Abstract
Purpose
The need for chronic systemic immunosuppression, which presents a host of acute risks to transplantation patients, remains the primary limitation for the translation of many cell therapies, such as insulin secreting cells for the treatment of type 1 diabetes. Trophoblasts are the professional tolerogenic cells of the placenta, and they secrete a range of soluble factors to induce antigen specific tolerance toward allogeneic fetal tissue during pregnancy, including extracellular vesicles. Here we develop a trophoblast extracellular vesicle-delivering hydrogel designed for sustained, localized tolerogenic factor delivery within a transplant site to induce localized tolerance toward cell grafts.
Methods
We engineer a synthetic poly(ethylene glycol)-based hydrogel system to tether extracellular vesicles for sustained delivery, and compare this system to passive vesicle entrapment within an alginate hydrogel system. We characterize trophoblast extracellular vesicles for size and morphology, and evaluate vesicle tolerogenic protein content via proteomic analysis. We validate the retention and tethering of extracellular vesicles within the hydrogel systems via scanning electron and stimulated emission depletion microscopy, and measure vesicle release rate over time. Finally, we evaluate trophoblast extracellular vesicle influence on natural killer cell activation in vitro.
Results
We isolated trophoblast extracellular vesicles and proteomics confirmed the presence of tolerogenic factors. We confirmed the presence of extracellular vesicles within hydrogel delivery vehicles, and synthetic hydrogels extended extracellular vesicle release relative to a passive hydrogel system. Finally, extracellular vesicles reduced natural killer cell activation in vitro, confirming the tolerogenic potential of hydrogel-delivered extracellular vesicles.
Conclusions
This tolerogenic extracellular vesicle-delivering hydrogel platform designed for delivery within a transplant site could serve as an alternative to systemic immunosuppression in cell transplantation, potentially reducing the risks associated with cell therapies and widening the eligible patient population.
Keywords: Extracellular vesicles, Hydrogels, Drug delivery, Immune tolerance, Immune engineering, Cell therapy, Trophoblasts
Introduction
Cell transplantation holds great promise as a therapeutic drug delivery approach to treat an array of diseases, with current clinical trials in the applications of type 1 diabetes [1, 2] and hemophilia [3] among many others. With the growth of cell product manufacturing for therapies, such as stem cell-derived and genetically modified cell products, the accessibility and availability of allogeneic cell therapies to patients is expanding rapidly [4, 5]. One major limitation in the use of allogeneic cell therapies is the requirement of systemic immunosuppression to prevent immune rejection, which poses acute risks to patients such as infections and malignancies [6–8]; these acute risks can limit the eligible cell therapy patient population to the critically ill, despite the significant benefits of cell-derived therapeutics. As such, alternative means to immunosuppression are required to eliminate these risks and widen the population of cell therapy-eligible patients. Examples of leading approaches to eliminate systemic immunosuppression include localized immunomodulation within the cell graft site and the induction of localized tolerance toward cell grafts [9–11]. Typically, the delivery of a single immunomodulatory factor, such as PD-L1 [11], FasL [10], or TGF-β [9], results in moderate graft survival, indicating that more complex, multifactorial approaches are necessary to fully induce tolerance toward the allogeneic graft.
Outside of central and peripheral tolerance mechanisms, where self-reactive lymphocytes are eliminated either in primary or secondary lymphatic organs [12], there are limited biological mechanisms available to induce de novo tolerance toward allogeneic cell therapies. One promising model of de novo tolerance in adult organisms is placental pregnancy [13]. This tolerance is mediated by the professional tolerogenic cells of the placenta, the trophoblasts, of which there are three primary phenotypes: the uninucleate cytotrophoblasts (CT), the multinucleate syncytiotrophoblasts (ST) that contact the maternal decidua, and the extravillous trophoblasts (EVT) that actively invade and remodel the maternal decidua [14]. During pregnancy, trophoblasts establish antigen specific tolerance toward fetal antigen by several multilayered mechanisms, including expression of non-classical human leukocyte antigen (HLA) [15], delivery of soluble tolerogenic molecules [16–18], and release of extracellular vesicles [18–21].
Extracellular vesicles (EV) are phospholipid-bound structures released by a variety of cell types, and are involved in cell communication and modulation of the immune system [22]. EVs modulate cell function via the delivery of intracellular signals (RNA, lipids, and proteins) from the originating cell, delivered via endocytosis by, or fusion with, target cells [23, 24]. They are classified based on their biogenesis pathway as exosomes (50–150 nm diameter) and ectosomes (100–1000 nm); exosomes arise from multivesicular body formation while ectosomes arise from external budding of the phospholipid layer [23, 25, 26]. The role of trophoblast EVs in modulating fetal–maternal tolerance has become evident over the past decade and reports have shown that trophoblast EVs are loaded with soluble tolerogenic factors and paternal antigen [27]. Trophoblast EVs, derived from the ST phenotype and present in high concentrations in maternal blood throughout pregnancy [28], promote tolerogenic phenotypes in maternal immune cells such as natural killer cells and T lymphocytes both locally at the site of implantation as well as in draining lymph nodes [19, 29–32], through potential mechanisms such as endocytosis and immune cell cross-dressing [33–37]. Placental EVs are characterized by high concentrations of tolerogenic immune factors, as well as characteristic molecules such as placental alkaline phosphatase (PLAP), placental growth factor (PlGF), and major histocompatibility complex class I G (HLA-G) [28]. We hypothesize that localized delivery of tolerogenic trophoblast-derived EVs within an allogeneic cell transplant site could shift local infiltrating immune cells toward a tolerogenic phenotype, similarly to the local immune dynamics at the maternal-fetal interface, and eliminate host immune responses toward the allogeneic graft.
There has been substantial recent interest in using EVs as therapeutics, particularly sourced from cells with anti-inflammatory functions such as mesenchymal stem cells [38]. Given the small size of EVs and their rapid clearance, interest has increased in the development of hydrogel delivery systems to both localize and retain EVs within a site of interest for improved therapeutic benefit [39]. Typical hydrogel matrices developed to deliver EVs include collagen, alginate, and poly(ethylene glycol) (PEG) hydrogels, often relying on passive entrapment of EVs within the matrix where EV delivery over time is governed by matrix mesh size; typical delivery time scales with these methods are on the order of 1–2 weeks [39]. For many applications, this time scale may be insufficient to achieve the desired therapeutic effect; as such, alternative biomaterial methods may be employed to achieve a longer sustained release, such as covalent immobilization of EVs within a hydrogel matrix [40].
To implement EVs as a therapeutic, careful consideration of cell source is necessary to ensure robust and reproducible production of EVs [41], and in sufficient quantities to generate clinically relevant doses [42]. For trophoblast-derived EVs in particular, primary trophoblast EVs are largely limited to term placenta sources, which may be limited in potency and exhibit high batch-to-batch variability between donors [29]. Stem cell-derived trophoblast EVs may have similar batch-to-batch variability as primary trophoblast EVs, may have limited homology to primary trophoblast EVs, and may present issues with scalability of production [43]. Alternatively, human trophoblast cell lines, such as the choriocarcinoma lines JEG-3 and JAR, represent plentiful cell sources of trophoblast EVs that may allow scalable production of clinically relevant EV doses; however, homology to primary trophoblast EVs must be elucidated [44, 45].
