Abstract
The zygote has a daunting task ahead of itself; it must develop from a single cell (fertilized egg) into a fully functioning adult with a multitude of different cell types. In the beginning, the zygote has help from its mother, in the form of gene products deposited into the egg, but eventually, it must rely on its own resources to proceed through development. The transfer of developmental control from the mother to the embryo is called the maternal-to-zygotic transition (MZT). All animals undergo this transition, which is defined by two main processes—the degradation of maternal RNAs and the synthesis of new RNAs from the zygote's own genome. Here, we review the regulation of the MZT in Drosophila, but given the broad conservation of this essential process, much of the regulation is shared among metazoans.
Keywords: FlyBook, Drosophila, MZT, zygotic genome activation, ZGA, embryo, gene expression, FlyBookg
Graphical Abstract
Graphical Abstract.
Coordinated events drive the MZT
During the maternal-to-zygotic transition (MZT), the fertilized egg transitions from two specified germ cells to the totipotent cells of the early embryo that subsequently gives rise to an entirely new organism. Thus, in a short time, the zygote generates thousands of cells, pattern the embryonic axes, and prepare for gastrulation. For this to occur, multiple processes must be precisely coordinated, including the lengthening of the division cycle, degradation of maternally deposited products, transcriptional activation of the zygotic genome, and reorganization of the chromatin of the embryonic genome (Fig. 1).
Fig. 1.
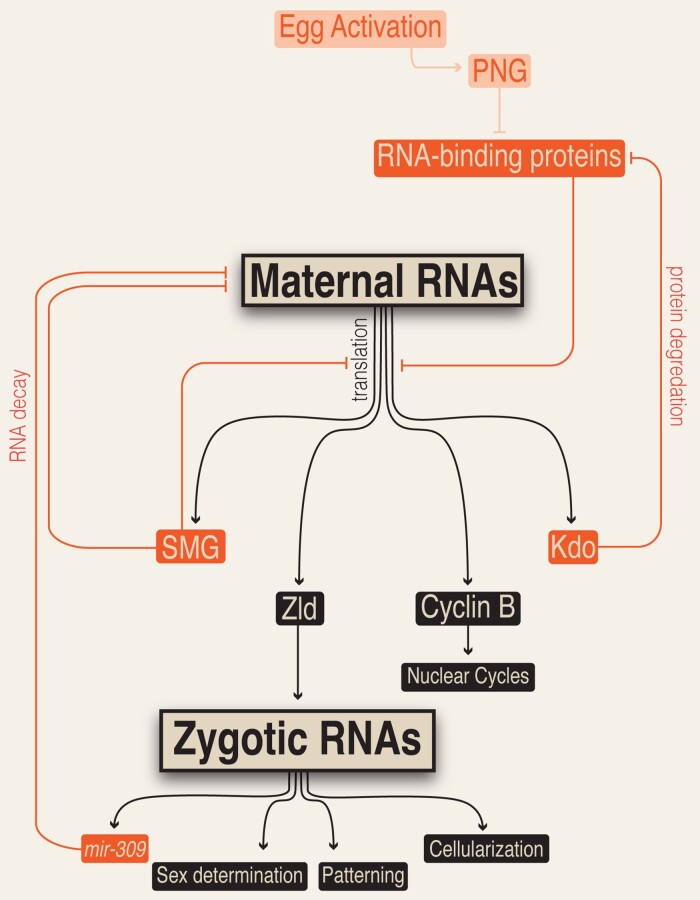
Handoff of developmental control is coordinated between maternal and zygotic programs. Egg activation triggers PNG kinase formation, which mediates phosphorylation events that result in translation of maternally provided mRNAs. This release of translational repression mediated by RNA-binding proteins results in the increase in protein levels of maternally encoded factors required for early and late-stage decay of maternal mRNA and proteins (denoted by orange boxes and lines), activation of the zygotic genome, and initiation of NCs. This allows multiple processes required for successful transition through the MZT to be coordinated and linked with egg activation.
After fertilization, the zygote undergoes a series of rapid and semisynchronous mitotic divisions that occur without cytokinesis, giving rise to a syncytial blastoderm (Zalokar and Erk 1976; Foe and Alberts 1983). These rapid cycles lack gap phases and are therefore a series of repeating rounds of DNA synthesis and mitotic division. The first 9 of these mitotic or nuclear cycles (NCs) last about 8 min each (at 25°C). At the ninth NC, the primordial germ cells form from nuclei that bud from the posterior and are cellularized during the subsequent division cycle (NC10). For the remaining nuclei, the division cycle gradually lengthens to about 60 min for NC14, due largely to increasing variability in the timing of DNA replication initiation and the addition of the first gap phase. During the extended interphase of NC14, the plasma membrane invaginates around the approximately 6,000 syncytial nuclei to form cells. Although, in this cellular blastoderm, all cells, with the exception of the future germ cells, appear nearly morphologically identical, they are molecularly distinct. By the end of NC14, cells have acquired positional information, and the fate map has been established (Lohs-Schardin, Sander, et al. 1979; Lohs-Schardin, Cremer, et al. 1979). Subsequent mitoses are asynchronous, occurring in mitotic domains determined by this fate map, and directly after cellularization, gastrulation movements begin (Foe and Alberts 1983; Foe 1989).
During the initial rapid divisions, the zygotic genome is largely transcriptionally quiescent with development controlled by maternally provided products, predominantly RNA. Impressively, over half of the protein-coding genome is encoded by these maternal RNAs, which are transcribed during oogenesis by nurse cells and the oocyte itself (>7,000 genes) (Tadros et al. 2003; De Renzis et al. 2007; Lecuyer et al. 2007; Thomsen et al. 2010). As development progresses, transcription of the zygotic genome is initiated, and this is coordinated with the degradation of the maternally provided products. Zygotic genome activation (ZGA) occurs gradually with a minor wave initiating at approximately NC8 and a major wave with many more genes becoming actively transcribed at NC14 (Edgar and Schubiger 1986; De Renzis et al. 2007; Lott et al. 2011; Chen et al. 2013). Together, these processes of transcriptional initiation and messenger RNA (mRNA) degradation result in a monumental shift in the transcriptome of the early embryo.
At NC14, the division cycle slows dramatically, transcription is broadly activated, the nuclei are cellularized, and morphological movements are initiated. The coordination of these processes marks the midblastula transition (MBT). This term was first used to describe the developmental transition in Xenopus embryos where similar cellular changes occurred, including lengthening of the cell cycle, asynchronous divisions, acquisition of cell motility, increased RNA synthesis, and dependence on the zygotic genome (Newport and Kirschner 1982a, 1982b). While the MBT takes place during the MZT, these terms refer to two distinct transitions. Here, we focus on the MZT: the process by which control is transferred from mother to offspring. Because the MZT is intimately linked with changes to the division cycle and to the chromatin template, we will also briefly discuss how these aspects are remodeled during early development and the tight regulation that couples these events together.
Degradation of maternally provided products
While approximately half of protein-coding genes are maternally contributed as mRNA to the future embryo, 60% of these transcripts will be degraded by the end of the MZT (Tadros et al. 2003; De Renzis et al. 2007; Lecuyer et al. 2007; Thomsen et al. 2010). Regulation of maternal RNAs occurs at the posttranscriptional level in which RNA-binding proteins mediate the stability, translation, and eventual decay of maternal transcripts. Simultaneously, 2% of the maternal proteome is also degraded through this period (Cao et al. 2020). Together, these networks of decay tightly coordinate the lifespan of maternal factors to ensure proper relinquishing of maternal control of the genome to the zygote.
Clearance of maternal mRNAs is regulated through both maternal and zygotically controlled mechanisms (Bashirullah et al. 1999). Maternally driven degradation of RNA is triggered at egg activation and proceeds regardless of whether an oocyte is fertilized. A principal regulator of this RNA decay pathway is the maternally encoded RNA-binding protein Smaug (Smg), which is responsible for the destabilization of an estimated two-thirds of maternally encoded mRNAs through the MZT (Tadros et al. 2007; Chen et al. 2014). Smg binds a stem-loop structure formed by smaug recognition elements located within the 3′UTRs of mRNAs (Smibert et al. 1996). This in turn recruits the CCR4-NOT-deadenylase complex to catalyze polyA-tail deadenylation, a mark important for mRNA stability (Semotok et al. 2005). Recruitment of the CCR4-NOT complex for downstream degradation is not exclusively dependent upon Smg. The RNA-binding proteins Brain Tumor (Brat) and Pumilio (Pum) recruit the CCR4-NOT complex to 3′UTRs to initiate mRNA decay (Gerber et al. 2006; Laver et al. 2015). In contrast to the maternal RNA degradation pathway, zygotic-mediated RNA decay requires fertilization and transcriptional activation of the zygotic genome. As such, this process begins approximately 2 h after egg laying (Bashirullah et al. 1999). This pathway is partially driven by expression of the mir-309 cluster, which contains 8 zygotically expressed microRNA (miRNA) genes that collectively code for 5 distinct seed sequences used to recognize and target maternal mRNA for depletion. Homology to these seed sequences is specifically enriched in maternal transcripts, suggesting the mir-309 locus has evolved over time to precisely target maternal rather than zygotic RNAs for degradation (Bushati et al. 2008). Interestingly, Smg is required for the activation of hundreds of zygotic loci, including the mir-309 cluster, suggesting a positive feedback loop to increase maternal RNA decay (Benoit et al. 2009). Although the mir-309 cluster is not deeply conserved, similar mechanisms of miRNA-mediated decay have been identified outside of Drosophila, including the mir-430 and mir-427 loci found in zebrafish and Xenopus, respectively (Giraldez et al. 2006; Lund et al. 2009).
Independent of RNA decay mechanisms, RNA-binding proteins like Smg, Pum, and Brat regulate translation of maternally provided transcripts to control protein expression. This balance between translational repression and RNA degradation is, in part, controlled by Pan Gu (Png) (Kronja, Whitfield, et al. 2014). In a complex with Plutonium (Plu) and Giant Nuclei (Gnu), Png reverses translational inhibition by repressors like Pum exclusively during the oocyte-to-embryo transition (Lee et al. 2003; Tadros et al. 2007; Hara et al. 2017) (Fig. 1). This activity is regulated by the ability of Png to directly phosphorylate proteins involved in translation, including Pum, Trailer Hitch (Tral), and MEI31B (Hara et al. 2018). A complex including Tral, Cup, and the deadbox helicase ME31B binds widely to maternal RNAs and inhibits translation within the first hour after fertilization (Nakamura et al. 2001; Tritschler et al. 2008). Png activation results in the decrease of the protein levels for all 3 factors over the span of the MZT and results in a shift from translational repression to RNA degradation of ME31B-bound mRNAs (Wang et al. 2017). While it is clear that Png activity regulates the transition from translational repression to RNA degradation, the direct mechanisms remain unclear.
During the MZT, a subset of the maternal proteome is degraded. While this functions on a smaller scale than maternal mRNA decay, it is still important for regulating this transition. Clearance of maternal proteins is partially dependent upon the E2 conjugating enzyme Marie Kondo (Kdo) and the E3 C-terminal to Lis1 Homology (CTLH) ligase complex, which promotes degradation of translational repressors such as ME31B, Cup, and Tral (Cao et al. 2020; Zavortink et al. 2020). Like maternal RNA clearance, degradation of maternal proteins is dependent upon the Png kinase, as upregulation of Kdo protein is impeded in png mutants (Zavortink et al. 2020) (Fig. 1). Conversely, removal of Smg occurs at the end of the MZT and requires the E3 Skp/Cullin/F-box-containing (SCF) ubiquitin ligase complex (Cao et al. 2020). Targeting of Smg for degradation is specifically mediated through an interaction of the F-box containing protein Bard with the C-terminus of Smg (Cao et al. 2022). Proper clearance of maternal repressor proteins during the MZT is important for an error-proof transition to zygotic control. Failure to degrade Smg by the end of the MZT results in Smg-mediated degradation of zygotically expressed targets that are necessary for developmental progression.
