A biobank of colorectal cancer organoids with matched cancer-associated fibroblasts was generated and serves as a preclinical model that can determine the impact of the tumor microenvironment on transcriptomic subtypes and sensitivity to therapeutic drugs.
Abstract
In colorectal cancers, the tumor microenvironment plays a key role in prognosis and therapy efficacy. Patient-derived tumor organoids (PDTO) show enormous potential for preclinical testing; however, cultured tumor cells lose important characteristics, including the consensus molecular subtypes (CMS). To better reflect the cellular heterogeneity, we established the colorectal cancer organoid–stroma biobank of matched PDTOs and cancer-associated fibroblasts (CAF) from 30 patients. Context-specific phenotyping showed that xenotransplantation or coculture with CAFs improves the transcriptomic fidelity and instructs subtype-specific stromal gene expression. Furthermore, functional profiling in coculture exposed CMS4-specific therapeutic resistance to gefitinib and SN-38 and prognostic expression signatures. Chemogenomic library screening identified patient- and therapy-dependent mechanisms of stromal resistance including MET as a common target. Our results demonstrate that colorectal cancer phenotypes are encrypted in the cancer epithelium in a plastic fashion that strongly depends on the context. Consequently, CAFs are essential for a faithful representation of molecular subtypes and therapy responses ex vivo.
Significance:
Systematic characterization of the organoid–stroma biobank provides a resource for context dependency in colorectal cancer. We demonstrate a colorectal cancer subtype memory of PDTOs that is independent of specific driver mutations. Our data underscore the importance of functional profiling in cocultures for improved preclinical testing and identification of stromal resistance mechanisms.
This article is featured in Selected Articles from This Issue, p. 2109
INTRODUCTION
Colorectal cancer belongs to the three most frequent types of cancer in both men and women and is the second most diagnosed cancer and the second leading cause of cancer-related death worldwide. Despite improvements in screening and therapy over the last years, incidence, prevalence, and mortality remain high in industrialized countries (1). Patients with colon cancer diagnosed with stage 4 disease have only an 11% survival rate at 5 years due to the ineffectiveness of current treatment regimens, strongly highlighting the need for new strategies for cancer screening and therapy that go beyond current approaches. Colorectal cancer should not be considered as one single disease, but rather as a heterogeneous complex of diseases. It would be too simplistic to reduce colorectal cancer growth to an accumulation of genetic alterations that change the behavior of tumor cells in a cell-autonomous manner. Instead, colorectal cancer depends on an intricate reciprocal interplay between tumor cells and their surrounding stroma and relies greatly on the plasticity of both tumor and surrounding cells within the tumor microenvironment (TME; ref. 2). The consensus molecular subtype (CMS) classification led to the identification of four subtypes (CMS1–4) and supports the clinical relevance of stromal cells for colorectal cancer (3). CMS1 represents hypermutated, microsatellite instable (MSI) tumors that are highly immunogenic and display immune cell infiltration and responsiveness to checkpoint inhibition (4). CMS2 tumors are associated with favorable prognosis and responsiveness to anti-EGFR therapy (5). Their gene expression is dominated by tumor-intrinsic features that are driven by specific DNA copy-number gains (6). CMS3 tumors are enriched for oncogenic KRAS variants and display metabolic changes. In contrast, CMS4 tumors, which have the worst prognosis of all subtypes, are characterized by abundant mesenchymal stroma and strong TGFβ signaling profiles (7, 8). Deregulated TGFβ signaling induces cancer-associated fibroblasts (CAF) that control local invasion, metastasis, and immune evasion, thereby driving tumor growth and progression (9). Importantly, inhibition of TGFβ signaling in fibroblasts enables T-cell infiltration and responsiveness to checkpoint inhibition in advanced, metastatic non-MSI tumors (10). Development of genetically engineered mouse colorectal cancer models that resemble clinical features of CMS4 tumors has allowed the study of therapeutic strategies such as inhibition of neutrophils or Notch signaling (11, 12). However, cross-species differences complicate the translation, and preclinical models that accurately reflect human colorectal cancer subtypes remain limiting.
Patient-derived tumor organoids (PDTO) have emerged as an excellent preclinical model for colorectal cancer. The technology is based on the expansion of primary epithelial cells under the control of defined growth factors and in a 3D extracellular matrix (ECM). Originally developed for the mouse small intestine (13), protocols have been adapted for efficient organoid derivation from normal and tumor tissues from the human colon (14) and other tumor entities (15–19). Several studies have demonstrated that living organoid biobanks can accurately represent the tumor heterogeneity among and within patients with colorectal cancer (20–22). More recently, coclinical trials have shown a high predictive value of colorectal cancer organoids for the individual patient response to chemo- and radiotherapy (23–26), demonstrating a great potential for personalized testing. However, PDTOs can only inadequately recapitulate the cellular contexture of the various molecular subtypes. Considering the importance of stromal cells, particularly for the prognosis of CMS4 tumors, PDTO monocultures are most likely limited to predict clinical responses. Thus, we aimed to establish a tumor organoid–stroma biobank to better reflect molecular subtype dependencies and to improve drug testing.
RESULTS
Development of a Colorectal Cancer Organoid–Stroma Biobank
To generate the colorectal cancer organoid–stroma biobank comprising PDTOs and CAFs, 30 tumors from various locations and stages of disease were included to reflect clinical heterogeneity in colorectal cancer (Fig. 1A and B; Supplementary Table S1). Organoids displayed diverse morphologies (Supplementary Fig. S1A), whereas fibroblasts were more uniform in adhesive culture (Supplementary Fig. S1B). Immunofluorescence analysis showed stable expression of α-SMA, VIM, and PDPN after initial and prolonged culture of CAFs (Supplementary Fig. S2A and S2B). Subsequently, established fibroblasts from tumor and matched adjacent normal tissues were analyzed by RNA sequencing (Supplementary Fig. S2C and S2D). Principal component analysis (PCA) showed distinct clustering and gene set enrichment analysis (GSEA) showed upregulation of inflammation and contractility signatures in CAFs as well as cell growth and translation signatures in normal fibroblasts, indicating that classic CAF features (27) are maintained in vitro.
Figure 1.
Clinical and genetic features of the colorectal cancer organoid–stroma biobank. A, Collected biomaterials from primary tumors (T), matched organoids (O), and fibroblasts (F). CRC, colorectal cancer; NA, data not available; TCGA, The Cancer Genome Atlas; TIS, tumor in situ; TMA, tissue microarray. B, Summary of clinical parameters in the experimental cohort (also see Supplementary Table S1) and public data (TCGA; ref. 28). C, Total detected alterations (SNPs and indels; log scale) in T and O. D, Mutational concordance. Note that private mutations are more frequent in organoids reflecting increased detection sensitivity in the absence of stroma. E, Recurrent mutations in cancer driver genes (based on OncoKB). Variant allele frequencies (VAF) are color coded. Average mutation frequencies (right) in this cohort reflect public data for colorectal cancer (28). Also see Supplementary Table S4. F, Mean copy-number changes in T, O (n = 30 each), and TCGA data (n = 319); also see Supplementary Fig. S3.
In parallel, the original tumor samples were analyzed on the molecular and histologic level using tissue microarrays (TMA; see inventory in Supplementary Table S2). Somatic variants in tumors and organoids were identified by whole-exome sequencing (WES) in comparison with matched normal genomic DNA (Supplementary Tables S3 and S4). Total mutation count showed a hypermutation phenotype in three of 30 cases (Fig. 1C). Concordance analysis showed a 63% ± 19% overlap between tumors and organoids (Fig. 1D). Private mutations were more abundant in organoids (24% ± 1%) compared with tumor tissues (13% ± 10%), indicating increased detection sensitivity in organoids that lack normal stroma. Inspection of recurrent mutations confirmed the presence of known driver mutations (Fig. 1E; Supplementary Table S4) in similar frequencies as reported previously (28). Hypermutated cases showed alterations in MSH3, MSH6, and/or POLE that co-occurred with mutations in BRAF, PIK3CA, and FBXW7. Copy-number variations (CNV) were preserved in organoids (Supplementary Fig. S3) and average CNVs resembled data reported for colorectal cancer (Fig. 1F), validating the established cohort.
PDTOs Remanifest Their Original Molecular Subtypes in a Stromal Context
To assess how stable tumor-specific gene expression is maintained in vitro, RNA sequencing was performed using tumor tissue, organoids, and xenotransplants (Fig. 2A). PCA showed distinct clustering between colorectal cancer tissues and corresponding organoids that were overall more homogenous (Fig. 2B). Gene set variation analysis (GSVA) showed downregulation of stromal and inflammatory signatures in organoids and upregulation of proliferative and metabolic genes compared with primary tumor samples (Fig. 2C). To model the stromal influence, organoids were systematically xenotransplanted into NOD-scid IL2Rgammanull (NSG) mice. Tumors from 13 models were analyzed by RNA sequencing, followed by species deconvolution of mRNA reads to distinguish gene expression in tumor (human) and stroma (mouse). In the tumor compartment, xenotransplantation increased the global similarity to clinical tissues (Fig. 2B) and partially restored the gene signatures that were most differentially regulated in PDTOs (Fig. 2C).
Figure 2.
Microenvironmental context is essential for the manifestation of transcriptomic subtypes. A, Schematic strategy for RNA sequencing in matched samples from primary tumors and organoids (n = 30 each), and following subcutaneous PDTO transplantation in NSG mice. An arbitrary subset of xenotransplants (n = 13) was analyzed by RNA sequencing, and human reads (h) were studied. For information on xenotransplant growth, see Supplementary Table S2. B, PCA shows transcriptomic differences among tumors and organoids and partial normalization in xenografts. C, Single-sample GSVA. Unsupervised clustering of tumors and organoids; the most differentially regulated HALLMARK signatures are shown. Note the restoration of tumor-specific signatures in xenografted organoids. EMT, epithelial-to-mesenchymal transition; OX., oxygen; Xeno, xenotransplant. D, CMS classification in clinical samples (Guinney et al.; ref. 3) and experimental models (see Supplementary Table S5). E, Sankey plots show weak overlap between tumors and organoids and increased concordance upon organoid xenotransplantation. See Supplementary Figs. S4 and S5.
Next, we classified the CMS of all samples (Supplementary Table S5). In tumor tissues, a faithful representation of all subtypes was found (Fig. 2D). PCA identified the CMS as the strongest source of biological variation (Supplementary Fig. S4A) compared with other clinical parameters (sex, age, location, prior antitumor treatment, mutation count, CNV status; Supplementary Fig. S4B–S4E). Most organoids were classified as of “mixed or indeterminate subtype” (Fig. 2D), likely due to the strong impact of the stroma on CMS assignment, and only 16.6% concordance was observed to matched tumors (Fig. 2E). Moreover, PDTOs did not cluster by PCA according to the original CMS or other clinical or pathologic parameters (Supplementary Fig. S4F–S4J). After xenotransplantation, the CMS concordance was increased to 46.6% (Fig. 2D and E) and for CMS1/2/3 tumors an accurate subtype was recovered in eight of 10 models. Although classification using the colorectal cancer–intrinsic subtypes (CRIS; ref. 8) should be less sensitive to stromal gene expression, high concordance was again observed only after transplantation (76.9% compared with 26.7% after culture in vitro; Supplementary Fig. S5A and S5B and Supplementary Table S6). Thus, PDTOs lose their original subtype under standard culture conditions, but they can be partially restored in a stromal context. In contrast, CMS4 tumors were not recovered (in five cases analyzed), most likely due to the absence of fibroblast transcripts in the human reads.
Cancer Cell–Dependent Programming of the TME
To explore whether cancer cells can instruct a subtype-specific stromal response, we examined mouse mRNA reads in xenotransplants. We compared organoids derived from CMS4 and CMS2, which are characterized by high and low CAF content, respectively (3). Differential analysis showed increased expression of fibroblast/muscle genes (Myh1, Ttn, and Ryr1) in CMS4-derived models, and inflammatory gene expression (Ccl6, Cd209f, and Ifi27l2a) in CMS2-derived models (Fig. 3A). GSEA confirmed induction of fibroblast and CMS4 signatures and repression of inflammatory signatures in CMS4-derived models (Fig. 3B and C), demonstrating that transplanted PDTOs induce a subtype-specific stromal response, which represents an unappreciated level of tumor self-organization. To further characterize the cellular microenvironment, we analyzed a TMA of tumors by multifluorescent immunostaining (Supplementary Fig. S6A and S6B). We used antibody panels specific to tumor cells (CK20), CAFs (α-SMA and VIM), and diverse immune cell populations (CD45, CD4, CD8, and CD163). Automatic image analysis and comparison between CMS2 (n = 9) and CMS4 (n = 7) models showed a significantly increased frequency of α-SMA+/VIM+ CAFs and smooth muscle cells in CMS4 tumors (Fig. 3D). In CMS2 tumors, a higher tumor cell percentage was found, resulting in a significantly increased tumor–stroma ratio. In contrast, the overall presence of immune cells (CD45) was not significantly altered. We then studied if the TME is recapitulated in PDTO xenografts. Immunofluorescent analysis of human E-Cadherin (tumor cells) and mouse Vimentin showed a markedly similar histologic pattern when compared with the original tumors (Fig. 3E).
