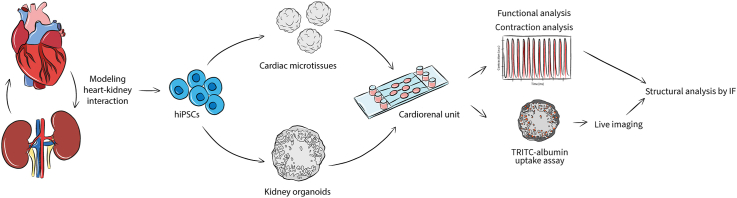
Abstract
Heart and kidney communicate with one another in an interdependent relationship and they influence each other's behavior reciprocally, as pathological changes in one organ can damage the other. Although independent human in vitro models for heart and kidney exist, they do not capture their dynamic crosstalk. We have developed a microfluidic system which can be used to study heart and kidney interaction in vitro. Cardiac microtissues (cMTs) and kidney organoids (kOs) derived from human induced pluripotent stem cells (hiPSCs) were generated and loaded into two separated communicating chambers of a perfusion chip. Static culture conditions were compared with dynamic culture under unidirectional flow. Tissue viability was maintained for minimally 72 h under both conditions, as indicated by the presence of sarcomeric structures coupled with beating activity in cMTs and the presence of nephron structures and albumin uptake in kOs. We concluded that this system enables the study of human cardiac and kidney organoid interaction in vitro while controlling parameters like fluidic flow speed and direction. Together, this “cardiorenal-unit” provides a new in vitro model to study the cardiorenal axis and it may be further developed to investigate diseases involving both two organs and their potential treatments.
Keywords: Cardiac microtissues, Kidney organoids, Microfluidics, Organ-on-a-chip, Cardiorenal disease, Multi-organ in vitro system
Graphical abstract
Credit author statement
Beatrice Gabbin; Conceptualization; Methodology; Software; Validation; Formal Analysis; Investigation; Data Curation; Writing Original Draft; Writing Review & Editing; Visualization, Viviana Meraviglia; Conceptualization; Validation; Data Curation; Writing Original Draft; Writing Review & Editing; Supervision, Maricke L. Angenent; Methodology; Writing Review & Editing, Dorien Ward-van Oostwaard; Resources; Writing Review & Editing, Wendy Sol; Resources; Writing Review & Editing, Christine L. Mummery; Writing Review & Editing; Funding Acquisition, Ton J. Rabelink; Writing Review & Editing; Funding Acquisition, Berend J. van Meer; Conceptualization; Data Curation; Writing Original Draft; Writing Review & Editing; Supervision; Funding Acquisition, Cathelijne W. van den Berg; Conceptualization; Data Curation; Writing Original Draft; Writing Review & Editing; Supervision; Funding Acquisition, Milena Bellin; Conceptualization; Data Curation; Writing Original Draft; Writing Review & Editing; Supervision; Project Administration; Funding Acquisition.
1. Introduction
In the human body, cardiovascular homeostasis, hemodynamic stability as well as control of fluid and nutrient perfusion of organs are regulated by the highly interdependent relationship between heart and kidney [1]. Dysfunction or disease of one organ may lead to disease of the other and vice versa [2] as a variety of pathways and paracrine signaling are involved in the communication of heart and kidney [3]. The complex, multifactorial, and dynamic mechanisms influencing the interaction and the reciprocal dysfunction of the two organs are not fully understood. Even though models for the heart and kidney have been created separately in vitro, most so far have not been designed to capture their dynamic interaction. The study of heart-kidney interaction would benefit from multi-organ in vitro models [4].
Human cell-based in vitro models, particularly of human pluripotent stem cells (hPSC) and organoids, have already increased our ability to recapitulate organ development, structure and function in health and disease. The study of more complex, multi-organ systems often relies on three-dimensional (3D) organization, co-culture of multiple cell types, extracellular matrix, and application of microfluidics (micro physiological systems or Organ-on-Chip; OoC). This can lead to the creation of controlled environments more closely representative of disease and thus better suited to development of future therapies [5,6]. The combination of an engineered environment and ability to manipulate cells is also required to understand the mechanisms underlying the heart-kidney interaction [4]. Microfluidic devices may enable the study of in vitro organ-organ interaction [7] as recently shown by studies which combined several organs on-chip [[8], [9], [10], [11], [12], [13], [14]].
In the present work, we describe the design and development of a multi-organ platform where hiPSC-derived cardiac and kidney organoids are combined on a microfluidic chip to form a cardiorenal unit. Static and flow culture conditions preserved structure- and function-specific properties of both human organoids in vitro, allowing the study of heart and kidney interaction in the future.
2. Material and methods
2.1. Cell lines
The results shown here were obtained using the hiPSC lines LUMC0020iCTRL-06 (https://hpscreg.eu/cell-line/LUMCi028-A, [[15], [16], [17]]) and LUMC0072iCTRL-01 (https://hpscreg.eu/cell-line/LUMCi029-A, [18]) generated by the hiPSC core facility of the LUMC. Protocols for generation of and research with hiPSCs were approved by the medical ethical committee at Leiden University Medical Center.