Dynamics of genome activation
Genome-wide RNA profiling assays were used to characterize the transcriptomes of early embryos and, importantly, to distinguish maternally loaded RNAs from newly transcribed zygotic RNAs (see Box 1). Tens of genes (∼100) become transcriptionally active in 1–2 h after egg laying (NC8–13), while a few thousand genes are activated at 2–3 h (NC14). While activation is a gradual process, these are referred to as the minor and major waves of ZGA, respectively (Tadros et al. 2003; Pilot et al. 2006; De Renzis et al. 2007; Thomsen et al. 2010; Lott et al. 2011; Chen et al. 2013). Genes activated during the minor wave typically contain TATA boxes in their promoters, a feature associated with high rates of RNA polymerase II initiation and lack of polymerase pausing (Chen et al. 2013; Pimmett et al. 2021). Such strong promoter activity is in accordance with the high-level expression observed for these genes and their roles in subsequent developmental processes including cellularization (e.g. halo, bottleneck [bnk], and slow as molasses [slam]), sex determination and dosage compensation (e.g. Sex-lethal [Sxl] and sisterless [sis] genes), mitotic-cycle lengthening (e.g. frühstart [frs] and tribbles [trbl]), chromatin organization (Elba genes), and patterning (e.g. zerknüllt [zen], snail [sna], and hunchback [hb]) (Liang et al. 2008). As identified for genes expressed in the minor wave in many species, these genes tend to be short and lack introns (De Renzis et al. 2007; Chen et al. 2013; Heyn et al. 2014).
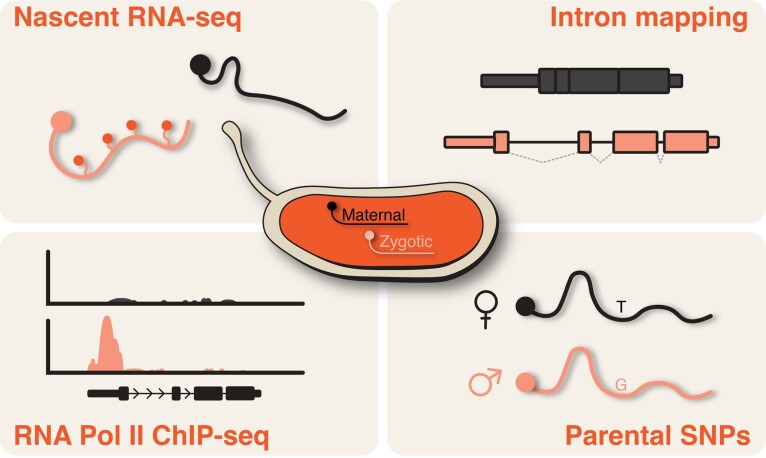
Box 1. Methods to identify zygotic gene expression (see Fig. 3).
The large quantity of maternally provided RNA that drives early embryonic development presents a unique challenge in identifying transcription from the zygotic genome. To address this challenge, multiple different strategies have been developed to elucidate the dynamics of zygotic genome activation. Early analysis relied on injections of radioactive nucleotide followed by identification of transcripts based on molecular weight to determine when large classes of transcripts became detectable (Zalokar 1976; Anderson and Lengyel 1979; Edgar and Schubiger 1986). Subsequently, nascent transcription was determined at the level of single genes using in situ hybridization to detect dots within the nucleus (Shermoen and O’Farrell 1991). With the advent of microarrays, thousands of transcripts could be detected simultaneously. Profiling transcript levels over development enabled changes in expression over time to be determined and used as a method to identify zygotically expressed genes as those whose levels increased over the first few hours of development (Arbeitman et al. 2002; Tadros et al. 2003; Pilot et al. 2006). Nonetheless, maternal transcripts could mask zygotic expression of the same gene. To avoid this complication, expression levels were determined over the MZT in embryos lacking specific chromosomal arms, allowing gene expression to be directly correlated with the presence or absence of a specific locus (De Renzis et al. 2007). In addition, comparing the transcriptomes of unfertilized eggs with those of fertilized embryos from multiple time points through development revealed maternal vs zygotic specific transcripts (Tadros et al. 2003; Thomsen et al. 2010). The increased sensitivity provided by high-throughput sequencing enabled transcriptional profiling in single embryos. Using polymorphic differences between two wild-type strains of flies allowed assignment of transcripts to either the paternal or maternal chromosome. Because any transcripts from the paternal chromosome must be expressed from the zygotic genome, thousands of zygotically expressed genes could be identified with high-temporal resolution (Lott et al. 2011; Ali-Murthy et al. 2013). Additional strategies have taken advantage of distinctive features of active transcripts, including the binding profile of the RNA polymerase II enzyme across the gene (Chen et al. 2013; Blythe and Wieschaus 2015), the ability to selectively identify RNA engaged by an actively transcribing polymerase (GRO-seq) (Saunders et al. 2013), the incorporation of an exogenously provided base analog that allows specific isolation of recently transcribed RNAs (Kwasnieski et al. 2019), and intron-mapping reads (Riemondy et al. 2023). Together, these diverse strategies have taken advantage of technology as it is developed to sensitively identify the dynamics of zygotic genome activation.
Genes expressed during the major wave include many housekeeping genes, which are also maternally expressed. In addition, expression of hundreds of patterning genes is activated by feed-forward transcriptional and signaling networks established by maternal and minor-wave zygotic factors (Nien et al. 2011; Chen et al. 2013). These genes exhibit RNA polymerase II pausing about 50-bp downstream of the transcription start site, a feature frequently found at developmental regulatory genes. In contrast to genes expressed during the minor wave, the promoters of genes expressed during the major wave of ZGA are enriched for motifs other than TATA, for example, pause button (PB), Initiator (Inr), and downstream promoter element (DPE) (Zeitlinger et al. 2007; Chen et al. 2013). They typically contain cis-regulatory modules (CRMs), or enhancers, that interact with specific trans-activators and repressors, which together direct spatiotemporal expression patterns (see more below). In many cases, genes are regulated by multiple enhancers. These can be referred to as primary and shadow enhancers or proximal and distal enhancers, depending on whether one enhancer was identified first or if one is closer to the transcription start site than the other (Hong et al. 2008; Dunipace et al. 2013). In some cases, multiple enhancers regulate transcription in a similar manner and function redundantly to provide resilience to environment or genetic perturbation. In other cases, enhancers act additively or synergistically to elaborate the pattern and can affect polymerase kinetics differently (Perry et al. 2010, 2012; Dunipace et al. 2013, 2019; Staller et al. 2015; Wunderlich et al. 2015; Scholes et al. 2019; Whitney et al. 2022).
As compared to the somatic cells of the embryo, transcriptional dynamics in the germline cells of the blastoderm are delayed. The germ “pole” cells remain quiescent until after gastrulation with activation as early as 3–5 h after fertilization (Williamson and Lehmann 1996; Seydoux and Dunn 1997; Siddiqui et al. 2012). This repression is dependent on posterior determinants, such as nanos and germ cell-less (gcl) as mutations in these genes activate the sis genes precociously (Deshpande et al. 1999; Leatherman et al. 2002). However, it is unclear how they function, as neither encodes a transcriptional repressor. The product of the gene polar granule component (pgc) may be actively involved in repressing transcription in the germline through inhibition of RNA polymerase II phosphorylation (Martinho et al. 2004; Hanyu-Nakamura et al. 2008), and this mechanism has similarities to repression of gene expression in the primordial germline in Caenorhabditis elegans (Seydoux and Dunn 1997). Thus, genome activation in primordial germ cells is regulated distinctly from that of the somatic cells, resulting in a delay in activation that is likely essential for proper germ cell specification. However, there is evidence that this control is not purely autonomous and that regulators of somatic ZGA also influence primordial germ cell development (Colonnetta et al. 2023).
Regulating genome activation
The dynamic activation of the zygotic genome is precisely controlled by multiple interdependent mechanisms. Changes in the length of the division cycle, the ratio of nuclear content to cytoplasm, and the levels of specific transcriptional activators all influence the timing of genome activation.
Division cycle
As discussed above, the nuclear division cycles are highly abbreviated, consisting of only a synthesis (S) phase and mitosis (M) with no gap phases. Because transcription is suppressed during mitosis (Shermoen and O’Farrell 1991), the 5-min S phases preceding NC12 provide little time for transcriptional elongation. In the early embryo, RNA polymerase II elongation rates have been estimated to be ∼1.5–3.0 kb/min (Ardehali and Lis 2009; Garcia et al. 2013; Fukaya et al. 2017), suggesting an upper limit on the length of transcripts that can be expressed during these early division cycles. Supporting the model that S-phase length may constrain zygotic transcription, early transcripts tend to be short and lack introns (De Renzis et al. 2007; Chen et al. 2013). Furthermore, aborted transcripts, as identified by 5′ read biases in RNA sequencing (RNA-seq) data, are more abundant during the highly abbreviated division cycles of NC7–9 than in NC14, comprising ∼60% of the identified zygotic transcripts (Kwasnieski et al. 2019). Some of these aborted transcripts may be actively generated and, if translated, produce truncated protein products with unique functions (Sandler et al. 2018). Thus, division cycle dynamics shape the transcriptome of the early embryo.
Multiple mechanisms ensure the progressive slowing of the division cycle as the MZT progresses. Because mitosis continues to take approximately 5 min in all the early divisions, the lengthening of the division cycle is largely due to the extension of S phase (Fig. 2). In the very early cycles, replication initiates nearly simultaneously throughout the genome (Blumenthal et al. 1974; Shermoen et al. 2010). Later in development, this synchronicity is lost. Late replication occurs predominantly at silenced satellite regions. The binding of Rif1 to these satellite sequences selectively delays their replication (Seller and O’Farrell 2018). In addition to the lengthening of S phase, a gap phase (G2) is added to the division cycle at NC14. This is controlled through the downregulation of maternally provided Cdc25 phosphatase, which results in the inhibitory phosphorylation of the mitotic kinase Cdk1 (Farrell et al. 2012; Di Talia et al. 2013; Farrell and O’Farrell 2013). Additionally, replication rates are controlled by the availability of the dNTPs, the building blocks of the replicated genomes. While the mother deposits sufficient supplies of many of the molecules required for early development, the egg contains only about a third of the required dNTPs (Song et al. 2017). Thus, the embryo must produce the additional dNTPs required to replicate the genome. This process is tightly regulated as dNTPs negatively regulate the enzyme required for their production (Song et al. 2017; Djabrayan et al. 2019). The decreasing levels of dNTPs also result in replication stress that feeds into the slowing of the division cycle (Liu et al. 2019).
Fig. 2.
