Abstract
Rare germline pathogenic variants in cancer‐predisposing genes have a high impact and potential for clinical utility. In the last 30 years, based on evidence of cancer risk associated with germline pathogenic variants, several measures have been suggested for personalized medicine, including the development of novel treatments, treatment stratification, risk reduction by surgical measures, chemoprevention, removal of environmental factors, and surveillance for early detection among specific high‐risk individuals. However, this evidence is mainly based on evaluations of European populations. Our large‐scale analyses of more than 100,000 individuals, including 14 disease cases and non‐cancer controls in the Japanese population, suggest some discrepancies in the associations between cancer‐predisposing genes and diseases, expansion of the targeted diseases of BRCA1 and BRCA2, and a potential novel risk‐reduction measure for gastric cancer. They are likely to be explained by population and region variations; therefore, more population‐wide and region‐wide research could provide improved personalized medicine as well as a better understanding of disease mechanisms. This review summarizes current personalized medicine and discusses the potential use of germline pathogenic variants.
Keywords: cancer, germline pathogenic variant, personalized medicine, risk estimation, risk reduction
We have analyzed germline pathogenic variants in 27 cancer‐predisposing genes among more than 100,000 Japanese individuals, including 14 disease cases and non‐cancer controls. This review summarizes current personalized medicine and discusses the potential use of germline pathogenic variants.
Abbreviations
- ACMG/AMP
American College of Medical Genetics and Genomics and Association for Molecular Pathology
- BRCA1/2
BRCA1 and BRCA2
- ENIGMA
Evidence‐based Network for the Interpretation of Germline Mutant Alleles
- GPV
germline pathogenic variant
- H. pylori
Helicobacter pylori
- NCCN
National Comprehensive Cancer Network
- PARP
poly(ADP‐ribose)polymerase
- VUS
variant of unknown significance
1. INTRODUCTION
In the past 30 years, there have been significant improvements in cancer management, particularly in the treatment of certain types of cancer. 1 This management promotes pairing individuals with cancer with drugs that target specific somatic variants in their tumor, aiming to produce long‐lasting remission and extend their survival. 2 In addition, germline variants, naturally possessing individual variations, also play essential roles in cancer management. Developing risk reduction and surveillance strategies based on germline variants for both those who have not yet developed the disease and those for which it has been developed would improve cancer management. 1
Many genetic analyses have identified thousands of germline variants associated with cancer; however, most variants with common allele frequencies confer relatively small increments in risk (1.1‐ to 1.5‐fold). 3 By contrast, rare germline variants have a high impact (two‐fold or more) and potential for high clinical utility. 3 In this review, we summarize current personalized medicine and discuss the potential use of GPVs.
2. CURRENT PERSONALIZED MEDICINE THROUGH GPVs
The first step toward personalized medicine based on germline variants is to identify genes that are associated with the onset of disease and to clarify the risk, prevalence, and clinical characteristics of GPV carriers in cancer‐predisposing genes. Based on this evidence, further studies have been performed to develop novel treatment strategies and risk‐reduction methods using preventive surgery, chemoprevention, removal of environmental factors, and surveillance. Some measures have already been established with sufficient evidence and clinically used, whereas evidence is still accumulating for other strategies.
2.1. Estimation of cancer risk and GPV prevalence
Most cancer‐predisposing genes, such as BRCA1/2, have been identified by linkage analyses, a traditional genome‐wide scan with many families, including affected individuals, and functional candidate gene studies from decades ago. The risk of disease has been estimated from prospective cohort studies that follow individuals with GPVs, and case–control studies that compare the prevalence of GPVs among patients and individuals without the disease. In 1994, BRCA1/2 were identified as breast cancer susceptibility genes. 4 , 5 , 6 Relationship between BRCA1/2 and the risk of breast cancer have received widespread attention because of the celebrity work done in 2013. 7 The latest established evidence was reported as follows: in 2017, from a prospective cohort study including ~10,000 GPV carriers in BRCA1/2 of mainly European populations, the cumulative risks for breast cancer to age 80 were 72% for BRCA1 and 69% for BRCA2. 8 In 2021, two population‐based studies, containing approximately 30,000 9 and 60,000 10 women with breast cancer in mainly European populations, provided prevalence and genetic risk in BRCA1/2 and other cancer‐predisposing genes (ATM, BARD1, CDH1, CHEK2, RAD51C, RAD51D, PALB2, and TP53). 9 , 10 In an analysis of 10,000 patients with cancer across 33 diseases, 8% had GPVs and 21 genes were found to be associated with one or multiple diseases. 11 Evidence has been still accumulating over the past 30 years since the use of genetic identification, even in well known relationships, such as that of breast cancer, although BRCA1/2 is already in clinical use as described below.
