The presence of cell-free tumor DNA was reported in breast milk from patients with breast cancer, and its use as a screening method surpassed plasma liquid biopsy in terms of detecting and molecularly profiling early-stage tumors, even prior to diagnosis.
Abstract
Breast cancer occurring during pregnancy (PrBC) and postpartum (PPBC) is usually diagnosed at more advanced stages compared with other breast cancer, worsening its prognosis. PPBC is particularly aggressive, with increased metastatic risk and mortality. Thus, effective screening methods to detect early PrBC and PPBC are needed. We report for the first time that cell-free tumor DNA (ctDNA) is present in breast milk (BM) collected from patients with breast cancer. Analysis of ctDNA from BM detects tumor variants in 87% of the cases by droplet digital PCR, while variants remain undetected in 92% of matched plasma samples. Retrospective next-generation sequencing analysis in BM ctDNA recapitulates tumor variants, with an overall clinical sensitivity of 71.4% and specificity of 100%. In two cases, ctDNA was detectable in BM collected 18 and 6 months prior to standard diagnosis. Our results open up the potential use of BM as a new source for liquid biopsy for PPBC detection.
Significance:
For the first time, we show that BM obtained from patients with breast cancer carries ctDNA, surpassing plasma-based liquid biopsy for detection and molecular profiling of early-stage breast cancer, even prior to diagnosis by image.
See related commentary by Cunningham and Turner, p. 2125.
This article is featured in Selected Articles from This Issue, p. 2109
INTRODUCTION
Breast cancer is the most common malignancy diagnosed and most common cancer-related death during pregnancy and lactation. Two entities are distinguished according to diagnosis momentum: breast cancer diagnosed during pregnancy (PrBC) and up to 5 to 10 years postpartum (PPBC; refs. 1, 2). Both subtypes encompass up to 55% of breast cancers diagnosed in women <45 years old (3). Considering that aging increases breast cancer risk and the tendency to delay pregnancy in developed countries, new cases are expected to rise in the years to come (4).
PrBC and PPBC are independently associated with poor survival but for different reasons. In the case of PrBC, it is mainly due to a delay in diagnosis and presentation at advanced stages (5, 6). The inability to detect tumors early during pregnancy results from the limited sensitivity of standard diagnostic imaging tools to reliably detect tumors amidst the morphologic changes that mammary glands undergo, as well as an underestimation of symptoms by patients and health care professionals during this period (6). After adjusting for clinicopathologic factors, prognosis of PrBC patients was indistinguishable from non-PrBC patients. On the contrary, PPBC has been shown not to present with a different stage than nulliparous or later parous young women but has a poorer prognosis and worse survival rates (7) and double metastatic risk (3, 8, 9), potentially linked to mammary gland involution after weaning (10). In stages I and II or estrogen receptor (ER)–positive subtype malignancies, usually considered as having good outcomes, a postpartum diagnosis has been revealed to be a dominant feature associated with increased risk for metastasis and death (9). Biological causes underlying their distinct clinical presentation were mainly studied in rodents and healthy human mammary tissue, pointing to the breast involution program. The programmed death of the alveolar epithelium mirrors a wound-healing scenario generating a protumorigenic stromal environment (11). Moreover, luminal PPBC aggressiveness in human tumors has recently been associated with a distinct gene expression signature (12) and increased risk of presenting liver metastases (9, 13).
Patients with PPBC represent a population in dire need of novel, sensitive, noninvasive early detection screening methods to reduce mortality. Moreover, the young age of these patients excludes them from population-based early diagnosis programs, starting in most countries from the age of 50 onward. Blood liquid biopsy, involving genetic and epigenetic analysis of cell-free tumor DNA (ctDNA) in patients’ plasma, represents a promising approach. In patients with metastatic breast cancer, ctDNA is detected in up to 85% of the blood samples, and this clinical relevance is well established (14, 15). In contrast, localized breast cancer releases comparatively little ctDNA in blood, complicating its use for early breast cancer detection (14, 16, 17). Using an alternative body fluid in intimate contact with the tumor may provide an alternative approach, as it has been successfully applied using urine for bladder malignancies or saliva for oral cancer (18).
In the present work, given the unique physiologic condition of the study population, we hypothesized that breast milk (BM) could represent a reliable source of ctDNA that may be used as a strictly noninvasive method for early PPBC detection. We demonstrate for the first time the presence of ctDNA in the BM collected from women diagnosed with early PrBC or PPBC, even in samples collected before diagnosis and whose solid tumors were genomically profiled in parallel.
RESULTS
Study Cohorts and Tumor Genomic Profiling
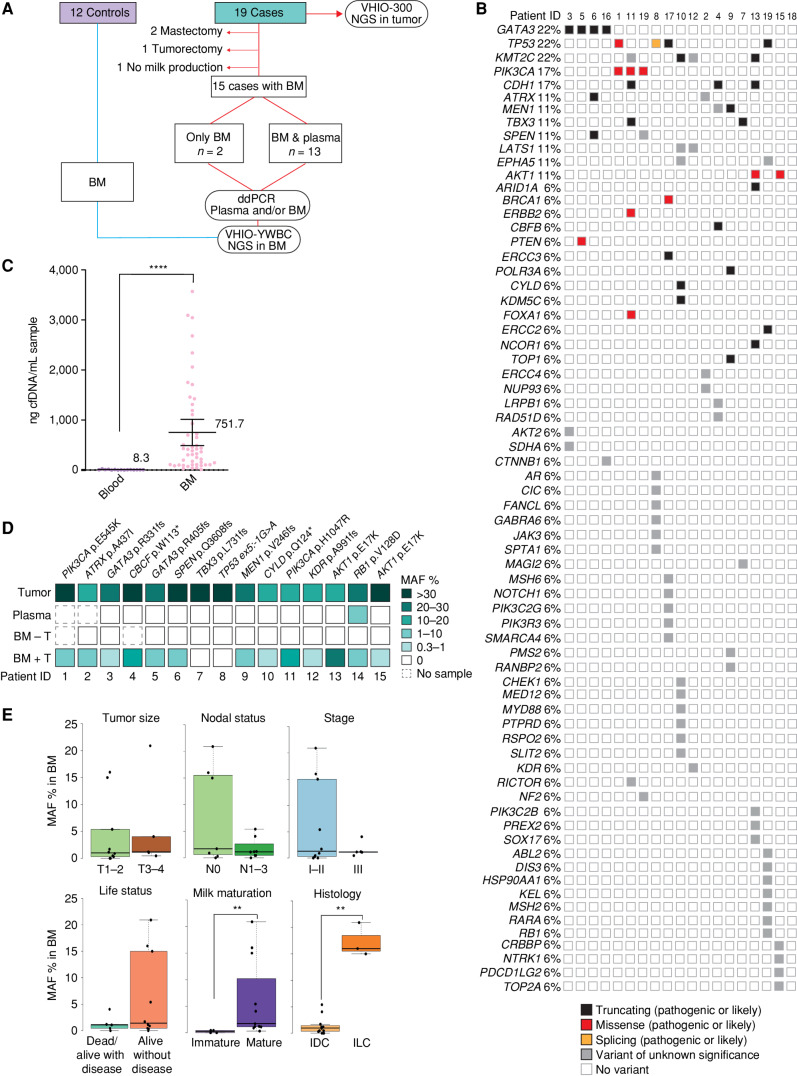
To address our hypothesis, we assembled two cohorts: a case group of patients (n = 19) and a control group of healthy donors with a minimum of 1 year of medical follow-up (n = 12; Fig. 1A). The case cohort included 19 patients with available tumor tissue prospectively collected in a single institution during 40 months: 10 women diagnosed with breast cancer during pregnancy (PrBC) and nine diagnosed during breastfeeding (PPBC; Table 1). The mean age of patients included was 36.2 years (range, 25–48), 74% of patients were diagnosed with stage I–II breast cancer, and 79% of patients had luminal tumors. Matched tumor samples were profiled by next-generation sequencing (NGS) using a custom 432-gene hybrid capture–based panel (VHIO-300; Supplementary Table S1) in all cases except BC-01, whose primary tissue was analyzed with a similar panel after enrollment in the AURORA (NCT02102165) clinical trial (ref. 19; 411 genes; Supplementary Table S2; a list of samples analyzed by NGS is available in Supplementary Excel Table S1). The variants detected were consistent with the young median age of the patients (20) and a majority of luminal subtypes, showing a 22% prevalence of GATA3, TP53, and KMT2C mutations, followed by alterations in PIK3CA and CDH1 in 17% of the samples (Fig. 1B; for list of variants detected in the tumors, see also Supplementary Excel Table S2).
