Abstract
The conserved chromosomal passenger complex (CPC) consists of Ipl1Aurora-B, Sli15INCENP, Bir1Survivin, and Nbl1Borealin, and localizes at the kinetochore/centromere to correct kinetochore attachment errors and to prevent checkpoint silencing. After anaphase entry, the CPC moves from the kinetochore/centromere to the spindle. In budding yeast, CPC subunit Sli15 is phosphorylated by both cyclin-dependent kinase (CDK) and Ipl1 kinase. Following anaphase onset, activated Cdc14 phosphatase reverses Sli15 phosphorylation imposed by CDK to promote CPC translocation. Although abolished Sli15 phosphorylation imposed by Ipl1 also causes CPC translocation, the regulation of Ipl1-imposed Sli15 phosphorylation remains unclear. In addition to Sli15, Cdc14 also dephosphorylates Fin1, a regulatory subunit of protein phosphatase 1 (PP1), to enable kinetochore localization of Fin1-PP1. Here, we present evidence supporting the notion that kinetochore-localized Fin1-PP1 likely reverses Ipl1-imposed Sli15 phosphorylation to promote CPC translocation from the kinetochore/centromere to the spindle. Importantly, premature Fin1 kinetochore localization or phospho-deficient sli15 mutation causes checkpoint defects in response to tensionless attachments, resulting in chromosome missegregation. In addition, our data indicate that reversion of CDK- and Ipl1-imposed Sli15 phosphorylation shows an additive effect on CPC translocation. Together, these results reveal a previously unidentified pathway to regulate CPC translocation, which is important for accurate chromosome segregation.
INTRODUCTION
Before mitosis, microtubules (MTs) polymerize from spindle pole bodies for the capture of the kinetochores (KTs) of chromosomes. Bipolar attachment is established when sister KTs are attached by MTs from opposite spindle poles. This attachment generates tension, which is essential for sister chromatid segregation. Defective or tensionless KT attachments activate the spindle assembly checkpoint (SAC), which delays anaphase entry. The SAC consists of multiple proteins that congregate at the KT to monitor KT attachment; among them are Bub1, Bub3, Mad1, Mad2, and Mps1 kinase (Hoyt et al., 1991; Li and Murray, 1991; Hardwick et al., 1996). The activity of SAC proteins relies on a balance of phosphorylation and dephosphorylation at the KT (Sherwin and Wang, 2019). In budding yeast, Ipl1 kinase, the homologue of Aurora B in mammalian cells, together with Sli15INCENP, Bir1Survivin, and Nbl1Borealin, constitutes the chromosomal passenger complex (CPC), which localizes at the KT and centromere before anaphase entry to regulate KT–MT interaction and SAC activity (Wang et al., 2014).
Previous work from our lab showed that Ipl1 kinase prevents SAC silencing in the presence of tensionless attachments through the phosphorylation of KT protein Dam1 (Jin and Wang, 2013). Ipl1 and Dam1, along with centromere-associated protein Sgo1, make up the tension checkpoint, which not only prevents anaphase entry in response to tensionless attachments, but also destabilizes chromosome–MT attachment for error correction (Biggins et al., 2001; Indjeian et al., 2005; Jin and Wang, 2013; Jin et al., 2017). Dam1 phosphorylation by Ipl1 reduces the MT affinity of KTs, which destabilizes KT–MT attachments (Cheeseman et al., 2002; Tanaka et al., 2002; Tien et al., 2010; Jin et al., 2017). Thus, the subsequently detached KTs may activate the SAC to delay anaphase entry and allow for corrections (Pinsky et al., 2006b). Upon tension generation on chromosomes, Ipl1-dependent phosphorylation of Dam1 is reversed by protein phosphatase I (PP1), which strengthens KT–MT interactions to create more stable KT attachments (Cheeseman et al., 2002; Jin et al., 2017). Therefore, Ipl1-dependent phosphorylation of KT proteins regulates the stability of KT–MT interaction and SAC activity. Moreover, we have shown that KT recruitment of Fin1-PP1 removes SAC proteins Bub1/Bub3 from the KT to prevent SAC reactivation (Bokros et al., 2016; Bokros et al., 2021).