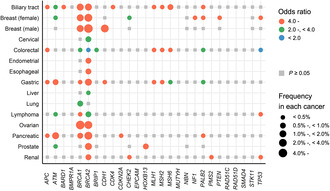
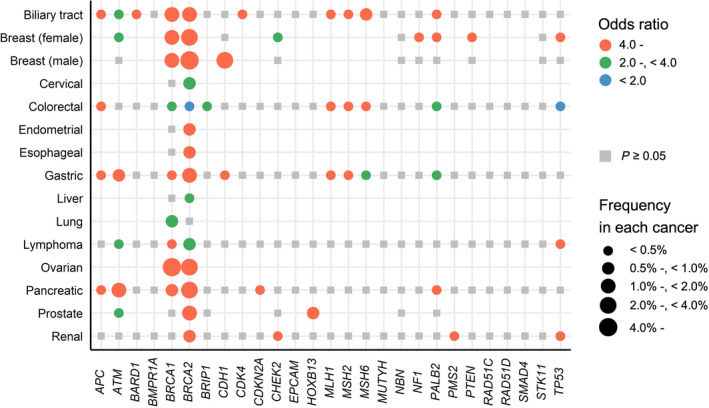
These reports are based mainly based on data from European populations in the USA and Europe, and it is difficult to apply them without precise examination to Japan because of the differences in GPV prevalence across populations 12 and incidence rates of the disease across regions. 13 Therefore, we started a comprehensive new project in 2015 to reveal the associations between cancer risk and GPVs in cancer‐predisposing genes in more than 100,000 Japanese individuals, including 14 disease cases and non‐cancer controls. 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 The summary of the associations in our previous study is shown in Figure 1. The prevalence of GPVs in BRCA1/2 was especially large, and the odds ratios for many diseases were mostly 4.0 or more. To compare our results with global evidence, the current evidence of NCCN guidelines 23 , 24 , 25 in May 2023 is summarized in Figure 2. We observed only half of the associations in the Japanese population among those with strong or very strong evidence according to the NCCN guidelines and vice versa. This can be explained by two main reasons. First, the associations described in the NCCN guidelines are biologically true; however, their prevalence is very low in patients. CDH1 and STK11 cause syndromes whose GPVs show a higher incidence of breast cancer. 26 , 27 However, the prevalence of GPVs was very low in the Japanese population (0.03% and 0.01%) 14 among breast cancer patients, and association analysis did not show significant differences. This is also applicable to European populations (0.02% and 0.01%). 10 Second, the associations may be identified in diseases that are common in a specific population or region. We found that GPVs in BRCA1 and/or BRCA2 were associated with biliary tract, esophageal, and gastric cancer risk, which have several‐fold higher incidence rates in East Asian countries. 13 , 19 These results suggest that information on the prevalence and risk observed in each population and region is indispensable for better personalized medicine.
FIGURE 1.
Summary of associations of germline pathogenic variants in 27 cancer‐predisposing genes among BioBank Japan. The associations of germline pathogenic variants in 27 cancer‐predisposing genes and cancer risk identified in our previous studies (breast cancer, 14 prostate cancer, 15 colorectal cancer, 16 pancreatic cancer, 17 renal cancer, 18 lymphoma, 20 biliary tract cancer, 21 gastric cancer, 22 and 14 diseases for BRCA1/2 19 ) from BioBank Japan are shown. Unanalyzed associations are indicated by empty fields and associations that did not reach P < 0.05 are indicated by gray squares.
FIGURE 2.
Summary of evidence of germline pathogenic variants in 27 cancer‐predisposing genes in the National Comprehensive Cancer Network guidelines in May 2023. The evidence of germline pathogenic variants in 27 cancer‐predisposing genes has been extracted from the National Comprehensive Cancer Network (NCCN) guidelines. 23 , 24 , 25 The evidence level in the NCCN guidelines is as follows: very strong: prospective cohort studies in a population‐based setting have demonstrated risk; strong: traditional case–control studies or more than three case–control studies including those with cases ascertained by commercial laboratories or those without controls from the same population (traditional case–control study: a retrospective study that compressed patients with a disease or specific outcome with patients without the disease or outcome). 23 , 24 , 25
2.2. Changing treatment strategies
Identifying the association between the onset of disease and GPVs has also changed treatment strategies. By the beginning of the 21st century, BRCA1/2 was shown to play a key role in homologous recombination repair. 28 Following studies demonstrated the synthetic lethality of PARP inhibitors in homologous recombination‐deficient cells. 29 , 30 , 31 When homologous recombination is disrupted and PARP is inhibited, replication fork collapse cannot be repaired, and the cell undergoes cell death. 29 , 30 , 31 These findings led to a novel paradigm shift in cancer treatment. After the first clinical trials, 32 many clinical trials have reported the efficacy of PARP inhibitors and widely recommend the treatment in a variety of neoplasms—breast, ovarian, pancreatic, and prostate cancer. 24 This is a great example of how the identification of cancer‐risk genes could clarify the pathogenesis and introduce a novel treatment agent.
In addition to the development of therapeutic agents, treatment stratification may change based on the clinical characteristics of GPV carriers. Prostate cancer risk is associated with GPVs in homologous recombination genes. 15 Patients with GPVs in BRCA1/2 showed more aggressive prostate cancer phenotypes with a higher probability of nodal involvement and distant metastasis. 33 Regarding the Japanese population, GPV carriers in homologous recombination genes were also associated with a short time to castration resistance and poor overall survival. 34 These studies indicate that GPVs may also have potential as prognostic factors, not only as cancer‐predisposing risk factors. Depending on the prognosis, better timing for genetic testing, more optimal treatment intensity, and more optimal post‐treatment follow‐up can occur.
Thus, the accumulation of clinical characteristics of GPV carriers might lead to further development of personalized medicine in terms of novel treatments and treatment stratification.
