The neurotransmitter GABA was identified as a mediator of functional communication within the melanoma microenvironment, which highlights the role of electrical activity in melanoma initiation and suggests GABA as a potential therapeutic target in this disease.
Abstract
Oncogenes can initiate tumors only in certain cellular contexts, which is referred to as oncogenic competence. In melanoma, whether cells in the microenvironment can endow such competence remains unclear. Using a combination of zebrafish transgenesis coupled with human tissues, we demonstrate that GABAergic signaling between keratinocytes and melanocytes promotes melanoma initiation by BRAFV600E. GABA is synthesized in melanoma cells, which then acts on GABA-A receptors in keratinocytes. Electron microscopy demonstrates specialized cell–cell junctions between keratinocytes and melanoma cells, and multielectrode array analysis shows that GABA acts to inhibit electrical activity in melanoma/keratinocyte cocultures. Genetic and pharmacologic perturbation of GABA synthesis abrogates melanoma initiation in vivo. These data suggest that GABAergic signaling across the skin microenvironment regulates the ability of oncogenes to initiate melanoma.
Significance:
This study shows evidence of GABA-mediated regulation of electrical activity between melanoma cells and keratinocytes, providing a new mechanism by which the microenvironment promotes tumor initiation. This provides insights into the role of the skin microenvironment in early melanomas while identifying GABA as a potential therapeutic target in melanoma.
See related commentary by Ceol, p. 2128.
This article is featured in Selected Articles from This Issue, p. 2109
INTRODUCTION
Melanoma arises at the dermal–epidermal junction, commonly harboring mutations in genes such as BRAF or NRAS (1). These same mutations occur in benign nevi, raising the question of why some melanocytes, but not others, are competent to form melanoma. Previous work has shown that the developmental state of the cell plays a dominant role in such competence because more neural crest–like melanocytes have a chromatin landscape that makes them permissive for oncogenesis (2). Melanocytes in the skin are encased in a dense network of microenvironmental cells, including keratinocytes, which make up the majority of the skin surface. We and others have previously shown that keratinocytes can have both protumorigenic (3–8) and antitumorigenic (9–13) roles in melanoma. Whether these abundant microenvironmental cells such as keratinocytes play a role in melanoma initiation remains unclear. Here, we identify a specific protumorigenic keratinocyte population in direct communication with melanoma cells and identify the pathways mediating this communication.
RESULTS
A Reporter System to Detect Melanocyte/Keratinocyte Communication
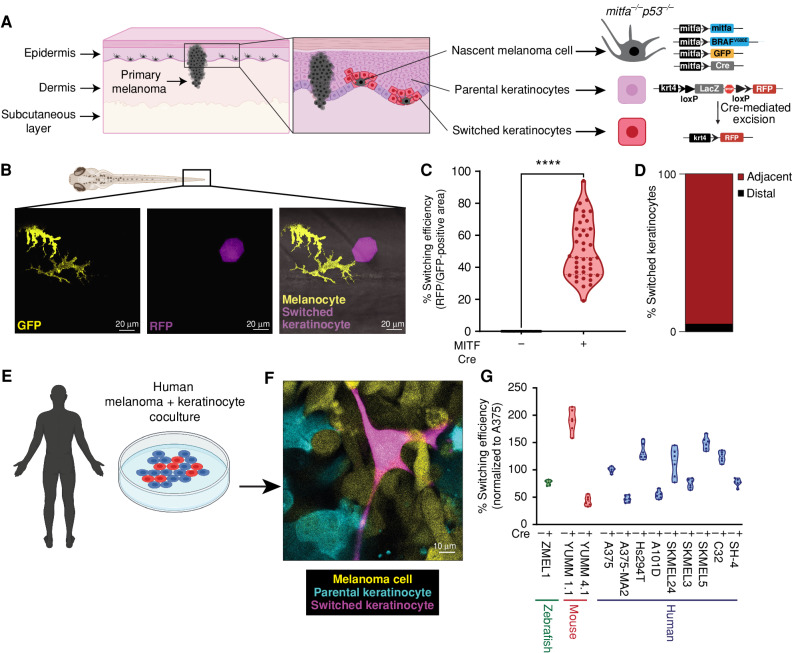
In normal physiology, melanocytes are connected to keratinocytes through dendrites. These dendrites allow for the export of a pigment-containing organelle called a melanosome into the surrounding keratinocytes forming the epidermal melanin unit (14–16). This unusual organelle transfer between cell types is responsible for skin coloration, as keratinocytes make up the vast majority of the skin surface. In addition to melanosomes, melanocytes also export smaller extracellular vesicles such as exosomes that contain RNA and proteins (17). We took advantage of this normal physiologic mechanism to develop a genetic reporter of melanocyte/keratinocyte communication. We engineered transgenic zebrafish to express Cre under the melanocyte-specific mitfa promoter, along with a floxed lacZ or GFP to RFP reporter under the keratinocyte-specific krt4 promoter. Both the mitfa and krt4 promoters drive specific expression in their respective cell types when assayed using the zebrafish melanoma model (Supplementary Fig. S1A and S1B). This approach has been previously used to study vesicular communication between different cell types, both during normal development and in cancer (18, 19). In this system, any keratinocyte that takes up Cre from the melanocyte will switch to express RFP fluorescence. To put this in the context of melanoma, we used the previously described MiniCoopR transgenic system (20), in which the melanocytes were further engineered to express BRAFV600E in the context of p53−/− along with a palmitoylated GFP fluorophore (Fig. 1A). The MiniCoopR transgenic system is engineered such that expression of the MiniCoopR construct rescues melanocytes in a Tg(mitfa:BRAFV600E);p53(lf);mitfa(lf) background while also driving expression of a candidate gene (like Cre) in the rescued melanocytes. Upon transgene injection, control animals without Cre in the melanocytes had no RFP-positive keratinocytes, as expected. In contrast, animals in which the melanocytes expressed Cre had, on average, 55% RFP-positive switched keratinocytes (Fig. 1B and C). We observed that RFP-positive switched keratinocytes were frequently directly adjacent to melanocytes in the zebrafish embryos. To quantify this phenomenon, we performed live confocal imaging of injected zebrafish embryos, and using Imaris, a 3D reconstruction software, we were able to quantify direct cell–cell contacts between nascent melanoma cells and switched keratinocytes. We call these melanocytes “nascent” melanoma cells because they possess the BRAFV600E and p53−/− genetic alterations, which are necessary but not sufficient to initiate melanoma. In 92% of cases (Fig. 1D), the RFP-positive keratinocytes were directly adjacent to the BRAFV600E, p53−/− melanocytes (marked by palmGFP fluorescence), suggesting it was only the subset of keratinocytes in physical contact with the nascent melanoma cells that exhibited such communication (Supplementary Fig. S1C–S1G; Supplementary Video S1).
Figure 1.
Nascent melanoma cells are in direct communication with keratinocytes. A, Schematic representation of the genetic reporter system to identify melanoma/keratinocyte communication (created with BioRender.com). Left, the epidermal melanin unit is disrupted in primary melanoma; zoomed-in image shows nascent melanoma cells and keratinocytes in direct physical contact. Right, the genetic reporter system for detecting melanoma keratinocyte communication in zebrafish casper F0 embryos with the genotype p53−/−mitfa:BRAFV600E injected with the indicated melanocyte and keratinocyte reporter constructs (± mitfa:Cre). B, Representative image of an F0 zebrafish embryo with nascent melanoma cells overexpressing Cre and palmGFP in direct communication with a switched keratinocyte overexpressing RFP. Individual cells are pseudocolored as indicated. C, Percentage switching efficiency calculated as percentage of RFP-positive area normalized to GFP-positive area in 3 days post-fertilization (dpf) zebrafish embryos. Data represent n = 40 control fish (negative for mitfa-Cre) and n = 40 switch fish (positive for mitfa-Cre) pooled from 3 biological replicates. Error bars, SD; P values generated by two-tailed unpaired t test; ****, P < 0.0001. D, Bar plot showing percentage of switched keratinocytes (RFP-positive) in direct contact (adjacent) or not in direct contact (distal) with a nascent melanoma cell (palmGFP-positive). Data are pooled from 3 biological replicates (n = 60). E, Schematic representation of human melanoma/keratinocyte coculture (schematic created with BioRender.com). F, Representative confocal image of human melanoma/keratinocyte coculture with nonswitched keratinocytes, melanoma cells, and switched keratinocytes pseudocolored as indicated. G, Percentage switching efficiency in human keratinocytes when cocultured with zebrafish, mouse, or human melanoma cell lines calculated as the number of switched keratinocytes per well, normalized to the human melanoma cell line A375 (ratio: 1:3, keratinocyte:melanoma). No switching was observed in the absence of Cre-expressing melanoma cell lines. Data represent n = 6 for control cocultures (no Cre) and n = 6 for switched cocultures (+ Cre) pooled from 3 biological replicates for each cell line indicated. Error bars, SD.
Human and Mouse Melanoma Cells Communicate with Keratinocytes
To test the relevance of these findings in human and mouse melanoma progression, we developed a similar system using human or mouse melanoma cells in coculture with human keratinocytes (Supplementary Fig. S2A). As a positive control, we first engineered a zebrafish melanoma cell line (ZMEL1) to express Cre and cocultured it with HaCaT keratinocytes expressing a floxed dsRED to GFP reporter. Similar to what we saw in vivo in the zebrafish, we found that a consistent percentage of cells (1.5% switching efficiency) in vitro also underwent Cre-mediated switching at 48 hours. We observed both single (GFP)- and double (dsRED/GFP)-positive switched cells in our assay because each cell receives multiple copies of the lentiviral construct. Because this in vitro rate was lower than what we saw in vivo, we monitored switching efficiency over time and found that this increased to 30% when the cells were cocultured for extended periods of time of 21 days (Supplementary Fig. S2B and S2C). This suggests a time dependency on the interaction between the cells, the nature of which remains to be investigated. We then tested a panel of mouse (YUMM cells) and human lines (A375, Hs294T, etc.) in the same fashion. Although there was some variation from line to line, as expected, all the tested lines exhibited Cre-mediated fluorescent switching (Fig. 1E–G), and this again primarily occurred when melanoma cells and keratinocytes were physically adjacent to each other (Supplementary Fig. S2D–S2I). Consistent with this, when the cells were separated by a Transwell membrane, the keratinocytes failed to undergo fluorescent switching, confirming that they require direct physical contact (Supplementary Fig. S2J and S2K).
To eliminate the possibility of cell–cell fusion between melanoma cells and keratinocytes, resulting in the formation of GFP-positive keratinocytes, we performed karyotypic analysis of individual cell populations as well as immunofluorescent studies for melanoma (SOX10) and keratinocyte (KRT14) markers. Karyotypic analysis of individual keratinocyte populations (dsRED-positive and GFP-positive) indicated that GFP-positive keratinocytes were karyotypically identical to dsRED-positive keratinocytes (Supplementary Fig. S3A). Further, we noted that GFP-positive keratinocytes were negative for the melanoma marker, SOX10, thus eliminating the possibility of melanoma cell and keratinocyte fusion (Supplementary Fig. S3B). Prior studies using this system have reported that this form of communication is dependent on several cell-intrinsic factors, such as growth rate, vesicle content, and target cell uptake, which might explain the variability we observe between different cell lines (21). To ensure that this was not a unique feature associated with HaCaT cells only, we tested this in a second human keratinocyte cell line, Ker-CT. Coculture of Ker-CT keratinocytes with human melanoma cell lines showed similar robust Cre-mediated switching, again observed only when melanoma cells and keratinocytes were physically touching each other (Supplementary Fig. S3C–S3G). We also wanted to know if this form of communication was used by melanoma cells to communicate with each other (melanoma/melanoma cross-talk). To test this, we engineered a similar cassette as above with melanoma cells expressing a floxed dsRED to GFP switch reporter and found no evidence of such communication between melanoma cells alone (Supplementary Fig. S3H–S3J). In addition, increasing the ratio of melanoma cells to keratinocytes increased switching efficiency suggesting that this mode of communication was dependent upon the density of melanoma cells (Supplementary Fig. S4A). Because the original studies using this Cre-based system were based on vesicle delivery, we wanted to confirm if this was the case here using both pharmacologic and genetic loss-of-function approaches. Treatment of melanoma/keratinocyte cocultures with GW4869, which is an inhibitor of exosome biogenesis, or genetic knockdown in melanoma cells of nSMase2, which is involved in exosome biogenesis (22), substantially reduced keratinocyte switching efficiency (Supplementary Fig. S4B–S4D), independent of any proliferation associated changes (Supplementary Fig. S4E and S4F), highlighting the role of exosome-like vesicles in melanoma/keratinocyte communication. Further, upon staining the melanoma cells only with MemBright, a dye that selectively stains vesicle-like structures in live cells (23), we find internalization of melanoma vesicles in switched keratinocytes (Supplementary Fig. S4G–S4I; Supplementary Video S2). These data indicate that the vesicle-mediated communication we report is a unique property only in the context of melanoma/keratinocyte communication.
Melanocyte/Keratinocyte Communication Is Required for Melanoma Initiation
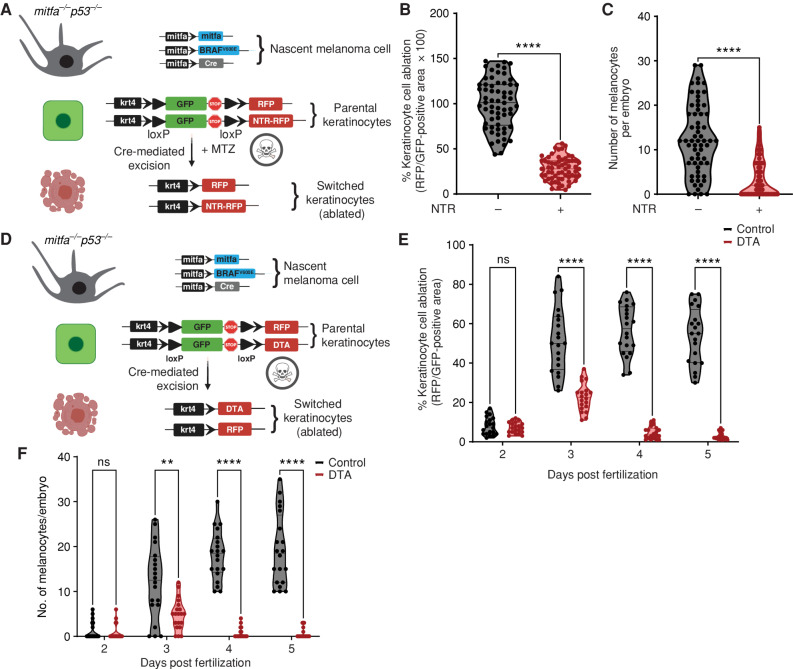
This interaction between the keratinocytes and melanocytes could have been either protumorigenic or antitumorigenic. We tested this using a previously established nitroreductase-mediated (NTR) inducible cell-ablation strategy in zebrafish (24, 25). This cell-ablation method was recently improved, such that it requires 100-fold lower concentrations of the prodrug due to improved catalytic efficiency of the nitroreductase enzyme (NTR2.0), hence providing a superior strategy to ablate specific cell types in the zebrafish without prodrug-associated off-target effects. Using the same melanoma/keratinocyte reporter as above, we reengineered the keratinocyte cassette to express a floxed GFP to nitroreductase (NTR2.0) transgene, such that any keratinocyte that receives Cre from the melanocyte would undergo cell death in the presence of the prodrug metronidazole (MTZ). This would help us understand if switched keratinocytes played a role in melanoma initiation, using our MiniCoopR melanocyte rescue model. We also coexpressed a GFP to RFP switch cassette in the keratinocytes to mark the switched cells over time (Fig. 2A). As a control, we found a significant average decrease (70%) in the RFP-positive area, which labels switched keratinocytes in animals expressing the NTR2.0 cassette in the presence of MTZ, validating that we could ablate keratinocytes in this setting (Fig. 2B). We then measured the number of pigmented BRAFV600E-positive melanocytes. Although control animals not expressing keratinocyte-NTR2.0 had, on average, 12 rescued nascent melanoma cells, in the presence of keratinocyte-NTR2.0, this was reduced to an average of less than four (Fig. 2C). We further tested this using another method of cell ablation in the zebrafish, which is based on the constitutive expression of diphtheria toxin (DTA) in specific cell types and does not require the use of a prodrug. Using the same strategy as above (Fig. 2D), we found that upon prolonged ablation of switched keratinocytes over a period of 5 days (ablation efficiency, 67% at day 3 to 95% at day 5; Fig. 2E), there is a stronger decrease in the rescue of nascent melanoma cells (97% decrease, Fig. 2F). This suggests that switched keratinocytes comprise a protumorigenic keratinocyte population and are critical for the rescue of transformed melanocytes in the early developmental window between 3 and 5 days after fertilization. Because in the MiniCoopR system only these MITF-positive pigmented melanocytes are capable of giving rise to melanomas that form later in life, this indicates that communication with these keratinocytes is important for melanoma initiation by BRAFV600E.