Extensive studies show several mechanisms for the association of CPC with KTs and centromeres in yeast and mammalian cells (Broad and DeLuca, 2020). One mechanism is that phosphorylation of histone H3 by Haspin kinase recruits CPC to centromeres via H3-Bir1Survivin interaction in both yeast and higher eukaryotic cells (Wang et al., 2010; Edgerton et al., 2016). Interestingly, Bir1-dependent CPC centromere localization is dispensable for error correction and checkpoint activity in budding yeast (Campbell and Desai, 2013). Moreover, H2A-associated Sgo1 recruits CPC to the centromere region through Sgo1-Nbl1 interaction (Kawashima et al., 2007; Yamagishi et al., 2010). In addition, the Sli15/Ipl1 complex in budding yeast directly binds to inner KT protein Ctf19 of the COMA complex; and disruption of this interaction causes a defect in chromosome biorientation (Fischbock-Halwachs et al., 2019; Garcia-Rodriguez et al., 2019). Therefore, multiple mechanisms ensure the KT/centromere (KT hereafter to simplify) localization of CPC. Before anaphase entry, Sli15 localizes at the KT and its phosphorylation by both cyclin-dependent kinase (CDK) and Ipl1 facilitates this localization. CDK phosphorylates Sli15 at six residues within its MT-binding domain, which prevents CPC from association with MTs; however, reversal of CDK-mediated phosphorylation during anaphase induces CPC dissociation from KTs and translocation to the spindle (Mackay et al., 1993; Kang et al., 2001; Pereira and Schiebel, 2003). Additionally, CDK-dependent Sli15 phosphorylation is required for cell-cycle arrest in response to loss of cohesion, suggesting the role of Sli15 phosphorylation in SAC function when tensionless KT attachment occurs (Mirchenko and Uhlmann, 2010). Ipl1-dependent phosphorylation of Sli15 was shown to prevent premature CPC spindle binding as well. Interestingly, the Ipl1 consensus sites on Sli15 also exist within Sli15’s MT-binding domain, suggesting that two distinct pathways regulate CPC translocation (Nakajima et al., 2011; Makrantoni et al., 2014).
After anaphase entry, cells change the balance of kinases/phosphatases to facilitate mitotic exit. In budding yeast, Cdc14 phosphatase inactivates CDK and dephosphorylates its substrates during anaphase. Before anaphase entry, nucleolar proteins Net1/Cfi1 and Tof2 retain Cdc14 within the nucleolus (Shou et al., 1999; Visintin et al., 1999; Waples et al., 2009). The dephosphorylation of Net1 by protein phosphatase 2A (PP2ACdc55) strengthens Net1-Cdc14 interaction to prevent release of Cdc14 (Queralt et al., 2006; Wang and Ng, 2006). The activation of the FEAR (Cdc14 early anaphase release) pathway promotes the release of a small amount of Cdc14 from the nucleolus to the nucleus (Stegmeier et al., 2002). Further activation of the mitotic exit network allows for Net1 phosphorylation and a more robust Cdc14 release for mitotic exit (Bardin et al., 2000; Mohl et al., 2009).
Cdc14-dependent dephosphorylation of S-phase CDK substrates is necessary for efficient spindle elongation and anaphase progression (Jin et al., 2008). One important substrate of Cdc14 phosphatase in this transition is Sli15. As mentioned above, Sli15 dephosphorylation by Cdc14 promotes CPC translocation from the KT to the spindle in anaphase (Pereira and Schiebel, 2003). This translocation serves two functions. First, it removes CPC from the KT region, thereby preventing Ipl1 from phosphorylating KT substrates. As a consequence, it decreases the phosphorylation of KT proteins imposed by Ipl1, which stabilizes KT–MT interaction and facilitates SAC silencing (Pinsky et al., 2006b; Jin and Wang, 2013; Jin et al., 2017). Second, CPC spindle localization in early anaphase enables spindle stabilization and elongation (Rozelle et al., 2011). Subsequently, CPC concentrates at the spindle midzone to assist in MT depolymerization just before mitotic exit (Buvelot et al., 2003).