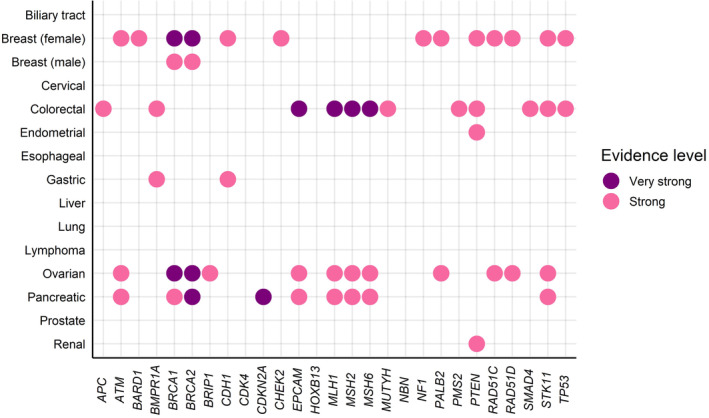
2.3. Risk reduction
Based on risk estimation among GPV carriers, some surgical measures are being considered for GPV carriers of some genes, including bilateral mastectomy for breast cancer, 24 salpingo‐oophorectomy for ovarian cancer risk, 24 colectomy for colorectal cancer, 25 and gastrectomy for gastric cancer 23 with various evidence for levels of recommendation (Table 1). Such surgical measures reduce the risk of developing cancer in the relevant organs and might improve prognosis. 35 However, several points should be considered. First, there is insufficient evidence on whether such surgical measures can improve the prognosis of some cancers. Even in breast cancer, a meta‐analysis showed that risk‐reducing bilateral mastectomy reduces the development of breast cancer; however, it was not clearly shown to reduce all‐cause mortality. 36 This might be due to limited long‐term follow‐up or evaluation. Second, careful decisions must be made regarding the appropriate timing and side effects. Regarding salpingo‐oophorectomy, it induces premature menopause with potential short‐term effects (hot flushes and impaired sexual functioning) 37 and long‐term effects (risk of osteoporosis, cardiovascular disease, and cognitive decline). 38 Fertility preservation is also an important theme. 39 Finally, it is also important to address the psychosocial effect and quality‐of‐life aspects of undergoing such risk‐reducing surgical measures. A previous study in the USA reported that 70% of women were satisfied with risk‐reducing mastectomy and would choose it again; by contrast, 11% of women were neutral in their response, and 19% of women were dissatisfied with this procedure. 40 Healthcare professionals need to provide GPV carriers with the available information and encourage them to take time to consider all options currently available. 40
TABLE 1.
Evidence of surgical measures for risk reduction in the National Comprehensive Cancer Network guidelines in May 2023.
| Cancer | Gene | Measures | Evidence level | Age |
|---|---|---|---|---|
| Breast | BRCA1 | Mastectomy | Discuss | – |
| Breast | BRCA2 | Mastectomy | Discuss | – |
| Breast | CDH1 | Mastectomy | Discuss | – |
| Breast | PALB2 | Mastectomy | Discuss | – |
| Breast | STK11 | Mastectomy | Discuss | – |
| Ovarian | BRCA1 | Salpingo‐oophorectomy a | Recommend | 35–40 years old b |
| Ovarian | BRCA2 | Salpingo‐oophorectomy a | Recommend | 35–45 years old b |
| Ovarian | BRIP1 | Salpingo‐oophorectomy a | Recommend | 45–50 years old |
| Ovarian | PALB2 | Salpingo‐oophorectomy a | Consider | >45 years old |
| Ovarian | RAD51C | Salpingo‐oophorectomy a | Recommend | 45–50 years old |
| Ovarian | RAD51D | Salpingo‐oophorectomy a | Recommend | 45–50 years old |
| Colorectal | APC | Colectomy | Consider–Recommend | – |
| Gastric | CDH1 | Gastrectomy | Recommend | 18–40 years old |
Risks and benefits of premature surgical menopause versus the risk of cancer and family history should be carefully considered, and the guidelines recommend that patients seek expert care. 24
Salpingo‐oophorectomy is typically recommended at 35–40 years of age and upon completion of childbearing. Because ovarian cancer onset in patients with pathogenic variant in BRCA2 is an average of 8–10 years later than that in patients with pathogenic variant in BRCA1, it is reasonable to delay salpingo‐oophorectomy for the management of ovarian cancer risk until the age of 40–45 years in patients with pathogenic variant in BRCA2 unless the age at diagnosis in the family warrants earlier age for consideration of surgery. 24
Another approach is chemoprevention. A double‐blinded, randomized controlled trial reported that individuals with Lynch syndrome who received a daily 600 mg aspirin developed lower rates of colorectal cancer than ones who received a placebo. 41 Therefore, in the NCCN guidelines, all individuals with Lynch syndrome who have a risk of future colorectal cancer are recommended to consider using aspirin daily. 25 There may be some other candidates such as a selective estrogen receptor modulator for invasive breast cancer in postmenopausal individuals with GPVs in BRCA1/2 and non‐steroidal anti‐inflammatory drugs for polyp regression in individuals with GPVs in APC although the evidence is insufficient. 24 , 25 The accumulation of evidence about effectiveness among GPV carriers and long‐term evaluation is also required for application in clinical settings.
