Abstract
Transparent and FAIR disclosure of meta-information about healthcare data and infrastructure is essential but has not been well publicized. In this paper, we provide a transparent disclosure of the process of standardizing a common data model and developing a national data infrastructure using national claims data. We established an Observational Medical Outcome Partnership (OMOP) common data model database for national claims data of the Health Insurance Review and Assessment Service of South Korea. To introduce a data openness policy, we built a distributed data analysis environment and released metadata based on the FAIR principle. A total of 10,098,730,241 claims and 56,579,726 patients’ data were converted as OMOP common data model. We also built an analytics environment for distributed research and made the metadata publicly available. Disclosure of this infrastructure to researchers will help to eliminate information inequality and contribute to the generation of high-quality medical evidence.
Subject terms: Epidemiology, Outcomes research, Databases, Epidemiology
Introduction
Numerous studies using routinely collected large healthcare data have provided invaluable evidence representing routine clinical practice1,2. Administrative data representing the nationwide population have been used for secondary analysis in healthcare research for various purposes, including consecutive monitoring of disease and medical expenditure, comparative effectiveness of medical interventions, and even machine learning3–6. The Korean National Health Insurance system is a single public insurance system for all citizens, and all medical institutions are applied as mandatory designation systems. The Health Insurance Review and Assessment Service (HIRA) establishes health insurance reimbursement criteria and reviews all medical claims for reimbursement. Therefore, the HIRA has accumulated a vast amount of claims data at the national level, and it can be used as a secondary data source for high-quality real-world evidence7. For example, statistics from the HIRA database are used in OECD statistics as representative statistics for Korea.
Administrative data, despite being a commonly used source for research, has drawn significant criticism predominantly due to concerns over the validity of its coded information. For instance, coding practices like “upcoding” can lead to inaccuracies; this is where providers code for a more severe illness than the patient actually has to receive higher reimbursement8,9. While the debate on coded information’s validity continues, less attention is being directed towards the stewardship of this extensive healthcare data. Chief among these are issues including: 1. Non-scalability and non-interoperability; 2. Ignored reproducibility; and 3. Protection of privacy of the national population. Such areas might pose even more profound implications for the utility and reliability of large healthcare datasets.
A distributed research system based on a common data model has emerged as a promising alternative to address the concerns surrounding the use of large healthcare datasets10. The Observational Medical Outcome Partnership Common Data Model (OMOP-CDM) is a standardized data model maintained by Observational Health Data Sciences and Informatics (OHDSI), which is a global, multi-stakeholder, interdisciplinary community. The OMOP-CDM was designed to enable the systematic analysis of large observational datasets from multiple data sources by providing a common structure and vocabulary for observational data. In response to the urgent requirement for coronavirus disease-2019 (COVID-19) research, the HIRA was the first institution in the world to standardize the data of patients with COVID-19 into OMOP-CDM, providing access to international researchers without compromising patient privacy11. This approach inspired other database owners, enabling researchers to conduct multiple high impact studies using the multi-national database in a timely manner12. However, thus far, the HIRA database has been standardized to OMOP-CDM for individual studies, and standardized data have not been maintained13.
We aimed to standardize HIRA data into OMOP-CDM, build infrastructure providing scalable accessibility and a flexible data analysis environment with privacy-by-design protection, and verify whether the infrastructure guarantees the reproducibility of research. The aim of this study was to enhance the FAIRness of the national healthcare database, which refers to its ability to be easily Findable, Accessible, Interoperable, and Reusable (FAIR)14. Specifically, in this study, the process of converting national claims data into research data to establish research infrastructure, mapping local code to standard vocabulary system, verifying data through type 2 diabetes mellitus (T2DM) cases and replicating previously published COVID-19 prediction study. In addition, external disclosure of the infrastructure by the FAIR principle was reviewed.
Results
Basic statistics of HIRA CDM
We extracted, transformed, and loaded (ETL) the HIRA database into the OMOP-CDM version 5.3.1. All tables specified by the OMOP-CDM conversion specifications were created. The number of converted claims specification and number of patients included were 10,098,730,241 and 56,579,726, respectively (Table 1). Among the converted data, the number of males and females was 28,439,311 (50.3%) and 28,140,325 (49.7%), respectively. All records of the source database were converted into CDM format without errors in classification by year, type of visit, and type of claiming medical institution (Table S1 in the Supplements). Among the CDM tables, the death table contained information of 3,804,948 people who had died over 11 years, accounting for 6.7% of the total population (Table 1). The condition, drug, and procedure tables, which are the main clinical information of the OMOP-CDM, included more than 99.0% of patients, and devices and measurements included more than 90.0% of patients (Table 1).