Figure 1.
Genomic profile of tumor tissue and matched plasma and BM ctDNA. A, Cohorts included in the study, samples, and workflow. ddPCR, droplet digital PCR; NGS, next-generation sequencing. B, Oncoprint of all solid tumors analyzed by NGS from the case cohort (n = 19). C, cfDNA concentration purified from blood samples of the case group (n = 12) and compared with BM samples collected from the control and case groups (n = 49). Individual values, mean, and 95% confidence interval are included. Nonparametric two-sided Mann–Whitney–Wilcoxon test was performed (****, P < 0.0001). D, Targeted detection of selected clonal variants by ddPCR in the parallel tumor, plasma, and BM samples from the affected breast (BM + T n = 15; BM – T n = 13) from the case cohort. E, Association between MAF percentage and tumor size, nodal status, disease stage, current life status, milk maturation, and histology (Mann–Whitney–Wilcoxon test; **, P < 0.01). IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma.
Table 1.
Clinical–pathologic characteristics of the cohorts
| Cohort | Classifier | Subtype | N | % |
|---|---|---|---|---|
| Healthy | Age | 34.2 years (28–40) | 12 | 100% |
| Milk stagea | Colostrum | 0 | 0% | |
| Transitional | 0 | 0% | ||
| Mature | 12 | 100% | ||
| Breast cancer cases | Age | 36.2 years (25–48) | 19 | 100% |
| pT | 1 | 5 | 26% | |
| 2 | 8 | 42% | ||
| 3 | 5 | 26% | ||
| 4 | 1 | 5% | ||
| pN | 0 | 9 | 47% | |
| ≥1 | 10 | 53% | ||
| M | 0 | 18 | 95% | |
| 1 | 1 | 5% | ||
| Stage | I | 2 | 11% | |
| II | 11 | 58% | ||
| III | 5 | 26% | ||
| IV | 1 | 5% | ||
| HR | Positive | 16 | 84% | |
| Negative | 3 | 16% | ||
| HER2 | Positive | 1 | 5% | |
| Negative | 18 | 95% | ||
| Molecular subtypeb | Luminal A | 6 | 32% | |
| Luminal B | 9 | 47% | ||
| HER2+ | 1 | 5% | ||
| TNBC | 3 | 16% | ||
| Diagnosis timec | PrBC | 10 | 53% | |
| PPBC | 9 | 47% | ||
| Histologic subtype | IDC | 15 | 79% | |
| ILC | 4 | 21% | ||
| Milk stagea | Colostrum | 3 | 21% | |
| Transitional | 1 | 7% | ||
| Mature | 11 | 79% |
NOTE: Healthy and breast cancer–diagnosed individuals are described in detail.
Abbreviations: HR, hormone receptor; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; PR, progesterone receptor; TNBC, triple-negative breast cancer.
aMilk stage is defined as colostrum: 0–5 days after childbirth; transitional: 6–14 days after childbirth; and mature: after 14 days from childbirth.
bMolecular subtype is based on IHC: luminal A is defined as ER- or PR-positive, HER2-negative, and Ki-67 <20%; luminal B is defined as ER- or PR-positive, HER2-negative, and Ki-67 ≥20%; and HER2+ is defined as HER2-positive regardless of ER, PR, and Ki-67 status. TNBC is defined as ER-, PR-, and HER2-negative.
cDiagnosis time refers to breast cancer diagnosed during pregnancy (PrBC) or postpartum (PPBC).
The control group consisted of healthy pregnant women under 40 years of age (34.2, range, 28–40) who voluntarily donated BM samples during breastfeeding (Table 1). Individuals of the healthy cohort were free of breast cancer until the present publication, with a minimum follow-up of 1 year.
BM Contains ctDNA and Surpasses Plasma in Sensitivity of Breast Cancer Detection
Out of the 19 patients from the case cohort, four were excluded from further analysis [three cases were discarded due to tumor surgery before childbirth (patients 16, 18, and 19)]. In one case, there was no production of BM, probably due to neoadjuvant chemotherapy treatment (BC-17); Fig. 1A; Supplementary Table S3). From all the controls and the remaining 15 patients, the total cell-free DNA (cfDNA) concentration extracted from BM of both breasts was, on average, 90-fold higher than from blood (751.7 ng per mL for any BM vs. 8.3 ng per mL of blood, P < 0.0001; Fig. 1C). Mean concentrations of cfDNA in BM did not differ between controls and cases (Supplementary Fig. S1A) or between affected versus unaffected breasts of cases (Supplementary Fig. S1A and S1B). Of note, a donor age ≥35 years, but not the presence of a tumor, significantly increased the abundance of cfDNA in BM (Supplementary Fig. S1C; Supplementary Table S4). BM cfDNA integrity was also superior to that in blood, with fragments ranging from 30 to >10,380 base pairs (bp) versus mono- (167 bp) and polynucleosome (n ×167 bp) fractions routinely observed in plasma-derived cfDNA (Supplementary Fig. S1D). We next assessed whether ctDNA was present in BM from patients. To this end, we tested paired samples of BM from both breasts and plasma by droplet digital PCR (ddPCR) except for patients BC-01 and -02, who lacked synchronous blood samples. We designed a specific ddPCR assay per case based on NGS data obtained from tumor tissue. Analysis was positive in 13 of 15 BM samples from the affected breasts for the patient-specific tumor variant, whereas synchronous plasmas yielded negative results except in BC-14 (Supplementary Table S3). In parallel, the BM of unaffected breasts was all negative (Fig. 1D). The total number of genome equivalents (GE) analyzed by ddPCR in BM and plasma reinforced the superior concentration of total cfDNA in BM seen in cfDNA quantification (Supplementary Fig. S1E) and points to a higher prevalence of ctDNA compared with plasma upon normalization of GEs analyzed (Supplementary Fig. S1F). As nothing is known regarding the biology underlying ctDNA release into the BM, we sought to explore whether ctDNA abundance in BM correlates with disease biomarkers, considering the relatively small number of samples in the study. Interstingly, mutant allele fraction (MAF) levels in BM did not show a significant correlation with disease burden biomarkers such as tumor size, stage, nodal status, or progression of disease (Fig. 1E). Instead, we detected striking differences in the median percentage of MAF in BM depending on its maturation stage (21). Immature samples, including colostrum (BM collected in the first 5 days of lactancy) and transitional specimens (from 6 to 14 days), had almost 7 times lower variant frequencies detected compared with those collected after 14 days of lactancy and considered mature samples (median 0.25 vs. 1.7, respectively; P = 0.0059). We also observed a strong correlation between MAF in BM and the lobular or ductal subtype of the tumor. The MAF percentage (%) in BM from patients diagnosed with invasive lobular carcinoma (ILC) was far superior when compared with those from patients with invasive ductal carcinoma (IDC; median 1.0 vs. 15.0, respectively; P = 0.0044; Fig. 1E).