Risk reduction can also be achieved by removing certain environmental factors, due to the excess risk from the interaction between germline variants and environmental factors. One instance is that of alcohol consumption and an East Asian specific common variant (rs671) in ALDH2 showing heterozygotic (Glu/Lys) and homozygotic (Lys/Lys) individuals of the derived allele have lower than 50% and <1–4% of the wild‐type enzymatic activity. 42 Alcohol consumption and rs671 in ALDH2 interact with upper aerodigestive tract cancer and lead to excess risk; estimated cumulative risks through 80 years among individuals with heavy drinking habits were 20.2% in Glu/Lys genotype and 2.9% in the wild‐type; by contrast, they were less than 3% among individuals with moderate drinking habits regardless of genotype status. 43 This suggests that modification of alcohol consumption according to the ALDH2 genotype has a large impact on upper aerodigestive tract cancer‐risk reduction. 43 Another recent instance we reported is H. pylori infection and GPVs in homologous recombination genes in gastric cancer. 22 H. pylori infection is a well known gastric cancer risk. In our recent study, GPVs in homologous recombination genes interacted with H. pylori infection to excessively increase the risk of gastric cancer. 22 The estimated cumulative risks through 85 years among individuals with H. pylori infection were 45.5% in GPV carriers in homologous recombination genes and 14.4% in GPV non‐carriers, while they were less than 5% among individuals without H. pylori infection regardless of GPV carrier status, indicating that GPVs in homologous recombination genes seems to boost the risk of H. pylori infection‐related gastric cancer. 22 This is probably due to genome instability caused by H. pylori infection that contributes to gastric carcinogenesis. 22 , 44 By contrast, there was no apparent interaction of GPVs with other environmental factors such as smoking habits or sodium intake. 22 These indicate that GPV carriers in homologous recombination genes need more attention, especially for H. pylori infection. The above excess risks could have been clarified because of evaluations in East Asia, where the fact that the genetic variation largely influencing acetaldehyde‐oxidizing capacities is polymorphic 45 contributes to the former and a region‐specific highly virulent type of H. pylori 46 to the latter. These examples illustrate the importance of population‐ and region‐wide analyses to provide better personalized medicine and to elucidate the novel pathogenesis of the disease.
It is crucial to accumulate evidence regarding risk‐reduction measures and share information with the individuals concerned so that they can make informed decisions.
2.4. Surveillance
Surveillance is considered for early detection among specific high‐risk individuals. There is accumulating evidence concerning the surveillance of GPV carriers. First, GPV carriers in BRCA1/2 are recommended for surveillance of breast cancer for early detection using MRI. 24 This recommendation is based on the largely increased risk of breast cancer and the sensitivity with MRI that is significantly higher than mammography in females with GPVs in BRCA1/2. 47 Second, GPV carriers in MLH1, MSH2, MSH6, and PMS2 are recommended regular colonoscopies from young ages. 25 In particular, GPV carriers in MLH1 or MSH2 are considered for high‐quality colonoscopies from their twenties because of their high risk. 25 In these ways, it is necessary to accumulate evidence on the sensitivity and specificity of surveillance that is established in the general population for high‐risk individuals, and evidence on risk and age of presentation according to gene should be accumulated.
Although many evaluations have attempted to improve survival rates through early detection, there are some cautions to consider in surveillance. Even if surveillance improves survival rates, it should be carefully evaluated to determine whether it truly reduces excess mortality. 48 , 49 This is because it can be biased by earlier diagnosis without postponing the time of death (lead‐time bias) or increased detection of indolent cancers (length bias), although the survival rate is undoubtedly an essential parameter of prognosis. 48 , 49 Furthermore, excessive surveillance recommendations may not only add to the burden in terms of cost but may also exacerbate the physical and psychological burden of GPV carriers. Thus, it is essential to remember, “All screening programs do harm; some do good as well, and, of these, some do more good than harm at a reasonable cost.” 50 Balanced discussions based on evidence should be continued.
3. CHALLENGES FOR BROADENED PERSONALIZED MEDICINE
Here, we describe the use of personalized medicine in GPVs. Expansion of the evaluation for each population or region would provide more insight into personalized medicine. Several remaining challenges need to be addressed, especially from the perspective of author expertise, genetics, expansion of other genes that establish evidence to be associated with cancer risk, better annotation of the clinical importance of variants, and optimization of target individuals who would benefit from genetic testing.
3.1. Identification of further cancer‐risk genes
Only a small number of genes among ~20,000 genes are still being utilized in personalized medicine. These genes were mainly identified using traditional linkage analyses with limited statistical power and subsequent functional candidate gene studies. Because the prevalence of GPVs is very low, GPVs cannot be identified by genome‐wide association studies, and sequencing analyses are needed. In addition, ~18,000 and 10,000 cases are required to detect an odds ratio of 2.0 and 4.0, respectively (simulated: a case–control ratio 1:3, power 0.80, a prevalence of GPVs in the controls 0.1%, and P = 1.0 × 10−4 in burden tests). 51 Therefore, whole‐genome or whole‐exome sequencing with tens of thousands of samples is required to identify new genes. However, only a few diseases have been analyzed on this scale. 52 An alternative efficient approach would be to analyze only candidate genes found in genome‐wide association studies in a higher number of samples because the enrichment of rare variants is mainly identified in such genes, although whole‐exome sequencing is conducted. 53 Also, because of founder variants or gene–environmental interactions, there may be a possibility that population‐wide and region‐wide evaluation would increase detection power and identify important genes, as the importance of CHEK2 was raised by a European‐specific founder pathogenic variant (c.1100delC).