Table 1.
Number of records, number of persons, and their ratio in HIRA CDM database.
| OMOP-CDM tables | Records (n) | Person (n) | Person/total person (%) |
|---|---|---|---|
| CARE SITE | 213 758 | 0 | 0.0 |
| CONDITION ERA | 12 600 281 758 | 56 536 873 | 99.9 |
| CONDITION OCCURRENCE | 26 798 208 704 | 56 536 873 | 99.9 |
| COST | 76 131 071 211 | 0 | 0.0 |
| DEATH | 3 804 948 | 3 804 948 | 6.7 |
| DEVICE EXPOSURE | 892 251 206 | 51 913 063 | 91.8 |
| DRUG ERA | 15 052 413 048 | 56 363 138 | 99.6 |
| DRUG EXPOSURE | 28 732 916 071 | 56 389 812 | 99.7 |
| MEASUREMENT | 10 190 277 150 | 53 603 975 | 94.7 |
| OBSERVATION | 499 711 003 | 40 949 894 | 72.4 |
| OBSERVATION PERIOD | 56 579 726 | 56 579 726 | 100.0 |
| PAYER PLAN PERIOD | 56 579 726 | 56 579 726 | 100.0 |
| PERSON | 56 579 726 | 56 579 726 | 100.0 |
| PROCEDURE OCCURRENCE | 27 683 994 844 | 56 547 684 | 99.9 |
| VISIT OCCURRENCE | 10 098 730 241 | 56 579 728 | 100.0 |
HIRA: Health Insurance Review and Assessment Service; OMOP: Observational Medical Outcome Partnership; CDM: common data model; n: number.
The results of vocabulary mapping from the Electronic Data Interchange (EDI) codes of Korea to the OMOP standardized vocabulary are shown in Table 2. Table 2 lists the number of EDI codes according to the OMOP domain, ratio of codes mapped to standard terminologies, and number of mapped records per source record. Regarding the ratio of mapped codes to source codes, condition (99.1%), drug (100.0%), observation (99.97%), and procedure (84.5%) were high, however, device (10.8%) and measurement (31.0%) were relatively low. However, the ratio of mapped records (mapped records per source records) was over 85.0% in all domains including device (87.6%) and measurement (91.5%).
Table 2.
Status of vocabulary mapping in the converted HIRA CDM.
| Contents | OMOP-CDM tables | |||||
|---|---|---|---|---|---|---|
| CONDITION OCCURRENCE | PROCEDURE OCCURRENCE | DRUG EXPOSURE | DEVICE EXPOSURE | MEASUREMENT | OBSERVATION | |
| Source code, n | 19 084 | 322 136 | 63 095 | 20 082 | 22 765 | 3 481 |
| Mapped code, n | 18 910 | 272 163 | 63 095 | 2 159 | 7 049 | 3 480 |
| Mapped code ratio, % | 99.1 | 84.5 | 100.0 | 10.8 | 31.0 | 99.97 |
| Source records, n | 26 798 208 704 | 27 683 994 844 | 28 732 916 071 | 892 251 206 | 10 190 277 150 | 499 711 003 |
| Mapped records, n | 26 599 002 701 | 26 111 671 178 | 28 692 327 376 | 781 209 308 | 9 326 266 819 | 499 596 519 |
| Mapped records ratio, % | 99.3 | 94.3 | 99.9 | 87.6 | 91.5 | 99.98 |
HIRA: Health Insurance Review and Assessment Service; CDM: common data model; OMOP: Observational Medical Outcome Partnership;
Data quality and reliability
We compared the amount of original (source) and converted data for the condition/drug/procedure/device codes. The number of records from the source and converted data and their differences from the top 10 codes in each domain are presented in the Tables S2–S6 in the Supplements. The differences were due to (1) the multiple mapping of the source code, (2) the assignment to a different domain table from the source table, and (3) the absence of mapping to OMOP standardized vocabulary.