Figure 2.
Melanoma/keratinocyte communication drives melanoma initiation. A, Schematic representation of the zebrafish genetic reporter system to detect melanoma/keratinocyte communication and specifically ablate melanoma-associated keratinocytes (RFP-positive, switched) using the transgenic expression of NTR2.0 and MTZ treatment (created with BioRender.com). B, Switched keratinocyte ablation calculated as percentage of RFP-positive area normalized to GFP-positive area in 3 days post-fertilization (dpf) zebrafish embryos (± NTR2.0 expression) in keratinocytes. Absolute values were normalized to no NTR control to calculate cell-ablation efficiency. Data represent n = 60 control fish (negative for NTR) and n = 60 NTR2.0 fish (positive for NTR) pooled from 3 biological replicates. Error bars, SD; P values generated by two-tailed unpaired t test; ****, P < 0.0001. C, Number of pigmented melanocytes per embryo in ± NTR conditions. Data represent n = 60 control fish and n = 60 NTR fish pooled from 3 biological replicates. Error bars, SD; P values generated by two-tailed unpaired t test; ****, P < 0.0001. D, Schematic representation of the zebrafish genetic reporter system to detect melanoma keratinocyte communication and specifically ablate melanoma-associated keratinocytes (RFP-positive, switched) using the transgenic expression of DTA (diphtheria toxin gene A chain; created with BioRender.com). E, Switched keratinocyte ablation calculated as percentage of RFP-positive area normalized to GFP-positive area in 2, 3, 4, and 5 dpf zebrafish embryos (± DTA expression) in keratinocytes. Absolute values were normalized to no DTA control to calculate cell-ablation efficiency. Data represent n = 20 control fish per time point (negative for mitfa-Cre) and n = 20 DTA fish per time point (positive for mitfa-Cre) pooled from 3 biological replicates. Error bars, SD; P values generated by multiple two-tailed paired t test; ****, P < 0.0001. ns, not significant. F, Number of pigmented melanocytes per embryo in 2, 3, 4, and 5 dpf embryos ± DTA expression. Data represent n = 20 control fish per time point (negative for mitfa-Cre) and n = 40 DTA fish per time point (positive for mitfa-Cre) pooled from 3 biological replicates. Error bars, SD; P values generated by multiple two-tailed paired t test; **, P < 0.01; ****, P < 0.0001.
A Screen for Melanoma/Keratinocyte Communication Reveals a Role for GABA
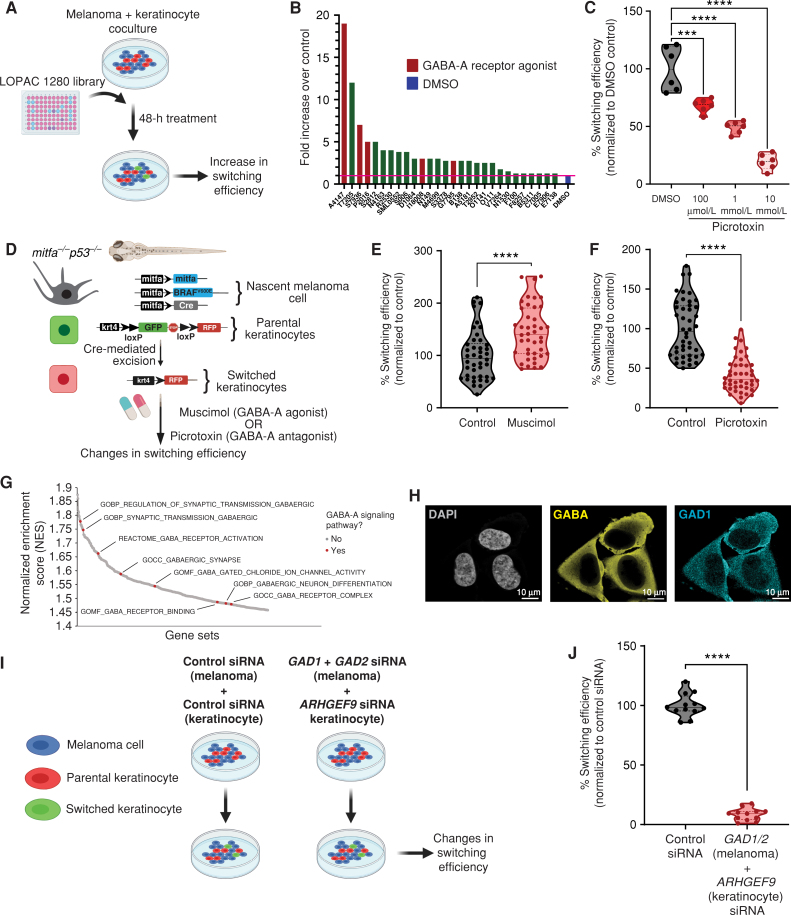
The mechanisms regulating this melanoma/keratinocyte cross-talk are unknown but could represent a means for abrogating melanoma initiation. To address this, we performed a small-molecule screen to identify pathways that mediated this communication. We used human A375 melanoma cells expressing Cre along with HaCaT keratinocytes expressing the floxed dsRED to GFP reporter, and then used fluorescent imaging to calculate the number of switched cells after applying the LOPAC 1280 small-molecule library, which contains a diverse set of chemicals affecting well-defined biological pathways (Fig. 3A). Overall, we found 28 molecules that increased switching above the DMSO control wells (Supplementary Table S1). Among the top 10 hits, we found three molecules (30%) that were all involved in GABAergic signaling. For example, the top hit from the screen was the GABA-A receptor agonist homotaurine (3-Amino-1-propanesulfonic acid sodium), which caused a nearly 20-fold increase in keratinocyte switching compared with control. Other hits included the GABA-A–positive allosteric modulators SB205384 and tetrahydrodeoxycorticosterone (THDOC; Fig. 3B). To validate results from our screen, we tested switching efficiency in cocultures with GABA, a natural agonist of GABA-A receptors, and muscimol, a previously validated, ionotropic GABA-A receptor agonist (26), and found significant increases in switching efficiency in melanoma/keratinocyte cocultures (34% and 57% increase, respectively; Supplementary Fig. S5A). We further tested the GABA-A receptor antagonist picrotoxin and found that it significantly decreased Cre-mediated recombination and switching efficiency in keratinocytes in a dose-dependent manner, further confirming the role of the GABA-A receptor in this communication (Fig. 3C). To test whether GABAergic signaling–mediated melanoma/keratinocyte communication occurred in vivo, we used the transgenic zebrafish system described above to calculate switching efficiency (Fig. 1C). We bathed the fish in either a GABA-A agonist (muscimol) or a GABA-A antagonist (picrotoxin) and measured the number of keratinocytes that had switched to RFP fluorescence (Fig. 3D). Consistent with the in vitro results of the screen, we found that GABA-A agonist activity increased the number of RFP-positive cells (44% increase; Fig. 3E), whereas GABA-A antagonist activity strongly reduced the number of RFP-positive cells (59% decrease; Fig. 3F; Supplementary Fig. S5B–S5D). These data support the notion that GABA is a specific mediator of melanoma/keratinocyte communication.
Figure 3.
GABAergic signaling drives melanoma/keratinocyte communication. A, Schematic representation of the LOPAC small-molecule library screen in human melanoma/keratinocyte cocultures treated with control (DMSO) or LOPAC 1280 library compounds (10 μmol/L each, indicated by their Sigma library identifiers) for 48 hours and quantified for an increase in switching efficiency (created with BioRender.com). B, Fold change over control (DMSO) in switching efficiency in the top 28 hits of the LOPAC small-molecule library screen. Red bars indicate compounds that are agonists or allosteric modulators of the GABA-A receptor; blue bar represents DMSO control. C, Percentage switching efficiency calculated as number of switched cells per well normalized to control (DMSO) upon treatment with the GABA antagonist picrotoxin (100 μmol/L, 1 mmol/L, 10 mmol/L) in melanoma/keratinocyte cocultures for 48 hours pooled from 6 biological replicates (n = 12; ratio: 1:3, keratinocyte:melanoma). Error bars, SD; P values generated by one-way ANOVA with multiple comparisons; ****, P < 0.0001. D, Schematic representation of the F0 zebrafish genetic reporter assay to quantify changes in keratinocyte switching efficiency in zebrafish embryos treated with a GABA-A agonist (muscimol) or a GABA-A antagonist (picrotoxin). Created with BioRender.com. E and F, Percentage switching efficiency calculated as percentage of RFP-positive area normalized to GFP-positive area, normalized to control in 3 days post-fertilization zebrafish embryos treated with muscimol (10 μmol/L; E) or picrotoxin (100 μmol/L; F). Data represent n = 44 DMSO-treated fish, n = 42 muscimol-treated fish, and n = 44 picrotoxin-treated fish pooled from 3 biological replicates. Error bars, SD; P values generated by two-tailed unpaired t test; ****, P < 0.0001. G, Waterfall plot of enriched pathways from GSEA of switched vs. parental keratinocytes. GABA-A receptor pathways are highlighted in red. H, Immunostaining for GAD1 (enzyme) and GABA in A375 melanoma cells. Individual cells are pseudocolored as indicated. I, Schematic representation of the human in vitro switch reporter assay in melanoma/keratinocyte cocultures with genetic loss of function in GABA pathway components. Created with BioRender.com. J, Percentage switching efficiency calculated as number of switched cells per well normalized to control siRNA when cocultures were treated with a combination of GAD1/2 (melanoma cells) and ARHGEF9 (keratinocytes) targeting siRNA pooled from 3 biological replicates (n = 12; ratio: 1:3, keratinocyte:melanoma). Error bars: SD; P values generated by two-tailed unpaired t test; ****, P < 0.0001.
GABAergic Genes Are Differentially Expressed in Keratinocytes versus Melanoma Cells
GABAergic signaling has mainly been studied in the context of neuronal communication, but our data suggested it may unexpectedly play an analogous role in melanoma/keratinocyte communication. In neurons, GABA is synthesized in presynaptic neurons via GAD1 or GAD2 (27, 28) and is exported into the synapse where it binds to GABA-A receptors on postsynaptic neurons. In addition, GABA-A receptors can also be activated extrasynaptically in neurons to drive a persistent and noisy “tonic” activation (29). We hypothesized that components of this machinery might also be expressed by melanoma cells and keratinocytes, since previous studies have reported the presence of certain GABAergic signaling components in the skin (30–33). To test this, we performed RNA sequencing (RNA-seq) of the keratinocyte populations that had undergone Cre-mediated switching. Consistent with our chemical screen, gene set enrichment analysis (GSEA) demonstrated that switched keratinocytes had a marked enrichment for pathways related to activation of the GABA-A receptor (Fig. 3G; Supplementary Table S2), with upregulation of individual genes including GABA-A receptor subunits such as GABRA3, GABRB3, and GABRG2 as well as ARHGEF9 (collybistin), a gene encoding an assembly protein that ensures proper focal membrane organization of the GABA-A receptor at both synaptic and extrasynaptic sites (refs. 34–37; Supplementary Fig. S5E). We also analyzed publicly available gene expression data (from the Cancer Cell Line Encyclopedia and Wistar melanoma cell lines; ref. 38) and found that melanoma cells (but not mature melanocytes) express high levels of the GABA synthesizing enzyme GAD1 (Supplementary Fig. S5F). Moreover, GAD1 expression is induced upon oncogene (BRAFV600E) expression and correlated with melanoma oncogenic competence in our previously developed human pluripotent stem cell–derived melanocyte model (refs. 2, 39; Supplementary Fig. S5G). To further test this, we performed immunofluorescence studies and were able to detect the presence of GABA itself and GAD1 in melanoma cells (Fig. 3H; Supplementary Fig. S6A–S6E). To genetically test the role of GABA-A signaling, we knocked down the GABA synthesis enzymes GAD1 and GAD2 only in melanoma cells, and the GABA-A receptor organizer ARHGEF9 (collybistin) only in keratinocytes, and then measured Cre-mediated fluorescent switching (Fig. 3I). Although individual knockdowns showed a partial decrease in switching (Supplementary Fig. S7A–S7D), double knockdown of GAD1 and GAD2 in melanoma cells, and ARHGEF9 in keratinocytes, showed a much larger decrease (92%; Fig. 3J), highlighting the primary role of the GABAergic pathway in this form of melanoma/keratinocyte communication.
Melanoma Cells Form Specialized Inhibitory Electrochemical Cell–Cell Junctions with Keratinocytes
In the adult nervous system, GABAergic signaling is primarily involved in inhibitory neurotransmission via an influx of chloride ions into the postsynaptic cell, resulting in a decreased likelihood of a postsynaptic action potential (40, 41). Recent studies using melanocyte/keratinocyte cocultures have demonstrated the presence of calcium spike–based electrical activity between these cell types during normal development (42). Based on this, we hypothesized that because physical contact was essential for switching (Fig. 1D), melanoma cells were reviving this developmental mechanism to promote tumor initiation by forming inhibitory GABAergic cell–cell junctions with keratinocytes.
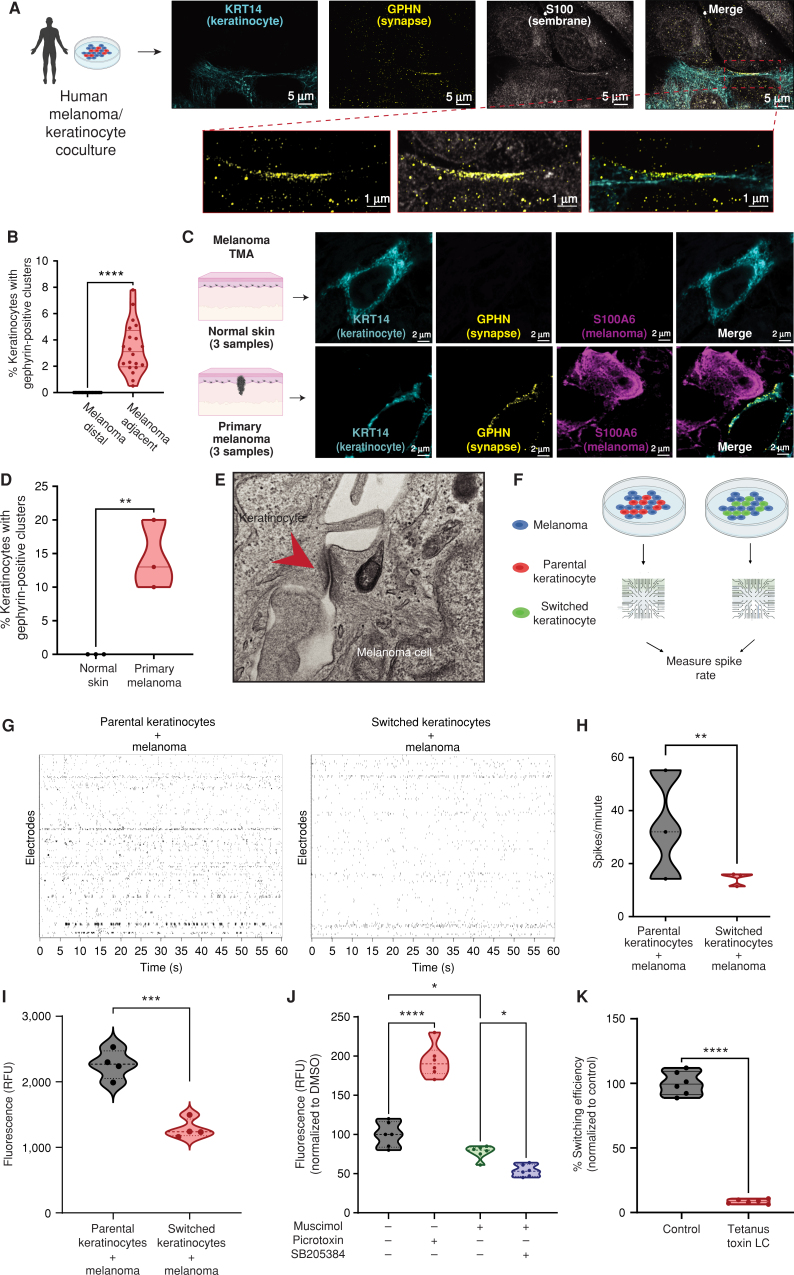
Studies in neurons have shown that both synaptic and extrasynaptic GABA-A receptor activation is regulated posttranscriptionally and requires the clustering of assembly proteins such as gephyrin, which ensures proper stabilization of membrane GABA-A receptors (42–44). Further, gephyrin clustering is highly dependent on the activity of collybistin (ARHGEF9; ref. 34), which we previously found was upregulated in switched keratinocytes (Supplementary Fig. S5E). To test whether melanoma/keratinocyte cocultures expressed such markers of GABA-A receptor activation, we performed immunofluorescence studies using gephyrin as a GABA-A receptor marker. We detected gephyrin positivity in keratinocytes upon confocal imaging, which we define as a high density of 3D gephyrin-positive signal aggregation in focal regions along the membrane. We observe this gephyrin cluster positivity specifically in keratinocytes proximal to melanoma cells in coculture (either in direct contact or adjacent), highlighting the activation of the GABA-A receptor machinery in keratinocytes in close proximity to melanoma cells (Fig. 4A and B; Supplementary Video S3). To further test this in melanoma patient samples, we stained a series of in situ melanomas in a tumor microarray (TMA) to look for the presence of the GABA-A receptor machinery in the keratinocytes directly adjacent to the tumor cells. We marked melanoma cells with S100A6 and stained for gephyrin (as a marker of the GABA-A receptor), and found that the tested melanoma patient samples contained gephyrin-positive clusters in keratinocytes directly adjacent to melanoma cells, suggesting activation of the GABAergic machinery in melanoma-associated keratinocytes, but not in normal skin (Fig. 4C and D).
