Abstract
Lipids are a major component of extracellular vesicles; however, their significance in tumorigenesis and progression has not been well elucidated. As we previously found that lipid profiles drastically changed in breast tumors upon progression, we hypothesized that lipid profiles of plasma‐derived extracellular vesicles could be utilized as breast cancer biomarkers. Here, we adopted modified sucrose cushion ultracentrifugation to isolate plasma‐derived extracellular vesicles from breast cancer (n = 105), benign (n = 11), and healthy individuals (n = 43) in two independent cohorts (n = 126 and n = 33) and conducted targeted lipidomic analysis. We established a breast cancer diagnostic model comprising three lipids that showed favorable performance with the area under the receiver operating characteristic curve of 0.759, 0.743, and 0.804 in the training, internal validation, and external test sets, respectively. Moreover, we identified several lipids that could effectively discriminate breast cancer progression and subtypes: phosphatidylethanolamines and phosphatidylserines were relatively higher in Stage III, whereas phosphatidylcholines and sphingomyelins were higher in Stage IV; phosphatidylcholines and ceramides were correspondingly concentrated in HER2‐positive patients, while lysophosphatidylcholines and polyunsaturated triglycerides were concentrated in the triple‐negative breast cancer subtype. Lipid profiling of plasma‐derived extracellular vesicles is a non‐invasive and promising approach for diagnosing, staging, and subtyping breast cancer.
Keywords: biomarker, breast cancer, extracellular vesicles, lipidomics, liquid biopsy
This study investigated the use of lipid profiles in plasma‐derived extracellular vesicles as non‐invasive biomarkers for diagnosing, staging, and subtyping breast cancer. Lipid profiling effectively discriminated breast cancer progression and subtypes, suggesting its potential as a promising approach to breast cancer management.
Abbreviations
- AUC
area under the ROC curve
- BC
breast cancer
- Cers
ceramides
- CUC
cushion ultracentrifugation
- EVs
extracellular vesicles
- FAs
fatty acids
- LPA
lysophosphatidic acid
- LysoPCs
lysophosphatidylcholines
- NTA
nanoparticle tracking analysis
- PCs
phosphatidylcholines
- PEs
phosphatidylethanolamines
- PIs
phosphatidylinositols
- PSs
phosphatidylserines
- QC
quality control
- ROC
receiver operating characteristic
- SEC
size exclusion chromatography
- SMs
sphingomyelins
- TEM
transmission electron microscopy
- TGs
triacylglycerols
- THCs
trihexosylceramides
- UC
ultracentrifugation
- WB
western blotting
1. INTRODUCTION
Over the past decades, early screening and precision medicine have improved the survival of patients with BC. 1 Although mammography and ultrasonography are commonly used for the early detection of BC, their accessibility and accuracy vary with region, population, and age distribution; thus, there is a high demand for more accessible and cost‐effective options. 2 , 3 , 4 Tumor biopsy is essential for precision treatment, but liquid biopsy based on biofluid samples, such as circulating tumor cells, DNA, or EVs, is gaining popularity because of its good compliance, easy specimen acquisition, and dynamic monitoring capabilities. 5
EVs, also referred to as “exosomes,” are 30–150 nm vesicles formed by the inward budding of late endosomes and released into the extracellular environment upon fusion with the plasma membrane. 6 EVs can also be released into blood vessels, carrying many molecules that can regulate the biological activities of cells at distant sites. 7 Several studies have confirmed that proteins, miRNAs, and long RNAs in blood‐derived EVs could be biomarkers for early diagnosis, therapeutic efficacy prediction, and prognosis evaluation in BC patients. 8 , 9 , 10 However, whether lipid profiles of blood‐derived EVs can be used for these purposes remains unclear. Most importantly, standardized methods for analyzing lipids in EVs have not been well established.
We previously demonstrated that phospholipids in human breast tumors alter their fatty acid (FA) composition, probably due to the dysregulation of tumor lipid metabolism. This alteration was particularly evident in phosphatidylinositols (PIs), and we identified the enrichment of PI_C38:3 in tumor cells as a potential biomarker for BC immune status. 11 Based on this observation, we hypothesized that lipid dysregulation in tumor cells could be transferred to those of tumor‐derived EVs and that they could be used for diagnosing or differentiating BC.
2. MATERIALS AND METHODS
2.1. Sample collection
In total, 159 blood samples from two cohorts were used in this experiment, comprising 126 cases in Cohort 1 and 33 cases in Cohort 2. Samples for both cohorts were obtained from the same biobank in the Kyoto University Hospital. The samples were selected in a consecutive manner: Cohorts 1 and 2 comprised samples primarily collected during 2016 and 2017, respectively. The inclusion criteria in the study were as follows: (1) Histologically proven BC with surgically operable or recurrent/metastatic lesions; (2) Mammography or ultrasonography screening confirmed‐BC‐free individuals. Patients with a history of other tumors or dyslipidemia were excluded. The study was approved by the Ethics Committee of Kyoto University Hospital, and written informed consent was obtained from all participants before sample collection. Blood samples were collected before cancer‐related treatment with a definite pathological diagnosis at the Breast Cancer Unit, Kyoto University Hospital. Plasma was separated by centrifugation at 1800 × g for 10 min at room temperature (RT, 23°C) in EDTA vacutainer tubes, aliquoted, and stored at −80°C.
2.2. EV isolation
The plasma sample (500 μL) was thawed and centrifuged (4000 × g, 15 min, RT). The supernatant was collected to isolate EVs using the following three methods with some adjustments. 12 , 13 , 14
2.2.1. Protocol 1: Ultracentrifugation
Plasma was diluted 1:2 in PBS and centrifuged (20,000 × g, 60 min, 4°C). The supernatant was centrifuged (120,000 × g, 90 min, 4°C) using an ultracentrifuge (Optima XE‐100, Beckman Coulter) in 4 mL tubes (#344062, Beckman Coulter) with an SW60Ti rotor. The pellets were washed with PBS and centrifuged again (120,000 × g, 70 min, 4°C). The purified EVs were resuspended in 100 μL of PBS and stored at −80°C until use.
2.2.2. Protocol 2: Sucrose cushion ultracentrifugation
Plasma was diluted 1:2 in PBS and centrifuged (20,000 × g, 60 min, 4°C). The supernatant was then centrifuged (120,000 × g, 90 min, 4°C) on 375 μL of Tris/sucrose/D2O solution (30% sucrose cushion). Next, 300 μL of sucrose cushion was collected, diluted with PBS, and centrifuged again (120,000 × g, 70 min, 4°C). The EVs were stored as described in protocol 1.