NGS-Based Approach for ctDNA Detection in BM
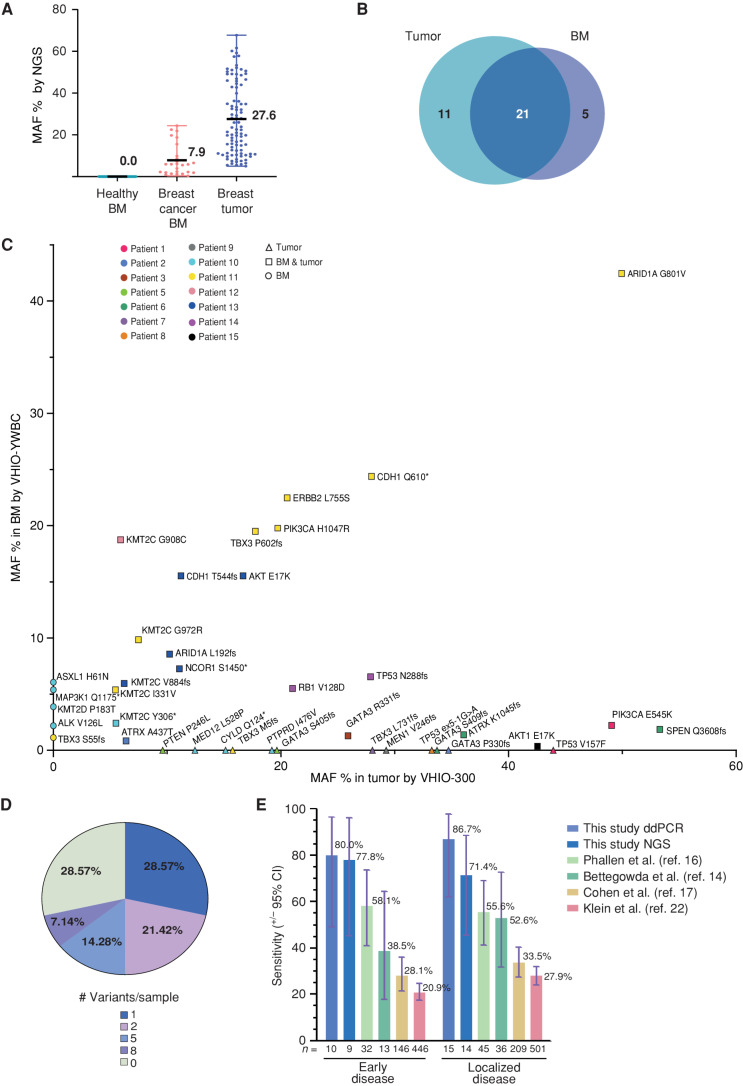
Although ddPCR is exquisitely sensitive, it requires prior knowledge of mutations and the design of variant-specific assays, which limits its application as a population-based potential future screening assay. Hence, we sought to adopt an NGS approach and designed a unique molecular index (UMI) hybrid-capture test that could be implemented as a prospective, mutation-agnostic BM-based PPBC screening assay. The custom-designed panel targets 54 genes frequently mutated in young women with breast cancer up to 45 years old (see Methods for the specific design, VHIO-YWBC; Supplementary Table S5). Analytical validation of the panel was performed using as a reference a commercial mixture of human genomic DNA spiked with synthetic controls in two different conditions at 0.5% MAF and 1% MAF (Supplementary Table S6 and Supplementary Fig. S2A). High accuracy of the three triplicates was detected per condition (Supplementary Fig. S2B). The test reported an average analytic sensitivity of 93.3% for 0.5% MAF and 100% for 1% MAF, whereas analytical specificity, positive predictive value, and negative predictive value were all 100% for both conditions (Supplementary Fig. S2C). Then, BM cfDNA from the cases and the healthy control subjects was reanalyzed with VHIO-YWBC except for BC-04, due to sample exhaustion. In total, the VHIO-YWBC panel detected 26 variants in the affected BM, with a mean MAF % of 7.9 (range, 0.4–24.4), compared with 96 variants with a mean MAF % of 27.6 (range, 5–67.7) in the tumor tissues using the VHIO-300 panel. VHIO-YWBC reported no variants in the BM samples analyzed from the healthy cohort (Fig. 2A). Comparing only genes detected in both panels, up to 21 of the 26 variants detected in the affected BM were coincident with the matched tumor tissue, whereas five were BM-specific. Eleven variants were detected only in the tumor but not in the BM (Fig. 2B and C; for a detailed list of variants detected in BM, see Supplementary Excel Table S3). Overall, by applying the VHIO-YWBC panel, we were able to detect at least one disease variant present in tumor tissue in 71.4% of BM samples and more than one in 42.9% of specimens, ranging from two to eight variants per sample (Fig. 2D). Focusing only on early disease, the panel increased to a sensitivity of 77.8% in stages I–II (Fig. 2E). Compared with ddPCR (sensitivity of 86.7%), two variants in GATA3 and MEN1 (patients BC-05 and BC-09) were not detected by NGS in BM samples, likely due to the low MAF in BM (detected by ddPCR) and their large indel size (≥10 bp; Supplementary Table S3; Supplementary Excel Table S2). In order to set our findings in the context of liquid biopsy, we compared the performance of the VHIO-YWBC panel in BM-derived ctDNA genotyping with previous reports applying NGS protocols for localized breast cancer detection in plasma through diverse methods such as ctDNA detection alone (14), targeted error correction sequencing (16), ctDNA detection combined with other biomarkers (17), or even analysis of DNA-methylation signatures (22). Our approach confirmed superior sensitivity to all the other protocols for both early (stages I–II) and localized (I–III) disease detection (Fig. 2E; Supplementary Table S7).
Figure 2.
NGS-based detection of breast cancer in BM using a targeted sequencing assay. A, Variants detected by the NGS panel VHIO-YWBC in healthy BM (n = 0) and breast cancer BM (n = 26), as well as in tumor tissue with the VHIO-300 NGS panel (n = 94), represented as % of MAF. The mean and range are depicted in the chart. B, Number of variants detected in common between BM and matched tumor tissue from the case cohort, focusing only on genes captured by both the VHIO-YWBC and VHIO-300 panels. C, Variants detected by NGS in BM samples by VHIO-YWBC (y-axis) vs. variants detected by the VHIO-300 panel in formalin-fixed, paraffin embedded (FFPE) solid tumor biopsies (x-axis) only in genes common to both panels. Data are represented as MAF %. Color coding is used to discriminate patients. The shape of the symbols represents variants detected only in BM (circles), only in FFPE (triangles), or in both (squares). D, Percentage of cases according to the number of variants detected in BM by NGS (n = 14). E, Sensitivity of our ddPCR and NGS approaches for the detection of ctDNA in early (stages I–II) and localized (stages I–III) disease and compared with four reported methods for early breast cancer detection from plasma; data are represented as a percentage, with exact mean numbers and plus/minus a 95% confidence interval (CI).