2.2. Cell differentiation and generation of cardiac microtissues
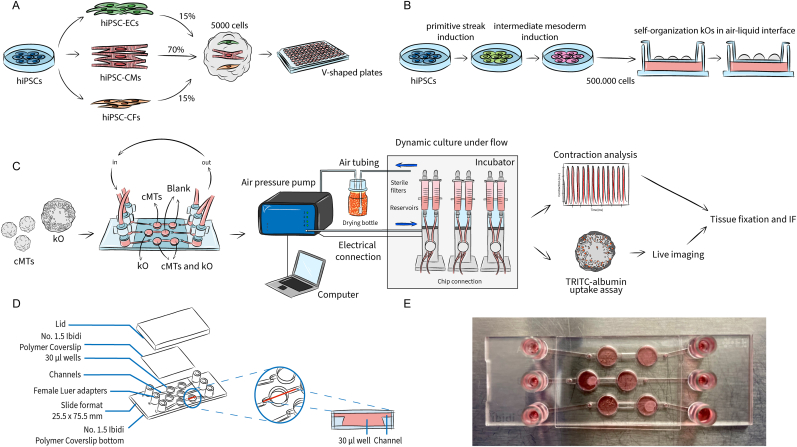
Cardiac microtissues (cMTs) were generated from cardiomyocytes (CMs), cardiac fibroblasts (CFs) and cardiac endothelial cells (ECs), all derived from hiPSCs, as previously described [17]. Briefly, differentiated and cryopreserved cardiac cells were thawed 3–5 days before cMT formation and then combined to a total of 5 × 103 cells in 50 μl in LI-BPEL medium supplemented with 50 ng/ml VEGF (Miltenyi Biotec, Cat. No: 130-109-386) and 5 ng/ml FGF-2 (Miltenyi Biotec, Cat. No: 130-093-842) using a defined ratio of 70:15:15 (CMs:CFs:ECs) in V-shaped culture microplates (Greiner Bio-one, Cat. No: 651161). (Fig. 1A). Microplates were centrifuged at 300 for 10 min at room temperature (RT) to facilitate the aggregation of cells at the bottom of the plates. Plates were gently placed in an incubator at 37 °C, 5% CO2 at day 0 of cMT formation. To allow proper aggregation of the cells, plates were not moved for 3 days. From day 3 until day 18, cMTs were refreshed every 3 days with a partial medium change of 25 μl LI-BPEL supplemented with 50 ng/ml VEGF and 5 ng/ml FGF-2.
Fig. 1.
Formation of organoid models and microfluidic setup. (A) cMTs were generated using hiPSC-derived cardiomyocytes (CMs), cardiac endothelial cells (ECs) and cardiac fibroblasts (CFs). After mixing them in defined ratios, the cardiac cells self-aggregated to form cMTs and were cultured in V-shaped 96 wells plates. (B) kOs were generated from undifferentiated hiPSCs. After one week of differentiation in a monolayer, cells were detached and subjected to microcentrifugation. Pellets of 500.000 cells were then pipetted onto a membrane and cultured in an air-liquid interface. (C) Schematic representation of the workflow: after the organoid generation, the tissues were loaded into the chip and kept in dynamic culture conditions for 72 h. The integrated Ibidi setup comprises a pump system which is, on one hand, connected to the computer and controlled through the PumpControl software; inside the incubator, on the other hand, the pumps are connected to 3 FUs, where the black perfusion set is mounted and whose tubing is in turn attached to each channel of the μ-Slide III 3D Perfusion chip. Organoids were retrieved and used for functional downstream analyses, i.e. contraction analysis (for cMTs), TRITC-albumin uptake assay (for kOs) and immunostaining (both). (D) The geometry of the μ-Slide III 3D Perfusion chip is characterized by three channels where two wells are interconnected, allowing the co-culture of the two organoids while keeping them separated; the perfusion set tubing of each FU is connected to the reservoirs of the channels through its end luers; an adhesive coverslip allows for the sealing of the on-chip culture chambers; the lid aids the transportation of the loaded chip before its connection and after retrieval. (E) Image of a loaded chip.
2.3. Cell differentiation and generation of kidney organoids
kOs were generated as previously described from the adaptation of the protocol by Takasato et al. using hiPSCs [18,19]. In summary, 1.5 × 105 cells/cm2 hiPSCs were plated on vitronectin-coated culture dishes in E8 medium (Thermo Fisher Scientific, Cat. No: A1517001) supplemented with RevitaCell (Thermo Fisher Scientific, Cat. No: A2644501). Differentiation was initiated after 24 h, when cell confluency had reached 10%–20%. Cells were cultured for 4 days in 8 μM CHIR99021 (R&D Systems, Cat. No: 4423) in STEMdiff APEL-2 medium (APEL-2, Cat. No: 05275) supplemented with 1% Protein Free Hybridoma Medium II (PFHMII, Thermo Fisher Scientific, Cat. No: 12040077) and Antibiotic-Antimycotic solution (Thermo Fisher Scientific, Cat. No: 15240062). On day 4, culture medium was replaced by APEL-2 medium containing 200 ng/ml rhFGF9 (R&D Systems, Cat. No: 273-F9) and 1 mg/ml heparin (Sigma-Aldrich, Cat. No: 9041-08-1). On day 7 of differentiation cells were switched from monolayer to 3D culture on Transwell 0.4 μm pore polyester membranes in the same medium after a 1 h pulse with 5 μM CHIR99021 (Fig. 1B). On day 7 + 5, growth factors were removed and a 1.2 ml APEL-2 medium change was performed every 2 days. Organoids were cultured at 37 °C, 5% CO2 until day 7 + 18.
2.4. Loading of the chip
The uncoated μ-Slide III 3D Perfusion chip (Ibidi, Cat. No: 80376) was selected for this study. The alignment in time of the protocols allowed us to use both organoids for microfluidic experiments between 18 and 25 days in vitro after their formation. Microfluidic experiments were performed using LI-BPEL as common media, without any added growth factor.
Chips were pre-warmed for about 1 h before loading. 10 cMTs were collected in a 1.5 ml tube and allowed to sediment. Culture media was replenished with 30 μl of LI-BPEL and cMTs were transferred into the designated chamber of the chip. The chamber containing the kO was pre-filled with 30 μl LI-BPEL and the tissue was then transferred with a metal spatula from the membrane of the Transwell. After all the chambers had been filled, medium was added to the microfluidic channels and reservoirs. Lastly, the chip was sealed with the adhesive Ibidi Polymer Coverslip avoiding bubble formation.
Control tissues were kept in static culture conditions following the culture maintenance schedule.
2.5. Setup of the microfluidic system and chip connection
The integrated pump system used (Ibidi, Cat. No: 10906) allows for the application of continuous, unidirectional recirculatory flow designed for the Ibidi chips. The system runs through the PumpControl software provided only. The complete setup consists of the pump system, the required number of Fluidic Units (FUs), the perfusion set of choice (Black, Ibidi, Cat. No: 10966) and the microfluidic chip. 3 FUs were connected to the pump to allow the simultaneous perfusion of the microfluidic lanes and testing of multiple experimental conditions at the same time.