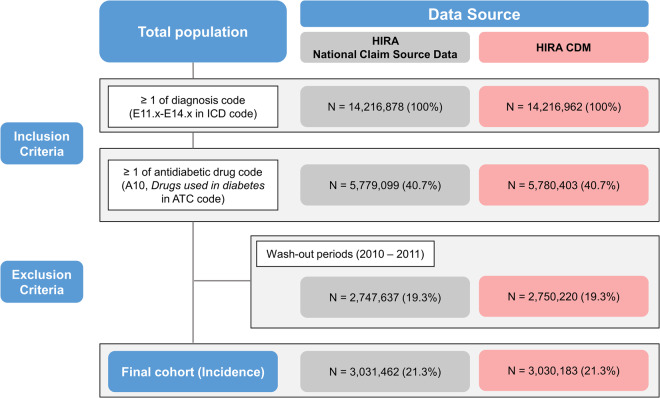
The number of patients with T2DM was extracted according to the same definition from the source and converted data, and the numbers of patients were 3,031,462 (21.3%) and 3,030,183 (21.3%), respectively (Fig. 1). The incidence of T2DM per 100,000 patients ranged from 550.1 to 650.9 and 549.9 to 649.7 in the source and converted HIRA CDM database, respectively (Table 3). In 2012, the difference in the number of patients with T2DM between the source and converted data was 590, and the difference in the incidence rate was the largest at 1.2 per 100,000 patients. The difference in the number of patients was 14, and the difference in the incidence rate was 0.0 in 2020. In addition, there were no differences in T2DM incidence by year-gender and year-age groups (Tables S7, S8 in the Supplements).
Fig. 1.
Flow chart of type 2 diabetes mellitus phenotype and comparison of incidences from the source and converted CDM databases. CDM: common data model.
Table 3.
Incidence and difference of type 2 diabetes mellitus phenotype by year between source and converted HIRA CDM data.
| Year | Population size at the middle of each year | HIRA Source data | HIRA CDM | Differences | |||
|---|---|---|---|---|---|---|---|
| N | Incidence* | N | Incidence* | N | Incidence* | ||
| (A) | (B) | (C) | (D) | (A)-(C) | (B)-(D) | ||
| 2012 | 50 199 853 | 326 732 | 650.9 | 326 142 | 649.7 | 590 | 1.2 |
| 2013 | 50 428 893 | 304 986 | 590.4 | 304 594 | 589.7 | 392 | 0.7 |
| 2014 | 50 746 659 | 291 837 | 550.1 | 291 750 | 549.9 | 87 | 0.2 |
| 2015 | 51 014 947 | 304 215 | 558.7 | 304 784 | 559.8 | −569 | −1.1 |
| 2016 | 51 217 803 | 337 933 | 607.7 | 337 561 | 607.0 | 372 | 0.7 |
| 2017 | 51 361 911 | 350 511 | 617.0 | 350 327 | 616.7 | 184 | 0.3 |
| 2018 | 51 585 058 | 356 570 | 615.1 | 356 420 | 614.8 | 150 | 0.3 |
| 2019 | 51 764 822 | 374 174 | 631.1 | 374 115 | 631.0 | 59 | 0.1 |
| 2020 | 51 836 239 | 384 504 | 636.6 | 384 490 | 636.6 | 14 | 0.0 |
*Age/sex-standardized incidence rate per 100 000 standard fixed population. Fixed population was referenced from KOSIS, Statistics Korea. HIRA: Health Insurance Review and Assessment service; CDM: common data model, N: number.
In the HIRA CDM database, by 2020, 32,633 outpatients were diagnosed with COVID-19. We could validate a previously published COVID-19 prediction model (COVER model) which developed based on the OMOP-CDM15. The performance of the COVER models to predict hospitalization for pneumonia, admission to the ICU or death from pneumonia, and all-cause death were 0.816, 0.891, and 0.892, respectively (Table S9 in the Supplements). We also tried to validate the models using newly updated sampled database. The HIRA 20% sample database until April 2022, 1,530,350 outpatients were diagnosed with COVID-19, and the performance of the model was 0.748 (hospitalization for pneumonia), 0.879 (admission to the ICU with pneumonia or death due to pneumonia), and 0.891 (all-cause mortality). Through version control of the database, we confirmed that predictive models developed earlier could be easily applied to databases of different versions with different periods.
Data analytic environment and open policy
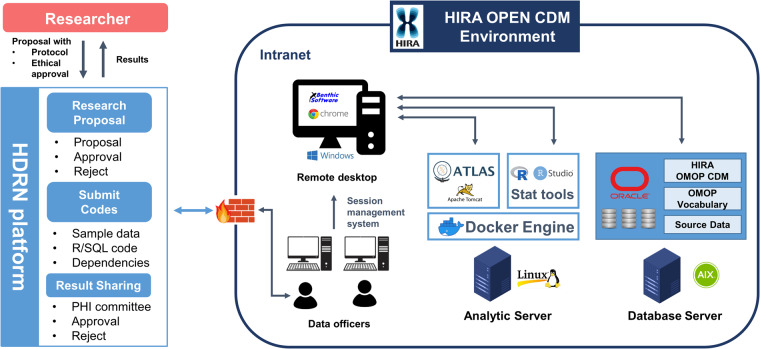
We built a Docker-based analytic environment for the use of open-source tools even in an intranet environment (offline for Internet) of the HIRA and to enable the installation of statistical tools and frequently updated packages (Fig. 2)16. For data security, the data officer of the HIRA is responsible for managing access sessions and logs from database and analytic servers.