Evidence for Early Breast Cancer Detection in BM Liquid Biopsy
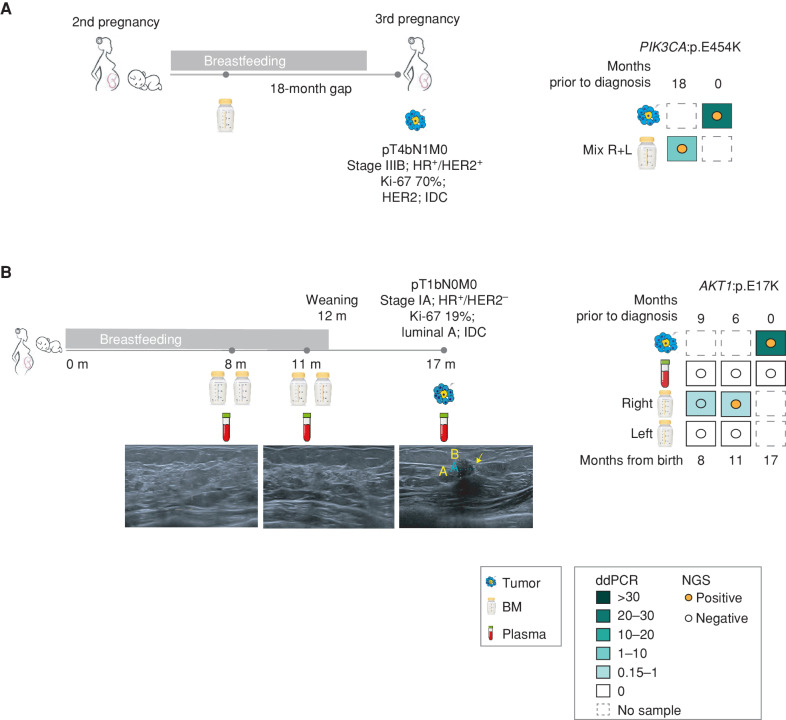
Although our case cohort is relatively small, two cases illustrate the potential use of ctDNA analysis in BM as a promising tool for early breast cancer detection. Patient BC-01 was diagnosed with breast cancer during her third pregnancy. Coincidentally, the patient had a frozen BM sample (pooled from both breasts) that had been collected during the lactation of her second child (and 18 months prior to breast cancer diagnosis). Strikingly, a PIK3CA p.E545K mutation was already present in the frozen BM sample, which was also identified in the tumor diagnosed 1.5 years later (Fig. 3A). A second patient (BC-15) volunteered after 8 months of childbirth for close monitoring in the context of a local project to follow-up on high-risk patients due to her age at first pregnancy (47 years). She received ultrasound follow-ups at 8, 11, and 17 months after delivery, along with plasma and BM collection (at the 17-month time point, BM was not collected due to weaning at month 12). The first two ultrasounds were negative. However, the third ultrasound revealed a 6.7 × 7 mm lesion in the right breast, which was further biopsied and diagnosed as a stage IA, luminal A IDC, making her eligible for the present study (Fig. 3B; Supplementary Fig. S3A–S3E; Supplementary Table S3). Retrospective ddPCR analysis of the BM cfDNA collected at time points 8 and 11 (6 and 9 months prior to diagnosis) identified an AKT1:p.E17K clonal variant in the tumor tissue. Only BM derived from the affected breast tested positive for ctDNA, and plasma samples were negative for all time points tested prior to and after diagnosis. NGS analysis, less sensitive than ddPCR, detected the variant at the 6-month time point prior to tumor radiologic and pathologic diagnosis. The earlier 9-month time point revealed two consensus reads carrying AKT1:p.E17K, a result under the threshold of the analysis pipeline (see Methods) to consider it informative (Fig. 3B).
Figure 3.
Early detection of breast cancer through BM ctDNA analysis. A, Description of case #1, timeline, sample collection, and test results by ddPCR and NGS. Patient 1 was diagnosed with PrBC during her third pregnancy. The patient provided a mixture of both breasts from the lactancy of a previous pregnancy and collected 18 months prior to diagnosis. HR, hormone receptor; R + L, right and left. B, Description of positive high-risk case BC-15, timeline (m = months from childbirth), sample collection, and the corresponding right breast ultrasound images. The third image taken at a 17-month time point corresponds to the time of clinical diagnosis. The yellow arrow points to the malignant lesion observed and its measurement by image (A = 7.4 mm; B = 6.4 mm). The different samples collected and test results obtained by ddPCR and NGS. Colored squares represent the MAF % of the pathogenic variant detected by ddPCR. Colored dots represent the detection of the same variant by NGS.
DISCUSSION
Overall, our data robustly demonstrate for the first time the existence of ctDNA in the BM of patients with breast cancer diagnosed while pregnant or postpartum. Given the aggressiveness of PPBC, the lack of population-wide breast cancer screening protocols in young women, and the tendency to postpone the first pregnancy in developed countries, researchers have high hopes in plasma-based liquid biopsy to allow early cancer detection, but sensitivity is a challenging issue in breast cancer. Previous reports explored diverse cancer biomarkers in alternative fluids in close contact with the breast tumor. This is the case for nipple aspirate fluid (NAF), ductal lavage (DL), or BM. So far, NAF has been tested for proteins and miRNAs, and DL has been used for cytomorphologic analysis (23, 24). Theoretically, NAF may be obtained from any woman except those pregnant or lactating, but its general utility for biomarker assessment is hampered by its low yield (on average, 20–50 μL). DL is a more invasive procedure, requiring local anesthesia to introduce a microcatheter to flush every duct independently (the average number of ducts in the breast is 9.4 ± 2.9), thereby obtaining nonphysiologic fluid that may or may not drag epithelial cells. This procedure has considerable rates of failure in addition to causing high discomfort; hence, alternative procedures need to be explored for breast cancer screening (25). Like NAF, BM is produced by the lobular tissue distributed throughout the ducts, in intimate contact with the tumor tissue, but in large quantities and only in pregnant and lactating women. BM has been used as a noninvasive surrogate of the mature mammary gland cellularity and its microenvironment (26), cellular dynamics during human lactation (27), promoter methylation of specific CpGs (28), cancer-associated miRNAs (29), or detection of protein biomarkers of breast cancer risk (30). However, these studies were conducted mainly in healthy individuals or women with noncancerous breast disease. So far, one study reported the enrichment of cancer stem–like cells (CSC) in BM, reporting genomic alterations on the mammospheres derived from BM-isolated CSCs upon five passages of in vitro culturing. CSCs were obtained from only two of 10 healthy donors and a single case of breast cancer (31). No comparison with the patient's tumor was performed for this last individual. Hence, it is unclear whether the CSCs were or were not of tumor origin. These results might be of interest and deserve further exploration, but CSC isolation and culturing require quick processing of a fresh sample, with a complex translation into the clinical setting and a long time frame to obtain results. On the contrary, our work not only demonstrates the presence of ctDNA in the BM of patients with localized breast cancer but also shows that its genomic profiling recapitulates the variants detected in the tumors analyzed by ddPCR or NGS, with a protocol closer to the clinical routine.
We also show that BM surpasses plasma regarding cfDNA abundance and integrity as well as ctDNA shedding. As mentioned before, low rates of ctDNA in plasma are expected in breast cancer early disease, in which sensitivity by NGS ranges from 21% to 58% depending on the approach (14, 16, 17, 22). That said, 8% of plasma ctDNA–positive samples of women diagnosed with PrBC and PPBC are scarce. Lower levels of ctDNA shedding to the blood of lactating women might be explained by the blood–milk barrier formed by breast epithelial cells during pregnancy. Such cell remodeling reduces permeability and increases tight junctions to prevent blood leakage of milk components upon lactation (32). Obviously, the higher cfDNA concentration in BM results in more available genomic equivalents (GE) to be analyzed and enhances the chance for variant detection. We also observed that upon in silico normalization of BM GEs with the lower plasma GEs analyzed by ddPCR, the number of ctDNA-positive samples in BM would still remain unchanged. This observation aligns with a higher ratio of ctDNA/cfDNA in BM compared with blood in PrBC and PPBC.
The highest abundance of ctDNA in BM belonged to the three patients with ILC. All cases carried a loss-of-function variant of the E-cadherin gene (CDH1), a hallmark of lobular carcinomas (33). Altered CDH1 decreases tumor cell junction tightness, possibly explaining the ∼16-fold higher ctDNA shedding in ILC. Early diagnosis of ILC is particularly interesting because it is challenging to detect using standard radiologic imaging, whereas BM genotyping enabled the detection of a tumor as small as a T1a (BC-04). As the three ILC samples were collected after 15 days postpartum and thus were all mature, we wondered whether the molecular subtype biased the positive correlation with maturity. Importantly, mature milk's median percentage of variant alleles was still significantly higher than immature specimens when ILC samples were excluded from the analysis (median 0.25 vs. 1.2, P = 0.0162; Supplementary Fig. S1G). The lowest MAF % of ctDNA among all BM samples belonged to immature milk specimens, including two BM samples corresponding to colostrum that were ctDNA negative by ddPCR and NGS. Although the cohort size is small, this result suggests that BM maturation might affect ctDNA abundance. In this regard, systematic collection of mature BM samples (>15 days after birth) should improve PPBC screening sensitivity in future studies.