Figure 3.
Organoid xenotransplantation elicits a subtype-specific stromal response. A, Differential analysis of stromal gene expression after xenotransplantation of CMS2-derived (n = 4) and CMS4-derived (n = 5) organoids in NSG mice. Volcano plot of mouse transcripts (base mean expression ≥10). Significantly changed genes (P < 0.05) are colored. B and C, GSEA of differential expression in CMS2 vs. CMS4 tumors and corresponding PDTO xenografts (mouse reads). MSigDB signatures for fibroblasts and inflammation (B) and the CMS4 signature (C) were studied. Normalized enrichment score (NES) and P values are shown. D, Multifluorescent analysis of the TME of matched TMA. Quantification of diverse cell populations. CAFs were defined by VIM expression and grouped into α-SMA–positive and α-SMA–negative cells. Significant changes between CMS2 (n = 9) and CMS4 tumors (n = 7) are labeled (*, P < 0.05; **, P < 0.01; Mann–Whitney U test). The tumor–stroma ratio was determined after image-based tissue segmentation (mean ± SD). E, Histologic analysis of TMA stained for pan-cytokeratin and Vimentin (top) and tumors obtained after xenotransplantation of matched PDTOs (bottom) stained for E-Cadherin and Vimentin. Staining was replicated in ≥2 animals each. All scale bars are 200 μm. See Supplementary Fig. S6.
In Vitro Modeling of Colorectal Cancer Subtype Determination
Our results indicate that both cell-autonomous and nonautonomous traits are encrypted in the tumor cell compartment. Niche factor dependency was expectedly linked to specific somatic alterations, including positive correlation of SMAD4 and KRAS/BRAF mutations with requirement for Noggin and EGF, respectively (Supplementary Fig. S7A and Supplementary Table S7; ref. 21). However, we did not detect any significant association with CMS (Supplementary Fig. S7B), and we reasoned that organoid–CAF cocultures could help to understand subtype determination, in particular for CMS4.
To address compartment specificity, we first tested differences between autologous and heterologous CAFs. Luciferase/GFP-transgenic organoids derived from two CMS2 and two CMS4 tumors were combined in Matrigel with matched or unmatched fibroblasts. To allow paracrine signaling, cells were cultured in a growth factor–reduced medium (Fig. 4A). We then studied the influence on colony number (Fig. 4B), organoid size (Fig. 4C), and tumor cell viability (luciferase activity; Fig. 4D). Characteristic differences between organoid lines were observed that were largely independent of the source of CAFs. The transcriptional response in organoids was then assessed after Transwell coculture (Fig. 4E). In a comparable setup, we had previously shown that Matrigel-embedded CAFs induce epithelial-to-mesenchymal transition (EMT) gene expression in mouse tumor organoids (29). We observed organoid-specific induction of EMT markers (LUM, SNAIL2, SPARC, and VIM) independently of the CAF origin. A weaker response was induced after the transfer of the CAF-conditioned medium (Fig. 4F), indicating reciprocal signaling activity. RNA sequencing of organoids (Fig. 4G) showed similar global transcriptomic effects by autologous and heterologous coculture. Thus, the presence of CAFs per se causes profound cellular and transcriptional changes that were largely defined by the responsiveness of tumor cells. RNA sequencing of CAFs showed that coculture with autologous or heterologous organoids induced a similar gene expression pattern (Fig. 4H) that was characterized by interferon and TGFβ signaling (Fig. 4I and J). qPCR analysis of four top-induced genes (NPR3, IFI6, ID1, and ANO1) showed a similar expression in CAFs and matched normal fibroblasts upon coculture, demonstrating an instructive role of tumor-derived signals (Fig. 4K). PDTO-conditioned medium, however, only partially recapitulated the effect of continuous coculture, suggesting the requirement for direct cell–cell contact and/or reciprocal signaling.
Figure 4.
Coculture restores the subtype-specific colorectal cancer phenotype that is determined by the tumor cell compartment. A, Direct coculture: luciferase-transgenic PDTOs were single-cell dispersed and co-embedded with autologous or heterologous CAFs in Matrigel followed by culture for 6 days in the reduced medium. B–D, Swapping experiment shows the comparable effect of autologous or heterologous CAFs on organoid number (B), area (C; both in n = 4 wells each), and viability (D; luciferase assay, n = 12 wells each). The cells were derived from CMS2 (blue) or CMS4 (green) tumors. Significant changes compared with organoids alone were determined by two-sided t tests (unequal variance, unpaired, FDR 1%). Experiments show mean ± SD and were repeated twice independently. ***, P < 0.001; **, P < 0.01; *, P < 0.05; n.s., P > 0.05. a.u., arbitrary units; Rel., relative. E, EMT gene expression in tumor organoids after Transwell coculture. qPCR analysis in n = 3 wells each. Shown is relative expression (log2-fold) compared with monoculture. The response depends on the tumor organoid rather than the source of fibroblasts. The experiment was repeated twice independently. F, EMT gene expression in tumor organoids (O11) after transfer of conditioned media (CM) from CAFs. Experiments in 3 wells each, and Transwell coculture was performed as control. Pooled data from two independent experiments are shown. G and H, RNA sequencing (RNA-seq) of organoids (G) and CAFs (H) after Transwell coculture. PCA plots show that tumor organoids and CAFs are influenced in a similar fashion by autologous and heterologous coculture. Note that the magnitude of PC1 (variation between individuals) exceeds PC2 (separating culture conditions). I and J, Differential expression analysis in CAFs after Transwell coculture (F11 and F17 ± autologous PDTOs). I, Volcano plot shows significantly changed genes (P < 0.05, colored). J, GSEA of HALLMARK signatures. Normalized enrichment score (NES) and P values are shown. K, qPCR analysis of gene expression in CAFs and matched normal fibroblasts (NAF). Relative expression (compared with monocultures) after transwell coculture with autologous PDTOs (left) or transfer of PDTO CM (right). Pooled data from two independent experiments are shown. L, Organoid/fibroblast coculture (O+F) affects organoid number, area, and viability (luciferase assay). Measured in n = 29 models (color indicates original CMS). Significance was analyzed by Wilcoxon matched-pairs signed rank test. ****, P < 0.0001. See also Supplementary Table S7. M, Spearman correlation of phenotypes (from L) to original tumor subtype. Organoids with >1,000 alterations were defined as mismatch repair deficient (dMMR). For cocultures, relative values compared with monoculture were studied. Significance was analyzed by Mann–Whitney U test; n.s., P > 0.05. Luc, luciferase; Rel., relative. N, CAFs induce subtype-specific transcriptomic features. Single-sample GSVA in tumors and in vitro contexts. Unsupervised clustering of differentially regulated HALLMARK signatures between CMS2- and CMS4-derived tumors (n = 9 each). Corresponding organoids (shown in the same order) lose subtype-specific signatures in full medium. Differences are recovered upon culture in reduced (red.) medium and Transwell coculture with fibroblasts. DN, down; HOMEOST., homeostasis; OX, oxidative; RESP., response. O, Pearson correlation of MSigDB signatures (GSEA) compared with tumors. Normalized enrichment scores between CMS2 (n = 9) and dMMR (n = 3), CMS3 (n = 6), or CMS4 (n = 9) models. CAF coculture selectively induces similarity in CMS4-derived PDTOs. Conditions as in N. P, Concordance of CMS and CRIS of PDTOs in different contexts and matched tumors. See Supplementary Fig. S8.
Next, we tested the generalized influence of CAFs on PDTO phenotypes. Cocultures of all biobank models (n = 29) resulted in significantly increased organoid number, size, and viability (Fig. 4L; Supplementary Table S7). Between individual lines, profound differences were observed that did not correlate with the original molecular subtype (Fig. 4M). We then explored whether modulation of the culture environment affected subtype fidelity in vitro. RNA sequencing and GSVA were performed either under regular PDTO culture, after growth factor reduction, or after additional Transwell coculture with CAFs in comparison with primary tumor tissues (Fig. 4N). In each condition, we focused on the changes between models originating from CMS2 and CMS4 tumors (n = 9 each). This comparison emphasized the differences in microenvironment-induced gene expression (CMS4) and in proliferation-associated signatures (CMS2) in primary tumor tissues that were essentially lost under regular PDTO culture conditions (Fig. 4N). In contrast, a stepwise restoration of the subtype-specific gene expression was observed after growth factor reduction and CAF coculture, which was further confirmed by a global correlation of enrichment scores for the MSigDB signatures (Fig. 4O). To evaluate if CAFs also regulate other subtypes, a similar comparison was performed for the mismatch repair–deficient (dMMR; n = 3) and CMS3 (n = 6) models. Although reduced medium had a moderate effect, presence of CAFs did not further improve similarity in dMMR and CMS3 models (Fig. 4O), indicating CMS4-selective responsiveness to coculture. The overall concordance of the resulting CMS and CRIS only moderately improved (Fig. 4P; Supplementary Fig. S8A and S8B), arguing that additional stromal signals may be required for increased similarity to matched tumors. Together, our results indicate that the manifestation of individual colorectal cancer characteristics strongly depends on the culture context and that a growth factor–rich medium perturbs subtype-specific expression.
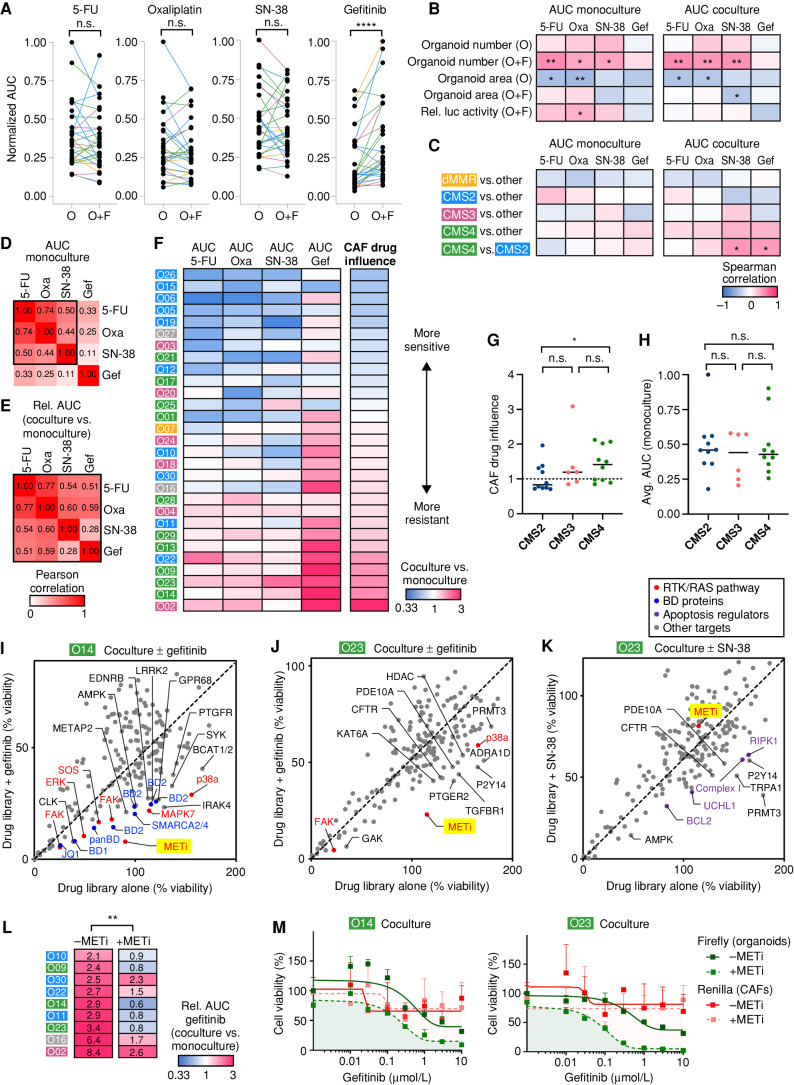
Manifestation of Subtype-Specific Responses to Chemotherapy Depends on CAFs
Given their increased transcriptomic fidelity, we reasoned that organoid–stroma cocultures could provide improved preclinical models for therapy. To address this hypothesis, we developed a platform for drug testing in cocultures. We noted that sensitivity to the EGFR inhibitor gefitinib, which correlates with response to the anti-EGFR antibody cetuximab (20), strongly depends on tumor cell density (Supplementary Fig. S9A and S9B). Therefore, we first normalized the seeding for each model to achieve a comparable colony number (Supplementary Fig. S9C and S9D). To faithfully determine tumor cell viability in cocultures, Firefly luciferase–transgenic organoids were co-embedded with CAFs in Matrigel in a 96-well format (Fig. 5A; Supplementary Fig. S9E). For efficient single-cell outgrowth, cells were seeded in a regular medium followed by drug treatment in the presence of a reduced medium to foster tumor–stroma interactions (Supplementary Fig. S9F–S9H). Using this setup, dose responses to the chemotherapeutic drugs 5-Fluorouracil (5-FU), oxaliplatin, SN-38, and gefitinib were determined (Supplementary Fig. S9I and S9J). CAFs had a significant impact on the drug response and could render PDTOs either more resistant or more sensitive. Mix-and-match experiments with autologous and heterologous CAFs demonstrated that the impact on the therapy response was PDTO-specific and not determined by the molecular subtype from which the CAFs originated (Fig. 5B and C; Supplementary Table S8). Given the growth-promoting effect of CAFs, drug sensitivity in CAFs may indirectly affect organoids. To monitor both compartments in cocultures, we developed a dual luciferase assay by combining Firefly luciferase–positive organoids with matched Renilla luciferase–positive CAFs (Supplementary Fig. S10A and S10B). CAFs generally showed reduced drug sensitivity compared with organoids (Fig. 5D; Supplementary Fig. S10C and S10D). Thus, indirect effects on CAF survival are not likely the major cause for the differential drug sensitivity of organoids in cocultures.