3.2. Better annotation of the clinical importance of variants
Sequencing analysis identifies genetic variants, but not all of them are GPVs. It is critical to identify GPVs; however, many germline variants remain VUS. One of the greatest challenges in personalized medicine for germline variants is the annotation of non‐synonymous variants. In addition, loss‐of‐function variants are not always GPVs (ex. c.4068_4071dupGATT in MSH6), nor all ClinVar interpretations were not true (ex. c.1744C>G in MLH1), which we have experienced in our study with Japanese population. 16 Even among the GPVs, there are variants with ambiguous functional defects that show intermediate cancer risk (ex. c.5096G>A in BRCA1) 54 and variants in a specific location of the amino acid sequence showing different characteristics of the disease (ex. promoter 1B in APC). 25 , 55
To overcome these challenges, functional and in silico assays have been conducted to evaluate VUS, 56 , 57 , 58 , 59 , 60 , 61 and guidelines or consensuses have been developed for determining the clinical interpretation of variants. According to the American College of Medical Genetics and Genomics and Association for Molecular Pathology guidelines, which provides interpretative categories of sequence variants and an algorithm for interpretation. 62 In addition, there are classification criteria developed by members of the consortium according to the Clinical Genome Resource, 63 such as the Evidence‐based Network for the Interpretation of Germline Mutant Alleles Consortium for BRCA1/2. 64 These guidelines or criteria allow us to separate variants into clinical significance (Figure 3A,B). Nevertheless, most germline variants remain uncertain (among 4804 variants in 27 cancer‐predisposing genes in approximately 36,000 individuals: 84.8% 16 ; among 1810 variants in BRCA1/2 in approximately 100,000 individuals: 54.4%). 19
FIGURE 3.
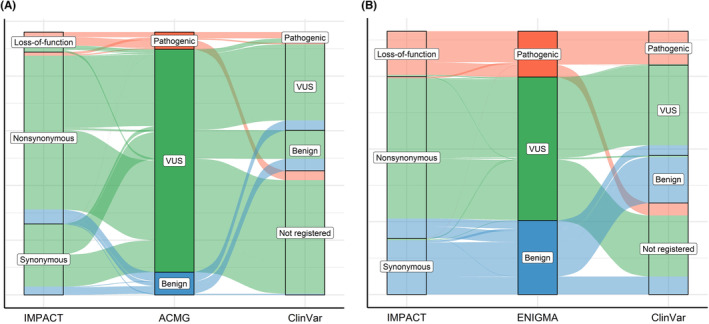
Difference in clinical importance detected by three methods. The annotation of germline variants in our previous studies is shown. 16 , 19 (A) In total, 4804 variants in 27 cancer‐predisposing genes in ~36,000 individuals were annotated using ACMG/AMP guidelines, information on ClinVar interpretation, and IMPACT of amino acid sequences. (B) In total, 1810 variants in BRCA1/2 in ~100,000 individuals were annotated using the ENIGMA consensus criteria, information on ClinVar interpretation, and IMPACT of the amino acid sequence. ACMG/AMP, American College of Medical Genetics and Genomics and Association for Molecular Pathology; ENIGMA, Evidence‐based Network for the Interpretation of Germline Mutant Alleles.
Any ambiguity or misclassification of the significance of variants may lead to incorrect medical care. Further evaluation of these variants will provide valuable information for personalized medicine.
3.3. Optimization of target individuals benefit from genetic testing
In the United States Consensus Conference, germline testing has been expanding for metastatic prostate cancer or for all patients with a personal history of breast cancer. 65 , 66 These expansions might be extended to individuals unaffected by the disease. In addition, risk elaboration combining GPVs with other factors, such as the polygenic risk score, family history, and environmental factors, has also been considered. 67 , 68 This will allow for more detailed risk estimation of the disease (Figure 4). The average risk estimated for all GPV carriers may divide individuals into relatively high‐risk and low‐risk groups by risk elaboration. CanRisk, 69 an online tool that enables healthcare professionals to calculate an individual's future risk of breast and ovarian cancers, has been developed for use in clinical research, although the current version is only applicable to European populations. Thus, personalized medicine that considers GPVs is being optimized for individuals who can benefit from it and will continue to expand.
FIGURE 4.
