Abstract
miRNA is a noncoding RNA found in recent years and more than one third of human genes are the target of miRNAs. miR-624, located on human chromosome 14, is associated with tumorigenesis. However, the role of miR-624 in human hepatocarcinogenesis is still unclear. Herein, our results indicate that miR-624 accelerates the growth of liver cancer cells in vivo and in vitro. Moreover, the modification distribution of H3K9me1 on chromosomes is different between rLV group and rLV-miR-624 group. miR-624 affects epigenetic regulation of several genes in human liver cancer cells, such as RAB21, SMARCD3, MAPK6,PRRX1, ZFHX3, EMC3 (TMEM111). Furthermore, miR-624 affects transcriptome of some genes in liver cancer, including RAB21, UBE2N, PPP1CC,KPNA3, RAB7A,CPEB2,KLF4, MARK2, JUN, ARF6, TMEM39A. On the other hand, miR-624 affects proteome of several genes in liver cancer, such as, RBM5,PTK2, KDM2A,POLR2H, POLR2G,CDK6,KIF15,CUL2,FKBP2,ErbB-3,JUN, PKM2, CyclinE,PLK1, mTOR, PPARγ, Rab7A,ARAF, UPF3B ,PTEN, SUZ12, GADD45, H3.3, CUL5, ARF6,EMC3,ATG4B,ATG14,CALR. Interestingly, miR-624 affects the RAB7A interaction network in liver cancer cells, involving in CLTC,ITGB1,HNRNPU, DARS1, RPS16, CTPS1,H3–3B,JUN,MYH10, CUL5, CPSF7. Strikingly, excessive MEC3 abrogates the carcinogenic functions of miR-624. Importantly, our findings indicate that miR-624 affects some signaling pathway in liver cancer, including Wnt signaling pathway,Hippo signaling pathway,mTOR signaling pathway, Ras signaling pathway,MAPK signaling pathway,PI3K-Akt signaling pathway, erbB signaling pathway. These results provide a basis for the treatment of human liver cancer.
Keywords: miR-624, Liver cancer, EMC3, Transcriptome, Proteome, Transcriptomics, Proteomics
miR-624, located on human chromosome 14, is associated with prognosis in several diseases [1,2]. Recently, inhibiting the expression of miR-624 promotes the Warburg effect and malignancy of hepatocellular carcinoma cells through increasing the expression of H3F3B [3]. miR-624 promotes the generation and migration of human osteosarcoma cells by inhibiting the Hippo signaling pathway [4]. However, the exact molecular mechanism of miR-624 in human hepatocarcinogenesis is still unclear. EMC4 (ER membrane protein complex subunit 4) is also called transmembrane protein 111 (TMEM111) as a member of the endoplasmic reticulum associated secret pathway [[5], [6], [7]]. Recently, a study suggests that EMC3 is one of the characteristic markers of several diseases [8,9]. EMC3 could be a potential therapeutic target in melanoma and basal cell carcinoma [10]. However, the role of EMC3 in human hepatocarcinogenesis is still unclear. In this study, we clearly demonstrate that miR-624 affects transcriptome and proteome, and accelerates the growth of liver cancer cells by reducing EMC3. These results are of great significance for the prevention and treatment of human liver cancer.
