SUMMARY
Clinical evidence points to a function for B cell-activating factor (BAFF) in pregnancy. However, direct roles for BAFF-axis members in pregnancy have not been examined. Here, via utility of genetically modified mice, we report that BAFF promotes inflammatory responsiveness and increases susceptibility to inflammation-induced preterm birth (PTB). In contrast, we show that the closely related A proliferation-inducing ligand (APRIL) decreases inflammatory responsiveness and susceptibility to PTB. Known BAFF-axis receptors serve a redundant function in signaling BAFF/APRIL presence in pregnancy. Treatment with anti-BAFF/APRIL monoclonal antibodies or BAFF/APRIL recombinant proteins is sufficient to manipulate susceptibility to PTB. Notably, macrophages at the maternal-fetal interface produce BAFF, while BAFF and APRIL presence divergently shape macrophage gene expression and inflammatory function. Overall, our findings demonstrate that BAFF and APRIL play divergent inflammatory roles in pregnancy and provide therapeutic targets for mitigating risk of inflammation-induced PTB.
Graphical abstract
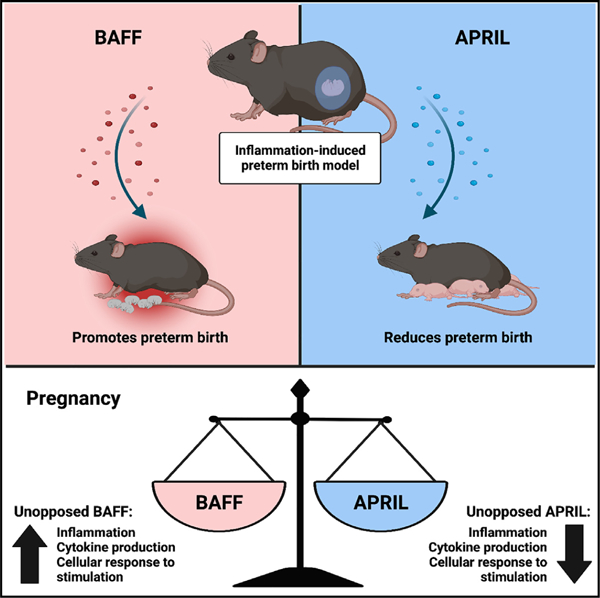
In brief
Doll et al. demonstrate that B cell-activating factor (BAFF) promotes inflammatory responsiveness and susceptibility to preterm birth while the closely related a proliferation-inducing ligand (APRIL) mediates the opposite effect. Notably, BAFF and APRIL play divergent roles in pregnancy and could be therapeutically targeted for mitigating risk of inflammation-induced preterm birth.
INTRODUCTION
Preterm birth (PTB), a leading cause of neonatal morbidity and mortality, affects nearly 500,000 neonates annually in the United States.1–3 Despite the clinical significance, the etiology of PTB is not well understood.4 Inflammation is the dominant driver of PTB, with both innate and adaptive immune cells implicated as important mediators.5,6 Hence, identification of the cross-talk between key immune axes holds therapeutic promise. Regulation of cytokine production, especially within the milieu of hemochorial placentas where maternal blood comes in contact with fetal structures, is essential for normal pregnancy.7,8 The role of tumor necrosis factor (TNF) and interleukin (IL)-6 in promotion of timely parturition and PTB is well established.9–13 However, therapies targeted at blocking TNF and IL-6 have not yielded expected outcomes.14,15 Thus, identification of additional factors that modify inflammatory responses and immune regulation during pregnancy are needed.
B cell-activating factor (BAFF), also known as tumor necrosis factor ligand superfamily member 13B (TNFSF13B), is essential for B cell differentiation and survival.16 BAFF also enhances natural killer (NK) cell activity, decreases survival of regulatory T cells, and activates monocytes.17–19 Physiologically, both BAFF20 and the close homolog of BAFF, A proliferation-inducing ligand (APRIL), play a role in autoimmunity21 and regulation of weight gain.22 BAFF and APRIL are produced by diverse myeloid, lymphoid, and non-hematopoietic cell types.23–27 BAFF binds to BAFF receptor (BAFF-R), B cell maturation antigen (BCMA), and T cell activator and calcium modulating ligand interactor (TACI).28 APRIL binds to BCMA, TACI, and heparan sulfate proteoglycans.20,29 Cumulatively, the overlapping receptor engagement of BCMA and TACI has propagated the idea that APRIL mimics BAFF inflammatory functions.30,31
In humans, serum BAFF levels decline over the course of pregnancy32,33 but increase during parturition.34 Structural mutation of BAFF is a risk factor for preeclampsia,35 and elevated serum BAFF levels correlate with induction of spontaneous abortion.33,36 Further, both BAFF and APRIL are expressed at a high level in chorionic villi,37 and their expression is increased in choriodecidual epithelial and stromal cells of mothers with preterm labor.38 How the BAFF axis affects systemic and local immune responses in pregnancy and PTB remains unknown. Additionally, the cellular source of BAFF/APRIL at the maternal-fetal interface in inflammation-induced PTB is unclear.
In this study, we examined the role of the BAFF axis in shaping susceptibility to inflammation-induced PTB in mouse models of pregnancy and correlate such findings with rhesus macaque pregnancy and primary human samples. We show that loss of BAFF resulted in decreases in inflammatory responsiveness and decreased susceptibility to PTB, while elevated BAFF levels increased susceptibility to PTB. In contrast, loss of APRIL correlated with increased overall inflammatory responsiveness and susceptibility to PTB. Deletion of single BAFF-axis receptors did not modify PTB outcome. Therapeutic administration of anti-BAFF/APRIL monoclonal antibody or BAFF/APRIL recombinant protein was sufficient to manipulate susceptibility to PTB. BAFF and APRIL divergently influenced macrophage gene expression and inflammatory function. Together, our findings uncover a counterregulatory role for BAFF and APRIL signaling in pregnancy that has high potential for therapeutic exploitation in inflammation-driven PTB.
RESULTS
BAFF/APRIL axis is activated in inflammation-induced PTB
Previous work has captured diverse cell types at the human maternal-fetal interface and established a publicly accessible database of the characterized gene expression at the single-cell level (http://placenta.grid.wayne.edu).39 Within this database, increased expression of BAFF (TNSF13B) is observed predominantly in myeloid cells in the basal plate, placental villi, and chorioamniotic membranes from both term and preterm labor.39 Hence, we examined BAFF and APRIL transcript expression in a rhesus macaque model of intrauterine inflammation in pregnancy (Figure 1A).40 Akin to human samples, upregulation of BAFF (p < 0.01) and APRIL (p < 0.05) transcript expression was observed in pregnant macaques following lipopolysaccharide (LPS) challenge (Figure 1B).
Figure 1. BAFF/APRIL axis is activated in experimental models of inflammation-induced preterm birth.
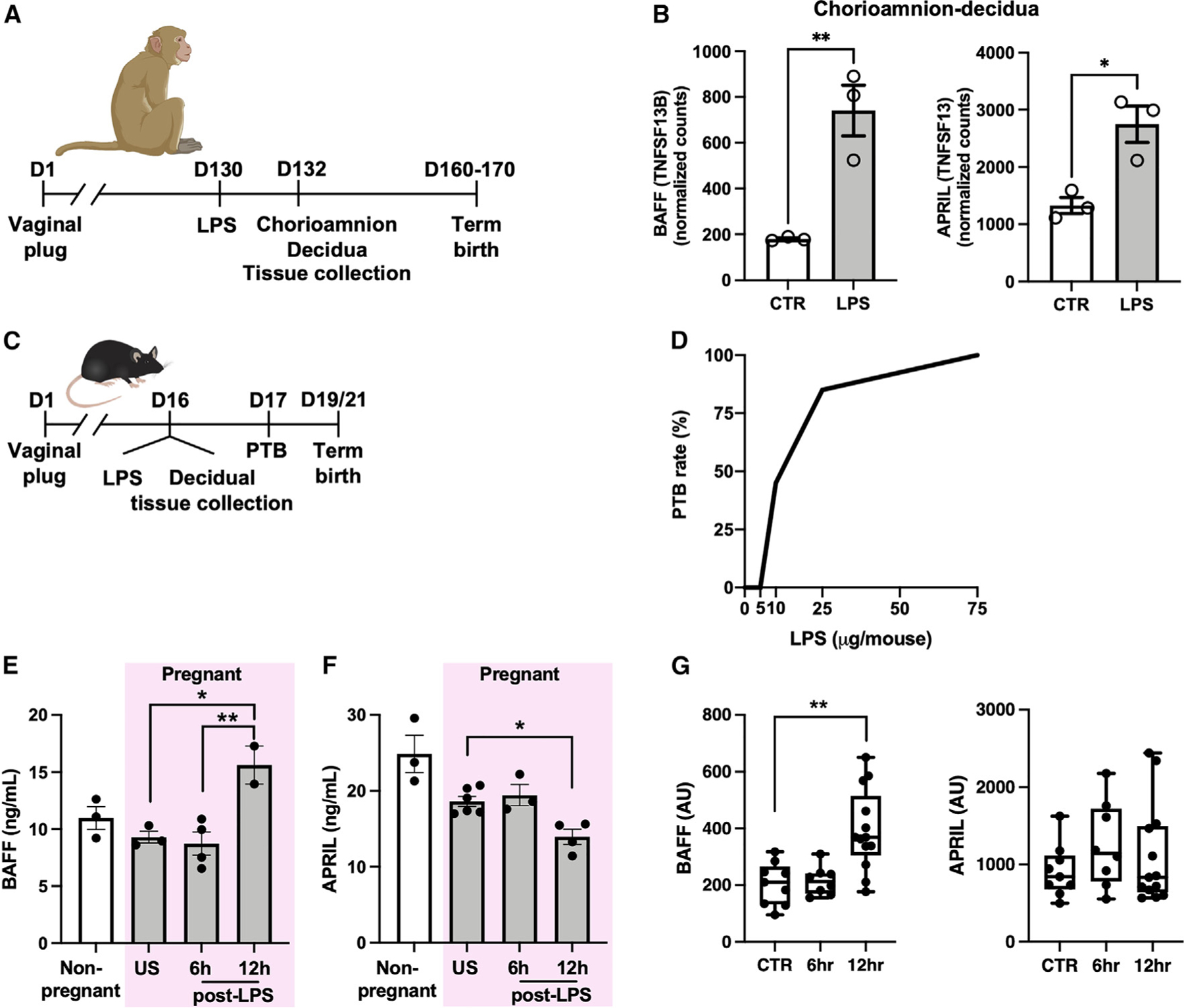
(A) Approach used to study gene expression in gravid rhesus macaques with or without intraamniotic LPS challenge.
(B) mRNA expression in chorioamnion-decidua of rhesus macaques with or without LPS challenge (n = 3/condition).
(C) Approach used to study PTB in gravid mice following LPS challenge.