Fig. 2.
HIRA CDM analytic environment and data open process. Researchers can request the use of the HIRA CDM through the HDRN platform, which is an open public healthcare data platform. HDRN, Healthcare Distributed Research Network; PHI, personal health information; HIRA, Health Insurance Review and Assessment Service; CDM, common data model; OMOP, Observational Medical Outcome Partnership.
By implementing the open policy of the HIRA CDM, researchers can apply for research requests through the healthcare distributed research network (HDRN) platform operated by the Korean government (https://hcdl.mohw.go.kr/). The specific application method is as follows: (1) The researcher must request a review of their research hypotheses and plan for ethical feasibility through an institutional or public review board. (2) The research must submit an approval letter from the review board and the research protocol to the HDRN platform. (3) The HIRA reviews the appropriateness of research/data provision and decides whether to provide it. (4) The researcher writes an analysis query, code, or package based on the open sample data and environment and sends it to the HIRA. (5) The HIRA reviews queries and expected results and derives results by running queries/codes/packages. (6) After the results are reviewed and the protected health information checked for infringement, the results are exported to the researcher.
We followed all FAIR principles, and the results of applying each principle to the HIRA CDM are shown in Table S10 in the Supplement. Metadata, disclosure policy, and sample data of HIRA CDM have been made available to the public online (https://opendata.hira.or.kr/op/opb/selectNotice.do?sno=13906&ntfcIteDivCd=&searchCnd=&searchWrd=cdm&pageIndex=1).
Discussion
The HIRA CDM database is a useful national resource that encompasses abundant medical information of virtually all citizens and institutions in the Republic of Korea. An open research analysis system that complies with the FAIR principle was established to transparently utilize it for biomedical and healthcare research. While increasing researchers’ access to data resources, a distributed research system with privacy by design was established, such that national claims data across the country can be safely disclosed to external researchers without access to patient-level data. The established database and environment demonstrated the reproducibility and scalability of the research through comparative verification with source data and previously developed predictive models.
The Data Quality Dashboard17, the official quality assessment tool of OMOP CDM, was not performed because of limited hardware resources. This was because it was expected to take several months to be running to HIRA CDM, such a large size of data. In the comparison of T2DM incidence performed for quality assessment, there were differences between the original data and the CDM, however, which were attributed to changes in disease coverage during code conversion (source code to OMOP standardized vocabulary). This is an issue of mapping to different vocabulary systems and is not a data quality issue, however, researchers should be aware of such cases.
Facilitating transparent and reproducible research
The retraction of COVID-19 research from high-profile journals underscores the necessity for open and reproducible science in healthcare18, which is particularly important for promoting confidence in science during the global health crisis. The current scientific landscape relies heavily on researchers’ reliability and trustworthiness. However, significantly high rates of data fabrication, falsification, and false-positive findings occur in healthcare research using big data for secondary use, further highlighting the necessity for more transparent research practices19,20. The usual policy of ‘sharing data upon request’ may not be optimal as it may limit the accessibility and usability of the data21. The common challenge against open science in healthcare is that patient-level data are inherently highly sensitive, making it difficult to share such data while preserving privacy. This challenge must be addressed by developing innovative approaches and technologies that can ensure safe and secure sharing of patient data while promoting open and reproducible science. Distributed research based on standardized data and vocabulary may guarantee reproducibility of research while preventing p-hacking.
Scalable accessibility with privacy-by-design protection
Distributed research systems aimed toward data standardization guarantee scalable accessibility without privacy concerns because they enable privacy-by-design protection. Researchers cannot access patient-level data, and only anonymized data can be exported from the system to researchers. Despite being an internal environment with no Internet access, we utilized several open-source tools (most are Internet-dependent) to build our analytic environment. This unique approach uses the analytic codes or programs to perform the analysis instead of providing data to external researchers. Analytic queries, codes, and even a Docker-based analytic environment can be applied, enabling researchers to conduct reproducible analyses in the same local environment.
Interoperability across countries
HIRA data can be used as a common data model such as OMOP-CDM in various approaches. Depending on the characteristics of the claims data, they include the life cycle information of the entire population; thus, expansion into various fields, such as the calculation of national statistics, research for clinical effectiveness, health care policy, and AI algorithms, is possible. The Republic of Korea is in the process of introducing OMOP-CDM to 57 medical institutions through past large national funding, suggesting that HIRA data can be utilized in association with the EHR-based databases of medical institutions using various methods. In addition, internationally, it is possible to cooperate with large-scale projects such as OHDSI, N3C22, EHDEN23, and DARWIN-EU24 based on the OMOP-CDM. Furthermore, as a national data infrastructure, it is possible to promote data harmonization with other data standards such as Fast Healthcare Interoperability Resources (e.g., http://omoponfhir.org/).