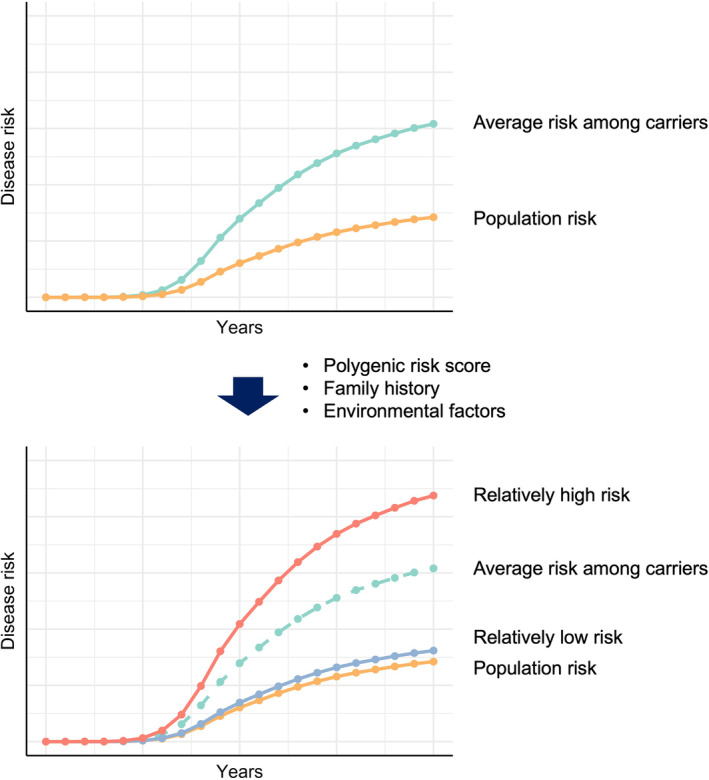
Scheme of detailed risk stratification among germline pathogenic variant carriers. The average risk estimated for all germline pathogenic variant carriers may divide individuals into relatively high‐risk and low‐risk groups by risk elaboration.
Several ethical, social, and legal challenges remain to be addressed. When receiving the genetic result, concerns and potential harms include increased emotional burden on the individual and family, information complexity, and difficulty communicating probabilistic information. 70 Moreover, we should focus on the proper and just use of genetic data for both the individual and society, recognizing how the data might be used to discriminate unfairly now or in the future. 71 Counseling and education systems that enable people to return genomic information are also desirable. It is necessary to establish a system to ensure that benefits are widely and equally available, rather than only to a limited number of individuals.
4. POSSIBILITY FOR FURTHER PERSONALIZED MEDICINE
Here, we discussed current and future topics about personalized medicine from the perspective of common cancers from GPVs. Genetic testing and risk reduction measures have received widespread attention because of the celebrity work done in 2013. 7 However, most of the current genetic evidence has been established mainly in European populations. Expansion of the evaluation for each population or region may clarify novel associated diseases and risk‐reduction measures. 19 , 22 Although caution should be exercised in dealing with a harmful response to GPVs overreaction, germline evaluation could be an effective approach to developing further personalized medicine.
FUNDING INFORMATION
This research was supported by Japan Agency for Medical Research and Development (AMED) under Grant No. JP19kk0305010, JP22ama121015, and JP23ck0106805.
CONFLICT OF INTEREST STATEMENT
The author declares no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by an Institutional Reviewer Board: N/A.
Informed Consent: N/A.
Registry and the Registration No. of the study/trial: N/A.
Animal Studies: N/A.
ACKNOWLEDGMENTS
We acknowledge the staff of the Laboratory for Genotyping Development at RIKEN for their helpful comments and discussions.
Usui Y, Momozawa Y. Personalized medicine with germline pathogenic variants: Importance of population‐ and region‐wide evidence. Cancer Sci. 2023;114:3816‐3824. doi: 10.1111/cas.15922
REFERENCES
- 1. Turnbull C, Sud A, Houlston RS. Cancer genetics, precision prevention and a call to action. Nat Genet. 2018;50:1212‐1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prasad V. Perspective: the precision‐oncology illusion. Nature. 2016;537:S63. [DOI] [PubMed] [Google Scholar]
- 3. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wooster R, Neuhausen SL, Mangion J, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12‐13. Science. 1994;265:2088‐2090. [DOI] [PubMed] [Google Scholar]
- 5. Miki Y, Swensen J, Shattuck‐Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66‐71. [DOI] [PubMed] [Google Scholar]
- 6. Futreal PA, Liu Q, Shattuck‐Eidens D, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120‐122. [DOI] [PubMed] [Google Scholar]
- 7. Desai S, Jena AB. Do celebrity endorsements matter? Observational study of BRCA gene testing and mastectomy rates after Angelina Jolie's New York times editorial. BMJ. 2016;355:i6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402‐2416. [DOI] [PubMed] [Google Scholar]
- 9. Hu C, Hart SN, Gnanaolivu R, et al. A population‐based study of genes previously implicated in breast cancer. N Engl J Med. 2021;384:440‐451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dorling L, Carvalho S, Allen J, et al. Breast cancer risk genes ‐ association analysis in more than 113,000 women. N Engl J Med. 2021;384:428‐439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang KL, Mashl RJ, Wu Y, et al. Pathogenic germline variants in 10,389 adult cancers. Cell. 2018;173:355‐370.e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoshida R. Hereditary breast and ovarian cancer (HBOC): review of its molecular characteristics, screening, treatment, and prognosis. Breast Cancer. 2021;28:1167‐1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209‐249. [DOI] [PubMed] [Google Scholar]
- 14. Momozawa Y, Iwasaki Y, Parsons MT, et al. Germline pathogenic variants of 11 breast cancer genes in 7,051 Japanese patients and 11,241 controls. Nat Commun. 2018;9:4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Momozawa Y, Iwasaki Y, Hirata M, et al. Germline pathogenic variants in 7636 Japanese patients with prostate cancer and 12 366 controls. J Natl Cancer Inst. 2020;112:369‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fujita M, Liu X, Iwasaki Y, et al. Population‐based screening for hereditary colorectal cancer variants in Japan. Clin Gastroenterol Hepatol. 2022;20:2132‐2141.e2139. [DOI] [PubMed] [Google Scholar]