(D) PTB rate of WT mice after intraperitoneal LPS injection at indicated doses on day 16 gestation (n = 5–24/condition).
(E and F) Quantification of serum (E) BAFF and (F) APRIL in WT nonpregnant and gravid mice after 75-µg LPS challenge. US, unstimulated.
(G) mRNA expression in mouse decidua/myometrium after 75-µg LPS challenge or saline control (CTR) (n = 8–13/condition).
(B) Student’s t test. *p < 0.05, **p < 0.01. (D) Chi-square (2 × 5 matrix), p < 0.0001. (E–G) Data represent average ±SEM. One-way ANOVA with Tukey’s multiple comparison test. *p < 0.05, **p < 0.01.
We next used a mouse model to study the role of BAFF in induction of inflammation-induced PTB9,41 (Figure 1C). LPS induces PTB in a dose-dependent manner in this system (Figure 1D), thus allowing for delineation of the inflammatory response sufficient to induce PTB. Following LPS challenge of gravid wild-type (WT) C57BL/6 mice, elevated BAFF levels were detected in maternal serum (Figure 1E) (p < 0.01), which correlated with a decline in systemic APRIL levels (Figure 1F) (p < 0.05). Decidua and myometrium had increased transcript expression of Tnfsf13b (BAFF) (p < 0.01) at 12 h post LPS challenge, although no significant change in Tnfsf13 (APRIL) transcript expression was seen (Figure 1G). Thus, consistent with observations in human pregnancy and macaque PTB, the BAFF/APRIL axis is activated in the mouse model of PTB.
BAFF and APRIL differentially modulate susceptibility to inflammation-induced PTB
The correlation between BAFF expression and induction of PTB suggests that BAFF-axis signaling may regulate both inflammation and parturition. The necessity of BAFF for induction of PTB was examined in BAFF-deficient mice. BAFF-deficient mice were less susceptible to LPS-induced PTB compared with WT C57BL/6 mice challenged with the same dose (p = 0.0292 at 5 µg of LPS), while induction of PTB was not mitigated in these mice at the 75-µg LPS challenge (Figure 2A). The ability of BAFF to enhance susceptibility to PTB using BAFF transgenic mice (BAFF-Tg), which express high levels of BAFF,22 was analyzed next. BAFF-Tg mice were more susceptible to LPS-induced PTB (Figure 2B) (p = 0.0278 at 5 µg of LPS). Strikingly, APRIL-deficient mice were also found to be more susceptible to LPS-induced PTB (p = 0.0104 at 1 µg of LPS), and 100% of APRIL-deficient mice had PTB after challenge with 1 µg of LPS (Figure 2C). These data invoke a notion that APRIL, unlike BAFF, may have anti-inflammatory properties in pregnancy.
Figure 2. BAFF/APRIL axis modulates susceptibility to PTB.
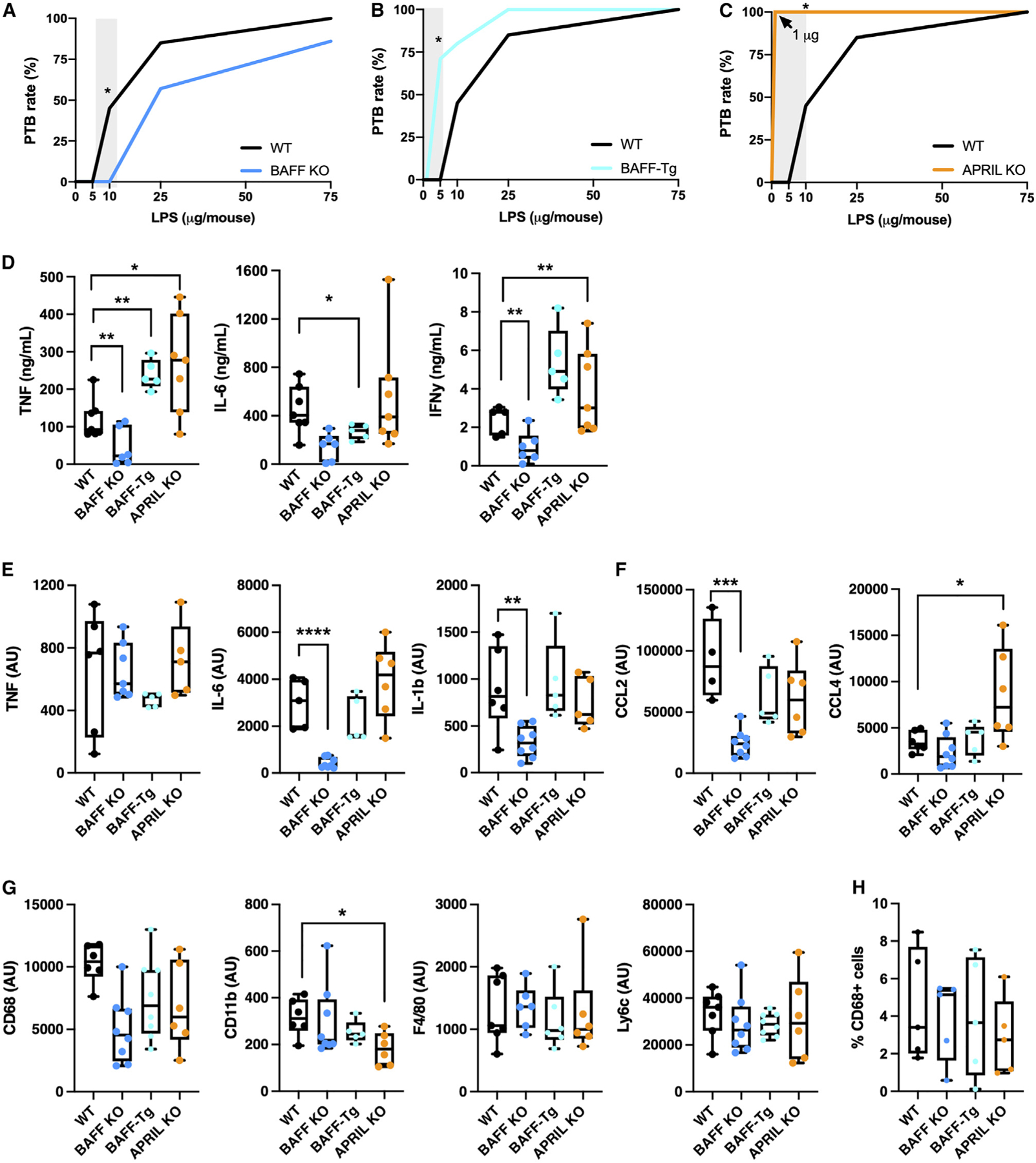
(A–C) PTB rate of WT and (A) BAFF-deficient, (B) BAFF-transgenic (BAFF-Tg), or (C) APRIL-deficient mice after intraperitoneal LPS injection on day 16 gestation (n = 5–24/condition).
(D) Serum TNF, IL-6, and IFNγ determined by in vivo cytokine capture assay (IVCCA) in WT, BAFF-deficient, BAFF-Tg, and APRIL-deficient mice following LPS challenge (n = 5–8/condition).
(E–G) Decidua/myometrium mRNA expression of (E) Tnf, Il6, Il1b, (F) Ccl2, Ccl4, (G) Cd68, CD11b, Adgre1 (F4/80), and Ly6c in WT, BAFF-deficient, BAFF-Tg, and APRIL-deficient mice at 6 h following 75-µg LPS challenge (n = 4–8/condition).
(H) Quantification of CD68-positive cells out of total nuclei by immunohistochemistry in WT, BAFF-deficient, BAFF-Tg, and APRIL-deficient mouse myometrium/decidua/placenta at 6 h following 75-µg LPS challenge (n = 5 fields/condition using 20 × objective).
(A–C) Fisher’s exact test p = 0.0292, 0.0278, 0.0104 respectively. (D–H) Student’s t test *p < 0.05, **p < 0.01, ***p < 0.001.
Cytokine production is central to induction of PTB.42 Accordingly, systemic levels of TNF (p < 0.01), IL-6 (p < 0.05), and interferon gamma (IFNγ) (p < 0.01) post LPS challenge were diminished in BAFF-deficient mice, while TNF (p < 0.01) and IFNγ (p < 0.01) were elevated in BAFF-Tg mice (Figure 2D). APRIL-deficient mice exhibited enhanced systemic TNF (p < 0.05) levels following LPS exposure, compared with WT controls (Figure 2D). To investigate differential inflammatory pathway activation, expression of genes corresponding to proinflammatory cytokines known to drive PTB (e.g., Tnf, Il6, Il1b)10–12,41,43 was evaluated in the decidua/myometrium. Compared with WT controls, tissue transcript expression of Il6 and Il1b is reduced (p < 0.0001, p < 0.01) in BAFF-deficient mice (Figure 2E). BAFF-Tg and APRIL-deficient mice did not have significant changes in tissue inflammatory mediator transcript expression relative to WT mice post LPS challenge (Figure 2E).
Given the relevance of myeloid cells in induction of parturition,41,44,45 as well as the myeloid-specific BAFF transcript expression identified in human studies, transcript expression of chemokines driving macrophage infiltration and activation was next quantified. Reduced expression of Ccl2 (p < 0.001), but not Ccl4, was observed in BAFF-deficient mice (Figure 2F). Although Ccl2 and Ccl4 expression was similar in BAFF-Tg decidua/myometrium compared with WT (Figure 2F), the uterine tissue from APRIL-deficient mice showed a significant increase in Ccl4 transcript expression (p < 0.05) (Figure 2F). To further explore changes in the decidua/myometrium, the transcript expression of various immune cell-associated markers (e.g., Cd68, Cd11b, Adgre1 [F4/80], Ly6c, and Ciita [MHC II]) was measured. Expression of Cd68, Adgre1, Ly6c, or Ciita was not different between WT, BAFF-Tg, BAFF-deficient, or APRIL-deficient mice (Figures 2G and S1A). However, a decrease in Cd11b (p < 0.05) transcript expression was noted in APRIL-deficient mice (Figure 2G).
We next analyzed transcript expression of Ptgs2 (Cox-2), a mediator involved in production of prostaglandins, which cause myometrial contractions critical for induction of labor.46,47 Ptgs2 expression was decreased in BAFF-deficient mice compared with WT controls (p < 0.05) (Figure S1B). Notably, in our studies, all mice that did not have PTB had normal gestational length (data not shown), indicating that BAFF is not causative in modifying the timing of parturition/gestation length, but alters susceptibility to PTB-inducing stimuli. Together, these findings demonstrate that BAFF modifies responsiveness to LPS challenge and PTB outcome.
To investigate whether observed findings in uterine tissue inflammation are consistent with changes in macrophage-like cell absolute number, presence of CD68+ cells was quantified in sections of myometrium/decidua/placenta by immunohistochemistry. WT, BAFF-deficient, BAFF-Tg, and APRIL-deficient mice had similar numbers of CD68+ cells at the maternal-fetal interface at baseline (following saline treatment) (Figure S1C) as well as following LPS challenge (Figures 2H and S1D). Together, these data suggest that BAFF and APRIL likely do not modify CD68+ cell tissue presence but rather differentially regulate immune cell function (see Figure 4).