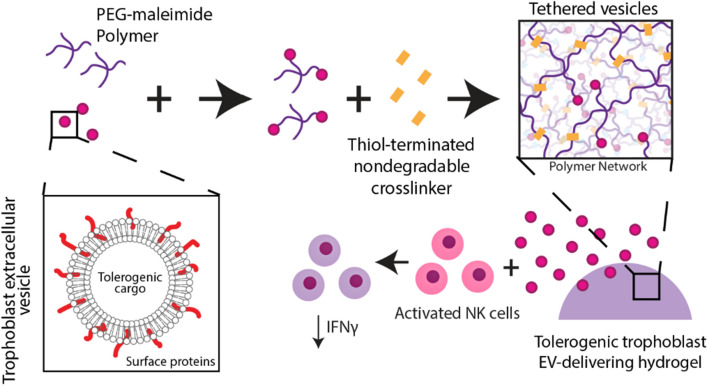
Here we aim to develop a trophoblast EV-delivering hydrogel engineered for sustained, localized tolerogenic factor delivery within a transplant site to induce tolerance toward allogeneic cell grafts (Fig. 1). We engineer a synthetic PEG-based hydrogel system to tether EVs for sustained delivery and compare the release of this system against passive EV entrapment within alginate hydrogels. We characterize trophoblast EVs for size and morphology, and evaluate tolerogenic protein content via proteomic analysis. We validate the retention and tethering of EVs within our synthetic hydrogel system via scanning electron and stimulated emission depletion microscopy, and measure vesicle release rate over time. Finally, we evaluate hydrogel-delivered trophoblast EV influence on activated natural killer cells in vitro.
Fig. 1.
Schematic illustrating design of trophoblast EV-delivering hydrogel to induce tolerance in immune cells
Materials and Methods
Materials
All chemical reagents were obtained from Sigma Aldrich and all cell culture reagents obtained from ThermoFisher unless otherwise noted.
Isolation of Extracellular Vesicles from JAR and JEG-3 Cell Lines
Human choriocarcinoma cell lines, JAR and JEG-3, were obtained from ATCC. Cell passages were maintained at no more than twelve past thawing from storage in − 80 °C with initial seeding densities within the range of 2–3 million cells into T-175 treated cell culture flasks (VWR). Culture conditions were under standard conditions (37 °C, 20% O2, 5% CO2). The recommended media for these cell lines were RPMI-1640 (Gibco) for JAR and MEM (Gibco) for JEG-3. These were supplemented with 10% exosome depleted fetal bovine serum (Thermo Fisher Scientific), 1% penicillin streptomycin, 1% sodium pyruvate, 1% MEM NEAA and 10 mM HEPES buffer solution. For extracellular vesicles isolation, media was harvested when confluency reached 75–85%. Culture volume per flask was maintained at 20 mL and media harvest and passage was performed at 3 day intervals. Harvested media was centrifuged at 2000×g for 30 min and stored in − 80 °C until use for extracellular vesicle collection. Two methods of isolation were investigated: reagent-based isolation using Total Exosome Isolation Reagent (Thermo Fisher Scientific) and size exclusion chromatography based iZON qEV columns. As per manufacturer’s protocol, the harvested media was centrifuged at 2000×g for 30 min and Total Exosome Isolation Reagent (Thermo Fisher Scientific) was introduced at recommended volumes and allowed to incubate overnight at 4 °C. The following day, the media was centrifuged at 10000×g for 1 h at 4 °C and the resulting pellet of extracellular vesicles were resuspended in Dulbecco’s phosphate buffer saline (Thermo Fisher Scientific) at recommended volumes and stored at − 80 °C for further applications. For column based isolation, as per recommendation, media was centrifuged at 1500×g for 10 min initially followed by 10000×g for 10 min prior to introduction into the column. Column elutes were collected at recommended volumes and extracellular vesicles were further filtered using Amicon 15 mL ultra-centrifugal filter unit (Sigma-Aldrich) at 10 kDa cutoff.
Dynamic Light Scattering (DLS) Analysis
Extracellular vesicles from JAR and JEG-3 suspended in DPBS were transferred into 2.5 mL PMMA cuvettes (BrandTech) for analysis. DLS was run with particle size characterization SOP with Delsa Nano Submicron Particle Size and Zeta Potential with Delsa Nano UI software.
Nanoparticle Tracking Analysis (NTA)
Quantification and size characterization of extracellular vesicles was performed using NanoSight NS300 using standard measurement parameters. Resulting extracellular vesicle fractions isolated from cell lines were diluted to appropriate loading capacity of the instrument. 30–60 s video segments with five replicates per sample was collected. Nanosight NTA 3.2 software was used for all analysis with standard settings.
Transmission Electron Microscopy (TEM) Imaging
Extracellular vesicle samples from JAR and JEG-3 were adhered to glow-discharged, carbon-formvar for 2 min for negative stain analysis, coated 400 mesh copper grids, then washed 2× with deionized water. They were stained with two consecutive drops of 2% aqueous uranyl acetate. Imaging was performed on Philips CM 12 TEM. All sample prep and imaging was performed by the Eyring Materials Center at Arizona State University. Image analysis was performed using ImageJ on n = 3 images per group, with n = 23–33 vesicles measured per group.
Western Blotting
JAR cells were cultured under above specified conditions. The cell pellet was lysed with 1× RIPA lysis buffer (Millipore Sigma) diluted with DI water and 2× Halt Protease and Phosphatase Inhibitor cocktail for 10 min on ice. The samples were then centrifuged at 14,000×g for 10 min for supernatant collection. EVs were isolated from JAR under TER conditions and resulting EV pellet fractions were subjected to similar lysis conditions. Samples were stored in − 80 °C until ready to use. Pierce BCA Protein Assay kit and Micro BCA Protein Assay kit were used to determine protein concentrations for cell lysate and EV lysate respectively. 10 µg of protein samples were mixed with Bolt LDS sample buffer (4×) and Bolt Reducing Agent (10×) and vortexed as per manufacturer’s recommendations. Resulting samples were incubated in a 100 °C water bath for 10 min to denature proteins. 30 µL of samples were loaded into the wells of Bolt Bis-Tris 4–12% 1 mm gels and electrophoresis was run under MOPS SDS Running buffer for 32 min at 200 V in Invitrogen Mini Gel Tank. Electrotansfer of proteins was performed using iBlot 2 onto mini nitrocellulose 0.2 µm membranes using the P0 preset method. The membrane was then washed with TBST buffer and then blocked with TBST + 5% (w/v) nonfat dry milk for 1 h on a rocker at room temperature. The membrane was again washed with TBST and then incubated at 4 °C overnight with the primary antibody cocktail (TBST + 5% (w/v) non-fat dry milk + CD63 + TSG 101 + CD81 (Cell Signalling Human Reactive Exosome Marker Antibody Sampler Kit #80610) + PCNA (Cell Signalling #13110S) at 1:1000 dilution). After overnight incubation, the membrane was washed with TBST and incubated with the secondary antibody cocktail for 1 h at room temperature (TBST + 5% (w/v) non-fat dry milk + anti-rabbit IgG HRP-linked antibody at 1:1000). The membrane was developed using Bio-Rad Clarity Western ECL substrate in dark prior to imaging with Analytik Jena™ UVP Chemstudio and exposure was optimised using VisionWorks software. All reagents were purchased from Thermo Fisher Scientific unless otherwise specified.
Mass Spectrometry and Proteomics
Samples were processed using the Protifi S-trap Micro Columns as per manufacturer instructions (using S-trap Ultra High Recovery Protocol). Samples were solubilized in SDS/TEAB and 50 mM dithiothreitol was added, vortexed, and then incubated for 10 min at 95 °C. Proteins were alkylated with approximately 40 mM final concentration freshly prepared iodoacetamide and incubated at 20 °C for 30 min in the dark (Pierce). Samples were then acidified and 2.0 μg of trypsin was introduced. 7× S-trap buffer (was added to samples and allowed to permeate into the S-Trap columns and were washed 3× with S-trap buffer. Samples were eluted off the S-trap columns using 50 mM TEAB, 0.2% formic acid in water, and 50% acetonitrile/50% water + 0.2% formic acid and dried down via speed vac and resuspended in formic acid. All LC–MS analyses were performed at the Biosciences Mass Spectrometry Core Facility at Arizona State University. All data-dependent mass spectra were collected in positive mode using an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific) coupled with an UltiMate 3000 UHPLC (Thermo Scientific).