1.5 ml of medium per reservoir was used to operate each reservoir during the experiments. After filling the reservoirs and tubing with pre-warmed medium, degassing and equilibrating volumes, the chip was connected. Experiments ran up to 72 h per round at 37 °C, 5% CO2. After 72 h of dynamic culture conditions, the pump was stopped and the chip disconnected. The adhesive coverslip sealing the chip was removed and the tissues were retrieved. 5 cMTs for each condition (i.e. static control, 72 h flow monoculture and 72 h flow co-culture) were transferred individually into a V-shaped culture microplate for MUSCLEMOTION analysis of contraction. 1 kO per condition was transferred using a metal spatula to a 24-well plate for the staining with TRITC-albumin. The remaining tissues were fixed for immunostaining.
2.6. Immunostaining
2.6.1. Immunostaining of cardiac microtissues
cMTs were stained as previously described [17]. Briefly, cMTs were fixed using 4% paraformaldehyde (PFA, Merck, Cat. No: 104005) for 1 h at 4 °C. Permeabilization was performed using 0.2% Triton X-100 (Sigma-Aldrich, Cat. No: T8787) in PBS+/+ (Thermo Fisher Scientific, Cat. No: 14040-091) for 30 min at RT. Following a washing step in PBS+/+, a blocking solution containing 10% fetal bovine serum (FBS Sigma-Aldrich, Cat. No: T8787) in PBS+/+ was added for 2 h at RT. Tissues were then incubated overnight in blocking solution with primary antibodies for ACTN2 (mouse monoclonal, Sigma-Aldrich Cat. No: A7811 dilution 1:1000) and TNNT2 (rabbit polyclonal Abcam Cat. No: ab45932 dilution 1:1500). The next day, tissues were washed 3 times for 5 min in PBS+/+ and incubated in blocking solution with Alexa-Fluor 594 and 488 secondary antibodies (Thermo Fisher Scientific, Cat. No: A-21203; Thermo Fisher Scientific, Cat. No: A-21206) for 2 h at room temperature on a rocker. Hoechst solution in PBS+/+ was used for nuclear staining (Invitrogen, Cat. No: H3570, dilution 1:1000). Lastly, cMTs were resuspended in ProLong Gold Antifade Mountant (Thermo Fisher Scientific, Cat. No: P36930) and mounted onto a glass slide. Imaging was performed with Dragonfly 200 Series high-speed confocal imaging platform (Oxford Instruments).
2.6.2. Immunostaining of kidney organoids
kOs were fixed and stained as previously described [17,19]. Tissues were fixed in 4% PFA for 30 min at 4 °C. Permeabilization/blocking of kOs was performed using 0.3% Triton X-100 with 10% donkey serum in PBS+/+ for 2 h at RT. Tissues were then incubated overnight in blocking solution with primary antibodies for NPHS1 (sheep polyclonal, R&D Systems, Cat. No: AF4269, dilution 1:100) and FITC-conjugated lotus tetragonolobus lectin (LTL, Fluorescein Vector Laboratories, Cat. No: FL-1321, dilution 1:300). The next day, tissues were washed 3 times for 5 min in 0.3% Triton X-100 in PBS+/+ and incubated in blocking solution with Alexa-Fluor 647 secondary antibody (Thermo Fisher Scientific, Cat. No: A21448) for 2 h at RT on a rocker. Hoechst solution in PBS+/+ was used for nuclear staining (dilution 1:1000). kOs were mounted using the ProLong Gold Antifade Mountant onto 35 mm glass bottom dishes (MatTek, 14 mm microwell No. 1.5 coverglass, Cat. No: P35G-1.5-14-C). Imaging was performed using the SP8WLL Inverted fluorescent confocal laser-scanning microscope (Leica Microsystems, Model No: TCS SP8 X equipped with White Light Laser).
2.7. MUSCLEMOTION analysis of cardiac microtissues
Contraction video analysis was performed using MUSCLEMOTION software as previously described [20]. Briefly, videos of cMTs were recorded for 10 s at acquisition speed of 100 frames per second using a 10× objective (Nikon Eclipse Ti inverted microscope, ThorLabs DCC3240 M camera) in a controlled environment at 37 °C and 5% CO2. Tissues were stimulated through electrical pacing at 1 Hz (20 V, 3 ms) using custom-made electrodes fitting the V-shaped wells [17]. Videos from the cMTs not following the pacing were excluded from the analysis. Contraction duration, time to peak, relaxation time, 50 to 50 transient and contraction amplitude were quantified. The triangulation of the contraction shape (similar to triangulation of action potential) was calculated by dividing the 10 to 10 transient by the 90 to 90 transient value before averaging.
2.8. TRITC-albumin live imaging of kidney organoids
For albumin uptake assays, kOs were incubated overnight in 300 μl LI-BPEL, 10 μg/ml TRITC-albumin (10 mg/ml stock, Sigma Aldrich, Cat. No: A2289) [21] and 1:300 FITC-conjugated LTL. After overnight incubation at 37 °C and 5% CO2, organoids were thoroughly washed using PBS+/+ 3 times and transferred onto 35 mm glass bottom dishes for live imaging using the SP8WLL Inverted fluorescent confocal laser-scanning microscope.
2.9. Dissociation of kidney organoids
Dissociation of kOs was performed by placing one organoid per condition in collagenase buffer. Collagenase buffer was prepared by mixing 600 U/ml collagenase type I (Worthington Biochemicals, Cat. No: LS004196) and 0.75 U/ml DNAse I (Sigma Aldrich, Cat. No: D4527, dilution 1:100) in HBSS+/+ (Thermo Fisher Scientific, Cat. No: 14025092). Organoids were then incubated for 40 min at 37 °C with repeated pipetting. Cells were then centrifuged at 300 for 7 min and the pellet resuspended in 2.5 ml of TrypLE buffer, containing 80% 10x TypLE (Thermo Fisher Scientific, Cat. No: A1217702), 755 U/ml DNAse I and 0.05 μg/ml Heparin in PBS−/-. The cell suspension was incubated for 5 min at 37 °C followed by pipetting to single cells. The dissociation was stopped by adding cold HBSS+/+ supplemented with 10% FBS and further diluted with cold HBSS+/+. Tubes were centrifuged at 300 for 5 min, the supernatant was aspirated, cells resuspended in cold PBS−/- with 0.1% BSA and filtered through a 30 μm filter. Tubes were kept on ice while performing antibody staining.