Figure 4. Macrophages produce BAFF and are differentially responsive to BAFF and APRIL.
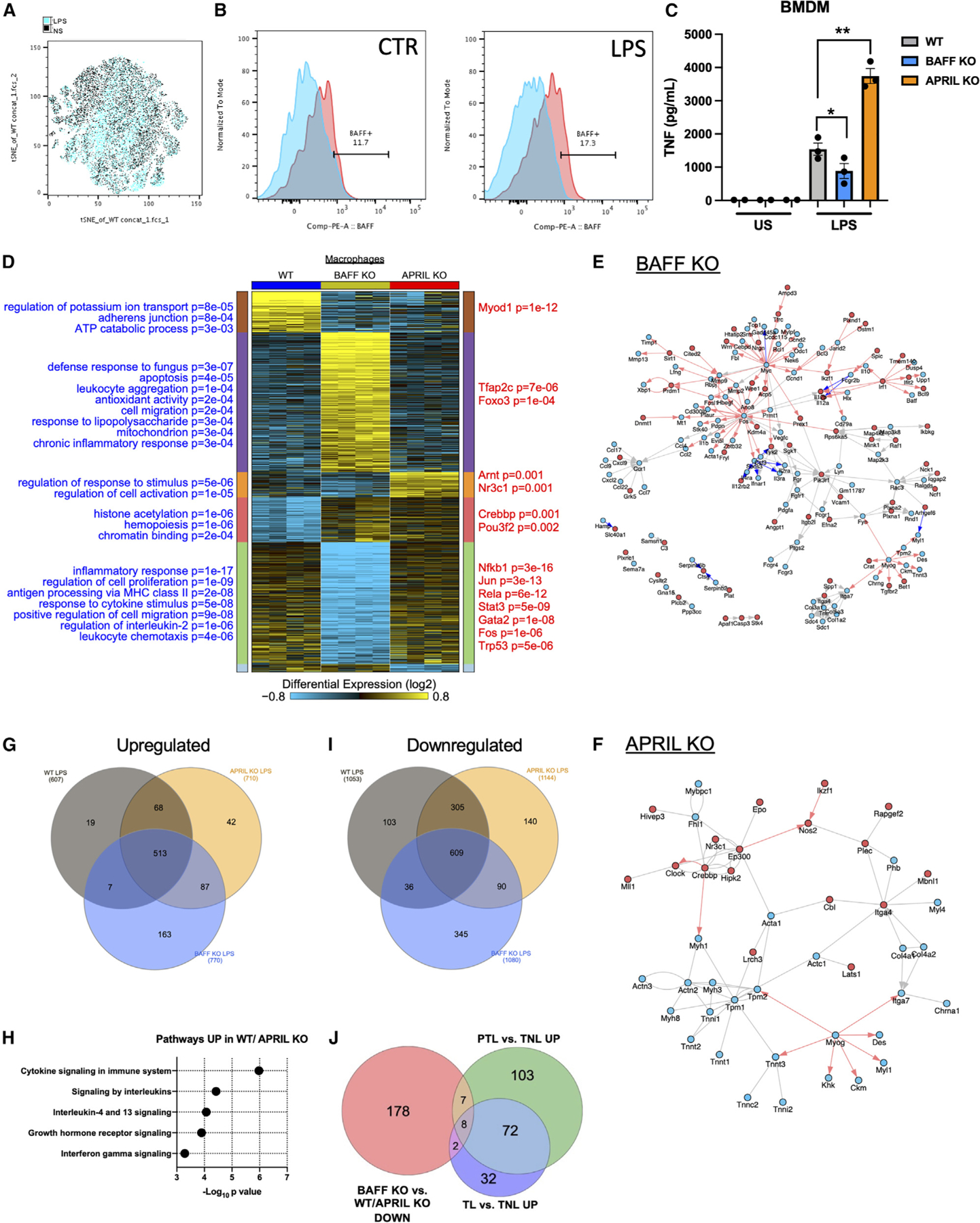
(A) t-distributed stochastic neighbor embedding (tSNE) analysis of flow cytometry on WT mouse decidua/myometrium following 6-h LPS challenge (blue) or saline (black) injection (n = 2/condition).
(B) BAFF expression in WT (blue) and BAFF-RFP+ (red) decidua/myometrium macrophage (CD45+F4/80+CD11b+) populations collected 6 h following saline (CTR) or LPS challenge.
(C) Supernatant TNF production from WT, BAFF-deficient, and APRIL-deficient mouse BMDMs, unstimulated (US) or stimulated with 10 ng LPS for 4 h.
(D) Heatmap of differentially expressed genes (FDR significant), organized by their predominant expression patterns (MarkerFinder algorithm in AltAnalyze) in WT, BAFF-deficient, and APRIL-deficient macrophages following LPS stimulation. Statistically enriched Gene Ontology (GO) terms are shown on the left of each associated cluster and putative transcriptional regulators derived from PAZAR annotated transcriptional targets on the right, from the software GO elite.
(E and F) Predicted interaction network of differentially expressed genes (up- or downregulated >1.5-fold) in (E) BAFF KO or (F) APRIL KO, with a moderated t test p < 0.05 (FDR corrected) in the NetPerspective module of AltAnalyze. Red arrows indicate putative transcriptional target genes from PAZAR or Amadeus, red nodes indicate upregulation, and blue nodes indicate downregulation. Blue arrows indicate inhibitory interactions.
(G) Venn diagram depicting overlap of upregulated genes (>1.5-fold) following LPS challenge of WT, APRIL-deficient, and BAFF-deficient macrophages.
(H) Top enriched biological pathways (ToppFun) from 68 genes upregulated in both WT and APRIL-deficient macrophages.
(I) Venn diagram depicting overlap of genes that were downregulated (>1.5-fold) following LPS challenge of WT, APRIL-deficient, and BAFF-deficient macrophages.
(J) Venn diagram of fold >1.2 and p < 0.05 (FDR) gene expression of macrophages from preterm labor (PTL) versus term no labor (TNL) and term labor (TL), from the study by Pique-Regi et al.39 compared with genes that were <1.5-fold in BAFF-deficient macrophages versus APRIL-deficient macrophages in the mouse.
(C) One-way ANOVA with Tukey’s multiple comparison test. *p < 0.05, **p < 0.01, ****p < 0.0001.
Redundancy of BAFF and APRIL receptors nullifies susceptibility to PTB
To investigate the role of BAFF-axis receptors in BAFF- and APRIL-mediated changes in inflammation and susceptibility to PTB, transcript expression of Tnfrsf13c (BAFF-R), Tnfrsf17 (BCMA), and Tnfrsf13b (TACI) was measured in uterine tissue. Expression of Tnfrsf13c was not significantly changed in the uterus of gravid WT mice following LPS challenge, while the expression of Tnfrsf17 and Tnfrsf13b was downregulated (p < 0.05) (Figure S1E). To test whether a single receptor signaling pathway was necessary for BAFF-axis-mediated inflammatory responsiveness, receptor knockout (KO) mice were challenged with LPS and PTB susceptibility was evaluated. At both high and low doses of LPS, the single-receptor-deficient mice exhibited similar PTB outcomes to WT controls (Figure S1F). Additionally, mice with genetic deletion of all three receptors (BAFF, TACI, and BCMA triple KO) exhibited susceptibility to LPS-induced PTB comparable with WT mice (Figure S1F).
Therapeutic potential for manipulation of BAFF/APRIL in inflammation-induced PTB
We next examined the contribution of BAFF to inflammatory responsiveness and susceptibility to LPS-induced PTB via pharmacological manipulation of BAFF in WT mice. Anti-BAFF treatment 4 h prior to LPS challenge was sufficient to reduce PTB occurrence in gravid WT mice (Figure 3A) (p = 0.0043). Transcript expression of inflammatory cytokines Tnf, Il6, and Il1b was not significantly decreased (Figure S2A). Similarly, transcript levels of immune-cell-recruiting chemokines Ccl2 and Ccl4 were not different between anti-BAFF-treated LPS-challenged mice and control mice (Figure S2B). Anti-BAFF-treated LPS-challenged mice had reduced uterine transcript expression of Cd68 (p < 0.001), while Cd11b, Adgre1 (F4/80), Ly6c, and Ciita (MHC II) transcript expression was unchanged compared with control LPS-challenged mice (Figure S2C). Consistent with the reduction in PTB occurrence following anti-BAFF treatment prior to LPS challenge, anti-BAFF-treated LPS-challenged mice had reduced transcript expression of Ptgs2 (Cox-2) (p < 0.05) in uterine tissue (Figure S2D). The ability of anti-BAFF to protect from PTB in the context of elevated inflammation was further examined in gravid APRIL-deficient mice and was found to reduce PTB occurrence (Figure S2E) (p = 0.0014). These findings support the proinflammatory function of BAFF and highlight the therapeutic potential of anti-BAFF treatment for reducing susceptibility to PTB.
Figure 3. Pharmacologic manipulation of BAFF/APRIL modifies susceptibility to PTB.
