollination mode in Thalictrum
To test the degree to which floral morphology reflects the different pollination modes (wind, insect, or both) in Thalictrum, we adopted clustering-based methods that use predictions to fill gaps in natural history data and are therefore ideally suited for systems with a paucity of empirical data (van der Niet, 2021). Initially, using both standard and phylogenetically informed multivariate analyses to visualize divergence across morphospace, we showed that continuous floral traits are able to discriminate, in part, between wind- and insect-pollinated taxa, with a less distinct morphospace that includes a mix of ambophilous, wind- and insect-pollinated taxa. Next, we used these data to test the predictive power of a synthetic PI that summarizes seven key traits as a proxy for pollination mode.
Based on combined evidence from both these approaches, we set more precise ranges for using PI as an indicator of pollination mode when empirical data are not available and applied a clustering approach to increase predictability within the intermediate range. Partly overlapping boundaries in the intermediate PCA cluster are consistent with a more generalist, opportunistic pollination mode in Thalictrum (Robertson, 1928; Kaplan and Mulcahy, 1971; Melampy and Hayworth, 1980; Motten, 1986; Steven, 2003). It thus appears that neither PI nor continuous floral traits can accurately discriminate between pollination modes within the species cluster with intermediate PI values. Pollen release biomechanics is another, functional trait that discriminates wind- from insect-pollinated species at the ends of the distribution but is less accurate at determining the pollination mode for those with intermediate PI values (e.g. T. alpinum; Timerman and Barrett, 2019). Interestingly, those species are newly classified as ‘ambiguous’ in our adjusted PI boundary system (PI 1.43–2.43) because they fall in our intermediate cluster, where predictions based on morphology are less accurate and empirical studies are most needed. Whether based on quantification and evolutionary analyses of flower morphology (this study) or on the biomechanical aspects of pollen release (Timerman and Barrett, 2019), there seems to be a strong signal at the extremes of the wind–insect pollination distribution and a ‘grey’ area of mixed features in the middle. We suggest that this intermediate zone of floral morphospace, currently supported by two independent studies (Timerman and Barrett, 2019; this study), should not be discounted as a lack of the power of floral morphometrics to detect pollination syndromes, but should be embraced as representative of a plastic space leading to the use of either or both pollination vectors, depending on environmental circumstances.
Overall, floral morphology is a better predictor of the pollination mode for more specialized Thalictrum, towards the extremes of the insect–wind adaptation spectrum. We hope that our study contributes to efforts towards improving pollination mode predictions based on floral morphology in Thalictrum. Other floral traits, such as flower scent, mild overall in Thalictrum but richer in volatile compound diversity in the insect-pollinated species (Wang et al., 2019), and inflorescence architecture, known to be labile in other ranunculids (Zhao et al., 2012), might also contribute to better discrimination of the pollination mode for species with intermediate flower morphologies. Complex functional traits, such as the ability of stamens to release pollen, provide a promising avenue (Timerman and Barrett, 2021) but will be harder to dissect at the genetic level, given that they are likely to result from multiple underlying developmental processes. The female side of wind pollination, pollen reception, is another key component that emerges as having an evolutionary signal from our analyses, in the form of style (and stigma) length, warranting further investigation.
Intermediate floral morphologies and ambophily as a step in the evolution of more specialized pollination modes
This study identified a floral morphotype in multivariate space, the intermediate cluster, that comprises all three pollination modes and thus appears more generalist than those in the other four PCA clusters. These less showy flowers are consistent with morphologies found in other ambophilous taxa (reviewed by Abrahamczyk et al., 2023). Two other morphotypes, consisting of petaloid sepals or showy stamens, appear to have converged on insect pollination via different morphological expressions of insect-attracting features, evolving in parallel at least twice in the genus (Fig. 2D). The last two morphotypes, consisting of small unisexual flowers (staminate and carpellate) with elongated sexual organs and small green sepals, are strongly associated with wind pollination, which has evolved independently at least eight times (Wang et al., 2019). Phylogenetic reconstruction of these four more specialized flower morphotypes suggests that they are more likely to derive from an intermediate morphology within clade II. We have previously shown that insect pollination is ancestral not only for clade II but for the entire genus (Soza et al., 2012; Wang et al., 2019). Here, we propose that a generalist and, potentially, more plastic ancestral trait space, the intermediate cluster, enabled the subsequent evolution of a more specialized wind pollination mode, or a reversal to insect pollination in clade II of Thalictrum. Additional investigation of the pollination biology of species falling within the intermediate flower morphotype and the characterization of other discriminating traits are needed to test further the hypothesis that the intermediate morphotype represents an evolutionarily transitional state. At the genus level, our analysis was able to limit the probable ancestral states to two floral morphotypes (showy stamens or intermediate morphologies), while excluding the other two (petaloid sepals or small unisexual flowers). An expansion of flower morphotype analyses to other Ranunculaceae is needed to inform these hypotheses further.