Figure 4.
Melanoma cells form specialized cell–cell junctions with keratinocytes. A, Representative image of immunostaining for KRT14 (keratinocyte marker), S100 (membrane marker), and gephyrin (postsynapse GABAergic marker) in human melanoma/keratinocyte cocultures. Gephyrin-positive clusters are observed in keratinocytes only at sites of melanoma cell contact (zoomed in). Individual cells are pseudocolored as indicated. Left schematic created with BioRender.com. B, Quantification of gephyrin-positive clusters in melanoma/keratinocyte cocultures. Each datapoint represents a microscopic field quantified for the presence of keratinocyte gephyrin-positive clusters pooled from 4 biological replicates (n = 20). P values generated by unpaired t test; ****, P < 0.0001. C, Representative images of a patient malignant melanoma in situ and normal skin sample with immunostaining for KRT14 (keratinocyte marker), S100A6 (melanoma marker), and gephyrin (postsynapse GABAergic marker). Individual cells are pseudocolored as indicated. Left schematic created with BioRender.com. D, Violin plots showing percentage of keratinocytes with gephyrin-positive clusters in melanoma patient samples and normal skin. Data represent samples from n = 3 melanoma in situ patients, n = 3 normal skin; P values generated by unpaired t test; **, P < 0.01. E, Transmission electron microscopy of melanoma/keratinocyte cocultures with specialized cell–cell junction structures indicated with red arrowhead. A representative image is shown. F, Schematic representation for MEA experimental setup in human melanoma cocultures with parental or switched keratinocytes for 48 hours. Created with BioRender.com. G, Representative spike rastergrams of 1 minute of activity on MEA chip in 48-hour melanoma/keratinocyte cocultures. H, Quantification of MEA activity in 48-hour melanoma/keratinocyte cocultures calculated as spikes per minute (ratio: 1:5, melanoma:keratinocyte). Data represent 3 biological replicates per condition, with P values calculated using multiple unpaired t test using the Holm–Šídák method for multiple comparisons; **, P < 0.01. I, Calcium spike activity in cocultures of melanoma cells with switched or parental keratinocytes. RFU is relative fluorescence units measured using the calcium dye Rhod-4 (ratio: 1:5, melanoma:keratinocyte). Data represent 3 biological replicates per condition calculated using unpaired t test; ***, P < 0.001. J, Calcium spike activity in melanoma/keratinocyte cocultures upon the addition of a GABA-A agonist (muscimol), a GABA-A antagonist (picrotoxin), and a GABA-A allosteric modulator (SB205384; ratio: 1:5, melanoma:keratinocyte). RFU is a relative fluorescence unit measured using the calcium dye Cal-520. Data represent 6 biological replicates per condition calculated using one-way ANOVA with multiple comparisons; *, P < 0.05; ****, P < 0.0001. K, Percentage switching efficiency calculated as number of switched cells per well normalized to control upon melanoma-specific expression of tetanus toxin light chain (LC) in melanoma/keratinocyte cocultures for 48 hours pooled from 6 biological replicates (n = 12; ratio: 1:3, keratinocyte:melanoma). Error bars, SD; P values generated by unpaired t test; ****, P < 0.0001.
In addition, we looked at differentially upregulated pathways in switched keratinocytes using GSEA. We found a strong enrichment of pathways related to specialized cell–cell junctions including synapse formation, particularly associated with the post-synapse (Supplementary Fig. S8A). This suggests that cells in the melanoma microenvironment transcriptionally upregulate some but not all components of the synapse-like machinery, revealing a novel mode of direct contact–mediated cell–cell communication in the skin. To further test whether the melanoma cells and keratinocytes were forming such specialized cell–cell junctions, we performed electron microscopy on melanoma/keratinocyte cocultures, which revealed strong, localized electron-dense sites of cell–cell contact between melanoma cells and keratinocytes, with an average cleft distance of 21 nm (± 3 nm), smaller than the typical epithelial cell junction cleft size of 35 nm. In addition, we saw specialized bands of extracellular material between the membranes, but we did not see an electron-dense midline that would be more characteristic of an epithelial cell–cell junction (Fig. 4E; Supplementary Fig. S8B–S8D; refs. 45–47). To functionally test the properties of this specialized GABAergic cell–cell junction, we performed extracellular electrophysiology recordings of melanoma/keratinocyte cocultures. This allowed us to quantify fast changes in membrane voltage. We plated melanoma cells with parental keratinocytes or Cre-recombined switched keratinocytes on a multielectrode array (MEA) system to record the electrical spikes of both cell types together (Fig. 4F). Whereas melanoma/parental keratinocytes had substantial spiking activity (33 spikes/minute), this was greatly diminished with the melanoma/switched keratinocytes (14 spikes/minute), consistent with an inhibitory effect of GABAergic signaling present in those keratinocytes (Fig. 4G and H; Supplementary Video S4). To further test this, we performed calcium imaging using fluorescent calcium indicators (48), an important marker of calcium spike activity in neurons (49). We loaded melanoma cells and either parental or switched keratinocytes with the calcium dye Rhod-4. Consistent with the switched keratinocytes having more GABA signaling resulting in a hyperpolarizing effect, we found decreased calcium activity in these cells compared with parental cocultures, suggesting that GABA-A receptor–mediated chloride influx resulted in a calcium ion efflux, thus effectively lowering the calcium spike activity in switched keratinocytes (35% less activity; Fig. 4I). We then tested the effect on calcium currents after treatment with the GABA-A agonist muscimol, the GABA-A antagonist picrotoxin, and a GABA-A receptor–positive allosteric modulator, SB205384, either alone or in combination. Although treatment with picrotoxin had little to no effect on monocultures (Supplementary Fig. S8E), we found that treatment with a GABA-A agonist decreased calcium activity slightly (23%), which was further reduced upon addition of an allosteric modulator (47%), whereas a GABA-A antagonist substantially increased calcium activity in the melanoma/keratinocyte cocultures, indicative of an increase in calcium spike activity upon loss of inhibitory GABAergic signaling between melanoma/keratinocytes (Fig. 4J). This is consistent with previous studies that suggest that positive allosteric modulators, which have binding sites distinct from the GABA-A receptor agonists, increase the amplitude of the inhibitory signal at the GABA-A receptors, thus further reducing calcium activity when applied in combination with a GABA-A receptor agonist (50).
In neurons, GABA-A receptor–mediated inhibitory neurotransmission is associated with an inward flux of chloride ions, which ensures rapid hyperpolarization and decreased action potential in the postsynaptic cell. To test whether the chloride ion itself was involved in melanoma/keratinocyte communication, we used a reduced chloride medium to decrease the extracellular and intracellular chloride concentration and block the GABA-A receptor–mediated chloride influx in keratinocytes, as described earlier (51, 52). We found that decreasing intracellular chloride ions substantially decreased switching efficiency in cocultures and disrupted melanoma/keratinocyte communication, without any effect on cell proliferation, highlighting the critical role of the chloride influx in this form of communication (Supplementary Fig. S8F and S8G).
To further understand whether this melanoma/keratinocyte interaction was dependent on the SNARE-dependent vesicular pathways regulating GABA release (53), we overexpressed the tetanus toxin light chain selectively in human melanoma cells. Tetanus toxin light chain blocks the exocytosis of neurotransmitters such as GABA from inhibitory neurons via SNARE proteins and blocks communication at neuronal as well as immunologic synapses (54, 55). We found a striking loss in melanoma/keratinocyte communication (80% decrease) upon expression of the tetanus toxin light chain in melanoma cells, suggesting that the GABAergic signaling activity in melanoma/keratinocyte cocultures is dependent on the SNARE-mediated exocytosis of GABA at specialized cell–cell junctions (Fig. 4K; Supplementary Fig. S8H). To investigate whether this specialized GABAergic cell–cell junction regulates melanoma initiation in vivo, we overexpressed a dominant-negative form of the GABA-A receptor subunit gabrg2, driven by the keratinocyte-specific krt4 promoter, which disrupts GABAergic communication, specifically in keratinocytes (56). This resulted in a significant decrease in the rescue of nascent melanoma cells in our previously described zebrafish melanoma model, suggesting a significant role of microenvironmental components of the GABAergic signaling pathway in melanoma initiation in vivo (Supplementary Fig. S8I).
Collectively, these data indicate that melanoma cells and keratinocytes form specialized cell–cell junctions with some characteristics of both synaptic and extrasynaptic GABAergic signaling (57) and that GABAergic signaling from melanoma cells acts as an inhibitor of electrical activity in melanoma/keratinocyte cocultures. Finally, we observed the presence of exosome-like vesicles close to the specialized GABAergic cell–cell junctions in both our light and fluorescent microscopy studies (Supplementary Fig. S9A–S9C). We hypothesize that this might be a mechanism to ensure polarized delivery of exosomes at specific sites of cell–cell contact between melanoma cells and keratinocytes, as has been observed previously in T-cell immune synapses (58–60).
GABA Signaling Promotes Melanoma Initiation
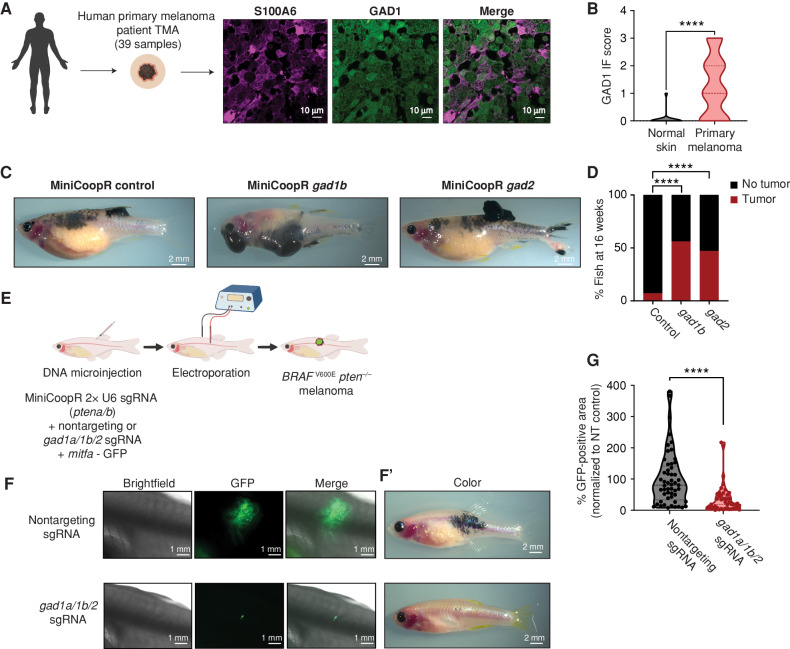
The above data suggested that melanoma/keratinocyte communication is important in melanoma initiation and that this communication is mediated by GABA. Based on this, we wanted to genetically test the effects of GABA on melanoma initiation in vivo. To identify which GABA-related proteins were most relevant, we analyzed components of the GABAergic signaling pathway in human melanoma tissue samples. We performed immunofluorescence for GAD1 (as a marker of GABA synthesis) and S100A6 (as a melanoma marker) on a human tissue microarray containing both normal skin as well as primary melanomas (n = 39 samples). Whereas nearly all normal skin samples (n = 9) were negative for GAD1, 40% of primary melanoma samples (n = 30, Fig. 5A and B) were positive for GAD1. Further, we used the zebrafish to test the effect of GAD1 (and its related protein GAD2) on melanoma initiation. We used the MiniCoopR system described above to activate BRAFV600E in melanocytes (along with germline p53 loss) but sensitized the system by injecting low doses of the rescue plasmids, such that most of the fish would not develop melanoma on their own (Supplementary Fig. S10A). As expected, only 10% of control fish receiving MiniCoopR alone developed tumors at the 16-week time point. In contrast, transgenic fish overexpressing either gad1b (n = 53) or gad2 (n = 63) in melanocytes had a higher rate of melanoma initiation by BRAFV600E (Fig. 5C and D). Furthermore, we noted that the gad-overexpressing fish tended to have multiple tumors, suggesting that GAD/GABA signaling lowers the threshold for tumor initiation in this model, with no significant difference in metastasis formation (Supplementary Fig. S10B–S10D). To further test this, we then performed loss-of-function experiments. Because zebrafish melanomas express multiple GAD genes (gad1a/gad1b/gad2; ref. 61; Supplementary Fig. S11A–S11D), we took advantage of the Transgene Electroporation in Adult Zebrafish (TEAZ) model of transgenic melanoma, in which multiplexed CRISPR cassettes can be directly electroporated into the skin of an adult fish (62). We designed single-guide RNAs (sgRNA) against all three zebrafish gad genes (gad1a/gad1b/gad2) and then initiated tumors using melanocyte-specific expression of BRAFV600E along with loss of pten and p53 (Fig. 5E; Supplementary Fig. S12A and S12B). Although we do not see a difference in the fluorescence area of the mitfa-positive cells, 1 week after electroporation (Supplementary Fig. S12C), we found that CRISPR-mediated deletion of the gad genes resulted in a significant decrease in tumor size at both 6 weeks (Fig. 5F, F’, and G) and 10 weeks (Supplementary Fig. S12D) after electroporation in the gad knockouts compared with the nontargeting control animals (65% decrease).
Figure 5.