CPC translocation is regulated by reversal of both CDK- and Ipl1-dependent Sli15 phosphorylation. After anaphase onset, Cdc14 reverses the Sli15 phosphorylation imposed by CDK, but it is unclear which phosphatase reverses Ipl1-imposed Sli15 phosphorylation. Previous data show that PP1 is responsible for the reversal of Ipl1-dependent phosphorylation (Pinsky et al., 2006a), and our lab has recently characterized the role of PP1 regulator Fin1 in cell-cycle progression. Fin1 is a CDK/Cdc14 substrate, and its phosphorylation by CDK prevents KT association of Fin1-PP1 because of the sequestration of phosphorylated Fin1 by Bmh1/Bmh2 (Loog and Morgan, 2005; Akiyoshi et al., 2009). Active Cdc14 dephosphorylates Fin1 to allow its release from Bmh1/Bmh2 for KT binding (Bremmer et al., 2012; Bokros et al., 2016). We further showed that Fin1 KT localization promotes dephosphorylation of KT protein Ndc80, which subsequently removes checkpoint proteins Bub1/Bub3 from the KT (Bokros et al., 2021). Untimely Ndc80 dephosphorylation contributes to checkpoint defects in response to tensionless syntelic attachment, indicating the need for proper Fin1-PP1 activity at the KT in faithful chromosome segregation.
In this study, we uncovered a new pathway that promotes CPC translocation through Fin1-PP1 dependent dephosphorylation of Sli15. It has been shown that at anaphase onset, activated Cdc14 dephosphorylates CDK substrate Sli15 to remove CPC from the KT for spindle translocation. In addition, Cdc14 dephosphorylates Fin1 to promote the KT localization of Fin1-PP1. We found that KT-localized Fin1-PP1 likely reverses Ipl1 kinase-imposed phosphorylation of Sli15, which also promotes CPC’s KT delocalization and spindle association in anaphase. In addition, either enforced KT localization of Fin1-PP1 or mutations of the CDK/Ipl1 phosphorylation sites in Sli15 causes premature removal of CPC from KTs. This leads to defective checkpoint response to tension defects as well as chromosome missegregation in the presence of syntelic attachment, wherein two sister KTs are attached by MTs from the same spindle pole. We further showed that the effects of Sli15 dephosphorylation by Cdc14 and Fin1-PP1 are additive. Therefore, this study reveals an additional, previously unknown layer of regulation for CPC translocation via Sli15 dephosphorylation by Fin1-PP1.
RESULTS
Fin1-PP1 partially reverses Sli15 phosphorylation
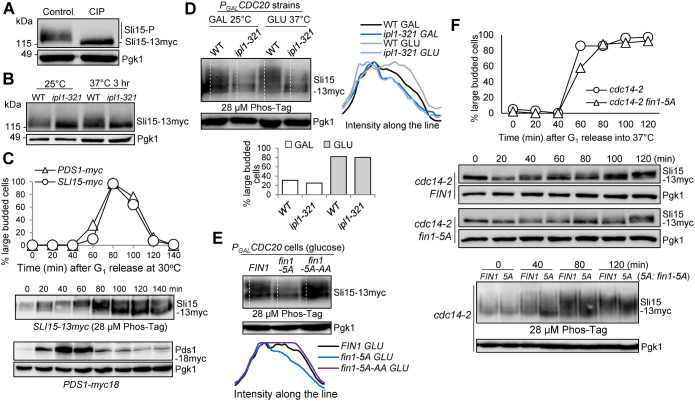
Sli15 is phosphorylated by both Ipl1 and CDK kinases during the cell cycle (Kang et al., 2001; Pereira and Schiebel, 2003). We examined the bandshift of Sli15 caused by phosphorylation. Protein samples were prepared from asynchronous yeast cells expressing 13myc-tagged Sli15, and slow-migrating forms of Sli15 were detected. After treating the protein samples with calf-intestinal alkaline phosphatase, the slow-migrating forms largely disappeared (Figure 1A), indicating that the bandshift is a result of phosphorylation. Ipl1 kinase phosphorylates Sli15 on 17 consensus sites located within the MT-binding domain to prevent its translocation to the spindle (Nakajima et al., 2011). To visualize Ipl1-dependent Sli15 phosphorylation, we observed a strong decrease in slow-migrating forms of Sli15 in temperature-sensitive ipl1-321 mutants compared with wild-type (WT) cells after 3 h incubation at 37°C (Figure 1B). Decreased Sli15 phosphorylation was also observed in ipl1-321 cells at 25°C, indicating comprised Ipl1 kinase activity at permissive temperature (Biggins and Murray, 2001; Jin et al., 2012). To clearly visualize Sli15 phosphorylation, we used Phos-tag SDS–PAGE (Bokros et al., 2021). We first measured Sli15 phosphorylation in synchronized cells, and 18myc-tagged Pds1 was used as a cell cycle marker for anaphase entry (Cohen-Fix et al., 1996). Figure 1C shows an increase in Sli15 phosphorylation beginning at 40 min as indicated