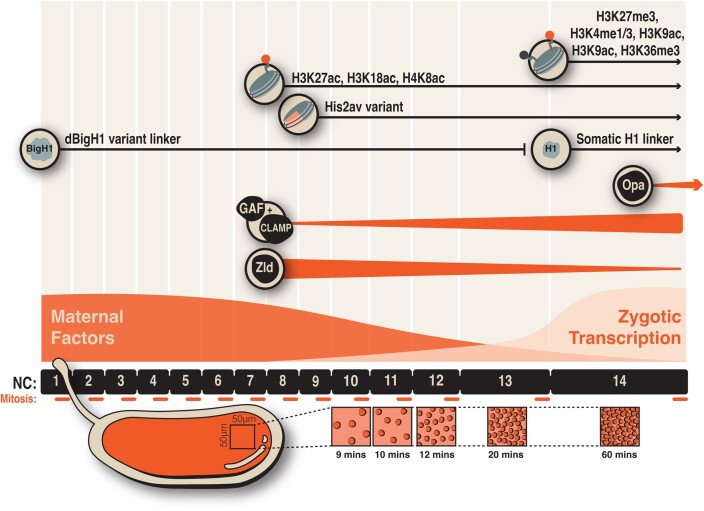
The early zygotic genome is reprogrammed during the MZT. Timeline depicting the initiation and longevity of processes defining the MZT. Within 3 h, the MZT undergoes 14 nuclear division cycles (NC) that are a series of alternating S and M phases. These begin to slow at NC10. Cycle slowing is largely achieved through longer S phases, as mitosis occurs in approximately 5 min regardless of division cycle. Because these nuclear division cycles occur in a shared cytoplasm, nuclear density increases, resulting in an increase in the N/C ratio. Transcriptional activation is mediated by pioneer factors Zld, GAF, Clamp, and Opa, which preferentially function at different time points over this developmental transition (denoted by triangles). Simultaneously, chromatin is restructured through the addition of posttranslational modifications and incorporation of histone variants (presence of protein or modification represented by a line).
Fig. 3.
Multiple methods can be used to classify maternal and zygotic transcripts in the Drosophila embryo on the genome-wide scale (see Box 1). Maternal transcripts are represented in black. Zygotic transcripts are depicted in pink.
In addition, zygotic transcriptional activation shapes the division cycle. Global inhibition of RNA polymerase II transcription leads to additional mitotic divisions and precocious transcription can halt the division cycle (Edgar et al. 1986; Sung et al. 2013). Transcription directly affects the slowing of the cycle through the expression of specific zygotic genes required for slowing the division cycle, such as frs and trbl (Großhans and Wieschaus 2000; Mata et al. 2000; Seher and Leptin 2000; Großhans et al. 2003; Farrell and O’Farrell 2013). In addition, during NC13 conflict between RNA polymerase II binding and DNA replication activates a replication checkpoint to slow the division cycle (Sibon et al. 1997; Blythe and Wieschaus 2015). This interdependence ensures the coordination of genome activation with the dramatic changes in division-cycle dynamics.
N/C ratio
Seminal experiments in Xenopus demonstrated a role for the ratio of the nuclear content to the cytoplasm (nuclear-to-cytoplasmic [N/C] ratio) as regulating MZT dynamics (Newport and Kirschner 1982a, 1982b). In eggs from oviparous organisms, the cytoplasmic volume remains constant during early development while the number of nuclei increases exponentially with each division. Thus, the ratio of cytoplasm to both nuclear volume and nuclear content (DNA) decreases over early development. In frogs, both the addition of exogenous DNA and decreasing cytoplasmic volume could initiate precocious transcription (Newport and Kirschner 1982b). Together, these and other data led to a model whereby titration of a maternally supplied repressor, by the increasing DNA content generated during each division cycle, resulted in progressive activation of the zygotic genome. More recently, studies in both frogs and zebrafish have implicated core histones as maternally supplied repressors that may be either titrated by the increasing DNA content or by changes in their concentration due to changes in nuclear volume (Amodeo et al. 2015; Joseph et al. 2017). Of note, the role of histones may not be directly related to their incorporation into chromatin. The unstructured tail of histone H3 is a substrate for the checkpoint kinase Chk1/grapes and acts as a competitive inhibitor during the early NCs when this histone is abundant (Shindo and Amodeo 2021). Thus, the titration of histones by DNA may function to regulate Chk1-dependent slowing of the division cycle.
In flies, the evidence for the role of the N/C ratio in directly controlling transcription is limited. Haploid embryos, which contain half the DNA content of wild-type embryos, fail to dramatically slow the division cycle at NC14 and undergo an additional round of division. Nonetheless, they attempt to undergo cellularization during the abbreviated NC14, demonstrating that zygotically expressed genes required for cellularization are transcribed. Furthermore, transcription is activated on a similar time frame in embryos arrested after NC10 as in dividing embryos, suggesting that time, and not nuclear content or volume, is regulating activation (Edgar et al. 1986; Strong et al. 2020). Genomic and imaging experiments on haploid embryos have identified the transcriptional activation of a subset of genes that is sensitive to DNA content (Lu et al. 2009; Syed et al. 2021). Nonetheless, many genes assayed in these haploid embryos show a contribution of both developmental time and DNA content to the dynamics of activation (Lu et al. 2009). These data suggest that the while N/C ratio has substantial effects on timing of the division cycle, the direct effects on transcriptional activation are more subtle and likely connected with the effects on division-cycle dynamics.
It is increasingly evident that the slowing of the division cycle, the increased N/C ratio, and transcriptional activation of the zygotic genome are interconnected. Transcription initiates broadly in embryos arrested at interphase of NC12 (Strong et al. 2020). The coupling of widespread ZGA with the lengthening of S phase and the robust transcriptional initiation in embryos arrested with a low N/C ratio together suggest that the N/C ratio is primarily regulating the slowing of the division cycle. It is this slowing, along with the translation of transcriptional activators, that ultimately enables ZGA. This may be a conserved feature of the MZT as ZGA occurs in zebrafish embryos arrested at low N/C ratios. Like Drosophila, transcriptional activation in zebrafish is largely dependent on developmental time and translation of transcriptional activators (Chan et al. 2019).
Regulation of transcriptional activators
While original models for the timing of genome activation focused on a maternally deposited repressor, the regulated expression of essential transcriptional activators also controls the timing of gene expression (Almouzni and Wolffe 1995). These activators are maternally deposited as mRNAs and are subsequently translated, driving transcriptional activation. This has been demonstrated for TATA-binding protein (TBP) in frogs, a histone acetyltransferase in zebrafish, and a transcriptional activator in Drosophila (Veenstra et al. 1999; Chan et al. 2019; Larson et al. 2022). The first indication of the existence of a sequence-specific DNA-binding protein that could initiate genome activation was based on work in Drosophila. A set of related sequence motifs were identified enriched in the regulatory regions of the earliest expressed genes (ten Bosch et al. 2006). The discovery of the protein Zelda (Zld; also known as Vielfaltig [Vfl]) that could bind to these sequences identified the first genome activator in any species (Staudt et al. 2006; Liang et al. 2008).
Discovery of Zld as a master regulator of the Drosophila MZT
The discovery of Zld was predicated on the identification of CAGGTAG and related sequence elements upstream of genes involved in sex determination and dosage compensation (sisA, sisB/scute [sc], and Sxl) (Erickson and Cline 1998; Wrischnik et al. 2003). These were hypothesized to be numerator sequences specific to the sex determination pathway. With the publication of the full genome sequence, it became clear that these sequence elements were enriched not only in the promoters of genes involved in sex determination, but more broadly in the promoters of multiple genes expressed during the earliest stages of development (ten Bosch et al. 2006; De Renzis et al. 2007). Transgenic analysis showed that CAGGTAG and related sequences were required to rescue mutations in sisB/sc and drive expression from the Sxl promoter, demonstrating the necessity of these motifs for promoting gene expression (ten Bosch et al. 2006). Furthermore, addition of CAGGTAG elements to regulatory regions could promote precocious gene expression in a manner dependent on the number of motifs, demonstrating the sufficiency of these motifs in driving early gene activation (ten Bosch et al. 2006). Additional support for the importance of these sequence motifs came from the identification of their enrichment at genomic loci bound by multiple transcription factors involved in early embryonic patterning (Li et al. 2008). The identification of sequence elements that could regulate early zygotic gene expression suggested the existence of a sequence-specific transcription factor with a broad role in genome activation. In 2008, Zld was isolated through its recognition of these sequence elements in a yeast one-hybrid screen and shown to be an essential master regulator of ZGA (Liang et al. 2008).
Zld is encoded by a maternally supplied mRNA and is translated within the first hour of embryogenesis (Nien et al. 2011; Larson et al. 2022). Zld binds the genome and activates the minor wave of transcription as early as NC8 (Harrison et al. 2011). This binding is driven largely by sequence; 64% of Zld-binding motifs are occupied at NC8–10. This sequence-driven binding may be unique to the embryo. In neuroblasts of the larval brain, Zld binding is not dependent on the CAGGTAG motif, suggesting that distinct features that remain to be elucidated regulate Zld binding outside the early embryo (Larson, Komori, et al. 2021). Not all Zld-bound targets activate transcription upon binding. Some early Zld-bound sites do not drive expression until the major wave of ZGA at NC14. Thus, Zld marks these regions for subsequent activation (Harrison et al. 2011). Embryos lacking maternally contributed zld fail to complete the MZT with morphological defects during cellularization (Liang et al. 2008). Furthermore, the loss of maternally encoded Zld results in the downregulation of thousands of genes by the end of the MZT (Liang et al. 2008; Nien et al. 2011; McDaniel et al. 2019). Among these genes are a set of miRNAs that target maternal transcripts for decay, including the mir-309 cluster discussed above (Liang et al. 2008; Fu et al. 2014). Thus, Zld-mediated activation contributes to the degradation of maternal mRNAs and in so doing helps coordinate the reprogramming of the embryonic transcriptome (Fig. 1).
The open reading frame of zld codes for a 1,596 amino acid protein containing 6 C2H2 zinc fingers and a large intrinsically disordered activation domain. Zld is expressed across multiple tissue types, including the embryo, nervous system, larval imaginal discs, and brain (Staudt et al. 2006; Liang et al. 2008; Pearson et al. 2012; Giannios and Tsitilou 2013). Additionally, Zld is present in multiple isoforms, of which the full-length protein is dominant during the MZT (Pearson et al. 2012; Giannios and Tsitilou 2013; Hamm et al. 2015; Reichardt et al. 2018). Mutation to the 4 zinc fingers clustered in the C-terminus fails to activate transcription in a sequence-dependent manner while mutation to the conserved second zinc finger near the N-terminus results in a hyperactive mutant (Hamm et al. 2015, 2017). Embryos either lacking maternally encoded zld or supplied with hyperactive Zld fail to complete the MZT, indicating a delicate balance of Zld levels must be maintained for the embryo to reach gastrulation (Hamm et al. 2017).
Zld is a pioneer factor
The finding that Zld targets are bound before they are transcribed, along with the observed enrichment of CAGGTAG motifs at regions bound by multiple unrelated transcription factors, suggested Zld may function as a pioneer factor (Li et al. 2008; Harrison et al. 2011; Nien et al. 2011; Satija and Bradley 2012). Pioneer factors are particularly well suited to drive developmental transitions, like ZGA, through their ability to bind and displace nucleosomes to increase local chromatin accessibility. This induces a permissible environment in which transcription factors can access binding motifs to upregulate target gene expression (Zaret 2020; Larson, Marsh, et al. 2021). Zld overcomes nucleosomal barriers by bindingDNA wrapped around histones and promoting chromatin accessibility at hundreds of cis-regulatory regions (Schulz et al. 2015; Sun et al. 2015; McDaniel et al. 2019). This Zld-mediated accessibility facilitates the binding of multiple additional transcription factors (Yanez-Cuna et al. 2012; Foo et al. 2014; Xu et al. 2014; Schulz et al. 2015; Li and Eisen 2018).