Evolutionary integration of floral traits: the role of selection and structural constraint
Floral integration tends to be higher in species with specialist pollinators than in those with more generalist pollination modes (Pérez-Barrales et al., 2007; Rosas-Guerrero et al., 2011; Gómez et al., 2014). Typically, floral integration is calculated from the variance in a trait correlation matrix for a given species (Wagner, 1984; Cheverud et al., 1989). Fewer studies have modelled trait integration explicitly within a phylogenetic context (Joly et al., 2018; Kriebel et al., 2020), which requires the fitting of multivariate models of trait evolution in order to test for evolutionary correlation (Harmon, 2018). As opposed to most other plants whose pollination biology has been placed in phylogenetic context (e.g. Smith et al., 2008; Lagomarsino et al., 2017; Reich et al., 2020), Thalictrum flowers lack organ fusion and whorled phyllotaxy (the latter often being considered a morphological precursor of fusion; Endress, 2016), both symplesiomorphic character states within angiosperms (Sauquet et al., 2017). Although apetalous, nectarless Thalictrum flowers are pollinated by a variety of generalist insects (Kaplan and Mulcahy, 1971) and are not expected to exhibit specialized adaptations, morphological specialization to wind pollination has evolved repeatedly in the genus. In fact, our analyses identified traits that exhibit a strong positive evolutionary correlation within the genus: between anther, style and stigma lengths but not between stamen filament and ovary length, highlighting the modularity of stamens and carpels. The lengths of anthers, styles and stigmas also appear to play an important role in segregating floral morphotypes in multivariate space based on the shared direction of their vectors in the biplots. The association between long, exserted stamens, styles and stigmas and wind pollination is often cited (Friedman and Barrett, 2009), but it had not been tested explicitly within a phylogenetic framework. Given that we do not observe a positive evolutionary correlation between all aspects of stamen and carpel morphology, e.g. filament length is not positively correlated with any carpel-related traits and ovary length is not positively correlated with staminate traits, we propose that these reproductive evolutionary correlations are more likely to result from the opposing selective pressures favouring abiotic vs. biotic pollination than from structural constraints (such as allometry).
We also identified a positive evolutionary correlation between the number of stamens and sepal size in the staminate dataset, probably driven by sexual dimorphism in unisexual flowers, whereby staminate flowers tend to have bigger sepals than carpellate flowers. This sexual dimorphism has previously been postulated to result from an ectopic role of certain B-class genes, which are floral organ identity genes that specify petal and stamen identity, in sepals (Di Stilio et al., 2005; LaRue et al., 2013). In particular, certain Thalictrum homologues of the APETALA3 lineage are differentially expressed in the perianth of staminate flowers, presumably making their sepals look larger and more petaloid than those of their carpellate counterparts (Galimba et al., 2018). Alternatively, this might be attributable to broader structural constraints, in that flower meristem size is known to dictate the number and initial size of floral organ primordia (Moyroud and Glover, 2017).
Our study offers new insight into the common and difficult problem of assigning the pollination mode when there are gaps in empirical data and into understanding whether and how suites of correlated floral characters evolve in concert in one of the few groups where wind pollination has evolved repeatedly within a genus. Although a multivariate phylogenetic approach alone does not identify the ultimate causal processes underlying the observed correlations (Boucher et al., 2018), distinguishing convergent floral morphologies to specific pollination vectors from those attributable to shared descent brings us closer to that goal. In future, it would be desirable to achieve an evolutionary synthesis of all available sources of floral quantitative morphology and functional data to identify further tractable developmental and genetic indicators along the biotic–abiotic pollination spectrum.
Multiple paths towards wind-pollinated morphologies are more likely than one
Given that wind pollination has evolved from insect pollination at least eight times in Thalictrum (Soza et al., 2012; Wang et al., 2019), we did not expect to find a single pathway from a flower morphology perspective. Evidence presented here supports the more likely scenario that Thalictrum species have used various paths emerging from an evolutionarily plastic, in flux floral morphospace associated with mixed pollination (whether stable or temporary) as a strategy to exploit readily available wind for sexual reproduction under a putative shortage of insect pollinators. Thus, we find instances of multiple floral morphologies sharing the same pollination mode in what is best described as a morphotype continuum between insect and wind pollination.
SUPPLEMENTARY DATA
Supplementary data are available at Annals of Botany online and consist of the following. Table S1: PCR and sequencing primers. Table S2: phylogenetic models. Table S3: voucher information. Figure S1: representative flower scans and traits. Figure S2: maximum likelihood Thalictrum phylogeny. Figure S3: Bayesian Thalictrum phylogeny. Figure S4: variation in Thalictrum floral traits. Figure S5: validation of principal component analysis of Thalictrum floral traits. Figure S6: principal component analysis of pollination index in Thalictrum. Video S1: first three dimensions of the PCA of Thalictrum floral traits.
ACKNOWLEDGEMENTS
We thank Simra Zahid for assistance with flower scans and measurements, University of Washington Biology Greenhouse staff for plant care, Dr Sungwon Son for photographing T. coreanum flowers, and the Cornell University Statistical Consulting Unit for advice on statistical analysis. J.M.-G. thanks Elizabeth Mahood for the discussion on data dimensionality reduction and clustering methods. Two anonymous reviewers provided helpful comments that greatly improved the original manuscript.We thank anonymous reviewers for helpful comments that improved an earlier version of the manuscript.
APPENDIX
Thalictrum (Ranunculaceae) and outgroup samples used in this study for DNA extraction and phylogenetic analysis
Taxon, voucher (Herbarium), GenBank accessions: ndhA intron, ndhF, rbcL, rpl32-trnL intergenic spacer, trnL intron and trnL-trnF intergenic spacer. Asterisks are voucher information for the trnL-trnF region.