by the band shift. However, Sli15 dephosphorylation was detected at 80 min, which concurs with Pds1 degradation, indicating that Sli15 dephosphorylation happens after anaphase entry. We further used Phos-tag to compare Sli15 phosphorylation in WT and ipl1-321 cells arrested in metaphase using PGALCDC20 strains (Figure 1D). After 2-h incubation in glucose for Cdc20 depletion, we noticed a significant decrease in phosphorylated species of Sli15 in ipl1-321 mutants at both 25°C and 37°C. This is likely attributed to reduced enzyme activity of Ipl1 even at the permissive temperature as mentioned above. Overall, these results suggest the significant contribution of Ipl1 kinase to Sli15 phosphorylation.
FIGURE 1:
Fin1-PP1 promotes Sli15 phosphorylation. (A) Sli15 is phosphorylated in vivo. Asynchronous WT cells (781-2-2) containing Sli15-13myc were grown in YPD medium at 25°C to log phase. Protein samples were collected and half of the protein sample was treated with calf intestinal alkaline phosphatase (CIP). Western blotting was performed with anti-myc antibody. Pgk1, loading control. (B) Sli15 is phosphorylated by Ipl1 kinase. WT (781-2-2) and ipl1-321 (4175-1-1) cells with Sli15-13myc were grown in YPD medium at 25°C to log phase then shifted to 37°C for 3 h. Protein samples were collected before and after temperature shift. Western blotting was performed with anti-myc antibody. Pgk1, loading control. (C) Sli15 phosphorylation during cell cycle. G1-arrested SLI15-13myc (781-2-2) and PDS1-18myc (JBY649) cells were released into 30°C YPD medium. α-factor was added back after 40-min release to block the following cell cycle. Cells were collected every 20 min to prepare protein samples and to count budding index (N = 100 cells). Phos-tag SDS–PAGE was performed to visualize the bandshift of Sli15 during cell cycle using anti-myc antibody. Regular SDS–PAGE was used to examine Pds1 protein level during cell cycle. Pgk1, loading control. (D) Analyze Sli15 phosphorylation in WT and ipl1-321 cells arrested at metaphase using Phos-tag SDS–PAGE. Asynchronous SLI15-13myc PGALCDC20 (4595-2-1) and ipl1-321 SLI15-13myc PGALCDC20 (4600-2-2) cells were grown overnight in galactose media at 25°C. Glucose was added for Cdc20 depletion and cells were shifted to 37°C at the same time. Cells were collected before and after temperature shift (2 h) to prepare protein samples and to count budding index (N = 100 cells). Phos-tag SDS–PAGE was performed with anti-myc antibody. Pgk1, loading control. Quantifications of the Sli15 protein band-shift (phosphorylation) from the western blot were performed using ImageJ. GLU: glucose; GAL: galactose. (E) Examination of Sli15 dephosphorylation in different fin1 mutants using Phos-tag SDS–PAGE. Asynchronous SLI15-13myc PGALCDC20 (4595-2-1) cells with either FIN1 (pSB1252), phospho-deficient fin1-5A (pSB1359), or phospho- and PP1 binding-deficient fin1-5A-AA (pSB1361) plasmids were grown overnight in galactose media at 30°C. Glucose was added for Cdc20 depletion and metaphase arrest. After 2 h, cells were collected to prepare protein samples and Phos-tag SDS–PAGE was performed to detect Sli15 bandshift with anti-myc antibody. Pgk1, loading control. Quantifications of the Sli15 protein bandshift (phosphorylation) from the Western blot were performed using ImageJ. (F) Premature Fin1-PP1 KT localization promotes Sli15 dephosphorylation in cdc14-2 cells. G1-arrested cdc14-2 SLI15-13myc (4048-1-3) cells containing either FIN1 (pMB6) or fin1-5A (pMB7) plasmids were released into YPD at 37°C. Samples were collected every 20 min to prepare protein samples and to count budding index (N = 100 cells) (top). Regular western blotting was performed to examine Sli15 band shift. Pgk1, loading control. The samples from the indicated time points were also subjected to Phos-tag SDS–PAGE to show Sli15 phosphorylation (bottom). Pgk1, loading control.