2.2.3. Protocol 3: Size exclusion chromatography
Plasma was centrifuged (20,000 × g, 60 min, 4°C) without dilution. The qEV 70 nm column (iZON Science) was equilibrated with PBS before use. Then, 500 μL of plasma was loaded on the top of the column; the first 3.0 mL of the elute was discarded. EV‐rich fractions (1.5 mL, 7–9 fractions) were collected and concentrated (7500 × g, 30 min, RT) using Amicon Ultra‐2 10 kDa centrifugal filters (Merck Millipore, Co. Cork). EVs were collected and stored as previously described.
2.3. Transmission electron microscopy
In total, 10 μL of EVs were placed on 200‐mesh copper grids (#6511, Nisshin EM) and allowed to settle for 10 min. The EVs were blotted and negatively stained with three successive drops of 1.0% uranyl acetate for 10 s‐10 s‐60 s. The grids were air dried and imaged using a TEM (H‐7650, Hitachi) operating at 80 kV.
2.4. Nanoparticle tracking analysis
For size and particle concentration analysis, EVs were diluted with PBS at a ratio of 1:100 and examined via NTA using a NanoSight NS300 system (Malvern Instruments) configured with a 488 nm laser as per the manufacturer's instructions. Data were analyzed using NTA 3.4 software.
2.5. Western blotting
EV quantification was performed using the Qubit™ Protein Assay Kit (#Q33211, Thermo Fisher Scientific) and a Qubit® 3.0 Fluorometer (Thermo Fisher Scientific). Equal amounts of EV proteins were separated by SDS‐PAGE and transferred onto PVDF membranes. The membranes were blocked with 5% nonfat dry milk for 1 h at RT and incubated with antibodies overnight at 4°C. The membranes were then incubated with secondary antibodies for 1 h at RT. The following antibodies were used: CD9 (#AHS0902, Invitrogen), TSG101 (#ab125011, Abcam), Hsp90α (#CA1023, Sigma‐Aldrich), ApoB (#20578‐1‐AP, Proteintech), ApoE (#66830‐1‐lg, Proteintech), Mouse IgG (#31430, Invitrogen), and Rabbit IgG (#31460, Invitrogen).
2.6. Lipid extraction
Lipids were extracted from EVs (5 μg) using a modified Bligh‐Dyer method as follows. 15 EVs were incubated in an extraction solution of chloroform and methanol (1:2) for 10 min at RT. Chloroform and 1% acetic acid were then added sequentially at a 1:1 ratio. The mixture was centrifuged (2000 × g, 10 min, RT), and an equal volume of the lower organic phase was vacuum‐dried for 1 h using a SpeedVac system. The lipid extracts were re‐dissolved in a methanol:chloroform:water (8:4:1) solution and injected for LC/MS analysis.
2.7. LC/MS analysis
LC analysis was performed on a Nextera (Shimadzu) ultra‐high‐performance liquid chromatography system with a TSKgel Amide‐80 column (150 × 2.0 mm, 3 μm, Tosoh) in hydrophilic interaction liquid chromatography mode. The mobile phase comprised A = ultrapure water (Milli‐Q; Millipore) solution and B = acetonitrile: water = 95:5 (10 mM ammonium formate +0.12% formic acid, pH 3) solution. The gradient program was as follows: 96%B (0 min), 96%B (3 min), 70%B (15 min), 96%B (15.01 min), and stop (20 min). The flow rate was set at 0.2 mL/min, and the column temperature was maintained at 40°C. A triple–quadruple mass spectrometer (LCMS‐8060, Shimadzu) coupled with an LC system was operated in positive electron spray ionization in multiple reaction monitoring (MRM) mode for targeted lipidomics analysis. Lipid detection was performed using an in‐house MRM library (Shimadzu). Table S1 summarizes the list of targeted lipids and MRM transitions. LabSolutions LCMS software (version 5.53, Shimadzu) was used to acquire data and analyze the peaks automatically.
2.8. Data processing and statistical analyses
Equal volumes of lipid extracts from EVs were pooled as QC samples, which were measured intermittently to monitor the performance of the LC/MS system. Systematical error removal using random forest was applied to reduce the system error and correct the signal drift (Figure S1). 16 Data were processed with MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/). Briefly, lipids were removed with >80% missing values in all samples; the missing values of residual lipids were replaced with 1/5 of the minimum positive value. Lipids with a relative standard deviation over 30% in QC samples were filtered, and samples were normalized by sum. Finally, the average of repeated measurements of the same specimen was used for subsequent analyses.
Statistical analyses were performed using GraphPad version 8.0.2, SPSS Statistics v26, and R version 4.2.0. Principal component analysis (PCA), partial least squares discriminant analysis (PLS‐DA), and heatmaps were generated using MetaboAnalyst 5.0. Candidate biomarkers were screened by variable importance projection (VIP) > 1, |log2 fold change| > log2 1.2, and p‐value < 0.05 (Wilcoxon Mann–Whitney U test). A volcano plot was constructed using Metware Cloud (https://cloud.metware.cn). The least absolute shrinkage and selection operator (LASSO) regression was adopted to narrow down the lipid biomarkers (glmnet, R package) and create a predictive model based on the training set. The ROC curve and AUC were used to evaluate the performance of the diagnostic panel (pROC, R package). The independent samples t test, Mann–Whitney U test, one‐way ANOVA, or Kruskal–Wallis test was used for comparison between groups.