Figure 5.
CAFs’ influence on drug responses is dependent on the tumor cell compartment. A, Experimental setup for drug testing in cocultures using luciferase/GFP transduced organoids. B and C, Sensitivity to 5-FU, oxaliplatin, SN-38, and gefitinib in monoculture and coculture with autologous or heterologous CAFs. Tumor cell viability was measured by luciferase activity (mean ± SD in triplicate wells). Representative dose–response curves (B) and heat map of drug sensitivity [relative (rel.) AUC compared with monoculture; C]. Experiments were repeated twice. D, Measurement of organoid- and CAF-specific toxicity by dual luciferase assay. Coculture was performed using organoids expressing Firefly luciferase and matched CAFs expressing Renilla luciferase. Heat map shows normalized (norm.) AUC. CAFs show reduced sensitivity to all treatments compared with organoids. See Supplementary Figs. S9 and S10 and Supplementary Table S8.
Subsequent pharmacotyping of all biobank models (n = 29) in the presence and absence of CAFs revealed a heterogeneous spectrum of drug sensitivity, as determined by the normalized area under the curve (AUC). Although dose responses to 5-FU, oxaliplatin, and SN-38 were nonuniformly affected by the presence of CAFs (Fig. 6A; Supplementary Table S9), CAFs induced generalized resistance to gefitinib. Spearman analysis was performed to study how individual growth characteristics are linked (Fig. 6B). The organoid area was correlated with sensitivity to 5-FU and oxaliplatin in accordance with vulnerability of proliferating tumor cells. In contrast, the effect of CAFs on colony number was correlated with chemotherapy resistance. However, both associations were observed in monocultures and cocultures and therefore cannot explain the differential influence of CAFs. In monocultures, no subtype-specific drug sensitivity was observed (Fig. 6C). In contrast, coculture caused significantly higher resistance to SN-38 and gefitinib in CMS4- compared with CMS2-derived organoids. The responses to the three chemotherapeutic drugs were correlated, reflecting generalized drug sensitivity and resistance phenotypes as noted before (Fig. 6D; refs. 25, 26). Yet, we observed that the differential drug sensitivity in the presence/absence of CAFs [relative AUC (rel. AUC)] showed an even stronger correlation for all four drugs (Fig. 6E). Based on this finding, we established a CAF drug influence score for each patient. Comparison with the original CMS (Fig. 6F and G) showed that CAFs induced drug sensitization in CMS2 PDTOs. In contrast, CMS4 PDTOs were rendered more resistant to the therapeutic compounds. Importantly, such association was not observed when PDTOs were treated in monoculture (Fig. 6H). Taken together, our results emphasize the requirement of cocultures to faithfully reflect subtype-specific responses to therapy.
Figure 6.
Coculture exposes subtype-specific therapy resistance and individualized drug vulnerabilities. A, Pharmacotyping of four clinical drugs in biobank (n = 29, color indicates original CMS). Tumor cell viability was assessed in monocultures (O) and cocultures (O+F) by luciferase measurement. Normalized AUC was calculated by dividing the maximum AUC value for each drug. ****, P < 0.0001; n.s.,P > 0.05 (Wilcoxon matched-pairs signed rank test). B and C, Spearman correlation between drug responses and growth characteristics (B; data from Fig. 4) or original tumor subtypes (C). Organoids with >1,000 somatic alterations were defined as dMMR. Significant changes are labeled: *, P < 0.05; **, P < 0.01 (Mann–Whitney U test). Gef, gefitinib; Oxa, oxaliplatin; Rel luc, relative luciferase. D and E, Pearson correlation between different drug treatments. AUC in monoculture (D) and the relative change of drug sensitivity in the presence of CAFs (E; AUCco/AUCmono) are shown. F, Heat map of differential drug sensitivity in the presence of CAFs (AUCco/AUCmono). Data are sorted according to the CAF drug influence, representing the average relative change of all four treatments. CMS of original tumors are labeled. G and H, Subtype comparison between CAF drug influence, (G) and the average (Avg.) AUC in monoculture (H). Medians are marked. Coculture induces significantly higher resistance in CMS4- compared with CMS2-derived models. Mann–Whitney U test (*, P = 0.029; n.s., P > 0.05). I–K, Pharmacologic screens show patient- and treatment-specific resistance mechanisms. A chemogenomic library containing 186 drugs was tested in O14 and O23 in coculture with F14. Stroma-induced resistance was analyzed by comparison of the library alone or in combination with a sublethal concentration of gefitinib (I/J) or SN-38 (K). Tumor cell viability was assessed by luciferase measurement in transgenic organoids. Mean data from two experimental replicates are shown. Top hits comprise MET inhibitor (METi; BAY-474) and other RTK/RAS pathway–associated proteins (red), BD protein inhibitors (blue), and apoptosis regulators (violet). L, MET inhibitor treatment to overcome gefitinib resistance. Heat map shows differential gefitinib response in resistant cocultures (AUCco/AUCmono). Addition of 1 μmol/L BAY-474 restores sensitivity in eight of nine tested cocultures. **, P < 0.01 (Wilcoxon matched-pairs signed rank test). M, Dual luciferase assay in cocultures (O14 and O23); 1 μmol/L BAY-474 induces vulnerability of tumor cells (Firefly, green) but not of CAFs (Renilla, red). Mean viability (+SD in triplicate wells) relative to DMSO alone. Experiments were repeated twice independently. See Supplementary Fig. S11 and Supplementary Tables S9 and S10.
Chemogenomic Screening Identifies Patient- and Drug-Specific Mechanisms of Stromal Resistance
To explore how CAFs modulate therapy responses, we performed pharmacologic library screens. We analyzed cocultures of O14 and O23 that showed an exceptional increase of resistance to gefitinib (both) and SN-38 (O23; see Fig. 6F). The library contained 186 active and 106 negative control compounds that had been previously characterized within the Donated Chemical Probes (DCP) and Structural Genomics Consortium (SGC) programs or had been assembled as part of the EUbOPEN chemogenomic libraries (30–32). Apart from kinases, targets included epigenetic proteins and solute carriers (Supplementary Table S10). To identify drugs that resensitize the coculture to therapy, we performed the compound screen at a single concentration alone or in the presence of a sublethal dose of gefitinib (Fig. 6I and J) or SN-38 (Fig. 6K). In O14 and O23, inhibition of MET (BAY-474) was identified as the top sensitizer for gefitinib together with other drugs targeting the RTK/RAS pathway, including FAK, SOS, p38a/MAPK14, and MAPK7. In contrast to O23, O14 also showed a pronounced therapy-induced vulnerability to bromodomain (BD)-containing proteins (inhibitors for BD1, BD2, as well as a pan-BD BET inhibitor and an inhibitor for the BDs of SMARCA2/4 and PB1 BD5; Fig. 6I). To overcome SN-38 resistance in O23, different targets were identified, including apoptosis regulators (BCL2, RIPK1) or the respiratory chain complex 1 (Fig. 6K). Interestingly, inhibition of the purinergic receptor P2Y14 sensitized O23 organoids, to both gefitinib and SN-38, suggesting that targeting affected CAFs rather than tumor organoids, in line with a negligible expression of P2Y14 on colorectal tumor cells (33). Dose titration in resistant cocultures validated that combination with a MET inhibitor causes a significant resensitization to gefitinib (Fig. 6L; Supplementary Fig. S11). Dual luciferase assays, moreover, showed a direct effect of BAY-474 on tumor cells and not on CAFs (Fig. 6M). Collectively, our results highlight that CAFs confer therapy resistance by general mechanisms but also in a patient- and therapy-dependent manner. Thus, organoid–stroma models represent a promising translational tool to exploit strategies for individualized treatment.
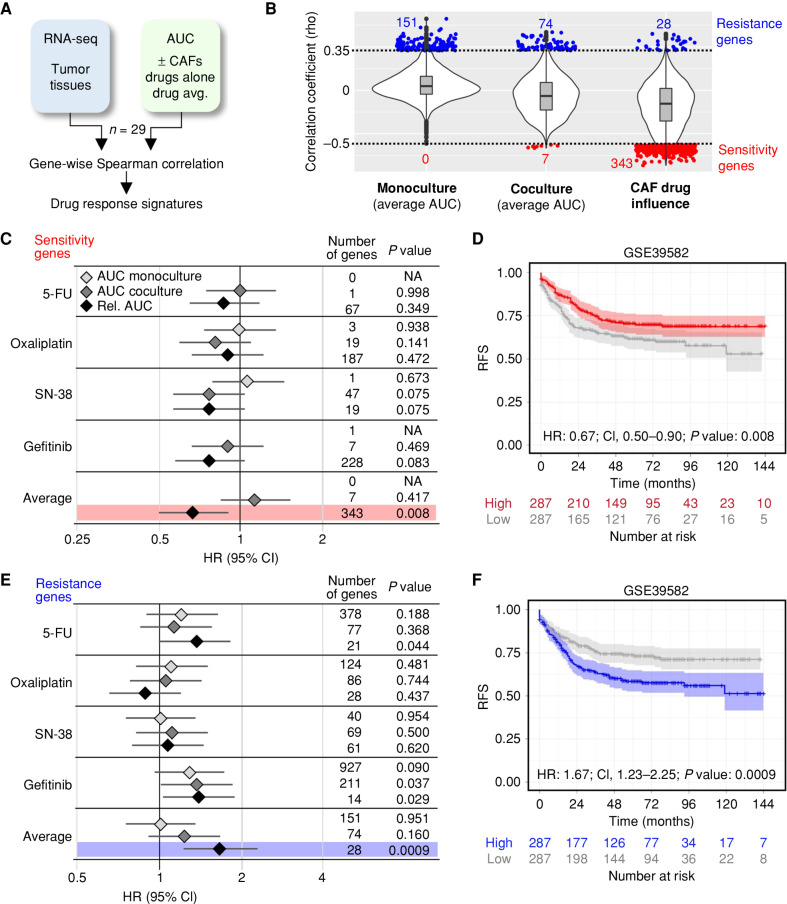
Pharmacotranscriptomic Signatures in Cocultures Allow Prognostic Colorectal Cancer Stratification
To evaluate if the divergent drug responses in organoid–stroma models are linked with prognosis of patients with colorectal cancer, we derived transcriptomic signatures. Spearman correlations were computed between gene expression in tumor tissues and the drug responses in corresponding PDTOs in the presence or absence of CAFs (Fig. 7A; n = 29). For the average AUC in monocultures and cocultures, only a few significant sensitivity genes were identified (Fig. 7B; rho < −0.5, P < 0.05). In contrast, the CAF drug influence was correlated with 343 sensitivity genes (Supplementary Table S11). Genes correlating with drug resistance (rho >0.35, P < 0.05) were observed in all conditions, and, subsequently, we tested the prognostic value of each gene signature for relapse-free survival (RFS) using a public gene expression cohort (GSE39582; ref. 34). The sensitivity signatures for individual drugs were not linked to an altered hazard ratio (HR). In contrast, expression of sensitivity genes based on the CAF drug influence identified patients with significantly improved RFS [Fig. 7C and D; HR: 0.67; 95% confidence intervals (CI), 0.50–0.90, P = 0.008]. A corresponding resistance signature identified patients with significantly worse RFS (Fig. 7E and F; HR:1.67; 95% CI, 1.23–2.25, P = 0.0009), highlighting that drug testing in organoid–stroma models shows improved potential for prognostic stratification compared with monocultures.