2.10. FACS analysis of kidney organoids
Cells were fixed in 4% PFA for 15 min at 4° C and then washed 3 times using FACS buffer, containing PBS−/- with 0.1% BSA and 2 mM EDTA (Thermo Fisher Scientific, Cat. No: AM9912). Cells were stained for LTL (FITC-conjugated, Vector Laboratories, Cat. No: FL-1321) positive populations in 100 μl FACS buffer for 20 min in the dark. Cells were washed and resuspended in 100 μl FACS buffer and kept on ice until analysis. FACS was performed using the Miltenyi Biotec MACSQuant® VYB flow cytometer and data analyzed in FlowJo (BD Life Sciences).
2.11. Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.2.0 and RStudio. Kruskal-Wallis one-way analysis of variance followed by multiple comparisons was applied. Chi-square test was used to analyze the observed frequencies of cMTs following the pacing. Results with p-values p < 0.05 were considered statistically significant. Data are expressed as the Mean ± SD as indicated in figure legends. The sample size used in each experiment is indicated in the figure legends.
3. Results
3.1. Selection of the chip and establishment of the microfluidic setup
A schematic overview of the workflow and of the microfluidic setup is shown in Fig. 1C. Three FUs were connected to the pump for the simultaneous perfusion of the microfluidic channels and the testing of three experimental conditions at the same time. The first two were used for the monoculture of cMTs and a kO, respectively. The third channel was used for the co-culture of the tissues. Because of the difference in size of the two biological models and on-chip space limitations, a ratio of 10 cMTs to 1 kO was selected. In parallel, control tissues were kept in static culture conditions: cMTs in a V-shaped 96 wells plate and kOs in air-liquid interface on a Transwell membrane, respectively.
The Ibidi μ-Slide III 3D Perfusion chip consists of three on-chip channels of 130 μl volume and a total of 6 wells of 30 μl volume. Each channel connects two chambers. Channels have a rectangular cross-section of 0.5 mm height and 1 mm width. Each on-chip reservoir contains 60 μl, reaching 250 μl per channel (Fig. 1D). Fig. 1E shows a chip loaded with either cMTs (upper channel) or a kO only (middle channel), or with both tissues (lower channel).
To keep the working volume low, the smallest type of compatible reservoirs was used, reaching a total volume of 3 ml per channel. The pump system can generate constant flow of medium by supplying the reservoirs of each FU with constant air pressure (mbar) which is set by the software based on the required flow rate. Each FUs features two integrated valves that are continuously switched between two possible states and that ensure unidirectional flow over the chip. The valve switching time was set to 60 s. The system ran with a positive air pressure, perfusing the channel from left to right. Medium viscosity was set to 0.0072 Pa s and calibration factor to 1.
The flow rate selected for the experiments was of 100 μl/min. The choice was made upon simulation (Fig. S1) where it is shown that fluid velocity inside the organ chamber is relatively low. Further validation will be needed to achieve the best conditions for a facilitated organ crosstalk, adequate nutrition and gas exchange.
Each cMT was formed from 5 × 103 cells and reaching a diameter of approximately 300 μm at maturation. kOs were instead developed from 5 × 105 cells and their final size was approximately 2 mm in diameter (Table 1). Because of these size differences, 10 cMTs were linked to one kO in the culture setup.
Table 1.
Overview of cMTs and kOs parameters.
| Tissue type | Cell number | Cell types | Diameter |
|---|---|---|---|
| Cardiac microtissues | 5000 | 70% hiPSC-cardiomyocytes | 200–300 μm |
| 15% hiPSC-endothelial cells | |||
| 15% hiPSC-cardiac fibroblasts | |||
| Kidney organoids | 500000 | hPSCs-derived nephrons, | 2–3.5 mm 500 μm height |
| collecting ducts, | |||
| endothelial cells, | |||
| interstitial cells |
The required volume to completely fill the microfluidic channels and chambers limits the total volume of an OoC system. The total on-chip volume of a single lane on the μ-Slide III 3D Perfusion chip is equal to 250 μl, which increases when connected to the external pumps for recirculation. In vivo values for organ mass, density and blood flow rates were used to determine their relative ratios (Table 2) [[22], [23], [24], [25]]. An overview of the volumetric data of in vivo tissues compared to on-chip tissue and medium volumes is available Table 3. A volumetric approximation was made to determine tissue-to-volume ratios. The tissue volume was calculated based on the organoid size, where kO was approximated to have a disc-like shape.
Table 2.
Physical parameters used to determine an estimation of relevant scaling ratios for experimental design. Values based on a 70 kg human [26].
| Organ | In vivo organ mass [g] | Specific density [g/cm3] | Relative organ weight [%] |
|---|---|---|---|
| Heart | 330a | 1.03 | 0.47 |
| Kidney | 310 | 1.05 | 0.44 |
| Blood | 5500 | 1.06 | 7.9 |
tissue only.
Table 3.
Volumetric data of in vivo tissues compared to on-chip tissues and medium volumes.
| Tissue type | In vivo volume [ml] | Relative in vivo ratio | On-chip volume [μl] | Achieved on-chip relative ratio | Scaling factor |
|---|---|---|---|---|---|
| Cardiac microtissues | 320 | 1 | 0.14 | 1 | 2286000 |
| Kidney organoids | 295 | 0.9 | 4.8 | 34 | 61500 |
| Blood | 5200 | 16 | 250 | 1790 | 20800 |
For the purpose of the experiments, the organoids were transferred onto the chip once they had formed and reached the required day in culture. As cellular behavior of kOs in LI-BPEL remained comparable to that of the golden standard APEL medium (data not shown), LI-BPEL without any added growth factors was used for on-chip experiments and co-culture purposes. The chip geometry in combination with the low constant flow rate of 100 μl/min allowed the cultured organoids to remain at the bottom of the chambers. The culture was subjected to unidirectional recirculating medium for 72 h before the cMTs and kOs were retrieved from the chip for morphological and functional analyses.