GABAergic signaling is protumorigenic in melanoma. A, Representative image of a patient primary melanoma sample from a primary melanoma TMA with immunostaining for S100A6 (melanoma marker) and GAD1 (GABA marker). Individual cells are pseudocolored as indicated. Left schematic created using BioRender.com. B, Violin plots of immunofluorescence (IF) score and quantification of GAD1 immunostaining in primary melanoma tumor samples and normal skin. Data represent samples from n = 30 primary melanoma patients and n = 9 normal skin; P values generated by unpaired t test with Welch correction; ****, P < 0.0001. C, Representative images of 16-week-old zebrafish with the genotype (mitfa−/−p53−/−mitfa:BRAFV600E) in the casper background injected with MiniCoopR rescue plasmids showing control (GFP) or gad1b- and gad2-overexpressing tumors. D, Quantification of melanoma incidence expressed as percentage of fish with tumors in 16-week-old zebrafish overexpressing GFP or gad1b or gad2 under a melanocyte-specific promoter. Data represent n = 55 control (GFP) fish, n = 53 gad1b, and n = 63 gad2 fish pooled from 3 biological replicates. P values generated by the chi-squared test; ****, P < 0.0001. E, Schematic representation of the TEAZ-based loss-of-function system in zebrafish to knockout GABA synthesis genes. Plasmids expressing MiniCoopR, mitfa:Cas9, and sgRNAs targeting pten were coelectroporated with plasmids expressing nontargeting or gad-targeting sgRNAs to generate control (nontargeting) or gad knockout BRAFV600Epten−/− melanomas in vivo. (Created using BioRender.com.) F and F’, Representative images of four different transgenic fish electroporated with melanocyte-specific Cas9 and a nontargeting sgRNA or gad1a/gad1b/gad2 sgRNAs, showing differences in both fluorescence area (F) and pigmented melanocyte area (F’) 6 weeks after electroporation. G, Quantification of tumor area calculated as GFP-positive area and normalized to the nontargeting (NT) control plasmid group. Data represent 53 nontargeting sgRNA fish and 46 gad1a/1b/2 sgRNA fish pooled from 3 biological replicates. Error bars, SD; P values generated by Mann–Whitney test; ****, P < 0.0001.
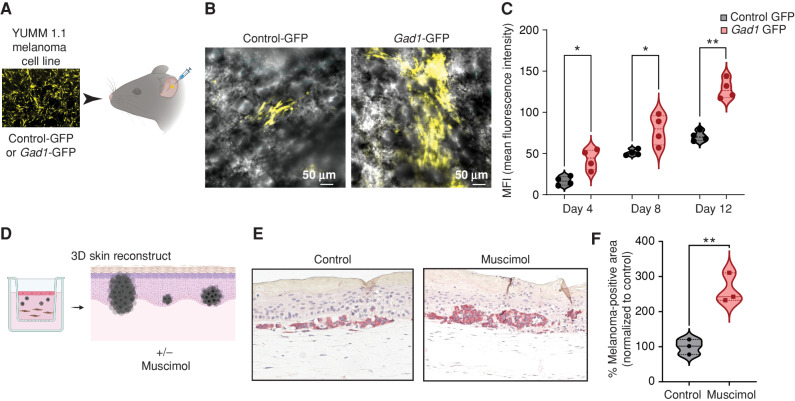
To test this in a mammalian system, we overexpressed Gad1 in the mouse melanoma cell line YUMM 1.1 and transplanted these cells into the mouse ear, which is rich in microenvironmental keratinocytes, thus providing an ideal model to test melanoma/microenvironment interactions (Fig. 6A). Although we did not observe any difference in the proliferation and invasion potential of the Gad1-overexpressing cells in monoculture in vitro (Supplementary Fig. S13A and S13B), we found a marked increase in tumor growth using this system, suggesting that Gad1 expression is protumorigenic in a mouse melanoma model (Fig. 6B and C). We next wanted to test this in a human context. We created 3D skin melanoma reconstructs, widely used to understand tumor/stromal interactions in melanoma, using human fibroblasts, keratinocytes, and melanoma cells together in a collagen matrix (63, 64). We found that treatment with the GABA-A agonist muscimol strongly increases the growth of initiated melanoma cells in a keratinocyte-rich microenvironment in this model, thus suggesting that activation of the GABA-A receptor in keratinocytes is protumorigenic in zebrafish, mouse, as well as human melanoma models (Fig. 6D–F; Supplementary Fig. S13C).
Figure 6.
GABA-A receptor activation drives melanoma growth in mouse models and human 3D skin reconstructs. A, Schematic representation of the mouse ear transplant experiment in which YUMM 1.1 cells expressing control-GFP or Gad1-GFP are injected into mouse ears and imaged at 4, 8, and 12 days after injection. Created using BioRender.com. B, Representative images of mouse ears transplanted with control- or Gad1-GFP–expressing YUMM 1.1 mouse melanoma cells, 4 days after injection. C, Mean fluorescence intensity (MFI) in control- or Gad1-GFP–expressing mouse melanoma cells transplanted into mouse ears and imaged at 4, 8, and 12 days after injection in 4 biological replicates. Error bars, SD; P values generated by two-tailed unpaired t test; *, P < 0.05; **, P < 0.01. D, Schematic representation of 3D melanoma skin reconstruction experiments, in which the constructs were treated with the GABA-A receptor agonist muscimol. Created using BioRender.com. E, Representative images of IHC performed on 3D melanoma skin reconstructs treated with control media or muscimol-containing media, with melanoma cells stained with an anti-BRAFV600E antibody, after 24 days in coculture with keratinocytes and fibroblasts in 3D reconstruct media. F, Percentage melanoma-positive area (BRAFV600E-positive) in 3D skin reconstructs treated with control media or muscimol-containing media (100 μmol/L). Data are pooled from 3 biological replicates, normalized to control media. Error bars, SD; P values generated by two-tailed unpaired t test; **, P < 0.005.
Because GABA/GABA-A agonists could be acting in a cell-autonomous manner on the melanoma cells themselves (rather than on the keratinocytes), we excluded this possibility by treating melanoma cells in vitro with GABA or picrotoxin and found no change in melanoma proliferation rate (Supplementary Fig. S13D and S13E). Similarly, the knockdown of GAD1/2 in melanoma cells in culture also did not affect melanoma proliferation (Supplementary Fig. S13F), indicating that the effect in vivo is not due to cell-autonomous effects on the melanoma cells themselves. Collectively, these data strongly implicate GABA signaling as a key factor in melanoma initiation and growth in vivo via an interaction with keratinocytes.
GABA-A Receptor–Positive Keratinocytes Promote Melanoma Growth via the MYCN/LIF Axis
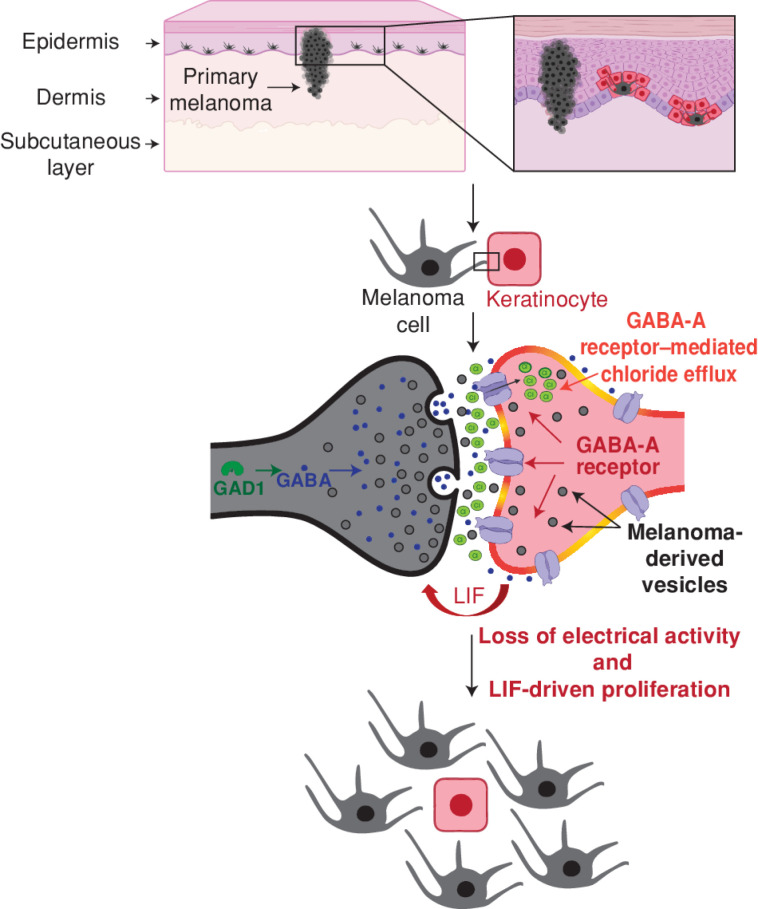
The above data suggested that the GABAergic keratinocytes were promoting the growth of the melanoma cells in vivo. Keratinocytes have previously been shown to suppress melanoma formation through physical tethering, which acts to restrain the growth of the nascent melanoma (13). In part, this is controlled through expression of Par3 on the keratinocytes, and its loss then allows the nascent melanoma cells to “decouple” or “escape” from the growth control of the keratinocytes (10). In addition to physical decoupling, the keratinocytes can also express secreted factors such as EDN1, EDN3, or FGFs, which can promote melanoma growth by binding to EDNRB or FGFR receptors (65, 66). Our data indicate that another way that melanoma cells could escape from keratinocyte growth control is through the activation of inhibitory GABAergic signaling, but whether these GABAergic keratinocytes promote the growth of melanoma through similar secreted factors remained unclear. To address this, we first cocultured our human melanoma cells with parental keratinocytes or Cre-recombined switched keratinocytes and monitored the proliferation of the melanoma cells over time (Supplementary Fig. S14A). Coculture with switched keratinocytes increased melanoma cell proliferation over a period of 48 hours (Supplementary Fig. S14B–S14D). Further, the increase in proliferation was also seen when melanoma cells were treated only with conditioned media from the switched keratinocytes (Supplementary Fig. S14E and S14F), suggesting that part of the protumorigenic effect could be mediated by a secreted factor from switched keratinocytes. We analyzed our RNA-seq data of the Cre-switched keratinocytes compared with the parental keratinocytes to find putative secreted ligands that would promote melanoma proliferation. Although we saw no significant increase in expression of EDN1 or FGF1/2, we found a significant elevation of FGF5, BMP6, and LIF, factors known to promote melanoma growth (refs. 67–70; Supplementary Fig. S15A). Based on this, we then tested whether inhibition of these pathways would slow the proliferation of the melanoma cells grown with conditioned media from keratinocytes. This revealed that only inhibition of LIF receptor signaling by EC330 decreased melanoma cell proliferation predominantly when treated with conditioned media from switched keratinocytes, though there is a slight growth disadvantage in the parental cell conditioned media as well (Supplementary Fig. S15B). Furthermore, targeting LIFR in melanoma cells using siRNA approaches decreased melanoma proliferation (Supplementary Fig. S15C), consistent with recent data that expression of the LIF receptor in melanoma is associated with poor prognosis in this disease (71). We detected the presence of transcripts for LIF/GABA-associated components in our zebrafish melanoma tumors as well, suggesting that LIF-mediated melanoma signaling is prevalent in vivo in melanoma (Supplementary Fig. S16A–S16D). Interestingly, LIF expression has been widely reported in the nervous system and is associated with increased growth, survival, and neuroprotection (72, 73). Taken together, this suggests that GABA-A receptor–positive keratinocytes mediate their protumorigenic effect by secreting LIF into the melanoma microenvironment, which increases melanoma cell proliferation independent of cell–cell contact (Fig. 7).
Figure 7.
Model for GABAergic signaling–driven melanoma/keratinocyte communication. Created using BioRender.com.
We further wanted to identify the upstream regulators of LIF in the switched keratinocytes. Our RNA-seq studies detect a strong upregulation of MYCN in switched keratinocytes (Supplementary Fig. S17A), which is a known upstream activator of LIF expression (74). We analyzed existing ChIP-seq datasets and found that MYCN was bound to several sites at the LIF gene locus in neuroblastoma cells (Supplementary Fig. S17B). To understand whether MYCN is involved in the upstream regulation of LIF, we perturbed MYCN expression in keratinocytes using siRNA approaches and found a marked reduction in LIF expression (Supplementary Fig. S17C and S17D). It has been previously shown that MYCN is associated with early skin development in mammals and is expressed in keratinocytes (75, 76), suggesting that GABAergic signaling in melanoma cells may activate an early developmental program in keratinocytes that supports melanoma growth.
DISCUSSION
In this study, we identify a novel mode of communication between melanoma cells and keratinocytes in the melanoma microenvironment. Although melanocyte and keratinocyte cells are closely intertwined in normal skin physiology, they typically become decoupled from each other during the early stages of melanoma development (77–79). Although physical decoupling is one such mechanism, we found that keratinocytes and melanoma cells form specialized GABAergic inhibitory electrochemical cell–cell junctions not previously reported in skin. Given that electrical activity is increasingly recognized to play a role in tumorigenesis (80–82), it is likely that this type of GABAergic signaling mechanism may be true in other epithelial tissues, which awaits further study.
Although it is well understood that electrical activity primarily regulates cell–cell communication in excitable cells such as neurons, nonexcitable cells can also be regulated by electrical activity–based communication (83, 84). In the skin, for example, certain ion channels regulating electrical activity and membrane potential play an important role in establishing skin pigmentation patterns (85, 86). In tumors, some of the earliest studies looking at electrical activity found that nascent tumor cells show loss of electrical activity upon transformation (87). Further studies highlighted that the loss of electrical activity was specifically between “transformed” cancer cells and “nontransformed” healthy cells, suggesting a functional loss of communication between tumor cells and the “nontransformed” healthy cells in their microenvironment (88, 89). Our data indicate that there is electrical signaling between melanoma cells and keratinocytes (based on MEAs) and that this communication is dependent upon extracellular chloride. Our present study suggests that one of the mechanisms that could drive such loss of electrical activity–based communication in melanoma is the activation of GABAergic inhibitory signaling in the tumor microenvironment, promoting increased proliferation of nascent tumor cells via the secretion of protumorigenic factors. We do not yet know whether tumor initiation depends on this inhibition of electrical activity per se, but we find that GABA is both necessary and sufficient for tumor initiation, suggesting it may play a role. It also remains to be understood how loss of electrical activity directly contributes to tumorigenesis in this context and whether loss of electrical activity also invokes LIF-independent, nonsecreted mechanisms of communication. We find a strong upregulation of GAD1 in our zebrafish as well as human patient samples, and this upregulation correlates well with primary tumor–forming ability but not metastasis in melanoma. This suggests that GAD1 might be a useful prognostic marker to predict early tumor initiation during routine melanoma screening of pretumorigenic moles that might progress to primary melanoma tumors.
Recent studies in brain tumors have shown that functional synapses can form between a tumor cell and a neuron, which aids in tumor progression (90–92). These studies suggest that the formation of a tumor cell to neuron synapse is protumorigenic both via increased neuronal activity due to stronger synaptic connections and via secreted paracrine factors that promote tumor proliferation. In our study, we show for the first time that specialized inhibitory GABAergic cell–cell junctions between skin cells can be formed in primary melanoma, independent of any input from neurons. We do not yet know the precise nature of this junction, and it will be important to explore whether it has a resemblance to the synaptic connections or whether it is extrasynaptic in nature, because our current assays cannot distinguish between the phasic and tonic forms of inhibition, which primarily distinguish these two forms of signaling (29). It also remains to be tested whether this unique communication pathway between melanoma cells and keratinocytes is a part of the normal physiology of melanocyte/keratinocyte communication during development. Interestingly, a recent study using GCaMP-based calcium imaging in skin showed the presence of calcium spikes and “electric-like” activity in melanocyte/keratinocyte cocultures, suggesting that melanocytes and keratinocytes might communicate via electrical signals (42). Further, a recent single-cell RNA-seq study in epidermal melanocyte populations at different stages of development found that “synapse formation” is a highly upregulated pathway during melanocyte development, in addition to canonical melanocyte-specific pathways such as pigmentation and organelle maturation (93). The above two studies suggest that electrical activity–based communication and the ability to form synapse-like structures may be closely intertwined with melanocyte development. Combined with our findings, this might indicate that nascent melanoma cells induce GAD1 expression and use their preexisting trafficking machinery to activate specialized inhibitory GABAergic cell–cell junctions in keratinocytes, effectively decoupling themselves from the stringent growth control of the skin keratinocytes. It remains to be identified whether additional cell types in the melanoma microenvironment can form such specialized structures with melanoma cells as the tumor cells progress and metastasize to different organs. This might be of particular importance in the melanoma immune microenvironment, because several immune cell types, including T cells, express GABA-A receptors and form cell–cell junctions with tumor cells called the “immunologic synapse” (55, 94).