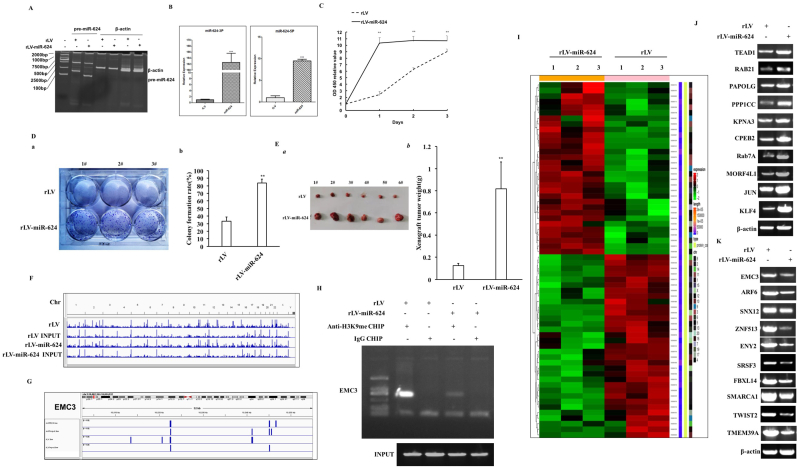
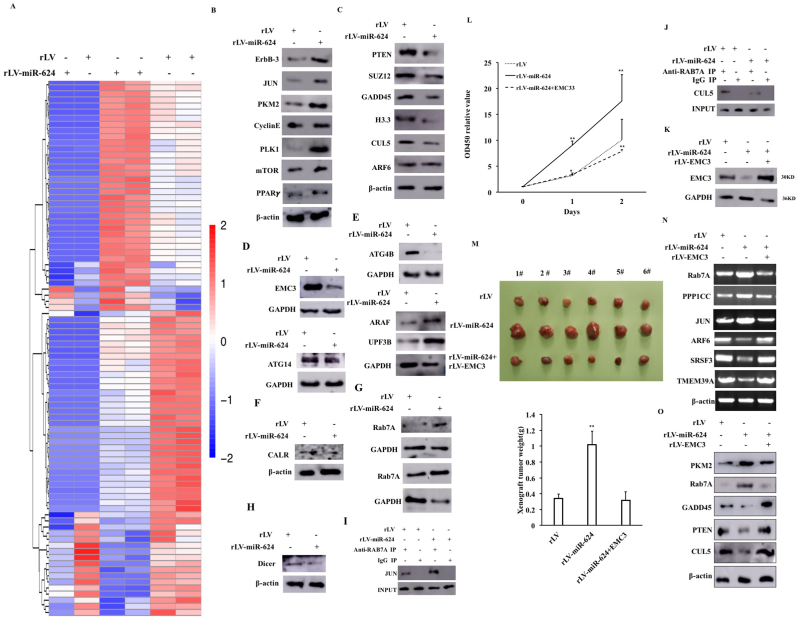
To investigate the effect of miR-624 on liver cancer cells, we cloned the miR-624 (MI0003638) precursor sequence (AATGCTGTT TCAAGGTAGTACCAGTACCTTGTGTTCAGTGGAACCAAGGTAAACACAAGGTATTGGTATTACCTTGAGATAGCATTACACCTAAGTG) into the lentiviral vector pLVX-ZsGreen-miRNA-Puro (pLVX-miR-624) and prepared rLV-miR-624 lentivirus. Next, rLV and rLV-miR-624 were infected liver cancer cell Hep3B (Fig. S1A, Fig. 1A–B). The proliferation ability (Fig. 1C), the cell colony formation ability (Fig.1Da-b), the cell injury repair ability (Fig. S1Ba-b), the weight of transplanted tumors (xenograft) (Fig. 1E), the poorly differentiated cells and PCNA positive rate (Fig. S2A–B) were significantly increased in the rLV-miR-624 group compared to rLV group. Chromatin immunoprecipitation sequencing (Chip-Seq) with anti-H3K9me1 high-throughput analysis shows that miR-624 affects epigenetic regulation in human liver cancer cells (Fig. 1F, Fig. S3A-H). In particular, the loading of H3K9me1 on EMC3 promoter region was significantly decreased compared to rLV group (Fig. 1G–H). RNA sequencing showed that miR-624 affects transcriptome in human liver cancer cells, including TEAD1,ARCN1,RAB21,VIM,ACTN1,PAPOLG,HNRNPU,ARID1A,UBE2N,YWHAQ,PURA,PPP1CC,KPNA3,PRCC,SMARCD3,UBE2M,MORF4L1,HMGB1,FAR1,RAB7A,CPEB2,KLF4,GNAQ,BZW1,MARK2,PCDH7,JUN,SUMO2,HNRNPA3, EXT1,PABPC1,MAP4K4,TWIST2,ARF6,AZIN1,MTDH,ZNF513,FAM78B,DCTN1,PRRX1,TMSB4X,ENY2,RAD23B,TMEM39A,BTG1,SRSF3,PDIK1L,EEF1A1,GNAS,SMARCA1,ZFHX3,PHF21A,EIF1,EIF4E2,SNX12,MAPK6,FL2,EIF3E,WAPL (Fig. 1I-K, Fig. S4A-G). The proteolytic peptides were analyzed by label free mass spectrometry and showed that miR-624 alters proteome in liver cancer(Fig. 2A, Fig. S5A-H).In particular, as shown in Fig. 2B-G, the expression of ErbB-3,JUN,PKM2,CyclinE,PLK1,mTOR,PPARγ,Rab7A,ARAF, UPF3B were significantly enhanced in rLV-miR-624 group compared to rLV group and the expression of PTEN,SUZ12, GADD45, H3.3, CUL5, ARF6,EMC3, ATG4B, ATG14,CALR, Dicer were significantly reduced in rLV-miR-624 group compared to rLV group. The immunoprecipitation with anti-RAB7A-mass spectrometry of proteolytic peptides analysis shows that miR-624 affects the RAB7A interaction network in liver cancer cells (Fig. 6A-F). Immunoprecipitation analysis showed that the interaction between RAB7A and JUN was significantly increased in rLV-miR-624 group compared with rLV group (Fig. 2I),and the interaction between RAB7A and CUL5 was significantly reduced in rLV-miR-624 group compared with rLV group (Fig. 2J). Strikingly, although the cell proliferation ability, the colony forming ability, the weight of transplanted tumors was increased in the rLV-miR-624 group, it was not altered in rLV-miR-624+rLV-EMC3 group compared with rLV group (Fig. 2K-M, Figs. S7A–C). In particular, the expression of PKM2 and Rab7A were significantly increased and the expression of GADD45, PTEN, CUL5 were significantly reduced in the rLV-miR-624 group, however, it was not changed in rLV-miR-624+ rLV-EMC3 group compared with rLV group (Fig. 2N–O).