Once established, Zld remains bound to largely the same genomic regions throughout the MZT despite the rapid division cycles (Harrison et al. 2011). While many transcription factors are not bound to the condensed mitotic chromosomes, a subset of pioneer factors can remain bound and promote gene expression following division, suggesting a possible mechanism for the stable Zld occupancy (Caravaca et al. 2013). However, live imaging demonstrated that Zld is not bound to chromatin during mitosis and, instead, rapidly reengages the genome following division (Dufourt et al. 2018; Mir et al. 2018). While some genes retain a “memory” of their prior transcriptional state through the disruptive process of mitosis, Zld is not important in this process. Instead, Zld functions to decrease the delay in transcriptional reactivation following mitosis and, in so doing, accelerate transcriptional kinetics (Dufourt et al. 2018). Thus, Zld is a pioneer factor that promotes chromatin accessibility, transcription-factor binding, and transcriptional activation.
Zld pioneer activity modulates the activity of morphogens essential for patterning in the embryo
Polarity of the Drosophila body plan is initiated in the early embryo by morphogen gradients. Bicoid (Bcd) and Dorsal (Dl) are maternally encoded transcription factors that are expressed in gradients across the early embryo (Johnston and Nüsslein-Volhard 1992; Irizarry and Stathopoulos 2021). Bcd is concentrated in the anterior to define the developing head and thorax. Dl localization is graded from the ventral side of the embryo, where it functions to specify the dorsal–ventral axis. Zld is not a morphogen. Instead, it is uniformly expressed in nuclei throughout the embryo. Loss of Zld function through either knockout or deletion of its binding motif affects the temporal and spatial expression of morphogen target genes, delaying and, in some cases, narrowing the expression domain (Nien et al. 2011; Kanodia et al. 2012; Foo et al. 2014). Zld therefore acts upstream of Bcd and Dl as a uniform effector of their delineated morphogen gradients, ensuring the proper timing and spatial expression of their targets. Zld-mediated pioneer activity is essential for the robust recruitment of Bcd, Dl, and Twist to target enhancers (Yanez-Cuna et al. 2012; Foo et al. 2014; Xu et al. 2014; Schulz et al. 2015; Sun et al. 2015). As Zld binding can be used to identify early embryonic enhancers, Zld-mediated chromatin accessibility may be generally required for the binding of patterning factors, both transcriptional activators and repressors (Nien et al. 2011; Fu et al. 2014). Thus, although Zld itself is not a morphogen, it potentiates morphogen activity.
The effect Zld has on morphogen-mediated activation is particularly relevant in areas of low concentration (Mir et al. 2018; Yamada et al. 2019). In the embryo, Zld forms subnuclear hubs that are required for the formation of hubs of other transcription factors, such as Bcd and Dl (Dufourt et al. 2018; Mir et al. 2018; Yamada et al. 2019). Thus, Zld may function at the top of developmental patterning networks by creating transcription-factor hubs that result in increased accessibility at cis-regulatory regions, permitting the recruitment of additional transcription factors required for specifying the Drosophila body plan.
Multiple pioneer factors are required to initiate ZGA
Although Zld is a master regulator of ZGA and required for development beyond NC14, additional factors function with Zld to reprogram the early embryonic transcriptome. Initial studies suggested roles for additional pioneer factors as not all Zld-bound regions required Zld for chromatin accessibility. Zld-bound regions that remained accessible in the absence of Zld were enriched for promoters containing GA-dinucleotide repeats (Schulz et al. 2015; Sun et al. 2015; Blythe and Wieschaus 2016; Moshe and Kaplan 2017), a motif recognized by at least two maternally encoded proteins, GAGA-factor (GAF)/Trithorax-like (Trl) and Clamp (Biggin and Tjian 1988; Soruco et al. 2013). Similar to the role of Zld in pioneering chromatin accessibility and gene expression in the early embryo, both GAF and Clamp are required for early embryonic viability, chromatin accessibility, and transcriptional activation (Duan et al. 2021; Gaskill et al. 2021). Together, these pioneer factors progressively establish chromatin accessibility during the MZT. Accessibility is initially evident over Zld-bound enhancers. Subsequently, GAF-bound, GA-rich promoters gain accessibility (Blythe and Wieschaus 2016; Gaskill et al. 2021). Supporting this sequential role for GAF and Zld, genes that require Zld alone are enriched for the earliest expressed genes, while those that require GAF are expressed later during the major wave of ZGA at NC14 (Gaskill et al. 2021). Thus, Zld is the first pioneer factor to initiate transcription and is subsequently followed by GAF and Clamp to further amplify transcription into the major wave. These proteins access the genome largely independently, although Zld and Clamp increase the occupancy of the other at a subset of regions (Duan et al. 2021; Gaskill et al. 2021). Gene expression is progressively controlled by a series of pioneer factors. Following NC14, the zygotically expressed Odd-paired (Opa) is required for accessibility of regulatory elements that control gene expression changes during gastrulation (Koromila et al. 2020; Soluri et al. 2020). In contrast to Zld, GAF and Clamp, Opa is not maternally encoded but is expressed during NC14 and functions to specify transcriptional networks associated with body segmentation (Benedyk et al. 1994). Together, it is evident that early embryonic reprogramming requires the sequential coordination of multiple transcription factors that function to activate transcription from the zygotic genome (Fig. 2).
Zld serves as a model to study pioneer factor function during ZGA
While the MZT is evolutionarily conserved, Zld is only conserved within the Pancrustacea lineage of arthropods (Ribeiro et al. 2017). As in Drosophila melanogaster, maternally encoded Zld drives activation of the zygotic genome in more distantly related species of cockroach and short-germ beetle (Ribeiro et al. 2017; Ventos-Alfonso et al. 2018). Despite the lack of conservation at the sequence level, since the original identification of Zld in Drosophila, pioneer factors have been identified as activators of the zygotic genome in frogs, fish, mice, and humans. Zebrafish and Xenopus rely on orthologs of the Yamanaka reprogramming factors Oct4, Sox2, and Nanog for accessibility (Lee et al. 2013; Leichsenring et al. 2013; Gentsch et al. 2019; Larson, Marsh, et al. 2021). In zebrafish, multiple pioneer factors (Pou5f3, Sox19b, and Nanog) function in concert to activate the developmental program required for progression through the MZT. By contrast, Pou5f3 and Sox3 are individually essential for early Xenopus development (Gentsch et al. 2019). Additional pioneer factors are also important for ZGA in Xenopus, including Foxh1 (Charney et al. 2017; Blitz and Cho 2021). In both mice and humans, the DUX family of factors is important for activating early embryonic gene expression but is not required for development (De Iaco et al. 2017; Hendrickson et al. 2017). More recently, Nr5a2 has been identified as an essential pioneer factor for early mouse development (Gassler et al. 2022). Regardless of the conserved nature of specific factors, the mechanism by which maternally provided pioneer factors overcome nucleosomal barriers to kick-start zygotic transcription is conserved, highlighting the power of Drosophila as a model to understand this essential developmental transition.
Chromatin structure during genome activation
Within the nucleus, DNA is organized by wrapping around histone octamers to form nucleosomes that can then be further compacted. Posttranslational modifications to the histones, variants in histones, nucleosome positioning, and 3D chromatin structure all impact genome organization and gene expression. During the MZT, the genomes of the specified gametes must unite and be reprogrammed to totipotency. This complex process requires dramatic changes to the genomic structure, including rewriting of histone marks, positioning of nucleosomes, and organizing the 3D chromatin structure, and this widespread genomic reprogramming is coordinated with the activation of the zygotic genome.
Histone marks are reestablished during ZGA
Posttranslational modifications to the unstructured tails of histones incorporated into chromatin can change the structure of the chromatin itself and provide a platform for binding by proteins that affect gene expression. These histone modifications are highly correlated with specific genomic features, such as enhancers or promoters, and individual modifications are correlated with the activity of these elements. Profiling of posttranslational modifications to the histone tails (H4K5ac, H4K8ac, H3K9ac, H3K18ac, H3K27ac, H3K4me1, H3K4me3, H3K27me3, and H3K36me3) on tightly staged embryos at multiple stages spanning the MZT (NCs 8, 12, 14a, and 14c) demonstrated a widespread shift in the modification state as embryos developed (Li et al. 2014) (Fig. 2). All of the modifications (except for the replication-associated mark H4K5ac) increased dramatically over the stages assayed. Indeed, both H3K4me3, associated with transcriptional activity, and H3K27me3, associated with Polycomb-mediated repression, are largely absent during the MZT prior to NC14, suggesting transcriptional activity is not dependent on these modifications (Chen et al. 2013; Li et al. 2014). Additional studies identified 32 repressive H3K27me3 domains that are inherited from the oocyte and persist through the MZT (Zenk et al. 2017). In addition to evidence of maternal inheritance of H3K27me3, the histone mark H4K16ac is also maintained from oocytes to embryos, and failure to inherit this modification leads to defects in genome activation and 3D genome organization (Samata et al. 2020). Thus, while the extent of inherited histone modifications remains unclear, it is evident that following fertilization parental histones are diluted through the multiple replication cycles, and these marks must be robustly reestablished.
The chromatin modification state of the early embryo is also actively reorganized to allow the partitioning of the active and silent genomes. Histone acetylation on H4K8, H3K18, and H3K27ac arises on promoters and enhancers concomitantly with gene expression, and H3K18ac at enhancers is at least partially dependent on Zld (Li et al. 2014), suggesting Zld binding or transcriptional activity might help direct this modification. However, while there is a correlation between histone modifications and transcriptional activity, the causative relationship is less clear. While the histone acetyltransferase Nejire (Drosophila CBP) is required for ZGA, this activity may not be absolutely dependent on its enzymatic activity (Ciabrelli et al. 2023). Concomitant with the establishment of the active genome, silent heterochromatin is progressively established. H3K9me3, a mark associated with constitutive heterochromatin, is robustly evident at NC14 (Yuan and O’Farrell 2016). The methyltransferase Eggless helps establish this mark at specific chromatin domains, and this activity may require the slowing of the cell cycle for the robust establishment of H3K9me3 (Seller et al. 2019).
In addition to posttranslational modifications on histone tails, chromatin structure can also be modulated by incorporation of variant histone isoforms. In flies, an embryonic variant of the linker histone H1, dBigH1, is specifically expressed during the MZT and is replaced by the somatic variant (dH1) during genome activation. Indeed, loss of dBigH1 results in precocious elongating RNA polymerase and increased gene expression, suggesting a role in limiting gene expression during the MZT (Pérez-Montero et al. 2013). This is likely a conserved function of variant linker histones, as embryo-specific H1 proteins have been identified in mammals, frogs, zebrafish, and other invertebrates (Godde and Ura 2009). Variants also exist in the core histones. Indeed, the H2A variant H2A.Z (His2Av) is deposited at transcription start sites prior to ZGA and is required for activation of housekeeping genes at NC14 (Ibarra-Morales et al. 2021). Thus, histone variants may function along with transcription factors to structure the early embryonic genome and mediate gene expression (Fig. 2).
Changes in chromatin accessibility
Nucleosome occupancy and organization help to define chromatin structure and its accessibility to chromatin-binding factors, including the transcription factors that drive changes in gene expression. In turn, transcription-factor binding can also influence the accessibility landscape. Thus, chromatin accessibility is both a determinant of transcription-factor occupancy and a reflection of the regulatory potential of a locus. Determining chromatin accessibility at 3-min intervals spanning NC11–13 using the assay for transposase-accessible chromatin sequencing (ATAC-seq) identified the sequential establishment of chromatin accessibility over the MZT (Blythe and Wieschaus 2016). These detailed assays revealed that accessibility was maintained through mitosis and was transiently disrupted but rapidly reestablished during the early replication cycles. Initially accessible regions were enriched for Zld-bound enhancers with GAF-enriched promoter regions gaining accessibility during NC13. Studies using orthogonal methods to identify accessible chromatin domains in embryos depleted for maternal zld (micrococcal nuclease sequencing [MNase-seq] or formaldehyde-assisted isolation of regulatory elements [FAIRE-seq]) demonstrated a requirement for Zld to establish accessibility at hundreds of loci but also showed a role for additional factors, including the ubiquitously expressed protein GAF (Schulz et al. 2015; Sun et al. 2015). Indeed, the capacity of Zld to bind to nucleosomes has been proposed to enable it to facilitate the rapid reestablishment of accessibility following replication (McDaniel et al. 2019). While Zld-mediated chromatin accessibility influences binding of the additional transcription factors, these factors may also influence the accessibility landscape (Hannon et al. 2017; Haines and Eisen 2018; Brennan et al. 2022). Furthermore, the relationship between accessibility and gene expression is not straightforward. Promoters of many genes are accessible throughout the blastoderm embryo despite expression in limited domains. By contrast, enhancers show differences in accessibility that reflect anterior–posterior biases in the expression patterns of the genes they drive (Haines and Eisen 2018). Thus, similar to the widespread increase in histone modifications over the MZT, the genome gains specific regions of accessibility that reflect the regulatory landscape of promoters and enhancers.