Finally, our study suggests that melanoma/keratinocyte communication is mediated by exosome-like vesicles. Our loss-of-function studies indicate that perturbation of exosome-like vesicle machinery in melanoma cells by pharmacologic (GW4869 treatment) or genetic approaches (nSMase2 knockdown) is sufficient to block vesicle-mediated communication between melanoma cells and keratinocytes (Supplementary Fig. S4B and S4C). Because most vesicle-based cargo is targeted for degradation in the recipient cell (95, 96), GABAergic signaling may be one such mechanism that increases the amplitude of vesicle-based communication between different cell types. In accordance with this, we note that disrupting the GABA-A receptor–mediated chloride efflux by blocking the chloride channel or by lowering the intracellular chloride concentration (Fig. 3C and F; Supplementary Fig. S8F) is sufficient to disrupt this vesicle-based communication. It remains to be identified which components in the melanoma vesicle induce the MYCN/LIF axis in the switched keratinocytes, but several recent studies suggest a role for noncoding RNAs such as miRNAs or long noncoding RNAs in extracellular vesicle–mediated reprogramming of recipient cells (97, 98). Interestingly, drugs inducing chloride accumulation in the cell and facilitating “endosomal escape” are being increasingly studied for the functional delivery of extracellular vesicle cargo (99, 100). Future identification of the role of chloride in this process may pave the way for a new generation of therapeutic targets to disrupt tumor/microenvironment communication.
METHODS
Zebrafish
Zebrafish Husbandry
All zebrafish experiments were approved by and carried out in accordance with institutional animal protocols from the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Animal Care and Use Committee (IACUC), protocol number 12-05-008. Fish stocks were kept under standard conditions at 28.5°C under 14-hour light/10-hour dark cycles, with salinity and pH (7.4) controlled conditions. Animals were fed a standard zebrafish diet consisting of brine shrimp followed by Zeigler pellets. Embryos were collected from natural mating and incubated in E3 buffer (5 mmol/L NaCl, 0.17 mmol/L KCl, 0.33 mmol/L CaCl2, 0.33 mmol/L MgSO4) at 28.5°C. All anesthesia was performed using Tricaine-S (MS-222, Syndel USA) with a 4 g/L, pH 7.0 stock. Sex determination in embryos is not possible at 3 to 5 days post-fertilization (dpf). Power analysis was not performed to determine the sample size. For all zebrafish experiments, a large sample size was maintained and estimated from similar experiments in previous publications from the group. For all adult zebrafish animal experiments to determine tumor initiation and progression, more than 40 animals per condition were used. No data exclusions were performed in any of the datasets.
In Vivo Switch Assay in Zebrafish Embryos
For the in vivo switch assay in zebrafish, one-cell stage casper (101) mitfa:BRAFV600E p53−/− embryos (30 embryos per condition) were injected with the following reporter constructs at 5 ng/μL with tol2 mRNA at 20 ng/μL:
krt4-loxP-lacZ-loxP-tdTomato OR krt4-loxP-GFP-loxP-tdTomato
MiniCoopR-palmGFP OR MiniCoopR-empty
mitfa-cre OR mitfa-empty
Embryos were screened for GFP fluorescence and imaged at 3 dpf to measure GFP and tdTomato (RFP) fluorescence. To calculate switching efficiency in embryos, we used GFP fluorescence from the krt4-loxP-GFP-loxP-tdTomato construct to mark all rescued keratinocytes in the embryo and tdTomato (RFP) fluorescence to mark switched keratinocytes only. For the NTR experiments, DMSO or 100 μmol/L MTZ was added directly to the fish water at 2 dpf and quantifications were performed after 24 hours, at 3 dpf. For quantifying switching efficiency, background autofluorescence was subtracted and the rescued keratinocyte area was quantified by uniformly thresholding the GFP intensity using the default ImageJ segmentation algorithm, followed by quantifying the switched keratinocyte area by uniformly thresholding the tdTomato (RFP) intensity across all animals. The percentage of switching efficiency was calculated as tdTomato (RFP)-positive area/GFP-positive area times 100 using ImageJ (NIH; RRID:SCR_003070). All images were acquired on a Zeiss AxioZoom V16 fluorescence microscope.
Pharmacologic Treatment of Zebrafish Embryos
Zebrafish melanoma-prone embryos (casper mitfa:BRAFV600E p53−/−) were injected with the following constructs: krt4-loxP-GFP-loxP-tdTomato + MiniCoopR-Cre (20 embryos per condition) at the one-cell stage. At 8 hours post-fertilization (hpf), embryos were placed in a 40-μm cell strainer in a 6-well dish in 6 mL of E3 water. Each well was treated with the following compounds: DMSO or muscimol (Sigma #M1523) at 10 μmol/L, or picrotoxin (Sigma #P1675) at 100 μmol/L. The compounds were added at 8 hpf and reapplied at 1 dpf and 2 dpf. Embryos were imaged at 3 dpf using the same imaging and quantification protocol as described above to calculate percentage switching efficiency. Individual animals were randomly allocated into control or experimental groups for pharmacologic treatment.
Generation of gad1b- and gad2-Overexpressing MiniCoopR Transgenic Fish
Transgenic zebrafish were generated by injection of 5 ng/μL of MiniCoopR-GFP and empty vector or MiniCoopR-GFP; mitfa-gad1b or MiniCoopR-GFP; mitfa-gad2 and tol2 mRNA at 20 ng/μL in melanoma-prone casper mitfa:BRAFV600E p53−/− embryos. Embryos were screened for melanocyte rescue at 5 dpf. Embryos with successful melanocyte rescue were grown to adulthood and scored for the emergence of tumors at 10 and 16 weeks post-fertilization (wpf). For quantifying kidney metastasis, individual animals were screened for GFP-positive areas in the kidney marrow combined with verification using IHC staining.
Generation of gabrg2 Dominant-Negative Transgenic Fish
The gabrg2 gene containing the premature stop codon resulting in a dominant-negative mutation was generated as described previously (102) and cloned into a middle-entry gateway compatible vector. The final construct, krt4:gabrg2-DN-394, was generated using gateway cloning.
TEAZ Electroporation
TEAZ was utilized to generate melanomas as previously described (62). The casper mitfa:BRAFV600E p53−/− transgenic animals (5–6 months old) were electroporated with plasmids to generate BRAFV600E, p53−/−, pten−/− tumors expressing either a nontargeting sgRNA or sgRNAs against gad1a, gad1b, and gad2. Plasmids electroporated per animal included MiniCoopR 2× U6 sgRNAs-pten, mitfa:Cas9 (300 ng; ref. 103) to induce melanocyte rescue and loss of pten, mitfa:GFP (125 ng), Tol2 (58 ng), and 100 ng of nontargeting or gad sgRNAs. Briefly, adult male fish were anesthetized with 0.2% tricaine and injected with 1 μL of plasmid mix described above into the skin below the dorsal fin. Fish were electroporated and allowed to recover in fresh system water. Electroporation was performed using a CM830 Electro Square Porator from BTX Harvard Apparatus and Genepaddles 3 × 5 mm, with a voltage of 45 V, 5 pulses, 60 ms pulse length, and 1 s pulse interval. Fish were imaged every week using brightfield and fluorescent imaging at 25× and 10× using a Zeiss AxioZoom V16 fluorescence microscope. Tumor area was quantified by GFP fluorescence using ImageJ.
All gad sgRNAs for this experiment were designed using CHOPCHOP (104) and GuideScan (105). The sgRNA sequences are outlined below:
Nontargeting: AACCTACGGGCTACGATACG
ptena: GAATAAGCGGAGGTACCAGG
ptenb: GAGACAGTGCCTATGTTCAG
gad1a: TGACGTCACCTATGACACGG
gad1b: TACGACAACCTGCCACAAGT
gad2: GTAGAGATCCGAAAAGCACG
Plasmid Construction
For melanocyte-specific constructs, the previously developed mitfa promoter (106) in a gateway-compatible 5′ entry vector was used. For keratinocyte-specific constructs, we used the previously described krt4 promoter, which labels differentiated keratinocytes in the zebrafish skin (107). The keratinocyte-specific switch construct was generated by modifying the previously generated ubiquitin switch construct in zebrafish, ubb-loxP-GFP-loxP-tdTomato (108). The following plasmids were constructed using the Gateway Tol2 kit (109):
krt4:loxP-lacZ-loxP-tdTomato-394
krt4:loxP-GFP-loxP-tdTomato-394
krt4:loxP-GFP-loxP-DTA-394
krt4:loxP-GFP-loxP-NTR2-394
krt4:gabrg2-DN-394
MiniCoopR:palmGFP
MiniCoopR: Cre
mitfa:palmGFP-394
mitfa:cre-394
ubb:cre-394bsd (blasticidin-resistance cassette in 394 backbone)
mitfa:gad1b-394
mitfa:gad2-394
For cloning gad1a-, gad1b-, and gad2-targeting sgRNAs into a plasmid backbone, we used three different U6 promoters as described previously (110) and cloned all the sgRNAs (nontargeting or gad) in a single gateway-compatible 394 backbone with tol2 arms. Guide RNA cutting efficiency was validated using the Surveyor mutation kit (IDT #706020). MiniCoopR 2× U6:gRNA, mitfa:Cas9 (MAZERATI; ref. 103) was a gift from Leonard Zon, Harvard Medical School, Harvard University (Addgene plasmid #118844, RRID:Addgene_118844).
Quantification of Cell–Cell Contacts
For cell–cell direct contact measurements in Fig. 1 and Supplementary Fig. S1, confocal imaging was performed on live zebrafish embryos injected with the above constructs. Three dpf, live zebrafish embryos were mounted in 96-well, glass-bottomed dishes with low melting agarose and tricaine to restrict the mobility of the fish. Following this, the embryos were imaged on a Leica SP5 inverted confocal microscope (20× objective) using the xyz scan mode. Imaging was performed by focusing on a switched keratinocyte (RFP-positive) cell with 40 μm depth and a step size of 0.54 μm. Contacts were scored as adjacent only if there was a GFP-positive signal in direct continuity with an RFP-positive signal without any empty area (no fluorophore signal) per embryo. This was verified using the surfaces program from Imaris, a 3D rendering software to assess cell–cell contacts (RRID:SCR_007370). For in vitro cell–cell direct contact measurements, widefield microscopy images were acquired on a Zeiss AxioObserver fluorescence microscope and cells were scored to be in direct contact if a GFP-positive cell had a direct contacting azurite-positive cell without any fluorophore-negative area in between in the cell monolayer of melanoma/keratinocyte cocultures. To visualize points of direct contact between melanoma cells and keratinocytes, cocultures were grown on 35-mm glass bottom dishes and z-stack images were acquired on the Zeiss LSM880 microscope using AiryScan mode (63× objective) with 20 μm depth and a step size of 1 μm.
Cell Culture
Melanoma Cell Culture
Human melanoma cell lines A375 (RRID:CVCL_6233), A375-MA2 (RRID:CVCL_X495), Hs294T (RRID:CVCL_0331), A101D (RRID:CVCL_1057), SKMEL24 (RRID:CVCL_0599), SKMEL3 (RRID:CVCL_0550), SKMEL5 (RRID:CVCL_0527), C32 (RRID:CVCL_1097), SH-4 (RRID:CVCL_1692), and BJ (RRID:CVCL_3653) were obtained from ATCC (August 2013) where routine cell line authentication is performed using short tandem repeat profiling. The zebrafish melanoma cell line (ZMEL1) was derived from a tumor in a mitfa-BRAFV600E p53−/− zebrafish as described previously (111). Mouse melanoma cell lines, YUMM 1.1 and YUMM 4.1 (112), were a kind gift from the Neal Rosen lab, Molecular Pharmacology Program, MSKCC (December 2019). Human melanoma cell lines, A375, A375-MA2, Hs294T, A101D, and SH-4, were maintained in DMEM (Gibco #11965) supplemented with 10% FBS (Gemini Bio) and 1× penicillin/streptomycin (Gibco #15140122); SKMEL24 and C32 were maintained in EMEM (ATCC #30–2003) supplemented with 10% FBS (Gemini Bio) and 1× penicillin/streptomycin (Gibco #15140122); and SKMEL3 was maintained in McCoy's 5a Medium Modified (ATCC #30–2007) supplemented with 10% FBS (Gemini Bio) and 1× penicillin/streptomycin (Gibco #15140122). Mouse melanoma lines, YUMM 1.1 and YUMM 4.1, were maintained in DMEM-F12 (ATCC #30–2006) supplemented with 10% FBS (Gemini Bio), 10% Nonessential Amino Acids (Gibco #11440–076), and 1× penicillin/streptomycin (Gibco #15140122). The zebrafish melanoma cell line ZMEL1 was maintained in DMEM (Gibco #11965) supplemented with 10% FBS (Gemini Bio), 1× penicillin/streptomycin/glutamine (Gibco #10378016), and 1× GlutaMAX (Gibco #35050061) and grown at 28°C with 5% CO2 in a humidified incubator. All cells were passaged less than 20 times before a low-passage batch was thawed. They were routinely tested for Mycoplasma using a luminescence-based Mycoplasma detection kit (MycoAlert Mycoplasma Detection Kit, Lonza #LT07-318, last tested December 2022).
Keratinocyte Cell Culture
The HaCaT keratinocyte cell line (113) was obtained from Addexbio and authenticated at the MSKCC Molecular Cytogenetics Core. Ker-CT (RRID:CVCL_S877) is an hTERT-immortalized keratinocyte cell line (114) and was obtained from ATCC. HaCaT cells were maintained in DMEM (Gibco #11965) supplemented with 10% FBS (Gemini Bio) and 1× penicillin/streptomycin (Gibco #15140122). Ker-CT cells were maintained in KGM-Gold Keratinocyte growth medium supplemented with the KGM-Gold BulletKit (Lonza #00192060). Cells were split when confluent, approximately 2× per week, and were used directly in cocultures. For melanoma/keratinocyte cocultures, 50:50 media from melanoma cells and keratinocytes were used and cocultures were maintained for 48 hours to 3 weeks. For low-chloride media experiments, Na-gluconate and K-gluconate were used as a substitute for chloride as described in ref. 51 to maintain isotonicity.
Cell Line Generation
All human and mouse melanoma lines were engineered to overexpress Cre under the UBC promoter modified from Addgene plasmid #65727 (RRID:Addgene_65727) and subjected to puromycin selection as described previously (115). Zebrafish ZMEL1-Cre–overexpressing lines were generated using electroporation with the plasmid ubb:Cre-394Bsd (Cre expression driven by the ubb promoter) using the Neon transfection system (Thermo Fisher). All human and mouse melanoma lines were selected with puromycin (1 μg/mL), whereas the zebrafish ZMEL1 line was selected with blasticidin (4 μg/mL) for 3 weeks. For switch cell line generation in melanoma cells (A375) and keratinocytes (HaCaT and Ker-CT), the plasmid pLV-CMV-LoxP-DsRed-LoxP-eGFP (Addgene plasmid #65726, RRID:Addgene_65726) was used for lentiviral infection. All switch lines were subjected to puromycin selection followed by two rounds of FACS to eliminate any double-positive (dsRED/GFP-positive) cells.
In Vitro Switch Assay in Melanoma/Keratinocyte Cocultures
HaCaT or Ker-CT reporter keratinocytes expressing the switch cassette and all human/mouse/zebrafish melanoma cell lines expressing Cre were seeded in 1:3 ratios in 6-well/48-well/96-well plates with 50/50 keratinocyte/melanoma cell media. For zebrafish melanoma (ZMEL1) and human keratinocyte (HaCaT) cocultures, cells were maintained in a 28°C humidified incubator with 5% CO2. For all other cocultures, cells were maintained in a 37°C with 5% CO2 humidified incubator. After 48 hours of coculture, conditions were blinded and each well was manually scored for the number of switched keratinocytes (GFP-positive) per condition. Control cocultures with no Cre melanoma cells were scored in parallel to eliminate the possibility of background switching in each condition. For MemBright staining, melanoma cells were incubated with the MemBright probe (23) for 20 minutes, washed, and incubated with the reporter keratinocytes for 48 hours. All imaging was performed using either a Zeiss AxioObserver fluorescence microscope or a Zeiss LSM880 confocal microscope. Z-stacks were acquired at a depth of 20 μm with 1 μm step size. Experiments were repeated using more than 3 biological replicates with different seeding densities and different cell ratios by three independent investigators.
siRNA Treatment
For siRNA studies, Dharmacon ON-TARGETplus SMARTpool siRNAs were used to knockdown individual human genes and cells were treated based on the manufacturer's instructions. Briefly, melanoma cells or keratinocytes were seeded overnight in regular media with no antibiotics in 6-well dishes as monocultures and transfected with the indicated SMARTpool siRNAs and DharmaFECT 1 transfection reagent (Horizon #T-2005-01) in serum-free media. Seventy-two hours after transfection, siRNA-treated cells (melanoma cells or keratinocytes) were treated with a second dose of siRNA, followed by a 48-hour incubation, and then cocultured with the corresponding nontargeting siRNA-treated melanoma cells or keratinocytes and incubated for an additional 48 hours. For calculating switching efficiency, the number of switched cells per condition (3 technical replicates) was counted 48 hours after coculture and 7 days after transfection. Knockdown efficiency was validated using qPCR at 5 days and Western blot at 7 days after transfection. All siRNA-treated cells were monitored for signs of toxicity or changes in proliferation rate using the Cyquant Cell Proliferation Assay (see below), and no toxicity was observed for the indicated siRNA treatments.