1 μL of peptides were fractionated using an Easy-Spray LC column (25 cm Å–75 μm ID, PepMap C18, 2 μm particles, 100 pore size, Thermo Scientific). The mass spectra were collected using the “Universal” method optimized for peptide analysis provided by Thermo Scientific. Raw spectral files were imported into Proteome Discoverer v2.5 using standard processing and consensus methods as provided by Thermo. A minimum peptide length was set to 6aa and up to two missed cleavage sites allowed. Sequest HT was used to identify peptide spectral masses (PSMs) and a fixed-value PSM validation method employed. Parameters were set as follows: Database used, Uniprot Homo sapiens (Tax ID 9606), precursor mass tolerance set to 20 ppm and fragment mass tolerance 0.5 Da, static modifications used were carbamidomethyl on cysteines (+ 57.021 Da). Protein FDR confidence levels set to 0.01 (strict) and 0.05 (relaxed). Identified PSMs, peptides and proteins were exported to Excel for further analysis. Further analysis involved submitting the generated list of proteins to Enrichr database to categorise proteins based on biological processes. In addition, to generate immunomodulatory proteins, list of trophoblast specific proteins was generated using the Human Gene Atlas database. In addition, literature review was performed to generate an additional list of trophoblast specific proteins, chemokines and immunomodulators for comparison studies. Samples prep and analysis was performed by the Bioscience Mass Spectrometry core at Arizona State University.
Fluorescent Labelling of JAR Isolated Extracellular Vesicles
Quantification of extracellular vesicles was performed as total protein content and analyzed with Nanodrop (Fisher Scientific) measuring absorbance at 280 nm. Fluorescent labelling of extracellular vesicles was performed with the addition of Alexa fluor 647 NHS-Ester dye (Thermo Fisher Scientific) at 1% v/v and incubated for 30 min while gently vortexing in dark. To remove unbound dye, ultrafiltration was performed using Amicon 0.5 mL ultra-centrifugal filter unit (Sigma-Aldrich) at 10 kDa cutoff thrice to retain labelled extracellular vesicles.
Scanning Electron Microscopy (SEM) and Cryo-SEM of Hydrogels
Scanning electron microscopy of hydrogel samples was performed through incubation of hydrogels in 2.5% glutaraldehyde/DPBS for 30 min and washed three times with DPBS. This was followed by incubation in 1% osmium tetroxide/DPBS for 30 min. Samples were then washed twice in deionized water for 10–15 min and gold coated for imaging. For cryo-SEM, hydrogel samples were mounted into a slotted sample stub and plunge frozen in a liquid nitrogen bath prior to a quick loading into Quorum PP3010 cryo system to stabilize. The samples were then moved into the Helios 5UX (Thermo Scientific) chamber and visualized. All sample prep and imaging were performed by the Eyring Materials Center at Arizona State University.
Release Kinetics of Fluorescently Labelled Extracellular Vesicles from PEG and Alginate Hydrogels
20 kDa 4-arm PEG-maleimide hydrogel macromers (Laysan Bio) were functionalized with RGD adhesion ligand (1 mM) (GRGDSPC, Genscript) and crosslinked with DTT (3470 µg/mL) for a final gel concentration of 5% w/v PEG. Slow-gelling alginate hydrogels (2% w/v, UP-MVG, Novamatrix) were made by adding 30 mM calcium carbonate to 4% w/v alginate and mixing with 60 mM glucono-d-lactone in a 1:1 ratio. The hydrogel volumes used within the study were 20 µL and labelled extracellular vesicles were encapsulated within the PEG (7.944 µg, n = 18 gels/group) and alginate (9.93 µg, n = 15 gels/group) matrices and placed within Dulbecco’s phosphate buffer saline (Thermo Fisher Scientific). The gels were incubated under physiological conditions (37 °C) throughout the duration of study. Gel buffer was replaced every 24 h for a course of 14 days and the release of extracellular vesicles were measured through change in fluorescence intensity of buffer using a BioTek Synergy H1 plate reader. PEG-maleimide and alginate hydrogels encapsulated with fluorescent label alone served as controls.
Fluorescence Microscopy Imaging of Hydrogels
PEG and alginate hydrogels encapsulated with fluorescently labelled extracellular vesicles or fluorescent dye alone were imaged using EVOS FL Auto Live Cell Imaging System on days 1, 4, 7, 10 and 14 post-encapsulation. Tile images of gels were generated and analysis of change in fluorescence was performed using ImageJ software. N = 3 per group from three independent experiments.
Natural Killer Cell Activation and Suppression by EVs
NK-92 cells (ATCC) were cultured with Myelocult H5100 (Stemcell technology) complete media supplemented with 100 units/mL of IL-2 and 12.6% of horse serum (Gibco) under standard conditions (37 °C, 20% O2, 5% CO2). 100,000 cells were seeded onto 48-well non-treated well plates (VWR) with 400 µL media volume with JAR extracellular vesicles (5 µg/mL and 20 µg/mL) or TGF-β (50 ng/mL) for 6 h. After EV or TGF-β preconditioning, the activation groups were activated for 2 h with phorbol 12-myristate 13-acetate (PMA, 10 ng/mL) and ionomycin (1 µg/mL). The media was collected and centrifuged at 180×g for 10 min. Supernatants were collected to measure IFNγ secretion utilizing ELISA Max deluxe set Human IFNγ kit (Biolegend). N = 6 per group from two independent experiments.
Activated Natural Killer Cell Suppression by EVs Delivered by PEG and Alginate Hydrogels
NK-92 cells were cultured as described above. JAR EVs were encapsulated within PEG or alginate hydrogels as described above. 100,000 cells were seeded onto 48-well non-treated plates (VWR) with 400 µL media volume and cultured with 20 µL PEG or alginate hydrogels (20 µg EV/gel or blank) or TGF-β (50 ng/mL) for 24 or 72 h. After EV or TGF-β preconditioning, the activation groups were activated for 2 h with phorbol 12-myristate 13-acetate (PMA, 10 ng/mL) and ionomycin (1 µg/mL). The media was collected and centrifuged at 180×g for 10 min. Supernatants were collected to measure IFNγ secretion utilizing ELISA Max deluxe set Human IFNγ kit (Biolegend). N = 6 per group from two independent experiments. Data was normalized to activated NK cell control.
Activation of NK-92 cells was also tested with EVs released from hydrogels. EV-containing or blank PEG or alginate hydrogels were incubated in 400 µL of NK-92 media for 72 h. 100,000 NK-92 cells were seeded onto 48-well non-treated plates (VWR) and incubated with pre-conditioned media for 24 h. After 24 h, the activation groups were activated for 2 h with phorbol 12-myristate 13-acetate (PMA, 10 ng/mL) and ionomycin (1 µg/mL). The media was collected and centrifuged at 180×g for 10 min. Supernatants were collected to measure IFNγ secretion utilizing ELISA Max deluxe set Human IFNγ kit (Biolegend). N = 6 per group from two independent experiments. Data was normalized to activated NK cell control after normalization to NK cell DNA per well using Picogreen assay.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software (version 9.1). Comparison of TER and SEC isolation method evaluated by Mann–Whitney test. EV release from hydrogels was compared using a two-way ANOVA with Sidak’s multiple comparisons test. JAR EV impact on NK cell IFN-γ secretion was evaluated using a two-way ANOVA with Dunnett’s multiple comparisons test.