3.2. Cardiac microtissues maintain their structural and contractile properties on-chip
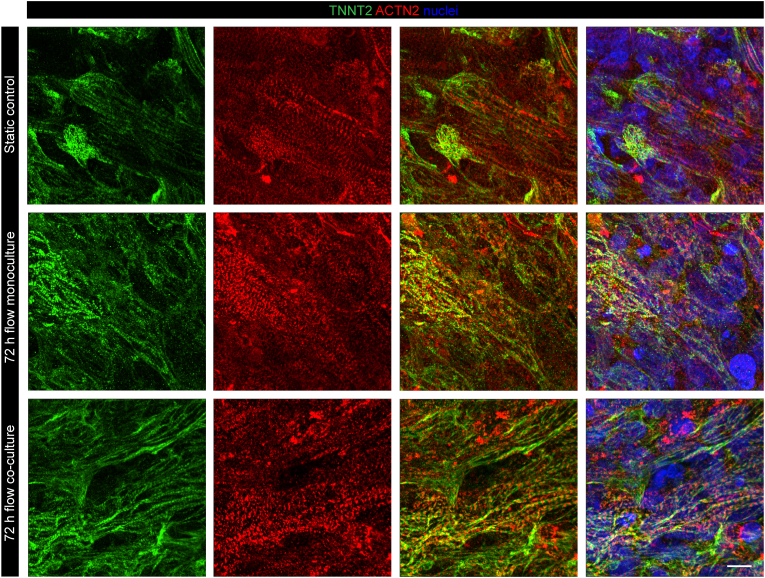
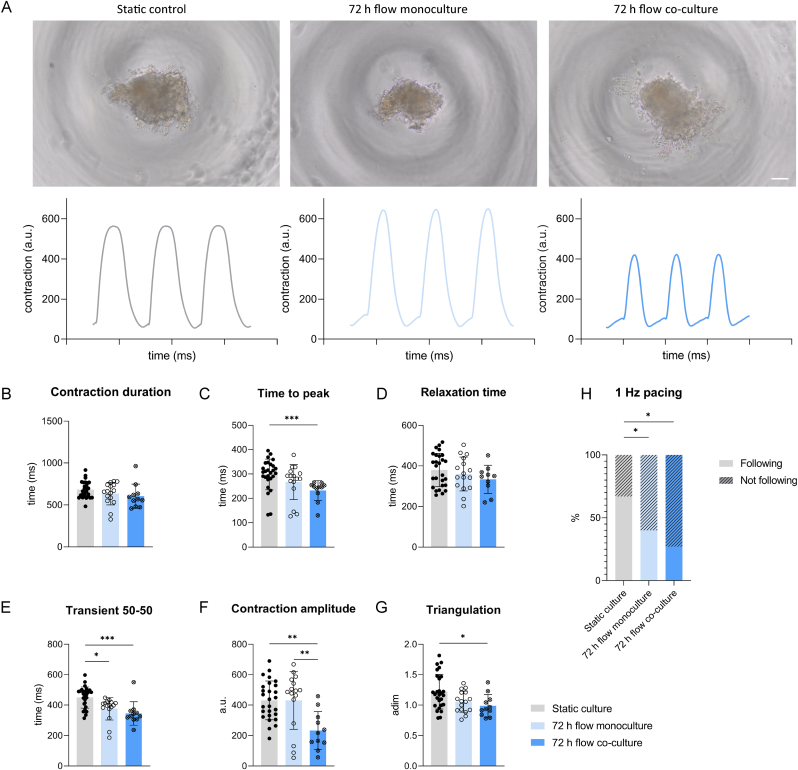
We first assessed the maintenance of structural and functional properties of cMTs. Immunofluorescent staining confirmed the presence of sarcomeric structures in all three conditions: static culture, 72 h flow monoculture and 72 h flow co-culture (Fig. 2). Contractile properties were also investigated. Right before and after retrieval from the chip and transfer to a 96-well plate, cMTs exhibited spontaneous contractile activity. Upon electrical stimulation, cMTs in all three conditions followed the pacing, confirming the retain of their functionality. Representative images and contraction recordings of cMTs at 1 Hz are shown in Fig. 3A. Contraction duration in the three groups showed no significant difference (Fig. 3B). A change in time-to-peak was detected when comparing the static control to the 72 h flow co-culture condition, while relaxation times did not differ among groups (Fig. 3C and D). The contraction time at 50% of the amplitude was not significantly different between the two on-chip conditions but both were lower than in static culture (Fig. 3E). Contraction amplitude was unchanged between static culture and 72 h flow monoculture cMTs, yet lower for cMTs in co-culture with a kO (Fig. 3F). The shape of contraction peaks can be described by the triangulation, a parameter similar to that of an action potential; among the groups, triangulation was statistically different only between the control tissues and those kept in co-culture for 72 h (Fig. 3G). Finally, we observed a significant difference between the static and on-chip conditions with regards to percentages of cMTs following the pacing (Fig. 3H).
Fig. 2.
Sarcomeric structures in cMTs. Immunofluorescence analysis of the sarcomeric markers TNNT2 (green) and ACTN2 (red) of cMTs in static control, 72 h flow monoculture and 72 h flow co-culture. Nuclei are stained in blue. Representative images from n = 4 independent experiments. Scale bar: 10 μm.
Fig. 3.
Contraction analysis of cMTs. (A) Brightfield images of cMTs from static control, 72 h flow monoculture and 72 h flow co-culture and their respective contraction plots upon electrical pacing at 1 Hz. Scale bar: 100 μm. Contraction amplitude is indicated as arbitrary unit (a.u.). (B–G) Contraction duration (B), time to peak (C), relaxation time (D), contraction duration at 50% of the amplitude (E), contraction amplitude (F) and triangulation indicated as adimensional (adim) (G) of cMTs paced at 1 Hz; n ≥ 11 from 4 independent batches; p < 0.0001, ANOVA results of Kruskal-Wallis test, followed by multiple comparisons. Data are shown as mean ± SD. (H) Percentage of cMTs following and not following pacing at 1 Hz; *p < 0.05, Chi-square test.
