STRUCTURED ABSTRACT
INTRODUCTION:
The ketogenic diet (KD) is an intriguing therapeutic candidate for Alzheimer’s disease (AD) given its protective effects against metabolic dysregulation and seizures. Gut microbiota are essential for KD-mediated neuroprotection against seizures as well as modulation of bile acids, which play a major role in cholesterol metabolism. These relationships motivated our analysis of gut microbiota and metabolites related to cognitive status following a controlled ketogenic diet intervention compared with a low-fat diet intervention.
METHODS:
Pre-diabetic adults, either with mild cognitive impairment (MCI) or cognitively normal (CN), were placed on either a low-fat American Heart Association Diet or high-fat Modified Mediterranean Ketogenic Diet for six weeks, then after a six week washout period, crossed over to the alternate diet. We collected stool samples for shotgun metagenomics and untargeted metabolomics at five timepoints to interrogate individuals’ microbiome and metabolome throughout the dietary interventions.
RESULTS:
Participants with MCI on the MMKD had lower levels of GABA-producing microbes Alistipes sp. CAG:514 and GABA, and higher levels of GABA-regulating microbes Akkermansia muciniphila. MCI individuals with curcumin in their diet had lower levels of BSH-containing microbes and an altered bile acid pool, suggesting reduced gut motility.
DISCUSSION:
Our results suggest that the MMKD may benefit adults with MCI through modulation of GABA levels and gut-transit time.
Keywords: Alzheimer’s disease, mild cognitive impairment, ketogenic diet, microbiome, metabolome, Low-Fat American Heart Association Diet (AHAD)
BACKGROUND
Despite decades of research, the pathophysiologic events that result in cognitive impairment are not fully understood, delaying the development of effective therapeutic options for neurodegenerative diseases such as Alzheimer’s disease (AD) (1, 2). However, it is now appreciated that a variety of systemic changes occur throughout the course of AD, including metabolic dysregulation (3, 4), mitochondrial dysfunction (5, 6), inflammation (7), and body compositional changes (8, 9).
AD-related metabolic dysregulation has been interrogated by comparing AD metabolomic profiles to those of unaffected individuals. However, identifying metabolites universally associated with AD or cognitive decline is not straightforward. Both the Alzheimer’s Disease Neuroimaging Initiative’s (ADNI) studies (10, 11) and mouse models (12) have demonstrated that metabolic signatures for AD are sex and APOE genotype-dependent. Though study of the UK Biobank cohort showed that AD incidence was correlated with metabolites including the ketone body β-hydroxybutyrate, acetone, and valine, these metabolites were also associated with age and APOE genotype, complicating interpretation (13). Triglycerides (particularly polyunsaturated serum triglycerides) are also broadly associated with mild cognitive impairment (MCI) and AD onset (14, 15). In terms of symptom-specific associations, both short- and medium/long-chain acylcarnitines were associated with episodic memory scores and AD severity in the ADNI cohort (16), while C3 (propionylcarnitine) was associated with decreased amyloid-β accumulation and higher memory scores (17). These observations have provided new directions for AD therapeutics.
In parallel, research into the gut-microbiota brain axis has spearheaded investigation into an altered gut microbiome in AD (18). It is hypothesized that gut microbiome disruptions may lead to local gastrointestinal inflammation or disturbance of the intestinal mucosal barrier, resulting in dysregulation of the signaling pathways between the gut and brain (19). Another hypothesis implicating the gut microbiome in AD comes from the connection between cholesterol, bile acids, and the gut microbiota. Cholesterol metabolism is consistently reported to be dysregulated in AD and AD-related diseases (20–22). Cholesterol accumulation in the brain contributes to hepatic encephalopathy, a chronic liver disease that leads to neuron loss and increased risk for AD, through bile acid mediated effects on the farnesoid X receptor (23). This observation is significant because bile acids, the main source of cholesterol elimination in the brain, are transformed by the gut microbiota (24). One preliminary study into this link showed that bile acids in serum were associated with “A/T/N’’ AD biomarkers, while another demonstrated that AD-related cognitive impairment is tied to higher levels of bacterially-modified secondary bile acids and lower levels of primary bile acids (25, 26). Similarly, another group reported that the deoxycholate/cholate (DCA/CA) ratio and primary bile acid synthesis were among the metabolites and metabolic pathways that most differentiated MCI and cognitively normal (CN) groups (27).
A significant factor uniting the gut microbiome, inflammation, and neurocognitive status with metabolic disorders is their relationship with diet (28–31). The ketogenic diet (KD) is a candidate therapeutic for AD because of its ability to improve mitochondrial function and cerebral bioenergetics, enhance autophagy, and reduce oxidative stress (30, 31). It also reduces neuronal hyperexcitability and leads to improved amyloid and tau regulation, substantiating its potential use for cognitive impairment (32).
We previously performed a pilot trial of a low carbohydrate modified Mediterranean ketogenic diet (MMKD) in patients at-risk for AD. We found that the MMKD increased levels of Aβ42 and decreased levels of tau in the cerebral spinal fluid (CSF) as well as improved peripheral lipid and glucose metabolism (i.e. reduction of Hb1Ac, insulin, triglyceride levels) in patients with mild cognitive impairment (MCI) (32). Patients on the MMKD also showed increased cerebral ketone body uptake and cerebral perfusion, important metrics of brain function (32). Microbially, the MMKD was associated with increased relative abundances of the Enterobacteriaceae, Akkermansia, Slackia, Christensenellaceae, and Erysipelotrichaceae taxa and decreases in the relative abundances of Bifidobacterium and Lachnobacterium (33). Metabolically, the MMKD was associated with increased propionate and butyrate and decreased lactate and acetate in stool (33). Interrogation into the mycobiome, or fungal microbiome, demonstrated that MMKD increased the relative abundance of Agaricus and Mrakia and decreased the relative abundance of Saccharomyces and Claviceps in patients with MCI (34).
Although thought-provoking, previous analyses relied on amplicon sequencing (of the 16S ribosomal RNA gene and internal transcribed spacer regions). In the present study, we sought to further characterize the complex relationship between diet, cognitive status, and the human microbiome and metabolome using shotgun metagenomic sequencing and untargeted metabolomics on the same samples. Furthermore, we employed a novel reference data-driven tool to read out dietary metabolites empirically and retrospectively from our samples (35). By updating our methods and integrating multiple data types, we identified key modulations to the gut microbiome and metabolome related to diet alone, cognitive status alone, or both.
METHODS
Participant Information:
Participants were deemed at risk for AD based on their systemic metabolic dysfunction and cognitive dysfunction or subjective/objective memory complaints at study onset. Participants were diagnosed by a physician or neuropsychologist as either cognitively normal with subjective memory complaints (CN) using Alzheimer’s Disease Neuroimaging Initiative (ADNI) criteria or mild cognitively impaired (MCI) using NIA-AA guidelines for clinical diagnoses without reference to biomarker status. Exclusion criteria included statin use and neurological conditions other than MCI. See the Methods Supplement for full inclusion and exclusion criteria for this study. The protocol was approved by the Wake Forest Institutional Review Board (ClinicalTrials.gov Identifier: NCT02984540), and written informed consent was obtained from all participants and/or their study partners. Participants were medically supervised by clinicians, with safety monitoring overseen by the Wake Forest Institutional Data and Safety Monitoring Committee.