3. RESULTS
3.1. Comparison of the yield and purity of plasma‐derived EVs isolated by UC, CUC, and SEC
Plasma samples contained various nanoparticles, including lipoproteins that are similar in size to the EVs. The lipid profiles of these lipoproteins may be influenced by dietary fat intake and interfere with the subsequent lipidomic analysis of plasma‐derived EVs. Thus, minimizing lipoprotein contamination to ensure accurate results is mandatory. We compared three methods to isolate plasma‐derived EVs: UC‐based, CUC‐based, and SEC‐based methods. The detailed procedure for each method is shown in Figure 1A. In TEM images, EVs typically appeared as cup‐shaped vesicles (Figure 1B, red arrows). Although we could detect EVs using all three methods, the SEC method contained many small spherical particles in the background, which indicated lipoprotein contamination (Figure 1B, blue arrows). Compatible with the observation in TEM, NTA analysis showed that the SEC method was greater in particle concentration (71.83 × 109/mL in SEC, 8.437 × 109/mL in CUC, and 3.683 × 109/mL in UC) and smaller in particle size (83.07 nm in SEC, 115.9 nm in CUC and 117.9 nm in UC) (Figure 1C). The minimal information for studies of EVs 2018 guidelines was used for EV identification, where CD9, TSG101, and Hsp90α were recommended as specific EV markers. Additionally, the apolipoproteins ApoB and ApoE, which commonly occur in lipoprotein particles, were used as markers for lipoprotein contamination. 17 While WB for CD9, TSG101, and Hsp90α confirmed the presence of EVs in all three methods, high ApoB and ApoE expression in the SEC method supported the massive contamination of lipoprotein (Figure 1D). The yield and purity of EVs tended to be higher in CUC than in UC, suggesting that the CUC method is the best way to isolate plasma‐derived EVs for lipidomics with minimal lipoprotein contamination and excellent efficacy. Thus, we decided to employ the CUC method to purify the EVs in subsequent experiments.
FIGURE 1.
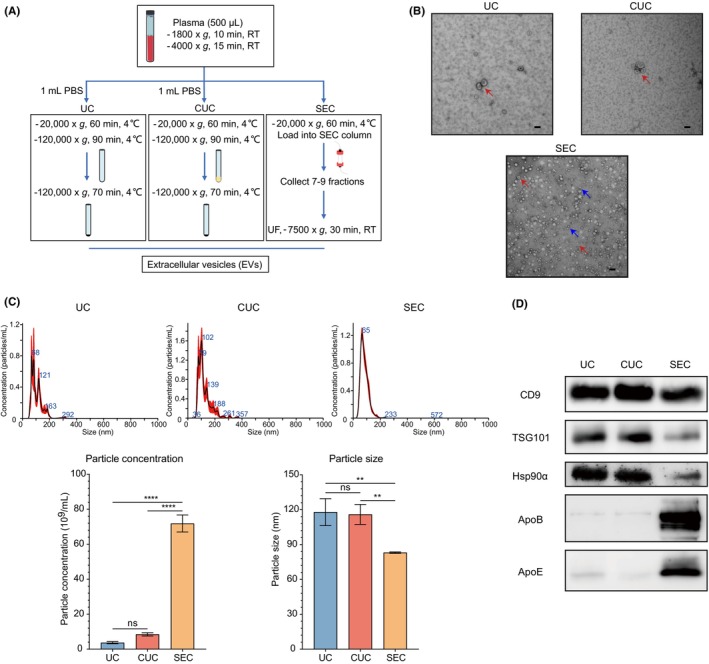
Comparison of the yield and purity of plasma‐derived extracellular vesicles (EVs) isolated by ultracentrifugation (UC), cushion ultracentrifugation (CUC), and size exclusion chromatography (SEC). (A) Workflow for isolating plasma‐derived EVs by UC, CUC, and SEC. (B) Representative transmission electron microscopy images of plasma‐derived EVs isolated by UC, CUC, and SEC. Scale bar: 100 nm. The red and blue arrows indicate EVs and lipoproteins, respectively. (C) Distribution of particle concentration and size of plasma‐derived EVs isolated by UC, CUC, and SEC. (D) Western blotting of EV‐related proteins (CD9, TSG101, and Hsp90α) and lipoproteins (ApoB and ApoE). Numerical data are shown as means ± SD of three independent experiments. ns, not significant. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
3.2. Effects of different postprandial times on characteristics of plasma‐derived EVs
Although research on the effects of dietary intake on EV concentration in humans is limited, plasma lipoprotein levels increase slightly upon the dietary fat intake. 18 , 19 Therefore, to explore the effect of dietary intake on the stability of plasma‐derived EVs, we compared the characteristics of plasma‐derived EVs using blood collected at three different time points from a healthy volunteer, at fasting, 3 h, and 6 h after a meal. TEM showed that the shape of the EVs was intact without large variations across three different time points and that no lipoproteins appeared in the background (Figure 2A). The concentration and size of plasma‐derived EVs showed no significant differences across the three time points (fasting, 3 h, and 6 h) (Figure 2B). Moreover, ApoB and ApoE were hardly detected in the WB, which confirmed that there was no fluctuation in contamination due to changes in lipoproteins and postprandial times in the plasma‐derived EVs (Figure 2C). The above results demonstrate that the effect of dietary fat intake on plasma‐derived EVs was minimal as long as we used the CUC method for EV purification.
FIGURE 2.
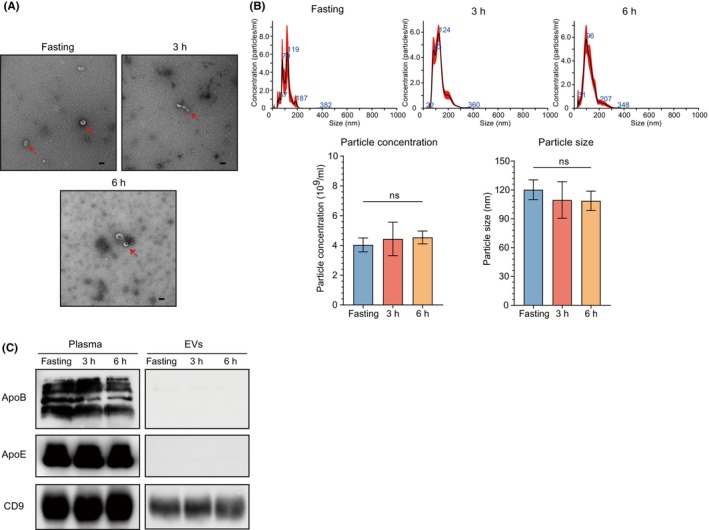
Effects of different postprandial times on characteristics of plasma‐derived extracellular vesicles (EVs). (A) Representative transmission electron microscopy images of plasma‐derived EVs at different time points (fasting, 3 h, and 6 h after a meal). Scale bar: 100 nm. The red arrows indicate EVs. (B) Distribution of particle concentration and size of plasma‐derived EVs at different time points. (C) Western blotting for assessment fluctuation of EV‐related protein (CD9) and lipoproteins (ApoB and ApoE) at three different time points. Numerical data are shown as means ± SD of three independent experiments. ns, not significant. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
3.3. Lipid profiles of plasma‐derived EVs can discriminate BC patients from benign and healthy individuals although the difference is unclear
To discover the different lipid profiles of plasma‐derived EVs between BC patients and controls (benign and healthy individuals), we performed targeted lipidomics covering 380 lipids using plasma‐derived EVs collected from 126 cases (33 healthy volunteers, 11 benign tumor patients, and 82 BC patients) (Cohort 1). The BC patients included 15 cases of ductal carcinoma in situ (DCIS), 35 cases of Stage I/II, 17 cases of Stage III, and 15 cases of Stage IV. The detailed clinicopathological information is shown in Table S2. The study design and data processing procedures are illustrated in Figure 3A. As a result, 186 lipids were retained for subsequent analysis.