FAIR research stewardship
As a custodian of nationwide healthcare data, the HIRA builds infrastructure for better research and data stewardship. Although data disclosure is important, the FAIR principle has rarely been applied to large-scale healthcare databases, owing to the sensitivity of personal data. In addition, the nature of the healthcare data provision process, in which researchers must rigorously vet data providers, often means that they do not provide sufficient information about the data. Providing metadata in accordance with FAIR can be part of a culture that improves access to information, and thus address information inequalities. For example, the structure of the database, original source of the data, time period of data, vocabulary, and application process for data access, etc.
Methods
Data source
HIRA claims data include complete information about medical services, such as patients’ visits to medical institutions, demographic information, medical service use, cost, disease conditions, and treatments including medications and procedures. The Republic of Korea introduced a mandatory national health insurance service to manage eligible citizens for health insurance from birth to death. In addition, a computerized system that enables the real-time linkage of medical records generated by medical institutions with the HIRA has been established. This study used the national claims data of the HIRA, which cover approximately 97% of the total population of the Republic of Korea (https://www.mohw.go.kr/eng/hs/hs0110.jsp?PAR_MENU_ID=1006&MENU_ID=100610). Furthermore, the HIRA data were linked to the national death registry of Statistics Korea; therefore, they were also included in this study. Data conversion and analyses were performed according to local laws and regulations and with approval from the respective scientific and ethics committees (Health Insurance Review and Assessment Institutional Review Board: 2022-014-001).
Mapping to standardized vocabulary
Health insurance details (for diagnoses, medical fees, medication, and therapeutic materials) are reimbursed using the EDI code system in Korea; therefore, all details in the HIRA database are stored as EDI codes. We established a standard dictionary for the EDI code to construct the OMOP-CDM and integrated the EDI system into the OMOP standardized vocabulary through previous research25. Vocabulary mapping was conducted from terms for the reimbursement/non-reimbursement list of the EDI to the standard concepts for each domain according to OHDSI standardized vocabulary, e.g., diagnostic codes were mapped to SNOMED-CT, medication codes were mapped to RxNorm system (https://github.com/OHDSI/Vocabulary-v5.0/wiki/General-Structure-and-Use). Two or more healthcare experts independently conduct vocabulary mapping, and in case where their results differ, a third-party review makes the final decision. The final mapping list has been transparently disclosed online (Basic medical examination and diagnosis fee: https://opendata.hira.or.kr/op/opb/selectRfrm.do?rfrmTpCd=&searchCnd=&searchWrd=%EC%9A%A9%EC%96%B4&sno=13305&pageIndex=1 and Operation and Procedure fee: https://opendata.hira.or.kr/op/opb/selectNotice.do?searchCnd=&searchWrd=%EC%9A%A9%EC%96%B4&sno=13603&pageIndex=1).
Because standardized analysis using the OMOP-CDM is based on a standard vocabulary, if the ratio of unmapped records is high, information loss may occur because it cannot be used in the analysis. Code mapping and mapping record rates were checked to evaluate the possible information loss according to the vocabulary dictionary.
Data conversion and quality assessment
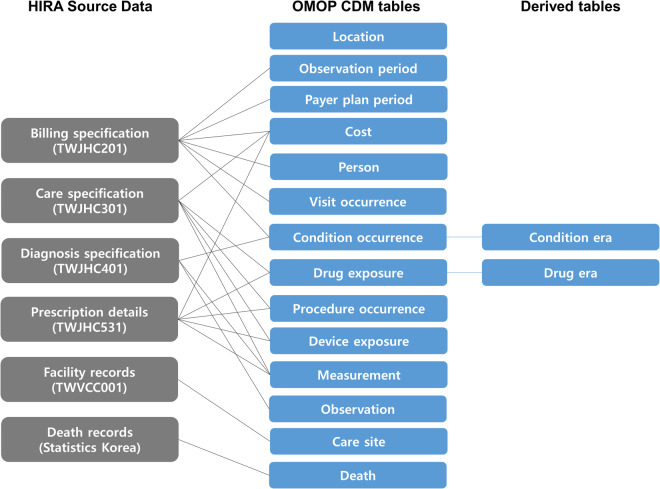
In this study, approximately 10 billion claim specifications for 56 million patients from 2010 to 2020 were converted into the OMOP-CDM. The data included information on healthcare institutions and death registry data, as well as general information, diagnosis, care, and prescription details of billing specifications. The source data of HIRA was converted by referring to the specification of OMOP-CDM version 5.3.1 (https://ohdsi.github.io/CommonDataModel/cdm53.html). Six types of source data were converted into 25 data tables of five table domains (clinical data, health system data, health economics data, standardized derived elements, metadata) and the data loaded with OMOP standardized vocabulary tables (Fig. 3). HIRA data were linked to the national death registry of Statistics Korea by national identification number. Under the current OMOP-CDM 5.3 convention, the death table was populated with the date of death and only one representative cause of death (underlying antecedent cause of death) for deceased patients. The pseudonymized patient identifiers and visit identifiers in the source data are maintained for consistency of the future conversion.