Procedure:
The primary pilot trial was a randomized crossover design in which participants consumed either a Modified Mediterranean-Ketogenic Diet (MMKD) or the control American Heart Association Diet (AHAD) for 6 weeks, followed by a 6-week washout with their pre-study diet, after which the second diet was consumed for 6 weeks (Figure S1). For further information on diet information and education, see the Methods Supplement.
Data Collection and Processing:
Stool samples were collected from participants at five study timepoints: at the beginning and end of each diet intervention, and 6 weeks after washout of the second diet (Figure S1). Methods for collection were adapted from the Manual of Procedures for the Human Microbiome Project (NIH, Version Number 12.0). We extracted DNA for metagenomic sequencing and metabolites for untargeted MS/MS analysis. For more information on metagenomics, metabolomics, and food-omics data processing and analysis, see the Methods Supplement.
Data Availability:
Metagenomic sequencing data are available in Qiita under study ID 13662 (36). Mass spectrometry data (.mzXML format) are available in MassIVE under ID MSV000087087. The classical molecular networking job is available in GNPS at the following link: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=90f1ba6e1e4d4d89b75b9017a0631983 (37). The feature-based molecular networking job is available in GNPS at the following link: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=0b828a2a6f224fb293de1fe39bee5fc5 (38). The code utilized for these analyses are available at https://github.com/ahdilmore/BEAM_MultiOmics.
RESULTS
Twenty-three adults were enrolled in the study and twenty completed the full intervention. Participant characteristics by AD risk group are in Table 1.
Table 1. Participant characteristics for full sample and by AD risk group at study baseline.
Table values are mean (SD) or N (%). +Denotes a difference between CN and MCI, p<0.05, using t-test for continuous and Fisher’s exact test for categorical variables. ÂPOE genotype information unavailable for one participant.
| Variable | All (n=20) | CN (n=11) | MCI (n=9) |
|---|---|---|---|
| Age, years | 64.3 (6.3) | 64.9 (7.9) | 63.4 (4.0) |
| Female sex | 15 (75%) | 9 (81.8%) | 6 (66.7%) |
| Black race | 7 (35%) | 2 (18.2%) | 5 (55.6%) |
| APOE4-positive | 6 (31.6%) | 2 (20%)^ | 4 (44.4%) |
| Education, years | 16.1 (2.5) | 16.5 (2.3) | 15.7 (2.9) |
| BMI, kg/m2 | 28.4 (5.7) | 26.9 (6.2) | 30.3 (4.7) |
| MMSE | 28.7 (1.1) | 28.9 (1.0) | 28.3 (1.2) |
| Glucose, mg/dL | 97.6 (18.3) | 93.9 (11.4) | 102.1 (24.4) |
| BHB, mmol/L | 0.23 (0.27) | 0.35 (0.31)+ | 0.1 (0.14)+ |
| Insulin, μIU/mL | 8.3 (6.1) | 5.2 (3.4)+ | 12.0 (6.8)+ |
| HbA1c, % | 5.9 (0.3) | 5.9 (0.2) | 6.1 (0.4) |
| Total Chol, mg/dL | 215.2 (43.7) | 200.5 (42.1) | 233.2 (40.6) |
| HDL Chol, mg/dL | 67.4 (25.8) | 69.5 (25.8) | 64.8 (27.1) |
| VLDL Chol, mg/dL | 20.0 (11.1) | 15.2 (6.2)+ | 25.8 (13.2)+ |
| LDL Chol, mg/dL | 121.5 (41.2) | 104.1 (45.6)+ | 142.7 (24.2)+ |
| Triglycerides, mg/dL | 99.8 (55.7) | 75.5 (30.5)+ | 129.4 (66.4)+ |
Dimensionality Reduction of Microbiome, Food-omics, and Metabolome datasets:
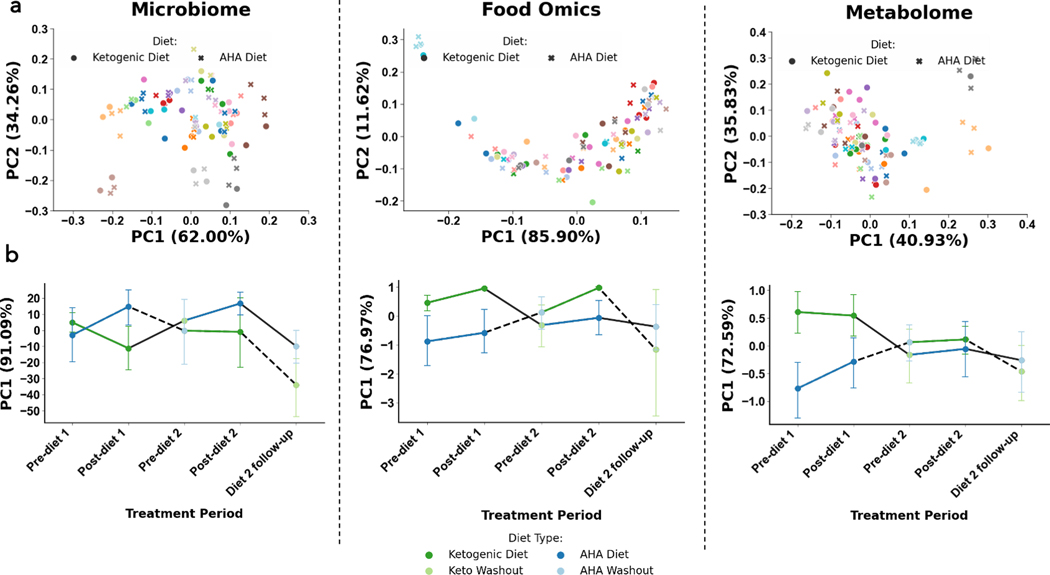
Each of the three data layers collected is high-dimensional (i.e. hundreds to thousands of microbes, foods features, and metabolites are quantified in each assay), making it difficult to draw direct comparisons between individuals. One strategy to mitigate this challenge is to reduce the high-dimensional data into a few informative dimensions. We did so with both the Robust Aitchison Principal Component Analysis (RPCA) and Compositional Tensor Factorization (CTF) metrics, then computed the effect size of variables of interest [diet, cognitive status, diet order, and subject identity] in the low-dimensional space using permutational multivariate analysis of variance (PERMANOVA) tests (39, 40; Figure 1). RPCA is a distance metric that accounts for the sparse, compositional nature of microbiome data, but does not perform any correction when a given subject is measured multiple times (39). CTF, on the other hand, represents multiple samples from a given host as a tensor to account for the fact that the microbiome and metabolome of a given individual correlated over time (40). Therefore, we hypothesized that subject identity would have the highest effect size with RPCA, while other variables of interest would have a larger effect size with CTF. This was not the case; subject identity only had the largest effect size by RPCA for metabolomics (F=29.68, padj=4.33e-6). No variables of interest were significant by RPCA for metagenomics nor food-omics. However, diet was significant by CTF in all groups (metagenomics: F=10.17, padj=0.012; food-omics: F=41.99, padj=1.07e-7, metabolomics: F=42.73, padj=8.64e-8). This suggests that, once repeated measured are accounted for, diet has a larger effect on gut microbiome and metabolome than cognitive status. Results from all PERMANOVAs performed after RPCA and CTF are available in Table S1.