Transwell Assay
A Transwell assay was performed to detect the requirement of cell–cell contact for switching in keratinocytes as described previously (115). Briefly, 20,000 HaCaT keratinocytes expressing the switch construct were plated alone or in combination with 60,000 A375 melanoma cells expressing Cre per well of a 24-well plate. Twenty-four hours after plating, 20,000 A375 melanoma cells (± Cre) were seeded into the upper Transwell chamber (400 nm size) with a layer of keratinocytes seeded previously at the bottom. Forty-eight hours after seeding the transwell chamber, switched GFP-positive cells were counted per condition. Cells were incubated for an additional 2 weeks and scored for GFP-positive switched keratinocytes in the bottom well.
Sigma LOPAC Library Small-Molecule Screen
For the small-molecule screen, we wanted to identify enhancers of switching efficiency, which is a readout of melanoma/keratinocyte communication, in cocultures, using the in vitro switch assay outlined above. To do this, human melanoma/keratinocyte cocultures (HaCaT and A375 cells) were seeded in 96-well plates at 1:3 ratios on day 0. Twenty-four hours after seeding, cocultures were treated with 10 μmol/L chemicals from the Sigma LOPAC 1280 library. All chemicals were prepared from 10 mmol/L stocks and diluted in a cell growth medium. Eight DMSO-only wells were included in each plate to quantify basal switching efficiency. Cocultures were incubated for an additional 48 hours after chemical addition, and switching efficiency was quantified 3 days after seeding and 48 hours after adding chemicals. Each well was manually screened and the number of GFP-positive switched keratinocytes was counted. Fold change was calculated by dividing the number of switched keratinocytes per well to the average number of switched keratinocytes per well in DMSO controls. After the identification of positive hits, a majority of which were modulators of the GABA-A signaling pathway, a second round of validation was performed in a 6-well format, using bona fide GABA-A receptor agonists such as GABA (Sigma #A2129) and muscimol (Sigma #M1523), as well as GABA-A receptor antagonists such as bicuculline methbromide (Sigma #B7561) and picrotoxin (Sigma #P1675), followed by in vivo validation in zebrafish embryos.
Tetanus Toxin Overexpression in Melanoma Cells
A plasmid encoding the tetanus toxin light chain was obtained from Addgene (#166603, RRID:Addgene_166603) and cloned into a lentiviral backbone with a blasticidin-resistance cassette. Empty vector or tetanus toxin–encoding plasmids were electroporated into melanoma-Cre lines and selected with blasticidin (10 μg/mL) for 10 days. Following this, the control vector or tetanus toxin light chain–overexpressing melanoma-Cre lines were cocultured with the previously mentioned loxP reporter keratinocyte lines and switching efficiency was measured as described above. Tetanus toxin overexpression was verified using qPCR, and proliferation assays were performed to account for any toxicity associated with tetanus toxin expression in melanoma cells, which showed no differences in melanoma cell proliferation upon tetanus toxin light chain expression.
RNA-seq of Keratinocytes
The HaCaT keratinocyte cell line expressing the switch construct was grown either in monoculture or in coculture with A375 melanoma cells expressing Cre in a 1:3 keratinocyte:melanoma ratio for 21 days in complete DMEM. Cocultures were split 1× per week upon reaching confluence. Monoculture keratinocytes (dsRED-positive) and coculture keratinocytes, both switched and nonswitched (GFP-positive and dsRED-positive), were then isolated using FACS and plated on 6-well dishes for recovery following the FACS procedure. Total RNA was extracted from the FACS-isolated keratinocytes after recovery using the Quick-RNA Miniprep Kit (Zymo), and purified RNA was delivered to GENEWIZ for mRNA preparation with the TruSeq RNA V2 kit (Illumina) and 150-bp paired-end sequencing on the Illumina HiSeq2500. Quality control of the raw reads from RNA-seq fastq files was performed using FASTQC (Babraham Bioinformatics), and trimming was performed using TRIMMOMATIC (115, 116). Trimmed reads were mapped to the human (hg38) genome using STAR (117). Gene counts of aligned reads were performed using the feature counts algorithm (118), followed by differential gene expression analysis using DeSeq2 (119). Pathway and gene ontology analysis was performed using GSEA (120). All data have been deposited in the Gene Expression Omnibus (GEO) database (GSE236806).
Chromatin Immunoprecipitation Sequencing Data Analysis
Existing chromatin immunoprecipitation sequencing (ChIP-seq) datasets for MYCN from neuroblastoma cell lines (NB1643, Kelly, SHEP21) with previously analyzed data were downloaded from ChIP-Atlas (http://chip-atlas.org/peak_browser), and visualization was performed using IGV.
3D Skin Reconstructs
3D skin reconstructs were generated using the protocol developed by the Herlyn lab (63). Briefly, an acellular collagen layer was created and incubated for 30 minutes at room temperature (RT). Following this, 7.5 × 104 fibroblasts (BJ fibroblast cell line) were seeded on top of the acellular layer and incubated at 37°C for 45 minutes, followed by the addition of DMEM containing 10% FBS. After incubation for 4 days, the inserts were washed and seeded with melanoma cells (A375) and keratinocytes (Ker-CT) at a ratio of 1:5 melanoma cells:keratinocytes and incubated for an additional 20 days with regular change in media every 48 hours. Control media or muscimol (100 μmol/L) containing media was added starting from day 10 to day 24 of cultures. On day 24, the 3D reconstructs were fixed in 4% formalin for 24 hours, sectioned, and stained with an antibody for BRAFV600E (Abcam, ab228461) using IHC. Melanoma cell area was quantified using ImageJ software.
Mouse
Mouse Ear Transplants
YUMM 1.1 cells expressing control-GFP or Gad1-GFP were grown for 48 hours. Tumor cells (5 × 104) resuspended in 20 μL PBS were injected subcutaneously on the dorsal side of the ear of C57BL/6J wild-type mice using 31G 6 mm BD Ultra-Fine insulin syringes. Mice were anesthetized with 2% isoflurane during tumor transplantation. Tumor cells expressing Gad1-GFP were injected into the right ear; control tumor cells expressing GFP only were injected into the contralateral ear. Mice were sacrificed at indicated time points after tumor transplantation. Ears were removed for downstream imaging analyses. Both female and male mice were used for studies. Mice were bred and maintained in the animal facility at MSKCC. Experiments were approved by and performed in compliance with the MSKCC IACUC. The tumor area was monitored over a period of 2 to 12 days after transplantation. The GFP-positive area was imaged on a Zeiss LSM880 confocal microscope and quantified using ImageJ software.
Immunostaining
Immunofluorescence Staining of Cultured Cells
HaCaT/A375 cocultures were grown on 35-mm glass-bottom dishes (Thermo Fisher #150682) for 48 hours. After 48 hours of incubation, growth media were aspirated from the dishes and cells were fixed with 4% PFA for 20 minutes (except for GABA staining where cells were fixed using 4% PFA + 0.25% glutaraldehyde) at RT and washed 3× with PBS. Following this, cells were permeabilized for 15 minutes in 0.1% Triton X-100 (diluted from 10% stock, Sigma #93443) in 1× PBS, followed by 3× washes with PBS. Blocking was performed using 10% normal goat serum (Thermo Fisher #50062Z) for 1 hour at RT, followed by overnight incubation with primary antibodies at 4°C. Cells were subsequently washed 3× with PBS and incubated with secondary antibodies diluted in blocking solution for 1.5 hours at RT, followed by 3× PBS washes. For fluorophore-conjugated secondary antibodies, samples were individually treated with the specific conjugated antibodies for 2 hours at RT after secondary antibody incubation, followed by 3× washes with PBS. Finally, cells were counterstained using Hoechst 33343 (Invitrogen #H3570) in PBS for 20 minutes, followed by 3× PBS washes. Fresh PBS was added after the final wash to the cells, and the cells were imaged on an LSM880 high-resolution confocal microscope with a 63× objective using AiryScan imaging. The following primary antibodies were used: rabbit anti-human GPHN (Thermo Fisher #PA529036, RRID AB_2546512); rabbit anti-human SOX10 (Thermo Fisher #PA5-84795, RRID AB_2791945); rabbit anti-human KRT17 conjugated to Alexa Fluor 546 (Santa Cruz Biotechnology #sc-393002 AF546, RRID AB_2893006); and rabbit anti-GABA (Sigma #A2052, RRID AB_477652). The following secondary antibodies were used: AlexaFluor 488 anti-mouse IgG (Cell Signaling Technology #4408S, RRID AB_10694704) and AlexaFluor 647 anti-rabbit IgG (Cell Signaling Technology #4414S, RRID AB_10693544).
Immunofluorescence Staining of Patient Samples
Malignant melanoma in situ samples and TMAs were obtained from the MSKCC TMA Database and US Biomax (ME208). Paraffin sections of all tumors and normal skin were deparaffinized, and antigen retrieval was performed using the heat-induced epitope retrieval method with 1× Antigen Retrieval solution (Invitrogen #00-4955-58) in a pressure cooker at 95°C for 10 minutes. Slides were washed 3× with PBS and blocked with 10% normal goat serum (Thermo Fisher #50062Z) for 2 hours at RT and incubated with primary antibodies overnight at 4°C. Slides were washed with PBS (3×), followed by incubation with secondary antibodies for 2 hours at RT and 3× washes with PBS after secondary antibody incubation. For fluorophore-conjugated secondary antibodies, samples were individually treated with the specific conjugated antibodies for 2 hours at RT after secondary antibody incubation, followed by 3× washes with PBS. After the final PBS wash, slides were counterstained with Hoechst 33342 (Invitrogen #H3570) and mounted in ProLong Glass Antifade Mountant (Fisher Scientific #P36984). The following primary antibodies were used: mouse anti-human GAD1 (Santacruz #sc28376, RRID AB_627650), rabbit anti-human GPHN (Thermo Fisher #PA529036, RRID AB_2546512), mouse anti-human KRT14 (Abcam #ab7800, RRID AB_306091), and rabbit anti-human S100A6 conjugated to Alexa Fluor 647 (Abcam #ab204028). The following secondary antibodies were used: AlexaFluor 488 anti-mouse IgG (Cell Signaling Technology #4408S, RRID AB_10694704) and Alexa Fluor 555 anti-rabbit IgG (Cell Signaling Technology #4413S, AB_10694110). For calculating the GAD1 immunofluorescence score, we probed the intensity (score 0–2) and the coverage (score 1–4) of each individual sample from melanoma patients and normal skin and assigned the immunofluorescence score by multiplying the intensity score with the coverage score. All sections were imaged on an LSM880 high-resolution confocal microscope with a 63× objective using AiryScan imaging.
Proliferation/Invasion Assays
A375 Phospho-Histone H3 Immunostaining
For proliferation studies, A375 melanoma cells (Azurite fluorophore-positive) were cocultured either with switched (GFP-positive) or parental (dsRED-positive) keratinocytes in low-serum (2%) media, in which melanoma cells and keratinocytes were plated in a 1:5 ratio for 48 hours. Following 48 hours of incubation, cells were fixed in 4% PFA and stained with a phospho-Histone H3 primary antibody (1:1,000; Millipore #05-806) overnight at 4°C. Cells were counterstained with AlexaFluor 647 anti-mouse IgG (Cell Signaling Technology #4410), and more than 10 images per condition were acquired in a Zeiss AxioObserver fluorescence microscope. The number of mitotic cells was quantified by calculating double-positive cells (Azurite+, AlexaFluor647+) as a fraction of the total number of Azurite+ cells in each field.
CellTiter-Glo Assay in Melanoma Cells Treated with Keratinocyte Conditioned Media
For conditioned media experiments, switched and parental keratinocytes were grown in 2% serum-containing media for 24 hours and collected in 50 mL Falcon tubes followed by centrifugation at 500 × g for 5 minutes to remove any dead cells or debris. To measure cell proliferation, monocultures of A375 Azurite-positive cells were plated at a density of 5,000 cells per well in a 96-well plate in 100 μL of conditioned media from switched or parental keratinocytes. After 48 hours of conditioned media treatment, CellTiter-Glo reagent (Promega #G7570) was added to the cells as per the manufacturer's instructions and luminescence was read using a BioTek Synergy 96-well plate reader. All values were normalized to the A375 cells treated with parental keratinocytes conditioned media, done in quadruplicate for all cell lines. For the antagonist treatments, the same protocol as above was used except cells were treated with the following antagonists, which were directly added to the conditioned media: AZD4547 (FGF receptor, Fisher Scientific #NC0660421, 100 ng/mL), DMH1 (BMP receptor, Sigma #D8946, 0.5 μmol/L), or EC330 (LIF receptor, Fisher #501871773, 30 nmol/L).
Cyquant Proliferation Assay in Monocultures and Cocultures
For proliferation experiments upon treatment with GABA/picrotoxin, 2,000 or 5,000 cells/well of melanoma cells were plated in 96-well plates. Twenty-four hours after plating, GABA/picrotoxin was added to the media followed by a 48-hour incubation. Proliferation was measured using the Cyquant Cell Proliferation Assay as per the manufacturer's instructions, and fluorescence was read using a BioTek Synergy 96-well plate reader. All values were normalized to the control conditions.
Invasion Assay
For invasion assays, 20,000 YUMM 1.1 cells/well expressing control-GFP or Gad1-GFP were plated on Biocoat Matrigel Invasion chambers (Corning #354481). After 48 hours, cells were fixed and migrated cells in the bottom chamber were stained with Hoechst 33343, 1:2,000 (Invitrogen #H3570), for 20 minutes and 6 to 8 fields per condition were imaged. The number of migrated cells was quantified using ImageJ.
Transmission Electron Microscopy
Melanoma/keratinocyte cocultures were incubated for 48 hours and fixed using 4% paraformaldehyde, 2.5% glutaraldehyde, and 0.002% picric acid in 0.1 mol/L sodium cacodylate buffer, pH 7.3, for transmission electron microscopy (12,000–15,000×). Imaging was performed on the JEOL JSM 1400, operated at 100 kV. Images were captured on a Veleta 2K × 2K CCD camera (EM-SIS).
Electrophysiology Studies
MEA
For extracellular recordings, melanoma cell and keratinocyte cocultures were seeded onto complementary metal oxide semiconductor MEA probes (3Brain AG) coated with poly d-lysine (Sigma #P6407) overnight in a 37°C incubator. A 100-μL droplet of cell suspension was placed directly on the probe recording area. After 1 hour of incubation at 37°C, 1.5 mL of medium was added to the probe and replaced daily. Recordings were performed 48 hours days after plating. Two minutes of spontaneous activity was sampled from the 4,096 electrodes in each probe using the BioCAM X system. Recordings were analyzed using the BrainWave 4 software. Spike detection was performed using a Precise Timing Spike Detection (121) algorithm by applying a threshold of 8 SDs to each channel trace. Average firing rates were calculated from a subset containing the 64 most active electrodes in each recording.
Calcium Activity Assay
Calcium activity was measured using the Cal-520 (Abcam ab171868) and Rhod-4 (Abcam ab112156) dyes following the manufacturer's instructions. Briefly, melanoma and keratinocyte monocultures and cocultures were plated in black-bottom, 96-well plates (Fisher #07-200-566) and incubated for 48 hours. Following incubation, cells were treated with Cal-520 or Rhod-4 dyes at 37°C, followed by RT incubation. For Cal-520 measurements, picrotoxin (100 μmol/L, Sigma #P1675), muscimol (100 μmol/L, Sigma #M1523), and SB205384 (10 μmol/L, Sigma #7936) were added to monocultures and cocultures following RT incubation. Readings were measured using a BioTek Synergy 96-well plate reader at 488 and 540 nm wavelengths, respectively.