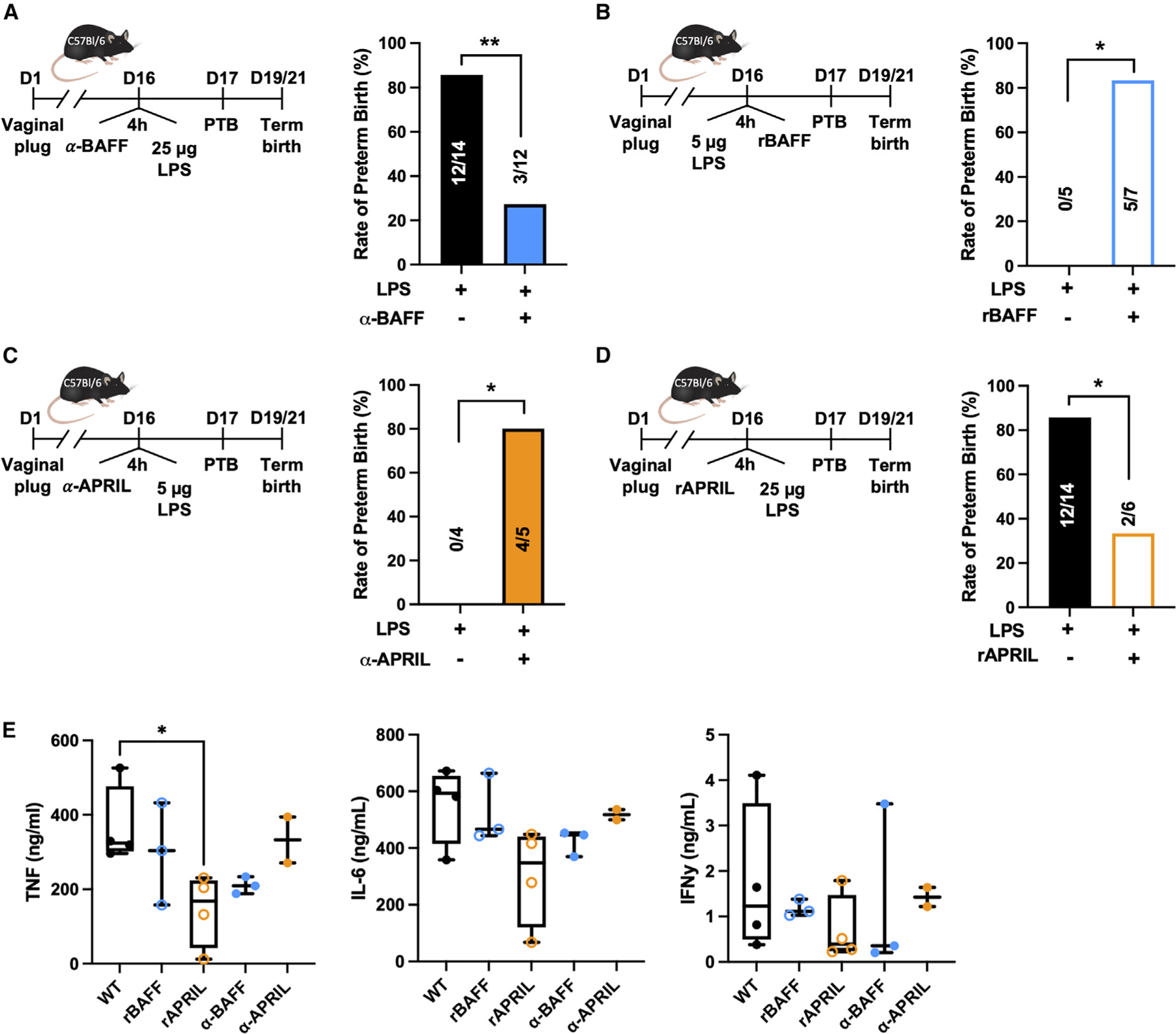
(A–D) PTB rate of gravid WT mice after intraperitoneal injection with (A) anti-BAFF (2 µg/g), then 25 µg LPS; (B) 5 µg LPS, then 2 µg recombinant BAFF; (C) anti-APRIL (2 µg/g), then 5 µg LPS; or (D) recombinant APRIL (2 µg), then 25 µg LPS; on day 16 gestation.
(E) Serum TNF, IL-6, and IFNγ determined by IVCCA in WT mice treated with recombinant BAFF (2 µg), recombinant APRIL (2 µg), anti-BAFF (2 µg/g), or anti-APRIL (2 µg/g) and challenged with LPS 4 h later (n = 2–4/condition). One-way ANOVA with Tukey’s multiple comparison test. *p < 0.05.
(A–D) Fisher’s exact test p = 0.0043, 0.0278, 0.0476, 0.0374 respectively.
The sufficiency of BAFF to enhance susceptibility to PTB was next examined by exogenous addition of recombinant BAFF. Injection of recombinant BAFF 4 h after LPS challenge increased PTB occurrence in gravid WT mice (Figure 3B) (p = 0.0278). Given the opposing phenotypes observed in BAFF-deficient and APRIL-deficient mice, the sufficiency of APRIL to alter susceptibility to LPS-induced PTB was similarly addressed in WT mice. Neutralization of APRIL in gravid WT mice with anti-APRIL antibody injected 4 h prior to LPS challenge increased PTB occurrence (Figure 3C) (p = 0.0476), in direct contrast to what was observed when BAFF was neutralized. Further, administration of recombinant APRIL 4 h prior to LPS challenge protected against PTB in WT mice (Figure 3D) (p = 0.0374). The impact of these treatments on systemic cytokine production was evaluated next. Consistent with the reduction in inflammation-induced PTB, anti-BAFF treatment showed a trend in reduction of systemic cytokine production, while recombinant APRIL treatment was sufficient to significantly reduce systemic TNF levels (p < 0.05) when exposed to 25-µg LPS challenge (Figure 3E). At the 25-µg LPS challenge, recombinant BAFF or anti-APRIL administration did not further exacerbate the inflammatory response (Figure 3E). Together, these data suggest that BAFF and APRIL are important contributors to PTB and that biological manipulation of BAFF or APRIL levels can effectively modulate induction of inflammation-induced PTB.
BAFF axis modulates macrophage inflammatory function
To identify cellular sources capable of producing BAFF during inflammation, uterine immune cells were analyzed in gravid WT and BAFF-reporter mice.48 Changes in uterine immune cell composition were observed at 6 h post challenge (Figure 4A), and, notably, the population of inflammatory macrophages (F4/80+CD11b+Ly6c+) was increased (Figure S3A). In fact, increased BAFF expression following LPS challenge was primarily detected in uterine macrophage populations (Figures 4B and S3B).
Macrophages are identified as key producers of proinflammatory cytokines and regulators of PTB outcome.41,44,45 Bone marrow-derived macrophages (BMDMs) isolated from WT, BAFF-deficient, and APRIL-deficient mice were stimulated with LPS and inflammatory responsiveness was measured. Similar to previous experiments measuring systemic cytokine levels, macrophages from BAFF-deficient mice produced less TNF (p < 0.05), while APRIL-deficient macrophages produced more TNF (p < 0.01) compared with WT controls (Figure 4C). To identify gene expression changes associated with these observed differences, we performed RNA sequencing (RNA-seq) on macrophages from APRIL-deficient and BAFF-deficient mice following LPS stimulation. We identified numerous differentially expressed genes between LPS-challenged WT and APRIL-deficient macrophages (mice susceptible to LPS-driven PTB) compared with BAFF-deficient macrophages (mice resistant to LPS-driven PTB) (Figures 4D, S3C, and S3D; Table S1). BAFF-deficient macrophages display induction of genes associated with apoptosis, cell migration, inflammation/defense response, and response to LPS, while similar and opposing Gene Ontology terms were downregulated, including proliferation, response to cytokine stimulation, chemotaxis, and regulation of IL-2 (Table S2). Network analysis of genes significantly differentially expressed in BAFF-deficient versus WT macrophages additionally show downregulation of Myc and Fos and their target genes, downregulation of Ccr1 and its ligands, downregulation of extracellular matrix components (e.g., Itga7, Col1a2), and upregulation of If1 and its target genes (Figure 4E). We also noted downregulation of these same extracellular matrix genes in the APRIL-deficient macrophages, suggesting these are common responses to loss of either APRIL or BAFF (Figure 4F). Downregulated genes in BAFF-deficient macrophages were enriched in transcriptional regulators of macrophage inflammation, including Nfkb1, Rela, Stat3, and Jun (Table S2). Upregulation of genes in APRIL-deficient and downregulation in BAFF-deficient macrophages were associated with response to stimulus and cell activation, including Tnfsf15, Tlr8, Notch1, Rara, Epo, and Ptgs2.
Gene transcripts significantly upregulated in both WT and APRIL-deficient macrophages are involved in cytokine signaling (Figures 4G and 4H), and all three genotypes revealed induction of inflammatory response pathways (Figure S4A), as anticipated following LPS challenge. There is significant divergence between groups in genes that are downregulated following LPS challenge (Figures 4I and S4B). Given the robust outcomes of BAFF-axis manipulation and our unbiased recognition of underlying pathways in mouse models of PTB, we next compared these findings with human pregnancy. To better assess the impact of different macrophage subsets on human pregnancy, potentially associated with APRIL and BAFF expression, we re-analyzed existing human patient placental single-cell RNA-seq39 (Table S3). Cells assigned a macrophage identity, based on a published comprehensive placental reference compendium (cellHarmony), identified 190 genes differentially expressed in the placental macrophages of preterm labor versus term not in labor (fold > 1.2 and moderated t test p < 0.05, false discovery rate [FDR] corrected). Of these genes, 15 were downregulated in our BAFF-deficient macrophages (CD74, OLR1, KDM68, MAP2K3, HLA-DQB1, FOSL2, MYADM, IL1RN, MT2A, PLAUR, CD83, IER3, C15ORF48, SAMSN1, IL1B). Placental cells from individuals with term labor versus term not in labor39 had up-regulated expression of 114 genes in macrophages, of which 10 were downregulated in our BAFF-deficient macrophages (IL1RN, MT2A, PLAUR, CD83, IER3, C15ORF48, SAMSN1, IL1B, MAP3K8, CCL4) (Figures 4J and S4C; Table S3). Of the commonly upregulated genes, MT2A was the most significantly altered. Notably, MT2 plays a significant role throughout pregnancy and can influence myo-endometrial contractility.49 Mt2 (mouse equivalent) is the second most differentially expressed gene in BAFF-deficient macrophages compared with APRIL-deficient macrophages as well as BAFF-deficient macrophages compared with WT macrophages, consistent with a role for BAFF in regulation of labor induction.
Previous studies have identified NK cells, macrophages, and CD8+ T cells as dominant immune cell types at the maternal-fetal interface and established the importance of these cells to pregnancy maintenance as well as parturition.42,50–53 BAFF and APRIL expression was identified in CD8+ T cells, NK cells, and macrophages in the decidua of first-trimester pregnant women,51–53 with the most robust expression detected in macrophages (Figure S4D) (p < 0.0001). In support of our findings, another study also identified BAFF, and to a lesser extent APRIL, in macrophages of the decidua from first-trimester pregnancies (http://data.teichlab.org [maternal-fetal interface]).54 Together, our data support a role for macrophages in production of BAFF and suggest an important role for the BAFF/APRIL axis in regulating inflammatory responses during pregnancy and parturition.
DISCUSSION
Targeting inappropriate inflammatory responses while preserving the pregnancy and fetal viability is of great clinical significance. However, current knowledge of mediators that facilitate cross-talk between branches of the immune system to modulate the overall response to inflammatory stimuli in pregnancy remains limited. Our study identified elevated BAFF levels in the pregnant mouse following inflammatory challenge. This is in agreement with published reports showing that BAFF levels decrease during human pregnancy32,55 but elevate on the day of delivery.34 Together, these data invoke the potential role for BAFF as a mediator of parturition. While the clinical value of BAFF level modulation in autoimmune disease is appreciated,56 therapeutic management of BAFF for treatment of other inflammatory conditions, including PTB, represent a new possibility.