Figure 1. Microbiome (left column), food-omics (middle column), and metabolome (right column), separate by subject and by diet across time.
Robust Aitchison Principal Component Analysis (RPCA) PC1 (x-axis) and PC2 (y-axis) colored by subject identification with marker shape by diet across study, clusters by subject. Compositional tensor factorization, method accounting from repeated measures across time (x-axis) PC1 (y-axis) colored by diet types, follows crossover study design in each data modality.
Relative Abundance Analyses:
After performing a broad assessment of the microbiome, food-ome, and metabolome’s associations with diet and cognitive status via dimensionality reduction, we utilized Bayesian Inferential Regression to identify particular microbes, foods, and metabolites associated with diet and/or cognitive status. Specifically, we modeled differential abundances using a Negative Binomial Linear Mixed Effect model designed specifically for crossover trials, then identified features that were most associated with the MMKD or AHAD and CN or MCI. The results are detailed below:
Features Associated with Cognitive Status Independent of Diet:
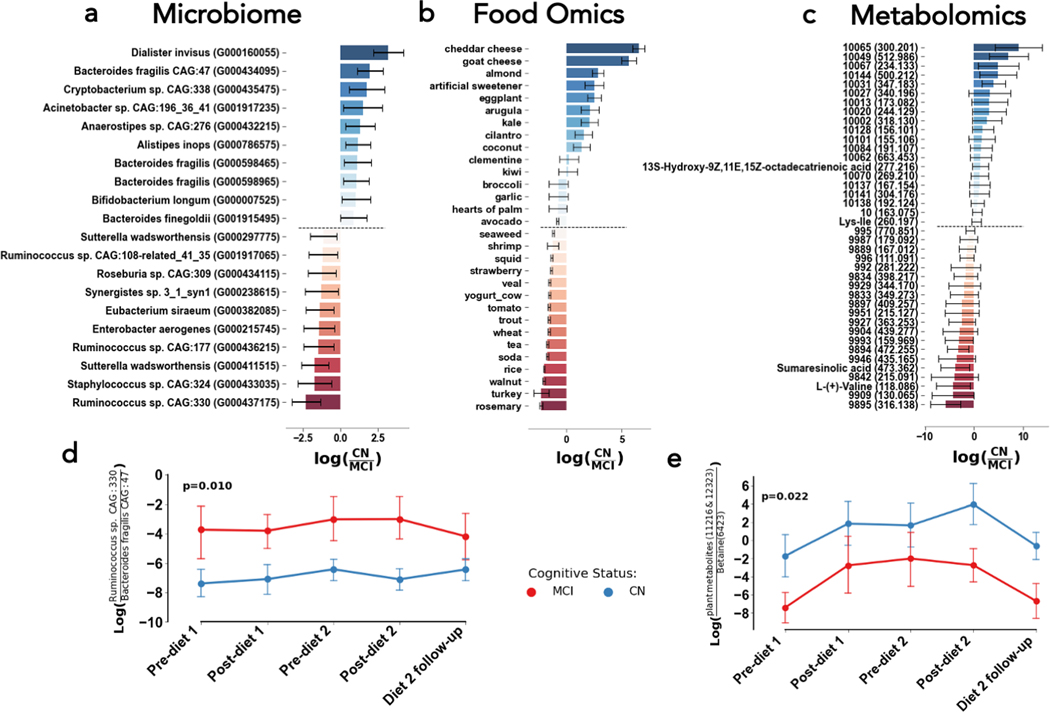
The top features associated with cognitive status independent of diet were identified by sorting the values in the cognitive status column of our Bayesian Inferential Regression results; more positive values represented stronger association with CN while more negative values indicated stronger association with MCI. The top ten microbial, food-derived metabolite, and overall metabolite features associated with normal cognition or mild cognitive impairment are displayed in Figure 2A–C. Notably, Ruminococcus sp. CAG:330 is associated with MCI while Bacteroides fragilis and Dialister invisus are associated with CN independently of diet or timepoint. Figure 2D illustrates that the ratio of Ruminococcus sp. CAG:330 to Bacteroides fragilis CAG: 47 is consistently elevated in MCI at each timepoint of the study, regardless of each participant’s diet order (p=0.016). Interestingly, cognitively normal individuals had higher levels of dietary metabolites that matched to cheddar cheese, goat cheese, and almonds while individuals with MCI had relatively higher loads of dietary metabolites that matched to turkey, smelt, and almonds (Figure 2B). All these foods are likely to be consumed on a ketogenic diet, reflecting that individuals of both cognitive status categories consumed both diets. The metabolites 13S-hydroxy-9Z,11E,15Z-octadecatrienoic acid (a polyunsaturated fat related to many cooking oils) and Ile-Lys were associated with CN, while sumaresinolic acid and L-Valine were associated with MCI. The role that these metabolites may or may not have in disease pathology are unknown, though it is interesting to see a polyunsaturated fat (PUFA) correlated with CN, since consumption of fats overall is increased on the MMKD. Furthermore, we found that the ratio of two particular plant-associated metabolites (IDs 11216 and 12323) to betaine, an osmoregulatory downstream of choline, was significantly higher in CN than MCI (p=0.022). However, it is unclear whether this reflects higher consumption of plants or lower amounts of betaine, which could be related to bile acid metabolism.
Figure 2. Microbial strains (left), foods (middle), and metabolites (right) related to cognitive status, but not necessarily diet type.
The differential abundances for the top 10 and bottom 10 with mean (bar) and model 95% confidence interval (error bars) associated with cognitive normal (more positive, blue) and mild cognitive impairment (more negative, red) (A, B, C). The log-ratio (y-axis) of the top and bottom bacterial strains (D&E) plotted across time (x-axis) and colored by cognitive status. Presented cognitive status p-values in log-ratio plots are determined from linear mixed effects models with fixed effects of diet, cognitive status, period, and diet sequences with random effect of subject.