Fig. 1.
miR-624 promotes the growth of liver cancer cells in vitro and affects proteomics in liver cancer. A. The precursor of miR-624 was detected by reverse transcription polymerase chain reaction (RT-PCR). β-actin was used as internal reference gene.B. Quantitative RT-PCR was used to detect the mature miR-624. U6 was used as internal reference gene. The values of each group were expressed as mean ± SD (n = 3), * *, P < 0.01, and *, P < 0.05. C. CCK8 method was used to determine the cell proliferation ability. The values of each group were expressed as mean ± SD (n = 3), * *, P < 0.01, and *, P < 0.05. D. The colony forming ability of cells was measured. a. photos of plate colonies. b. the analysis of cell colony formation ability. The values of each group were expressed as mean ± standard deviation (bar ± SD, n = 3), * *, P < 0.01, *, P < 0.05. E. the xenograft tumor was dissected. B. Comparison of tumor size (g). The values of each group were expressed as mean ± SD (n = 6), * *, P < 0.01, and *, P < 0.05, respectively. F. IGV browser interface (Demo): visualization of the reads of the modification distribution of H3K9me1 on 23 pairs of chromosomes in rLV group and rLV-miR-624 group. G. IGV browser interface (Demo): visualization of the reads of modification distribution of H3K9me1 on gene region. H. Chromatin immunocoprecipitation (CHIP) analysis was performed by anti-H3K9me1. The PCR amplification was carried out by using primers designed according to the DNA of EMC3 promoter region. IgG CHIP was used as the negative control. I. Heat map analysis (cluster) of all gene expression in the two groups. D. Up-regulated genes and down-regulated genes. J. RT-PCR for several up-regulated genes. β-actin was used as internal reference gene. K. RT-PCR for several down-regulated genes. β-actin was used as internal reference gene.
Fig. 2.
miR-624 affects proteomics, RAB7A-proteins interaction network in liver cancer and excessive MEC3 abrogates the carcinogenic functions of miR- 624 in liver cancer. A. Differential protein cluster Heatmap. The vertical is the clustering of samples and the horizontal is the clustering of proteins. B–H. Several proteins were detected by Western blotting. β-actin was used as internal reference gene. I. Co-immunoprecipitation analysis with anti-RAB7A and Western blot analysis with anti-JUN. Western blot analysis with anti-RAB7A as INPUT. J. Co-immunoprecipitation analysis with anti-RAB7A and Western blot analysis with anti-CUL5. Western blot analysis with anti-RAB7A as INPUT. K. The Western blot was used to detect the EMC3. GAPDH was used as the internal reference gene. L. cell proliferation ability assay. M. Photograph of xenograft tumor and comparison of the size (g) of transplanted tumors. The values of each group are expressed as mean ± standard deviation (mean ± SEM, n = 6), **, P < 0.01, *, P < 0.05).N. RT-PCR for several genes. β-actin was used as internal reference gene. O. Western blot for several proteins with related antibodies. β-actin was used as internal reference gene.
In conclusions, miR-624 accelerates the growth of liver cancer cells in vivo and in vitro. Moreover, miR-624 affects epigenetic regulation of several genes, transcriptome, proteome in liver cancer. Evidently, our present observations clearly demonstrate that miR-624 affects the RAB7A interaction network in liver cancer cells. In particular, excessive MEC3 abrogates the carcinogenic functions of miR-624. Importantly, miR-624 affects several signaling pathway in human liver cancer cell Hep3B. These results provide a basis for the treatment of human liver cancer.