Fig. 3.
Data mapping to OMOP-CDM from HIRA source claims database. OMOP: Observational Medical Outcome Partnership; CDM: common data model; HIRA: Health Insurance Review and Assessment Service.
After the ETL process, we evaluated the quality of the HIRA CDM by assessing the concordance of descriptive statistics from the source and converted data. Statistical concordance between the source and HIRA CDM was evaluated. We compared the size of the data (by year and by type of medical service), number of medical institutions, and number of records with frequent codes within each domain. In addition, the number of patients with T2DM and its incidence in the middle of the year were calculated using the source and CDM databases. The digital phenotyping of T2DM was defined as those that had corresponding codes to E11-E14 of the International Classification of Disease (ICD-10) and A10 (‘Drug used in diabetes’) of Anatomical Therapeutic Chemical (ATC) Classification system26.
A previously published clinical prediction model was applied to corroborate the usability of the database and infrastructure established in this study. The COVER model was developed in the 2020 OHDSI COVID-19 study-a-thon, and the subset of HIRA database has already been used for the model validation study15. In the previous study, HIRA data included information of the patients with COVID-19 from 1 January to 4 April, 2020; however, in this study, we re-validated using data from two different databases: (1) the HIRA CDM database; 1 January, 2020, to 31 December, 2020, (2) 20% sampled database which newly updated information of the patients with COVID-19 until 30 April, 2022.
The target population was patients with COVID-19 infection and was defined as COVID-19 diagnosis or severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) virus positive through the reverse transcription polymerase chain reaction (RT-PCR) test. The population was limited to adults (age ≥ 18) and without flu symptoms and pneumonia diagnosis within the previous 60 days. The outcomes to be predicted were as follows: (1) hospitalization for pneumonia within 30 days, (2) hospitalization for pneumonia requiring intensive care service or death after hospitalization for pneumonia from an index up to 30 days after the index, and (3) death within 30 days. The detailed model development process and evaluation method were performed in the same manner as described as in the previous publication.
Infrastructure and data open policy
To utilize the HIRA CDM as a national data infrastructure, we established an open analytic environment and data access process for external researchers. To establish the analytic environment, our aim was to ensure that the analytic package developed by an external researcher using open-source tools (e.g., R) was sufficiently run, even in the closed intranet network of HIRA. We established a data acquisition process for external researchers, and the HIRA CDM data were disclosed according to the principle of distributed research using metadata and sample data. In all processes, we followed the FAIR principle, published the metadata online, and performed version control of the database.
Supplementary information
Acknowledgements
This work was supported by the Health Insurance Review and Assessment Service (HIRA).
Author contributions
J.W.K. and Y.L. collected data. J.W.K., C.K., R.W.P. and S.C.Y. conceptualized and designed the study. J.W.K., C.K., D.H.Y. and H.B. conducted data analyses including data conversion, developing analytic infrastructures, and proof-of-concept analyses. All authors interpreted the results. J.W.K. and C.K. drafted the manuscript and K.H.K., J.Y., R.W.P. and S.C.Y. made critical revisions to the manuscript. R.W.P. and S.C.Y. finalized the manuscript.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Data on vocabulary mapping were disclosed on the HIRA website (Basic medical examination and diagnosis fee: https://opendata.hira.or.kr/op/opb/selectRfrm.do?rfrmTpCd=&searchCnd=&searchWrd=%EC%9A%A9%EC%96%B4&sno=13305&pageIndex=1 and Operation and Procedure fee: https://opendata.hira.or.kr/op/opb/selectNotice.do?searchCnd=&searchWrd=%EC%9A%A9%EC%96%B4&sno=13603&pageIndex=1). According to Personal Information Protection Act in the Republic of Korea, HIRA does not permit us to share patient-level source data or data derivatives with individuals and institutions.