Features Associated with Diet Intervention Independent of Cognitive Status:
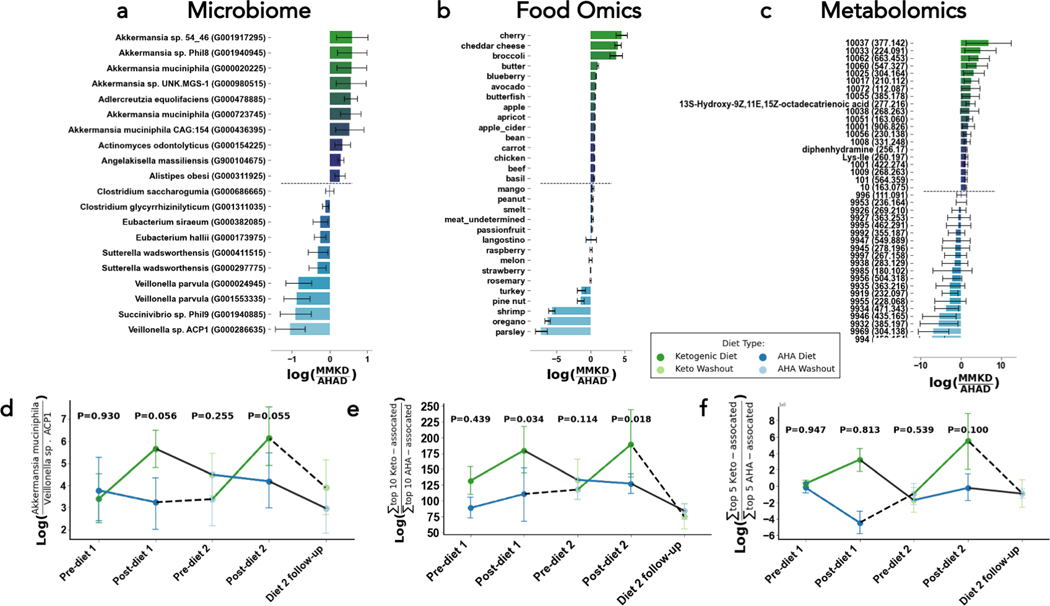
The top features associated with diet independent of cognitive status were identified by sorting the values in the diet column of our Bayesian Inferential Regression results; more positive values represented stronger association with the MMKD while more negative values indicated stronger association with the AHAD. The top ten microbial, food-derived metabolite, and overall metabolite features associated with the MMKD and AHAD are displayed in Figure 3A–C. Many of the top microbes associated with MMKD are of the Akkermansia taxa, while Veillonella, Sutterella, and Eubacteria are associated with AHAD. We consistently observed higher relative abundances of Akkermansia muciniphila on the MMKD, and higher relative abundances of Veillonella sp. ACP1 on the AHAD (Figure 3D). Foodwise, those on the MMKD had higher relative abundances of dietary metabolites found in high-fat dairy products including cheddar cheese, low-sugar fruits like cherries, and low-sugar vegetables including broccoli (Figure 3B). Those on the AHAD diet had higher relative abundances of dietary metabolites that matched to low-fat foods including parsley, oregano, and shrimp (Figure 3B). None of the top metabolites associated with the AHAD were annotated through FBMN, but the metabolites 13S-hydroxy-9Z,11E,15Z-octadecatrienoic acid, Ile-Lys, and diphenhydramine (an antihistamine) were associated with the MMKD. The former two were independently associated with CN, suggesting that particular PUFAs or conjugated amino acids may be implicated in the relationship between the MMKD and CN. However it is important to note that the log ratios of the sum of the top 10 most MMKD-associated to the sum of top 10 AHAD-associated dietary metabolites were not significantly different over the course of the study (Figure 3E–F).
Figure 3. Microbial strains (left), foods (middle), and metabolites (right) related to diet type, but not necessarily cognitive status.
The differential abundances for the top 10 and bottom 10 with mean (bar) and model 95% confidence interval (error bars) associated with cognitive Ketogenic (more positive, green) and low-fat / american heart association diets (more negative, blue) (A, B, C). The log-ratio (y-axis) of the top and bottom bacterial strains (D), and sum of the top and bottom 10 foods (E) or metabolites (F) plotted across time (x-axis) and colored by diet. Presented diet stage p-values in log-ratio plots are determined from linear mixed effects models with fixed effects of diet, cognitive status, period, and diet sequences with random effect of subject.
Microbes and Metabolites Jointly Associated with Cognitive Status and Dietary Intervention:
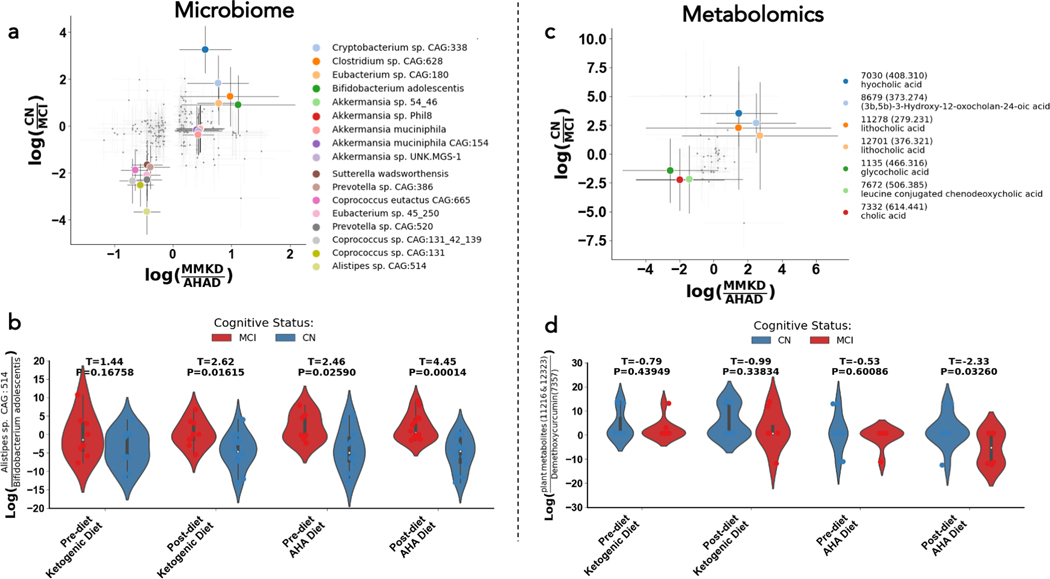
To identify microbes and metabolites that had strong diet- and cognitive status-specific associations, we plotted the log ratios for CN to MCI against the log ratios for MMKD over AHAD and examined features that fell in each corner of the plot (Figure 4A, 3C). Each corner corresponds to a different subpopulation: the top right corner represents microbes/metabolites associated with the MMKD and normal cognition, the top left corner contains the microbes/metabolites correlated with the AHAD and normal cognition, the bottom left corner shows microbes/metabolites associated with the AHAD and mild cognitive impairment, and the bottom right corner corresponds to microbes/metabolites correlated with the MMKD and MCI. These investigations allowed us to examine microbial and metabolite changes that were modulated by both diet and cognitive status.
Figure 4. Microbial strains (left), foods (middle), and metabolites (right) related to both cognitive status and diet.
Scatter plot of log ratios of differential abundances by diet (x-axes) and cognitive status (y-axes) for microbes, with the ten most differential microbes and metabolites in both axes colored (A). Violin plot of the log-ratio of Alistepes sp. CAG: 514 to Bifidobacterium adolescentis at four different timepoints in the study (B). Scatter plot of log ratios of differential abundances by diet (x-axes) and cognitive status (y-axes) for microbes, with the ten most differential microbes and metabolites in both axes colored (C). Violin plot of the log-ratio of plant metabolites to Diarylheptanoids at four different timepoints in the study (D). In the scatterplots shown in (A) and (C), each corner corresponds to a different subpopulation: the top right corner represents microbes/metabolites associated with the MMKD and CN, the top left corner contains the microbes/metabolites correlated with the AHAD and CN, the bottom left corner shows microbes/metabolites associated with the AHAD and mild cognitive impairment, and the bottom right corner corresponds to microbes/metabolites correlated with the MMKD and MCI. These analyses allowed us to examine microbial and metabolite changes that were modulated by both diet and cognitive status.