3.3. Kidney organoids maintain their structure and functional uptake properties on-chip
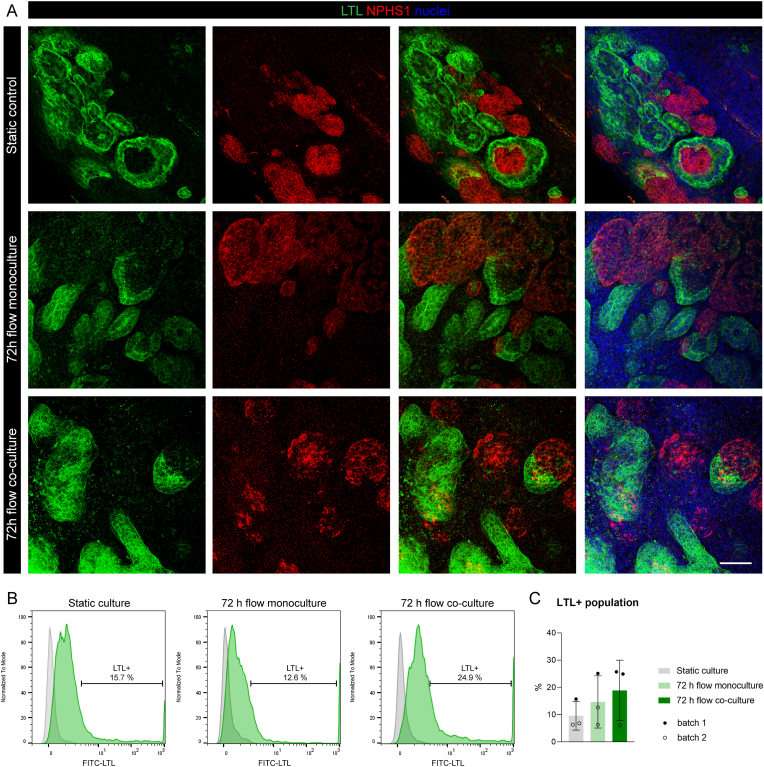
Immunofluorescence staining was performed to assess potential change of tissue morphology for kOs. Fig. 4A shows the presence of nephron structures in all groups, indicating that glomerular (marked by nephrin, NPHS1) and tubular (marked by lectin, LTL) structures were not altered by the 72 h in dynamic culture conditions. Fig. 4B and C shows the quantification of the proximal tubule cell population (LTL+) for each condition, confirming that on-chip culture do not have a negative impact on the overall presence of tubular structures.
Fig. 4.
Nephron structures and FACS analysis of proximal tubule cell population in kOs. (A) Immunofluorescence analysis of LTL (green) and NPHS1 (red) in static control, 72 h flow monoculture and 72 h flow co-culture kOs. Nuclei are stained in blue. Representative images from n = 4 independent experiments. Scale bar: 50 μm. (B) FACS analysis for the LTL+ cell population in kOs (green histograms). Gray histograms represent the unstained control for each condition. (C) Summary of the percentages of LTL+ cell population; n = 3 experiments from 2 independent batches; p < 0.0001, ANOVA results of Kruskal-Wallis test, followed by multiple comparisons. Data are shown as mean ± SD.
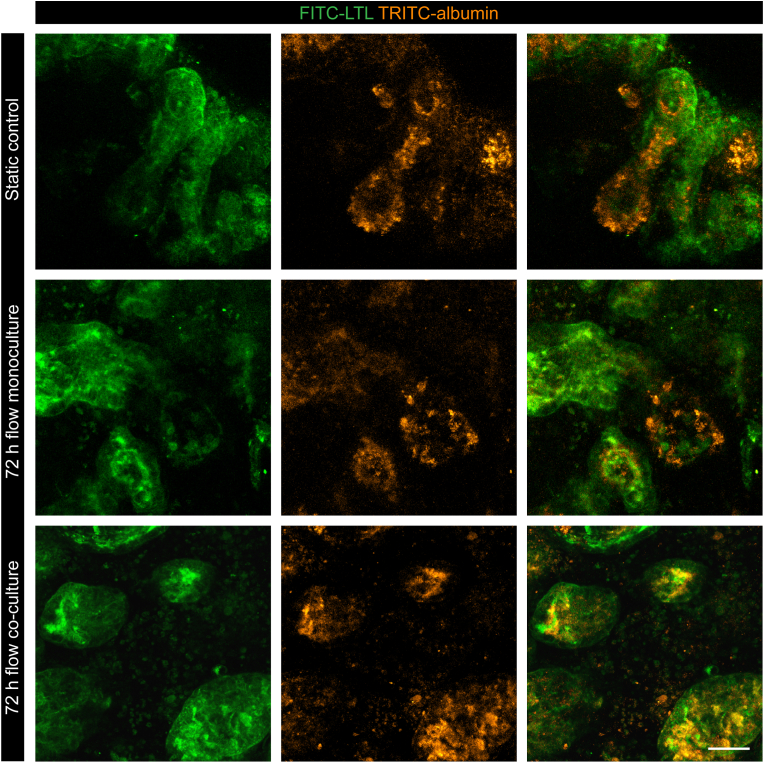
To characterize nephron functionality in the tissues, we examined the expression and presence of transporters by performing a substrate uptake assay specific for proximal tubules. kOs showed evidence of uptake of fluorescently labelled albumin (TRITC-albumin) among the three conditions, indicating the specificity of the receptors by the proximal tubules where the signal with LTL co-localized (Fig. 5).
Fig. 5.
Functional protein uptake properties in kOs. Uptake assay of kOs performed as live imaging of TRITC-albumin (orange); proximal tubules structures are marked by LTL (green). Representative images from n = 4 independent experiments. Scale bar: 50 μm.
4. Discussion
Our aim in this study was to develop a system in which human kOs and cMTs could be coupled and studied under the physiological reality of fluidic flow. The starting point of the development of a microfluidic setup is the selection of a suitable chip. We selected the Ibidi uncoated μ-Slide III 3D Perfusion chip for several reasons. Firstly, it is commercially available, and has previously undergone quality control, which decreases inter-chip variability. Secondly, the chip architecture consists of two on-chip chambers and allows fluidic connection of two types of tissue without them being in direct contact. Finally, another advantage of the two on-chip interconnected chambers, is the reduced tubing complexity compared with e.g. two chips connected in series, which limits dead-volume.
Although the intended use of the μ-Slide III 3D Perfusion chip is for either spheroids or organoid embedded within gel matrices, our data suggested that the chip could also be used by simply allowing the tissue to sediment at the bottom of the respective chamber, without the addition of scaffold material.