Chromatin structure arises over the MZT
Apart from DNA accessibility, the 3D structure of the genome may also influence gene expression. Briefly, the genome is organized into both active (A) and inactive (B) compartments that are further subdivided into smaller topologically associated domains (TADs) (for more information, see the FlyBook Chapter; Schwartz and Cavalli 2017). While the extent to which this 3D structure impacts gene expression remains debated, it is clear that much of this structure arises during the MZT (Hug et al. 2017). Given that TADs and compartments are lost during metaphase, it is possible that the rapid embryonic division cycles provide a barrier to the stable formation of 3D chromatin structure. Prior to ZGA, the genome is largely unstructured with TAD boundaries being established concomitant with recruitment of RNA polymerase II and transcriptional activation. Despite different gene expression profiles, TAD boundary structure is nearly identical between the anterior and posterior halves of the embryo (Stadler et al. 2017). Furthermore, RNA polymerase II activity is not required for TAD formation, suggesting active transcription is not required for boundary formation (Hug et al. 2017). Zld motifs are enriched at boundaries established during NC12–13, and Zld is required for TAD formation at a discrete number of loci. These data provide a connection between the local chromatin effects mediated by Zld and more global changes in genome organization (Hug et al. 2017; Ogiyama et al. 2018). Contact sites for later-established repressive domains are enriched for GAF-binding sites, implicating GAF in organizing the 3D genome following the MZT (Ogiyama et al. 2018). Contrary to this prediction, GAF is dispensable for TAD formation but has a role in facilitating the interactions between promoters and enhancers mediated by tethering elements (Gaskill et al. 2021; Batut et al. 2022; Levo et al. 2022; Li et al. 2023). Because TADs are largely maintained throughout development and between cell types, these data suggest that some 3D structure is organized concomitantly with the activation of the zygotic genome during the MZT.
Phase separation may facilitate the 3D organization of chromatin within the nucleus. Multivalent interactions between low-complexity protein domains have been shown to promote the condensation of proteins and nucleic acids to form membraneless organelles. A major component of constitutive heterochromatin, Heterochromatin Protein 1 (HP1a), can phase separate in vitro and forms phase separated foci in vivo along with the establishment of heterochromatin during NC13–14 (Strom et al. 2017). This is likely instructive in promoting the compartmentalization of chromatin as loss of HP1a leads to a decrease in segregation between the active A compartment and the inactive B compartment (Zenk et al. 2017). Nonetheless, not all foci are mediated through multivalent interactions by low-complexity domains. In addition to acting as a pioneer factor to activate the zygotic genome, GAF is instrumental in promoting silenced heterochromatin at highly abundant AAGAG simple satellite repeats. In this case, it is the DNA binding of GAF that localizes it to these repeats, rather than low-complexity regions of the protein (Gaskill et al. 2023. Together, these data demonstrate that multiple mechanistic features drive the segregation of the active and inactive genome during the MZT.
Outlook and future directions
Over the first few hours following fertilization, the embryo must rapidly transition from maternal-to-zygotic control of development. This rapid and efficient reprogramming requires the precise coordination of multiple processes, including the division cycle, maternal mRNA decay, degradation of maternally provided proteins, and activation of transcription from the zygotic genome. The robust regulation of these processes is, in part, mediated through their interdependence (Fig. 1). Additionally, in Drosophila, stabilization of the Png kinase links progression through the MZT to egg activation and helps ensure the coordinated timing of all the processes required for this essential transition. By phosphorylating proteins involved in translational regulation, the Png kinase promotes translation of a large number of genes, including cyclin B, smg, zld, and kdo, which are required for the division cycle, mRNA degradation, ZGA, and protein degradation, respectively (Tadros et al. 2007; Vardy and Orr-Weaver 2007; Kronja, Yuan, et al. 2014; Zavortink et al. 2020; Larson et al. 2022) (Fig. 1). Decades of study in Drosophila have demonstrated that slowing of the division cycle, degradation of maternal products, and transcriptional activation of the zygotic genome are each required for development and identified the importance of pioneer transcription factors in this reprogramming event. As in the past, it is evident that studies in Drosophila will continue to pave the way in understanding the processes that drive the MZT. Whether it is through the use of state-of-the-art imaging of transcriptional dynamics, determination of single-molecule dynamics, powerful gene-editing strategies, or detailed genomic methodologies, Drosophila will be at the forefront of determining the mechanisms of this essential, conserved developmental transition (see Box 2).
Box 2. Tools for studying genome activation (see Fig. 4).
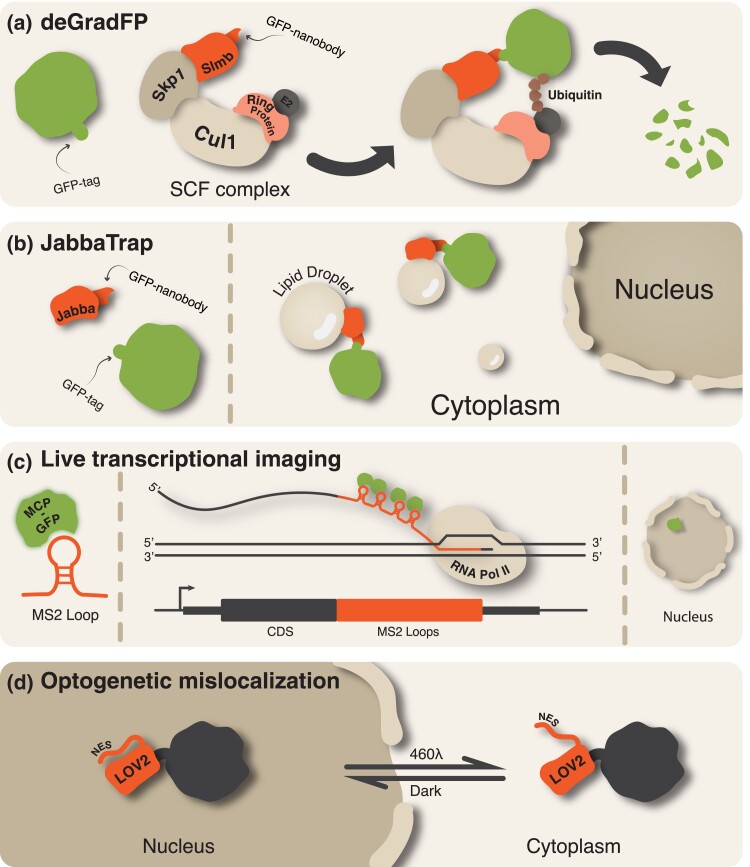
The essential nature of factors present during the MZT makes maternal depletion experiments a challenge in many model systems. Maintaining homozygous mutants is impossible as animals fail to survive to adulthood or result in female infertility. The Drosophila system is particularly suited to circumvent these problems through genetic tools that restrict maternal depletion to early embryo development. Adaption of the recombinase FLP/FRT system revolutionized early developmental genetics through the generation of germline mutant clones and has been used to study quintessential ZGA players like Zld (Theodosiou and Xu 1998). Knock-down of maternal mRNA via RNA interference has been optimized through the use of the GAL4/UAS system in which maternal GAL4 drivers restrict the expression of a short hairpin RNA to the oocyte for targeted depletion by the RNA-induced silencing complex (Staller et al. 2013). Maternal factors can also be knocked down at the protein level through maternal expression of deGradFP. This transgene codes for a F-box protein fused to a GFP nanobody that intercalates with the fly's endogenous E3 ubiquitin ligase complex to ubiquitinylate GFP-tagged proteins and mark them degradation (Caussinus et al. 2012; Gaskill et al. 2021) (Fig. 4a). A related strategy, called JabbaTrap, utilizes a GFP nanobody fused to Jabba to sequester GFP-tagged proteins to lipid droplets. This prevents their localization to the nucleus and results in a failure to activate targets (Seller et al. 2019) (Fig. 4b). In addition to maternal depletion, visualization and isolation of endogenous maternal factors have become relatively straightforward with Cas9-mediated genome-editing strategies, allowing researchers to tag seemingly any maternal protein of interest (Gratz et al. 2013). The lack of pigmentation in the Drosophila embryo makes live imaging of fluorescently tagged proteins easy. Expression of a fluorescently tagged histone can be used to measure nuclear density and therefore to precisely stage developmental timing (Fig. 2). Transcription can be imaged in live embryos using MS2-tagged reporters. In this approach, MS2 stem loops are added to the coding sequence of interest. When expressed as RNA, these loops are bound by transgenically expressed and fluorescently labeled MS2-coat protein (MCP). As RNA polymerase II transcribes the MS2 loops, MCP binds the secondary loop structure and serves as a visual readout of nascent transcription in vivo (Garcia et al. 2013; Gregor et al. 2014) (Fig. 4c). Finally, protein function can be manipulated with light-sensitive tags that act immediately upon stimulation and are not limited by the requirement for specific drivers like the previously mentioned depletion strategies. The plant derived CRY2 optogenetic tag has been used to inactivate endogenously tagged maternal proteins like Bcd and Zld during ZGA after blue-light treatment of embryos through ZGA (Huang et al. 2017; McDaniel et al. 2019). Optogenetic regulation of nuclear localization is a powerful strategy to control transcription factor activity (Kögler et al. 2021) (Fig. 4d). In addition, blue light can be used to induce release of a degradation signal with temporal precision (Irizarry et al. 2020). The ever-expanding toolkit for studies in Drosophila has ensured that the model remains an essential system for defining the mechanisms driving the MZT.
Fig. 4.
Genetic tools used to study factors involved in the MZT (see Box 2). a) Degradation of endogenously tagged GFP proteins through maternal expression of the deGradFP nanobody fusion. b) The JabbaTrap transgenic system sequesters GFP-tagged transcription factors outside the nucleus for loss of function studies. c) MS2 reporters enable visualization of nascent transcription in the living embryo. Transcription is visualized as a fluorescent spot in the nucleus (right). d) Reversible mislocalization of transcription factors to the cytoplasm can be achieved through the iLEXY optogenetic system. NES, nuclear export sequence.
Contributor Information
Melissa M Harrison, Department of Biomolecular Chemistry, University of Wisconsin-Madison, Madison, WI 53706 USA.
Audrey J Marsh, Department of Biomolecular Chemistry, University of Wisconsin-Madison, Madison, WI 53706 USA.
Christine A Rushlow, Department of Biology, New York University, New York, NY 10003 USA.
Funding
M.M.H. was supported by R35 GM136298 and C.A.R. by R35 GM148241 from the National Institutes of Health. A.J.M. was supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1747503.