Data Analysis
Analysis of Publicly Available GAD1 Expression Data
For GAD1 expression analysis, microarray expression data from Wistar melanoma cell lines were obtained from (122). The GAD1 expression value for each melanoma cell line (radial growth phase, vertical growth phase, and metastatic) and normal cell lines (melanocytes, keratinocytes, and fibroblasts) was plotted, and GAD1 positivity was assigned based on default analysis parameters used in the dataset.
Data and Software Availability
RNA-seq data have been deposited to the GEO database with the accession number GSE236806. All raw data files will be made available upon request. All transgenic zebrafish lines are available upon request from the authors or via the ZIRC zebrafish stock center (https://zebrafish.org/home/guide.php). Plasmids generated in this study will be made available upon request.
Statistical Analysis
All analysis and visualization of data and statistical tests were performed using GraphPad Prism 9. Detailed information about individual statistical tests, sample size, and P values is indicated in the figure legends. All analysis was performed on data pooled from 3 to 6 biological replicates per experiment.
Supplementary Material
LOPAC Screen results in melanoma keratinocyte co-cultures
RNA-seq and GSEA results in switched vs non-switched keratinocytes
Combined supplementary figures and figure legends. Supplementary Fig. 1: Switching requires direct contact between nascent melanoma cells and keratinocytes in vivo. Supplementary Fig. 2: Switching requires direct contact between melanoma cells and keratinocytes in vitro. Supplementary Fig. 3: Switching does not involve cell fusion between keratinocytes and melanoma cells. Supplementary Fig. 4: Exosome-like vesicles are transferred from melanoma cells to keratinocytes. Supplementary Fig. 5: GABAergic signaling is active in skin and melanoma cells. Supplementary Fig. 6: Melanoma cells express GAD1 and produce GABA. Supplementary Fig. 7: Disruption of GABA signaling blocks melanoma/keratinocyte communication. Supplementary Fig. 8: Specialized cell-cell junction signatures are enriched in melanoma cells and keratinocytes. Supplementary Fig. 9: Melanoma vesicles are present near specialized GABAergic cell-cell junctions in co-cultures. Supplementary Fig. 10: gad activation correlates with higher tumor burden in melanoma. Supplementary Fig. 11: GABA producing enzymes are expressed in zebrafish melanoma cells. Supplementary Fig. 12: Disruption of GABA synthesis blocks tumor initiation in melanoma. Supplementary Fig. 13: GAD expression/GABA treatment is pro-tumorigenic in a non cellautonomous manner. Supplementary Fig. 14: Switched keratinocytes are pro-tumorigenic in melanoma. Supplementary Fig. 15: LIF expression in keratinocytes drives melanoma cell proliferation. Supplementary Fig. 16: Zebrafish melanoma scRNAseq identifies GABA and LIF associated components in the TME. Supplementary Fig. 17: MYCN drives LIF expression in keratinocytes.
3D rendering of melanoma/keratinocyte contacts in zebrafish embryos
3D rendering of melanoma/switched keratinocyte contacts and stained vesicles in human melanoma/keratinocyte co-cultures
3D rendering of gephyrin positive clusters in melanoma/keratinocyte co-cultures
Representative trace videos of spike activity in melanoma/keratinocyte co-cultures +/- GABA signaling
Acknowledgments
The authors thank members of the White lab for helpful discussions and feedback on this project. We thank Jose A. Esteban for critical feedback on the manuscript. We also thank Ling Li and Meenhard Herlyn for technical help with 3D skin reconstruction experiments. We thank Lee-Cohen Gould and Juan Jimenez for their help with transmission electron microscopy and Chenura Jayewickreme for help with the optimization of immunofluorescence protocols. We also thank the MSKCC Molecular Cytology Core for imaging assistance, the Flow Cytometry Core for cell sorting assistance, and the Molecular Cytogenetics Core for karyotyping assistance. All schematic figures were created using BioRender.com with a paid BioRender license. This work was funded with support from the NIH/NCI Cancer Center Support Grant P30 CA008748, the Melanoma Research Alliance, The Debra and Leon Black Family Foundation, NIH Research Program Grants R01CA229215 and R01CA238317, the NIH Director's New Innovator Award DP2CA186572, the Pershing Square Sohn Foundation, The Mark Foundation, The Alan and Sandra Gerry Metastasis Research Initiative at MSKCC, The Harry J. Lloyd Foundation, Consano, the Starr Cancer Consortium, and the American Cancer Society RSG-19-024-01-DDC (all to R.M. White); the Melanoma Research Foundation and an MSKCC TROT fellowship (both to S. Suresh); a Marie-Josée Kravis Women in Science Endeavor Postdoctoral Fellowship and the Experimental Immuno-Oncology Scholars Fellowship (to M. Tagore); and The Cancer Cell Map Initiative NIH/NCI U54 CA209891 (to T. Ideker).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
M. Tagore reports a Marie-Josée Kravis Women in Science Endeavor Postdoctoral Fellowship, MSKCC, and the Center for Experimental Immuno-Oncology Scholars Fellowship, MSKCC, during the conduct of the study. S. Melendez reports other support from the MSKCC SCORE Program and the McNulty Scholars Program during the conduct of the study. T.J. Hollmann reports that this work was completed while he was an employee of MSKCC; his current affiliation is Bristol Myers Squib, Princeton, NJ. L. Studer reports grants from the National Institute of Neurological Disorders and Stroke/NIH and the Melanoma Research Association during the conduct of the study, as well as grants and personal fees from BlueRock Therapeutics and Dacapo Brainsciences outside the submitted work. R.M. White reports grants from the NIH/NCI (Cancer Center Support Grant P30 CA008748), the Melanoma Research Alliance, The Debra and Leon Black Family Foundation, the NIH (Research Program Grants R01CA229215 and R01CA238317, as well as the NIH Director's New Innovator Award DP2CA186572), the Pershing Square Sohn Foundation, The Mark Foundation, The Alan and Sandra Gerry Metastasis Research Initiative at MSKCC, The Harry J. Lloyd Foundation, Consano, and the Starr Cancer Consortium during the conduct of the study, as well as personal fees from N-of-One, a subsidiary of Qiagen, outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
M. Tagore: Conceptualization, resources, data curation, software, formal analysis, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. E. Hergenreder: Investigation, visualization, methodology, writing–review and editing. S.C. Perlee: Investigation, methodology, writing–review and editing. N.M. Cruz: Investigation, methodology. L. Menocal: Investigation, methodology. S. Suresh: Investigation, visualization, methodology, writing–review and editing. E. Chan: Visualization, methodology. M. Baron: Investigation, visualization, methodology, writing–review and editing. S. Melendez: Investigation, methodology. A. Dave: Methodology. W.K. Chatila: Visualization. J. Nsengimana: Visualization. R.P. Koche: Visualization. T.J. Hollmann: Investigation. T. Ideker: Project administration. L. Studer: Project administration. A. Schietinger: Project administration. R.M. White: Conceptualization, resources, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 2015;161:1681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baggiolini A, Callahan SJ, Montal E, Weiss JM, Trieu T, Tagore MM, et al. Developmental chromatin programs determine oncogenic competence in melanoma. Science 2021;373:eabc1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Golan T, Messer AR, Amitai-Lange A, Melamed Z, Ohana R, Bell RE, et al. Interactions of melanoma cells with distal keratinocytes trigger metastasis via notch signaling inhibition of MITF. Mol Cell 2015;59:664–76. [DOI] [PubMed] [Google Scholar]
- 4. Kim IS, Heilmann S, Kansler ER, Zhang Y, Zimmer M, Ratnakumar K, et al. Microenvironment-derived factors driving metastatic plasticity in melanoma. Nat Commun 2017;8:14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Halaban R, Rubin JS, Funasaka Y, Cobb M, Boulton T, Faletto D, et al. Met and hepatocyte growth factor/scatter factor signal transduction in normal melanocytes and melanoma cells. Oncogene 1992;7:2195–206. [PubMed] [Google Scholar]
- 6. Kunisada T, Yamazaki H, Hirobe T, Kamei S, Omoteno M, Tagaya H, et al. Keratinocyte expression of transgenic hepatocyte growth factor affects melanocyte development, leading to dermal melanocytosis. Mech Dev 2000;94:67–78. [DOI] [PubMed] [Google Scholar]
- 7. Brenner M, Degitz K, Besch R, Berking C. Differential expression of melanoma-associated growth factors in keratinocytes and fibroblasts by ultraviolet A and ultraviolet B radiation. Br J Dermatol 2005;153:733–9. [DOI] [PubMed] [Google Scholar]
- 8. Chung H, Suh EK, Han IO, Oh ES. Keratinocyte-derived laminin-332 promotes adhesion and migration in melanocytes and melanoma. J Biol Chem 2011;286:13438–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ableser MJ, Penuela S, Lee J, Shao Q, Laird DW. Connexin43 reduces melanoma growth within a keratinocyte microenvironment and during tumorigenesis in vivo. J Biol Chem 2014;289:1592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mescher M, Jeong P, Knapp SK, Rübsam M, Saynisch M, Kranen M, et al. The epidermal polarity protein Par3 is a non-cell autonomous suppressor of malignant melanoma. J Exp Med 2017;214:339–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shih IM, Elder DE, Hsu MY, Herlyn M. Regulation of Mel-CAM/MUC18 expression on melanocytes of different stages of tumor progression by normal keratinocytes. Am J Pathol 1994;145:837–45. [PMC free article] [PubMed] [Google Scholar]
- 12. Hsu M, Andl T, Li G, Meinkoth JL, Herlyn M. Cadherin repertoire determines partner-specific gap junctional communication during melanoma progression. J Cell Sci 2000;113( Pt 9):1535–42. [DOI] [PubMed] [Google Scholar]
- 13. Hsu MY, Meier FE, Nesbit M, Hsu JY, Van Belle P, Elder DE, et al. E-cadherin expression in melanoma cells restores keratinocyte-mediated growth control and down-regulates expression of invasion-related adhesion receptors. Am J Pathol 2000;156:1515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ando H, Niki Y, Ito M, Akiyama K, Matsui MS, Yarosh DB, et al. Melanosomes are transferred from melanocytes to keratinocytes through the processes of packaging, release, uptake, and dispersion. J Invest Dermatol 2012;132:1222–9. [DOI] [PubMed] [Google Scholar]
- 15. Chen H, Weng QY, Fisher DE. UV signaling pathways within the skin. J Invest Dermatol 2014;134:2080–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fitzpatrick TB, Breathnach AS. [The epidermal melanin unit system]. Dermatol Wochenschr 1963;147:481–9. [PubMed] [Google Scholar]
- 17. Wäster P, Eriksson I, Vainikka L, Rosdahl I, Öllinger K. Extracellular vesicles are transferred from melanocytes to keratinocytes after UVA irradiation. Sci Rep 2016;6:27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015;161:1046–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ridder K, Keller S, Dams M, Rupp AK, Schlaudraff J, Del Turco D, et al. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol 2014;12:e1001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 2011;471:513–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Jong OG, Murphy DE, Mäger I, Willms E, Garcia-Guerra A, Gitz-Francois JJ, et al. A CRISPR-Cas9-based reporter system for single-cell detection of extracellular vesicle-mediated functional transfer of RNA. Nat Commun 2020;11:1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008;319:1244–7. [DOI] [PubMed] [Google Scholar]
- 23. Collot M, Ashokkumar P, Anton H, Boutant E, Faklaris O, Galli T, et al. MemBright: a family of fluorescent membrane probes for advanced cellular imaging and neuroscience. Cell Chem Biol 2019;26:600–14. [DOI] [PubMed] [Google Scholar]
- 24. Wang J, Panáková D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 2011;138:3421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharrock AV, Mulligan TS, Hall KR, Williams EM, White DT, Zhang L, et al. NTR 2.0: a rationally engineered prodrug-converting enzyme with substantially enhanced efficacy for targeted cell ablation. Nat Methods 2022;19:205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnston GAR. Muscimol as an ionotropic GABA receptor agonist. Neurochem Res 2014;39:1942–7. [DOI] [PubMed] [Google Scholar]
- 27. Asada H, Kawamura Y, Maruyama K, Kume H, Ding RG, Kanbara N, et al. Cleft palate and decreased brain gamma-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci U S A 1997;94:6496–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lazarus MS, Krishnan K, Huang ZJ. GAD67 deficiency in parvalbumin interneurons produces deficits in inhibitory transmission and network disinhibition in mouse prefrontal cortex. Cereb Cortex 2015;25:1290–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farrant M, Nusser Z. Variations on an inhibitory theme: phasic and tonic activation of GABA(A) receptors. Nat Rev Neurosci 2005;6:215–29. [DOI] [PubMed] [Google Scholar]
- 30. Denda M, Inoue K, Inomata S, Denda S. gamma-Aminobutyric acid (A) receptor agonists accelerate cutaneous barrier recovery and prevent epidermal hyperplasia induced by barrier disruption. J Invest Dermatol 2002;119:1041–7. [DOI] [PubMed] [Google Scholar]
- 31. Allen JR, Skeath JB, Johnson SL. Maintenance of melanocyte stem cell quiescence by GABA-A signaling in larval zebrafish. Genetics 2019;213:555–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pomeranz Krummel DA, Nasti TH, Kaluzova M, Kallay L, Bhattacharya D, Melms JC, et al. Melanoma cell intrinsic GABA receptor enhancement potentiates radiation and immune checkpoint inhibitor response by promoting direct and T cell-mediated antitumor activity. Int J Radiat Oncol Biol Phys 2021;109:1040–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ito K, Tanaka K, Nishibe Y, Hasegawa J, Ueno H. GABA-synthesizing enzyme, GAD67, from dermal fibroblasts: evidence for a new skin function. Biochim Biophys Acta 2007;1770:291–6. [DOI] [PubMed] [Google Scholar]
- 34. Kins S, Betz H, Kirsch J. Collybistin, a newly identified brain-specific GEF, induces submembrane clustering of gephyrin. Nat Neurosci 2000;3:22–9. [DOI] [PubMed] [Google Scholar]
- 35. Harvey K, Duguid IC, Alldred MJ, Beatty SE, Ward H, Keep NH, et al. The GDP-GTP exchange factor collybistin: an essential determinant of neuronal gephyrin clustering. J Neurosci 2004;24:5816–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Papadopoulos T, Korte M, Eulenburg V, Kubota H, Retiounskaia M, Harvey RJ, et al. Impaired GABAergic transmission and altered hippocampal synaptic plasticity in collybistin-deficient mice. EMBO J 2007;26:3888–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. George S, Chiou TT, Kanamalla K, De Blas AL. Recruitment of plasma membrane GABA-A receptors by submembranous gephyrin/collybistin clusters. Cell Mol Neurobiol 2022;42:1585–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hsu MY, Elder DE, Herlyn M. Melanoma: the Wistar melanoma (WM) cell lines. In: Masters JRW, Palsson B, editors. Human cell culture. Netherlands: Springer; 2002. p. 259–74. [Google Scholar]
- 39. Callahan SJ, Mica Y, Studer L. Feeder-free derivation of melanocytes from human pluripotent stem cells. J Vis Exp 2016;109:e53806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lefler Y, Yarom Y, Uusisaari MY. Cerebellar inhibitory input to the inferior olive decreases electrical coupling and blocks subthreshold oscillations. Neuron 2014;81:1389–400. [DOI] [PubMed] [Google Scholar]
- 41. Marlin JJ, Carter AG. GABA-A receptor inhibition of local calcium signaling in spines and dendrites. J Neurosci 2014;34:15898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Belote RL, Simon SM. Ca2+ transients in melanocyte dendrites and dendritic spine-like structures evoked by cell-to-cell signaling. J Cell Biol 2020;219:e201902014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Essrich C, Lorez M, Benson JA, Fritschy JM, Lüscher B. Postsynaptic clustering of major GABAA receptor subtypes requires the gamma 2 subunit and gephyrin. Nat Neurosci 1998;1:563–71. [DOI] [PubMed] [Google Scholar]
- 44. Danglot L, Triller A, Bessis A. Association of gephyrin with synaptic and extrasynaptic GABAA receptors varies during development in cultured hippocampal neurons. Mol Cell Neurosci 2003;23:264–78. [DOI] [PubMed] [Google Scholar]