In the field of liquid biopsy, plasma-derived ctDNA content correlates with disease burden, increasing with the stage (14). Blood-based biopsy is used for tumor profiling, to capture minimal residual disease, to study clonal evolution, or to detect resistance mechanisms before progression is diagnosed by image. In the case of BM, no correlation was found between ctDNA abundance and disease burden biomarkers. Lactation is suppressed upon breast cancer diagnosis, preventing repeated sample collection from monitoring disease evolution. But significantly enough, BM ctDNA reveals the mutational landscape of the tumor tissue and could become an excellent noninvasive biopsy for the most challenging application—early detection. For this purpose, an agnostic method is required, something that ddPCR cannot accomplish. Thus, we successfully reanalyzed ctDNA detection sensitivity in BM by NGS using the custom-designed YWBC-VHIO panel. Ten out of 14 BM samples were positive for variant detection. Twenty-one variants were commonly found in tumor tissue and BM, but some discordances were depicted. The case with the highest mutational burden in tissue (BC-10) encompasses seven of those nonoverlapping variants, most likely reflecting high tumor heterogeneity. Six frameshifts were also missed in BM, four with lengths superior to 10 bp, highlighting a commonly limited sensitivity for insertions/deletions in capture-based NGS panels.
The overall 77.8% and 80% sensitivity in early disease obtained by NGS and ddPCR (taking tumor samples as the reference), respectively, are auspicious results. But even more encouraging are the two cases whose BM was obtained far prior to diagnosis by image (BC-01 and BC-15) and even in parallel to negative radiologic controls (BC-15). Finally, but most importantly for early detection applicability, no variants were found in the healthy volunteers or the analyzed healthy breasts of patients diagnosed with breast cancer.
Notwithstanding these benefits, the study has some limitations. First, the size of the case cohort is relatively small but challenging to enlarge in a reasonable time frame. Collection of BM while the tumor is still in the breast, to ensure temporal coincidence of both entities, is a challenge. Surgery and chemotherapy can be performed during pregnancy (removing the tumor), and lactation is usually suppressed immediately after diagnosis of breast cancer (no BM available for study). Despite this, we report the largest collection published to date of BM from PPBC and PrBC patients. Second, the YWBC-VHIO panel missed two samples positive for ctDNA by ddPCR, with long frameshifts as the main drivers. To circumvent this weakness, enlargement of the panel size or combinations with PCR-based target amplification and capture-based enrichment chemistries should be tested.
As pregnant and lactating women lack specific early screening strategies—protocols are generally based on imaging and mainly performed in postmenopausal women—our study provides evidence that ctDNA analysis in mature BM might become an opportunity for detection of PPBC significantly sooner than standard imaging-based diagnosis. We demonstrate that BM is a superior source of ctDNA as compared with plasma in early-stage breast cancer and could be adapted for early cancer detection in a setting compatible with routine clinical practice. This approach would be focused on PPBC as long as malignant cells are already present during the lactating period. Even so, PPBC accounts for up to half of the diagnosed breast cancer cases in women up to 45 years old (11), a nonnegligible value for a tumor type of high aggressiveness. Similar to the neonatal heel prick for metabolic disease testing, routine NGS-based breast cancer screening using mature BM could dramatically improve the prognosis and quality of life of diagnosed mothers in the future.
METHODS
Patients’ Clinical Records and Samples
Individuals of both cohorts, healthy controls, and breast cancer–diagnosed case groups were recruited at the Breast Cancer Unit of the Vall d'Hebron University Hospital (HUVH) in Barcelona, Spain. Healthy individuals were women with a pregnancy under 40 years old and unknown carriers of germline mutations associated with breast cancer increased risk. The case group was women with a diagnosed breast cancer during any stage of pregnancy or breastfeeding. The HUVH Ethics Committee of Clinical Research approved the study according to local guidelines and regulations, with the reference PR(AG)197/2019. Informed written consent was obtained from all the patients included in this study. Our study was conducted in accordance with the ethical principles stated in the latest version of the Declaration of Helsinki, and genomic studies were performed in agreement with the Spanish Act 14/2007, July 3, on Biomedical Research.
Sample and Image Collection
Healthy individuals donated a BM sample from both breasts. In the case group, we collected synchronous tumor biopsies, blood, and BM samples of both breasts whenever possible. Specifically, BM, plasma, and solid biopsy were collected upon diagnosis in PPBC patients. For PrBC patients, plasma and formalin-fixed, paraffin embedded (FFPE) tissue were collected upon diagnosis and BM collection was postponed until live birth. All tumor specimens (breast biopsy, primary breast tumor, or metastatic sample) were stored as fresh-frozen FFPE material. Blood samples were collected in anticoagulant-treated tubes, and plasma fraction was isolated by double-step centrifugation at low speed to separate cellular fraction and at room temperature. For further purification, isolated plasma was aliquoted and stored at −80°C. Similarly, milk samples were fractionated by double-step low-speed and room temperature centrifugation to isolate the serum from the fat and cellular fraction. For further purification, isolated serum BM was aliquoted and stored at −80°C.
Isolation of cfDNA
cfDNA from BM was isolated using the QIAamp MinElute ccfDNA Midi Kit (cat no. 55284, Qiagen) or the QIAamp Circulating Nucleic Acid Kit according to the manufacturer's instructions. For plasma, cfDNA isolation was performed with the QIAamp Circulating Nucleic Acid Kit according to the manufacturer's instructions. Both cfDNAs were stored at −20°C and quantified by the Qubit dsDNA HS Assay Kit (Invitrogen). The quality and size of the cfDNA were measured by the High-Sensitivity 2100 Bioanalyzer Instrument (Agilent).
Isolation of FFPE-Derived Tumor DNA
DNA extraction of FFPE samples (ten 10-μm FFPE tissue sections) was performed with the automated system Maxwell16 FFPE Plus LEV DNA Purification Kit (Promega). DNA quality and concentration were measured with a NanoDrop 1000 spectrophotometer (Thermo Scientific).
VHIO-300 Panel NGS
Libraries were prepared according to the Agilent standard protocol (SureSelect XT Human, Agilent). Briefly, 500 ng of double-stranded DNA (dsDNA) was fragmented (S2, Covaris), end-repaired, A-tailed, and linked to adapters. After 10 cycles of PCR, 750 ng was used as input for capture, followed by an additional postcapture 12-cycle PCR step. For a detailed gene list of captured exons in the VHIO-300 panel, see Supplementary Table S1. Libraries were sequenced in a HiSeq2500 instrument (Illumina), 2 × 100 paired-end.
Sequencing reads were aligned (BWA v0.7.17, Samtools v1.9) against the hg19 reference genome, base recalibrated, realigned (ABRA2 v2-2.23), indel realigned [Genome Analysis Toolkit (GATK) v3.7.0], and variant called by two different tools (VarScan2 v2.4.3; GATK Mutect2 v4.1.0.0). The final average coverage depth per gene was ×1,118 (ranging from ×322 to ×1,859). A minimum of seven reads supporting the variant allele were required to call a mutation. Frequent single-nucleotide polymorphisms (SNP) in the population were filtered based on the gnomAD database (allele frequency ≤0.0001), and copy-number alterations were calculated (CNVkit v0.9.6.dev0). Clinical significance classification of the variants was performed using the following databases: COSMIC (https://cancer.sanger.ac.uk/cosmic), cBioPortal (http://www.cbioportal.org/), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), OncoKB (https://www.oncokb.org/), and VarSome (https://varsome.com/). Finally, manual data curation was performed among all the knowledge databases to harmonize their criteria.