Aquilegia buergeriana var. oxysepala (Trautv. & C.A.Mey.) Kitam., Park AF01 (YNUH), JX258647, KM206671, JX258432, JX258540, JQ691534. A. formosa L., Di Stilio 128 (WTU), MT427936, MT427953, MT427984, MT427968, MT428001/MT428018. Enemion raddeanum Regal., Park ER01 (YNUH), JX258645, KM206669, JX258430, JX258538, JQ691533. Isopyrum manshuricum Kom., Park IM02 (YNUH), JX258646, KM206670, JX258431, JX258539, JQ691532. Leptopyrum fumarioides (L.) Reichb., Zhang s.n. (KUN), JX258650, KM206674, JX258435, JX258543, JX573531. Paraquilegia microphylla (Royle) J.R.Drumm. & Hutch., Liang s.n. (KUN), JX258649, KM206673, JX258434, JX258542, JX573530. Semiaquilegia adoxoides (DC.) Makino., Park SE01 (YNUH), JX258648, KM206672, JX258433, JX258541, JX573529. Thalictrum actaeifolium Siebold & Zucc., Yamazaki 1104 (TI), JX258544, KM206569, JX258329, JX258436, JX573432. T. actaeifolium var. brevistylum Nakai, Park 029 (YNUH), JX258545, KM206570, JX258330, JX258437, JX573433. T. acutifolium (Hand.-Mazz.) B.Boivin, Mu 180 (KUN), JX258637, KM206662, JX258423, JX258529, JX573521. T. alpinum L., Tatewaki 1074 (TI), JX258546, KM206571, JX258331, JX258438, JX573434. T. alpinum var. elatum O.E.Ulbr., Boufford 28521 et al. (TI), JX258549, KM206574, JX258334, JX258441, JX573437. T. amurense Maxim., unvouchered, MT427937, MT427954, MT427985, MT427969, MT428002/MT428019. T. aquilegiifolium L., Kawahara 666 et al. (TI), JX258550, KM206575, JX258335, JX258442, JX573438. T. aquilegiifolium var. sibiricum Regel & Tiling, Park 12 (YNUH), JX258551, KM206576, JX258336, JX258443, JX573439. T. arkansanum B.Boivin, Carr 17995 (TEX), JX258552, KM206577, JX258337, JX258444, JX573440. T. arsenii B.Boivin, Barriga 4750 (TEX), JX258554, KM206579, JX258339, JX258446, JX573442. T. atriplex Finet & Gagnep., Ho 2594 et al. (TI), JX258638, KM206663, JX258424, JX258530, JX573522. T. baicalense Turcz. ex Ledeb., Quo 9156 (KUN)/Jeon s.n. (SKK)*, JX258555, KM206580, JX258341, JX258448, JQ691506. T. calabricum Spreng., Segelberg s.n. (S), JX258556, KM206581, JX258342, JX258450, JX573443. T. calcicola T.Shimizu, Shimizu 10034 et al. (TI), JX258644, KM206668, JX258429, JX258537, JX573528. T. chelidonii DC., Kanai 674743 et al. (TI), JX258639, KM206664, JX258425, JX258532, JX573523. T. clavatum DC., Kral 61853 (TEX), JX258557, KM206582, JX258343, JX258451, JX573444. T. confine Fernald, Fernald s.n. (TEX), JX258558, KM206583, JX258344, JX258452, JX573445. T. cooleyi H.E.Ahles, s.c. s.n. (OSC), MT427938, MT427955, MT427986, MT427970, MT428003/MT428020. T. coreanum H.Lév., Park 0501 (YNUH), JX258560, KM206585, JX258346, JX258454, JX573447. T. coriaceum (Britton) Small, Bozeman 10680 et al. (TEX), JX258561, KM206586, JX258347, JX258455, JX573448. T. cuernavacanum Rose, Floden s.n. (TENN), MT427939, MT427956, MT427987, MT427971, MT428004/MT428021. T. cultratum Wall., Qing 73-246 (KUN), JX258641, KM206665, JX258426, JX258534, JX573525. T. dasycarpum Fisch., C.A.Mey. & Avé-Lall., Carr 17991 (TEX), JX258563, KM206588, JX258349, JX258457, JX573450. T. decipiens B.Boivin, Dillon 3126 et al. (NY), JX258565, KM206590, JX258351, JX258459, JX573452. T. delavayi Franch., Boufford 27446 et al. (TI), JX258566, KM206591, JX258352, JX258460, JX573453. T. diffusiflorum C.Marquand & Airy Shaw, Liston 1161 (OSC), MT427940, MT427957, MT427988, MT427972, MT428005/MT428022. T. dioicum L., Wood 8903 and Wilson (TEX), JX258567, KM206592, JX258353, JX258461, JX573454. T. elegans Wall. ex Royle, Ludlow 20610 et al. (TI), JX258569, KM206594, JX258355, JX258463, JX573456. T. fargesii Franch. ex Finet & Gagnep., Boufford 26417 et al. (TI), JX258570, KM206595, JX258356, JX258464, JX573457. T. fendleri Engelm. ex A.Gray, Emily 5279 et al. (TEX), JX258571, KM206596, JX258357, JX258465, JX573458. T. filamentosum Maxim., Di Stilio 104 (WTU), MT427942, MT427959, MT427990, MT427973, MT428007/MT428024. T. finetii B.Boivin, Boufford 27614 et al. (TI), JX258642, KM206666, JX258427, JX258535, JX573526. T. flavum