FIGURE 3.
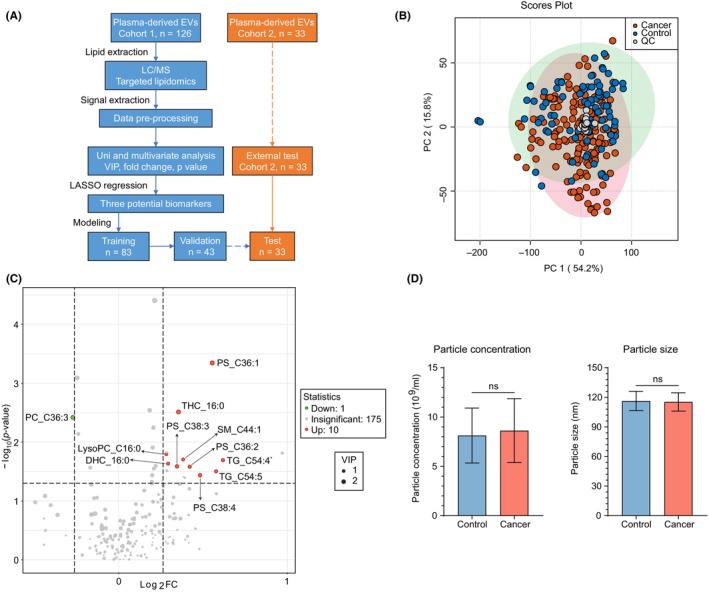
Lipid profiles of plasma‐derived EVs can discriminate BC patients from benign and healthy individuals although the difference is unclear. (A) Diagram of study design and data processing procedures. Cohort 1 (controls = 44, BC = 82) and Cohort 2 (healthy = 10, BC = 23) were two experiments performed independently. The blue text box refers to data from Cohort 1, and the orange text box refers to data from Cohort 2. (B) Unsupervised principal component analysis plot of lipids in Cohort 1 with quality control samples. LC/MS measurements were duplicated per sample. Each plot represents one measurement. (C) Volcano plot of lipids with differential concentrations between BC and control samples in Cohort 1. Red and green dots represent the upregulated and downregulated lipids that meet the criteria of variable importance projection > 1, |log2 fold change| > log2 1.2, and p‐value < 0.05, respectively. TG_C54:4* represents TG_C54:4 isomer‐1. (D) Distribution of particle concentration and size of plasma‐derived EVs isolated from BC and control samples in Cohort 1. Numerical data are shown as means ± SD of each group. ns, not significant. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
The QC samples were tightly clustered in the unsupervised PCA model, indicating the stability of the experimental system (Figure 3B). In the supervised PLS‐DA model, patients with BC were somewhat separated from controls, indicating different lipid profiles between the two groups (Figure S2a). Cross‐validation (10‐fold) and permutation tests (100 times) were used to prevent overfitting and ensure prediction accuracy. The optimal number of principal components was determined to be 1 (R 2 = 0.190 and Q 2 = 0.055) with p‐value < 0.01 (Figure S2b,c). As the PLS‐DA model showed suboptimal performance, we incorporated additional univariate analysis techniques, including fold change and U test, to further screen candidate biomarkers. The volcano plot identified 11 lipids with differential concentrations between BC patients and controls (Figure 3C), which comprised six glycerophospholipids, three sphingolipids, and two glycerolipids. Of these, 10 lipids showed higher concentrations and only one lipid showed a lower concentration in BC patients than in controls (Table S3). Although the concentration and size of EVs reportedly differ between patients with BC and healthy participants, 8 we found no significant difference in either the concentration or size of EVs between the BC patients and controls among the 126 cases (Cohort 1) (Figure 3D).
3.4. Lipid profiles of plasma‐derived EVs associated with clinicopathological characteristics of BC
We explored the association between lipid profiles of plasma‐derived EVs and disease progression, including subtypes, in the 126 cases. Disease progression was defined as control, DCIS, Stage I/II, Stage III, and Stage IV, according to clinicopathological characteristics. The heatmap shows the distribution of 53 lipids with statistically significant differences in concentration among the groups (Figure 4A). Most of these were higher in invasive diseases than in the control and DCIS, whereas Cer_d18:1/17:0, LysoPC_C20:0e, TG_C48:0, and TG_C50:0 had the highest concentrations in DCIS. Saturated or mono‐unsaturated PCs, PEs, and PSs were most abundant at Stage III. In contrast, the levels of polyunsaturated PCs, polyunsaturated TGs, LysoPCs, and SMs are elevated upon further progression. Interestingly, the polyunsaturated PIs, PI_C36:2, PI_C38:3, and PI_C38:4, showed similar trends to our previous observations in breast tumors, where they were lower in DCIS and higher in invasive diseases with metastasis (Figure S3a).
FIGURE 4.