PP1 regulatory subunit Fin1 is a KT-associated protein, and its KT localization is cell cycle regulated. Cdc14 phosphatase, upon release from the nucleolus triggered by active FEAR, reverses CDK-dependent phosphorylation of Fin1 to allow its KT binding (Woodbury and Morgan, 2007a; Akiyoshi et al., 2009; Bremmer et al., 2012). Because PP1 is responsible for the reversal of Ipl1-mediated phosphorylation and CPC localizes at the KT before anaphase entry, we suspected that KT recruitment of Fin1-PP1 might contribute to the dephosphorylation of Ipl1 sites on Sli15. To test this idea, we first followed Sli15 phosphorylation kinetics in synchronous WT and fin1Δ mutant cells. Surprisingly, fin1Δ cells did not show noticeable delay in Sli15 dephosphorylation and exhibited a relatively normal cell cycle progression (Supplemental Figure S1). We reasoned that this is likely because the contribution of Fin1-PP1 to Sli15 dephosphorylation is minor in the presence of active Cdc14 phosphatase. However, targeting Fin1-PP1 to the KT before Cdc14 activation may provide more insight into the extent of Sli15 dephosphorylation by Fin1-PP1 alone. To test this idea, we utilized a phospho-deficient mutant of Fin1, in which the five CDK consensus sites (S36A, S54A, T68A, S117A, and S148A) in Fin1 were mutated to alanine to generate a fin1-5A mutant (Woodbury and Morgan, 2007a, b). Fin1-5A protein exhibits premature KT localization even during metaphase, before Cdc14 activation (Bokros et al., 2021). PGALCDC20 strains were used to achieve metaphase arrest. After growing cells in glucose for 2 h for Cdc20 depletion and metaphase arrest, we examined Sli15 phosphorylation using Phos-tag gel in WT and fin1-5A cells. Hyper-phosphorylated Sli15 species were detected in WT cells (Figure 1E), but fin1-5A cells showed a clear reduction in phosphorylated Sli15 species. If prematurely targeting Fin1-PP1 to the KT is responsible for Sli15 dephosphorylation, the effect could be reversed by abrogating the PP1-binding motifs in a fin1-5A-AA mutant (Akiyoshi et al., 2009). Indeed, Sli15 hyper-phosphorylation was restored in fin1-5A-AA cells.
We further examined whether Fin1-PP1 KT localization was sufficient to induce Sli15 dephosphorylation in cells with inactive Cdc14. To this end, we utilized temperature-sensitive cdc14-2 mutant and monitored Sli15 phosphorylation in FIN1 and fin1-5A cells after G1 release into the nonpermissive temperature. Interestingly, we found decreased Sli15 phosphorylation in fin1-5A cells using regular SDS–PAGE (Figure 1F, top), as the majority of Sli15 protein was concentrated to the bottom band. Additionally, we utilized Phos-tag SDS–PAGE to examine Sli15 phosphorylation in the same strains following G1 release (Figure 1F, bottom). As expected, fin1-5A cells showed much slower migrating Sli15 bands than cells with FIN1 after G1 release for 40 min, indicating increased Sli15 dephosphorylation in fin1-5A mutants even when Cdc14 is inactive. Therefore, we concluded that premature Fin1 KT localization partially dephosphorylates Sli15, which is independent of Cdc14.