Similarity in function of BAFF and APRIL via utility of overlapping receptors is implicated in autoimmune diseases pathogenesis.21,30,31 Our data suggest opposite phenotypes in BAFF- and APRIL-deficient mice in inflammation-induced PTB. BAFF-deficient mice have dampened inflammatory responsiveness to LPS challenge and are less susceptible to PTB, albeit PTB was not mitigated in these mice under high-dose LPS challenge. In contrast, APRIL-deficient mice have exacerbated inflammatory responsiveness and are more susceptible to PTB. Interestingly, an anti-inflammatory role for APRIL was proposed in a study for treating multiple sclerosis57 and obesity.22 The overlapping receptor utilization of BAFF and APRIL may explain why single deletion of individual BAFF-axis receptors, or simultaneous deletion of all three receptors (BAFF-R, BCMA, TACI), did not alter inflammatory cytokine production or susceptibility to PTB, despite changes in immune profiles.29,58,59 Hence, studies utilizing double deletion of a combination of these three receptors are needed to further delineate signaling pathways whereby BAFF/APRIL promote/suppress inflammation and PTB.
Modulating BAFF/APRIL levels in pregnancy holds therapeutic implications. Administration of belimumab, a clinically available anti-BAFF antibody,56 to pregnant patients with autoimmune disease has not resulted in negative effects on the pregnancy.60,61 We found that administration of monoclonal anti-BAFF antibody decreased susceptibility to PTB in mice, which has implications for targeting at-risk human pregnancies. African American women are more likely to experience PTB1–3 and are more likely to develop autoimmune disease.62 Notably, African Americans have increased incidence of an SNP associated with higher BAFF levels and African Americans with systemic lupus erythematosus have higher BAFF levels compared with other races/ethnicities.63,64 Thus, as previous strategies for blocking TNF or IL-6 signaling were associated with an increased risk for pregnancy complications and/or birth defects,14,15 manipulation of BAFF may represent a safer target. In addition, the use of recombinant APRIL as a therapeutic should be investigated in further detail for safety and efficacy.
Within the immune compartment, we identify macrophages as a relevant BAFF-producing cell type in human pregnancy and in a mouse model of inflammation-driven PTB. Myeloid cells shape pregnancy outcome,5,41,65,66 and the divergent role of macrophage populations in the uterus is being increasingly appreciated.67 However, processes whereby macrophage populations exert pro- or anti-inflammatory effects in pregnancy remain elusive. Although our data suggest that modulation of the BAFF axis does not robustly affect the number of CD68-expressing cells present at the maternal-fetal interface, our findings do invoke that BAFF axis modifies macrophage function. These observations are further supported by a recent study demonstrating a role for BAFF in macrophage polarization and induction of a proinflammatory response.68 As we uncover BAFF as a product of the macrophage-dependent inflammatory responses, further characterization of pathway activation in BAFF-expressing and BAFF-responsive macrophages may reveal additional findings important in pregnancy.
In summary, we identified an immunomodulatory role for the BAFF/APRIL signaling axis in inflammation-induced PTB. We show that macrophages produce BAFF at the maternal-fetal interface and that BAFF enhances inflammatory responsiveness and increases susceptibility to PTB. In contrast, we show that APRIL has a counterregulatory function and is important for dampening inflammatory responses. Therapeutic manipulation of BAFF or APRIL is sufficient to augment susceptibility to PTB in mice. Inflammatory signaling pathways were significantly upregulated in APRIL-deficient macrophages but downregulated in BAFF-deficient macrophages. These pathways were similarly upregulated in human placental macrophages of preterm labor versus term not in labor. Together, our findings support the notion that therapeutic modulation of the BAFF/APRIL axis may hold promise to manage the risk of inflammation-driven PTB.
Limitations of the study
Although we identified similarities in BAFF and APRIL levels during pregnancy in humans, rhesus macaques, and mice, the models used had notable differences. Specifically, LPS was administered intraamniotically in macaques but was given intra-peritoneally in mice. Previous comparisons of intraamniotic and intraperitoneal injection routes in mice have not found significant differences in induction of inflammatory pathways and PTB outcome41; however, we have not specifically examined BAFF/APRIL in these settings. We measured serum levels of BAFF and APRIL in day 16 pregnant compared with nonpregnant mice, but protein levels were not temporally tracked to recapitulate multiple gestational time points examined in human pregnancy. We identified macrophages as producers of BAFF at the maternal-fetal interface; however, due to low recovery of uterine macrophages, the RNA-seq analysis was performed on BMDMs. Given the difference in cell function between different tissues, future studies are needed to formally examine the uterine macrophage populations and their gene expression patterns in inflammation-induced PTB. Our RNA-seq findings support differential signaling/function pathways being targeted by BAFF and APRIL; however, manipulation of these pathways is required to conclude that the observed opposing mechanisms of BAFF and APRIL action are due to targeting divergent signaling pathways.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Senad Divanovic, senad.divanovic@cchmc.org.
Materials availability
This study did not generate new unique reagents.
Data and code availability
RNA-seq data has been deposited at GEO and is publicly available. Macrophage bulk RNA-seq data is under accession number GEO: GSE202811. Previously deposited bulk RNA-seq data for rhesus macaque samples can be found under GEO: GSE181054.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Female C57BL/6J background, were mated with fertile male mice of the same strain beginning at 8 weeks of age. WT (Jackson), BAFF KO (Jackson), BAFF-Tg (Biogen Idec), APRIL KO (Jackson), BAFF-R KO (generously provided by Klaus Rajewsky71), BCMA KO (generously provided by Loren Erickson72), TACI KO (generously provided by Richard Bram73), Receptor TKO (in house), BAFF-reporter (generously provided by Daniela Giordano74) were bred at Cincinnati Children’s Hospital Medical Center (CCHMC). Animals were housed in a specific pathogen-free animal facility at CCHMC and handled in high-efficiency particulate-filtered laminar flow hoods with free access to food and water. For all pregnancy studies the presence of a vaginal plug was considered day 1 of pregnancy. Parturition events were monitored on days 17–21 and defined as complete delivery of pups.9,41 All murine studies were performed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals (National Academic Press, 2011) and approved by the CCHMC Institutional Animal Care and Use Committee.
Preterm birth
On day 16 gestation, gravid female mice were challenged intraperitoneally (i.p.) with ultrapure LPS (Invivogen) at the indicated concentrations. Where indicated, anti-BAFF and anti-APRIL (Adipogen; 2 µg/g) and recombinant BAFF and recombinant APRIL (R&D Systems; 2 µg/mouse) were administered i.p. Doses were selected based on previous evidence of a biological effect at the indicated concentration.22 PTB was defined as parturition within 24 h after LPS challenge (all pups deceased). Term birth was defined as parturition between days 19–21 (all pups alive).9,41
Macaque model
Normally cycling, adult female rhesus macaques were time mated. At ~130 d of gestation, the pregnant macaques received 1 mL saline solution or 1 mg LPS (Sigma-Aldrich). All injections were given by ultrasound-guided intraamniotic injection. Surgical deliveries were performed 48 h after injection. Chorioamnion-decidua was isolated as previously described.40 Total RNA was purified, treated with DNase using RNeasy mini kit according to manufacturer’s recommendation (Qiagen). RNA-library preparation was performed at DNA Core Facility at CCHMC. Single-end read sequencing by Illumina HiSeq2500 Ultra-High-throughput sequencing system (Illumina Inc.) was used at an average depth of 20 million reads per sample. Raw sequences were accepted once they passed the quality filtering parameters used in the Illumina GA Pipeline. The reads were mapped with STAR 2.5.3a to the Macaca mulatta genome (Mmul 8.0.1). The counts for each gene were obtained using quantMode GeneCounts in the STAR commands and only counts for the featured genes were reserved. Differential expression analyses were carried out using DESeq2. The animals used here were also reported in a previous study.40 All macaque studies were approved by the IACUC at the University of California Davis.
Human pregnancy tissues
As reported previously,51–53 first-trimester decidual samples (gestational age: 6–12 wk) were obtained from patients undergoing elective pregnancy termination at a women’s health clinic in Boston, MA. All human tissue was de-identified, discarded clinical material. The committee on the use of human subjects (Harvard Institutional Review Board) determined that this use of decidual material is not human subjects research. Leukocytes from decidual tissue were processed as previously described. Total RNA was isolated from sorted cells and subjected to one round of amplification and biotinylation using Ambion’s MessageAmp III RNA Amplification kit. Biotinylated amplified RNA was hybridized to human U133 Plus 2.0 chips. Chips were processed at Harvard University’s Microarray Core Facility and according to manufacturer’s protocols. Gene transcript levels were determined using algorithms in Microarray Suite 5.0 software (Affymetrix). All human studies were approved by the IRB at Boston Children’s Hospital and Harvard University.
METHOD DETAILS
Cell culture
Bone marrow cells were derived from the femurs of WT mice and cultured in complete culture medium (RPMI-1640 medium [Gibco] supplemented with 10% FCS [Gibco], 1% L-glutamine [Gibco], penicillin and streptomycin [Thermo Fisher Scientific]), and 10 ng/mL M-CSF (BioLegend) for macrophage differentiation. Media was added on day 3 and on day 6 macrophages and dendritic cells were collected. BMDMs (1 × 106 cells/well) were mock-stimulated (saline) or stimulated with ultrapure LPS (10 ng/mL) for 4 h and TNF production was determined by ELISA (BD).
Cytokine quantification
In vivo: Systemic IL-6, TNF, and IFNγ levels were quantified using in vivo cytokine capture assay (IVCCA).41,75,76 Briefly, biotinylated capture antibodies IL-6 (MQ2–13A5), TNF (TN3–19), and IFNγ (AN.18) (eBioscience) were injected i.p. 3 h prior to LPS (25 µg) challenge and serum cytokine levels were determined 4 h later.
Systemic BAFF (R&D Systems) and APRIL (MyBioSource) levels were quantified by ELISA per manufacturer instructions.
Gene expression by qRT-PCR
Uterine samples consisting of maternal decidua and myometrium were collected 6 h or 12h after ultrapure LPS challenge on d16 of gestation. For quantification of mRNA expression in murine samples, cells/tissues were homogenized in TRIzol (Invitrogen), RNA was extracted and cDNA was generated and quantified as previously described41 using Light Cycler 480 II (Roche Diagnostics). Single product qPCR was validated by melt curve analysis. Each reaction contained 2 µL of cDNA (25 ng/µL) and 8 mL of master mix consisting of SYBR Green PCR Master Mix (Life Technologies), 0.5 µM 5ʹ and 3ʹ primers, and ribonuclease-free water. MIQE compliant primer sets were obtained from BioRad for all targets. Data were normalized to β-actin mRNA expression and expressed as delta delta cycle threshold using the formula mRNA level = 1.8^(CtB-actin – Cttarget gene)*100000.
Histology and immunohistochemistry
Samples consisting of decidua, myometrium, and placenta from WT, BAFF deficient, BAFF-Tg, and APRIL deficient mice were collected 6 h after ultrapure LPS challenge (or saline) on d16 of gestation. In brief, samples were fixed in formalin, embedded in paraffin and cut into 10 µm sections. Slides were deparaffinized, blocked, and anti-CD68 antibody or isotype control was applied at a dilution of 1:25. Secondary anti-Rabbit HRP antibody was applied with hematoxylin used as a counterstain. Images were acquired on a Zeiss Axio Imager M1 widefield light microscope with an ICc 3 camera using AxioVision software. CD68+ cell percentage was quantified using the ImageJ Cell Counter plugin on 5 fields per condition and calculated as CD68+ cells/total nuclei per field.