- 45. Burette A, Collman F, Micheva KD, Smith SJ, Weinberg RJ. Knowing a synapse when you see one. Front Neuroanat 2015;9:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gray EG. Axo-somatic and axo-dendritic synapses of the cerebral cortex: an electron microscope study. J Anat 1959;93:420–33. [PMC free article] [PubMed] [Google Scholar]
- 47. Odland GF. The fine structure of the interrelationship of cells in the human epidermis. J Biophys Biochem Cytol 1958;4:529–38. [PMC free article] [PubMed] [Google Scholar]
- 48. Lock JT, Parker I, Smith IF. A comparison of fluorescent Ca2+ indicators for imaging local Ca2+ signals in cultured cells. Cell Calcium 2015;58:638–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang SJ, Jackson MB. GABA-activated chloride channels in secretory nerve endings. Science 1993;259:531–4. [DOI] [PubMed] [Google Scholar]
- 50. Meadows HJ, Kumar CS, Pritchett DB, Blackburn TP, Benham CD. SB-205384: a GABA(A) receptor modulator with novel mechanism of action that shows subunit selectivity. Br J Pharmacol 1998;123:1253–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heimlich G, Cidlowski JA. Selective role of intracellular chloride in the regulation of the intrinsic but not extrinsic pathway of apoptosis in Jurkat T-cells. J Biol Chem 2006;281:2232–41. [DOI] [PubMed] [Google Scholar]
- 52. Hull C, von Gersdorff H. Fast endocytosis is inhibited by GABA-mediated chloride influx at a presynaptic terminal. Neuron 2004;44:469–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McMahon HT, Ushkaryov YA, Edelmann L, Link E, Binz T, Niemann H, et al. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature 1993;364:346–9. [DOI] [PubMed] [Google Scholar]
- 54. Link E, Edelmann L, Chou JH, Binz T, Yamasaki S, Eisel U, et al. Tetanus toxin action: inhibition of neurotransmitter release linked to synaptobrevin proteolysis. Biochem Biophys Res Commun 1992;189:1017–23. [DOI] [PubMed] [Google Scholar]
- 55. Das V, Nal B, Dujeancourt A, Thoulouze MI, Galli T, Roux P, et al. Activation-induced polarized recycling targets T cell antigen receptors to the immunological synapse; involvement of SNARE complexes. Immunity 2004;20:577–88. [DOI] [PubMed] [Google Scholar]
- 56. Kang JQ, Shen W, Macdonald RL. The GABRG2 mutation, Q351X, associated with generalized epilepsy with febrile seizures plus, has both loss of function and dominant-negative suppression. J Neurosci 2009;29:2845–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brickley SG, Mody I. Extrasynaptic GABA(A) receptors: their function in the CNS and implications for disease. Neuron 2012;73:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mittelbrunn M, Vicente Manzanares M, Sánchez-Madrid F. Organizing polarized delivery of exosomes at synapses. Traffic 2015;16:327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2011;2:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Céspedes PF, Jainarayanan A, Fernández-Messina L, Valvo S, Saliba DG, Kurz E, et al. T-cell trans-synaptic vesicles are distinct and carry greater effector content than constitutive extracellular vesicles. Nat Commun 2022;13:3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Baron M, Tagore M, Hunter MV, Kim IS, Moncada R, Yan Y, et al. The stress-like cancer cell state is a consistent component of tumorigenesis. Cell Syst 2020;11:536–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Callahan SJ, Tepan S, Zhang YM, Lindsay H, Burger A, Campbell NR, et al. Cancer modeling by transgene electroporation in adult zebrafish (TEAZ). Dis Model Mech 2018;11:dmm034561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li L, Fukunaga-Kalabis M, Herlyn M. The three-dimensional human skin reconstruct model: a tool to study normal skin and melanoma progression. J Vis Exp 2011;54:2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kaur A, Webster MR, Marchbank K, Behera R, Ndoye A, Kugel CH 3rd, et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature 2016;532:250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hirobe T. Role of keratinocyte-derived factors involved in regulating the proliferation and differentiation of mammalian epidermal melanocytes. Pigment Cell Res 2005;18:2–12. [DOI] [PubMed] [Google Scholar]
- 66. Halaban R. Signal transduction in normal and malignant melanocytes. Pigment Cell Res 1994;7:89–95. [DOI] [PubMed] [Google Scholar]
- 67. Ghassemi S, Vejdovszky K, Sahin E, Ratzinger L, Schelch K, Mohr T, et al. FGF5 is expressed in melanoma and enhances malignancy in vitro and in vivo. Oncotarget 2017;8:87750–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Halaban R, Langdon R, Birchall N, Cuono C, Baird A, Scott G, et al. Basic fibroblast growth factor from human keratinocytes is a natural mitogen for melanocytes. J Cell Biol 1988;107:1611–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Venkatesan AM, Vyas R, Gramann AK, Dresser K, Gujja S, Bhatnagar S, et al. Ligand-activated BMP signaling inhibits cell differentiation and death to promote melanoma. J Clin Invest 2018;128:294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kuphal S, Wallner S, Bosserhoff AK. Impact of LIF (leukemia inhibitory factor) expression in malignant melanoma. Exp Mol Pathol 2013;95:156–65. [DOI] [PubMed] [Google Scholar]
- 71. Guo H, Cheng Y, Martinka M, McElwee K. High LIFr expression stimulates melanoma cell migration and is associated with unfavorable prognosis in melanoma. Oncotarget 2015;6:25484–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Blesch A, Uy HS, Grill RJ, Cheng JG, Patterson PH, Tuszynski MH. Leukemia inhibitory factor augments neurotrophin expression and corticospinal axon growth after adult CNS injury. J Neurosci 1999;19:3556–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li M, Sendtner M, Smith A. Essential function of LIF receptor in motor neurons. Nature 1995;378:724–7. [DOI] [PubMed] [Google Scholar]
- 74. Cotterman R, Knoepfler PS. N-Myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b. PLoS One 2009;4:e5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mill P, Mo R, Hu MC, Dagnino L, Rosenblum ND, Hui CC. Shh controls epithelial proliferation via independent pathways that converge on N-Myc. Dev Cell 2005;9:293–303. [DOI] [PubMed] [Google Scholar]
- 76. Silva-Vargas V, Lo Celso C, Giangreco A, Ofstad T, Prowse DM, Braun KM, et al. Beta-catenin and Hedgehog signal strength can specify number and location of hair follicles in adult epidermis without recruitment of bulge stem cells. Dev Cell 2005;9:121–31. [DOI] [PubMed] [Google Scholar]
- 77. Haass NK, Herlyn M. Normal human melanocyte homeostasis as a paradigm for understanding melanoma. J Investig Dermatol Symp Proc 2005;10:153–63. [DOI] [PubMed] [Google Scholar]
- 78. Lee JT, Herlyn M. Microenvironmental influences in melanoma progression. J Cell Biochem 2007;101:862–72. [DOI] [PubMed] [Google Scholar]
- 79. Li G, Satyamoorthy K, Herlyn M. Dynamics of cell interactions and communications during melanoma development. Crit Rev Oral Biol Med 2002;13:62–70. [DOI] [PubMed] [Google Scholar]
- 80. Barria A. Dangerous liaisons as tumour cells form synapses with neurons. Nature 2019;573:499–501. [DOI] [PubMed] [Google Scholar]
- 81. Rahrmann EP, Shorthouse D, Jassim A, Hu LP, Ortiz M, Mahler-Araujo B, et al. The NALCN channel regulates metastasis and nonmalignant cell dissemination. Nat Genet 2022;54:1827–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ganesh K. Uncoupling metastasis from tumorigenesis. N Engl J Med 2023;388:657–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. McCaig CD, Song B, Rajnicek AM. Electrical dimensions in cell science. J Cell Sci 2009;122:4267–76. [DOI] [PubMed] [Google Scholar]
- 84. Chernet B, Levin M. Endogenous voltage potentials and the microenvironment: bioelectric signals that reveal, induce and normalize cancer. J Clin Exp Oncol 2013; Suppl 1:S1–002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bellono NW, Kammel LG, Zimmerman AL, Oancea E. UV light phototransduction activates transient receptor potential A1 ion channels in human melanocytes. Proc Natl Acad Sci U S A 2013;110:2383–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Watanabe M, Iwashita M, Ishii M, Kurachi Y, Kawakami A, Kondo S, et al. Spot pattern of leopard Danio is caused by mutation in the zebrafish connexin41.8 gene. EMBO Rep 2006;7:893–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Loewenstein WR, Kanno Y. Intercellular communication and the control of tissue growth: lack of communication between cancer cells. Nature 1966;209:1248–9. [DOI] [PubMed] [Google Scholar]
- 88. Yamasaki H, Hollstein M, Mesnil M, Martel N, Aguelon AM. Selective lack of intercellular communication between transformed and nontransformed cells as a common property of chemical and oncogene transformation of BALB/c 3T3 cells. Cancer Res 1987;47:5658–64. [PubMed] [Google Scholar]
- 89. Enomoto T, Yamasaki H. Lack of intercellular communication between chemically transformed and surrounding nontransformed BALB/c 3T3 cells. Cancer Res 1984;44:5200–3. [PubMed] [Google Scholar]
- 90. Venkataramani V, Tanev DI, Strahle C, Studier-Fischer A, Fankhauser L, Kessler T, et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 2019;573:532–8. [DOI] [PubMed] [Google Scholar]
- 91. Venkatesh HS, Morishita W, Geraghty AC, Silverbush D, Gillespie SM, Arzt M, et al. Electrical and synaptic integration of glioma into neural circuits. Nature 2019;573:539–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zeng Q, Michael IP, Zhang P, Saghafinia S, Knott G, Jiao W, et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature 2019;573:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Belote RL, Le D, Maynard A, Lang UE, Sinclair A, Lohman BK, et al. Human melanocyte development and melanoma dedifferentiation at single-cell resolution. Nat Cell Biol 2021;23:1035–47. [DOI] [PubMed] [Google Scholar]
- 94. Tian J, Chau C, Hales TG, Kaufman DL. GABA(A) receptors mediate inhibition of T cell responses. J Neuroimmunol 1999;96:21–8. [DOI] [PubMed] [Google Scholar]
- 95. Hung ME, Leonard JN. A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery. J Extracell Vesicles 2016;5:31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kanada M, Bachmann MH, Hardy JW, Frimannson DO, Bronsart L, Wang A, et al. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc Natl Acad Sci U S A 2015;112:E1433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213–28. [DOI] [PubMed] [Google Scholar]
- 98. Dror S, Sander L, Schwartz H, Sheinboim D, Barzilai A, Dishon Y, et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nat Cell Biol 2016;18:1006–17. [DOI] [PubMed] [Google Scholar]
- 99. Heath N, Osteikoetxea X, de Oliveria TM, Lázaro-Ibáñez E, Shatnyeva O, Schindler C, et al. Endosomal escape enhancing compounds facilitate functional delivery of extracellular vesicle cargo. Nanomedicine 2019;14:2799–814. [DOI] [PubMed] [Google Scholar]
- 100. Sonawane ND, Szoka FC Jr, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem 2003;278:44826–31. [DOI] [PubMed] [Google Scholar]
- 101. White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2008;2:183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Rao MB, Didiano D, Patton JG. Neurotransmitter-regulated regeneration in the zebrafish retina. Stem Cell Rep 2017;8:831–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ablain J, Xu M, Rothschild H, Jordan RC, Mito JK, Daniels BH, et al. Human tumor genomics and zebrafish modeling identify loss as a driver of mucosal melanoma. Science 2018;362:1055–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res 2019;47:W171–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Perez AR, Pritykin Y, Vidigal JA, Chhangawala S, Zamparo L, Leslie CS, et al. GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol 2017;35:347–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol 2005;15:249–54. [DOI] [PubMed] [Google Scholar]
- 107. Gong Z, Ju B, Wang X, He J, Wan H, Sudha PM, et al. Green fluorescent protein expression in germ-line transmitted transgenic zebrafish under a stratified epithelial promoter from keratin8. Dev Dyn 2002;223:204–15. [DOI] [PubMed] [Google Scholar]
- 108. Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 2011;138:169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 2007;236:3088–99. [DOI] [PubMed] [Google Scholar]
- 110. Yin L, Maddison LA, Li M, Kara N, LaFave MC, Varshney GK, et al. Multiplex conditional mutagenesis using transgenic expression of Cas9 and sgRNAs. Genetics 2015;200:431–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Heilmann S, Ratnakumar K, Langdon E, Kansler E, Kim I, Campbell NR, et al. A quantitative system for studying metastasis using transparent zebrafish. Cancer Res 2015;75:4272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Meeth K, Wang JX, Micevic G, Damsky W, Bosenberg MW. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res 2016;29:590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol 1988;106:761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ramirez RD, Herbert B-S, Vaughan MB, Zou Y, Gandia K, Morales CP, et al. Bypass of telomere-dependent replicative senescence (M1) upon overexpression of Cdk4 in normal human epithelial cells. Oncogene 2003;22:433–44. [DOI] [PubMed] [Google Scholar]
- 115. Zomer A, Steenbeek SC, Maynard C, van Rheenen J. Studying extracellular vesicle transfer by a Cre-loxP method. Nat Protoc 2016;11:87–101. [DOI] [PubMed] [Google Scholar]
- 116. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 119. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Maccione A, Gandolfo M, Massobrio P, Novellino A, Martinoia S, Chiappalone M. A novel algorithm for precise identification of spikes in extracellularly recorded neuronal signals. J Neurosci Methods 2009;177:241–9. [DOI] [PubMed] [Google Scholar]
- 122. Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res 2006;19:290–302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LOPAC Screen results in melanoma keratinocyte co-cultures
RNA-seq and GSEA results in switched vs non-switched keratinocytes
Combined supplementary figures and figure legends. Supplementary Fig. 1: Switching requires direct contact between nascent melanoma cells and keratinocytes in vivo. Supplementary Fig. 2: Switching requires direct contact between melanoma cells and keratinocytes in vitro. Supplementary Fig. 3: Switching does not involve cell fusion between keratinocytes and melanoma cells. Supplementary Fig. 4: Exosome-like vesicles are transferred from melanoma cells to keratinocytes. Supplementary Fig. 5: GABAergic signaling is active in skin and melanoma cells. Supplementary Fig. 6: Melanoma cells express GAD1 and produce GABA. Supplementary Fig. 7: Disruption of GABA signaling blocks melanoma/keratinocyte communication. Supplementary Fig. 8: Specialized cell-cell junction signatures are enriched in melanoma cells and keratinocytes. Supplementary Fig. 9: Melanoma vesicles are present near specialized GABAergic cell-cell junctions in co-cultures. Supplementary Fig. 10: gad activation correlates with higher tumor burden in melanoma. Supplementary Fig. 11: GABA producing enzymes are expressed in zebrafish melanoma cells. Supplementary Fig. 12: Disruption of GABA synthesis blocks tumor initiation in melanoma. Supplementary Fig. 13: GAD expression/GABA treatment is pro-tumorigenic in a non cellautonomous manner. Supplementary Fig. 14: Switched keratinocytes are pro-tumorigenic in melanoma. Supplementary Fig. 15: LIF expression in keratinocytes drives melanoma cell proliferation. Supplementary Fig. 16: Zebrafish melanoma scRNAseq identifies GABA and LIF associated components in the TME. Supplementary Fig. 17: MYCN drives LIF expression in keratinocytes.
3D rendering of melanoma/keratinocyte contacts in zebrafish embryos
3D rendering of melanoma/switched keratinocyte contacts and stained vesicles in human melanoma/keratinocyte co-cultures
3D rendering of gephyrin positive clusters in melanoma/keratinocyte co-cultures
Representative trace videos of spike activity in melanoma/keratinocyte co-cultures +/- GABA signaling
Data Availability Statement
RNA-seq data have been deposited to the GEO database with the accession number GSE236806. All raw data files will be made available upon request. All transgenic zebrafish lines are available upon request from the authors or via the ZIRC zebrafish stock center (https://zebrafish.org/home/guide.php). Plasmids generated in this study will be made available upon request.