Literature cited
- Ali-Murthy Z, Lott SE, Eisen MB, Kornberg TB. An essential role for zygotic expression in the pre-cellular Drosophila embryo. PLoS Genet. 2013;9(4):e1003428. doi: 10.1371/journal.pgen.1003428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G, Wolffe AP. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. EMBO J. 1995;14(8):1752–1765. doi: 10.1002/j.1460-2075.1995.tb07164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodeo AA, Jukam D, Straight AF, Skotheim JM. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc Natl Acad Sci U S A. 2015;112(10):E1086-95. doi: 10.1073/pnas.1413990112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KV, Lengyel JA. Rates of synthesis of major classes of RNA in Drosophila embryos. Dev Biol. 1979;70(1):217–231. doi:0012-1606(79)90018-6. [DOI] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EEM, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297(5590):2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- Ardehali MB, Lis JT. Tracking rates of transcription and splicing in vivo. Nat Struct Mol Biol. 2009;16(11):1123–1124. doi: 10.1038/nsmb1109-1123. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J. 1999;18(9):2610–2620. doi: 10.1093/emboj/18.9.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut PJ, Bing XY, Sisco Z, Raimundo J, Levo M, Levine MS. Genome organization controls transcriptional dynamics during development. Science. 2022;375(6580):566–570. doi: 10.1126/science.abi7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedyk MJ, Mullen JR, DiNardo S. Odd-paired: a zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes Dev. 1994;8(1):105–117. doi: 10.1101/gad.8.1.105. [DOI] [PubMed] [Google Scholar]
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development. 2009;136(6):923–932. doi: 10.1242/dev.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin MD, Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53(5):699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Blitz IL, Cho KWY. Control of zygotic genome activation in Xenopus. Curr Top Dev Biol. 2021;145:167–204. doi: 10.1016/bs.ctdb.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38(0):205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Blythe SA, Wieschaus EF. Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell. 2015;160(6):1169–1181. doi: 10.1016/j.cell.2015.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, Wieschaus EF. Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. eLife. 2016;5:e20148. doi: 10.7554/eLife.20148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan KJ, Weilert M, Krueger S, Pampari A, Liu H-Y, Yang AWH, Hughes TR, Rushlow CA, Kundaje A, Zeitlinger J. Chromatin accessibility is a two-tier process regulated by transcription factor pioneering and enhancer activation. Dev Cell. 2023;58. doi:10.106/j.devcel.2023.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18(7):501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Cao WX, Kabelitz S, Gupta M, Yeung E, Lin S, Rammelt C, Ihling C, Pekovic F, Low TCH, Siddiqui NU, et al. Precise temporal regulation of post-transcriptional repressors is required for an orderly Drosophila maternal-to-zygotic transition. Cell Rep. 2020;31(12):107783. doi: 10.1016/j.celrep.2020.107783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao WX, Karaiskakis A, Lin S, Angers S, Lipshitz HD. The F-box protein Bard (CG14317) targets the Smaug RNA-binding protein for destruction during the Drosophila maternal-to-zygotic transition. Genetics. 2022;220(1):iyab177. doi: 10.1093/genetics/iyab177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 2013;27(3):251–260. doi: 10.1101/gad.206458.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E, Kanca O, Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol. 2012;19(1):117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- Chan SH, Tang Y, Miao L, Darwich-Codore H, Vejnar CE, Beaudoin J-D, Musaev D, Fernandez JP, Benitez MDJ, Bazzini AA, et al. Brd4 and P300 confer transcriptional competency during zygotic genome activation. Dev Cell. 2019;49(6):867–881.e8. doi: 10.1016/j.devcel.2019.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charney RM, Forouzmand E, Cho JS, Cheung J, Paraiso KD, Yasuoka Y, Takahashi S, Taira M, Blitz IL, Xie X, et al. Foxh1 occupies cis-regulatory modules prior to dynamic transcription factor interactions controlling the mesendoderm gene program. Dev Cell. 2017;40(6):595–607.e4. doi: 10.1016/j.devcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Dumelie JG, Li X, Cheng MH, Yang Z, Laver JD, Siddiqui NU, Westwood JT, Morris Q, Lipshitz HD, et al. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome Biol. 2014;15(1):R4. doi: 10.1186/gb-2014-15-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Johnston J, Shao W, Meier S, Staber C, Zeitlinger J. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife. 2013;2:e00861. doi: 10.7554/eLife.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciabrelli F, Rabbani L, Cardamone F, Zenk F, Löser E, Schächtle MA, Mazina M, Loubiere V, Iovino N. CBP And Gcn5 drive zygotic genome activation independently of their catalytic activity. Sci Adv. 2023;9(16):eadf2687. doi: 10.1126/sciadv.adf2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonnetta MM, Schedl P, Deshpande G. Germline/soma distinction in Drosophila embryos requires regulators of zygotic genome activation. eLife. 2023;12:e78188. doi: 10.7554/eLife.78188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, et al. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet. 2017;49(6):941–945. doi: 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, Wieschaus EF, Shu S. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo, (T. Kornberg, ed.). PLoS Biol. 2007;5(5):e117. doi:06-PLBI-RA-1961R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Yanowitz JL, Schedl PD. Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell. 1999;99(3):271–281. doi: 10.1016/s0092-8674(00)81658-x. [DOI] [PubMed] [Google Scholar]
- Di Talia S, She R, Blythe SA, Lu X, Zhang QF, Wieschaus EF. Posttranslational control of Cdc25 degradation terminates Drosophila's early cell-cycle program. Current Biology: CB. 2013;23(2):127–132. doi: 10.1016/j.cub.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djabrayan NJ-V, Smits CM, Krajnc M, Stern T, Yamada S, Lemon WC, Keller PJ, Rushlow CA, Shvartsman SY. Metabolic regulation of developmental cell cycles and zygotic transcription. Curr Biol. 2019;29(7):1193–1198.e5. doi: 10.1016/j.cub.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Rieder L, Colonnetta MM, Huang A, Mckenney M, Watters S, Deshpande G, Jordan W, Fawzi N, Larschan E. CLAMP And Zelda function together to promote Drosophila zygotic genome activation. eLife. 2021;10:e69937. doi: 10.7554/eLife.69937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourt J, Trullo A, Hunter J, Fernandez C, Lazaro J, Dejean M, Morales L, Nait-Amer S, Schulz KN, Harrison MM, et al. Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. Nat Commun. 2018;9(1):5194. doi: 10.1038/s41467-018-07613-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunipace L, Ákos Z, Stathopoulos A. Coacting enhancers can have complementary functions within gene regulatory networks and promote canalization. PLoS Genet. 2019;15(12):e1008525. doi: 10.1371/journal.pgen.1008525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunipace L, Saunders A, Ashe HL, Stathopoulos A. Autoregulatory feedback controls sequential action of cis-regulatory modules at the brinker locus. Dev Cell. 2013;26(5):536–543. doi: 10.1016/j.devcel.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Kiehle CP, Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 1986;44(2):365–372. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986;44(6):871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- Erickson JW, Cline TW. Key aspects of the primary sex determination mechanism are conserved across the genus Drosophila. Development. 1998;125(16):3259–3268. doi: 10.1242/dev.125.16.3259. [DOI] [PubMed] [Google Scholar]
- Farrell JA, O’Farrell PH. Mechanism and regulation of Cdc25/Twine protein destruction in embryonic cell-cycle remodeling. Current Biology: CB. 2013;23(2):118–126. doi: 10.1016/j.cub.2012.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JA, Shermoen AW, Yuan K, O’Farrell PH. Embryonic onset of late replication requires Cdc25 down-regulation. Genes Dev. 2012;26(7):714–725. doi: 10.1101/gad.186429.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 1989;107(1):1–22. doi: 10.1242/dev.107.1.1. [DOI] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61(1):31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Foo SM, Sun Y, Lim B, Ziukaite R, O’Brien K, Nien C-Y, Kirov N, Shvartsman SY, Rushlow CA. Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr Biol. 2014;24(12):1341–1346. doi: 10.1016/j.cub.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Nien C-Y, Liang H-L, Rushlow C. Co-activation of microRNAs by Zelda is essential for early Drosophila development. Development (Cambridge, England). 2014;141(10):2108–2118. doi: 10.1242/dev.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Lim B, Levine M. Rapid rates of pol II elongation in the Drosophila embryo. Curr Biol. 2017;27(9):1387–1391. doi: 10.1016/j.cub.2017.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia HG, Tikhonov M, Lin A, Gregor T. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr Biol. 2013;23(21):2140–2145. doi: 10.1016/j.cub.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskill MM, Gibson TJ, Larson ED, Harrison MM. GAF is essential for zygotic genome activation and chromatin accessibility in the early Drosophila embryo. eLife. 2021;10:e66668. doi: 10.7554/eLife.66668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskill MM, Soluri IV, Branks AE, Boka AP, Stadler MR,Vietor K, Huang HS, Gibson TJ, Mukherjee A, Mir M, et al. Localization of the pioneer factor GAF to subnuclear foci is driven by DNA binding and required to silence satellite repeat expression. Dev Cell. 2023;58. doi: 10.106/j.devcel.2023.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassler J, Kobayashi W, Gáspár I, Ruangroengkulrith S, Mohanan A, Gómez Hernández L, Kravchenko P, Kümmecke M, Lalic A, Rifel N, et al. Zygotic genome activation by the totipotency pioneer factor Nr5a2. Science. 2022;378(6626):1305–1315. doi: 10.1126/science.abn7478. [DOI] [PubMed] [Google Scholar]
- Gentsch GE, Spruce T, Owens NDL, Smith JC. Maternal pluripotency factors initiate extensive chromatin remodelling to predefine first response to inductive signals. Nat Commun. 2019;10(1):4269. doi: 10.1038/s41467-019-12263-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103(12):4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannios P, Tsitilou SG. The embryonic transcription factor Zelda of Drosophila melanogaster is also expressed in larvae and may regulate developmentally important genes. Biochem Biophys Res Commun. 2013;438(2):329–333. doi: 10.1016/j.bbrc.2013.07.071. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312(5770):75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Godde JS, Ura K. Dynamic alterations of linker histone variants during development. Int J Dev Biol. 2009;53(2–3):215–224. doi: 10.1387/ijdb.082644jg. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided cas9 nuclease. Genetics. 2013;194(4):1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Garcia HG, Little SC. The embryo as a laboratory: quantifying transcription in Drosophila. Trends Genet. 2014;30(8):364–375. doi: 10.1016/J.TIG.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Großhans J, Müller HAJ, Wieschaus E. Control of cleavage cycles in Drosophila embryos by frühstart. Dev Cell. 2003;5(2):285–294. doi: 10.1016/S1534-5807(03)00208-9. [DOI] [PubMed] [Google Scholar]
- Großhans J, Wieschaus E. A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila. Cell. 2000;101(5):523–531. doi: 10.1016/S0092-8674(00)80862-4. [DOI] [PubMed] [Google Scholar]