Ethics approval and consent to participate
All methods were carried out in “accordance” with the approved guidelines. All experimental protocols “were approved by” a Tongji university institutional committee. Informed consent was obtained from all subjects. The study was reviewed and approved by the China national institutional animal care and use committee.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Funding
This study was supported by grants from National Natural Science Foundation of China (NCSF No.82073130) and by grants from Science and Technology Commission of Shanghai Municipality Shanghai Science and Technology Plan Basic Research Field Project(20JC1411400).
Authors' contributions
Dongdong Lu conceived the study and participated in the study design, performance, coordination and manuscript writing. Xiaoxue Jiang, Yi Lu, Sijie Xie, Yingji Chen, Xinlei Liu, Shujie Li, Shuting Song, Liyan Wang performed the research. All authors have read and approved the final manuscripts.
Declaration of competing interest
The authors declare that they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2023.09.005.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Ji J., Yamashita T., Budhu A., Forgues M., Jia H.L., Li C., Deng C., Wauthier E., Reid L.M., Ye Q.H., Qin L.X., Yang W., Wang H.Y., Tang Z.Y., Croce C.M., Wang X.W. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50(2):472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaca-Paniagua F., Alvarez-Gomez R.M., Maldonado-Martínez H.A., Pérez-Plasencia C., Fragoso-Ontiveros V., Lasa-Gonsebatt F., Herrera L.A., Cantú D. Revealing the molecular portrait of triple negative breast tumors in an understudied population through omics analysis of formalin-fixed and paraffin-embedded tissues. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0126762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo Y., Liu W., Tang P., Jiang D., Gu C., Huang Y., Gong F., Rong Y., Qian D., Chen J., Zhou Z., Zhao S., Wang J., Xu T., Wei Y., Yin G., Fan J., Cai W. miR-624-5p promoted tumorigenesis and metastasis by suppressing hippo signaling through targeting PTPRB in osteosarcoma cells. J. Exp. Clin. Cancer Res. 2019;38(1):488. doi: 10.1186/s13046-019-1491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y., Song S., Zhang L., Zhang Y. Circular RNA hsa_circ_0091579 facilitates the Warburg effect and malignancy of hepatocellular carcinoma cells via the miR-624/H3F3B axis. Clin. Transl. Oncol. 2021;23(11):2280–2292. doi: 10.1007/s12094-021-02627-4. [DOI] [PubMed] [Google Scholar]
- 5.Ring G., Khoury C.M., Solar A.J., Yang Z., Mandato C.A., Greenwood M.T. Transmembrane protein 85 from both human (TMEM85) and yeast (YGL231c) inhibit hydrogen peroxide mediated cell death in yeast. FEBS Lett. 2008;582(17):2637–2642. doi: 10.1016/j.febslet.2008.06.042. [DOI] [PubMed] [Google Scholar]
- 6.Nayak R.R., Kearns M., Spielman R.S., Cheung V.G. Coexpression network based on natural variation in human gene expression reveals gene interactions and functions. Genome Res. 2009;19(11):1953–1962. doi: 10.1101/gr.097600.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pleiner T., Tomaleri G.P., Januszyk K., Inglis A.J., Hazu M., Voorhees R.M. Structural basis for membrane insertion by the human ER membrane protein complex. Science. 2020;369(6502):433–436. doi: 10.1126/science.abb5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z., Wang Z.X., Chen Y.X., Wu H.X., Yin L., Zhao Q., Luo H.Y., Zeng Z.L., Qiu M.Z., Xu R.H. Integrated analysis of single-cell and bulk RNA sequencing data reveals a pan-cancer stemness signature predicting immunotherapy response. Genome Med. 2022;14(1):45. doi: 10.1186/s13073-022-01050-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang X., Snowball J.M., Xu Y., Na C.L., Weaver T.E., Clair G., Kyle J.E., Zink E.M., Ansong C., Wei W., Huang M., Lin X., Whitsett J.A. EMC3 coordinates surfactant protein and lipid homeostasis required for respiration. J. Clin. Invest. 2017;127(12):4314–4325. doi: 10.1172/JCI94152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z., Wang Z.X., Chen Y.X., Wu H.X., Yin L., Zhao Q., Luo H.Y., Zeng Z.L., Qiu M.Z., Xu R.H. Integrated analysis of single-cell and bulk RNA sequencing data reveals a pan-cancer stemness signature predicting immunotherapy response. Genome Med. 2022;14(1):45. doi: 10.1186/s13073-022-01050-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.