Results and Discussion
Characterization of JAR and JEG-3 Trophoblast Cell Line Extracellular Vesicles
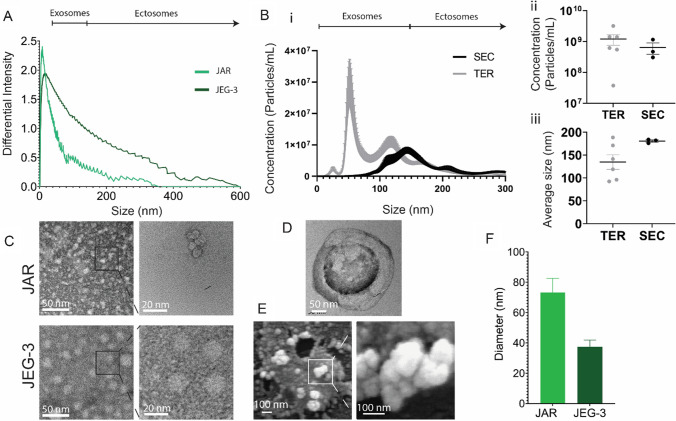
We first sought to isolate EVs from two cultured trophoblast cell lines, JEG-3 and JAR, and to characterize their size and quantity (Fig. 2). Both JEG-3 and JAR cell lines produced vesicles in the range of 0–600 nm and 0–375 nm diameter, respectively, which indicates the presence of vesicles in the size range of both exosomes and ectosomes, as measured via dynamic light scattering (Fig. 2A). We next sought to determine whether EV isolation method impacted EV quantity or size distribution. We evaluated the total exosome reagent (TER) and size exclusion chromatography (SEC) column methods, as opposed to the traditional ultracentrifugation technique, to limit damage to collected EVs and increase purity and reproducibility [41] (Fig. 2B). Nanoparticle tracking analysis demonstrates comparable quantity and size distribution between the TER method and the SEC purification method for JAR EVs (Fig. 2Bi–iii); given statistically comparable average yield (Fig. 2Bii) and size distribution (Fig. 2Biii), TER was used for subsequent studies as the TER method has a simpler workflow and faster yield time. Transmission electron microscopy (TEM) was used to verify the presence of vesicles in JEG-3 and JAR EV samples at high magnification (Fig. 2C), and additionally revealed the traditional “cup” morphology indicating spherical vesicles (Fig. 2D). Scanning electron microscopy (SEM) of JAR EV samples further demonstrated clusters of spherical vesicles (Fig. 2E), confirming the presence of EVs smaller than 100 nm diameter. Finally, we measured the average size of vesicles visualized with TEM, which demonstrated a larger average size for visualized JAR EVs relative to JEG-3 EVs (Fig. 2F). Altogether, this data demonstrates that EVs were successfully isolated from both JEG-3 and JAR cell lines, predominantly in the exosome size range. The size distribution of JEG-3 (0–600 nm) and JAR EVs (0–375 nm) aligns with previously reported size distribution of EVs isolated from primary trophoblast cell sources such as from perfused term placentas (50–500 nm) [20, 46], circulating EVs within the maternal plasma (0–300 nm) [47] as well as trophoblast stem cells (100–350 nm) [48, 49].
Fig. 2.
Characterization of JAR and JEG-3 isolated extracellular vesicles. A Dynamic light scattering analysis of JAR and JEG-3 EVs isolated via TER method. B Nanoparticle tracking analysis of JAR EVs isolated through SEC or TER methods showing (i) particle concentration relative to particle size, (ii) average concentration, and (iii) average particle size. C TEM imaging of JAR and JEG-3 EVs. D Magnified view of cup-like JAR EV morphology by TEM. E SEM of JAR EVs illustrating spherical morphology. F Quantification of EV diameter via TEM images. Data in F presented as mean ± SEM
Characterization of Trophoblast Cell Line Extracellular Vesicle Protein Composition and Tolerogenic Potential via Proteomics
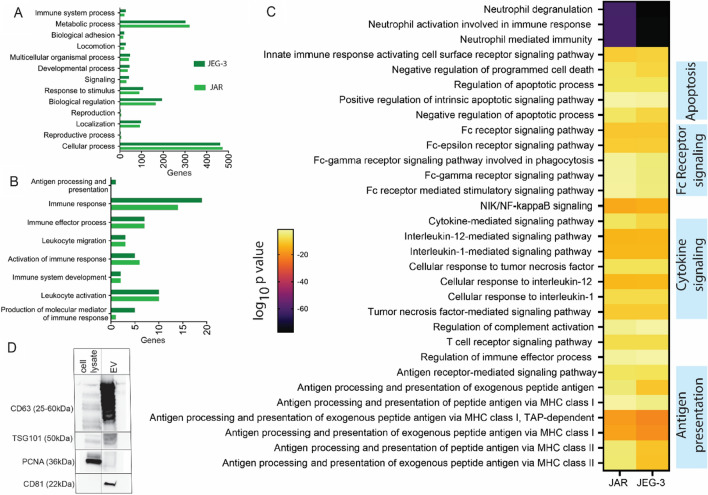
Given our interest in applying trophoblast EVs as a tolerogenic therapeutic, we next sought to characterize the composition of trophoblast cell line-derived EVs, with a focus on their immunomodulatory potential, and compare their composition to primary trophoblast EVs. We performed proteomic analysis on EVs isolated from both JEG-3 and JAR cell lines (Fig. 3), and identified a total of 1070 and 1072 proteins in in JAR and JEG-3 EVs, respectively. We first categorized these proteins based on biological processes through gene ontology enrichment analysis utilizing the Enrichr tool and PANTHER software (Fig. 3A), and found comparable protein expression between JEG-3 and JAR EVs across these broad biological processes. Overall, gene ontology enrichment analysis generated a list of 3845 and 3979 biological processes associated with JAR and JEG-3 vesicles, respectively. We next isolated processes related specifically to immune function (Fig. 3B) and found relatively comparable profiles between JEG-3 and JAR EVs, with the exception of JEG-3 expressing greater numbers of proteins related to antigen processing and presentation, immune response, and molecular mediators of immune response. A comparison between the significant biological processes between two cell types generated a list of 405 biological processes, and immune-specific pathways are highlighted in Fig. 3C with a heat map of adjusted p values for each cell line. The immune system processes identified are broadly involved in neutrophil activation, apoptosis signaling, Fc receptor signaling, cytokine signaling, and antigen presentation on MHC class I and II.
Fig. 3.
Proteomic characterization of JAR and JEG-3 isolated extracellular vesicles through liquid chromatography and mass spectrometry. A Gene ontology enrichment analysis performed on proteins identified in JAR and JEG-3 extracellular vesicles grouped based on biological processes. B Pathways involved in immune function identified in (A) in JAR and JEG-3 extracellular vesicles. C Immune system-related biological processes between JAR and JEG-3 isolated with adjusted p-values less than 0.05. D Western blot characterization of JAR extracellular vesicles and cell lysates for key exosome markers CD63, TSG101, and CD81, and non-extracellular vesicle marker nuclear antigen PCNA
We also looked for characteristic markers of placenta-derived EVs such as beta 2 microglobulin (a component of MHC class I molecules such as HLA-G), PlGF, and PLAP [28]. We identified beta 2 microglobulin in EVs from both JEG and JAR, PlGF in JEG only, and PLAP in JAR only. Both JAR and JEG-3 EVs expressed Annexin A1, CD63, and calreticulin, proteins identified in primary trophoblast EVs [16]; Annexin and calreticulin are “eat me” signals indicating these vesicles are likely to be phaged by recipient cells [16]. CD47 and CD31 have been used to identify macro and microvesicles isolated from placenta [48], and neither was present in EVs from either line, confirming a nanovesicle phenotype; these signals are also inhibitors of phagocytosis [48]. Similar to primary trophoblast EVs, JEG-3 and JAR EVs contained multiple complement factors, as well as CD55, a regulatory molecule that prevents complement activation at the maternal–fetal interface and regulates T cell function [48]. JEG-3 and JAR EVs were negative for some immunomodulatory markers observed in primary trophoblast EVs, such as MICA/B and ULBP1-5 [16], but did contain many other proteins identified in primary trophoblast EVs associated with tolerogenic immunomodulation [16, 31, 32, 48] (Table 1). Finally, we validated the presence of characteristic exosome markers CD63, CD81, and TSG101 within EV isolates via Western blot, and the lack of the non-extracellular vesicle protein marker proliferating cell nuclear antigen (PCNA) (Fig. 3D). We found that JAR EVs isolated via TER exhibited strong bands for EV markers and negative staining for nuclear protein PCNA, whereas cell lysates exhibited moderate staining for CD83 and TSG101 and strong presence of PCNA.
Table 1.