Microbially, we found that the ratio of Alistipes sp. CAG:514 to Bifidobacterium adolescentis was significantly different between MCI and CN individuals only after consuming the low-fat AHAD (Figure 4B; t=3.78, p=0.00082). This finding is significant because microbial strains that are similar to Alistipes sp. CAG:514 are known to produce GABA (45). This led us to re-investigate the relative relationship between Akkermansia muciniphila and Alistipes sp. CAG:514 and levels of the GABA metabolite in our population. We found that the ratio of Alistipes sp. CAG:514 to Akkermansia muciniphila is elevated in individuals with MCI relative to normal individuals only on the low-fat AHAD (Figure S3A). Furthermore, GABA levels are higher in the stool of cognitively-impaired individuals on the AHAD than on the MMKD, but there is no dietary-related difference in cognitively-normal individuals (Figure S3B). This trend was consistent in a separate dataset examining cerebrospinal fluid metabolites of the same individuals (Fig S3B). These observations led us to hypothesize that low-fat AHAD modulates key neurotransmitters, such as promoting GABA production, in individuals with cognitive impairment. This dysregulation may be related to increases in GABA-producing microbes such as Alistipes sp. CAG:514, while dietary interventions such as the MMKD may decrease GABA levels in the opposite direction through its associated increases in GABA-regulating microbes such as Akkermansia muciniphila.
Metabolite-wise, we found that the log ratio of two plant-derived metabolites to Diarylheptanoids, a class of plant secondary metabolites, was much lower in individuals with MCI only after consuming the AHAD (Figure 4D; t=−2.33, p=0.0326).This indicates that Diarylheptanoids, which most notably includes the turmeric-associated metabolite curcumin, may be selectively enriched in people with cognitive impairment after consuming a low-fat diet. This observation is interesting given curcumin’s previous implications with longer gut transit time and increased bile acid levels, which is particularly relevant to our work given the role of bile acids in cholesterol metabolism (41). Though curcumin was not annotated by FBMN, a nearest-neighbor suspect library suggested that metabolite 7357 in our dataset may be demethoxycurcumin (42).
Multi-omics Analyses:
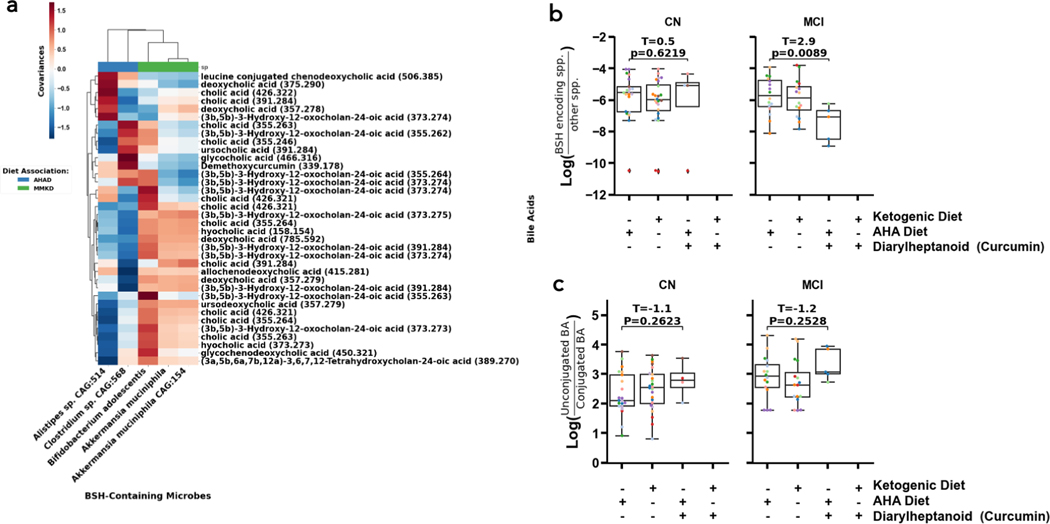
Given that cholesterol metabolism is consistently dysregulated in AD and MCI, we monitored bile acids in different subpopulations over the course of the dietary intervention. Bile acids are significant because they are the primary source of cholesterol elimination in the brain, and their chemistries are modified by microbes. Specifically, microbes can modulate the relative amounts of secondary and primary bile acids, as well as conjugate particular amino acids to bile acids (43). The former ratio is particularly relevant given that AD-related cognitive impairment was previously associated with a higher ratio of secondary bile acids to primary bile acids (26). The bacterial enzyme bile salt hydrolase (BSH) promotes deconjugation of conjugated bile acids and production of secondary bile acids, leading us to hypothesize that the abundance of BSH-containing microbes would positively co-vary with levels of secondary bile acids and deconjugated bile acids (41, 26). To test this hypothesis, we used MMvec to quantify covariances between bile acids and BSH-containing microbes (44). We also clustered relationships by diet, to determine if covariances between any particular bile acids and BSH-containing microbes were diet-specific. We found that Bifidobacterium adolescentis, Akkermansia muciniphila, and Akkermansia muciniphila CAG:154 were associated with several ion forms of cholic acid (CA), deoxycholic acid (DCA), (3a, 5b)-3-hydroxy-12-oxocholan-245-oic acid (3β-DCA), hyocholic acid,, ursodeoxycholic acid, and glycine-conjugated chenodeoxycholic acid in the MMKD while Alistipes sp. CAG:514 was associated with leucine-conjugated chenodeoxycholic acid, and a few ion forms of CA, DCA, and 3β-DCA and Clostridium sp. CAG:568 was associated with demethoxycurcumin, ursocholic acid, glycocholic acid, and a few ion forms of CA and 3β-DCA in the AHAD (Figure 5A). Each group contains all forms of bile acids (primary, secondary, conjugated, and unconjugated), so we could not draw conclusions on whether diet was associated with any particular shift in bile acids.
Figure 5. Strongest diet and cognitive status-linked differential abundances and co-occurrences linked to bile salt hydrolase (BSH) containing bacterial species, diarylheptanoids, and bile acids (BA).
The MMvec log cooccurrence probabilities (red-blue colors, colorbar) between differentially abundant and annotated metabolites (y-axis) and microbes (x-axis) colored by their association to diet (color bar top of x-axis, color legend) (A). The log-ratio of BSH containing microbes vs. those without BSH (y-axis) across diets and diarylheptanoids consumption (x-axis) (B). The log-ratio of unconjugated to conjugated BAs (y-axis) across diets and diarylheptanoids consumption (x-axis) (C). Annotations are Level II or Level III according to the Metabolomics Standards Initiative and significance was evaluated by a two-sided t-test. (50).
We were also interested in examining the metabolite demethoxycurcumin’s relationships with abundance of BSH-containing microbes as well as the ratio of unconjugated to conjugated bile acids. Turmeric is known to increase the levels of unconjugated bile acids specifically in hosts colonized with bacteria containing high levels of BSH (41). However, it is unknown if turmeric itself modulated levels of BSH-encoding microbes, and the effects of this system in inviduals experiencing cognitive impairment are unknown. Interestingly, we found that individuals with MCI had lower relative abundances of BSH-encoding microbes when their metabolome contained curcumin, while no difference was found in cognitively normal individuals (Figure 5B; t=2.9, p=0.0089; t=0.5, p=0.6308). On the other hand, there was no significant difference in the ratio of unconjugated bile acids to conjugated bile acids among the diet groups of either CN or MCI individuals (Figure 5C; t=−1.1, p=0.2623; t=−1.2, p=0.2528). The lower levels of BSH-encoding microbes and slightly elevated (though not significant) unconjugated to conjugated bile acid ratio in response to turmeric suggests that individuals with MCI may have decreased gut motility (increased gut transit time) relative to CN individuals (41, Figure S4).