Figure 7.
Drug response in the organoid–stroma biobank is linked to distinct colorectal cancer patient outcomes. A, Strategy for the identification of pharmacotranscriptomic signatures (see Methods). B, Violin plots show the transcriptome-wide distribution of Spearman correlation coefficients (rho). Sensitivity (red) and resistance (blue) genes were filtered (rho < −0.5, rho >0.35, and P < 0.05, respectively). Correlations to the absolute response in monocultures and in cocultures (average AUC of 5-FU, oxaliplatin, SN-38, and gefitinib) and the relative response (AUCco/AUCmono) defined as CAF drug influence. C and D, Prognostic value of signatures linked with drug sensitivity. A public cohort (GSE39582) was divided into high- and low-expression groups. C, Forest plot shows HR with 95% confidence interval (CI) for RFS in the high-expression group. Signatures were derived from AUC in monoculture, coculture, and relative change (AUCco/AUCmono) from each drug alone and from the average. For each signature, the number of genes and log-rank statistics (P value) are listed. NA, no correlated genes were present in GSE39582. D, Kaplan–Meier plot for high (red) and low (gray) expression of the sensitivity signature (CAF drug influence). E and F, Prognostic value of genes linked to drug resistance. Data are shown as above. F, Kaplan–Meier analysis with resistance signature derived from CAF drug influence. P values for C–F were calculated using a univariate log-rank test. See Supplementary Fig. S12 and Supplementary Table S11.
Next, we compared the expression of the signatures with the CMS classification system. In a clinical cohort, the identified sensitivity and resistance signatures showed a reciprocal pattern of expression in CMS2 and CMS4 tumors, respectively (GSE39582; Supplementary Fig. S12A and S12B), in support of the observed subtype dependency of the drug response (Fig. 6). As reported, patients with the CMS4 subtype displayed worse RFS compared with the other subtypes (Supplementary Fig. S12C; ref. 3). We therefore tested if the stromal drug signatures could further stratify the outcome in these patients. Indeed, high expression of the sensitivity and resistance signatures identified CMS4 patients with markedly increased or worsened prognosis, respectively (Supplementary Fig. S12D and S12E), whereas fewer differences were observed for the other subtypes. Collectively, our results demonstrate that functional profiling in a coculture context can provide additional prognostic value to traditional expression-based classification methods.
DISCUSSION
The TME has been recognized as a key determinant for tumorigenesis, therapy response, and prognosis (2). Although single-cell RNA sequencing greatly increases the ability to catalog cellular heterogeneity (35–38), which helps to determine associations, we lack mechanistic insights to functionally explain how cell-intrinsic and cell-extrinsic inputs regulate individual colorectal cancer phenotypes. Here, we have established an organoid–stroma biobank to demonstrate that individual tumor subtypes are encrypted within the cancer cell compartment yet critically depend on the cellular context. Coculture of PDTOs with CAFs reestablished transcriptional features observed in poorly prognosed primary CMS4 tumors and affected the response to clinical drugs. These results are in line with the recently established IMF [intrinsic epithelial subtype (I), microsatellite instability status (M), and fibrosis (F)] classification of colorectal cancer, representing a refined, epithelial cell–centered CMS classification (38), as we provide functional evidence that tumor cell–intrinsic mechanisms define the subtype classification. Yet, fibroblasts represent modulators of therapy response ultimately affecting patients’ overall survival and prognosis.
Using matched clinical samples as a reference, we demonstrate that standard culture conditions largely erase tumor-specific features including CMS and CRIS subtypes. Thus, a growth factor–rich medium, although required for efficient PDTO expansion, critically confounds the tumor cell phenotype. Such culture-induced biases are plausible given that the TME is characterized by limitations of niche factors and metabolites as has been recently noted in pancreatic cancer models (39). Indeed, our systematic analysis revealed that the reduction of growth factors and culture in the presence of CAFs suffice to restore tumor-intrinsic signaling processes, including proliferation-related signatures in CMS2-derived organoids and reexpression of characteristic EMT gene signatures in CMS4-derived organoids. This paradigm suggests that a less instructive environment results in more informative models. In this context, CAFs allow culture conditions that contain more physiologic levels of growth factors and cytokines, and more physiologic ECMs should be considered (40). Interestingly, although the phenotype of CMS2 tumors is more determined by tumor cell–intrinsic features (6), we did not observe a bias for niche factor dependency, arguing that the observed induction of CMS4 features does not merely reflect increased sensitivity to perturbation.
In addition, PDTOs showed the capacity to instruct subtype-specific stromal changes demonstrating an unappreciated level of tumor self-organization. We observed pronounced fibroblast recruitment and immunosuppressive signaling in xenografts from CMS4-derived PDTOs, two features that could be mechanistically linked, for example, via suppressive influence of fibroblast-derived TGFβ (7, 10). Retransplantation induced stromal features reminiscent of the original histology, suggesting that complex traits of the TME may be induced by coculture with other cells. Interestingly, we found that phenotypic responses were largely determined by the source of tumor cells and not by the origin of CAFs, strongly underscoring the high degree of stromal plasticity. For nonimmune-related phenotypes, our results indicate that heterologous CAFs or normal fibroblasts may be used in cases in which autologous CAFs are not available. Future experiments should systematically compare CAF plasticity in cocultures and matched tumors on a single-cell level and identify the cellular mediators that control interactions between both compartments.
Previous studies have reported that patients with mesenchymal CMS4 tumors show no benefit from anti-EGFR therapy when combined with oxaliplatin-based chemotherapy (5, 41). In contrast, when combined with irinotecan-based chemotherapy, all subtypes showed significant survival benefit from anti-EGFR (42). Although previous studies have demonstrated a high negative predictive value of PDTO models, the response to some drugs including oxaliplatin is not well reflected between patients and organoids (25). Here, we tested the hypothesis that the addition of an essential TME component known to be involved in poor prognosis (3) could provide more predictive models. Indeed, we observed that CMS-specific drug sensitivities are entirely lost in monocultures. Although we detected a general protective effect by CAFs on gefitinib, we found that CMS4 models display the most pronounced protective benefit from the stroma. This is in line with the recent finding that tumors with acquired resistance to cetuximab become enriched for the CMS4 phenotype and show increased expression of CAF-derived growth factors such as TGFβ, HGF, and FGF family ligands that can compensate for EGFR pathway blockade (43). Indeed, drug screening confirmed that stromal resistance to gefitinib creates a vulnerability for RTK/RAS signaling, most prominently affecting the HGF receptor MET. In addition, patient-specific sensitivity to BET protein inhibition was observed indicating that interference with epigenetic plasticity may represent a strategy to prevent CAF-mediated resistance. Our results also highlight that CAFs can affect therapy in various ways in a context-dependent manner. Most likely this is due to the high degree of CAF heterogeneity that depends in part on tumor cell–derived signals (44). Thus, our data underscore the utility of the organoid–stroma biobank to identify both general and individual mechanisms of therapy resistance.
A number of recent studies have reported the high prognostic and predictive value of the TME in primary colorectal cancer tissues (45). We postulate that functional characterization of organoid–stroma cocultures can provide an additional parameter that may be more independent of the high spatial variability of the TME (46–48). Intriguingly, testing in a stromal context clearly outperforms the response signatures established in monocultures. Coclinical studies should address the predictive and prognostic value following the perturbation of organoid–stroma models. Yet, CAFs represent only one aspect of the complex TME that is comprised of additional elements such as immune and vascular cells. Incorporation of immunomodulatory elements such as macrophages (49) could help to provide informative preclinical models, in particular for immunotherapy (50). We observed an unappreciated capacity of PDTOs to instruct subtype-specific stromal changes, indicating that self-organization processes in coculture may be exploited to unlock the phenotypic plasticity of cancers (51). However, until this complexity can be faithfully modeled and exploited ex vivo, our results provide evidence that organoid–CAF cocultures are instrumental in unleashing the full potential of PDTOs for personalized oncology.
METHODS
Collection of Clinical Samples and Data
Resection samples from 30 patients with colorectal cancer were provided by the University Cancer Center Frankfurt (UCT). All materials were collected as part of the interdisciplinary Biobank and Database Frankfurt after prior written informed consent, and the study was approved by the institutional review board of the UCT and the Ethical Committee at the University Hospital Frankfurt (ethics vote: 4/09; project numbers: SGI-06-2015 and SGI-12-2018). Samples were pseudonymized, and associated clinical data were obtained (age, sex, diagnosis; Supplementary Table S1). Fresh tissue samples were rapidly processed on ice and subjected to routine pathologic assessment for preparation of formalin-fixed, paraffin-embedded (FFPE) specimens. From tumor samples, necrotic regions were removed and tissues were cut with scalpels into pieces (∼1–2 mm diameter). In parallel, fresh tissue samples from tumor-adjacent normal regions were processed for calling somatic mutations. From each sample, three to five randomly chosen pieces were immediately frozen at −80°C for subsequent DNA/RNA isolation. The remaining tumor tissue was used for derivation of primary cells (see below). DNA/RNA extraction was performed by the AllPrep DNA/RNA Micro Kit (Qiagen) following the manufacturer's suggestions. Tissues were homogenized in lysis buffer using a scalpel and then passed on a QIAshredder microcentrifuge spin-column (Qiagen). DNA was eluted in 100 μL PCR-grade water and RNA in 40 μL RNAse-free water and stored at −80°C before analysis.
Derivation of the Colorectal Cancer Organoid–Stroma Biobank
Organoid cultures were established and maintained as described previously (20). Briefly, tissue pieces were collected in a cold washing medium (DMEM, Thermo Fisher Scientific; 0.4% BSA, Sigma-Aldrich; 1× penicillin/streptomycin, Thermo Fisher Scientific) and washed twice using cold PBS, followed by incubation for 30 minutes in 4 mmol/L EDTA/PBS. Every 10 minutes, the pieces were pipetted vigorously to release tumor cells. After the sedimentation of tissue pieces, the supernatant containing tumor cells was collected followed by the addition of 20 mL of cold washing medium and centrifugation. The cell pellet was washed twice using a cold washing medium and then resuspended in 75% Matrigel. Matrigel domes were incubated for 30 minutes at 37°C before the addition of the culture medium. Full tumor medium contained advanced DMEM/F12 supplemented with 10 mmol/L hepes, 1 × glutamax, 1 × penicillin/streptomycin, 2% B27, 12.5 mmol/L N-acetylcysteine, 500 nmol/L A83-01 (R&D Systems), 10 μmol/L SB202190 (Sigma-Aldrich), 20% R-spondin 1–conditioned medium, 10% Noggin-conditioned medium, and 50 ng/mL human EGF (PeproTech). Conditioned media were prepared as described (52). Y-27632 (10 μmol/L) was added to the medium for the first 3 days after seeding or passaging. Primocin (100 μg/mL; InvivoGen) was added to the culture medium for the first 2 passages. Organoids were passaged by applying mechanical force. Low passage number samples were cryopreserved as master aliquots, and cells were kept in culture for at most 10 passages before replacement from frozen stocks.
For the isolation of primary colon fibroblasts, the remaining tissue pieces were further incubated in fresh 10 mmol/L EDTA/PBS for 30 minutes on ice. After vigorous pipetting, the supernatant was discarded and the pieces were washed 3 times before seeding in DMEM (Thermo Fisher Scientific) including 1 × glutamax, 10% fetal bovine serum (Sigma-Aldrich), and 1× penicillin/streptomycin on a 10-cm tissue culture dish. Primocin (100 μg/mL) was added to the culture medium for the first 2 to 3 passages. After 24 hours of culture, the pieces were washed and plated in a fresh tissue culture dish that was coated for 30 minutes with 0.1% gelatin/PBS and incubated for 1 week without disturbance to promote attachment and migration of the fibroblasts. The medium was changed weekly, and the first passage was performed after 2 to 3 weeks. Cells were split at confluency in a 1:3 ratio after cell dissociation with TrypLE Express (Thermo Fisher Scientific). Low passage number samples were frozen as master aliquots.
Available materials of the colorectal cancer organoid–stroma cohort are listed in Supplementary Table S2. Cells from one case were excluded from the experiments because the patient had been diagnosed with HIV. Several CAF lines did not recover from liquid nitrogen or were lost due to a lack of cell proliferation. All fibroblasts were inspected after prolonged culture without passaging to detect potential tumor cell contamination visible by the presence of tumor cell colonies with cobblestone morphology in which case CAF lines were excluded. All cells were regularly tested for Mycoplasma.