- 17. Mizukami K, Iwasaki Y, Kawakami E, et al. Genetic characterization of pancreatic cancer patients and prediction of carrier status of germline pathogenic variants in cancer‐predisposing genes. EBioMedicine. 2020;60:103033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sekine Y, Iwasaki Y, Aoi T, et al. Different risk genes contribute to clear cell and non‐clear cell renal cell carcinoma in 1532 Japanese patients and 5996 controls. Hum Mol Genet. 2022;31:1962‐1969. [DOI] [PubMed] [Google Scholar]
- 19. Momozawa Y, Sasai R, Usui Y, et al. Expansion of cancer risk profile for BRCA1 and BRCA2 pathogenic variants. JAMA Oncol. 2022;8:871‐878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Usui Y, Iwasaki Y, Matsuo K, et al. Association between germline pathogenic variants in cancer‐predisposing genes and lymphoma risk. Cancer Sci. 2022;113:3972‐3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okawa Y, Iwasaki Y, Johnson TA, et al. Hereditary cancer variants and homologous recombination deficiency in biliary tract cancer. J Hepatol. 2023;78:333‐342. [DOI] [PubMed] [Google Scholar]
- 22. Usui Y, Taniyama Y, Endo M, et al. Helicobacter pylori, homologous‐recombination genes, and gastric cancer. N Engl J Med. 2023;388:1181‐1190. [DOI] [PubMed] [Google Scholar]
- 23. National Comprehensive Cancer Network . Gastric cancer. (Version 2) 2022. Accessed May 10, 2023. https://www.nccn.org/professionals/physician_gls/pdf/gastric.pdf
- 24. National Comprehensive Cancer Network . Genetic/familial high‐risk assessment: breast, ovarian, and pancreatic. (Version 2) 2023. Accessed May 10, 2023. https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf
- 25. National Comprehensive Cancer Network . Genetic/familial high‐risk assessment: colorectal (Version 2). 2022. Accessed May 10, 2023. https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf
- 26. Hansford S, Kaurah P, Li‐Chang H, et al. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1:23‐32. [DOI] [PubMed] [Google Scholar]
- 27. Hearle N, Schumacher V, Menko FH, et al. Frequency and spectrum of cancers in the Peutz‐Jeghers syndrome. Clin Cancer Res. 2006;12:3209‐3215. [DOI] [PubMed] [Google Scholar]
- 28. Venkitaraman AR. Functions of BRCA1 and BRCA2 in the biological response to DNA damage. J Cell Sci. 2001;114:3591‐3598. [DOI] [PubMed] [Google Scholar]
- 29. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917‐921. [DOI] [PubMed] [Google Scholar]
- 30. Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2‐deficient tumours with inhibitors of poly(ADP‐ribose) polymerase. Nature. 2005;434:913‐917. [DOI] [PubMed] [Google Scholar]
- 31. Curtin NJ, Szabo C. Poly(ADP‐ribose) polymerase inhibition: past, present and future. Nat Rev Drug Discov. 2020;19:711‐736. [DOI] [PubMed] [Google Scholar]
- 32. Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP‐ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123‐134. [DOI] [PubMed] [Google Scholar]
- 33. Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748‐1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kimura H, Mizuno K, Shiota M, et al. Prognostic significance of pathogenic variants in BRCA1, BRCA2, ATM and PALB2 genes in men undergoing hormonal therapy for advanced prostate cancer. Br J Cancer. 2022;127:1680‐1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eleje GU, Eke AC, Ezebialu IU, Ikechebelu JI, Ugwu EO, Okonkwo OO. Risk‐reducing bilateral salpingo‐oophorectomy in women with BRCA1 or BRCA2 mutations. Cochrane Database Syst Rev. 2018;8:CD012464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li X, You R, Wang X, et al. Effectiveness of prophylactic surgeries in BRCA1 or BRCA2 mutation carriers: a meta‐analysis and systematic review. Clin Cancer Res. 2016;22:3971‐3981. [DOI] [PubMed] [Google Scholar]
- 37. Vermeulen RFM, Beurden MV, Korse CM, Kenter GG. Impact of risk‐reducing salpingo‐oophorectomy in premenopausal women. Climacteric. 2017;20:212‐221. [DOI] [PubMed] [Google Scholar]
- 38. Rocca WA, Gazzuola‐Rocca L, Smith CY, et al. Accelerated accumulation of multimorbidity after bilateral oophorectomy: a population‐based cohort study. Mayo Clin Proc. 2016;91:1577‐1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang X, Niu J, Che T, Zhu Y, Zhang H, Qu J. Fertility preservation in BRCA mutation carriers‐efficacy and safety issues: a review. Reprod Biol Endocrinol. 2020;18:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frost MH, Schaid DJ, Sellers TA, et al. Long‐term satisfaction and psychological and social function following bilateral prophylactic mastectomy. JAMA. 2000;284:319‐324. [DOI] [PubMed] [Google Scholar]
- 41. Burn J, Sheth H, Elliott F, et al. Cancer prevention with aspirin in hereditary colorectal cancer (Lynch syndrome), 10‐year follow‐up and registry‐based 20‐year data in the CAPP2 study: a double‐blind, randomised, placebo‐controlled trial. Lancet. 2020;395:1855‐1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen CH, Ferreira JC, Gross ER, Mochly‐Rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev. 2014;94:1‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koyanagi YN, Ito H, Oze I, et al. Development of a prediction model and estimation of cumulative risk for upper aerodigestive tract cancer on the basis of the aldehyde dehydrogenase 2 genotype and alcohol consumption in a Japanese population. Eur J Cancer Prev. 2017;26:38‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Imai S, Ooki T, Murata‐Kamiya N, et al. Helicobacter pylori CagA elicits BRCAness to induce genome instability that may underlie bacterial gastric carcinogenesis. Cell Host Microbe. 2021;29(6):941‐958.e10. [DOI] [PubMed] [Google Scholar]