DISCUSSION
It is well-established that metabolic dysregulation (3, 4) and inflammation (7) occur in tandem with cognitive decline over the course of AD. Therefore, modulating host metabolism through specialized diets such as the ketogenic diet, is one strategy to mitigate the effects of AD-associated cognitive impairment. These diet-associated changes in cognitive status may be microbiota-dependent or microbiota-modulated. Previous work found that gut microbiota are required for the KD’s protection against seizures in a murine model of epilepsy (46). Furthermore, gut microbiota are known to modulate the bile acid pool; given that bile acids are the primary agent of cholesterol depletion in the brain, gut microbiota-induced changes to the bile acid pool may mitigate the dysregulation of cholesterol metabolism.
Broadly, our investigation demonstrated that controlled changes in diet led to widespread changes in the microbiome and metabolome over time. Our metagenomic results are consistent with those reported in a previous, preliminary analysis of the microbiome in this trial, but provide greater resolution since we utilized shallow shotgun sequencing over 16S rRNA gene amplicon sequencing (33). Furthermore, we utilized updated methods including Bayesian Inferential Regression with a model that was customized for the crossover nature of the study, while previous authors reported differences in relative abundances without accounting for the longitudinality of the data (33). Our studies made similar conclusions – we both reported an increase in Akkermansia on the MMKD relative to the AHAD and increases in Dialister and Bacteriodes in CN individuals relative to MCI individuals – but our study was able to identify several subspecies of Akkermansia muciniphila that were enriched on the MMKD and that Dialister invisus and multiple Bacteriodes fragilis strains that were enriched in cognitively normal individuals. Our observation of Akkermansia muciniphila’s enrichment on the MMKD is consistent with previous findings that the relative abundance of Akkermansia muciniphila greatly increases over the course of ketogenic dietary intervention in a mouse model of epilepsy (46). Interestingly, another study reported an increase in relative abundance of Akkermansia muciniphila in individuals who completed a calorie-restricted high-protein, moderate-fat (35% protein, 25% fat, 40% carbohydrate) diet (47). Both the calorie-restricted diet and our KD were observed to improve metabolic regulation and insulin sensitivity. It is possible, therefore, that Akkermansia muciniphila is involved in metabolic regulation in a manner that relates to effects such as improved insulin sensitivity rather than being tied specifically to dietary composition.
Our study also identified that the ratio of Alistipes sp. CAG:514 to Bifidobacterium adolescentis was significantly different between MCI and CN individuals only after consuming the low-fat AHAD. This finding is significant given Alistipes’ ability to produce GABA, which we validated by checking that GABA levels trended higher in the stool of cognitively-impaired individuals after consuming the AHAD but not the MMKD, while there is no such diet difference in cognitively-normal individuals (Fig S2B). We therefore hypothesize that the AHAD leads to increased GABA production in individuals with MCI through an increase in GABA-producing Alistipes. Inversely, the MMKD regulates GABA production in both MCI and CN individuals through its associated increase in production of GABA-regulating Akkermansia muciniphila. This possibility is inconsistent with some previous work relating GABA to AD; though dysfunction in GABA signaling has been hypothesized to play a role in AD, the strongest evidence shows lower levels of GABA in the post-mortem brains of patients with AD (48). However, recent studies have highlighted the multifaceted role GABA plays within the gut-brain-axis. GABA within the gut regulates local microbial populations (which may serve as growth factors), signals through the gastrointestinal tract impacting motility, regulates inflammation, and may impact GABA levels within other organs including the brain (49). Further, variable response of the GABAergic system to the KD has been observed in response to individual factors such as age (50). One such factor of relevance to AD is APOE genotype; our own data indicate that APOE4-positive individuals have higher loads of GABA-producing microbes in the gut and higher amounts of GABA in their cerebrospinal fluid (Fig S3). Therefore, it is possible that GABA dysfunction in the gut follows a different trend than GABA dysfunction in the brain, particularly at the earlier stages of disease we are observing in this study. Our results showing that GABA levels were higher after AHAD but not MMKD highlight that dietary intervention can alter the microbiome and downstream metabolites/signaling molecules. This was only seen in cognitively-impaired participants, suggesting that individuals throughout the cognitive spectrum may differentially respond to intervention impacting the microbiome. Nonetheless, our study was underpowered and the direct implications of these gut changes on the CNS remains unclear. Future studies assessing microbiome in patients with cognitive impairment and modification by intervention are needed.
Our study also addressed relationships between bile acids, diet, and cognitive status given bile acids role in cholesterol regulation. Though we found diet-specific relationships between BSH-containing microbes and particular bile acids, we did not find diet-specific relationships between more broad bile acid classifications. We found that differences in bile acid categories (unconjugated versus conjugated) were not differentiated by curcumin presence, dietary fat content, or cognitive status. This result is inconsistent with the previous finding that turmeric (which contains curcumin as an active ingredient) significantly increases the levels of unconjugated bile acids specifically in hosts colonized with high levels of bacteria containing BSH (41). However, our sample size is small, so it is possible that we lack the power to reliably detect this difference. Our study also did not investigate Bilophila wadsworthia, which has previously documented relationships to cognitive impairment in mice as well as dietary fat induced taurocholic acid (TCA) production (51, 52). We were unable to detect TCA through FBMN, but are interested in determining whether specific bile acid transformations by BSH (i.e. from conjugated TCA to unconjugated CA) are enriched in particular diets or in particular cognitive groups in future studies
Limitations & Further Work:
Our study is limited by its small size and relatively brief intervention period, as it is a pilot trial. We were also significantly limited by the low annotation rate of the untargeted metabolomics experiments, which left many of the key metabolites that were associated with cognitive status and diet unannotated. For this reason, much of our metabolomics analyses relied on metabolites that have previously been described in the Alzheimer’s disease and dietary metabolite literature. Additionally, as we did not run bile acid or neurotransmitter standards in these experiments, our annotations are Level II or Level III by the Metabolomics Standards Initiative; this could be a reason that we were unable to detect certain bile acids of interest, such as TCA (53). Since we know that bile acid metabolites and neurotransmitters such as GABA are potentially important AD-related mechanistic pathways, in the future we may choose to perform targeted metabolomics or at a minimum, run these compounds as standards in the same mass spectrometry run to increase the rigor of our annotations.
Similarly, while our food-omics tool provides a handy reverse method to readout food counts, it is not perfect in reproducing exact dietary features. Although it reproduces dietary patterns that are consistent with the ketogenic diet or a low-fat diet, the tool cannot be used to distinguish exact dietary conditions. As more reference foods are added to the FoodOmics database and the tool grows in power, its power will grow. In the future, we may also choose to utilize patient dietary logs to identify foods that are highly associated with changes in cognitive status.
An additional technical limitation is that the untargeted metabolomics platform does not provide the level of characterization of lipid metabolism and mitochondrial function that is available on targeted platforms. The current results should be interpreted as a snapshot of metabolism with more detailed analysis of lipid metabolism, ketogenesis, and other processes known both to be affected by diet and important for AD to come in future studies.