Sampling of tolerogenic proteins identified in JEG and JAR EVs via proteomics that correspond to primary trophoblast EV cargo [16, 48]
| Protein | Cell line | Function |
|---|---|---|
| Heat shock protein family E (Hsp10) member 1 (HSPE1) | JAR, JEG-3 | Regulate T cell expansion and Treg differentiation |
| CD276 | JAR, JEG-3 | Restrains T cell activation and proliferation |
| Programmed cell death protein 6 | JAR, JEG-3 | Apoptosis |
| Programmed cell death 6-interacting protein | JAR, JEG-3 | |
| CD81 antigen | JAR, JEG-3 | Regulation of complement cascade |
| Complement decay-accelerating factor (CD55) | JAR, JEG-3 | |
| Superoxide dismutase | JAR, JEG-3 | Anti-inflammatory enzyme |
| Galectin-1, 3 | JAR | Negative regulators of immune cell function |
| Galectin-7 | JAR, JEG-3 |
Engineering a Hydrogel Delivery Vehicle for Sustained Release of Tolerogenic Trophoblast Extracellular Vesicles
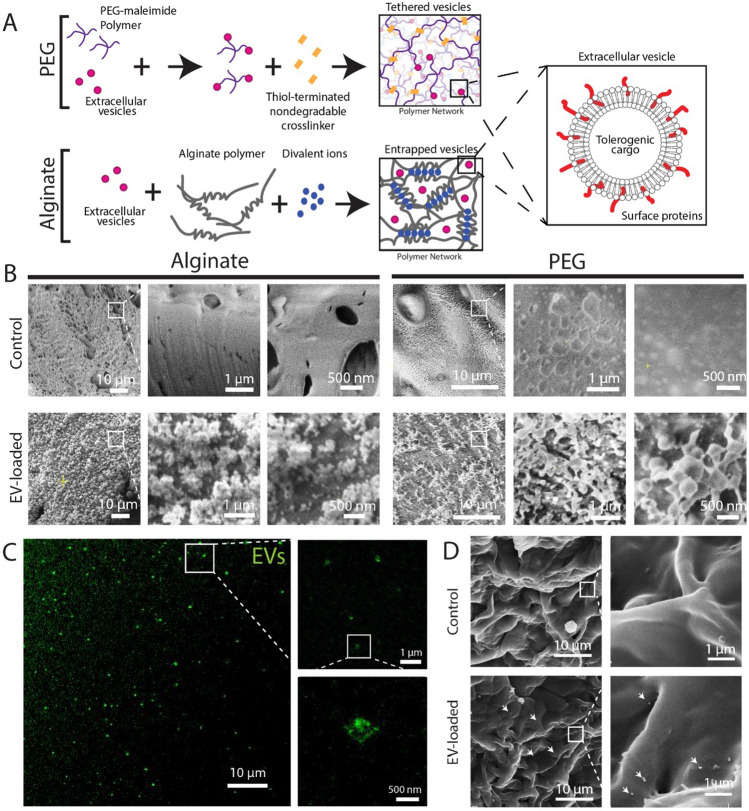
Given that proteomic characterization identified that JAR EV characteristics greater aligned with primary trophoblast extracellular vesicles (e.g. presence of placental alkaline phosphatase), JAR EVs were selected to engineer an EV-delivering hydrogel system (Fig. 4). To engineer a sustained release EV-delivering hydrogel system, we used a 4-arm synthetic poly(ethylene glycol) (PEG) hydrogel functionalized with maleimide end-groups, which enables rapid covalent linking with free thiol groups via a Michael-type addition reaction. EVs possess numerous surface proteins, which we hypothesized would contain free cysteines that would enable EV tethering to the PEG matrix; the hydrogel network was then crosslinked with a nondegradable synthetic dithiolated linker. As a control matrix, we used an alginate hydrogel to passively entrap EVs, as alginate polysaccharide polymers do not possess functional groups capable of linking EVs within the matrix (Fig. 4A). The hydrogels selected have comparable mesh sizes (5% w/v PEG = 15–35 nm [50], 2% w/v alginate = 10–50 nm [51]), which should result in comparable unbound EV diffusion kinetics from the matrix.
Fig. 4.
Imaging of EV-loaded hydrogel delivery systems. A Schematic illustrating EV tethering to PEG hydrogels and EV entrapment within alginate. B Cryo-scanning electron microscopy of blank (control, top row) and EV-loaded (bottom row) alginate (left) and PEG (right) hydrogels at increasing magnifications. C Stimulated emission depletion confocal microscopy of EV-loaded PEG hydrogels. D SEM imaging of PEG hydrogel surface. White arrows indicate EV location
We next sought to evaluate whether EVs could be visualized within the PEG and alginate matrices to confirm EV tethering or entrapment, respectively. We used cryo-scanning electron microscopy (cryo-SEM) to visualize nanoscale features of frozen, hydrated hydrogel-only (control) or freshly EV-loaded PEG and alginate matrices (Fig. 4B). Nanometer scale vesicles were visible for both alginate and PEG hydrogels in the EV-loaded group only. We further confirmed the presence of fluorescently-labeled nanoscale vesicles in PEG hydrogels using stimulated emission depletion confocal microscopy, which enables ~ 50 nm resolution fluorescence imaging (Fig. 4C), which revealed the presence of large EVs or clusters of EVs within the PEG hydrogel. Finally, we were also able to resolve EVs at the surface of PEG hydrogels using standard SEM imaging (Fig. 4D). Overall, this data demonstrates the presence of EV-like vesicles, in the size range characterized in Fig. 2, within or at the surface of EV-loaded hydrogels.
Extracellular Vesicle Delivery Kinetics from PEG and Alginate Hydrogels
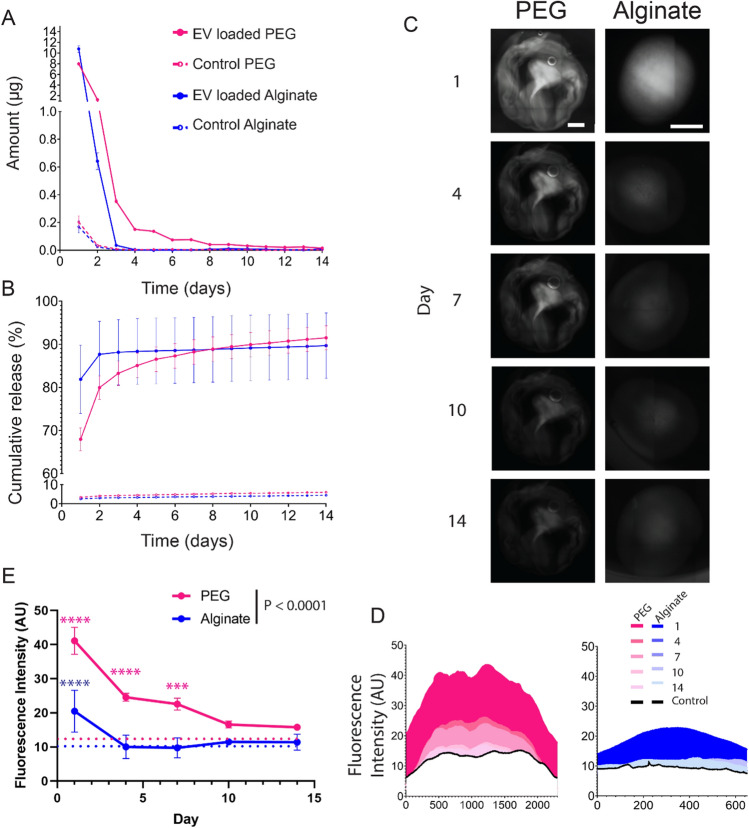
We next investigated the kinetics of extracellular vesicle release from PEG and alginate hydrogels. JAR EVs were labeled with a fluorescent tag and tethered to the PEG matrix or passively encapsulated within the alginate matrix; control hydrogels were loaded with an equivalent amount of fluorescent tag only. The quantity of released EVs was measured daily out to 14 days (Fig. 5A), where physically tethered PEG EVs exhibited sustained release over at least 14 days at approximately 100 ng/day between day 7 and 14. Conversely, EV entrapment within alginate led to a burst release completed within 3 days. Cumulatively, 68% and 82% of EVs were released from PEG and alginate hydrogels by day 1, and 91.5% and 90% on day 14, respectively (Fig. 5B), and cumulative daily release was not significant between alginate and PEG groups (P = 0.1388). Fluorescence microscopy was used to visualize EV retention within hydrogels over time (Fig. 5C), where quantification of fluorescence intensity across the hydrogels exhibited greater retention in PEG hydrogels out to day 14 (Fig. 5D). Time dependent analysis of fluorescent intensity demonstrated significantly retained fluorescence in EV-loaded PEG hydrogels relative to control PEG hydrogels out to day 7, whereas EV-loaded alginate hydrogel fluorescence did not significantly differ from alginate controls at any time point (Fig. 5E). PEG hydrogels exhibited significantly greater fluorescence relative to alginate throughout the time course (P = 0.0047). Overall, these results indicate that EV tethering within a PEG hydrogel matrix results in greater sustained release behavior, out to at least 2 weeks, relative to passive encapsulation within an alginate hydrogel matrix, where EV release is depleted within 2–4 days.