The CDM data converted in this study is available as a distributed research network way upon an application through an online web portal (https://hcdl.mohw.go.kr). HIRA CDM is updated on an annual basis. Researchers can apply for research requests through the healthcare distributed research network (HDRN) platform operated by the Korean government. The specific application method is as follows: (1) The researcher must request a review of their research hypotheses and plan for ethical feasibility through an institutional or public review board. (2) The research must submit an approval letter from the review board and the research protocol to the HDRN platform. (3) The HIRA reviews the appropriateness of research/data provision and decides whether to provide it. (4) The researcher writes an analysis query, code, or package based on the open sample data and environment and sends it to the HIRA. (5) The HIRA reviews queries and expected results and derives results by running queries/codes/packages. (6) After the results are reviewed and the protected health information checked for infringement, the results are exported to the researcher. Detailed application process for data use is descripted in https://hcdl.mohw.go.kr/static/data/dataApplyStep.
Code availability
We stored the CDM data using open-source codes of OHDSI for conforming to the database structure of OMOP CDM (https://github.com/OHDSI/CommonDataModel).
Competing interests
RWP reports grants from the Ministry of Trade, Industry & Energy (MOTIE, Korea) and the Ministry of Health & Welfare (Korea). SCY reports being CTO of PHI Digital Healthcare.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ji-Woo Kim, Chungsoo Kim.
These authors jointly supervised this work: Rae Woong Park, Seng Chan You.
Contributor Information
Rae Woong Park, Email: veritas@ajou.ac.kr.
Seng Chan You, Email: chandryou@yuhs.ac.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-023-02580-7.
References
- 1.Schneeweiss S. Learning from Big Health Care Data. New England Journal of Medicine. 2014;370:2161–2163. doi: 10.1056/NEJMp1401111. [DOI] [PubMed] [Google Scholar]
- 2.You SC, Krumholz HM. The Evolution of Evidence-Based Medicine: When the Magic of the Randomized Clinical Trial Meets Real-World Data. Circulation. 2022;145:107–109. doi: 10.1161/CIRCULATIONAHA.121.057931. [DOI] [PubMed] [Google Scholar]
- 3.Lo-Ciganic W-H, et al. Developing and validating a machine-learning algorithm to predict opioid overdose in Medicaid beneficiaries in two US states: a prognostic modelling study. The Lancet Digital Health. 2022;4:e455–e465. doi: 10.1016/S2589-7500(22)00062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nosrati E. Harnessing administrative data to study health inequality. The Lancet Public Health. 2022;7:e726–e727. doi: 10.1016/S2468-2667(22)00172-4. [DOI] [PubMed] [Google Scholar]
- 5.Portuondo JI, Harris AHS, Massarweh NN. Using Administrative Codes to Measure Health Care Quality. JAMA. 2022;328:825–826. doi: 10.1001/jama.2022.12823. [DOI] [PubMed] [Google Scholar]
- 6.Sarrazin MSV, Rosenthal GE. Finding Pure and Simple Truths With Administrative Data. JAMA. 2012;307:1433–1435. doi: 10.1001/jama.2012.404. [DOI] [PubMed] [Google Scholar]
- 7.Kim J-A, Yoon S, Kim L-Y, Kim D-S. Towards Actualizing the Value Potential of Korea Health Insurance Review and Assessment (HIRA) Data as a Resource for Health Research: Strengths, Limitations, Applications, and Strategies for Optimal Use of HIRA Data. J Korean Med Sci. 2017;32:718–728. doi: 10.3346/jkms.2017.32.5.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suissa S, Garbe E. Primer: administrative health databases in observational studies of drug effects—advantages and disadvantages. Nature Clinical Practice Rheumatology. 2007;3:725–732. doi: 10.1038/ncprheum0652. [DOI] [PubMed] [Google Scholar]
- 9.Steinbusch PJM, Oostenbrink JB, Zuurbier JJ, Schaepkens FJM. The risk of upcoding in casemix systems: A comparative study. Health Policy. 2007;81:289–299. doi: 10.1016/j.healthpol.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 10.You SC, Lee S, Choi B, Park RW. Establishment of an International Evidence Sharing Network Through Common Data Model for Cardiovascular Research. Korean Circ J. 2022;52:853–864. doi: 10.4070/kcj.2022.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rho Y, et al. COVID-19 International Collaborative Research by the Health Insurance Review and Assessment Service Using Its Nationwide Real-world Data: Database, Outcomes, and Implications. J Prev Med Public Health. 2021;54:8–16. doi: 10.3961/jpmph.20.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burn E, et al. Deep phenotyping of 34,128 adult patients hospitalised with COVID-19 in an international network study. Nature Communications. 2020;11:5009. doi: 10.1038/s41467-020-18849-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You SC, et al. Association of Ticagrelor vs Clopidogrel With Net Adverse Clinical Events in Patients With Acute Coronary Syndrome Undergoing Percutaneous Coronary Intervention. JAMA. 2020;324:1640–1650. doi: 10.1001/jama.2020.16167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson MD, et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams RD, et al. Seek COVER: using a disease proxy to rapidly develop and validate a personalized risk calculator for COVID-19 outcomes in an international network. BMC Medical Research Methodology. 2022;22:35. doi: 10.1186/s12874-022-01505-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merkel D. Docker: lightweight Linux containers for consistent development and deployment. Linux J. 2014;2014:Article 2. doi: 10.5555/2600239.2600241. [DOI] [Google Scholar]
- 17.Blacketer C, Defalco FJ, Ryan PB, Rijnbeek PR. Increasing trust in real-world evidence through evaluation of observational data quality. Journal of the American Medical Informatics Association. 2021;28:2251–2257. doi: 10.1093/jamia/ocab132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ledford, H. & Noorden, R. V. in Nature (Nature, 2020).