Despite these technical challenges, we performed a longitudinal trial and using statistical techniques that accounted for various challenging aspects of our data (sparsity, compositionality, and longitudinality). These statistical analyses allowed us to identify microbial and metabolite features that were specifically associated with both diet and cognitive status.
Supplementary Material
RESEARCH IN CONTEXT:
Systematic review:
The authors used PubMed to review literature related to the gut microbiome and metabolome’s association with either Alzheimer’s disease or the ketogenic diet. While literature relating the gut microbiome to Alzheimer’s is not well-established, there is significant literature examining ketogenic diet induced changes in the gut microbiome. We also reference previous work on this same study population.
Interpretation:
Our study suggested that the MMKD may benefit adults with MCI through modulation of GABA levels and gut-transit time.
Future directions:
Our work serves as a model for elucidating microbiome and metabolome alterations in a dietary intervention with advanced statistical techniques. In the future, we will use more precise metabolomics interrogations to elucidate how specific metabolites of interest are altered in this dietary intervention.
ACKNOWLEDGEMENTS / CONFLICTS / FUNDING SOURCES:
This work was supported by the Wake Forest Alzheimer’s Disease Research Center (P30-AG072947), the Hartman Family Foundation, and the Roena B. Kulynych Center for Memory and Cognition Research.
Funding for the AGMP (Alzheimer Gut Microbiome Project) and ADMC (Alzheimer’s Disease Metabolomics Consortium), led by Dr R.K.-D. at Duke University) was provided by the National Institute on Aging grants 5U19AG063744, 31U01AG061359–01, 1RF1AG059093, 1RF1AG057452, and R01AG046171, a component of the Accelerating Medicines Partnership for AD (AMP-AD) Target Discovery and Preclinical Validation Project (https://www.nia.nih.gov/research/dn/amp-ad-target-discovery-and-preclinical-validation-project) and the National Institute on Aging grant RF1 AG0151550.
R.K-D. is an inventor of key patents in the field of Metabolomics and holds equity in Metabolon, a biotech company in North Carolina. In addition, she holds patents licensed to Chymia LLC and PsyProtix with royalties and ownership, which is unrelated to this work. P.C.D. is on the scientific advisory board of Sirenas, Cybele Microbiome, Galileo, and founder and scientific advisor of Ometa Labs LLC and Enveda (with approval from UC San Diego).
Disclosures:
This work was supported by grants U19AG063744 (to R.K.D.) and P30AG072947 (to S.C.) from the National Institute of Health and a grant from the Hartman Family Foundation (to S.C.). PCD is a scientific co-founder of Ometa Labs and Enveta Biosciences with prior approval from UC San Diego, and is on the scientific advisory board of Cybele. R.K.D is an inventor on a series of patents on use of metabolomics for diagnosis and treatment of CNS diseases and which do not relate to the subject of this manuscript. R.K.D serves on the Metabolomics Society and the Metabolomics Association of North America, which does not interfere with the subject of this manuscript. R.K.D holds equity in Metabolon Inc., Chymia LLC and PsyProtix.
REFERENCES:
- 1.Yiannopoulou KG, Anastasiou AI, Zachariou V, Pelidou S-H. Reasons for Failed Trials of Disease-Modifying Treatments for Alzheimer Disease and Their Contribution in Recent Research. Biomedicines 2019;7. 10.3390/biomedicines7040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cummings J, Lee G, Zhong K, Fonseca J, Taghva K. Alzheimer’s disease drug development pipeline: 2021. Alzheimers Dement 2021;7:e12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pathak D, Berthet A, Nakamura K. Energy failure: does it contribute to neurodegeneration? Ann Neurol 2013;74:506–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Velpen V, Teav T, Gallart-Ayala H, Mehl F, Konz I, Clark C, et al. Systemic and central nervous system metabolic alterations in Alzheimer’s disease. Alzheimers Res Ther 2019;11:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swerdlow RH, Burns JM, Khan SM. The Alzheimer’s disease mitochondrial cascade hypothesis. J Alzheimers Dis 2010;20 Suppl 2:S265–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swerdlow RH, Burns JM, Khan SM. The Alzheimer’s disease mitochondrial cascade hypothesis: progress and perspectives. Biochim Biophys Acta 2014;1842:1219–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes C, Cunningham C, Zotova E, Woolford J, Dean C, Kerr S, et al. Systemic inflammation and disease progression in Alzheimer disease. Neurology 2009;73:768–74. 10.1212/wnl.0b013e3181b6bb95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchman AS, Wilson RS, Bienias JL, Shah RC, Evans DA, Bennett DA. Change in body mass index and risk of incident Alzheimer disease. Neurology 2005;65:892–7. [DOI] [PubMed] [Google Scholar]
- 9.Razay G, Vreugdenhil A, Wilcock G. Obesity, abdominal obesity and Alzheimer disease. Dement Geriatr Cogn Disord 2006;22:173–6. [DOI] [PubMed] [Google Scholar]
- 10.Arnold M, Nho K, Kueider-Paisley A, Massaro T, Huynh K, Brauner B, et al. Sex and APOE ε4 genotype modify the Alzheimer’s disease serum metabolome. Nat Commun 2020;11:1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang R, Trushina E, Zhu K, Zaidi SSA, Lau BM, Kueider-Paisley A, et al. Predictive metabolic networks reveal sex- and APOE genotype-specific metabolic signatures and drivers for precision medicine in Alzheimer’s disease. Alzheimers Dement 2022. 10.1002/alz.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao N, Ren Y, Yamazaki Y, Qiao W, Li F, Felton LM, et al. Alzheimer’s Risk Factors Age, APOE Genotype, and Sex Drive Distinct Molecular Pathways. Neuron 2020;106:727–42.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Amin N, Sproviero W, Arnold M, Batra R, Bonnechere B, et al. Longitudinal analysis of UK Biobank participants suggests age and APOE-dependent alterations of energy metabolism in development of dementia. bioRxiv 2022. 10.1101/2022.02.25.22271530. [DOI] [Google Scholar]
- 14.Huynh K, Lim WLF, Giles C, Jayawardana KS, Salim A, Mellett NA, et al. Concordant peripheral lipidome signatures in two large clinical studies of Alzheimer’s disease. Nat Commun 2020;11:5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernath MM, Bhattacharyya S, Nho K, Barupal DK, Fiehn O, Baillie R, et al. Serum triglycerides in Alzheimer disease: Relation to neuroimaging and CSF biomarkers. Neurology 2020;94:e2088–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horgusluoglu E, Neff R, Song W-M, Wang M, Wang Q, Arnold M, et al. Integrative metabolomics-genomics approach reveals key metabolic pathways and regulators of Alzheimer’s disease. Alzheimers Dement 2022;18:1260–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nho K, Kueider-Paisley A, Arnold M, MahmoudianDehkordi S, Risacher SL, Louie G, et al. Serum metabolites associated with brain amyloid beta deposition, cognition and dementia progression. Brain Commun 2021;3:fcab139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morais LH, Schreiber HL, Mazmanian SK. The gut microbiota–brain axis in behaviour and brain disorders. Nat Rev Microbiol 2020;19:241–55. [DOI] [PubMed] [Google Scholar]