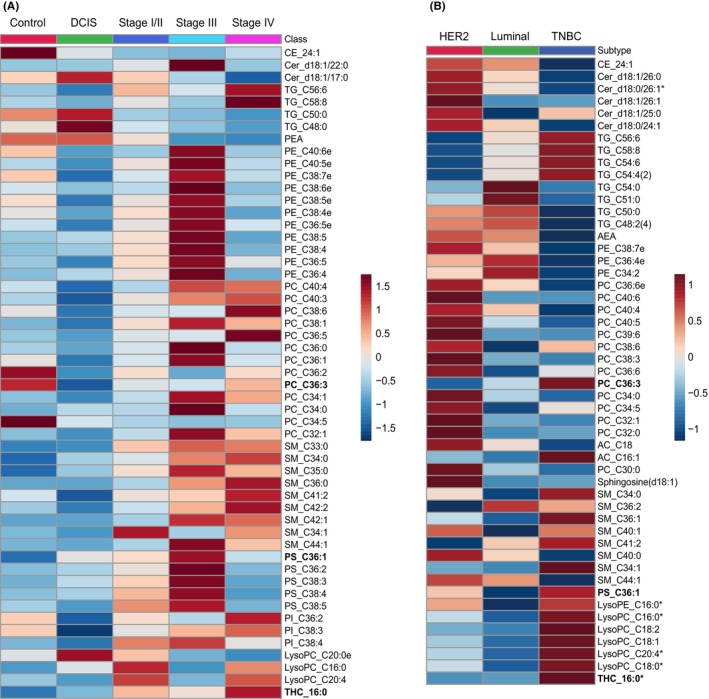
Lipid profiles of plasma‐derived extracellular vesicles (EVs) associated with clinicopathological characteristics of breast cancer (BC). (A) Heatmap of lipid concentrations with statistical significance among control, DCIS, Stage I/II, Stage III, and Stage IV samples in Cohort 1. (B) Heatmap of lipid concentrations between Luminal, HER2‐positive, and triple‐negative breast cancer (TNBC) subtypes in Cohort 1. Lipids significantly different among the groups were labeled with “*”; the top 50 lipids with the lowest p‐values and the lipids in the diagnostic model are used to generate the heatmap. PC_C36:3, PS_C36:1, and THC_16:0 are represented in bold. Each column represents the average concentration of specific lipids for a particular group.
The lipid profiles of plasma‐derived EVs vary according to their molecular subtype. Although a few lipids exhibited statistically significant differences, we could see the distribution patterns as follows: PEs and saturated‐TGs were relatively higher in the Luminal subtype, while PCs and Cers were higher in the HER2‐positive subtype, and polyunsaturated TGs, LysoPCs, and SMs were higher in the triple‐negative breast cancer (TNBC) subtype (Figure 4B). These findings imply that lipid metabolism is different between each molecular subtype of BC, which is reflected in the lipid profiles of EVs.
3.5. Discovery of lipid profiles in plasma‐derived extracellular vesicles as biomarkers for breast cancer diagnosis
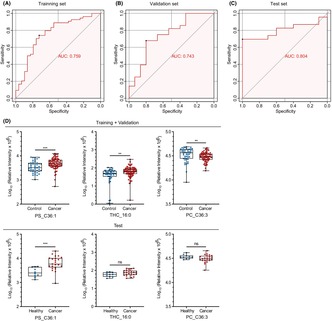
To establish a biomarker panel that could be utilized for BC screening, LASSO regression analysis was performed with 11 lipids in 126 cases (Cohort 1). We applied 10‐fold cross‐validation to evaluate the model's optimal parameters (i.e. lambda) and optimized the lipid panel. This analysis was repeated 25 times to ensure robustness and repeatability (Figure S4). Ultimately, three lipids were selected to generate a potential biomarker panel for BC screening, with strong statistical validity (Tables S4 and S5). The three lipids were PS_C36:1, THC_16:0, and PC_C36:3. Next, we divided the 126 cases into a training set (n = 83) and an internal validation set (n = 43) at a 2:1 ratio using a stratified random sampling method; another cohort (Cohort 2) of 33 cases (10 healthy volunteers and 23 BC patients) was used as an external test set (Figure S5; Figure 3A). The details of Cohorts 1 and 2 are described in Table S2. The AUC values in the training, internal validation, and external test sets were 0.759 (sensitivity, 0.741; specificity, 0.724), 0.743 (sensitivity, 0.679; specificity, 0.800), and 0.804 (sensitivity, 0.696; specificity, 1.000), respectively (Figure 5A–C). PS_C36:1 showed a significantly higher concentration in BC patients across the two cohorts (Figure 5D). As for the other two lipids, THC_16:0 and PC_C36:3, their differences in concentration within the test set did not reach statistical significance. However, the trends were similar between the two cohorts. Additionally, this diagnostic model exhibited promising discriminatory power between DCIS and controls with the AUC values of 0.711 (sensitivity, 0.733; specificity, 0.773), indicating its potential for early detection of BC (Table S6). Nevertheless, significant differences were not observed in the expression levels of PS_C36:1, THC_16:0, and PC_C36:3 across age groups and T1–4 stages among the population with cancer in Cohort 1 (Figure S3b). Some lipids that were not used in the model (PS_C36:2, PS_C38:3, PS_C38:4, DHC_16:0, and SM_C44:1) showed higher concentrations in patients with BC in both cohorts (Table S3). Isolation and lipidomic analysis of plasma‐derived EVs in Cohorts 1 and 2 were performed independently, which increased the reliability and validity of the results. Our findings suggest that the lipid profiles of plasma‐derived EVs can provide supportive information for BC screening.
FIGURE 5.
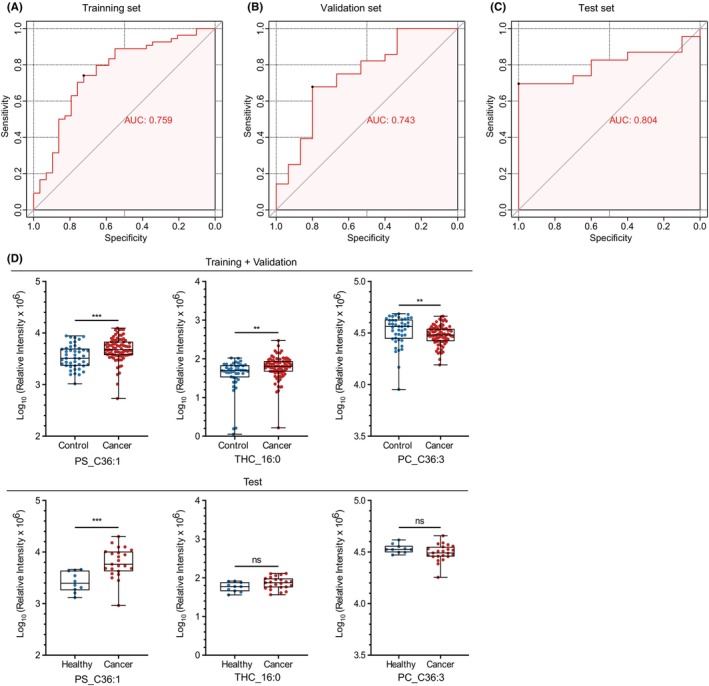
Discovery of lipid profiles in plasma‐derived extracellular vesicles as biomarkers for breast cancer diagnosis. (A–C) Receiver operating characteristic (ROC) curve and area under the ROC curve (AUC) of the diagnostic panel using the three lipid combination in the training, internal validation, and external test sets. (D) Boxplots depicting the concentrations of the three lipids, PS_C36:1, THC_16:0, and PC_C36:3, in the training + validation and external test sets. The box range represents the interquartile range, and the median is indicated by a line across the box. ns, not significant. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
4. DISCUSSION
To date, most studies on EV‐related biomarkers have focused on proteins and RNAs, and there is no standardized method for the lipidomic analysis of EVs. Here, we demonstrated that the modified CUC method, as described by Scott and Suchi, is appropriate for isolating plasma‐derived EVs for lipidomic analysis. 13 , 20 Furthermore, we highlight the importance of lipid profiles in understanding BC diagnosis, progression, and subtyping.