Fig. 5.
Extracellular vesicle delivery kinetics from PEG and alginate hydrogels. JAR EV release from PEG and alginate hydrogels measured as A amount release per day in µg and B cumulative release. C Fluorescent imaging of JAR EV loaded PEG and alginate hydrogels over 14 days, with maximum average fluorescent intensity quantified in (D). E Average fluorescent intensity values for EV-loaded PEG and alginate hydrogels (solid lines, n = 3 per group) relative to controls (dashed lines). Scale bar on all images = 1 mm. Two-way ANOVA with Sidak’s multiple comparisons test. ****P < 0.0001, ***P < 0.001. Pink and blue asterisk vs. PEG and alginate control hydrogels, respectively
With increasing interest in the use of EVs as localized therapeutics, several delivery systems have been designed for improved EV retention in vivo. Most previously developed EV delivery systems rely on passive encapsulation within a hydrogel matrix such as alginate [39]; however, passive encapsulation often results in poor control over EV delivery and rapid burst release, typicaly within 1 week [52, 53]. Synthetic hydrogel systems and chemistry techniques can be used to achieve more sustained EV delivery kinetics than passive entrapment, as we demonstrate (Fig. 5). One previous report demonstrated degradable PEG hydrogel-tethered mesenchymal stem cell-derived EVs achieved sustained delivery over 1 month [40]. Similarly, while our PEG-tethered EV system measured release out to 14 days, we calculate that the sustained release profile of 100 ng/day would continue up to approximately 28 days (Fig. 5B). It is possible that modifying our system with degradable crosslinkers would allow further modulation of EV release profile, and will be explored in future studies. Additionally, while passive systems have the disadvantage of less control over EV release kinetics, EVs covalently tethered within matrices may be impacted by physical tethering within the matrix, either by preventing whole EV release or alteration to surface proteins. These potential impacts to EVs by covalent immobilization within matrices should be investigated in further studies. Further, EV bioactivity stability over time is an important consideration, and EVs and their immunomodulatory component bioactivity should be verified as release profiles are extended further.
Tolerogenic Trophoblast EVs Reduce Activated Natural Killer Cell IFN-γ Secretion
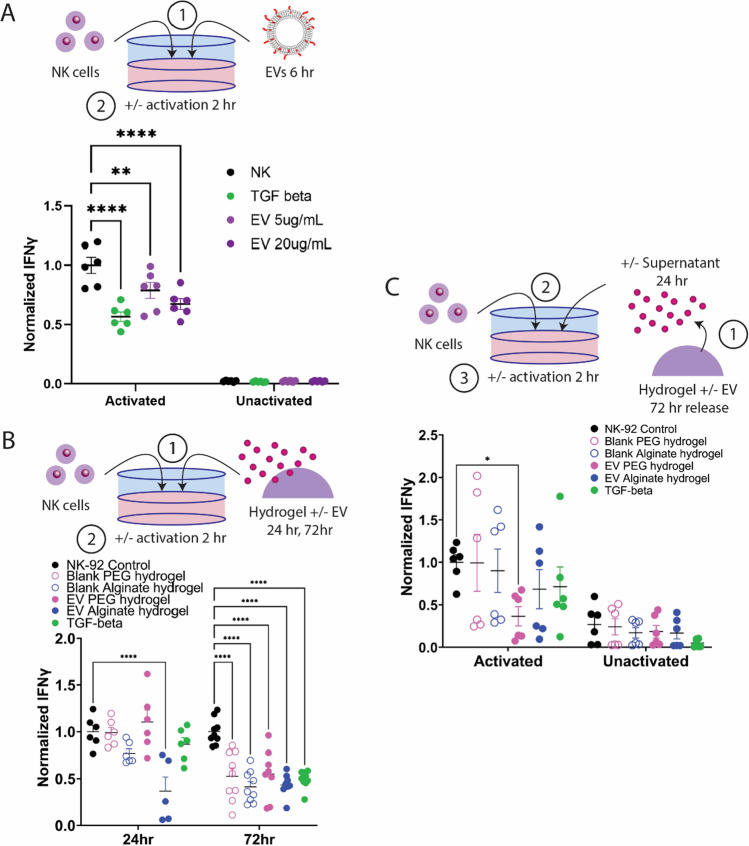
Finally, it is critical to determine the appropriate dose of EVs required to generate a tolerogenic response locally in immune cells within an allogeneic cell graft site (Fig. 6). We tested two dosages of JAR EVs, 5 and 20 µg/mL on activated natural killer (NK) cells to determine whether EVs are able to reduce NK cell activation comparably to known tolerogenic factors, such as high-dose TGF-β (50 ng/mL). Cancer-derived EVs and TGF-β have demonstrated the ability to reduce NK cell activation [54]. Human NK-92 cells were preconditioned with EVs or TGF-β for 6 h prior to activation with PMA/ionomycin for 2 h. EVs and TGF-β had no impact on unactivated NK cell IFN-γ secretion levels, whereas 5 and 20 µg/mL EVs significantly reduced activated NK cell IFN-γ secretion in a dose-dependent manner by 21% and 32%, respectively (Fig. 6A). TGF-β significantly decreased NK cell IFN-γ secretion by 43%, a level significantly lower than the 5 µg/mL EV dose, but not statistically different from the 20 µg/mL EV dose. Overall, this data demonstrates that a single dose of 5 or 20 µg/mL EVs can substantially reduce NK cell IFN-γ secretion, which may have downstream impacts to reduce the recruitment and activation of other immune cells in an in vivo environment.
Fig. 6.
Hydrogel-delivered trophoblast extracellular vesicles reduce NK cell activation. A JAR EVs exhibit dose-dependent decrease of activated NK cell IFN-γ secretion comparable to TGF-β control. B Activated NK cells co-cultured with EV-delivering or blank hydrogels for 24 or 72 h exhibited reduced IFN-γ secretion. C Supernatants from EV-loaded and blank hydrogels were collected after 72 h incubation in NK media and incubated with NK cells for 24 h prior to activation. Statistical comparison performed with two-way ANOVA with Dunnett’s multiple comparisons test. ****P < 0.0001, **P < 0.01, *P < 0.05
After establishing an effective EV dose to reduce activation in NK cells, we next incubated NK cells directly with blank or EV-loaded PEG and alginate hydrogels (20 µg EVs) for 24 or 72 h to determine whether our hydrogel systems could reduce NK cell activation (Fig. 6B). At 24 h incubation, only EV-loaded alginate hydrogels significantly reduced activated NK cell IFN-γ secretion (37%); however, at 72 h, both EV-loaded PEG and alginate hydrogels reduced activated NK cell IFN-γ secretion (55% and 43%, respectively), comparable to TGF-β controls (49%). Interestingly and unexpectedly, at 72 h of co-incubation, blank alginate and PEG hydrogel controls also significantly reduced activated NK cell IFN-γ secretion (41% and 53%, respectively). To determine whether NK cell activation was impacted by direct contact with hydrogels, and whether hydrogel-released EVs alone were capable of reducing NK cell activation, we next collected supernatants from blank and EV-loaded hydrogels after 72 h of release, and incubated these supernatants with NK cells for 24 h prior to activation (Fig. 6C). Supernatants containing hydrogel-delivered EVs resulted in significant reduction in activated NK cell IFN-γ secretion for EV-loaded PEG hydrogels only (37%). Altogether, this data indicates that direct incubation of NK cells with alginate and PEG hydrogels can reduce activated NK cell IFN-γ secretion, and that EVs released from PEG hydrogels are bioactive and capable of reducing activated NK cell IFN-γ secretion.