VHIO-YWBC Panel NGS
The panel comprises probes for coding regions of 54 cancer-related genes (Supplementary Table S5). The panel was designed based on three studies (34–36), using exclusively tissues from patients with breast cancer aged ≤45 years (n = 1,069). Only cancer-related genes by pathogenicity, druggability, and predictive/prognostic biomarker capacity were chosen. The minimum altered gene frequency of the published study cohort was 1% for the medium-size panels or 2% for whole-exome panels. Each gene was present in at least two of the three datasets. Libraries were prepared according to the Agilent standard protocol (SureSelect XT Human, Agilent). Briefly, ∼170 ng of dsDNA from BM were fragmented (S2, Covaris), end-repaired, A-tailed, and linked to adapters. After 10 cycles of PCR, ∼770 ng was used as input for capture, followed by an additional postcapture 10-cycle PCR step. In cfDNA from plasma, 16 ng of dsDNA was directly end-repaired, A-tailed, and linked to adapters. After 12 cycles of PCR, ∼750 ng was used as input for capture, followed by an additional postcapture 10-cycle PCR step. Adapters included 8 nt UMI from IDT for Illumina (UMI DNA/RNA UD Indexes, cat. #20034701). Libraries were sequenced in a HiSeq2500 instrument (Illumina), 2 × 100 paired-end, or NovaSeq 6000 (Illumina), 2 × 75 pair-ended. After quality assessment of the reads with the FastQC quality control suite, the UMI tags were extracted from the 5′ end of the reads, which were then aligned on the human reference genome GRCh37 from the 1000 Genomes Project with BWA-mem v0.7.17. Next, consensus sequences were generated from reads aligning on the same position and sharing the same UMI barcode with a minimum of three reads per consensus family using the Duplexseq tool, based on the protocol described by Kennedy and colleagues (37). Consensus sequences were aligned on the same genome reference and realigned with GATK v3.7.06. Variant calling was done with VarScan v2.4.37, with no filtering for minimum allele frequency. Frequent SNPs were filtered based on the gnomAD database (allele frequency ≤0.0001). The final consensus sequence average coverage depth per gene was ×2,427 (ranging from ×371 to ×12,340). Variant annotation was done with Annovar. Clinical significance classification of the variants was performed using the following databases: COSMIC (https://cancer.sanger.ac.uk/cosmic), cBioPortal (http://www.cbioportal.org/), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), OncoKB (https://www.oncokb.org/), and VarSome (https://varsome.com/). BM samples from cases #2 and #7 presented more than 75% of variants likely originating from oxidative damage (low frequency C>A and G>T). All variants with a frequency lower than 10% and a C>A or G>T change were excluded in these two cases. Stereotypical background errors across multiple genomic locations, repeated in at least 20% of the samples, or when detected only in the first or the last nucleotide of the consensus read according to the read coordinates were removed. The threshold to filter out false variants was P < 0.007. Variants with a P ≥ 0.007, classified as pathogenic or likely pathogenic and supported by at least four consensus families, were manually reviewed blinded to tissue sequencing data and sample type.
Analytic Validation of the VHIO-YWBC Panel
Analytic parameters of the VHIO-YWBC panel were assessed using two commercial reference DNAs with 1% allelic frequency (AF; SeraSeq ctDNA Mutation Mix v2 AF1%; cat. #0710-0140) and 0.5% AF (SeraSeq ctDNA Mutation Mix v2 AF0.5%; cat. #0710-0141). Three technical replicates were performed for each reference material. Libraries were prepared for BM samples but with 30 ng of starting material. The resulting libraries were captured and analyzed with our custom VHIO-YWBC panel and pipeline, respectively. Results obtained were compared with reference data from SeraSeq after capture with an Archer Reveal ctDNA 28 kit run on an Illumina MiSeq for all the exons of those genes present in both panels: AKT1, ALK, ERBB2, PIK3CA, ROS1, and TP53. True positives (TP) were considered those variants detected by the SeraSeq and VHIO-YWBC protocol; true negatives (TN) were considered those variants not detected by any method; false positives (FP) were those variants detected only by the VHIO-YWBC method; and false negatives (FN) were the variants detected only by SeraSeq. Sensitivity [TP/(TP + FN)], specificity [TN/(TN + FP)], positive predictive value [TP/(TP + FP)], and negative predictive value [TN/(TN + FN)] were calculated. The accuracy of the MAF % was analyzed as (X − X0/X0), where X is the detected value, X0 is the expected value (theoretical), and X − X0 is the absolute error.
ddPCR
The QX200 ddPCR System (Bio-Rad Laboratories) was used to confirm in plasma- and milk-derived cfDNA the presence of variants detected in tumor tissue by NGS. Genomic DNA from tumor tissue from the same patient was run as a positive control. TaqMan SNP genotyping assays for ddPCR were custom-designed to detect the mutations and purchased from Thermo Scientific (Supplementary Table S8). Assay design was chosen prioritizing pathogenicity and/or clonality.
Briefly, the 20 μL final volume of TaqMan PCR reaction mixture was assembled with 1× ddPCR Supermix for Probes (no dUTP), 900 nmol/L of each primer, 250 nmol/L of each probe and 8 μL of cfDNA from plasma, 15 ng of cfDNA from BM, or 30 ng of gDNA from FFPE per reaction (positive controls). Each assay was performed in duplicates or triplicates in separate mixtures and loaded in different wells for amplification. The thermal cycling program was performed according to the specifications of the manufacturer. After PCR, droplets were read in the Droplet Reader and analyzed with QuantaSoft version 1.7.4. Human reference genomic DNA was included as negative control and used to determine the cutoff for allele calling in each assay. ddPCR validations were carried out blinded to tumor genotype.
Statistical Methods
A descriptive analysis of all included variables in the study was performed. Continuous variables are expressed as median and range, and categorical variables are expressed as absolute values and percentages. Due to the limited number of cases, a nonparametric test was used to avoid the normality assumption. The nonparametric two-sided Mann–Whitney–Wilcoxon test was performed to compare the MAF % in BM serum across different patient characteristics. The threshold for statistical significance was defined as 0.05 (two-sided). No correction for multiplicity was performed due to the exploratory character of the study, and no data imputation was used.
Data and Material Availability
Patient-level somatic mutation data from BM, plasma, and FFPE solid tissue derived from ddPCR and the VHIO-YWBC NGS platform are available in Supplementary Table S3 and Supplementary Excel Tables S1 and S2. Due to patient privacy restrictions, raw NGS data will be shared with interested researchers upon submission of a proposal subject to approval by the Research Ethics Committee from Vall d'Hebron Hospital. The Methods sections “VHIO-300 Panel NGS” and “VHIO-YWBC Panel NGS” describe the implementation of established tools/pipelines; no custom software or computational code was created for this study. According to biological materials availability, samples are kept in the collection “Identification of diagnostic biomarkers, predictors of response and resistance to personalized therapies in patients with different types of cancer” (principal investigator: Josep Tabernero) registered at the National Registry of Biobanks (collection section), Instituto de Salud Carlos III with registration number C0003435.
Supplementary Material
VHIO-300 NGS panel gene list
AURORA NGS panel gene list.
Complete information of the Case cohort, with clinical features and results from the somatic mutation analysis
List of BM samples
VHIO-YWBC NGS panel gene list
Analytical validation of the VHIO-YWBC protocol
Sensitivity of the BM ctDNA detection assays.