Although UC and SEC methods are currently used extensively for isolating plasma‐derived EVs, the low UC yield and poor SEC purity have been widely concerned by scholars. 21 , 22 , 23 , 24 , 25 , 26 In line with this issue, we observed severe contamination of lipoprotein, especially in the SEC method, which probably affected downstream multi‐omics analysis. Furthermore, to assess the stability of plasma‐derived EVs and to exclude the effect of diet‐induced changes in lipoproteins, such as the rise in chylomicrons and VLDLs 3–6 h after meals, 27 , 28 we examined the properties and purity of EVs in plasma samples drawn at various intervals. However, similar to the conclusion reached by Simin, 18 we found no apparent changes in the morphology, concentration, or size of plasma‐derived EVs over time when using the CUC method. Additionally, there was no increase in lipoprotein contamination over time.
As a result, we generated a BC diagnosis model with three lipids and achieved favorable results. Unlike previous BC biomarker studies that included only patients with invasive carcinoma, 8 , 9 we enrolled patients with benign tumors and DCIS in Cohort 1 to develop the biomarker model, which made it possible to better detect BC at an earlier stage. However, including DCIS and benign tumors in Cohort 1 might have resulted in a lower diagnostic performance of the panel compared with Cohort 2, which did not include DCIS or benign tumors.
Among the three lipids in the panel, the elevation of PS_C36:1 and THC_16:0 has been reported in the blood or tumors of various cancers, such as bladder, kidney, lung, and breast cancers. 29 , 30 , 31 , 32 , 33 The association between elevated plasma THC_16:0 levels and cancer progression has been observed in prostate cancer and oral squamous cell carcinoma. 32 , 33 The increase of PS_C36:1 and THC_16:0 in cancers is reasonable as their structures contain saturated and mono‐unsaturated FAs synthesized through the de novo FA synthesis pathway, which is frequently elevated in cancer cells. 34 Additionally, the exposure of PS on the surface of cancer cells or cancer cell‐derived microvesicles suppresses immune reaction by inhibiting T‐cell activation via G protein‐coupled receptor 174 or diminishing T‐cell receptor signaling cascade of CD4+ and CD8+ T‐cells. 35 The reduction in PC_C36:3 has also been reported in the plasma of patients with kidney, prostate, and breast cancers, as well as in BC tissue. 36 , 37 The trends of these three lipids in our panel were similar to the results of previous studies, showing an increase in PS_C36:1 and THC_16:0 and a decrease in PC_36:3 in plasma‐derived EVs from BC patients.
Changes in lipid metabolism are intensified upon BC progression in response to an altered tumor microenvironment. In our previous study, we found that the composition of PI remained strictly constant across mammalian cells and increased markedly in BC tissues. Here, we noticed that the composition of 53 lipids in plasma‐derived EVs, including PIs, changed according to the tumor stage. Interestingly, the proportion of PI_C38:3 increased in the higher stages, which is consistent with our previous tissue‐based lipidomics results. In the current study, PSs and PEs showed the highest proportions at Stage III. This may reflect the high tumor burden of the primary site since apoptotic signaling activation and nutrient deprivation can trigger abnormal PS exposure and increase PE production. 38 , 39 , 40 In addition, we observed a higher proportion of SMs and a lower proportion of Cers in Stage IV. This inverse relationship is intriguing and is likely to result from the regulation of intracellular SMs and Cers by sphingomyelin synthase 2 that is often overexpressed in metastatic BC tissues. 41 Most lipids showed non‐linear trends with distinct increase or decrease in concentration at specific stages. When compared with that of the control group, the levels of PS_C36:1 significantly increased in Stage III (p = 0.004), THC_16:0 significantly increased in Stage I/II and Stage IV (p = 0.039 and 0.014, respectively), and PC_C36:3 significantly decreased in DCIS (p = 0.041). This unique non‐linear trend and stage specificity in the expression of lipid biomarkers is another significant discovery of this study.
Concerning tumor subtypes, we found that the proportions of PCs, PEs, TGs, and LysoPCs were substantially different among Luminal, HER2‐positive and TNBC subtypes. There are differences in lipid metabolism across molecular subtypes. For instance, the Luminal subtype generally relies on de novo lipogenesis, whereas the TNBC subtype relies on exogenous FAs. 42 , 43 In line with our observations, lysophospholipase D (LysoPLD) activation is consistently detected in TNBC MDA‐MB‐231 cells. LysoPLD can hydrolyze LysoPCs to produce LPA, which promotes tumor cell proliferation and contributes to the tumorigenesis, invasion, and metastasis of BC. 44 , 45 Over half of the patients with TNBC have elevated blood TG levels, which are associated with shorter survival. 46 , 47 , 48 PC metabolism plays an essential role in HER2‐positive BC, especially in tumor proliferation, making it a promising therapeutic target. 49 , 50 Therefore, we speculated that changes in the lipid metabolism of tumors translate into the lipid composition of plasma‐derived EVs, which warrants further investigation. Our study has some limitations:
(1) This was a single‐center study with a limited number of cases, and only a small number of lipid biomarkers reached statistical significance. Notably, all participants in this study were Japanese. Although some studies examining the lipids in the tissue or plasma from different race groups reported the same lipids as biomarkers for BC, caution is required while applying this model to populations from different races. 30 , 36 , 37 Therefore, to improve the performance of the diagnostic model, the sample size needs to be expanded to include multiple centers with different races and a broader range of lipids must be explored in future studies.
(2) EVs isolated using the CUC method were a mixture of EVs from cancerous and normal tissues. The limited distinguishing capability exhibited by PLS‐DA in our study could possibly be attributed to the sharing of a majority of EV lipid components between cancerous and non‐cancerous cells to sustain their structure and essential functions. Additionally, the higher number of EVs derived from non‐cancerous cells in the blood might also contribute to this limitation. 51 Nevertheless, although the PI distribution in the plasma‐derived EVs was consistent with our previous tissue lipidomics results, the absolute difference in this study was slightly smaller than that in the tissue lipidomics results. To effectively identify cancer‐derived EVs, we suggest incorporating advanced or targeted methods to optimize the purification process, such as using a specific antibody that recognizes cancer‐derived EVs.