- 19.Ioannidis JPA. Why Most Published Research Findings Are False. PLOS Medicine. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanelli D. How Many Scientists Fabricate and Falsify Research? A Systematic Review and Meta-Analysis of Survey Data. PLOS ONE. 2009;4:e5738. doi: 10.1371/journal.pone.0005738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyakawa T. No raw data, no science: another possible source of the reproducibility crisis. Molecular Brain. 2020;13:24. doi: 10.1186/s13041-020-0552-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haendel MA, et al. The National COVID Cohort Collaborative (N3C): Rationale, design, infrastructure, and deployment. Journal of the American Medical Informatics Association. 2021;28:427–443. doi: 10.1093/jamia/ocaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puttmann D, et al. Assessing the FAIRness of databases on the EHDEN portal: A case study on two Dutch ICU databases. International Journal of Medical Informatics. 2023;176:105104. doi: 10.1016/j.ijmedinf.2023.105104. [DOI] [PubMed] [Google Scholar]
- 24.Arlett P, Kjaer J, Broich K, Cooke E. Real-World Evidence in EU Medicines Regulation: Enabling Use and Establishing Value. Clinical pharmacology and therapeutics. 2022;111:21–23. doi: 10.1002/cpt.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seong Y, et al. Incorporation of Korean Electronic Data Interchange Vocabulary into Observational Medical Outcomes Partnership Vocabulary. Healthc Inform Res. 2021;27:29–38. doi: 10.4258/hir.2021.27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko SH, et al. Past and Current Status of Adult Type 2 Diabetes Mellitus Management in Korea: A National Health Insurance Service Database Analysis. Diabetes Metab J. 2018;42:93–100. doi: 10.4093/dmj.2018.42.2.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Data on vocabulary mapping were disclosed on the HIRA website (Basic medical examination and diagnosis fee: https://opendata.hira.or.kr/op/opb/selectRfrm.do?rfrmTpCd=&searchCnd=&searchWrd=%EC%9A%A9%EC%96%B4&sno=13305&pageIndex=1 and Operation and Procedure fee: https://opendata.hira.or.kr/op/opb/selectNotice.do?searchCnd=&searchWrd=%EC%9A%A9%EC%96%B4&sno=13603&pageIndex=1). According to Personal Information Protection Act in the Republic of Korea, HIRA does not permit us to share patient-level source data or data derivatives with individuals and institutions.
The CDM data converted in this study is available as a distributed research network way upon an application through an online web portal (https://hcdl.mohw.go.kr). HIRA CDM is updated on an annual basis. Researchers can apply for research requests through the healthcare distributed research network (HDRN) platform operated by the Korean government. The specific application method is as follows: (1) The researcher must request a review of their research hypotheses and plan for ethical feasibility through an institutional or public review board. (2) The research must submit an approval letter from the review board and the research protocol to the HDRN platform. (3) The HIRA reviews the appropriateness of research/data provision and decides whether to provide it. (4) The researcher writes an analysis query, code, or package based on the open sample data and environment and sends it to the HIRA. (5) The HIRA reviews queries and expected results and derives results by running queries/codes/packages. (6) After the results are reviewed and the protected health information checked for infringement, the results are exported to the researcher. Detailed application process for data use is descripted in https://hcdl.mohw.go.kr/static/data/dataApplyStep.
We stored the CDM data using open-source codes of OHDSI for conforming to the database structure of OMOP CDM (https://github.com/OHDSI/CommonDataModel).