L., Soza 1908 (WTU), MT427947, MT427964, MT427995, MT427978, MT428012/MT428029. T. foeniculaceum Bunge, Smith 6618 (S), JX258573, KM206598, JX258359, JX258467, JX573460. T. foetidum L., Klackenberg 820619-6 (S), JX258574, KM206599, JX258360, JX258468, JX573461. T. foliolosum DC., Kanai 672444 and Shakya (TI), JX258575, KM206600, JX258361, JX258469, JX573462. T. galeottii Lecoy., Lucia 1125 (TEX), JX258576, KM206601, JX258362, JX258470, JX573463. T. gibbosum Lecoy., Pedro 8954 (TEX), JX258577, KM206602, JX258363, JX258471, JX573464. T. grandiflorum Maxim., Tang 16 (PE), JX258578, KM206603, JX258364, JX258472, JX573465. T. grandifolium S.Watson, Hinton 24708 et al. (TEX), JX258579, KM206604, JX258365, JX258473, JX573466. T. guatemalense C.DC. & Rose, Elias 4989 (TEX), JX258580, KM206605, JX258366, JX258474, JX573467. T. heliophilum Wilken & DeMott, Waters s.n. (CS), MT427943, MT427960, MT427991, MT427974, MT428008/MT428025. T. henricksonii M.C.Johnst., Henrickson 13417 (RSA), MT427944, MT427961, MT427992, MT427975, MT428009/MT428026. T. hernandezii Tausch ex J.Presl, Pringle s.n. (S), JX258581, KM206606, JX258367, JX258475, JX573468. T. ichangense Lecoy. ex Oliv., Park 31 (YNUH), JX258582, KM206607, JX258368, JX258476, JX573469. T. isopyroides C.A.Mey., Di Stilio 111 (WTU), MT427945, MT427962, MT427993, MT427976, MT428010/MT428027. T. kiusianum Nakai, Brunet s.n. (OSC), MT427946, MT427963, MT427994, MT427977, MT428011/MT428028. T. lankesteri Standl., Williams 11399 (NY), JX258583, KM206608, JX258369, JX258477, JX573470. T. lecoyeri Franch., Boufford 35394 et al. (PE), JX258643, KM206667, JX258428, JX258536, JX573527. T. leuconotum Franch., Tang 0283 et al. (PE), JX258584, KM206609, JX258370, JX258478, JX573471. T. lucidum L., Barabas 1131294 (PE), JX258585, KM206610, JX258371, JX258479, JX573472. T. macrocarpum Gren., Schultz s.n. and Winter (RSA), JX258586, KM206611, JX258372, JX258480, JX573473. T. macrostylum Shuttlew. ex Small & A.Heller, unvouchered, MT427948, MT427965, MT427996, MT427979, MT428013/MT428030. T. minus var. hypoleucum (Siebold & Zucc.) Miq., Park 067 (YNUH)/Park 051 (YNUH)*, JX258587, KM206612, JX258373, JX258481, JQ691515. T. myriophyllum Ohwi, Mori s.n. (TI), JX258588, KM206613, JX258374, JX258482, JX573474. T. occidentale A.Gray, Karen 63 (TEX), JX258589, KM206614, JX258375, JX258483, JX573475. T. omeiense W.T.Wang & S.H.Wang, Wang 519 (PE), JX258591, KM206616, JX258377, JX258485, JX573477. T. peltatum DC., Soule 2679 and Loockerman (TEX), JX258592, KM206617, JX258378, JX258486, JX573478. T. petaloideum L., Park 72 (YNUH), JX258593, KM206618, JX258379, JX258487, JX573479. T. pinnatum S.Watson, Hemple 1067 and Jack (TEX), JX258594, KM206619, JX258380, JX258488, JX573480. T. podocarpum Kunth, Wiegend 2000/623 (OSC), MT427949, –, MT427997, MT427980, MT428014/MT428031. T. polycarpum (Torr.) S.Watson, Darin 0620 and Boyd (RSA), JX258596, KM206621, JX258382, JX258490, JX573482. T. pringlei S.Watson, Walker s.n. (NY), JX258598, KM206623, JX258384, JX258492, JX573484. T. przewalskii Maxim., Ho 2013 et al. (TI), JX258599, KM206624, JX258385, JX258493, JX573485. T. pubescens Pursh, Moldenke 30097 and Moldenke (TEX), JX258601, KM206625, JX258387, JX258495, JX573487. T. pubigerum Benth., Panero 4111 and Calzada (TEX), JX258603, KM206628, JX258389, JX258497, JX573489. T. punctatum H.Lév., Choi 60295 (YNUH), JX258604, KM206629, JX258390, JX258498, JQ691528. T. reniforme Wall., Naithani 37388 (TI), JX258605, KM206630, JX258391, JX258499, JX573490. T. reticulatum Franch., Boufford 42802 et al. (GH), MT427950, MT427966, MT427998, MT427981, MT428015/MT428032. T. revolutum DC., Fryxell 3756 (TEX), JX258606, KM206631, JX258392, JX258500, JX573491. T. rhynchocarpum Quart.