Flow cytometry on uterine tissues
Uterine samples consisting of maternal decidua and myometrium were collected 6 h after ultrapure LPS challenge (or saline) on d16 of gestation from WT and BAFF reporter (RFP48) mice. Immune cells were isolated using enzymatic digestion. Briefly, uterine tissue was dissected and finely minced. Tissue was digested using Liberase TM (21 µg/mL, Roche) and Dnase I (8.8 µg/mL, Roche) in DMEM containing HEPES and 2% BSA. Tissue was incubated at 37°C for 30 min at 220 RPM. After digestion, cells were filtered through a 100 µM strainer and centrifuged at 800 g for 5 min. Following red blood cell lysis, single cell suspensions were stained with Live/Dead (Zombie UV Dye: Biolegend) and directly conjugated monoclonal antibodies to CD45-PE-Dazzle594 (Biolegend, 104), F4/80-AF700 (Biolegend, BM8), TCRb-APC-ef780 (Invitrogen, H57–597), CD11b-ef450 (Biolegend, 17A2), CD8-BV510 (Biolegend, 53–6.7), B220-BV605 (Biolegend, RA3–6B2), NK1.1-FITC (Biolegend, PK136), CD11c-BV711 (Biolegend, N418), Ly6G-APC (Invitrogen, RB6–8C5), Ly6C-PerCP-Cy5.5 (eBioscience, HK1.4). Data was collected using LSR Fortessa (BD) and analyzed using FlowJo X software (vX10).
RNA-sequencing and gene expression quantification
RNA from murine bone marrow derived macrophages was isolated using the mirVANA RNA isolation kit. To measure the library concentration, NEBNext Ultra II Directional RNA Library Prep kit (New England BioLabs) was used. RNA-seq was performed using the Illumina NextSeq 550 with the sequencing setting of single read 85 bp to generate ~20 M reads per sample. RNA-seq data were aligned to the mouse genome (mm10) using the software STAR to produce BAM files. The BAM files were quantified as gene-level RPKMs using the software AltAnalyze version 2.1.4, with the Ensembl version 72 database.77 All differentially expressed genes were identified based on a +/− 1.5 fold change and empirical Bayes moderated t test p value <0.05, FDR corrected. Significantly differentially expressed genes were segregated into predominant patterns based on the software MarkerFinder in AltAnalyze. Pathway enrichment analysis was performed in AltAnalyze using the GO-Elite database (Gene Ontology and MergedTF-Target database) or from the software ToppFun78 as indicated. For Venn Diagram analyses, we additionally considered non-adjusted differentially expressed genes to assess potential overlaps. The RNA-seq data have been deposited in the Gene Expression Omnibus database (GEO: GSE202811). Human placenta single-cell RNA-Seq fastq files obtained from dbGAP (dbGaP: PRJNA560990) were aligned and initially processed using CellRanger 6.0.1. The resulting filtered count matrices were normalized, filtered and annotated using AltAnalyze and cellHarmony, against a prior published well-described placental reference atlas (Single-Cell Portal, accession SCP86054). The reference-aligned cell populations were used to identify and further analyze placental macrophages from women with premature labor, term labor and at term with a cesarean. Differential expression changes between these three patient groups were calculated in cellHarmony (fold >1.2 and empirical Bayes moderated t test p < 0.05, FDR corrected).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Any data outliers were detected by GROUT in Prism 8 (GraphPad Software, Inc.) and removed from datasets. All statistical comparisons were performed in Prism 8 (GraphPad Software, Inc.). Group comparisons were performed using Student’s t test for continuous variables, Fisher’s exact test for binary nominal variables, and chi-squared test for categorical nominal variables. Comparisons among multiple groups were performed using ANOVA with Tukey’s post-hoc test. The exact n for each experiment can be found in the Figure Legend corresponding to that dataset. A p value <0.05 was considered significant.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Zombie UV Dye | Biolegend | Cat# 423107 |
| anti-CD45-PE-Dazzle594 (clone 104) | Biolegend | Cat# 109845; RRID:AB_2564176 |
| anti-TCRβ-APC-ef780 (clone H57–597) | Invitrogen | Cat# 47–5961-82; RRID:AB_1272173 |
| anti-CD11b-ef450 (clone 17A2) | Invitrogen | Cat# 48–0112-82; RRID:AB_1582236 |
| anti-F4/80-AF700 (clone BM8) | Biolegend | Cat# 123130; RRID:AB_2293450 |
| anti-CD8-BV510 (clone 53–6.7) | Biolegend | Cat# 100752; RRID:AB_2563057 |
| anti-B220-BV605 (clone RA3–6B2) | Biolegend | Cat# 103244; RRID:AB_2563312 |
| Anti-NK1.1-FITC (clone PK136) | Biolegend | Cat# 108706; RRID:AB_313393 |
| Anti-CD11c-BV711 (clone N418) | Biolegend | Cat# 117349; RRID:AB_2563905 |
| Anti-Ly6G-APC (clone RB6–8C5) | Abcam | Cat# ab25273; RRID:AB_448758 |
| Anti-Ly6C-PerCP-Cy5.5 (clone HK1.4) | eBioscience | Cat# 45–5932-82; RRID:AB_2723343 |
| anti-TNFα purified tissue culture grade (clone TN3–19) | eBioscience | Cat# 16–7423-81; RRID:AB_469261 |
| anti-IFNγ purified tissue culture grade (clone AN.18) | eBioscience | Cat# 14–7313-85; RRID:AB_468472 |
| Anti-IL6 purified tissue culture grade (clone MP5–32C11) | eBioscience | Cat# 36–7062-85; RRID:AB_469761 |
| Anti-BAFF | Adipogen | Cat# AG-20B-0063PF-C500 |
| Anti-APRIL | Adipogen | Cat# AG-27B-0001PF-C500 |
| Anti-CD68 (polyclonal) | Abcam | Cat# 125212; RRID:AB_10975465 |
| Isotype IgG (for anti-CD68) | R&D Systems | Cat# AB-105-C; RRID:AB_354266 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Recombinant BAFF | R&D Systems | Cat# 8876-BF-010/CF |
| Recombinant APRIL | R&D Systems | Cat# 7907-AP-010/CF |
| LPS (E. coli 0111:B4) | Invivogen | Cat# vac-3pelps |
| M-CSF | Biolegend | Cat# 576406 |
| Fetal Bovine Serum (Seradigm Life Science) | VWR | Cat# 89510 |
| Percoll | Sigma-Aldrich | Cat# P1644 |
| TRizol reagent | Thermo Fisher Scientific | Cat# 15596–018 |
| Liberase | TM Roche | Cat# 05401127001 |
| DNase I | Roche | Cat# 10104159001 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat# A9647 |
| RPMI 1640 media | GIBCO | Cat# 11875–093 |
| DMEM (Dulbecco’s Modification of Eagle’s Medium) | Corning Cellgro | Cat# 10–013-CV |
| Penicillin and streptomycin | Thermo Fisher Scientific | Cat# 15240062 |
| Glutamax | GIBCO | Cat# 35050061 |
|
| ||
| Critical commercial assays | ||
|
| ||
| IL-6 ELISA kit, mouse | BD | Cat# 550950 |
| TNF ELISA kit, mouse | BD | Cat# 560478 |
| BAFF ELISA kit, mouse | R&D Systems | Cat# DY2106–05 |
| APRIL ELISA kit, mouse | MyBioSource | Cat# MBS738004 |
| Fixation/Permeabilization Solution Kit | eBioscience | Cat# 88–8824-00 |
| Quant Perfecta Sybr Green Supermix | QuantaBio | Cat# 95054–02K |
| Verso cDNA synthesis Kit | Thermo Fisher Scientific | Cat# AB-1453/B |
|
| ||
| Deposited data | ||
|
| ||
| Bulk RNA-seq, mouse macrophage | GEO | GEO: GSE202811 |
| Bulk RNA-seq, rhesus macaque | GEO | GEO: GSE181054 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6J CD45.2 WT mice | Jackson Laboratories | Cat# 000664 |
| BAFF transgenic mice | Biogen Idec | Biogen Idec |
| BAFF KO mice | Jackson Laboratories | Cat# 010572 |
| APRIL KO mice | Jackson Laboratories | Cat# 022971 |
| BAFF-R KO mice | Klaus Rajewsky | Harvard Medical School |
| BCMA KO mice | Loren Erickson | University of Virginia |
| BAFF-R/BCMA/TACI triple KO mice | CCHMC Transgenic Core | CCHMC |
| BAFF-reporter mice | Daniela Giordano | University of Washington |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S4 | BIO RAD | See Table S4 |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo, Version 10 | FlowJo | https://www.flowjo.com/ |
| GraphPad Prism version 7 | Prism | https://www.graphpad.com/ |
| ToppGene | CCHMC | https://toppgene.cchmc.org |
| Venn Diagrams | Bioinformatics & Evolutionary Genomics | http://bioinformatics.psb.ugent.be/webtools/Venn/ |
| AltAnalyze version 2.1.1, with ICGS version 1.0 | Olsson et al., 201669 | http://altanalyze.org/ |
| CellHarmony | DePasquale et al., 201970 | http://altanalyze.org/ |
| ImageJ | https://imagej.nih.gov/ij | RRID: SCR_003070 https://imagej.nih.gov/ij |
Highlights.