- 45. Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci U S A. 1984;81:258‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hayashi T, Senda M, Suzuki N, et al. Differential mechanisms for SHP2 binding and activation are exploited by geographically distinct helicobacter pylori CagA Oncoproteins. Cell Rep. 2017;20:2876‐2890. [DOI] [PubMed] [Google Scholar]
- 47. Passaperuma K, Warner E, Causer PA, et al. Long‐term results of screening with magnetic resonance imaging in women with BRCA mutations. Br J Cancer. 2012;107:24‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ellis L, Woods LM, Estève J, Eloranta S, Coleman MP, Rachet B. Cancer incidence, survival and mortality: explaining the concepts. Int J Cancer. 2014;135:1774‐1782. [DOI] [PubMed] [Google Scholar]
- 49. Welch HG, Schwartz LM, Woloshin S. Are increasing 5‐year survival rates evidence of success against cancer? JAMA. 2000;283:2975‐2978. [DOI] [PubMed] [Google Scholar]
- 50. Gray JA, Patnick J, Blanks RG. Maximising benefit and minimising harm of screening. BMJ. 2008;336:480‐483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Easton DF, Pharoah PD, Antoniou AC, et al. Gene‐panel sequencing and the prediction of breast‐cancer risk. N Engl J Med. 2015;372:2243‐2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Q, Dhindsa RS, Carss K, et al. Rare variant contribution to human disease in 281,104 UK biobank exomes. Nature. 2021;597:527‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Backman JD, Li AH, Marcketta A, et al. Exome sequencing and analysis of 454,787 UK biobank participants. Nature. 2021;599:628‐634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Spurdle AB, Whiley PJ, Thompson B, et al. BRCA1 R1699Q variant displaying ambiguous functional abrogation confers intermediate breast and ovarian cancer risk. J Med Genet. 2012;49:525‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li J, Woods SL, Healey S, et al. Point mutations in exon 1B of APC reveal gastric adenocarcinoma and proximal polyposis of the stomach as a familial adenomatous polyposis variant. Am J Hum Genet. 2016;98:830‐842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bouwman P, van der Gulden H, van der Heijden I, et al. A high‐throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013;3:1142‐1155. [DOI] [PubMed] [Google Scholar]
- 57. Takagi M, Yoshida M, Nemoto Y, et al. Loss of DNA damage response in Neuroblastoma and utility of a PARP inhibitor. J Natl Cancer Inst. 2017. doi: 10.1093/jnci/djx062 [DOI] [PubMed] [Google Scholar]
- 58. Findlay GM, Daza RM, Martin B, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217‐222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kleiblova P, Stolarova L, Krizova K, et al. Identification of deleterious germline CHEK2 mutations and their association with breast and ovarian cancer. Int J Cancer. 2019;145:1782‐1797. [DOI] [PubMed] [Google Scholar]
- 60. Ikegami M, Kohsaka S, Ueno T, et al. High‐throughput functional evaluation of BRCA2 variants of unknown significance. Nat Commun. 2020;11:2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Frazer J, Notin P, Dias M, et al. Disease variant prediction with deep generative models of evolutionary data. Nature. 2021;599:91‐95. [DOI] [PubMed] [Google Scholar]
- 62. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. ClinGen Clinical Genome Resource . Accessed May 10, 2023. https://www.clinicalgenome.org/
- 64. Spurdle AB, Healey S, Devereau A, et al. ENIGMA—Evidence‐based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat. 2012;33:2‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Giri VN, Knudsen KE, Kelly WK, et al. Implementation of germline testing for prostate cancer: Philadelphia prostate cancer consensus conference 2019. J Clin Oncol. 2020;38:2798‐2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Manahan ER, Kuerer HM, Sebastian M, et al. Consensus guidelines on genetic' testing for hereditary breast cancer from the American Society of Breast Surgeons. Ann Surg Oncol. 2019;26:3025‐3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nyberg T, Brook MN, Ficorella L, et al. CanRisk‐prostate: a comprehensive, externally validated risk model for the prediction of future prostate cancer. J Clin Oncol. 2023;41:1092‐1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee A, Mavaddat N, Wilcox AN, et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet Med. 2019;21:1708‐1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. University of Cambridge . Accessed May 10th, 2023. https://www.canrisk.org/
- 70. Hunter CL, Helft PR. Yes, we can, but should we? Ethical considerations in reporting germline findings from paired tumor‐Normal genomic testing in patients with advanced cancer. J Clin Oncol. 2023;41:1982‐1985. [DOI] [PubMed] [Google Scholar]
- 71. Seaver LH, Khushf G, King NMP, et al. Points to consider to avoid unfair discrimination and the misuse of genetic information: a statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2022;24:512‐520. [DOI] [PubMed] [Google Scholar]