-Dill. & A.Rich., Carvalho 3971 (NY), JX258608, KM206633, JX258394, JX258502, JX573493. T. rochebrunnianum Franch. et Sav., Park 106 (YNUH), JX258609, KM206634, JX258395, JX258503, JX573494. T. rochebrunnianum var. grandisepalum (H.Lév.) Nakai, Uchima s.n. (TI), JX258610, KM206635, JX258396, JX258504, JX573495. T. rostellatum Hook.f. & Thomson, Stainton 3392 and Williams (TI), JX258611, KM206636, JX258397, JX258505, JX573496. T. rotundifolium DC., Kanai 672698 and Shresta (TI), JX258612, KM206637, JX258398, JX258506, JX573497. T. rubescens Ohwi, Yamazaki 597 et al. (TI), JX258613, KM206638, JX258399, –, JX573498. T. rutifolium Hook.f. & Thomson, Boufford 29502 et al. (TI), JX258614, KM206639, JX258400, JX258507, JX573499. T. sachalinese Lecoy., Hara s.n. (TI), JX258615, KM206640, JX258401, JX258508, JX573500. T. saniculaeforme DC., Kanai 672443 and Shakya (TI), JX258616, KM206641, JX258402, JX258509, JX573501. T. simplex var. brevipes H.Hara, Park 56 (YNUH), JX258617, KM206642, JX258403, JX258510, JX573502. T. smithii B.Boivin, Boufford 28205 et al. (TI), JX258618, KM206643, JX258404, JX258511, JX573503. T. sparsiflorum Turcz.ex Fisch.& C.A.Mey., Jeon s.n. (SKK), JX258620, KM206645, JX258406, JX258513, JQ691522. T. squamiferum Lecoy., Boufford 29603 et al. (TI), JX258621, KM206646, JX258407, JX258514, JX573505. T. squarrosum Stephan ex Willd., Smith 7560 (S), JX258622, KM206647, JX258408, JX258515, JX573506. T. strigillosum Hemsl., Hinton 139 et al. (TEX), JX258623, KM206648, JX258409, JX258516, JX573507. T. tenue Franch., Smith 220 (S), JX258624, KM206649, JX258410, JX258517, JX573508. T. texanum (E.Hall ex A.Gray) Small, Carr 17939 (TEX), JX258625, KM206650, JX258411, JX258518, JX573509. T. thalictroides (L.) A.J.Eames & B.Boivin, Johm 37004 (TEX), JX258627, KM206652, JX258413, –, JX573511. T. trichopus Franch., Bartholomew 130 et al. (TI), JX258628, KM206653, JX258414, JX258520, JX573512. T. tripeltiferum B.Boivin, Detling 8788 (ORE), MT427951, N/A, MT427999, MT427982, MT428016/MT428033. T. tuberiferum Maxim., Park 055 (YNUH), JX258629, KM206654, JX258415, JX258521, JX573513. T. tuberosum L., Bremer 49 et al. (S), JX258630, KM206655, JX258416, JX258522, JX573514. T. uchiyamae Nakai, Park 156 (YNUH), JX258631, KM206656, JX258417, JX258523, JX573515. T. uncatum Maxim., Boufford 28389 et al. (TI), JX258632, KM206657, JX258418, JX258524, JX573516. T. uncinulatum Franch. ex Lecoy., Ho 1316 (PE), JX258633, KM206658, JX258419, JX258525, JX573517. T. urbainii Hayata, Liston 1162 (OSC), MT427941, MT427958, MT427989, –, MT428006/MT428023. T. venulosum Trel., Shirakashi 52 (TEX), JX258634, KM206659, JX258420, JX258526, JX573518. T. virgatum Hook.f. & Thomson, Boufford 30496 et al. (TI), JX258636, KM206661, JX258422, JX258528, JX573520. T. zernyi Ulbr., Gereau 3976 and Kayombo (MO), MT427952, MT427967, MT428000, MT427983, MT428017/MT428034.
Contributor Information
Jesús Martínez-Gómez, Department of Biology, University of Washington, PO Box 351800, Seattle, WA 98195, USA; School of Integrative Plant Sciences and L.H. Bailey Hortorium, Cornell University, Ithaca, NY 14853, USA.
Seongjun Park, Institute of Natural Science, Yeungnam University, Gyeongsan, Gyeongbuk, 38541, South Korea.
Samantha R Hartogs, Department of Biology, University of Washington, PO Box 351800, Seattle, WA 98195, USA.
Valerie L Soza, Department of Biology, University of Washington, PO Box 351800, Seattle, WA 98195, USA.
Seon Joo Park, Department of Life Sciences, Yeungnam University, Gyeongsan, Gyeongbuk, 38541, South Korea.
Verónica S Di Stilio, Department of Biology, University of Washington, PO Box 351800, Seattle, WA 98195, USA.
FUNDING
This work was supported by National Science Foundation, Division of Environmental Biology (Opportunities for Promoting Understanding through Synthesis—Mid-Career Synthesis) grant number [1911539] and The Fred C. Gloeckner Foundation [630550] to V.S.D.S., Mary Sue Ittner Pacific Bulb Society grant to S.R.H., and National Science Foundation Graduate Research Fellowship [1650441] to J.M.-G.
DATA AVAILABILITY
Floral measurements, scripts, R package citations, alignments and trees can be found at Zenodo (https://doi.org/10.5281/zenodo.6369383).