- Haines JE, Eisen MB. Patterns of chromatin accessibility along the anterior-posterior axis in the early Drosophila embryo. PLoS Genet. 2018;14(5):e1007367. doi: 10.1371/journal.pgen.1007367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm DC, Bondra ER, Harrison MM. Transcriptional activation is a conserved feature of the early embryonic factor Zelda that requires a cluster of four zinc fingers for DNA binding and a low-complexity activation domain. J Biol Chem. 2015;290(6):3508–3518. doi: 10.1074/jbc.M114.602292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm DC, Larson ED, Nevil M, Marshall KE, Bondra ER, Harrison MM. A conserved maternal-specific repressive domain in Zelda revealed by Cas9-mediated mutagenesis in Drosophila melanogaster, (M. B. Eisen, ed.). PLoS Genet. 2017;13(12):e1007120. doi: 10.1371/journal.pgen.1007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon CE, Blythe SA, Wieschaus EF. Concentration dependent chromatin states induced by the Bicoid morphogen gradient. eLife. 2017;6:e28275. doi: 10.7554/eLife.28275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451(7179):730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Lourido S, Petrova B, Lou HJ, Von Stetina JR, Kashevsky H, Turk BE, Orr-Weaver TL. Identification of PNG kinase substrates uncovers interactions with the translational repressor TRAL in the oocyte-to-embryo transition. eLife. 2018;7:e33150. doi: 10.7554/eLife.33150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Petrova B, Orr-Weaver TL. Control of PNG kinase, a key regulator of mRNA translation, is coupled to meiosis completion at egg activation. eLife. 2017;6:e22219. doi: 10.7554/eLife.22219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li X-YY, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition, (G. P. Copenhaver, ed.). PLoS Genet. 2011;7(10):e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim J-W, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet. 2017;49(6):925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn P, Kircher M, Dahl A, Kelso J, Tomancak P, Kalinka AT, Neugebauer KM. The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell Rep. 2014;6(2):285–292. doi: 10.1016/j.celrep.2013.12.030. [DOI] [PubMed] [Google Scholar]
- Hong J-W, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321(5894):1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A, Amourda C, Zhang S, Tolwinski NS, Saunders TE. Decoding temporal interpretation of the morphogen Bicoid in the early Drosophila embryo. eLife. 2017;6:e26258. doi: 10.7554/eLife.26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell. 2017;169(2):216–228.e19. doi: 10.1016/j.cell.2017.03.024. [DOI] [PubMed] [Google Scholar]
- Ibarra-Morales D, Rauer M, Quarato P, Rabbani L, Zenk F, Schulte-Sasse M, Cardamone F, Gomez-Auli A, Cecere G, Iovino N. Histone variant H2A.Z regulates zygotic genome activation. Nat Commun. 2021;12(1):7002. doi: 10.1038/s41467-021-27125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry J, McGehee J, Kim G, Stein D, Stathopoulos A. Twist-dependent ratchet functioning downstream from Dorsal revealed using a light-inducible degron. Genes Dev. 2020;34(13–14):965–972. doi: 10.1101/gad.338194.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry J, Stathopoulos A. Dynamic patterning by morphogens illuminated by cis-regulatory studies. Development. 2021;148(2):dev196113. doi: 10.1242/dev.196113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DS, Nüsslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68(2):201–219. doi: 10.1016/0092-8674(92)90466-P. [DOI] [PubMed] [Google Scholar]
- Joseph SR, Pálfy M, Hilbert L, Kumar M, Karschau J, Zaburdaev V, Shevchenko A, Vastenhouw NL. Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. eLife. 2017;6:e23326. doi: 10.7554/eLife.23326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanodia JS, Liang H-L, Kim Y, Lim B, Zhan M, Lu H, Rushlow CA, Shvartsman SY. Pattern formation by graded and uniform signals in the early Drosophila embryo. Biophys J. 2012;102(3):427–433. doi: 10.1016/j.bpj.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kögler AC, Kherdjemil Y, Bender K, Rabinowitz A, Marco-Ferreres R, Furlong EEM. Extremely rapid and reversible optogenetic perturbation of nuclear proteins in living embryos. Dev Cell. 2021;56(16):2348–2363.e8. doi: 10.1016/j.devcel.2021.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromila T, Gao F, Iwasaki Y, He P, Pachter L, Gergen JP, Stathopoulos A. Odd-paired is a pioneer-like factor that coordinates with Zelda to control gene expression in embryos. eLife. 2020;9:e59610. doi: 10.7554/eLife.59610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I, Whitfield ZJ, Yuan B, Dzeyk K, Kirkpatrick J, Krijgsveld J, Orr-Weaver TL. Quantitative proteomics reveals the dynamics of protein changes during Drosophila oocyte maturation and the oocyte-to-embryo transition. Proc Natl Acad Sci U S A. 2014;111(45):16023–16028. doi: 10.1073/pnas.1418657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I, Yuan B, Eichhorn SW, Dzeyk K, Krijgsveld J, Bartel DP, Orr-Weaver TL. Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Rep. 2014;7(5):1495–1508. doi: 10.1016/j.celrep.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasnieski JC, Orr-Weaver TL, Bartel DP. Early genome activation in Drosophila is extensive with an initial tendency for aborted transcripts and retained introns. Genome Res. 2019;29(7):1188–1197. doi: 10.1101/gr.242164.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson ED, Komori H, Fitzpatrick ZA, Krabbenhoft SD, Lee C-Y, Harrison M. Premature translation of the Drosophila zygotic genome activator Zelda is not sufficient to precociously activate gene expression. G3 (Bethesda). 2022;12(9):jkac159. doi: 10.1093/g3journal/jkac159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson ED, Komori H, Gibson TJ, Ostgaard CM, Hamm DC, Schnell JM, Lee C-Y, Harrison MM. Cell-type-specific chromatin occupancy by the pioneer factor Zelda drives key developmental transitions in Drosophila. Nat Commun. 2021;12(1):7153. doi: 10.1038/s41467-021-27506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson ED, Marsh AJ, Harrison MM. Pioneering the developmental frontier. Mol Cell. 2021;81(8):1640–1650. doi: 10.1016/j.molcel.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver JD, Li X, Ray D, Cook KB, Hahn NA, Nabeel-Shah S, Kekis M, Luo H, Marsolais AJ, Fung KYY, et al. Brain tumor is a sequence-specific RNA-binding protein that directs maternal mRNA clearance during the Drosophila maternal-to-zygotic transition. Genome Biol. 2015;16(1):94. doi: 10.1186/s13059-015-0659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman JL, Levin L, Boero J, Jongens TA. Germ cell-less acts to repress transcription during the establishment of the Drosophila germ cell lineage. Curr Biol. 2002;12(19):1681–1685. doi: 10.1016/s0960-9822(02)01182-x. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131(1):174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503(7476):360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LA, Van Hoewyk D, Orr-Weaver TL. The Drosophila cell cycle kinase PAN GU forms an active complex with PLUTONIUM and GNU to regulate embryonic divisions. Genes Dev. 2003;17(23):2979–2991. doi: 10.1101/gad.1132603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, Maes J, Mossner R, Driever W, Onichtchouk D. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 2013;341(6149):1005–1009. doi: 10.1126/science.1242527. [DOI] [PubMed] [Google Scholar]
- Levo M, Raimundo J, Bing XY, Sisco Z, Batut PJ, Ryabichko S, Gregor T, Levine MS. Transcriptional coupling of distant regulatory genes in living embryos. Nature. 2022;605(7911):754–760. doi: 10.1038/s41586-022-04680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-Y, Eisen MB. Zelda potentiates transcription factor binding to zygotic enhancers by increasing local chromatin accessibility during early Drosophila melanogaster embryogenesis. BioRxiv. 2018. doi: 10.1101/380857 [DOI] [Google Scholar]
- Li X-Y, Harrison MM, Villalta JE, Kaplan T, Eisen MB. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife. 2014;3:e03737. doi: 10.7554/eLife.03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Hendriks CLL, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6(2):e27. 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tang X, Bing X, Catalano C, Li T, Dolsten G, Wu C, Levine M. GAGA-associated factor fosters loop formation in the Drosophila genome. Mol Cell. 2023;83(9):1519–1526. doi: 10.1016/j.molcel.2023.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456(7220):400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Winkler F, Herde M, Witte C-P, Großhans J. A link between deoxyribonucleotide metabolites and embryonic cell-cycle control. Curr Biol. 2019;29(7):1187–1192.e3. doi: 10.1016/j.cub.2019.02.021. [DOI] [PubMed] [Google Scholar]
- Lohs-Schardin M, Cremer C, Nüsslein-Volhard C. A fate map for the larval epidermis of Drosophila melanogaster: localized cuticle defects following irradiation of the blastoderm with an ultraviolet laser microbeam. Dev Biol. 1979;73(2):239–255. doi: 10.1016/0012-1606(79)90065-4. [DOI] [PubMed] [Google Scholar]
- Lohs-Schardin M, Sander K, Cremer C, Cremer T, Zorn C. Localized ultraviolet laser microbeam irradiation of early Drosophila embryos: fate maps based on location and frequency of adult defects. Dev Biol. 1979;68(2):533–545. doi: 10.1016/0012-1606(79)90224-0. [DOI] [PubMed] [Google Scholar]
- Lott SE, Villalta JE, Schroth GP, Luo S, Tonkin LA, Eisen MB. Noncanonical compensation of the zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 2011;9(2):e1000590. doi: 10.1371/journal.pbio.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Li JM, Elemento O, Tavazoie S, Wieschaus EF. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development (Cambridge, England). 2009;136(12):2101–2110. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA. 2009;15(12):2351–2363. doi: 10.1261/rna.1882009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr Biol. 2004;14(2):159–165. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- Mata J, Curado S, Ephrussi A, Rørth P. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell. 2000;101(5):511–522. doi: 10.1016/S0092-8674(00)80861-2. [DOI] [PubMed] [Google Scholar]
- McDaniel SL, Gibson TJ, Schulz KN, Fernandez Garcia M, Nevil M, Jain SU, Lewis PW, Zaret KS, Harrison MM. Continued activity of the pioneer factor Zelda is required to drive zygotic genome activation. Mol Cell. 2019;74(1):185–195. doi: 10.1016/j.molcel.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M, Stadler MR, Ortiz SA, Hannon CE, Harrison MM, Darzacq X, Eisen MB. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife. 2018;7:e40497. doi: 10.7554/eLife.40497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshe A, Kaplan T. Genome-wide search for Zelda-like chromatin signatures identifies GAF as a pioneer factor in early fly development. Epigenetics Chromatin. 2017;10(1):33. doi: 10.1186/s13072-017-0141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128(17):3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982a;30(3):675–686. doi:0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982b;30(3):687–696. doi:0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7(10):e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiyama Y, Schuettengruber B, Papadopoulos GL, Chang J-M, Cavalli G. Polycomb-dependent chromatin looping contributes to gene silencing during Drosophila development. Mol Cell. 2018;71(1):73–88.e5. doi: 10.1016/j.molcel.2018.05.032. [DOI] [PubMed] [Google Scholar]
- Pearson JC, Watson JD, Crews ST. Drosophila melanogaster Zelda and single-minded collaborate to regulate an evolutionarily dynamic CNS midline cell enhancer. Dev Biol. 2012;366(2):420–432. doi: 10.1016/j.ydbio.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Montero S, Carbonell A, Morán T, Vaquero A, Azorín F. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev Cell. 2013;26(6):578–590. doi: 10.1016/j.devcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20(17):1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Bothma JP, Luu RD, Levine M. Precision of hunchback expression in the Drosophila embryo. Curr Biol. 2012;22(23):2247–2252. doi: 10.1016/j.cub.2012.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot F, Philippe J-M, Lemmers C, Chauvin J-P, Lecuit T. Developmental control of nuclear morphogenesis and anchoring by Charleston, identified in a functional genomic screen of Drosophila cellularisation. Development. 2006;133(4):711–723. doi: 10.1242/dev.02251. [DOI] [PubMed] [Google Scholar]