The hydrogel delivery systems herein were loaded with 20 µg EVs, resulting in approximately 13 and 17 µg EVs released by PEG and alginate hydrogels by 24 h, and 17 and 18 µg EVs by 72 h, respectively. NK cell activation data (Fig. 6A) demonstrates that a threshold of EVs is required to significantly suppress activated NK cell IFN-γ secretion, and that alginate hydrogel burst release achieves this level by 24 h, and sustained release from PEG hydrogels achieves this within 3 days (Fig. 6B). Future studies will evaluate the impact of higher EV loadings and larger hydrogel sizes on innate and adaptive immune cells in vitro and in vivo. Additionally, future studies will explore the impact of duration of release on immune cells, and subsequent impact on establishing immune tolerance in an in vivo study.
Conclusion
Here we demonstrate the isolation of human trophoblast cell line-derived EVs, and characterize their size and morphology, and tolerogenic potential via proteomics and in vitro assays with NK cells. We demonstrate the feasibility of tethering EVs within a synthetic PEG hydrogel matrix, which achieved a sustained release of at least 2 weeks relative to passive alginate entrapment methods where EV release is depleted within 2 days. We also demonstrate that hydrogels are capable of loading doses relevant to reducing NK cell activation in vitro. This tolerogenic extracellular vesicle-delivering hydrogel platform could serve as an alternative to systemic immunosuppression in cell transplantation, potentially reducing the risks associated with cell therapies and widening the eligible patient population.
Acknowledgements
This research was supported by the National Institutes of Autoimmunity and Infectious Diseases (R21AI151865). The confocal microscope used in these studies was acquired by an NIH SIG award (1 S10 RR027154-01A1) and is housed in the Regenerative Medicine Imaging Facility at Arizona State University. Data was generated using the facilities within the Eyring Materials Center at Arizona State University supported in part by NNCI-ECCS-2025490.
Funding
This research was supported by the Division of Extramural Research, National Institute of Allergy and Infectious Diseases (Grant No. R21AI151865).
Data Availability
Data is available for download at 10.5061/dryad.qfttdz0nb.
Declarations
Conflict of interest
Author S.C. Hiremath and J.D. Weaver have no conflicts to disclose.
Citation Diversity Statement
Recent work in several fields of science has identified a bias in citation practices such that papers from women and other minority scholars are undercited relative to the number of papers in the field. We recognize this bias and have worked diligently to ensure that we are referencing appropriate papers with fair gender and racial author inclusion.
Ethical Approval
No ethical approval was required.
Research Involving Human and/or Animal Participants
No animal or human subjects were used to conduct the studies in this manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ramzy A, et al. Implanted pluripotent stem-cell-derived pancreatic endoderm cells secrete glucose-responsive C-peptide in patients with type 1 diabetes. Cell Stem Cell. 2021;28(12):2047–2061.e5. doi: 10.1016/j.stem.2021.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro AMJ, et al. Insulin expression and C-peptide in type 1 diabetes subjects implanted with stem cell-derived pancreatic endoderm cells in an encapsulation device. Cell Rep. Med. 2021 doi: 10.1016/j.xcrm.2021.100466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapiro AD, et al. First-in-human phase 1/2 clinical trial of SIG-001, an innovative shielded cell therapy platform, for hemophilia Α. Blood. 2020;136:8. doi: 10.1182/blood-2020-141381. [DOI] [Google Scholar]
- 4.Jenkins MJ, Farid SS. Human pluripotent stem cell-derived products: advances towards robust, scalable and cost-effective manufacturing strategies. Biotechnol. J. 2015;10(1):83–95. doi: 10.1002/biot.201400348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pigeau GM, Csaszar E, Dulgar-Tulloch A. Commercial scale manufacturing of allogeneic cell therapy. Front. Med. 2018;5:233. doi: 10.3389/fmed.2018.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niethammer D, Kümmerle-Deschner J, Dannecker GE. Side-effects of long-term immunosuppression versus morbidity in autologous stem cell rescue: striking the balance. Rheumatology. 1999;38(8):747–750. doi: 10.1093/rheumatology/38.8.747. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz R, Kirk AD. Long-term toxicity of immunosuppressive therapy. Transplant. Liver. 2015 doi: 10.1016/B978-1-4557-0268-8.00097-X. [DOI] [Google Scholar]
- 8.Gallagher MP, et al. Long-term cancer risk of immunosuppressive regimens after kidney transplantation. J. Am. Soc. Nephrol. 2010;21(5):852–858. doi: 10.1681/ASN.2009101043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, et al. Immunosuppressive PLGA TGF-β1 microparticles induce polyclonal and antigen-specific regulatory T cells for local immunomodulation of allogeneic islet transplants. Front. Immunol. 2021;12:653088. doi: 10.3389/fimmu.2021.653088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Headen DM, et al. Local immunomodulation Fas ligand-engineered biomaterials achieves allogeneic islet graft acceptance. Nat. Mater. 2018;17(8):732–739. doi: 10.1038/s41563-018-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coronel MM, et al. Immunotherapy via PD-L1–presenting biomaterials leads to long-term islet graft survival. Sci. Adv. 2020;6(35):eaba5573. doi: 10.1126/sciadv.aba5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroemer G, Martínez-A C. Mechanisms of self tolerance. Immunol. Today. 1992;13(10):401–404. doi: 10.1016/0167-5699(92)90090-T. [DOI] [PubMed] [Google Scholar]
- 13.Trowsdale J, Betz AG. Mother’s little helpers: mechanisms of maternal-fetal tolerance. Nat. Immunol. 2006;7(3):241–246. doi: 10.1038/ni1317. [DOI] [PubMed] [Google Scholar]
- 14.Guttmacher AE, Maddox YT, Spong CY. The Human Placenta Project: placental structure, development, and function in real time. Placenta. 2014;35(5):303–304. doi: 10.1016/j.placenta.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tilburgs T, et al. Human HLA-G+ extravillous trophoblasts: immune-activating cells that interact with decidual leukocytes. Proc. Natl. Acad. Sci. U.S.A. 2015;112(23):7219–7224. doi: 10.1073/pnas.1507977112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mincheva-Nilsson L. Immunosuppressive protein signatures carried by syncytiotrophoblast-derived exosomes and their role in human pregnancy. Front. Immunol. 2021;12:717884. doi: 10.3389/fimmu.2021.717884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hönig A, Rieger L, Kapp M, Sütterlin M, Dietl J, Kämmerer U. Indoleamine 2,3-dioxygenase (IDO) expression in invasive extravillous trophoblast supports role of the enzyme for materno-fetal tolerance. J. Reprod. Immunol. 2004;61(2):79–86. doi: 10.1016/j.jri.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Southcombe J, Tannetta D, Redman C, Sargent I. The immunomodulatory role of syncytiotrophoblast microvesicles. PLoS ONE. 2011;6(5):e20245. doi: 10.1371/journal.pone.0020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atay S, Gercel-Taylor C, Suttles J, Mor G, Taylor DD. Trophoblast-derived exosomes mediate monocyte recruitment and differentiation. Am. J. Reprod. Immunol. 2011;65(1):65–77. doi: 10.1111/j.1600-0897.2010.00880.x. [DOI] [PubMed] [Google Scholar]