LITERATURE CITED
- Abrahamczyk S, Struck J-H, Weigend M.. 2023. The best of two worlds: ecology and evolution of ambophilous plants. Biological Reviews 98: 391–420. doi: 10.1111/brv.12911. [DOI] [PubMed] [Google Scholar]
- Berg RL. 1960. The ecological significance of correlation pleiades. Evolution 14: 171–180. doi: 10.2307/2405824. [DOI] [Google Scholar]
- Boucher FC, Démery V, Conti E, Harmon LJ, Uyeda J.. 2018. A general model for estimating macroevolutionary landscapes. Systematic Biology 67: 304–319. doi: 10.1093/sysbio/syx075. [DOI] [PubMed] [Google Scholar]
- Boyko JD, Beaulieu JM.. 2021. Generalized hidden Markov models for phylogenetic comparative datasets. Methods in Ecology and Evolution 12: 468–478. [Google Scholar]
- Charrad M, Ghazzali N, Boiteau V, Niknafs A.. 2014. NbClust: an R package for determining the relevant number of clusters in a data set. Journal of Statistical Software 61: 1–36. [Google Scholar]
- Cheverud JM, Wagner GP, Dow MM.. 1989. Methods for the comparative analysis of variation patterns. Systematic Biology 38: 201–213. [Google Scholar]
- Culley TM, Weller SG, Sakai AK.. 2002. The evolution of wind pollination in angiosperms. Trends in Ecology & Evolution 17: 361–369. [Google Scholar]
- Darwin C. 1862. On the various contrivances by which British and foreign orchids are fertilized by insects, and on the good effects of interbreeding. London: John Murray. [PMC free article] [PubMed] [Google Scholar]
- Davis SL. 1997. Stamens are not essential as an attractant for pollinators in females of cryptically dioecious Thalictrum pubescens Pursch. (Ranunculaceae). Sexual Plant Reproduction 10: 293–299. doi: 10.1007/s004970050101. [DOI] [Google Scholar]
- Dellinger AS, Chartier M, Fernández‐Fernández D, et al. 2019. Beyond buzz-pollination – departures from an adaptive plateau lead to new pollination syndromes. New Phytologist 221: 1136–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delph LF, Galloway LF, Stanton ML.. 1996. Sexual dimorphism in flower size. The American Naturalist 148: 299–320. doi: 10.1086/285926. [DOI] [Google Scholar]
- Di Stilio VS, Kramer EM, Baum DA.. 2005. Floral MADS box genes and homeotic gender dimorphism in Thalictrum dioicum (Ranunculaceae) – a new model for the study of dioecy. The Plant Journal 41: 755–766. doi: 10.1111/j.1365-313x.2005.02336.x. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A.. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biology 4: e88699–e88710. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A.. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. 2016. Development and evolution of extreme synorganization in angiosperm flowers and diversity: a comparison of Apocynaceae and Orchidaceae. Annals of Botany 117: 749–767. doi: 10.1093/aob/mcv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD.. 2004. Pollination syndromes and floral specialization. Annual Review of Ecology, Evolution, and Systematics 35: 375–403. doi: 10.1146/annurev.ecolsys.34.011802.132347. [DOI] [Google Scholar]
- Fenster CB, Reynolds RJ, Williams CW, Makowsky R, Dudash MR.. 2015. Quantifying hummingbird preference for floral trait combinations: the role of selection on trait interactions in the evolution of pollination syndromes. Evolution 69: 1113–1127. doi: 10.1111/evo.12639. [DOI] [PubMed] [Google Scholar]
- Fior S, Li M, Oxelman B, et al. 2013. Spatiotemporal reconstruction of the Aquilegia rapid radiation through next-generation sequencing of rapidly evolving cpDNA regions. New Phytologist 198: 579–592. doi: 10.1111/nph.12163. [DOI] [PubMed] [Google Scholar]
- Friedman J, Barrett SCH.. 2009. Wind of change: new insights on the ecology and evolution of pollination and mating in wind-pollinated plants. Annals of Botany 103: 1515–1527. doi: 10.1093/aob/mcp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimba KD, Martínez-Gómez J, Di Stilio VS.. 2018. Gene duplication and transference of function in the paleoAP3 lineage of floral organ identity genes. Frontiers in Plant Science 9: 334. doi: 10.3389/fpls.2018.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez JM, Perfectti F, Klingenberg CP.. 2014. The role of pollinator diversity in the evolution of corolla-shape integration in a pollination-generalist plant clade. Philosophical Transactions of the Royal Society B: Biological Sciences 369: 20130257. doi: 10.1098/rstb.2013.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán D. 2005. Funciones masculina y femenina de la reproducción en cinco especies de Thalictrum (Ranunculaceae) con diferentes vectores de polinización. Pirineos 160: 23–44. doi: 10.3989/pirineos.2005.v160.37. [DOI] [Google Scholar]
- Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. Journal of Statistical Software 33: 1–22. [PMC free article] [PubMed] [Google Scholar]
- Harmon L. 2018. Phylogenetic comparative methods: learning from trees. Minneapolis, MN: CreateSpace Independent Publishing Platform, 286. https://lukejharmon.github.io/pcm/ [Google Scholar]
- Humphrey RP. 2018. Sterile stamens do not enhance seed set in females of the cryptically dioecious Thalictrum macrostylum (Ranunculaceae). The Journal of the Torrey Botanical Society 145: 82–90. doi: 10.3159/torrey-d-16-00040.1. [DOI] [Google Scholar]