Altogether, we successfully discovered lipid biomarkers in plasma‐derived EVs using a modified EV isolation method, which could predict BC and its disease status. Importantly, these biomarkers exhibit different signatures across tumor stages and subtypes and reflect the known lipid profiles of tumors. Our findings show that lipid profiling of plasma‐derived EVs can be utilized as a liquid biopsy for BC. Further investigations with larger cohorts, broader coverage of lipid species, and improved EV isolation methods are warranted to optimize the model's efficacy and to gain deeper insights into tumor lipid metabolism.
AUTHOR CONTRIBUTIONS
L. L and M. K conducted the initial design of the study. L. L performed the experiments and analyzed the results. L. L, M. K, M. S, and K. S wrote and revised the manuscript. M. K and M. T supervised this study. K. K, M. K, F. P, W. L and M. S optimized the study design and guided the data processing. K. S and T. S performed the lipid extraction and LC/MS measurement. F. P helped to collect the plasma samples. M. T and T. G provided technical support in EV isolation and identification. All authors read and approved the final paper.
FUNDING INFORMATION
This study was conducted under the support of the Ministry of Education, Culture, Sports, Science and Technology KAKENHI (Project ID: 21365268), the JST FOREST Program (Grant Number: JPMJFR200K). It was also partly funded by the research support program of Kyowa Kirin Co., Shimadzu Co. and Nippon Kayaku Co. L. L was supported by the scholarship program from China Scholarship Council (Grant number: 202008050237).
CONFLICT OF INTEREST STATEMENT
Masakazu Toi is an editorial board member of Cancer Science. The authors have no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by an Institutional Reviewer Board: The study was approved by the Ethics Committee of Kyoto University Hospital (Authorization number: G0424) and performed in accordance with the Declaration of Helsinki.
Informed Consent: Informed consent was obtained from all participants.
Registry and the Registration No. of the study/trial: N/A.
Animal Studies: N/A.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
ACKNOWLEDGMENTS
Parts of this study were conducted through the Medical Research Support Center and Center for Anatomical Studies (Division of Electron Microscopic Study), Graduate School of Medicine, Kyoto University and the CORE Program of the Radiation Biology Center, Kyoto University. We appreciate Dr. Anfeng Mu, Prof. Minoru Takata and Prof. Hiroshi Harada, Radiation Biology Center, Kyoto University, for discussing the study and for their technical assistance. We also thank Mrs. Kayoko Koishihara for collecting clinical samples and the members of the Research Committee of the Kyoto Breast Cancer Research Network for the critical review of the study protocol.
Liu L, Kawashima M, Sugimoto M, et al. Discovery of lipid profiles in plasma‐derived extracellular vesicles as biomarkers for breast cancer diagnosis. Cancer Sci. 2023;114:4020‐4031. doi: 10.1111/cas.15935
REFERENCES
- 1. Plevritis SK, Munoz D, Kurian AW, et al. Association of Screening and Treatment with Breast Cancer Mortality by molecular subtype in US women, 2000‐2012. JAMA. 2018;319:154‐164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seely JM, Alhassan T. Screening for breast cancer in 2018‐what should we be doing today? Curr Oncol. 2018;25:S115‐S124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ohuchi N, Suzuki A, Sobue T, et al. Sensitivity and specificity of mammography and adjunctive ultrasonography to screen for breast cancer in the Japan strategic anti‐cancer randomized trial (J‐START): a randomised controlled trial. Lancet. 2016;387:341‐348. [DOI] [PubMed] [Google Scholar]
- 4. Lee JM, Arao RF, Sprague BL, et al. Performance of screening ultrasonography as an adjunct to screening mammography in women across the Spectrum of breast cancer risk. JAMA Intern Med. 2019;179:658‐667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lone SN, Nisar S, Masoodi T, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022;21:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao Y, Qin Y, Wan C, et al. Small extracellular vesicles: a novel avenue for cancer management. Front Oncol. 2021;11:638357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xie F, Zhou X, Fang M, et al. Extracellular vesicles in cancer immune microenvironment and cancer immunotherapy. Adv Sci (Weinh). 2019;6:1901779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vinik Y, Ortega FG, Mills GB, et al. Proteomic analysis of circulating extracellular vesicles identifies potential markers of breast cancer progression, recurrence, and response. Sci Adv. 2020;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Su Y, Li Y, Guo R, et al. Plasma extracellular vesicle long RNA profiles in the diagnosis and prediction of treatment response for breast cancer. NPJ Breast Cancer. 2021;7:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hannafon BN, Trigoso YD, Calloway CL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawashima M, Tokiwa M, Nishimura T, et al. High‐resolution imaging mass spectrometry combined with transcriptomic analysis identified a link between fatty acid composition of phosphatidylinositols and the immune checkpoint pathway at the primary tumour site of breast cancer. Br J Cancer. 2020;122:245‐257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andaluz Aguilar H, Iliuk AB, Chen IH, Tao WA. Sequential phosphoproteomics and N‐glycoproteomics of plasma‐derived extracellular vesicles. Nat Protoc. 2020;15:161‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Langevin SM, Kuhnell D, Orr‐Asman MA, et al. Balancing yield, purity and practicality: a modified differential ultracentrifugation protocol for efficient isolation of small extracellular vesicles from human serum. RNA Biol. 2019;16:5‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stranska R, Gysbrechts L, Wouters J, et al. Comparison of membrane affinity‐based method with size‐exclusion chromatography for isolation of exosome‐like vesicles from human plasma. J Transl Med. 2018;16:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911‐917. [DOI] [PubMed] [Google Scholar]
- 16. Fan S, Kind T, Cajka T, et al. Systematic error removal using random Forest for normalizing large‐scale untargeted Lipidomics data. Anal Chem. 2019;91:3590‐3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jamaly S, Ramberg C, Olsen R, et al. Impact of preanalytical conditions on plasma concentration and size distribution of extracellular vesicles using nanoparticle tracking analysis. Sci Rep. 2018;8:17216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langsted A, Freiberg JJ, Nordestgaard BG. Fasting and nonfasting lipid levels: influence of normal food intake on lipids, lipoproteins, apolipoproteins, and cardiovascular risk prediction. Circulation. 2008;118:2047‐2056. [DOI] [PubMed] [Google Scholar]