BAFF promotes susceptibility to preterm birth
APRIL reduces susceptibility to preterm birth
Targeting BAFF/APRIL axis has potential for therapeutic utility in preterm birth
BAFF and APRIL modulate macrophage gene expression and inflammatory vigor
ACKNOWLEDGMENTS
This work was supported in part by: Cincinnati Children’s Hospital Medical Center (CCHMC) Perinatal Institute Pilot and Feasibility Award, Burroughs Wellcome Fund Preterm Birth Research Grant 1015032, March of Dimes Prematurity Research Center Ohio Collaborative for an Innovation Catalyst Grant 22-FY16–125, and CCRF Endowed Scholar Award (to S.D.); The Arnold W. Strauss Award (to J.R.D.); American Diabetes Association 1–19-PMF-019 (to M.E.M.F.); National Institutes of Health U01ES029234 and U54 DK126108, and CCHMC Academic and Research Committee Grant (to C.A.C.); and CCHMC Pathology Research Core (RRID: SCR_022637) and Confocal Imag-ing Core (RRID: SCR_022628). We thank Dr. Daniel Lucas for assistance with immunohistochemistry of uterine tissues. The Graphical Abstract was created with BioRender.com (https://BioRender.com).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112352.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Hamilton BE, Martin JA, Osterman MJK, Driscoll AK, and Rossen LM (2019). Vital statistics rapid release. Birth 35, 1–10. [Google Scholar]
- 2.Martin JA, Hamilton BE, and Osterman MJK (2019). Births in the United States, 2018 (NCHS Data Brief), pp. 1–8. [PubMed]
- 3.Martin JA, and Osterman MJK (2018). Describing the Increase in Preterm Births in the United States, 2014–2016 (NCHS Data Brief), pp. 1–8. [PubMed]
- 4.Deshmukh H, and Way SS (2019). Immunological basis for recurrent fetal loss and pregnancy complications. Annu. Rev. Pathol 14, 185–210. 10.1146/annurev-pathmechdis-012418-012743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomez-Lopez N, StLouis D, Lehr MA, Sanchez-Rodriguez EN, and Arenas-Hernandez M (2014). Immune cells in term and preterm labor. Cell. Mol. Immunol 11, 571–581. 10.1038/cmi.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.PrabhuDas M, Bonney E, Caron K, Dey S, Erlebacher A, Fazleabas A, Fisher S, Golos T, Matzuk M, McCune JM, et al. (2015). Immune mechanisms at the maternal-fetal interface: perspectives and challenges. Nat. Immunol 16, 328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt JS, Pace JL, and Gill RM (2010). Immunoregulatory molecules in human placentas: potential for diverse roles in pregnancy. Int. J. Dev. Biol 54, 457–467. 10.1387/ijdb.082831jh. [DOI] [PubMed] [Google Scholar]
- 8.Maltepe E, Bakardjiev AI, and Fisher SJ (2010). The placenta: transcriptional, epigenetic, and physiological integration during development. J. Clin. Invest 120, 1016–1025. 10.1172/JCI41211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cappelletti M, Presicce P, Lawson MJ, Chaturvedi V, Stankiewicz TE, Vanoni S, Harley IT, McAlees JW, Giles DA, Moreno-Fernandez ME, et al. (2017). Type I interferons regulate susceptibility to inflammation-induced preterm birth. JCI Insight 2, e91288. 10.1172/jci.insight.91288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greig PC, Murtha AP, Jimmerson CJ, Herbert WN, Roitman-Johnson B, and Allen J (1997). Maternal serum interleukin-6 during pregnancy and during term and preterm labor. Obstet. Gynecol 90, 465–469. 10.1016/s0029-7844(97)00294-9. [DOI] [PubMed] [Google Scholar]
- 11.Gücer F, Balkanli-Kaplan P, Yüksel M, Yüce MA, Türe M, and Yardim T (2001). Maternal serum tumor necrosis factor-alpha in patients with preterm labor. J. Reprod. Med 46, 232–236. [PubMed] [Google Scholar]
- 12.Robertson SA, Christiaens I, Dorian CL, Zaragoza DB, Care AS, Banks AM, and Olson DM (2010). Interleukin-6 is an essential determinant of on-time parturition in the mouse. Endocrinology 151, 3996–4006. 10.1210/en.2010-0063. [DOI] [PubMed] [Google Scholar]
- 13.Romero R, Dey SK, and Fisher SJ (2014). Preterm labor: one syndrome, many causes. Science 345, 760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeltzenbein M, Beck E, Rajwanshi R, Gøtestam Skorpen C, Berber E, Schaefer C, and Østensen M (2016). Tocilizumab use in pregnancy: analysis of a global safety database including data from clinical trials and post-marketing data. Semin. Arthritis Rheum 46, 238–245. 10.1016/j.semarthrit.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Weber-Schoendorfer C, Oppermann M, Wacker E, Bernard N, Network of French Pharmacovigilance Centres, Beghin D, Cuppers-Maarschalkerweerd B, Richardson JL, Rothuizen LE, Pistelli A, et al. (2015). Pregnancy outcome after TNF-alpha inhibitor therapy during the first trimester: a prospective multicentre cohort study. Br. J. Clin. Pharmacol 80, 727–739. 10.1111/bcp.12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackay F, Schneider P, Rennert P, and Browning J (2003). Baff AND APRIL: a tutorial on B cell survival. Annu. Rev. Immunol 21, 231–264. 10.1146/annurev.immunol.21.120601.141152. [DOI] [PubMed] [Google Scholar]
- 17.Chang SK, Arendt BK, Darce JR, Wu X, and Jelinek DF (2006). A role for BLyS in the activation of innate immune cells. Blood 108, 2687–2694. 10.1182/blood-2005-12-017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stohl W, and Yu N (2020). Promotion of T Regulatory cells in mice by B cells and BAFF. J. Immunol 204, 2416–2428. 10.4049/jimmunol.1900057. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W, Wen L, Huang X, Liang J, Gao W, Zhang S, and Chen L (2008). hsBAFF enhances activity of NK cells by regulation of CD4(+) T lymphocyte function. Immunol. Lett 120, 96–102. 10.1016/j.imlet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Mackay F, Silveira PA, and Brink R (2007). B cells and the BAFF/APRIL axis: fast-forward on autoimmunity and signaling. Curr. Opin. Immunol 19, 327–336. 10.1016/j.coi.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Koyama T, Tsukamoto H, Masumoto K, Himeji D, Hayashi K, Harada M, and Horiuchi T (2003). A novel polymorphism of the human APRIL gene is associated with systemic lupus erythematosus. Rheumatology 42, 980–985. 10.1093/rheumatology/keg270. [DOI] [PubMed] [Google Scholar]
- 22.Chan CC, Harley ITW, Pfluger PT, Trompette A, Stankiewicz TE, Allen JL, Moreno-Fernandez ME, Damen MSMA, Oates JR, Alarcon PC, et al. (2021). A BAFF/APRIL axis regulates obesogenic diet-driven weight gain. Nat. Commun 12, 2911. 10.1038/s41467-021-23084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackay F, and Schneider P (2009). Cracking the BAFF code. Nat. Rev. Immunol 9, 491–502. 10.1038/nri2572. [DOI] [PubMed] [Google Scholar]
- 24.Alexaki VI, Notas G, Pelekanou V, Kampa M, Valkanou M, Theodoropoulos P, Stathopoulos EN, Tsapis A, and Castanas E (2009). Adipocytes as immune cells: differential expression of TWEAK, BAFF, and APRIL and their receptors (Fn14, BAFF-R, TACI, and BCMA) at different stages of normal and pathological adipose tissue development. J. Immunol 183, 5948–5956. 10.4049/jimmunol.0901186. [DOI] [PubMed] [Google Scholar]
- 25.Krumbholz M, Theil D, Derfuss T, Rosenwald A, Schrader F, Monoranu CM, Kalled SL, Hess DM, Serafini B, Aloisi F, et al. (2005). BAFF is produced by astrocytes and upregulated in multiple sclerosis lesions and primary central nervous system lymphoma. J. Exp. Med 201, 195–200. 10.1084/jem.20041674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Liu L, Moore DJ, Shen X, Peek RM, Acra SA, Li H, Ren X, Polk DB, and Yan F (2017). An LGG-derived protein promotes IgA production through upregulation of APRIL expression in intestinal epithelial cells. Mucosal Immunol 10, 373–384. 10.1038/mi.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundell AC, Nordström I, Andersson K, Lundqvist C, Telemo E, Nava S, Kaipe H, and Rudin A (2017). IFN type I and II induce BAFF secretion from human decidual stromal cells. Sci. Rep 7, 39904. 10.1038/srep39904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kampa M, Notas G, Stathopoulos EN, Tsapis A, and Castanas E (2020). The TNFSF members APRIL and BAFF and their receptors TACI, BCMA, and BAFFR in oncology, with a special focus in breast cancer. Front. Oncol 10, 827. 10.3389/fonc.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smulski CR, and Eibel H (2018). BAFF and BAFF-receptor in B cell selection and survival. Front. Immunol 9, 2285. 10.3389/fimmu.2018.02285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baert L, Manfroi B, Casez O, Sturm N, and Huard B (2018). The role of APRIL - a proliferation inducing ligand - in autoimmune diseases and expectations from its targeting. J. Autoimmun 95, 179–190. 10.1016/j.jaut.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 31.Vincent FB, Saulep-Easton D, Figgett WA, Fairfax KA, and Mackay F (2013). The BAFF/APRIL system: emerging functions beyond B cell biology and autoimmunity. Cytokine Growth Factor Rev 24, 203–215. 10.1016/j.cytogfr.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stohl HE, Lee RH, Manetta J, Kikly K, Korst LM, and Stohl W (2017). Maternal serum B-cell activating factor levels: candidate early biomarker for hypertensive disorders of pregnancy. Hypertension 70, 1007–1013. 10.1161/HYPERTENSIONAHA.117.09775. [DOI] [PubMed] [Google Scholar]
- 33.Stohl HE, and Stohl W (2023). Maternal and cord blood BAFF and APRIL levels during pregnancy. Am. J. Reprod. Immunol 89, e13654. 10.1111/aji.13654. [DOI] [PubMed] [Google Scholar]
- 34.Lima J, Cambridge G, Vilas-Boas A, Martins C, Borrego LM, and Leandro M (2019). Serum markers of B cell activation in pregnancy during late gestation, delivery and the postpartum period. Am. J. Reprod. Immunol 81, e13090. 10.1111/aji.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fenstad MH, Johnson MP, Roten LT, Aas PA, Forsmo S, Klepper K, East CE, Abraham LJ, Blangero J, Brennecke SP, et al. (2010). Genetic and molecular functional characterization of variants within TNFSF13B, a positional candidate preeclampsia susceptibility gene on 13q. PLoS One 5, e12993. 10.1371/journal.pone.0012993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stohl HE, Yu N, and Stohl W (2021). First-trimester serum BAFF:sFlt-1 ratio as a candidate early biomarker of spontaneous abortions. Am. J. Reprod. Immunol 86, e13428. 10.1111/aji.13428. [DOI] [PubMed] [Google Scholar]
- 37.Megli C, Morosky S, Rajasundaram D, and Coyne CB (2021). Inflammasome signaling in human placental trophoblasts regulates immune defense against Listeria monocytogenes infection. J. Exp. Med 218, e20200649. 10.1084/jem.20200649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang B, Faucette AN, Pawlitz MD, Pei B, Goyert JW, Zhou JZ, El-Hage NG, Deng J, Lin J, Yao F, et al. (2017). Interleukin-33-induced expression of PIBF1 by decidual B cells protects against preterm labor. Nat. Med 23, 128–135. 10.1038/nm.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pique-Regi R, Romero R, Tarca AL, Sendler ED, Xu Y, Garcia-Flores V, Leng Y, Luca F, Hassan SS, and Gomez-Lopez N (2019). Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife 8, e52004. 10.7554/eL-ife.52004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cappelletti M, Presicce P, Ma F, Senthamaraikannan P, Miller LA, Pellegrini M, Jobe AH, Divanovic S, Way SS, Chougnet CA, and Kallapur SG (2021). The intensity of the immune response to LPS and E. coli regulates the induction of preterm labor in Rhesus Macaques. Pre-print at bioRxiv 10.1101/2021.01.07.425700. [DOI] [PMC free article] [PubMed]