- 19.Sochocka M, Donskow-Łysoniewska K, Diniz BS, Kurpas D, Brzozowska E, Leszek J. The Gut Microbiome Alterations and Inflammation-Driven Pathogenesis of Alzheimer’s Disease—a Critical Review. Molecular Neurobiology 2019;56:1841–51. 10.1007/s12035-018-1188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baloni P, Funk CC, Yan J, Yurkovich JT, Kueider-Paisley A, Nho K, et al. Metabolic Network Analysis Reveals Altered Bile Acid Synthesis and Metabolism in Alzheimer’s Disease. Cell Rep Med 2020;1:100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wörheide MA, Krumsiek J, Nataf S, Nho K, Greenwood AK, Wu T, et al. An integrated molecular Atlas of Alzheimer’s disease. bioRxiv 2021. 10.1101/2021.09.14.21263565. [DOI] [Google Scholar]
- 22.Varma VR, Wang Y, An Y, Varma S, Bilgel M, Doshi J, et al. Bile acid synthesis, modulation, and dementia: A metabolomic, transcriptomic, and pharmacoepidemiologic study. PLoS Med 2021;18:e1003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMillin M, Grant S, Frampton G, Petrescu AD, Kain J, Williams E, et al. FXR-Mediated Cortical Cholesterol Accumulation Contributes to the Pathogenesis of Type A Hepatic Encephalopathy. Cell Mol Gastroenterol Hepatol 2018;6:47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MahmoudianDehkordi S, Arnold M, Nho K, Ahmad S, Jia W, Xie G, et al. Altered bile acid profile associates with cognitive impairment in Alzheimer’s disease-An emerging role for gut microbiome. Alzheimers Dement 2019;15:76–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nho K, Kueider-Paisley A, Ahmad S, MahmoudianDehkordi S, Arnold M, Risacher SL, et al. Association of Altered Liver Enzymes With Alzheimer Disease Diagnosis, Cognition, Neuroimaging Measures, and Cerebrospinal Fluid Biomarkers. JAMA Netw Open 2019;2:e197978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol 2014;30:332–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Wei R, Xie G, Arnold M, Kueider-Paisley A, Louie G, et al. Peripheral serum metabolomic profiles inform central cognitive impairment. Sci Rep 2020;10:14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, et al. Gut-microbiota-targeted diets modulate human immune status. Cell 2021;184:4137–53.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasior M, Rogawski MA, Hartman AL. Neuroprotective and disease-modifying effects of the ketogenic diet. Behav Pharmacol 2006;17:431–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maalouf M, Rho JM, Mattson MP. The neuroprotective properties of calorie restriction, the ketogenic diet, and ketone bodies. Brain Res Rev 2009;59:293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neth BJ, Mintz A, Whitlow C, Jung Y, Sai KS, Register TC, et al. Modified ketogenic diet is associated with improved cerebrospinal fluid biomarker profile, cerebral perfusion, and cerebral ketone body uptake in older adults at risk for Alzheimer’s disease: a pilot study. Neurobiology of Aging 2020;86:54–63. 10.1016/j.neurobiolaging.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagpal R, Neth BJ, Wang S, Craft S, Yadav H. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer’s disease markers in subjects with mild cognitive impairment. EBioMedicine 2019;47:529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagpal R, Neth BJ, Wang S, Mishra SP, Craft S, Yadav H. Gut mycobiome and its interaction with diet, gut bacteria and alzheimer’s disease markers in subjects with mild cognitive impairment: A pilot study. eBioMedicine 2020;59:102950. 10.1016/j.ebiom.2020.102950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gauglitz JM, Bittremieux W, Williams CL, Weldon KC, Panitchpakdi M, Di Ottavio F, et al. Reference data based insights expand understanding of human metabolomes. bioRxiv 2020:2020.07.08.194159. 10.1101/2020.07.08.194159. [DOI] [Google Scholar]
- 36.Gonzalez A, Navas-Molina JA, Kosciolek T, McDonald D, Vázquez-Baeza Y, Ackermann G, et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods 2018;15:796–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol 2016;34:828–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nothias L-F, Petras D, Schmid R, Dührkop K, Rainer J, Sarvepalli A, et al. Feature-based molecular networking in the GNPS analysis environment. Nat Methods 2020;17:905–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martino C, Morton JT, Marotz CA, Thompson LR, Tripathi A, Knight R, et al. A Novel Sparse Compositional Technique Reveals Microbial Perturbations. mSystems 2019;4. 10.1128/mSystems.00016-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martino C, Shenhav L, Marotz CA, Armstrong G, McDonald D, Vázquez-Baeza Y, et al. Context-aware dimensionality reduction deconvolutes gut microbial community dynamics. Nat Biotechnol 2021;39:165–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dey N, Wagner VE, Blanton LV, Cheng J, Fontana L, Haque R, et al. Regulators of gut motility revealed by a gnotobiotic model of diet-microbiome interactions related to travel. Cell 2015;163:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bittremieux W, Avalon NE, Thomas SP, Kakhkhorov SA, Aksenov AA, Gomes PWP, et al. Open Access Repository-Scale Propagated Nearest Neighbor Suspect Spectral Library for Untargeted Metabolomics. bioRxiv 2022:2022.05.15.490691. 10.1101/2022.05.15.490691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guzior DV, Quinn RA. Review: microbial transformations of human bile acids. Microbiome 2021;9:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morton JT, Aksenov AA, Nothias LF, Foulds JR, Quinn RA, Badri MH, et al. Learning representations of microbe–metabolite interactions. Nat Methods 2019;16:1306–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strandwitz P, Kim KH, Terekhova D, Liu JK, Sharma A, Levering J, et al. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol 2019;4:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell 2018;174:497. 10.1016/j.cell.2018.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 2016;65:426–36. [DOI] [PubMed] [Google Scholar]
- 48.Govindpani K, Calvo-Flores Guzmán B, Vinnakota C, Waldvogel HJ, Faull RL, Kwakowsky A. Towards a Better Understanding of GABAergic Remodeling in Alzheimer’s Disease. Int J Mol Sci 2017;18. 10.3390/ijms18081813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazzoli R, Pessione E. The Neuro-endocrinological Role of Microbial Glutamate and GABA Signaling. Front Microbiol 2016;7:1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dahlin M, Elfving A, Ungerstedt U, Amark P. The ketogenic diet influences the levels of excitatory and inhibitory amino acids in the CSF in children with refractory epilepsy. Epilepsy Res 2005;64:115–25. [DOI] [PubMed] [Google Scholar]
- 51.Olson CA, Iñiguez AJ, Yang GE, Fang P, Pronovost GN, Jameson KG, et al. Alterations in the gut microbiota contribute to cognitive impairment induced by the ketogenic diet and hypoxia. Cell Host Microbe 2021;29:1378–92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012;487:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007;3:211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metagenomic sequencing data are available in Qiita under study ID 13662 (36). Mass spectrometry data (.mzXML format) are available in MassIVE under ID MSV000087087. The classical molecular networking job is available in GNPS at the following link: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=90f1ba6e1e4d4d89b75b9017a0631983 (37). The feature-based molecular networking job is available in GNPS at the following link: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=0b828a2a6f224fb293de1fe39bee5fc5 (38). The code utilized for these analyses are available at https://github.com/ahdilmore/BEAM_MultiOmics.