- Hurvich CM, Tsai C-L.. 1989. Regression and time series model selection in small samples. Biometrika 76: 297–307. doi: 10.1093/biomet/76.2.297. [DOI] [Google Scholar]
- Jolliffe IT. 2002. Principal component analysis. New York: Springer. [Google Scholar]
- Joly S, Lambert F, Alexandre H, Clavel J, Léveillé‐Bourret E, Clark JL.. 2018. Greater pollination generalization is not associated with reduced constraints on corolla shape in Antillean plants. Evolution 72: 244–260. doi: 10.1111/evo.13410. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan SM, Mulcahy DL.. 1971. Mode of pollination and floral sexuality in Thalictrum. Evolution 25: 659–668. doi: 10.1111/j.1558-5646.1971.tb01923.x. [DOI] [PubMed] [Google Scholar]
- Kitazawa MS. 2021. Developmental stochasticity and variation in floral phyllotaxis. Journal of Plant Research 134: 403–416. doi: 10.1007/s10265-021-01283-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa MS, Fujimoto K.. 2014. A developmental basis for stochasticity in floral organ numbers. Frontiers in Plant Science 5: 545. doi: 10.3389/fpls.2014.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriebel R, Drew B, González‐Gallegos JG, et al. 2020. Pollinator shifts, contingent evolution, and evolutionary constraint drive floral disparity in Salvia (Lamiaceae): evidence from morphometrics and phylogenetic comparative methods. Evolution 74: 1335–1355. doi: 10.1111/evo.14030. [DOI] [PubMed] [Google Scholar]
- Lagomarsino LP, Forrestel EJ, Muchhala N, Davis CC.. 2017. Repeated evolution of vertebrate pollination syndromes in a recently diverged Andean plant clade. Evolution 71: 1970–1985. doi: 10.1111/evo.13297. [DOI] [PubMed] [Google Scholar]
- LaRue N, Sullivan A, Di Stilio V.. 2013. Functional recapitulation of transitions in sexual systems by homeosis during the evolution of dioecy in Thalictrum. Frontiers in Plant Science 4: 487. doi: 10.3389/fpls.2013.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melampy MN, Hayworth AM.. 1980. Seed production and pollen vectors in several nectar-less plants. Evolution 34: 1144–1154. doi: 10.1111/j.1558-5646.1980.tb04057.x. [DOI] [PubMed] [Google Scholar]
- Motten AF. 1986. Pollination ecology of the spring wildflower community of a temperate deciduous forest. Ecological Monographs 56: 21–42. doi: 10.2307/2937269. [DOI] [Google Scholar]
- Moyroud E, Glover BJ.. 2017. The evolution of diverse floral morphologies. Current Biology 27: R941–R951. doi: 10.1016/j.cub.2017.06.053. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Niet T. 2021. Paucity of natural history data impedes phylogenetic analyses of pollinator-driven evolution. New Phytologist 229: 1201–1205. [DOI] [PubMed] [Google Scholar]
- Ollerton J, Alarcón R, Waser NM, et al. 2009. A global test of the pollination syndrome hypothesis. Annals of Botany 103: 1471–1480. doi: 10.1093/aob/mcp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollerton J, Rech AR, Waser NM, Price MV.. 2015. Using the literature to test pollination syndromes — some methodological cautions. Journal of Pollination Ecology 16: 119–125. doi: 10.26786/1920-7603(2015)17. [DOI] [Google Scholar]
- Parachnowitsch AL, Kessler A.. 2010. Pollinators exert natural selection on flower size and floral display in Penstemon digitalis. New Phytologist 188: 393–402. doi: 10.1111/j.1469-8137.2010.03410.x. [DOI] [PubMed] [Google Scholar]
- Park S, Jansen RK, Park SJ.. 2015. Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biology 15: 40. doi: 10.1186/s12870-015-0432-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Barrales R, Arroyo J, Armbruster WS.. 2007. Differences in pollinator faunas may generate geographic differences in floral morphology and integration in Narcissus papyraceus (Amaryllidaceae). Oikos 116: 1904–1918. [Google Scholar]
- Phillips HR, Landis JB, Specht CD.. 2020. Revisiting floral fusion: the evolution and molecular basis of a developmental innovation. Journal of Experimental Botany 71: 3390–3404. doi: 10.1093/jxb/eraa125. [DOI] [PubMed] [Google Scholar]
- Plummer M, Best N, Cowles K, Vines K.. 2006. CODA: convergence diagnosis and output analysis for MCMC. R News 6: 7–11. [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA.. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67: 901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Berger A, Balthazar M von, et al. 2020. Modularity and evolution of flower shape: the role of function, development, and spandrels in Erica. New Phytologist 226: 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2009. Size-correction and principal components for interspecific comparative studies. Evolution 63: 3258–3268. doi: 10.1111/j.1558-5646.2009.00804.x. [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Revell LJ, Harmon LJ.. 2008. Testing quantitative genetic hypotheses about the evolutionary rate matrix for continuous characters. Evolutionary Ecology Research 10: 311–331. [Google Scholar]
- Robertson C. 1928. Flowers and insects; lists of visitors of four hundred and fifty-three flowers. Carlinville: n.p. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Guerrero V, Quesada M, Armbruster WS, Pérez-Barrales R, Smith SD.. 2011. Influence of pollination specialization and breeding system on floral integration and phenotypic variation in Ipomoea. Evolution 65: 350–364. doi: 10.1111/j.1558-5646.2010.01140.x. [DOI] [PubMed] [Google Scholar]