- 41.Cappelletti M, Doll JR, Stankiewicz TE, Lawson MJ, Sauer V, Wen B, Kalinichenko VV, Sun X, Tilburgs T, and Divanovic S (2020). Maternal regulation of inflammatory cues is required for induction of preterm birth. JCI Insight 5, e138812. 10.1172/jci.insight.138812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mor G, Aldo P, and Alvero AB (2017). The unique immunological and microbial aspects of pregnancy. Nat. Rev. Immunol 17, 469–482. 10.1038/nri.2017.64. [DOI] [PubMed] [Google Scholar]
- 43.Presicce P, Cappelletti M, Senthamaraikannan P, Ma F, Morselli M, Jackson CM, Mukherjee S, Miller LA, Pellegrini M, Jobe AH, et al. (2020). TNF-signaling modulates neutrophil-mediated immunity at the feto-maternal interface during LPS-induced intrauterine inflammation. Front. Immunol 11, 558. 10.3389/fimmu.2020.00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan H, Li H, Zhu L, Gao J, Li P, and Zhang Z (2019). Increased TLR4 and TREM-1 expression on monocytes and neutrophils in preterm birth: further evidence of a proinflammatory state. J. Matern. Fetal Neonatal Med 32, 2961–2969. 10.1080/14767058.2018.1452903. [DOI] [PubMed] [Google Scholar]
- 45.Yao Y, Xu XH, and Jin L (2019). Macrophage polarization in physiological and pathological pregnancy. Front. Immunol 10, 792. 10.3389/fimmu.2019.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, and Dey SK (2010). Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J. Clin. Invest 120, 803–815. 10.1172/JCI40051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Brien WF (1995). The role of prostaglandins in labor and delivery. Clin. Perinatol 22, 973–984. [PubMed] [Google Scholar]
- 48.Giordano D, Kuley R, Draves KE, Roe K, Holder U, Giltiay NV, and Clark EA (2020). BAFF produced by neutrophils and dendritic cells is regulated differently and has distinct roles in antibody responses and protective immunity against west nile virus. J. Immunol 204, 1508–1520. 10.4049/jimmunol.1901120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlomagno G, Minini M, Tilotta M, and Unfer V (2018). From implantation to birth: insight into molecular melatonin functions. Int. J. Mol. Sci 19, 2802. 10.3390/ijms19092802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crespo ÂC, van der Zwan A, Ramalho-Santos J, Strominger JL, and Tilburgs T (2017). Cytotoxic potential of decidual NK cells and CD8+ T cells awakened by infections. J. Reprod. Immunol 119, 85–90. 10.1016/j.jri.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Houser BL, Tilburgs T, Hill J, Nicotra ML, and Strominger JL (2011). Two unique human decidual macrophage populations. J. Immunol 186, 2633–2642. 10.4049/jimmunol.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kopcow HD, Eriksson M, Mselle TF, Damrauer SM, Wira CR, Sentman CL, and Strominger JL (2010). Human decidual NK cells from gravid uteri and NK cells from cycling endometrium are distinct NK cell subsets. Placenta 31, 334–338. 10.1016/j.placenta.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Zwan A, Bi K, Norwitz ER, Crespo ÂC, Claas FHJ, Strominger JL, and Tilburgs T (2018). Mixed signature of activation and dysfunction allows human decidual CD8(+) T cells to provide both tolerance and immunity. Proc. Natl. Acad. Sci. USA 115, 385–390. 10.1073/pnas.1713957115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polański K, Goncalves A, et al. (2018). Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353. 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muzzio DO, Soldati R, Ehrhardt J, Utpatel K, Evert M, Zenclussen AC, Zygmunt M, and Jensen F (2014). B cell development undergoes profound modifications and adaptations during pregnancy in mice. Biol. Reprod 91, 115. 10.1095/biolreprod.114.122366. [DOI] [PubMed] [Google Scholar]
- 56.Stohl W, Merrill JT, McKay JD, Lisse JR, Zhong ZJ, Freimuth WW, and Genovese MC (2013). Efficacy and safety of belimumab in patients with rheumatoid arthritis: a phase II, randomized, double-blind, placebo-controlled, dose-ranging Study. J. Rheumatol 40, 579–589. 10.3899/jrheum.120886. [DOI] [PubMed] [Google Scholar]
- 57.Baert L, Benkhoucha M, Popa N, Ahmed MC, Manfroi B, Boutonnat J, Sturm N, Raguenez G, Tessier M, Casez O, et al. (2019). A proliferation-inducing ligand-mediated anti-inflammatory response of astrocytes in multiple sclerosis. Ann. Neurol 85, 406–420. 10.1002/ana.25415. [DOI] [PubMed] [Google Scholar]
- 58.Seshasayee D, Valdez P, Yan M, Dixit VM, Tumas D, and Grewal IS (2003). Loss of TACI causes fatal lymphoproliferation and autoimmunity, establishing TACI as an inhibitory BLyS receptor. Immunity 18, 279–288. 10.1016/s1074-7613(03)00025-6. [DOI] [PubMed] [Google Scholar]
- 59.O’Connor BP, Raman VS, Erickson LD, Cook WJ, Weaver LK, Ahonen C, Lin LL, Mantchev GT, Bram RJ, and Noelle RJ (2004). BCMA is essential for the survival of long-lived bone marrow plasma cells. J. Exp. Med 199, 91–98. 10.1084/jem.20031330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Danve A, Perry L, and Deodhar A (2014). Use of belimumab throughout pregnancy to treat active systemic lupus erythematosus—a case report. Semin. Arthritis Rheum 44, 195–197. 10.1016/j.semarthrit.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 61.Góralczyk A, Kolossa K, Waszczak-Jeka M, Adamczak R, and Jeka S (2020). The exposure to biologic and targeted synthetic disease-modifying antirheumatic drugs in pregnancy and lactation. Postepy Dermatol. Alergol 37, 306–312. 10.5114/ada.2020.96294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams EM, Bruner L, Adkins A, Vrana C, Logan A, Kamen D, and Oates JC (2016). I too, am America: a review of research on systemic lupus erythematosus in African-Americans. Lupus Sci. Med 3, e000144. 10.1136/lupus-2015-000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar R, Williams LK, Kato A, Peterson EL, Favoreto S Jr., Hulse K, Wang D, Beckman K, Thyne S, LeNoir M, et al. (2012). Genetic variation in B cell-activating factor of the TNF family (BAFF) and asthma exacerbations among African American subjects. J. Allergy Clin. Immunol 130, 996–999.e6. 10.1016/j.jaci.2012.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ritterhouse LL, Crowe SR, Niewold TB, Merrill JT, Roberts VC, Dedeke AB, Neas BR, Thompson LF, Guthridge JM, and James JA (2011). B lymphocyte stimulator levels in systemic lupus erythematosus: higher circulating levels in African American patients and increased production after influenza vaccination in patients with low baseline levels. Arthritis Rheum 63, 3931–3941. 10.1002/art.30598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cappelletti M, Presicce P, and Kallapur SG (2020). Immunobiology of acute chorioamnionitis. Front. Immunol 11, 649. 10.3389/fimmu.2020.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pawelczyk E, Nowicki BJ, Izban MG, Pratap S, Sashti NA, Sanderson M, and Nowicki S (2010). Spontaneous preterm labor is associated with an increase in the proinflammatory signal transducer TLR4 receptor on maternal blood monocytes. BMC Pregnancy Childbirth 10, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gomez-Lopez N, Romero R, Varrey A, Leng Y, Miller D, Done B, Xu Y, Bhatti G, Motomura K, Gershater M, et al. (2021). RNA sequencing reveals diverse functions of amniotic fluid neutrophils and monocytes/macrophages in intra-amniotic infection. J. Innate Immun 13, 63–82. 10.1159/000509718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Zhang T, Zhang Z, Wang Z, Zhou YJ, and Wang Z (2021). B cell activating factor regulates periodontitis development by suppressing inflammatory responses in macrophages. BMC Oral Health 21, 426. 10.1186/s12903-021-01788-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, and Grimes HL (2016). Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702. 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DePasquale EAK, Schnell D, Dexheimer P, Ferchen K, Hay S, Chetal K, Valiente-Alandí Í, Blaxall BC, Grimes HL, and Salomonis N (2019). cellHarmony: cell-level matching and holistic comparison of single-cell transcriptomes. Nucleic Acids Res 47, e138. 10.1093/nar/gkz789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Otipoby KL, Sasaki Y, Schmidt-Supprian M, Patke A, Gareus R, Pasparakis M, Tarakhovsky A, and Rajewsky K (2008). BAFF activates Akt and Erk through BAFF-R in an IKK1-dependent manner in primary mouse B cells. Proc. Natl. Acad. Sci. USA 105, 12435–12438. 10.1073/pnas.0805460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang C, Loo WM, Greenley EJ, Tung KS, and Erickson LD (2011). B cell maturation antigen deficiency exacerbates lymphoproliferation and autoimmunity in murine lupus. J. Immunol 186, 6136–6147. 10.4049/jimmunol.1001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mantchev GT, Cortesão CS, Rebrovich M, Cascalho M, and Bram RJ (2007). TACI is required for efficient plasma cell differentiation in response to T-independent type 2 antigens. J. Immunol 179, 2282–2288. 10.4049/jimmunol.179.4.2282. [DOI] [PubMed] [Google Scholar]
- 74.Kuley R, Draves KE, Fuller DH, Giltiay NV, Clark EA, and Giordano D (2021). B cell activating factor (BAFF) from neutrophils and dendritic cells is required for protective B cell responses against Salmonella typhimurium infection. PLoS One 16, e0259158. 10.1371/journal.pone.0259158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Giles DA, Moreno-Fernandez ME, Stankiewicz TE, Graspeuntner S, Cappelletti M, Wu D, Mukherjee R, Chan CC, Lawson MJ, Klarquist J, et al. (2017). Thermoneutral housing exacerbates nonalcoholic fatty liver disease in mice and allows for sex-independent disease modeling. Nat. Med 23, 829–838. 10.1038/nm.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moreno-Fernandez ME, Giles DA, Stankiewicz TE, Sheridan R, Karns R, Cappelletti M, Lampe K, Mukherjee R, Sina C, Sallese A, et al. (2018). Peroxisomal beta-oxidation regulates whole body metabolism, inflammatory vigor, and pathogenesis of nonalcoholic fatty liver disease. JCI Insight 3, e93626. 10.1172/jci.insight.93626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, and Albrecht M (2010). AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res 38, W755–W762. 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen J, Bardes EE, Aronow BJ, and Jegga AG (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–W311. 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data has been deposited at GEO and is publicly available. Macrophage bulk RNA-seq data is under accession number GEO: GSE202811. Previously deposited bulk RNA-seq data for rhesus macaque samples can be found under GEO: GSE181054.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