WES Analysis
From each sample, genomic DNA from tumor, tumor organoids, and normal adjacent tissue were subjected to whole-exome analysis. Genomic DNA (450 ng) was used for library preparation using the Agilent Low Input Exome-Seq Human v6 kit. Indices were introduced using the Agilent SureSelect XT kit, and sequencing was performed on a HiSeq 4000 at the Genomics and Proteomics Core Facility at DKFZ Heidelberg. On average, 98 ± 15 million reads (100 bp, paired ends; Supplementary Table S2) were obtained and Q30 values were >95% and >93.7% for reads 1 and 2, respectively.
Pathologic variants were detected by following the GATK best practices (53) using Mutect2 (GATK3 v3.8). In addition, we used the variant callers Mutect1 v1.1.7 (54) and Varscan2 v2.4.3 (RRID:SCR_006849; ref. 55). Only variants with a variant allele frequency cutoff of 5% identified by Mutect2 and confirmed by one of the other two variant callers were considered and annotated using the Ensembl variant effect predictor (RRID:SCR_007931; ref. 56). Supplementary Table S3 lists the alterations that were included in the comparisons. For each model, the number of detected somatic alterations (shared and specific for tumors and organoids) is listed in Supplementary Table S4 and mutation frequencies were compared with published data (28). For recurrent mutations, the sum of allele frequencies of all identified variants of the indicated locus is shown. Tumor purity and ploidy were analyzed in tumor and organoid samples using the Sequenza package (RRID:SCR_016662; ref. 57). Heat map of individual CNVs was plotted using CNVkit v0.9.3 (RRID:SCR_021917; ref. 58). Average CNVs were displayed by “GenVisR” (59) and compared with 319 randomly selected samples from The Cancer Genome Atlas colorectal cancer cohort for which copy-number segments obtained through the DNAcopy R package (RRID:SCR_012560) were downloaded from the GDC Data Portal (RRID:SCR_014514; release 12.0).
RNA Sequencing Analysis
RNA concentration was measured using the Qubit RNA HS Assay Kit (Thermo Fisher Scientific). For clinical tissues, the RNA integrity was analyzed by RNA ScreenTape (Agilent). For library generation of matched tumors, organoids, and xenotransplants (Supplementary Table S2), the SMARTer Stranded Total RNA Sample Prep Kit–HI Mammalian kit (Takara Bio) was used following the manufacturer's recommendations. Total RNA (1 μg) was fragmentized in a bioruptor (Diagenode) for 5 minutes for samples with an RNA integrity number (RIN) >7, 4 minutes for RIN 4 to 7, 3 minutes for RIN 4 to 3, and 2 minutes for RIN ≤3 and chilled on ice water. Final libraries were measured by Qubit and ScreenTape analysis before multiplexing. RNA sequencing was performed using the TruSeq RNA 50 cycle kit (Illumina) and run on a HiSeq 2000 instrument (Illumina) at the Genomics and Proteomics Core Facility at DKFZ Heidelberg. On average, 29 ± 9 (SD) million reads per sample were measured. For in vitro experiments (Figs. 4 and 5), RNA was collected using the NucleoSpin RNA kit (Macherey-Nagel) according to the manufacturer's instructions and 500 ng RNA was used for sequencing (random priming, 50 bp single read) at the Integrative Genomics Core Unit of the University Medical Center Göttingen on a HiSeq 4000 instrument (Illumina), and on average 38 ± 9 (SD) million reads per sample were measured.
The quality of FASTQ files was checked using FastQC software (version 0.11.8) before they were preprocessed to trim adapters, and remove artifacts and low-quality reads using Fastp software (version 0.20.1). The reads were aligned using STAR version 2.7a (RRID:SCR_004463), and read counts were computed by the STAR function “quantMode” using reference genome GRCh38 for human and GRCm38 for mouse. Gene annotation and species conversion were done using the biomaRt R package (RRID:SCR_019214; version 2.38.0). Differential gene expression was performed in an unpaired manner, unless otherwise mentioned in the text, using the DESeq2 R package (RRID:SCR_000154; versions 1.12.4, 1.18.1, and 1.32.0). Genes with base mean expression <50 (or <10 for mouse stromal genes) were filtered out, and differential gene expression was considered significant if log2 fold change was ≥1 or ≤−1 and the adjusted P value was <0.05. For PCA, read counts were normalized using “vst” function of the DESeq2 package for the top 1,000 genes and plotted using the “autoplot” function of the ggplot2 package (version 3.3.5).
The Xenome tool was used to deconvolute human and mouse transcripts from xenograft bulk RNA sequencing data (60). In short, an index file was constructed from the human (graft) and mouse (host) reference genomes (GRCh38 and GRCm38, respectively) by converting the files from NCBI to FASTA format. The “classify” function was then used to sort the reads as “graft,” “host,” “both” (graft and host), “neither” (graft nor host), or “ambiguous,” and for each category, new FASTQ files were generated and subjected to differential gene expression analysis as above.
Gene Set Enrichment Analysis
GSEA was performed using the fgsea R package (RRID:SCR_020938; version 1.18.0) as previously reported (bioRxiv 060012). Genes with base mean <20 (or <10 for the mouse stroma) were removed before they were ranked ascendingly based on the log2 fold change. The ranked gene list was then compared against MSigDB (RRID:SCR_016863; version 7.4) using the “fgsea” function with the default settings. Normalized enrichment scores (NES) and adjusted P values were used for subsequent analyses. In Fig. 3C, the reported CMS4 signature (61) was filtered for robust expression using the GEO2R tool (RRID:SCR_016569; log2 expression >6; 398 genes) and analyzed using the preranked GSEA software (RRID:SCR_003199; version 4.04).
The GSVA R package (RRID:SCR_021058; version 1.40.1) was used to determine gene set enrichment at the single sample level (62). Briefly, raw read counts were converted to log2 RPKM using the “rpkm” function of the limma package (RRID:SCR_010943; version 3.48.3) and the “gsva” function was used to calculate enrichment of the HALLMARK signatures from MSigDB using the default settings. To determine the differentially expressed signatures, the “Bayes” function of the limma package was used. The top 20 signatures based on the adjusted P value were used to extract sample- or context-specific signatures and to create heat maps using the ComplexHeatmap R package (RRID:SCR_017270; version 2.8.0).
Colorectal Cancer Subtype Classification
For CMS classification, expression data were normalized using the “vst” function of the DESeq2 package. Gene names or Ensembl IDs were transformed to NCBI-Entrez IDs using the biomaRt R package. The data were then subjected to the CMSclassifier R package (version 1.0.0; to derive random forest and single-sample prediction output; Supplementary Table S5). In addition, all samples were analyzed using the R package CMScaller (63). For each sample, the subtype was assigned if at least two of three prediction methods were congruent or otherwise the sample was assigned as “mixed.” The CRIS classification was performed using the “predictCRISclassKTSP” function of the CRISclassifier R package (version 1.0.0; ref. 8) using vst-normalized expression data. Sankey diagrams were generated using the “sankeyNetwork” function of the networkD3 package (version 0.4).
TMA and Multiplex Immunofluorescent Analysis
FFPE tissues from 25 of 30 tumors were assembled into a TMA using TMA Grand Master (3DHISTECH). Regions were defined by expert pathologic assessment and for each tumor, one to three paraffin cores (1 mm diameter) were included. Paraffin sections (3-μm thick) were stained with Opal 7-Color Automation IHC Kits (Akoya Bioscience) in the BOND-RX Multiplex IHC Stainer (Leica) following established protocols (64). Each section was put through 6 sequential rounds of staining, which included blocking in 5% BSA, followed by incubation with primary antibodies of two panels that were used to study tissue architecture with a focus on fibroblast activation—PanCK, CD45, VIM, α-SMA, KI67, and PD-L1—or the immune microenvironment—CD3, CD4, CD8, CD163, FoxP3, and PD-1 (for staining conditions and RRIDs, see Supplementary Table S12). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) contained in the Opal 7-Color Automation IHC Kits, and slides were mounted with Fluoromount-G (SouthernBiotech). Imaging was performed with the VectraPolaris imaging system (Akoya Bioscience). Tumor regions of interest were defined manually, and out-of-focus images or high background signals were excluded from further analysis. Images were analyzed by using the phenotyping application of inForm software V2.54.10 (Akoya Bioscience; RRID:SCR_019155). Cell numbers of assigned phenotypes were then exported, and identifiers for each core were annotated using R. For tumors that were represented by multiple cores, cell numbers were summed over all replicates before calculating the ratios. P values for comparisons of indicated groups were calculated using the Mann–Whitney U test.
For immunofluorescence of fibroblasts, cells with low and high passage numbers were seeded in a 48-well plate coated with 0.1% gelatin/PBS. The cells were fixed using 4% PFA for 30 minutes at room temperature, followed by one wash with PBS. Cells were stored in PBST + 1% sodium azide at 4°C prior to staining. The following primary antibodies were diluted in PBST + 3% BSA: PDPN/VIM/α-SMA or CD31/EPCAM were coincubated overnight at 4°C. CD45-PE was incubated for 1 hour. Cells were washed 3 times with PBST and incubated for 1 hour at room temperature with the respective secondary antibodies. Finally, cells were washed 3 times with PBST and mounted with Prolong Gold Antifade with DAPI (Thermo Fisher Scientific), and confocal images were captured using a Cytation C10 Confocal Imaging Reader (Agilent; 10 × objective). A full list of antibodies, including staining conditions and RRIDs, is shown in Supplementary Table S12.
Transplantation Experiments and Tumor Histologic Analysis
All animal housing and experimental procedures were approved by the institutional animal care and the regional animal welfare office, Darmstadt (F123/1004 and F123/1038). For xenotransplantation, organoids were collected in a cold medium, mechanically dissociated, and centrifuged for 5 minutes at 1,200 RPM, and the pellet was resuspended in full tumor medium (see above) supplemented with 25% Matrigel and 10 μmol/L Y-27632. Per mouse, 100 μL organoid suspension representing two confluent wells (12-well plate) was injected subcutaneously into the right flank of NSG mice. Mouse cohorts were gender- and age-matched, and tumors were collected after 10 to 16 weeks and processed for RNA isolation (as above) or used for immunofluorescent analysis as described (29). Antibodies used were anti–E-Cadherin (BD Biosciences, cat. #610181; RRID:AB_397580) and anti-Vimentin (Abcam, cat. #ab92547; RRID:AB_10562134; Supplementary Table S12). Images were scanned using a Cytation C10 microscope (widefield optics; 10 × objective).
Niche Dependency and Coculture Experiments
For each tumor organoid, individual niche factor requirements were tested by weekly passage (1:4 split ratio) and culture in full medium and media lacking individual factors. After each passage, organoids were imaged to document dependency of the deprived factor and organoids were considered independent if they maintained growth for >5 passages (Supplementary Table S7). For culture in growth factor–reduced medium, organoids were kept in a medium containing advanced DMEM/F12 supplemented with 10 mmol/L hepes, 1 × glutamax, and 1 × penicillin/streptomycin.
For all cocultures, except in swapping experiments, autologous CAFs were preferentially used. When unavailable, heterologous CAFs were used that were derived from the tumors of the identical CMS (Supplementary Tables S2 and S9). All cocultures were performed in a reduced medium (as above). Transwell cocultures were performed as described (65) using 12-well tissue culture inserts (Transwell Corning; pore size 8 μm). In the lower compartment, 50,000 fibroblasts were embedded in 50 μL BME (Amsbio). In the top compartment, PDTOs were seeded after mechanical dissociation in 60 μL Matrigel and cells were cultured for 96 hours in the presence of a 2 mL reduced medium. As a control, each organoid was cultured in a reduced medium in the absence of fibroblasts. Organoid RNA was collected using the NucleoSpin RNA kit (Macherey-Nagel) and used for RNA sequencing (described above). For RT-qPCR analysis, cDNA was generated from 500 ng of RNA using random hexamers and M-MLV reverse transcriptase as described (29). Gene expression was measured using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) on a StepOneplus instrument (Applied Biosystems) using the primer pairs shown in Supplementary Table S12. The relative expression (normalized to HPRT) was calculated using the 2(−ΔΔCT) method.
For direct coculture, organoid cells after single-cell dissociation by 3 to 5 minutes of enzymatic digest with Accutase (Thermo Fisher Scientific) and filtration (40 μmol/L, Greiner) were seeded alone or with 5,000 fibroblasts in 15 μL 90% Matrigel in 96-well round-bottom plates (Sarstedt). Cells were overlaid with 100 μL/well medium and sealed with Breathe-Easy membranes (Sigma-Aldrich). The required number of organoid cells was determined for each line by a prior colony formation assay. For this, 500, 1,000, 2,000, and 5,000 live cells after trypan blue staining and manual counting were seeded per well and the colony number was determined after 9 days of culture in full organoid medium from images (EVOS FL; Thermo Fisher Scientific, 2 × objective) of triplicate wells using ImageJ software (RRID:SCR_003070). Linear regression analysis was performed with GraphPad Prism to calculate the number of organoids/2,000 input cells (Supplementary Table S7).