List of oligos and probes for each ddPCR assay
Table 1. Samples analyzed by NGS Table 2. VHIO-300 results from solid tumor Table 3. VHIO-YWBC results from BM positive for ctDNA
Breast milk charactherization as liquid biopsy specim and comparison to plasma
Analytical validation of the VHIO-YWBC protocol for NGS
Diagnostic images of the PPBC from case #15 right breast
Acknowledgments
We thank the patients who participated in the study and donated samples for analysis for their generous contribution, with particular thanks to the first patient, Maite, and her daughter Àneu, who inspired us to initiate this study (oral consent to name the patient and her daughter was provided by the patient, and her legal partner provided written consent after patient's exitus). We are grateful to Javier Carmona for his valuable contributions and support in the manuscript's conceptualization, preparation, and revision. VHIO would like to acknowledge the Cellex Foundation for providing research facilities and equipment and the CERCA Programme from the Generalitat de Catalunya for their support of this research. The authors from VHIO acknowledge the State Agency for Research (Agencia Estatal de Investigación) for the financial support as a Center of Excellence Severo Ochoa (CEX2020-001024-S/AEI/10.13039/501100011033). This research is financially supported by the “El paseíco de la mama” Foundation. C. Saura was the recipient of a II FERO-GHD grant from the FERO Foundation (FERO/5086), a Junior Clinical award from the Spanish Association Against Cancer Foundation (FAECC; CLJUN212026ORTI), and a SEOM-Daiichi Sankyo grant for its support on the Breast Cancer Research Projects 2021 (SEOM/FECMA2022) and received funding from the Department of Health (Generalitat de Catalunya SLT008/18/00198) and from the Instituto de Salud Carlos III (ISCIII) and Fondo Europeo de Desarrollo Regional (FEDER), cofunded by the European Union (PI21/01020). C. Ortiz was the recipient of a Junior Clinician award from the FAECC (CLJUN212026ORTI) and a SEOM-Daiichi Sankyo grant for its support on the Breast Cancer Research Projects 2021 (SEOM/FECMA2022), and received funding from the Department of Health (Generalitat de Catalunya SLT008/18/00198). N. Bayó-Puxan received funding from the Department of Health (Generalitat de Catalunya SLT008/18/00205), MCIN/AEI/10.13039/501100011033 (GPE2022-001029) and MCIN/AEI/10.130.39/501100011033, and the European Union “Next GenerationEU/PRTR” (ECT2020-000827). J.M. Miquel received funding from the Department of Health (Generalitat de Catalunya SLT008/18/00205), MCIN/AEI/10.130.39/501100011033, and the European Union “Next GenerationEU/PRTR” (ECT2020-000827). J. Arribas is funded by the Breast Cancer Research Foundation (BCRF-23-008), Instituto de Salud Carlos III (project reference numbers AC15/00062, CB16/12/00449, and PI22/00001), and the European Commission under the framework of the ERA-NET TRANSCAN-2 initiative cofinanced by FEDER and Asociación Española Contra el Cáncer. A. Vivancos was the recipient of a project award from the FAECC (AVP/18/AECC/3219) and received funding from the Advanced Molecular Diagnostic (DIAMAV) program from the FERO Foundation (8361) and from ISDIN for supporting the development of liquid biopsy applications at the Cancer Genomics Lab (1848). M. Sansó was the recipient of a II FERO-GHD grant from the FERO Foundation (FERO/5086) and an investigator award from the FAECC (INVES19056SANS), and received funding from the Health Research Institute of the Balearic Islands (IdISBa), the RADIX-Janssen program (RADIX/JANSSEN21/01), and the Miguel Servet Program funded by the ISCIII (CP22/00131).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
C. Saura reports nonfinancial support from the Cellex Foundation, the CERCA Programme from the Generalitat de Catalunya, and the State Agency for Research, and grants from Paseico de la Mama, the FERO-GHD Foundation, the Spanish Association Against Cancer Foundation (FAECC; Junior Clinical award), SEOM-Daiichi Sankyo, Department of Health-Generalitat de Catalunya, and Instituto de Salud Carlos III (ISCIII) and Fondo Europeo de Desarrollo Regional (FEDER; cofunded by the European Union) during the conduct of the study. V. Peg reports personal fees from Roche, AstraZeneca, Sysmex, and Merck Sharp & Dohme outside the submitted work. P. Nuciforo reports personal fees from Bayer, MSD Oncology, Novartis, and Targos Molecular Pathology GmbH outside the submitted work. M. Gómez-Rey reports nonfinancial support from the Cellex Foundation during the conduct of the study. G. Villacampa reports personal fees from MSD, Pfizer, AstraZeneca, Pierre Fabre, GSK, and Reveal Genomics outside the submitted work. M. Espinosa-Bravo reports grants from the Institute of Health Carlos III during the conduct of the study. R. Dienstmann reports personal fees from Roche, Boehringer Ingelheim, Ipsen, Amgen, Servier, Sanofi, Libbs, Merck Sharp & Dohme, Lilly, Janssen, and Takeda, grants and personal fees from AstraZeneca, and grants from Novartis, Daiichi Sankyo, and Pierre Fabre outside the submitted work. J. Arribas reports grants from the Breast Cancer Research Foundation, Instituto de Salud Carlos III, Asociación Española Contra el Cáncer, and Byondis during the conduct of the study; grants from Fundación Mutua Madrileña, Fundación La Caixa, and Worldwide Cancer Research outside the submitted work; and a patent for EP22382294.1 issued, a patent for EP20382457.8 licensed to Mnemo Therapeutics, and a patent for EP2360187B1 licensed to Innovent. J. Tabernero reports other support from Array Biopharma, AstraZeneca, Bayer, Boehringer Ingelheim, Cardiff Oncology, Chugai, Daiichi Sankyo, F. Hoffmann-La Roche, Genentech, HalioDX SAS, Hutchison MediPharma International, Ikena Oncology, Inspirna, IQVIA, Lilly, Menarini, Merck Serono, Merus, MSD, Mirati, Neophore, Novartis, Ona Therapeutics, Orion Biotechnology, Peptomyc, Pfizer, Pierre Fabre, Samsung Bioepis, Sanofi, Scandion Oncology, Scorpion Therapeutics, Seattle Genetics, Servier, Sotio Biotech, Taiho, TheraMyc, and Tolremo Therapeutics, and other support from Oniria Therapeutics, Imedex, Medscape Education, MJH Life Sciences, PeerView Institute for Medical Education, and Physicians’ Education Resource outside the submitted work. A. Vivancos reports personal fees from Bayer, Bristol Myers Squibb, and Roche, and grants from Bristol Myers Squibb, Roche, and Incyte during the conduct of the study; personal fees from Guardant Health, Merck, Roche, Bristol Myers Squibb, Incyte, Bayer, and Reveal Genomics (cofounder) outside the submitted work. M. Sansó reports grants from the Health Research Institute of the Baleric Islands and Janssen, Fundación Científica de la Asociación Española Contra el Cáncer, Fundacion FERO, and Instituto de Salud Carlos III, and nonfinancial support from the Cellex Foundation, the CERCA Programme from the Generalitat de Catalunya, and Agencia Estatal de Investigación during the conduct of the study, as well as a competitive starting package grant at the Health Research Institute of the Balearic Islands (IdISBa), funded by the institution as well as by Janssen Pharmaceuticals and for this reason declares it as a conflict of interest, since it might give the appearance of potentially influencing the present work. No disclosures were reported by the other authors.
Authors’ Contributions
C. Saura: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. C. Ortiz: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, writing–review and editing, co–first author with C. Saura. J. Matito: Conceptualization, resources, data curation, formal analysis, investigation, methodology, writing–review and editing. E.J. Arenas: Conceptualization, resources, investigation, methodology, writing–review and editing. A. Suñol: Resources, data curation, investigation, methodology, writing–review and editing. A. Martín: Resources, formal analysis, supervision, writing–review and editing. O. Córdoba: Conceptualization, formal analysis, investigation, methodology, writing–review and editing. A. Martínez-Sabadell: Resources, data curation, formal analysis, investigation, methodology, writing–review and editing. I. García-Ruiz: Resources, data curation, formal analysis, investigation, writing–review and editing. I. Miranda: Conceptualization, resources, funding acquisition, project administration, writing–review and editing. C. Morales-Comas: Resources, supervision, investigation, writing–review and editing. E. Carrasco: Conceptualization, resources, data curation, formal analysis, funding acquisition, investigation, project administration, writing–review and editing. C. Viaplana: Conceptualization, resources, data curation, formal analysis, funding acquisition, investigation, project administration, writing–review and editing. V. Peg: Conceptualization, resources, formal analysis, funding acquisition, investigation, project administration, writing–review and editing. P. Nuciforo: Conceptualization, resources, formal analysis, funding acquisition, investigation, project administration, writing–review and editing. N. Bayó-Puxan: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, project administration, writing–review and editing. A. Gonzalez-Medina: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. J.M. Miquel: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M. Gómez-Rey: Conceptualization, resources, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. G. Villacampa: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. S. Arévalo: Conceptualization, resources, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M. Espinosa-Bravo: Conceptualization, resources, formal analysis, supervision, investigation, writing–review and editing. J. Balmaña: Conceptualization, resources, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. R. Dienstmann: Conceptualization, resources, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. J. Arribas: Resources, methodology, writing–review and editing. J. Tabernero: Conceptualization, resources, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. A. Vivancos: Conceptualization, resources, data curation, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing, co–corresponding author with C. Saura and M. Sansó. M. Sansó: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing, co–corresponding author with C. Saura and A. Vivancos.