- 20.Cronqvist T, et al. Syncytiotrophoblast derived extracellular vesicles transfer functional placental miRNAs to primary human endothelial cells. Sci. Rep. 2017;7:4558. doi: 10.1038/s41598-017-04468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouyang Y, et al. Isolation of human trophoblastic extracellular vesicles and characterization of their cargo and antiviral activity. Placenta. 2016;47:86–95. doi: 10.1016/j.placenta.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 2016;36(3):301–312. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buzas EI. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 2022 doi: 10.1038/s41577-022-00763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Niel G, Carter DRF, Clayton A, Lambert DW, Raposo G, Vader P. Challenges and directions in studying cell–cell communication by extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2022;23(5):Art. no. 5. doi: 10.1038/s41580-022-00460-3. [DOI] [PubMed] [Google Scholar]
- 25.Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Meldolesi J. Exosomes and ectosomes in intercellular communication. Curr. Biol. 2018;28(8):R435–R444. doi: 10.1016/j.cub.2018.01.059. [DOI] [PubMed] [Google Scholar]
- 27.Morelli AE, Sadovsky Y. Extracellular vesicles and immune response during pregnancy: a balancing act*. Immunol. Rev. 2022;308(1):105–122. doi: 10.1111/imr.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kupper N, Huppertz B. The endogenous exposome of the pregnant mother: placental extracellular vesicles and their effect on the maternal system. Mol. Asp. Med. 2022;87:100955. doi: 10.1016/j.mam.2021.100955. [DOI] [PubMed] [Google Scholar]
- 29.Kshirsagar SK, et al. Immunomodulatory molecules are released from the first trimester and term placenta via exosomes. Placenta. 2012;33(12):982–990. doi: 10.1016/j.placenta.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petroff MG, Kharatyan E, Torry DS, Holets L. The immunomodulatory proteins B7-DC, B7-H2, and B7-H3 are differentially expressed across gestation in the human placenta. Am. J. Pathol. 2005;167(2):465–473. doi: 10.1016/S0002-9440(10)62990-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hedlund M, et al. Human placenta expresses and secretes NKG2D ligands via exosomes that down-modulate the cognate receptor expression: evidence for immunosuppressive function. J. Immunol. 2009;183(1):340–351. doi: 10.4049/jimmunol.0803477. [DOI] [PubMed] [Google Scholar]
- 32.Mincheva-Nilsson L, Baranov V. Review article: the role of placental exosomes in reproduction. Am. J. Reprod. Immunol. 2010;63(6):520–533. doi: 10.1111/j.1600-0897.2010.00822.x. [DOI] [PubMed] [Google Scholar]
- 33.Bracamonte-Baran W, et al. Modification of host dendritic cells by microchimerism-derived extracellular vesicles generates split tolerance. Proc. Natl. Acad. Sci. U.S.A. 2017;114(5):1099–1104. doi: 10.1073/pnas.1618364114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macedo C, et al. Donor-derived regulatory dendritic cell infusion results in host cell cross-dressing and T cell subset changes in prospective living donor liver transplant recipients. Am. J. Transplant. 2021;21(7):2372–2386. doi: 10.1111/ajt.16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mastoridis S, et al. Impact of donor extracellular vesicle release on recipient cell ‘cross-dressing’ following clinical liver and kidney transplantation. Am. J. Transplant. 2021;21(7):2387–2398. doi: 10.1111/ajt.16123. [DOI] [PubMed] [Google Scholar]
- 36.Squadrito ML, Cianciaruso C, Hansen SK, De Palma M. EVIR: chimeric receptors that enhance dendritic cell cross-dressing with tumor antigens. Nat. Methods. 2018;15(3):183–186. doi: 10.1038/nmeth.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakim LM, Bevan MJ. Cross-dressed dendritic cells drive memory CD8+ T-cell activation after viral infection. Nature. 2011;471(7340):629–632. doi: 10.1038/nature09863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiklander OPB, Brennan MÁ, Lötvall J, Breakefield XO, EL Andaloussi S. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 2019;11(492):eaav8521. doi: 10.1126/scitranslmed.aav8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chabria Y, Duffy GP, Lowery AJ, Dwyer RM. Hydrogels: 3D drug delivery systems for nanoparticles and extracellular vesicles. Biomedicines. 2021;9(11):Art no. 11. doi: 10.3390/biomedicines9111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mardpour S, et al. Hydrogel-mediated sustained systemic delivery of mesenchymal stem cell-derived extracellular vesicles improves hepatic regeneration in chronic liver failure. ACS Appl. Mater. Interfaces. 2019;11(41):37421–37433. doi: 10.1021/acsami.9b10126. [DOI] [PubMed] [Google Scholar]
- 41.Théry C, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Syromiatnikova V, Prokopeva A, Gomzikova M. Methods of the large-scale production of extracellular vesicles. Int. J. Mol. Sci. 2022;23(18):10522. doi: 10.3390/ijms231810522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaletka J, Lee KH, Altman J, Kanada M, Hardy JW. Listeria monocytogenes infection alters the content and function of extracellular vesicles produced by trophoblast stem cells. Infect. Immun. 2022;90(10):e00347–e422. doi: 10.1128/iai.00347-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conrad KP, Tuna KM, Mestre CT, Banwatt ES, Alli AA. Activation of multiple receptors stimulates extracellular vesicle release from trophoblast cells. Physiol. Rep. 2020;8(20):e14592. doi: 10.14814/phy2.14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godakumara K, et al. Trophoblast derived extracellular vesicles specifically alter the transcriptome of endometrial cells and may constitute a critical component of embryo-maternal communication. Reprod. Biol. Endocrinol. 2021;19(1):1–14. doi: 10.1186/s12958-021-00801-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villalobos-Labra R, et al. Placenta-derived extracellular vesicles from preeclamptic and healthy pregnancies impair ex vivo vascular endothelial function. Biosci. Rep. 2022;42(12):BSR20222185. doi: 10.1042/BSR20222185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, et al. Quantification of placental extracellular vesicles in different pregnancy status via single particle analysis method. Clin. Chim. Acta. 2023;539:266–273. doi: 10.1016/j.cca.2022.12.021. [DOI] [PubMed] [Google Scholar]
- 48.Tong M, et al. Proteomic characterization of macro-, micro- and nano-extracellular vesicles derived from the same first trimester placenta: relevance for feto-maternal communication. Hum. Reprod. 2016;31(4):687–699. doi: 10.1093/humrep/dew004. [DOI] [PubMed] [Google Scholar]
- 49.Cui J, et al. MiR-101-containing extracellular vesicles bind to BRD4 and enhance proliferation and migration of trophoblasts in preeclampsia. Stem Cell Res. Ther. 2020;11(1):231. doi: 10.1186/s13287-020-01720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weaver JD, Headen DM, Hunckler MD, Coronel MM, Stabler CL, García AJ. Design of a vascularized synthetic poly(ethylene glycol) macroencapsulation device for islet transplantation. Biomaterials. 2018;172:54–65. doi: 10.1016/j.biomaterials.2018.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Campbell KT, Wysoczynski K, Hadley DJ, Silva EA. Computational-based design of hydrogels with predictable mesh properties. ACS Biomater. Sci. Eng. 2020;6(1):308–319. doi: 10.1021/acsbiomaterials.9b01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shafei S, et al. Exosome loaded alginate hydrogel promotes tissue regeneration in full-thickness skin wounds: an in vivo study. J. Biomed. Mater. Res. A. 2020;108(3):545–556. doi: 10.1002/jbm.a.36835. [DOI] [PubMed] [Google Scholar]
- 53.Lv K, et al. Incorporation of small extracellular vesicles in sodium alginate hydrogel as a novel therapeutic strategy for myocardial infarction. Theranostics. 2019;9(24):7403–7416. doi: 10.7150/thno.32637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression1. J. Immunol. 2008;180(11):7249–7258. doi: 10.4049/jimmunol.180.11.7249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is available for download at 10.5061/dryad.qfttdz0nb.