Tumor cell viability in cocultures was measured by luciferase assay. For this purpose, tumor organoids (n = 29) were individually transduced with a Luciferase2-P2A-EGFP lentivirus as described (66) and stable expression was selected by expansion in the presence of 0.5 to 1 μg/mL puromycin. Monocultures and cocultures (as above) were then studied using the ONE-Glo EX assay (Promega) according to the manufacturer's instructions and luminescence was recorded on a SpectraMax iD3 Multimode Microplate Reader (Molecular Devices). Average results from triplicate wells were measured (Supplementary Table S7). For automated image morphometry, brightfield images from four independent wells were captured (EVOS, 2 × objective), and the organoid number and mean organoid area were measured using OrganoSeq software (ref. 67; Supplementary Table S7).
Drug Testing in Cocultures
The influence of cell density on drug sensitivity was tested by seeding 1,000 or 5,000 live cells in full medium in a 96-well format as above. After 4 days, the cells were exposed to different concentrations of gefitinib (Selleckchem). Medium containing drug was replenished after 3 days, and after 6 days of treatment, cell viability was assessed in triplicate wells using CellTiter-Glo assay (Promega) according to the manufacturer's instructions. In all subsequent experiments, the input organoid cell number was adjusted to obtain 150 organoids per well.
Organoid viability in cocultures was measured using Firefly luciferase/GFP transduced organoids (as above). Different media were tested to identify conditions that facilitate single-cell outgrowth and stromal interactions. Cells were cultured either for 9 days in full medium, reduced medium (both as above), or for 3 days in full medium, followed by two rounds of washes and culture for 6 days in reduced medium (mixed). Drug screening in monocultures and cocultures (n = 29) was performed in the mixed condition. On day 3, drugs were added to the reduced medium with a D300e digital dispenser (Tecan): 0.1 to 5 nmol/L SN-38 (Selleckchem), 0.5 to 60 μmol/L 5-FU (Sigma-Aldrich), 0.5 to 60 μmol/L oxaliplatin (Sigma-Aldrich), and 0.01 to 10 μmol/L gefitinib (Selleckchem) were administered in 7-point dilutions. Compounds were dissolved in DMSO, and DMSO content in all wells was normalized, not exceeding 1% final volume. The medium was replenished after 3 days, and cell viability was assessed in triplicates after 6 days of drug exposure using the ONE-Glo EX assay (as above).
For dual luciferase assays, CAFs were transduced with a Renilla luciferase-P2A-dsRED-T2A-Puro (Supplementary Fig. S10A). The vector was generated by the introduction of a Renilla luciferase-P2A-dsRED fragment into the BamHI and SgrDI sites of pCDH-CMV-Nluc-P2A-copGFP-T2A-Puro, a gift from Kazuhiro Oka (RRID:Addgene_73037), and stable expression was selected in the presence of 0.5 to 1 μg/mL puromycin. Cocultures were seeded and treated as described above, followed by Dual-Glo luciferase assay (Promega). For cell lysis, the medium was replaced by Dual-Glo Luciferase Reagent mixed with ONE-Glo EX Lysis Buffer at a 1:1 ratio, and the below steps were performed as suggested by the manufacturer. Staurosporine (0.001–1 μmol/L; Selleckchem) was used as a control.
Luciferase signals were normalized to the average of DMSO control. Individual values were excluded if cells were lost due to technical errors or if seeding was not uniform. Dose–response analysis was performed in GraphPad and curve fit (R2 value), IC50, and AUC values are listed in Supplementary Tables S8 and S9. The normalized AUC was obtained for each drug by dividing the AUC by the maximum AUC measured among all models. The range of the normalized AUC is between 0 and 1. Relative IC50 and AUC values were determined by division of monoculture and coculture values, and the CAF drug influence represents the mean of the relative AUC values of the four drugs.
Chemogenomic Drug Library Screen
For pharmacologic screening in resistant cocultures, luciferase-expressing organoids (O14 or O23) and nonmodified CAFs (F14) were seeded as described above in a rich medium. The library contained 186 test compounds and 106 corresponding inactive controls, which were assembled as part of the SGC Chemical Probe programs including the donated probe program and the EUbOPEN consortium (see Supplementary Table S10; refs. 30–32). For master plates, compounds were dissolved in DMSO at 1 mmol/L or 10 mmol/L. Three days after seeding, the culture medium was changed to a growth factor–reduced medium (as above) following cotreatment. The library was administered with an automatic multichannel pipette (Integra Mini 96) at a final concentration of 0.1 or 1 μmol/L (10,000-fold dilution) to cocultures in presence and absence of a sublethal dose of gefitinib (O14: 1 μmol/L and O23: 0.2 μmol/L) or SN-38 (O23: 1.36 nmol/L). Each experiment was performed in two replicates. The medium was renewed after 3 days, and treatment was stopped after 6 days, followed by measurement of organoid viability using ONE-Glo EX (Promega). Raw data were normalized to the average of the DMSO controls present on each plate, and the mean viability is listed in Supplementary Table S10. The effect of 1 μmol/L BAY-474 was validated in resistant cocultures as described above.
Pharmacotranscriptomic Analysis and Study of Prognostic Value
To derive pharmacotranscriptomic signatures, we adopted a strategy described recently in pancreatic cancer organoids (68). The Spearman rank correlation coefficients (rho) were calculated for the 10,000 most variably expressed genes in tumor tissues (Log2 count per million) and drug response in vitro. Specifically, we tested the AUC in monoculture and coculture and the relative drug sensitivity in the presence of CAFs (AUCco/AUCmono) for each drug alone and for the average of all four drugs (Supplementary Table S9). Genes were filtered by t test statistics using P < 0.05. Genes that correlated negatively (rho < −0.5) or positively (rho >0.35) with drug response were defined as sensitivity or resistance signatures, respectively. The signatures based on the CAF drug influence are shown in Supplementary Table S11.
For Kaplan–Meier analyses, the raw expression and phenotype data of GSE39582 was downloaded using the “getGEO” function of the GEOquery R package (RRID:SCR_000146). Affymetrix probe IDs were converted to Emsembl IDs using the biomaRt R package before Z-score normalization. For each signature, the sum of Z-scores was determined and a median split was performed to divide the cohort into two groups of high and low expression. Kaplan–Meier plots were generated using the “ggsurvplot” function of the survminer R package (RRID:SCR_021094). The “coxph” function of the same package was used to calculate the HR, P values (log-rank test), and 95% CIs displayed in Kaplan–Meier and Forest plots using default parameters.
Statistical Analysis
All error bars represent SD. Statistical significance was performed using GraphPad Prism 9.2.0 (RRID:SCR_002798). Two-tailed unpaired t tests were performed assuming unequal variance, with P < 0.05 considered statistically significant. In the case of multiple testing, P values were further investigated as indicated using the FDR approach with 1% FDR or by Bonferroni correction. The influence of coculture was assessed by paired nonparametric tests (Wilcoxon matched-pairs signed rank test). The average normalized AUC and average relative sensitivity values in different CMS models were compared by Mann–Whitney U tests. All the experiments were repeated at least two times independently unless mentioned in the legend.
For Pearson correlation of signature enrichment (NES from MSigDB) and drug response data, the “similarity matrix” function of the Morpheus software (Broad Institute; RRID:SCR_017386) was used. For correlation analysis of clinical, genetic transcriptomic and functional data, tumor location and T1 to T4 stages were converted to numerical values. P values for comparisons between two groups were calculated using the Fisher exact test for binary variables and the Mann–Whitney U test for nonbinary variables. Correlations between variables were calculated using Spearman rank correlation.
Data and Material Availability
The sequencing data have been deposited to the European Genome-phenome Archive (www.ebi.ac.uk/ega/) under accession numbers (EGAS00001007300; RNA sequencing data) and (EGAS00001007301; exome sequencing data). Raw drug screening data are available on Mendeley Data (doi: 10.17632/fypp6xhkjy.1). Patient-derived cells and materials can be provided upon reasonable request and following approval by the institutional review board of the UCT. DCP compounds can be ordered via https://www.sgc-ffm.uni-frankfurt.de/#!setorderview; SGC compounds can be ordered via https://www.thesgc.org/chemical-probes; and EUbOPEN chemogenomics compounds are accessible via contact information on the consortium homepage: https://www.eubopen.org/.
Supplementary Material
Supplementary Table S1 shows the clinical data of the CRC organoid-stroma cohort. Supplementary Table S2 shows the inventory of available materials and molecular analyses. Supplementary Table S3 shows the selected variant types for whole exome analysis. Supplementary Table S4 summarizes the genetic characterization of CRC models. Supplementary Table S5 shows the classification according to the consensus molecular subtypes (CMS). Supplementary Table S6 shows classification according to the CRC intrinsic subtypes (CRIS). Supplementary Table S7 summarizes the functional data in mono- and co-cultures. Supplementary Table S8 shows drug sensitivity of CRC organoids in co-culture with autologous and heterologous fibroblasts. Supplementary Table S9 show the drug sensitivity in all biobank models in mono- and co-culture. Supplementary Table S10 shows the cell viability data of the chemogenomic library screens in resistant co-culture models. Supplementary Table S11 shows the identified drug sensitivity and resistance signatures. Supplementary Table S12 lists all antibodies and primer sequences used in this study.
Supplementary Figure S1 shows light microscopic images of CRC organoids and CAFs. Supplementary Figure S2 shows immunostaining and RNA sequencing analysis of cultured CAFs. Supplementary Figure S3 shows chromosomal copy number changes in tumors and matched organoids. Supplementary Figure S4 shows the transcriptional variation among tumors and organoids. Supplementary Figure S5 shows the classification of cancer intrinsic subtypes (CRIS) in tumors, matched organoids and xenotransplants. Supplementary Figure S6 shows the tissue microarray analysis of the of tumor samples from the colorectal cancer organoid-stroma biobank cohort. Supplementary Figure S7 shows the association of growth characteristics with molecular features of colorectal cancer organoid-stroma biobank. Supplementary Figure S8 shows the shows CMS and CRIS classifications of tumors and of matched organoids in different contexts. Supplementary Figure S9 describes the establishment of a drug screening workflow in 3D organoid-stroma co-cultures. Supplementary Figure S10 describes the development of a dual luciferase assay to study cell viability simultaneously in organoids and CAFs. Supplementary Figure S11 demonstrates that the MET inhibitor BAY-474 sensitizes Gefitinib resistant co-cultures. Supplementary Figure S12 shows the association of identified sensitivity and resistance signatures with gene expression and prognosis in different CMS.
Acknowledgments
We thank all patients, families, and clinicians as well as the UCT Biobank for participating in this study. We thank Kathrin Hampel, Susanne Bösser, and Patrizia Schult-Dietrich for excellent technical support. The Genomics and Proteomics Core Facility, German Cancer Research Center (DKFZ), has provided excellent services for next-generation sequencing. Cell lines for the production of R-spondin 1 CM and Noggin CM were kind gifts from C. Kuo (Stanford University, CA) and H. Clevers (Hubrecht Institute, Utrecht, the Netherlands), respectively. This work was supported by institutional funds from the Georg-Speyer-Haus and the LOEWE Center Frankfurt Cancer Institute (FCI) funded by the Hessen State Ministry for Higher Education, Research and the Arts [III L 5-519/03/03.001-(0015)], the Deutsche Forschungsgemeinschaft (FOR2438: FA1334/2-2 and Gr1916/11-1; SFB1292-Project ID: 318346496-TP16; SFB1479-Project ID: 441891347-P02; GRK2336), the German Federal Ministry of Education and Research (BMBF; 01KD2206Q/SATURN3), and the European Research Council (Advanced Grant PLASTICAN-101021078). The Institute for Tumor Biology and Experimental Therapy, Georg-Speyer-Haus, is funded jointly by the German Federal Ministry of Health and the Ministry of Higher Education, Research and the Arts of the State of Hessen. H.F. Farin, F.R. Greten, S. Knapp, and S. Muller receive funding from the German Cancer Research Center (DKTK), the FCI, and the European Union (EU)/European Federation of Pharmaceutical Industries and Associations (EFPIA)/Ontario Institute for Cancer Research (OICR)/McGill/Royal Institute of Technology (KTH)/Diamond Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant no. 875510). S. Knapp and S. Muller are grateful for support from the Structural Genomics Consortium (SGC), a registered charity (no: 1097737) that also receives funds from Bristol Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute, Janssen, Merck KGaA, Pfizer, and Takeda.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
J. Bollrath reports personal fees from Boehringer Ingelheim, Roche, and Miltenyi Biotec outside the submitted work. T.O. Goetze reports grants from the German Research Foundation (DFG), Gemeinsamer Bundesausschuss, German Cancer Aid (Deutsche Krebshilfe), Lilly Pharma, AstraZeneca, and Incyte outside the submitted work. W.O. Bechstein reports personal fees from AOK Bundesverband, Astellas GmbH, Deutsche Gesellschaft für Chirurgie, Hessisches Ministerium für Soziales und Intergration, Landesäerztekammer Hessen, MCI Deutschland, Medupdate GmbH, Aventis, Charite Berlin, Deutsche Gesellschaft für Allgemein- und Viszeralchirurgie, Deutsche Gesellschaft für Serologie, Interplan, and Terumo outside the submitted work. S. Knapp reports grants from Innovative Medicines Initiative EUbOPEN during the conduct of the study. F.R. Greten reports personal fees from Amazentis outside the submitted work. No disclosures were reported by the other authors.