- Pimmett VL, Dejean M, Fernandez C, Trullo A, Bertrand E, Radulescu O, Lagha M. Quantitative imaging of transcription in living Drosophila embryos reveals the impact of core promoter motifs on promoter state dynamics. Nat Commun. 2021;12(1):4504. doi: 10.1038/s41467-021-24461-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt I, Bonnay F, Steinmann V, Loedige I, Burkard TR, Meister G, Knoblich JA. The tumor suppressor Brat controls neuronal stem cell lineages by inhibiting Deadpan and Zelda. EMBO Rep. 2018;19(1):102–117. doi: 10.15252/embr.201744188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro L, Tobias-Santos V, Santos D, Antunes F, Feltran G, de Souza Menezes J, Aravind L, Venancio TM, Nunes da Fonseca R. Evolution and multiple roles of the Pancrustacea specific transcription factor Zelda in insects, (C. Desplan, ed.). PLoS Genet. 2017;13(7):e1006868. doi: 10.1371/journal.pgen.1006868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemondy K, Henriksen JC, Rissland OS. Intron dynamics reveal principles of gene regulation during the maternal-to-zygotic transition. RNA. 2023;29(5):596–608. doi: 10.1261/rna.079168.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samata M, Alexiadis A, Richard G, Georgiev P, Nuebler J, Kulkarni T, Renschler G, Basilicata MF, Zenk FL, Shvedunova M, et al. Intergenerationally maintained histone H4 lysine 16 acetylation is instructive for future gene activation. Cell. 2020;182(1):127–144.e23. doi: 10.1016/j.cell.2020.05.026. [DOI] [PubMed] [Google Scholar]
- Sandler JE, Irizarry J, Stepanik V, Dunipace L, Amrhein H, Stathopoulos A. A developmental program truncates long transcripts to temporally regulate cell signaling. Dev Cell. 2018;47(6):773–784.e6. doi: 10.1016/j.devcel.2018.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Bradley RK. The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res. 2012;22(4):656–665. doi: 10.1101/gr.130682.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Sutcliffe C, Lis JT, Ashe HL. Extensive polymerase pausing during Drosophila axis patterning enables high-level and pliable transcription. Genes Dev. 2013;27(10):1146–1158. doi: 10.1101/gad.215459.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes C, Biette KM, Harden TT, DePace AH. Signal integration by shadow enhancers and enhancer duplications varies across the Drosophila embryo. Cell Rep. 2019;26(9):2407–2418.e5. doi: 10.1016/j.celrep.2019.01.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz KN, Bondra ER, Moshe A, Villalta JE, Lieb JD, Kaplan T, McKay DJ, Harrison MM. Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 2015;25(11):1715–1726. doi: 10.1101/gr.192682.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Cavalli G. Three-dimensional genome organization and function in Drosophila. Genetics. 2017;205(1):5–24. doi: 10.1534/genetics.115.185132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seher TC, Leptin M. Tribbles, a cell–cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation. Curr Biol. 2000;10(11):623–629. doi: 10.1016/S0960-9822(00)00502-9. [DOI] [PubMed] [Google Scholar]
- Seller CA, Cho C-Y, O’Farrell PH. Rapid embryonic cell cycles defer the establishment of heterochromatin by Eggless/SetDB1 in Drosophila. Genes Dev. 2019;33(7–8):403–417. doi: 10.1101/gad.321646.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seller CA, O’Farrell PH. Rif1 prolongs the embryonic S phase at the Drosophila mid-blastula transition. PLoS Biol. 2018;16(5):e2005687. doi: 10.1371/journal.pbio.2005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15(4):284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124(11):2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Shermoen AW, McCleland ML, O’Farrell PH. Developmental control of late replication and S phase length. Curr Biol. 2010;20(23):2067–2077. doi: 10.1016/j.cub.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, O’Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67(2):303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo Y, Amodeo AA. Excess histone H3 is a competitive Chk1 inhibitor that controls cell-cycle remodeling in the early Drosophila embryo. Curr Biol. 2021;31(12):2633–2642.e6. doi: 10.1016/j.cub.2021.03.035. [DOI] [PubMed] [Google Scholar]
- Sibon OC, Stevenson VA, Theurkauf WE. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388(6637):93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- Siddiqui NU, Li X, Luo H, Karaiskakis A, Hou H, Kislinger T, Westwood JT, Morris Q, Lipshitz HD. Genome-wide analysis of the maternal-to-zygotic transition in Drosophila primordial germ cells. Genome Biol. 2012;13(2):R11. doi: 10.1186/gb-2012-13-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert CA, Wilson JE, Kerr K, Macdonald PM. Smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10(20):2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- Soluri IV, Zumerling LM, Payan Parra OA, Clark EG, Blythe SA. Zygotic pioneer factor activity of Odd-paired/Zic is necessary for late function of the Drosophila segmentation network. eLife. 2020;9:e53916. doi: 10.7554/eLife.53916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Marmion RA, Park JO, Biswas D, Rabinowitz JD, Shvartsman SY. Dynamic control of dNTP synthesis in early embryos. Dev Cell. 2017;42(3):301–308.e3. doi: 10.1016/j.devcel.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soruco MML, Chery J, Bishop EP, Siggers T, Tolstorukov MY, Leydon AR, Sugden AU, Goebel K, Feng J, Xia P, et al. The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev. 2013;27(14):1551–1556. doi: 10.1101/gad.214585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MR, Haines JE, Eisen MB. Convergence of topological domain boundaries, insulators, and polytene interbands revealed by high-resolution mapping of chromatin contacts in the early Drosophila melanogaster embryo. eLife. 2017;6:e29550. doi: 10.7554/eLife.29550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller MV, Vincent BJ, Bragdon MDJ, Lydiard-Martin T, Wunderlich Z, Estrada J, DePace AH. Shadow enhancers enable Hunchback bifunctionality in the Drosophila embryo. Proc Natl Acad Sci U S A. 2015;112(3):785–790. doi: 10.1073/pnas.1413877112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller MV, Yan D, Randklev S, Bragdon MD, Wunderlich ZB, Tao R, Perkins LA, DePace AH, Perrimon N. Depleting gene activities in early Drosophila embryos with the “maternal-Gal4-shRNA” system. Genetics. 2013;193(1):51–61. doi: 10.1534/genetics.112.144915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt N, Fellert S, Chung HR, Jackle H, Vorbruggen G. Mutations of the Drosophila zinc finger-encoding gene vielfaltig impair mitotic cell divisions and cause improper chromosome segregation. Mol Biol Cell. 2006;17(5):2356–2365. doi: 10.1091/mbc.E05-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. Phase separation drives heterochromatin domain formation. Nature. 2017;547(7662):241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong IJT, Lei X, Chen F, Yuan K, O’Farrell PH. Interphase-arrested Drosophila embryos activate zygotic gene expression and initiate mid-blastula transition events at a low nuclear-cytoplasmic ratio. PLoS Biol. 2020;18(10):e3000891. doi: 10.1371/journal.pbio.3000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nien C-Y, Chen K, Liu H-Y, Johnston J, Zeitlinger J, Rushlow C. Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 2015;25(11):1703–1714. doi: 10.1101/gr.192542.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H, Spangenberg S, Vogt N, Grosshans J. Number of nuclear divisions in the Drosophila blastoderm controlled by onset of zygotic transcription. Curr Biol. 2013;23(2):133–138. doi: 10.1016/j.cub.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Syed S, Wilky H, Raimundo J, Lim B, Amodeo AA. The nuclear to cytoplasmic ratio directly regulates zygotic transcription in Drosophila through multiple modalities. Proc Natl Acad Sci U S A. 2021;118(14):e2010210118. doi: 10.1073/pnas.2010210118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG Is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12(1):143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Tadros W, Houston SA, Bashirullah A, Cooperstock RL, Semotok JL, Reed BH, Lipshitz HD. Regulation of maternal transcript destabilization during egg activation in Drosophila. Genetics. 2003;164(3):989–1001. doi: 10.1093/genetics/164.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133(10):1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- Theodosiou NA, Xu T. Use of FLP/FRT system to study Drosophila development. Methods. 1998;14(4):355–365. doi: 10.1006/meth.1998.0591. [DOI] [PubMed] [Google Scholar]
- Thomsen S, Anders S, Janga SC, Huber W, Alonso CR. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 2010;11(9):R93. doi: 10.1186/gb-2010-11-9-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V. Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol Cell Biol. 2008;28(21):6695–6708. doi: 10.1128/MCB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L, Orr-Weaver TL. The Drosophila PNG kinase complex regulates the translation of cyclin B. Dev Cell. 2007;12(1):157–166. doi: 10.1016/j.devcel.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Destree OH, Wolffe AP. Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol Cell Biol. 1999;19(12):7972–7982. doi: 10.1128/MCB.19.12.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventos-Alfonso A, Ylla G, Belles X. Zelda and the evolution of insect metamorphosis. bioRxiv. 2018. [Google Scholar]
- Wang M, Ly M, Lugowski A, Laver JD, Lipshitz HD, Smibert CA, Rissland OS. ME31B globally represses maternal mRNAs by two distinct mechanisms during the Drosophila maternal-to-zygotic transition. eLife. 2017;6:e27891. doi: 10.7554/eLife.27891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney PH, Shrestha B, Xiong J, Zhang T, Rushlow CA. Shadow enhancers modulate distinct transcriptional parameters that differentially effect downstream patterning events. Development. 2022;149(21):dev200940. doi: 10.1242/dev.200940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Lehmann R. Germ cell development in Drosophila. Annu Rev Cell Dev Biol. 1996;12(1):365–391. doi: 10.1146/annurev.cellbio.12.1.365. [DOI] [PubMed] [Google Scholar]
- Wrischnik LA, Timmer JR, Megna LA, Cline TW. Recruitment of the proneural gene scute to the Drosophila sex-determination pathway. Genetics. 2003;165(4):2007–2027. doi: 10.1093/genetics/165.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich Z, Bragdon MDJ, Vincent BJ, White JA, Estrada J, DePace AH. Krüppel expression levels are maintained through compensatory evolution of shadow enhancers. Cell Rep. 2015;12(11):1740–1747. doi: 10.1016/j.celrep.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Chen H, Ling J, Yu D, Struffi P, Small S. Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev. 2014;28(6):608–621. doi: 10.1101/gad.234534.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Whitney PH, Huang S-K, Eck EC, Garcia HG, Rushlow CA. The Drosophila pioneer factor Zelda modulates the nuclear microenvironment of a dorsal target enhancer to potentiate transcriptional output. Curr Biol. 2019;29(8):1387–1393.e5. doi: 10.1016/j.cub.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Cuna JO, Dinh HQ, Kvon EZ, Shlyueva D, Stark A. Uncovering cis-regulatory sequence requirements for context specific transcription factor binding. Genome Res. 2012;22(10):2018–2030. doi: 10.1101/gr.132811.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, O’Farrell PH. TALE-light imaging reveals maternally guided, H3K9me2/3-independent emergence of functional heterochromatin in Drosophila embryos. Genes Dev. 2016;30(5):579–593. doi: 10.1101/gad.272237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalokar M. Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Dev Biol. 1976;49(2):425–437. doi: 10.1016/0012-1606(76)90185-8. [DOI] [PubMed] [Google Scholar]
- Zalokar M, Erk I. Division and migration of nuclei during early embryogenesis of Drosophila melanogaster. J. Microbiol Cell. 1976;25:97–106. [Google Scholar]
- Zaret KS. Pioneer transcription factors initiating gene network changes. Annu Rev Genet. 2020;54(1):367–385. doi: 10.1146/annurev-genet-030220-015007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavortink M, Rutt LN, Dzitoyeva S, Henriksen JC, Barrington C, Bilodeau DY, Wang M, Chen XXL, Rissland OS. The E2 Marie Kondo and the CTLH E3 ligase clear deposited RNA binding proteins during the maternal-to-zygotic transition. eLife. 2020;9:e53889. doi: 10.7554/eLife.53889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong J-W, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39(12):1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenk F, Loeser E, Schiavo R, Kilpert F, Bogdanovic O, Iovino N. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science (New York, N.Y.). 2017;357(6347):212–216. doi: 10.1126/science.aam5339 [DOI] [PubMed] [Google Scholar]