The Ibidi pump system is easy to use and allowed the application of continuous, unidirectional recirculatory flow in our cardiac-renal model. Since the chip selected had three individual channels, it enabled three different experimental conditions to be tested at the same time, thus providing time-matched controls.
An inherent limitation of the Ibidi integrated pump system is the obligatory use of the dedicated software. The software provides automatic calculations of pressure, flow rate and shear stress once parameters such as perfusion, type of chip and medium viscosity are set and there is no guarantee of stability over time. As an example, flow rates might be affected by the presence of bubbles in the tubing or in the chambers. No real-time feedback loops based on measured data are provided by the PumpControl software. Moreover, calibration measurements should be performed prior to the initiation of any experiment. Reproducibility of the experiments may, as a result, be slightly altered and the exact flow rate is hard to monitor.
On the positive side, the minimal flow rate that can be achieved with the selected setup is 0.08 ml/min. The recommended flow rate to ensure an optimal supply of oxygen and nutrients to the culture chambers is reported to be in the range of 0.5–1 ml/min [27]. Usually, stringent requirements apply to flow rates in order to prevent exposure of tissues to high shear stress. Because the OoC model here involves organoids rather than cell monolayers, shear stress is not the main concern and there is no straightforward method to determine what the ideal flow rate should be.
When comparing the physiological tissue-to-volume ratio to the achieved on-chip relative ratio, a large discrepancy remains. This directly affects any molecules that may mediate crosstalk; they may be over-diluted which may impact the accuracy of read-outs as well as tissue interaction in the microscale. This limitation was acknowledged when interpreting results. Scaling is an important design consideration that will be implemented in future work to better render physiological dimensions.
The parallel execution of the protocols for the generation of the organoids allowed us to use both for microfluidic experiments as they reached sufficient levels of maturity, from day 18 to day 25 in vitro from first formation.
Results show that our tissue models maintained tissue morphology, structure and functions after 72 h on-chip dynamic culture conditions. In particular, contractile properties of cMTs were comparable among the study groups. Although time-to-peak decreased in co-culture conditions compared to control, the overall contraction duration showed no significant difference. This could be due to variability, as contraction duration is defined as the sum of time-to-peak and relaxation time.
On-chip monoculture of cMTs did not seem to alter functional performance compared to static culture, while the co-culture on-chip indicated some loss of function quality such as lower contraction amplitude. This might be an effect of the presence of the kO, either indirectly (e.g. through consumption of glucose or nutrients) or directly (e.g. through secreted factors). However, the proportion of cMTs following the pacing was affected in both the on-chip monoculture and co-culture. This might be due to the handling of the cMTs that need to be transferred from the chip to the 96-well plate to allow video recordings and mechanical damage could compromise their performance. Further experiments will need to be performed to confirm and improve this technical aspect; alternatively, on chip analysis may solve this issue.
kOs were also shown to maintain tissue morphology after 72 h on-chip dynamic culture conditions. The presence and quality of glomerular and proximal tubular structures was similar among groups. This is also true for the affinity of proximal tubule receptors for albumin, where the signal co-localized. Nevertheless, it is currently challenging to study functionality of glomeruli due to the absence of vascularization in the organoids. Statically controlled kOs were always kept at an air-liquid interface and did not lose directionality, in contrast to the 72 h flow monoculture and co-culture kOs, which were instead completely submerged in media.
Overall, the combination of cardiac and kidney organoids in a single microfluidic platform may allow the study the dialogue between of heart and kidney and possibly also the more complex responses which characterize dynamic crosstalk, where the functionality of one organoid may influence the response of the other. Integrating multiple functional human organoids on-chip in common media provides multiple capabilities to study physiological and pathological mechanisms that cannot be investigated in single tissue systems in vitro.
5. Conclusions
In this study we described the successful design and development of a microfluidic cardiorenal unit to study the interaction of heart and kidney in vitro. The hiPSC-derived cardiac and kidney organoids could be combined isogenically in a microfluidic chip and cultured under unidirectional flow in a closed perfusion system. We showed that cardiac and renal human organoids cultured in dynamic conditions were viable and retained their structural- and functional properties. Moreover, we demonstrated that organoids could be coupled in co-culture on-chip.
This microfluidic setup represents an in vitro platform to study the cardiorenal axis in humans. It establishes a baseline for on-chip experiments in the context of disease modeling and therapeutic discovery, since it is both reproducible and physiologically relevant.
Declaration of competing interest
C.L.M. is co-founder of Pluriomics (now Ncardia) and B.v.M. is co-founder and CTO of Demcon Biovitronix. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
We thank A. Verwey, R. van Nieuwland, E. Lievers and M. Mol for technical assistance. B. Gabbin is supported by the LUMC-PhD grant for the project UNIONS (Unified organoid system for modelling kidney and heart interaction in chronic disease and its treatment) and Eureka (Eurostars no. 113601). This work was supported by the European Research Council (ERC-CoG Mini-HEART no. 101001746), the Netherlands Organ-on-Chip Initiative, an NWO Gravitation project (no. 024.003.001) funded by the Ministry of Education, Culture, and Science of the government of the Netherlands, the Novo Nordisk Foundation Center for Stem Cell Medicine supported by Novo Nordisk Foundation grants (no. NNF21CC0073729) and The Netherlands Organisation for Health Research and Development ZonMW (PSIDER no. 10250022110004).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2023.100818.