- Rosas-Guerrero V, Aguilar R, Martén-Rodríguez S, et al. 2014. A quantitative review of pollination syndromes: do floral traits predict effective pollinators? Ecology Letters 17: 388–400. doi: 10.1111/ele.12224. [DOI] [PubMed] [Google Scholar]
- Sauquet H, von Balthazar M, Magallón S, et al. 2017. The ancestral flower of angiosperms and its early diversification. Nature Communications 8: 16047. doi: 10.1038/ncomms16047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9: 671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL.. 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany 94: 275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- Smith SD. 2016. Pleiotropy and the evolution of floral integration. New Phytologist 209: 80–85. [DOI] [PubMed] [Google Scholar]
- Smith SD, Kriebel R.. 2018. Convergent evolution of floral shape tied to pollinator shifts in Iochrominae (Solanaceae)*. Evolution 72: 688–697. doi: 10.1111/evo.13416. [DOI] [PubMed] [Google Scholar]
- Smith SD, Ané C, Baum DA.. 2008. The role of pollinator shifts in the floral diversification of Iochroma (Solanaceae). Evolution 62: 793–806. doi: 10.1111/j.1558-5646.2008.00327.x. [DOI] [PubMed] [Google Scholar]
- Soza VL, Brunet J, Liston A, Salles Smith P, Di Stilio VS.. 2012. Phylogenetic insights into the correlates of dioecy in meadow-rues (Thalictrum, Ranunculaceae). Molecular Phylogenetics and Evolution 63: 180–192. doi: 10.1016/j.ympev.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Soza VL, Haworth KL, Di Stilio VS.. 2013. Timing and consequences of recurrent polyploidy in meadow-rues (Thalictrum, Ranunculaceae). Molecular Biology and Evolution 30: 1940–1954. doi: 10.1093/molbev/mst101. [DOI] [PubMed] [Google Scholar]
- Statisticat LLC. 2021. LaplacesDemon: complete environment for Bayesian inference. Bayesian-Inference.com. R package version 16.1.6. https://web.archive.org/web/20150206004624/http://www.bayesian-inference.com/software [Google Scholar]
- Stebbins GL. 1951. Natural selection and the differentiation of Angiosperm families. Evolution 5: 299–324. doi: 10.2307/2405676. [DOI] [Google Scholar]
- Steven JC. 2003. Pollination and resource allocation in Thalictrum (Ranunculaceae): implications for the evolution of dioecy. Ph.D. thesis, University of Wisconsin; (Madison: ). https://www.proquest.com/openview/07b672c2cc03bf11b3edde164491530d/1?pq-origsite=gscholar&cbl=18750&diss=y [Google Scholar]
- Steven J, Waller DM.. 2004. Reproductive alternatives to insect pollination in four species of Thalictrum (Ranunculaceae). Plant Species Biology 19: 73–80. [Google Scholar]
- Tamura M. 1995. Ranunculaceae In: Hiepko P, ed. Die Naturlichen Pflanzenfamilien. Berlin: Duncker & Humblot, 223–497. [Google Scholar]
- Timerman D, Barrett SCH.. 2019. Comparative analysis of pollen release biomechanics in Thalictrum: implications for evolutionary transitions between animal and wind pollination. New Phytologist 224: 1121–1132. doi: 10.1111/nph.15978. [DOI] [PubMed] [Google Scholar]
- Timerman D, Barrett SCH.. 2021. The biomechanics of pollen release: new perspectives on the evolution of wind pollination in angiosperms. Biological Reviews 96: 2146–2163. [DOI] [PubMed] [Google Scholar]
- Tribble CM, Freyman WA, Landis MJ, et al. 2022. RevGadgets: an R package for visualizing Bayesian phylogenetic analyses from RevBayes. Methods in Ecology and Evolution 13: 314–323. [Google Scholar]
- Wagner GP. 1984. On the eigenvalue distribution of genetic and phenotypic dispersion matrices: evidence for a nonrandom organization of quantitative character variation. Journal of Mathematical Biology 21: 77–95. doi: 10.1007/bf00275224. [DOI] [Google Scholar]
- Walker JD, Geissman JW, Bowring SA, Babcock LE, comps. 2018. Geologic Time Scale v. 5.0: Geological Society of America. doi: 10.1130/2018.CTS005R3C. [DOI] [Google Scholar]
- Wang TN, Clifford MR, Martínez-Gómez J, Johnson JC, Riffell JA, Di Stilio VS.. 2019. Scent matters: differential contribution of scent to insect response in flowers with insect vs. wind pollination traits. Annals of Botany 123: 289–301. doi: 10.1093/aob/mcy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittall JB, Hodges SA.. 2007. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature 447: 706–709. doi: 10.1038/nature05857. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2011. ggplot2. Wiley Interdisciplinary Reviews: Computational Statistics 3: 180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- Wilke CO. 2020. cowplot: streamlined plot theme and plot annotations for ‘ggplot2’. R package version 1.1.1. https://CRAN.R-project.org/package=cowplot [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y.. 2017. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 8: 28–36. [Google Scholar]
- Zhao L, Bachelier JB, Chang H, Tian X, Ren Y.. 2012. Inflorescence and floral development in Ranunculus and three allied genera in Ranunculeae (Ranunculoideae, Ranunculaceae). Plant Systematics and Evolution 298: 1057–1071. doi: 10.1007/s00606-012-0616-6. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Floral measurements, scripts, R package citations, alignments and trees can be found at Zenodo (https://doi.org/10.5281/zenodo.6369383).