Disclaimer
This communication reflects the views of the authors, and the Joint Undertaking is not liable for any use that may be made of the information contained herein.
Authors’ Contributions
H.F. Farin: Conceptualization, formal analysis, supervision, funding acquisition, visualization, writing–original draft, writing–review and editing. M.H. Mosa: Formal analysis, investigation, visualization. B. Ndreshkjana: Formal analysis, investigation, visualization. B.M. Grebbin: Formal analysis, investigation. B. Ritter: Formal analysis, investigation. C. Menche: Formal analysis, investigation. K.B. Kennel: Formal analysis, investigation. P.K. Ziegler: Formal analysis. L. Szabó: Formal analysis, investigation. J. Bollrath: Formal analysis, investigation. D. Rieder: Formal analysis. B.E. Michels: Formal analysis. A. Kress: Formal analysis, investigation. M. Bozlar: Formal analysis, investigation. T. Darvishi: Investigation. S. Stier: Formal analysis, investigation. I.-M. Kur: Formal analysis, investigation. K. Bankov: Formal analysis. R. Kesselring: Resources. S. Fichtner-Feigl: Resources. B. Brune: Resources. T.O. Goetze: Resources. S.E. Al-Batran: Resources. C.H. Brandts: Resources. W.O. Bechstein: Resources. P.J. Wild: Resources. A. Weigert: Formal analysis, investigation. S. Muller: Resources. S. Knapp: Resources. Z. Trajanoski: Supervision. F.R. Greten: Conceptualization, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin 2022;72:409–36. [DOI] [PubMed] [Google Scholar]
- 2. Schmitt M, Greten FR. The inflammatory pathogenesis of colorectal cancer. Nat Rev Immunol 2021;21:653–67. [DOI] [PubMed] [Google Scholar]
- 3. Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trinh A, Trumpi K, De Sousa E Melo F, Wang X, de Jong JH, Fessler E, et al. Practical and robust identification of molecular subtypes in colorectal cancer by immunohistochemistry. Clin Cancer Res 2017;23:387–98. [DOI] [PubMed] [Google Scholar]
- 6. Berg KCG, Sveen A, Høland M, Alagaratnam S, Berg M, Danielsen SA, et al. Gene expression profiles of CMS2-epithelial/canonical colorectal cancers are largely driven by DNA copy number gains. Oncogene 2019;38:6109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calon A, Espinet E, Palomo-Ponce S, Tauriello DVF, Iglesias M, Céspedes MV, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 2012;22:571–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Isella C, Brundu F, Bellomo SE, Galimi F, Zanella E, Porporato R, et al. Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer. Nat Commun 2017;8:15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 2015;47:320–9. [DOI] [PubMed] [Google Scholar]
- 10. Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554:538–43. [DOI] [PubMed] [Google Scholar]
- 11. Jackstadt R, van Hooff SR, Leach JD, Cortes-Lavaud X, Lohuis JO, Ridgway RA, et al. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell 2019;36:319–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Varga J, Nicolas A, Petrocelli V, Pesic M, Mahmoud A, Michels BE, et al. AKT-dependent NOTCH3 activation drives tumor progression in a model of mesenchymal colorectal cancer. J Exp Med 2020;217:e20191515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009;459:262–5. [DOI] [PubMed] [Google Scholar]
- 14. Sato T, Stange DE, Ferrante M, Vries RGJ, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 2011;141:1762–72. [DOI] [PubMed] [Google Scholar]
- 15. Boj SF, Hwang CI, Baker LA, Chio IIC, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015;160:324–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 2015;148:126–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarró LM, Bradshaw CR, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 2018;172:373–86. [DOI] [PubMed] [Google Scholar]
- 19. Driehuis E, Kolders S, Spelier S, Lõhmussaar K, Willems SM, Devriese LA, et al. Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov 2019;9:852–71. [DOI] [PubMed] [Google Scholar]
- 20. van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015;161:933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 2016;18:827–38. [DOI] [PubMed] [Google Scholar]
- 22. Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 2018;556:457–62. [DOI] [PubMed] [Google Scholar]
- 23. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018;359:920–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ganesh K, Wu C, O'Rourke KP, Szeglin BC, Zheng Y, Sauvé C-EG, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med 2019;25:1607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ooft SN, Weeber F, Dijkstra KK, McLean CM, Kaing S, van WE, et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med 2019;11:eaay2574. [DOI] [PubMed] [Google Scholar]
- 26. Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell 2020;26:17–26. [DOI] [PubMed] [Google Scholar]
- 27. Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 2020;20:174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mosa MH, Michels BE, Menche C, Nicolas AM, Darvishi T, Greten FR, et al. A Wnt-induced phenotypic switch in cancer-associated fibroblasts inhibits EMT in colorectal cancer. Cancer Res 2020;80:5569–82. [DOI] [PubMed] [Google Scholar]
- 30. Müller S, Ackloo S, Arrowsmith CH, Bauser M, Baryza JL, Blagg J, et al. Donated chemical probes for open science. eLife 2018;7:e34311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu Q, Heidenreich D, Zhou S, Ackloo S, Krämer A, Nakka K, et al. A chemical toolbox for the study of bromodomains and epigenetic signaling. Nat Commun 2019;10:1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scheer S, Ackloo S, Medina TS, Schapira M, Li F, Ward JA, et al. A chemical biology toolbox to study protein methyltransferases and epigenetic signaling. Nat Commun 2019;10:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmitt M, Ceteci F, Gupta J, Pesic M, Böttger TW, Nicolas AM, et al. Colon tumour cell death causes mTOR dependence by paracrine P2X4 stimulation. Nature 2022;612:347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013;10:e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet 2017;49:708–18. [DOI] [PubMed] [Google Scholar]
- 36. Lee HO, Hong Y, Etlioglu HE, Cho YB, Pomella V, Van den Bosch B, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet 2020;52:594–603. [DOI] [PubMed] [Google Scholar]
- 37. Uhlitz F, Bischoff P, Peidli S, Sieber A, Trinks A, Lüthen M, et al. Mitogen-activated protein kinase activity drives cell trajectories in colorectal cancer. EMBO Mol Med 2021;13:e14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Joanito I, Wirapati P, Zhao N, Nawaz Z, Yeo G, Lee F, et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat Genet 2022;54:963–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raghavan S, Winter PS, Navia AW, Williams HL, DenAdel A, Lowder KE, et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell 2021;184:6119–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luo X, Fong ELS, Zhu C, Lin QXX, Xiong M, Li A, et al. Hydrogel-based colorectal cancer organoid co-culture models. Acta Biomater 2021;132:461–72. [DOI] [PubMed] [Google Scholar]
- 41. ten Hoorn S, Sommeijer DW, Elliott F, Fisher D, de Back TR, Trinh A, et al. Molecular subtype-specific efficacy of anti-EGFR therapy in colorectal cancer is dependent on the chemotherapy backbone. Br J Cancer 2021;125:1080–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aderka D, Stintzing S, Heinemann V. Explaining the unexplainable: discrepancies in results from the CALGB/SWOG 80405 and FIRE-3 studies. Lancet Oncol 2019;20:e274–83. [DOI] [PubMed] [Google Scholar]
- 43. Woolston A, Khan K, Spain G, Barber LJ, Griffiths B, Gonzalez-Exposito R, et al. Genomic and transcriptomic determinants of therapy resistance and immune landscape evolution during anti-EGFR treatment in colorectal cancer. Cancer Cell 2019;36:35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nicolas AM, Pesic M, Engel E, Ziegler PK, Diefenhardt M, Kennel KB, et al. Inflammatory fibroblasts mediate resistance to neoadjuvant therapy in rectal cancer. Cancer Cell 2022;40:168–84. [DOI] [PubMed] [Google Scholar]
- 45. Fridman WH, Miller I, Sautès-Fridman C, Byrne AT. Therapeutic targeting of the colorectal tumor stroma. Gastroenterology 2020;158:303–21. [DOI] [PubMed] [Google Scholar]
- 46. Alderdice M, Richman SD, Gollins S, Stewart JP, Hurt C, Adams R, et al. Prospective patient stratification into robust cancer-cell intrinsic subtypes from colorectal cancer biopsies. J Pathol 2018;245:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sirinukunwattana K, Domingo E, Richman SD, Redmond KL, Blake A, Verrill C, et al. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut 2021;70:544–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dunne PD, McArt DG, Bradley CA, O'Reilly PG, Barrett HL, Cummins R, et al. Challenging the cancer molecular stratification dogma: intratumoral heterogeneity undermines consensus molecular subtypes and potential diagnostic value in colorectal cancer. Clin Cancer Res 2016;22:4095–104. [DOI] [PubMed] [Google Scholar]
- 49. Khaliq AM, Erdogan C, Kurt Z, Turgut SS, Grunvald MW, Rand T, et al. Refining colorectal cancer classification and clinical stratification through a single-cell atlas. Genome Biol 2022;23:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dao V, Yuki K, Lo YH, Nakano M, Kuo CJ. Immune organoids: from tumor modeling to precision oncology. Trends Cancer 2022;8:870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov 2022;12:31–46. [DOI] [PubMed] [Google Scholar]
- 52. Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 2012;143:1518–29. [DOI] [PubMed] [Google Scholar]
- 53. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reble E, Castellani CA, Melka MG, O'Reilly R, Singh SM. VarScan2 analysis of de novo variants in monozygotic twins discordant for schizophrenia. Psychiatr Genet 2017;27:62–70. [DOI] [PubMed] [Google Scholar]
- 56. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol 2015;26:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016;12:e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Skidmore ZL, Wagner AH, Lesurf R, Campbell KM, Kunisaki J, Griffith OL, et al. GenVisR: genomic visualizations in R. Bioinformatics 2016;32:3012–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Conway T, Wazny J, Bromage A, Tymms M, Sooraj D, Williams ED, et al. Xenome–a tool for classifying reads from xenograft samples. Bioinformatics 2012;28:i172–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Michels BE, Mosa MH, Grebbin BM, Yepes D, Darvishi T, Hausmann J, et al. Human colon organoids reveal distinct physiologic and oncogenic Wnt responses. J Exp Med 2019;216:704–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Eide PW, Bruun J, Lothe RA, Sveen A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci Rep 2017;7:16618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Strack E, Rolfe PA, Fink AF, Bankov K, Schmid T, Solbach C, et al. Identification of tumor-associated macrophage subsets that are associated with breast cancer prognosis. Clin Transl Med 2020;10:e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 2017;214:579–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schnalzger TE, de Groot MH, Zhang C, Mosa MH, Michels BE, Röder J, et al. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J 2019;38:e100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Borten MA, Bajikar SS, Sasaki N, Clevers H, Janes KA. Automated brightfield morphometry of 3D organoid populations by OrganoSeg. Sci Rep. 2018;8:5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tiriac H, Belleau P, Engle DD, Plenker D, Deschênes A, Somerville TDD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov 2018;8:1112–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 shows the clinical data of the CRC organoid-stroma cohort. Supplementary Table S2 shows the inventory of available materials and molecular analyses. Supplementary Table S3 shows the selected variant types for whole exome analysis. Supplementary Table S4 summarizes the genetic characterization of CRC models. Supplementary Table S5 shows the classification according to the consensus molecular subtypes (CMS). Supplementary Table S6 shows classification according to the CRC intrinsic subtypes (CRIS). Supplementary Table S7 summarizes the functional data in mono- and co-cultures. Supplementary Table S8 shows drug sensitivity of CRC organoids in co-culture with autologous and heterologous fibroblasts. Supplementary Table S9 show the drug sensitivity in all biobank models in mono- and co-culture. Supplementary Table S10 shows the cell viability data of the chemogenomic library screens in resistant co-culture models. Supplementary Table S11 shows the identified drug sensitivity and resistance signatures. Supplementary Table S12 lists all antibodies and primer sequences used in this study.