References
- 1. Amant F, Lefrere H, Borges VF, Cardonick E, Lambertini M, Loibl S, et al. The definition of pregnancy-associated breast cancer is outdated and should no longer be used. Lancet Oncol 2021;22:753–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lyons TR, Schedin PJ, Borges VF. Pregnancy and breast cancer: when they collide. J Mammary Gland Biol Neoplasia 2009;14:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Callihan EB, Gao D, Jindal S, Lyons TR, Manthey E, Edgerton S, et al. Postpartum diagnosis demonstrates a high risk for metastasis and merits an expanded definition of pregnancy-associated breast cancer. Breast Cancer Res Treat 2013;138:549–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amant F, Loibl S, Neven P, Van Calsteren K. Malignancies in pregnancy 2 breast cancer in pregnancy. Lancet 2012;379:570–9. [DOI] [PubMed] [Google Scholar]
- 5. Amant F, Von Minckwitz G, Han SN, Bontenbal M, Ring AE, Giermek J, et al. Prognosis of women with primary breast cancer diagnosed during pregnancy: results from an international collaborative study. J Clin Oncol 2013;31:2532–9. [DOI] [PubMed] [Google Scholar]
- 6. Azim HA, Santoro L, Russell-Edu W, Pentheroudakis G, Pavlidis N, Peccatori FA. Prognosis of pregnancy-associated breast cancer: a meta-analysis of 30 studies. Cancer Treat Rev 2012;38:834–42. [DOI] [PubMed] [Google Scholar]
- 7. Shagisultanova E, Gao D, Callihan E, Parris HJ, Risendal B, Hines LM, et al. Overall survival is the lowest among young women with postpartum breast cancer. Eur J Cancer 2022;168:119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goddard ET, Bassale S, Schedin T, Jindal S, Johnston J, Cabral E, et al. Association between postpartum breast cancer diagnosis and metastasis and the clinical features underlying risk. JAMA Netw Open 2019;2:e186997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Z, Bassale S, Jindal S, Fraser A, Guinto E, Anderson W, et al. Young-onset breast cancer outcomes by time since recent childbirth in Utah. JAMA Netw Open 2022;5:E2236763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jindal S, Narasimhan J, Borges VF, Schedin P. Characterization of weaning-induced breast involution in women: implications for young women's breast cancer. npj Breast Cancer 2020;6:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Borges VF, Lyons TR, Germain D, Schedin P. Postpartum involution and cancer: an opportunity for targeted breast cancer prevention and treatments? Cancer Res 2020;80:1790–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jindal S, Pennock ND, Sun D, Horton W, Ozaki MK, Narasimhan J, et al. Postpartum breast cancer has a distinct molecular profile that predicts poor outcomes. Nat Commun 2021;12:6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bartlett AQ, Pennock ND, Klug A, Schedin P. Immune milieu established by postpartum liver involution promotes breast cancer liver metastasis. Cancers 2021;13:1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic— implementation issues and future challenges. Nat Rev Clin Oncol 2021;18:297–312. [DOI] [PubMed] [Google Scholar]
- 16. Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ponti G, Manfredini M, Tomasi A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit Rev Oncol Hematol 2019;141:36–42. [DOI] [PubMed] [Google Scholar]
- 19. Aftimos P, Oliveira M, Irrthum A, Fumagalli D, Sotiriou C, Gal-Yam EN, et al. Genomic and transcriptomic analyses of breast cancer primaries and matched metastases in AURORA, the Breast International Group (BIG) molecular screening initiative. Cancer Discov 2021;11:2796–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Azim HA, Nguyen B, Brohée S, Zoppoli G, Sotiriou C. Genomic aberrations in young and elderly breast cancer patients. BMC Med 2015;13:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ballard O, Morrow AL. Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am 2013;60:49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klein EA, Richards D, Cohn A, Tummala M, Lapham R, Cosgrove D, et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol 2021;32:1167–77. [DOI] [PubMed] [Google Scholar]
- 23. Patuleia SIS, Suijkerbuijk KPM, van der Wall E, van Diest PJ, Moelans CB. Nipple aspirate fluid at a glance. Cancers 2022;14:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patuleia SIS, Hagenaars SC, Moelans CB, Ausems MGEM, van Gils CH, Tollenaar RAEM, et al. Lessons learned from setting up a prospective, longitudinal, multicenter study with women at high risk for breast cancer. Cancer Epidemiol Biomarkers Prev 2021;30:441–9. [DOI] [PubMed] [Google Scholar]
- 25. Loud JT, Beckjord EB, Nichols K, Peters J, Giusti R, Greene MH. Tolerability of breast ductal lavage in women from families at high genetic risk of breast cancer. BMC Womens Health 2009;9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murphy J, Sherman ME, Browne EP, Caballero AI, Punska EC, Pfeiffer RM, et al. Potential of breastmilk analysis to inform early events in breast carcinogenesis: rationale and considerations. Breast Cancer Res Treat 2016;157:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Twigger AJ, Engelbrecht LK, Bach K, Schultz-Pernice I, Pensa S, Stenning J, et al. Transcriptional changes in the mammary gland during lactation revealed by single cell sequencing of cells from human milk. Nat Commun 2022;13:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong CM, Anderton DL, Smith-Schneider S, Wing MA, Greven MC, Arcaro KF. Quantitative analysis of promoter methylation in exfoliated epithelial cells isolated from breast milk of healthy women. Epigenetics 2010;5:645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qin W, Tsukasaki Y, Dasgupta S, Mukhopadhyay N, Ikebe M, Sauter ER. Exosomes in human breast milk promote EMT. Clin Cancer Res 2016;22:4517–24. [DOI] [PubMed] [Google Scholar]
- 30. Aslebagh R, Channaveerappa D, Arcaro KF, Darie CC. Proteomics analysis of human breast milk to assess breast cancer risk. Electrophoresis 2018;39:653–65. [DOI] [PubMed] [Google Scholar]
- 31. Bhat-Nakshatri P, Kumar B, Simpson E, Ludwig KK, Cox ML, Gao H, et al. Breast cancer cell detection and characterization from breast milk–derived cells. Cancer Res 2020;80:4828–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu F, Pawliwec A, Feng Z, Yasruel Z, Lebrun JJ, Ali S. Prolactin/Jak2 directs apical/basal polarization and luminal linage maturation of mammary epithelial cells through regulation of the Erk1/2 pathway. Stem Cell Res 2015;15:376–83. [DOI] [PubMed] [Google Scholar]
- 33. Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015;163:506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pereira B, Chin SF, Rueda OM, Vollan HKM, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun 2016;7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 2018;34:427–38.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ding L, Bailey MH, Porta-Pardo E, Thorsson V, Colaprico A, Bertrand D, et al. Perspective on oncogenic processes at the end of the beginning of cancer genomics. Cell 2018;173:305–20.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, et al. Detecting ultralow-frequency mutations by duplex sequencing. Nat Protoc 2014;9:2586–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
VHIO-300 NGS panel gene list
AURORA NGS panel gene list.
Complete information of the Case cohort, with clinical features and results from the somatic mutation analysis
List of BM samples
VHIO-YWBC NGS panel gene list
Analytical validation of the VHIO-YWBC protocol
Sensitivity of the BM ctDNA detection assays.
List of oligos and probes for each ddPCR assay
Table 1. Samples analyzed by NGS Table 2. VHIO-300 results from solid tumor Table 3. VHIO-YWBC results from BM positive for ctDNA
Breast milk charactherization as liquid biopsy specim and comparison to plasma
Analytical validation of the VHIO-YWBC protocol for NGS
Diagnostic images of the PPBC from case #15 right breast