Abbreviations
- 3D
Three-dimensional
- cMT
Cardiac microtissue
- CF
Cardiac fibroblast
- CM
Cardiomyocyte
- EC
Endothelial cell
- FBS
Fetal bovine serum
- FU
Fluidic unit
- hPSC
Human pluripotent stem cell
- hiPSC
Human induced pluripotent stem cell
- kO
Kidney organoid
- LTL
Lotus tetragonolobus lectin
- OoC
Organ-on-Chip
- PFA
Paraformaldehyde
- PFHMII
Protein Free Hybridoma Medium II
- RT
Room temperature
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Schefold J.C., Filippatos G., Hasenfuss G., Anker S.D., von Haehling S. Heart failure and kidney dysfunction: epidemiology, mechanisms and management. Nat. Rev. Nephrol. Oct. 2016;12(10):610–623. doi: 10.1038/nrneph.2016.113. [DOI] [PubMed] [Google Scholar]
- 2.Boudoulas K.D., Triposkiadis F., Parissis J., Butler J., Boudoulas H. The cardio-renal interrelationship. Prog. Cardiovasc. Dis. May 2017;59(6):636–648. doi: 10.1016/j.pcad.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Ronco C., Haapio M., House A.A., Anavekar N., Bellomo R. Cardiorenal syndrome. J. Am. Coll. Cardiol. Nov. 2008;52(19):1527–1539. doi: 10.1016/j.jacc.2008.07.051. [DOI] [PubMed] [Google Scholar]
- 4.Gabbin B., et al. Toward human models of cardiorenal syndrome in vitro. Frontiers in Cardiovascular Medicine. 2022;9 doi: 10.3389/fcvm.2022.889553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braam B., Joles J.A., Danishwar A.H., Gaillard C.A. Cardiorenal syndrome--current understanding and future perspectives. Nat. Rev. Nephrol. Jan. 2014;10(1):48–55. doi: 10.1038/nrneph.2013.250. [DOI] [PubMed] [Google Scholar]
- 6.Hoes M.F., Bomer N., van der Meer P. Concise Review: the current state of human in vitro cardiac disease modeling: a focus on gene editing and tissue engineering. Stem Cells Transl. Med. Jan. 2019;8(1):66–74. doi: 10.1002/sctm.18-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y., Kankala R.K., Wang S.-B., Chen A.-Z. Multi-Organs-on-Chips: towards long-term biomedical investigations. Molecules. Feb. 2019;24(4) doi: 10.3390/molecules24040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sung J.H., Shuler M.L. A micro cell culture analog (microCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip. May 2009;9(10):1385–1394. doi: 10.1039/b901377f. [DOI] [PubMed] [Google Scholar]
- 9.Bauer S., et al. Functional coupling of human pancreatic islets and liver spheroids on-a-chip: towards a novel human ex vivo type 2 diabetes model. Sci. Rep. Nov. 2017;7(1) doi: 10.1038/s41598-017-14815-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bovard D., et al. A lung/liver-on-a-chip platform for acute and chronic toxicity studies. Lab Chip. Dec. 2018;18(24):3814–3829. doi: 10.1039/c8lc01029c. [DOI] [PubMed] [Google Scholar]
- 11.Ong L.J.Y., et al. Self-aligning Tetris-Like (TILE) modular microfluidic platform for mimicking multi-organ interactions. Lab Chip. Jun. 2019;19(13):2178–2191. doi: 10.1039/c9lc00160c. [DOI] [PubMed] [Google Scholar]
- 12.Skardal A., et al. Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication. Feb. 2020;12(2) doi: 10.1088/1758-5090/ab6d36. [DOI] [PubMed] [Google Scholar]
- 13.Lee J., et al. A heart-breast cancer-on-a-chip platform for disease modeling and monitoring of cardiotoxicity induced by cancer chemotherapy. Small. Apr. 2021;17(15) doi: 10.1002/smll.202004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronaldson-Bouchard K., et al. A multi-organ chip with matured tissue niches linked by vascular flow. Nat. Biomed. Eng. Apr. 2022;6(4):351–371. doi: 10.1038/s41551-022-00882-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellin M., et al. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. Dec. 2013;32(24):3161–3175. doi: 10.1038/emboj.2013.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giacomelli E., et al. Human-iPSC-Derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell. Jun. 2020;26(6):862–879. doi: 10.1016/j.stem.2020.05.004. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campostrini G., et al. Generation, functional analysis and applications of isogenic three-dimensional self-aggregating cardiac microtissues from human pluripotent stem cells. Nat. Protoc. Apr. 2021;16(4):2213–2256. doi: 10.1038/s41596-021-00497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Berg C.W., et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Rep. Mar. 2018;10(3):751–765. doi: 10.1016/j.stemcr.2018.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takasato M., Er P.X., Chiu H.S., Little M.H. Generation of kidney organoids from human pluripotent stem cells. Nat. Protoc. Sep. 2016;11(9):1681–1692. doi: 10.1038/nprot.2016.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sala L., et al. MUSCLEMOTION: a versatile open software tool to quantify cardiomyocyte and cardiac muscle contraction in vitro and in vivo. Circ. Res. Feb. 2018;122(3) doi: 10.1161/CIRCRESAHA.117.312067. e5–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanslambrouck J.M., et al. Enhanced metanephric specification to functional proximal tubule enables toxicity screening and infectious disease modelling in kidney organoids. Nat. Commun. Oct. 2022;13(1):5943. doi: 10.1038/s41467-022-33623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies B., Morris T. Physiological parameters in laboratory animals and humans. Pharm. Res. (N. Y.) Jul. 1993;10(7):1093–1095. doi: 10.1023/a:1018943613122. [DOI] [PubMed] [Google Scholar]
- 23.Brown R.P., Delp M.D., Lindstedt S.L., Rhomberg L.R., Beliles R.P. Physiological parameter values for physiologically based pharmacokinetic models. Toxicol. Ind. Health. Jul-Aug 1997;13(4):407–484. doi: 10.1177/074823379701300401. [DOI] [PubMed] [Google Scholar]
- 24.Allen B.C., Hack C.E., Clewell H.J. Use of Markov chain Monte Carlo analysis with a physiologically-based pharmacokinetic model of methylmercury to estimate exposures in US women of childbearing age. Risk Anal. Aug. 2007;27(4):947–959. doi: 10.1111/j.1539-6924.2007.00934.x. [DOI] [PubMed] [Google Scholar]
- 25.Chen L., Yang Y., Ueno H., Esch M.B. Body-in-a-Cube: a microphysiological system for multi-tissue co-culture with near-physiological amounts of blood surrogate. Microphysiol Syst. 2020;4(Jun) doi: 10.21037/mps-19-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Report of the task group on reference man. Ann. ICRP. 1979;3(1–4):iii. doi: 10.1016/0146-6453(79)90123-4. [DOI] [PubMed] [Google Scholar]
- 27.μ-Slide III 3D Perfusion,” ibidi. [Online]. Available: https://ibidi.com/channel-slides/53--slide-iii-3d-perfusion.html. [Accessed: 24-April-2023].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.