Supplementary Figure S1 shows light microscopic images of CRC organoids and CAFs. Supplementary Figure S2 shows immunostaining and RNA sequencing analysis of cultured CAFs. Supplementary Figure S3 shows chromosomal copy number changes in tumors and matched organoids. Supplementary Figure S4 shows the transcriptional variation among tumors and organoids. Supplementary Figure S5 shows the classification of cancer intrinsic subtypes (CRIS) in tumors, matched organoids and xenotransplants. Supplementary Figure S6 shows the tissue microarray analysis of the of tumor samples from the colorectal cancer organoid-stroma biobank cohort. Supplementary Figure S7 shows the association of growth characteristics with molecular features of colorectal cancer organoid-stroma biobank. Supplementary Figure S8 shows the shows CMS and CRIS classifications of tumors and of matched organoids in different contexts. Supplementary Figure S9 describes the establishment of a drug screening workflow in 3D organoid-stroma co-cultures. Supplementary Figure S10 describes the development of a dual luciferase assay to study cell viability simultaneously in organoids and CAFs. Supplementary Figure S11 demonstrates that the MET inhibitor BAY-474 sensitizes Gefitinib resistant co-cultures. Supplementary Figure S12 shows the association of identified sensitivity and resistance signatures with gene expression and prognosis in different CMS.



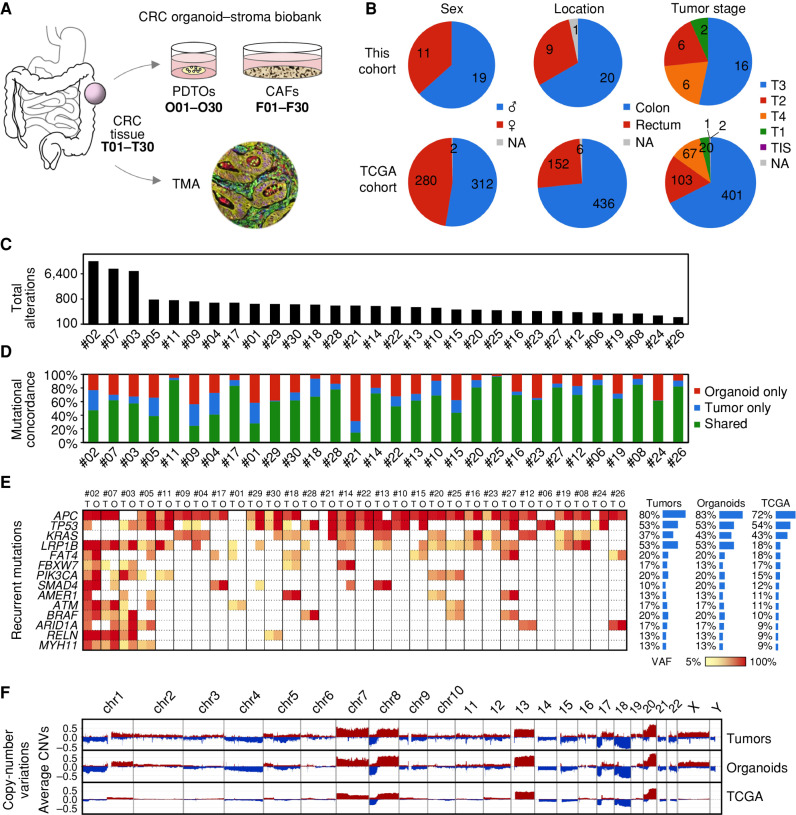
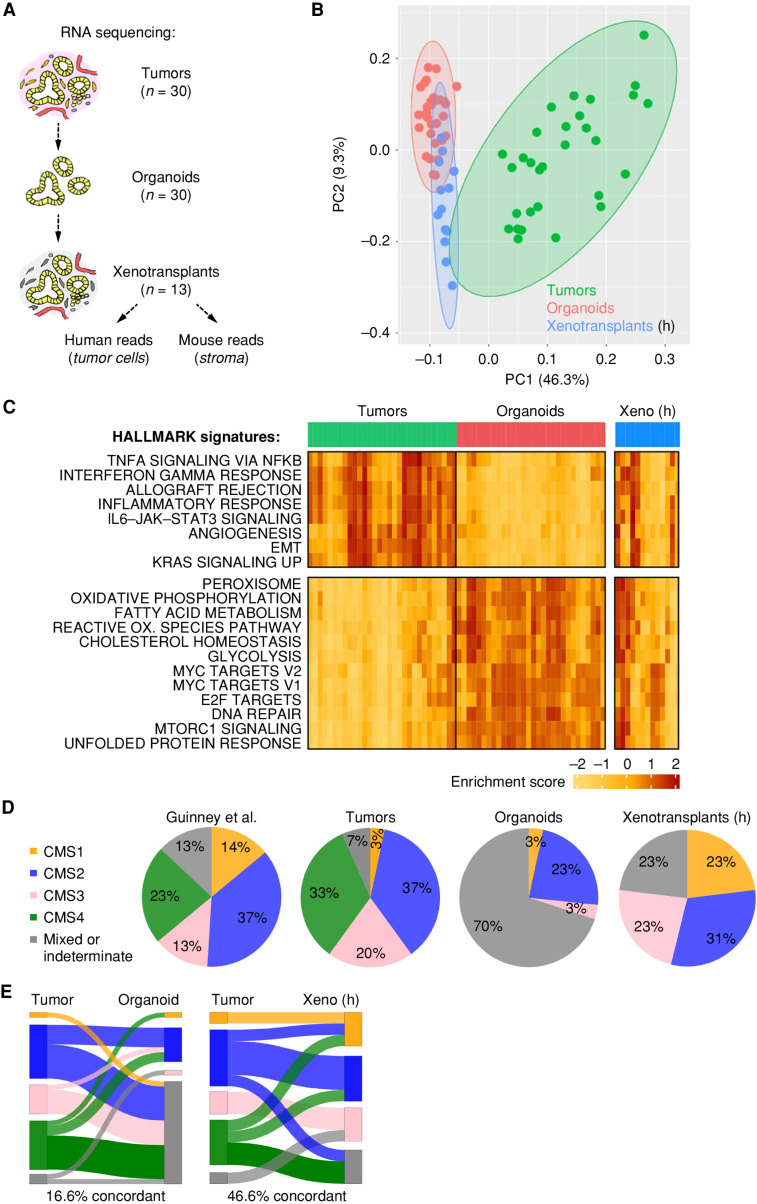
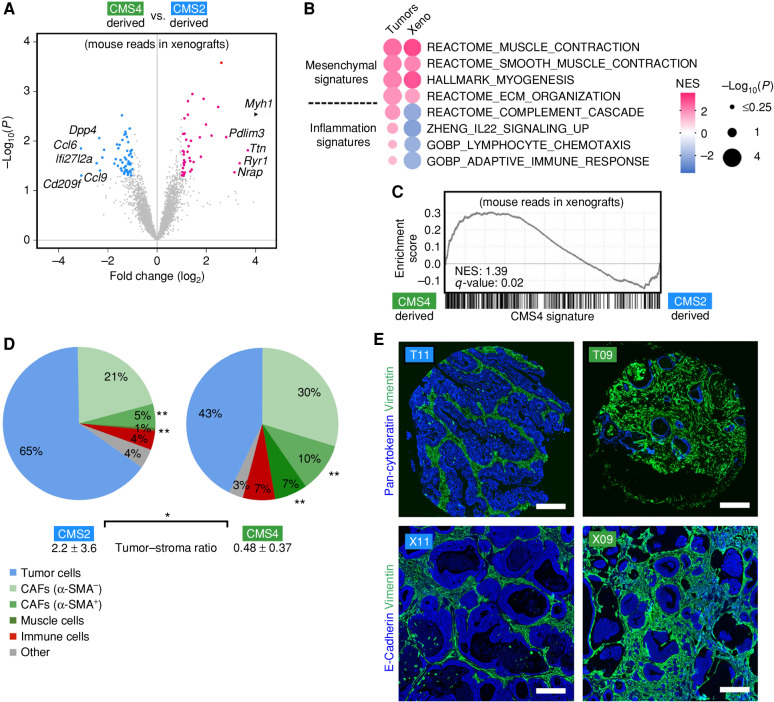
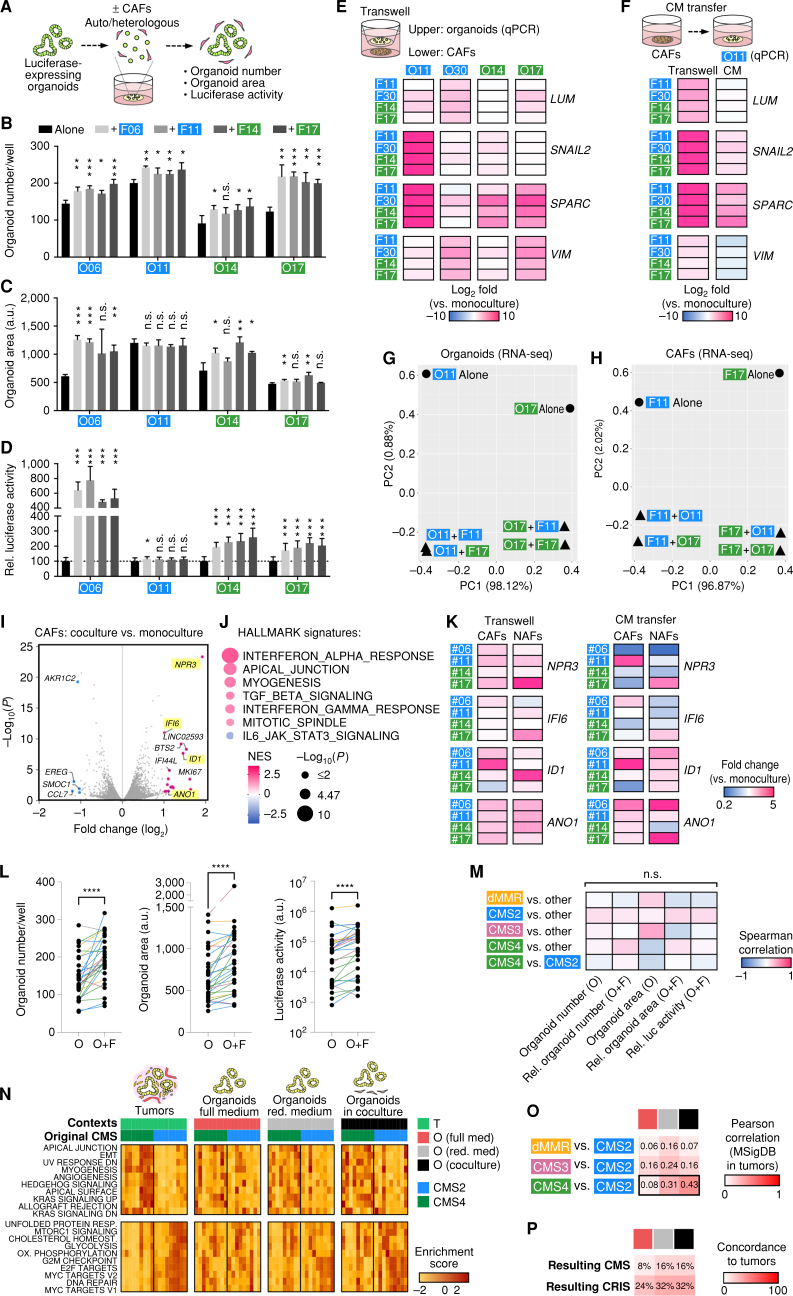
![Figure 5. CAFs’ influence on drug responses is dependent on the tumor cell compartment. A, Experimental setup for drug testing in cocultures using luciferase/GFP transduced organoids. B and C, Sensitivity to 5-FU, oxaliplatin, SN-38, and gefitinib in monoculture and coculture with autologous or heterologous CAFs. Tumor cell viability was measured by luciferase activity (mean ± SD in triplicate wells). Representative dose–response curves (B) and heat map of drug sensitivity [relative (rel.) AUC compared with monoculture; C]. Experiments were repeated twice. D, Measurement of organoid- and CAF-specific toxicity by dual luciferase assay. Coculture was performed using organoids expressing Firefly luciferase and matched CAFs expressing Renilla luciferase. Heat map shows normalized (norm.) AUC. CAFs show reduced sensitivity to all treatments compared with organoids. See Supplementary Figs. S9 and S10 and Supplementary Table S8.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/85ca/10551667/6cdb60a644b3/2192fig5.jpg)