- 20. Gupta S, Rawat S, Arora V, et al. An improvised one‐step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem Cell Res Ther. 2018;9:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dong L, Zieren RC, Horie K, et al. Comprehensive evaluation of methods for small extracellular vesicles separation from human plasma, urine and cell culture medium. J Extracell Vesicles. 2020;10:e12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brennan K, Martin K, FitzGerald SP, et al. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci Rep. 2020;10:1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hong CS, Funk S, Muller L, Boyiadzis M, Whiteside TL. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J Extracell Vesicles. 2016;5:29289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Y, Wang Y, Wei S, et al. Extracellular vesicles isolated by size‐exclusion chromatography present suitability for RNomics analysis in plasma. J Transl Med. 2021;19:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. An M, Wu J, Zhu J, Lubman DM. Comparison of an optimized ultracentrifugation method versus size‐exclusion chromatography for isolation of exosomes from human serum. J Proteome Res. 2018;17:3599‐3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tian Y, Gong M, Hu Y, et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano‐flow cytometry. J Extracell Vesicles. 2020;9:1697028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pirillo A, Norata GD, Catapano AL. Postprandial lipemia as a cardiometabolic risk factor. Curr Med Res Opin. 2014;30:1489‐1503. [DOI] [PubMed] [Google Scholar]
- 28. Lambert JE, Parks EJ. Postprandial metabolism of meal triglyceride in humans. Biochim Biophys Acta. 2012;1821:721‐726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wolrab D, Jirásko R, Chocholoušková M, Peterka O, Holčapek M. Oncolipidomics: mass spectrometric quantitation of lipids in cancer research. TrAC Trends Anal Chem. 2019;120:115480. [Google Scholar]
- 30. Santoro AL, Drummond RD, Silva IT, et al. In situ DESI‐MSI Lipidomic profiles of breast cancer molecular subtypes and precursor lesions. Cancer Res. 2020;80:1246‐1257. [DOI] [PubMed] [Google Scholar]
- 31. Hou Y, Li J, Xie H, et al. Differential plasma lipids profiling and lipid signatures as biomarkers in the early diagnosis of ovarian carcinoma using UPLC‐MS. Metabolomics. 2016;12:1‐12. [Google Scholar]
- 32. Wang L, Wang X, Li Y, et al. Plasma lipid profiling and diagnostic biomarkers for oral squamous cell carcinoma. Oncotarget. 2017;8:92324‐92332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Snider AJ, Seeds MC, Johnstone L, et al. Identification of plasma glycosphingolipids as potential biomarkers for prostate cancer (PCa) status. Biomolecules. 2020;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mashima T, Seimiya H, Tsuruo T. De novo fatty‐acid synthesis and related pathways as molecular targets for cancer therapy. Br J Cancer. 2009;100:1369‐1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang W, Fa H, Xiao D, Wang J. Targeting phosphatidylserine for cancer therapy: prospects and challenges. Theranostics. 2020;10:9214‐9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wolrab D, Jirasko R, Peterka O, et al. Plasma lipidomic profiles of kidney, breast and prostate cancer patients differ from healthy controls. Sci Rep. 2021;11:20322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chagovets VV, Starodubtseva NL, Tokareva AO, et al. Validation of breast cancer margins by tissue spray mass spectrometry. Int J Mol Sci. 2020;21:4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Belzile O, Huang X, Gong J, et al. Antibody targeting of phosphatidylserine for the detection and immunotherapy of cancer. Immunotargets Ther. 2018;7:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stoica C, Ferreira AK, Hannan K, Bakovic M. Bilayer forming phospholipids as targets for cancer therapy. Int J Mol Sci. 2022;23:5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu L, Bakovic M. Breast cancer cells adapt to metabolic stress by increasing ethanolamine phospholipid synthesis and CTP:ethanolaminephosphate cytidylyltransferase‐Pcyt2 activity. Biochem Cell Biol. 2012;90:188‐199. [DOI] [PubMed] [Google Scholar]
- 41. Zheng K, Chen Z, Feng H, et al. Sphingomyelin synthase 2 promotes an aggressive breast cancer phenotype by disrupting the homoeostasis of ceramide and sphingomyelin. Cell Death Dis. 2019;10:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gong Y, Ji P, Yang YS, et al. Metabolic‐pathway‐based subtyping of triple‐negative breast cancer reveals potential therapeutic targets. Cell Metab. 2021;33(51–64):e59. [DOI] [PubMed] [Google Scholar]
- 43. Simeone P, Tacconi S, Longo S, et al. Expanding roles of De novo lipogenesis in breast cancer. Int J Environ Res Public Health. 2021;18(7):3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Umezu‐Goto M, Kishi Y, Taira A, et al. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J Cell Biol. 2002;158:227‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu S, Umezu‐Goto M, Murph M, et al. Expression of autotaxin and lysophosphatidic acid receptors increases mammary tumorigenesis, invasion, and metastases. Cancer Cell. 2009;15:539‐550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maiti B, Kundranda MN, Spiro TP, Daw HA. The association of metabolic syndrome with triple‐negative breast cancer. Breast Cancer Res Treat. 2010;121:479‐483. [DOI] [PubMed] [Google Scholar]
- 47. Lofterod T, Mortensen ES, Nalwoga H, et al. Impact of pre‐diagnostic triglycerides and HDL‐cholesterol on breast cancer recurrence and survival by breast cancer subtypes. BMC Cancer. 2018;18:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dai D, Chen B, Wang B, et al. Pretreatment TG/HDL‐C ratio is superior to triacylglycerol level as an independent prognostic factor for the survival of triple negative breast cancer patients. J Cancer. 2016;7:1747‐1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Szumilo M, Rahden‐Staron I. Biological role of phosphatidylcholine‐specific phospholipase C in mammalian cells. Postepy Hig Med Dosw (Online). 2008;62:593‐598. [PubMed] [Google Scholar]
- 50. Pisanu ME, Ricci A, Paris L, et al. Monitoring response to cytostatic cisplatin in a HER2(+) ovary cancer model by MRI and in vitro and in vivo MR spectroscopy. Br J Cancer. 2014;110:625‐635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alberro A, Iparraguirre L, Fernandes A, Otaegui D. Extracellular vesicles in blood: sources, effects, and applications. Int J Mol Sci. 2021;22:8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6