Fin1-PP1 KT localization and Sli15 dephosphorylation cause premature CPC removal from the KT
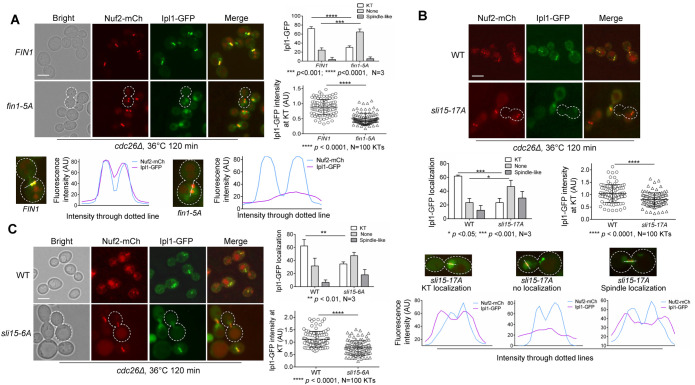
If premature KT localization of Fin1-PP1 promotes Sli15 dephosphorylation, then this might lead to untimely CPC’s KT delocalization and/or spindle binding. To test this idea, we analyzed the KT localization of CPC in FIN1 and fin1-5A cells with Ipl1-3GFP to mark the CPC and Nuf2-mCherry to mark KTs in a temperature-sensitive cdc26Δ background. Cdc26 is a subunit of the anaphase promoting complex and yeast cells lacking Cdc26 arrest at metaphase at high temperature (Zachariae et al., 1996; Hwang and Murray, 1997). After 2-h incubation at 36°C, we found that 72% of WT cells showed clear KT localization of Ipl1 (Figure 2A), as indicated by two distinct Ipl1 foci colocalizing with the two Nuf2-mCherry dots (Janke et al., 2001). However, in fin1-5A cells, the frequency of KT colocalization of Ipl1 was reduced to 30%, and the difference between FIN1 and fin1-5A cells is significant. In contrast, we observed a significant increase of fin1-5A cells with no detectable KT localization of Ipl1 (60%) compared with WT (30%), and many fin1-5A cells showed faint Ipl1-GFP signal throughout the entire nucleus. Furthermore, the overall intensity of Ipl1-GFP relative to 100 KTs (Nuf2-mCherry) was reduced in fin1-5A mutants compared with WT cells, and the reduction is significant. We further reasoned that the effects caused by premature KT localization of Fin1 could be reversed by a PP1-binding deficient fin1-5A-AA mutant. Indeed, Ipl1 KT localization was restored in cdc26Δ cells expressing fin1-5A-AA after G1 release at 36°C (Supplemental Figure S2A). Similar to WT cells (72%), we found that 73% of fin1-5A-AA cells showed clear Ipl1 KT localization (Supplemental Figure S2B). Additionally, no significant difference was detected for the intensity of Ipl1-GFP signal colocalizing with Nuf2-mCherry in WT and fin1-5A-AA cells (Supplemental Figure S2C), supporting the notion that KT recruitment of PP1 by Fin1 is required to dissociate CPC from the KT. Therefore, we conclude that premature KT localization of Fin1-PP1 is sufficient to dissociate CPC from the KT in metaphase, possibly through Sli15 dephosphorylation.
FIGURE 2:
Sli15 dephosphorylation promotes CPC disassociation from KTs in metaphase. (A) Premature KT localization of Fin1 removes CPC from KTs in metaphase cells. cdc26Δ IPL1-3GFP NUF2-mCherry (4421-3-2) cells containing either FIN1 (pMB6) or fin1-5A (pMB7) plasmids were grown to log phase in YPD at 25°C then shifted to 36°C for 2 h for metaphase arrest. Samples were collected for imaging. Ipl1 localization was categorized as no KT localization, KT localization, or spindle localization. A cell representing loss of Ipl1 KT localization is outlined with white dotted line. Scale bar, 5 μm. The experiment was repeated three times (N = 3), and 100 cells were counted for each experiment. The bars represent mean values ± SD (top right). p values were obtained by using a two-tailed unpaired t test, with asterisks indicating *p < 0.05, **p < 0.01, ***p < 0.001, and **** p < 0.0001. The intensity of Ipl1 relative to Nuf2 in 50 cells (100 KTs) was quantified using ImageJ as described in the Materials and Methods (bottom right). Statistical significance for relative Ipl1 intensity was analyzed by a two-tailed unpaired t test, with asterisks indicating *p < 0.05, **p < 0.01, ***p < 0.001, and **** p < 0.0001. Colocalization of Ipl1 with Nuf2 in representative cells expressing FIN1 or fin1-5A was quantified and plotted using ImageJ (bottom). (B) Abolishment of Ipl1-dependent phosphorylation in Sli15 reduces KT localization of CPC in metaphase cells. cdc26Δ IPL1-3GFP NUF2-mCherry (4421-3-2) and sli15-17A cdc26Δ IPL1-3GFP NUF2-mCherry (4437-1-3) cells were subjected to the same experiment as in panel A. A cell representing loss of Ipl1 KT localization is outlined with white dotted line. Scale bar, 5 μm. The pictures are representative of three experimental repeats where 100 cells were counted for each experiment. The statistical analysis of Ipl1 KT localization and Ipl1 intensity at KTs is the same as described in panel A. Representative cells for each of the three Ipl1 localization categories in WT and sli15-17A strains were quantified and plotted using ImageJ (bottom). (C) Abolishment of CDK-dependent phosphorylation in Sli15 reduces KT localization of CPC in metaphase cells. cdc26Δ IPL1-3GFP NUF2-mCherry (4421-3-2) and sli15-6A cdc26Δ IPL1-3GFP NUF2-mCherry (4426-5-3) cells were subjected to the same experiment as in panel A. A cell representing loss of Ipl1 KT localization was outlined with white dotted line. Scale bar, 5 μm. The pictures are representative of three experimental repeats. See above for the statistical analysis for the KT localization and intensity of Ipl1.
If Sli15 dephosphorylation by Fin1-PP1 promotes CPC removal from the KT, then the phospho-deficient mutant sli15-17A, in which the Ipl1 phosphorylation sites in Sli15 are mutated to alanine (Nakajima et al., 2011), should also exhibit CPC removal from the KT in cells arrested at metaphase with cdc26Δ. As expected, in sli15-17A cells, KT localization of Ipl1 was significantly lower (23%) compared with WT (61%) (Figure 2B). Furthermore, the overall intensity of Ipl1 in 50 cells (100 KT clusters) relative to Nuf2-mCherry was significantly decreased in sli15-17A cells compared with WT cells. Interestingly, not only did sli15-17A exhibit a higher frequency of cells lacking KT localization of Ipl1 (47%) than in WT (23%), but we also noticed a strong increase in premature spindle association of Ipl1 (30% in sli15-17A vs. 12% in WT cells). Spindle association was evidenced by even distribution of Ipl1-GFP along a bar structure, indicative of the metaphase spindle (Figure 2B, bottom). This is in clear contrast to WT cells at metaphase, which show two distinct Ipl1-GFP dots associating with KTs. However, we cannot exclude the possibility that these mutant cells show mixed KT and spindle association. We also examined Ipl1 KT localization in sli15-3 mutants to determine whether compromised Ipl1 activity causes CPC dissociation from the KT in metaphase, because sli15-3 mutants show impaired Ipl1 activation. Following 80 min after G1 release at the restrictive temperature (36°C), the intensity of Ipl1-GFP relative to 100 KTs was reduced in sli15-3 cells, and this reduction was significant (Supplemental Figure S3). We therefore conclude that abolishment of Sli15 phosphorylation by Ipl1 is sufficient to remove CPC from the KT in cells at metaphase.
In addition to Ipl1, Sli15 is also phosphorylated by CDK, which is reversed by Cdc14 phosphatase. Because mutation of the Sli15 phosphorylation sites by CDK to alanine (sli15-6A) leads to CPC spindle binding even in the absence of Cdc14 activity (Pereira and Schiebel, 2003), we also examined CPC localization in cdc26Δ sli15-6A cells (Figure 2C). Like sli15-17A mutants, sli15-6A cells showed a significant drop in frequency of KT localization of Ipl1 (35%) compared with WT (62%), and an increase in cells with uniform nuclear Ipl1 distribution (47% in sli15-6A and 31% in WT). Additionally, the average intensity of Ipl1 at 100 KTs was also reduced in sli15-6A. sli15-6A cells also showed a slight increase in Ipl1 spindle association (18%) compared with WT (6%), although not as pronounced as sli15-17A mutant cells. Altogether, these results support the conclusion that either premature Fin1-PP1 KT localization or abolished Sli15 phosphorylation by CDK or Ipl1 kinase can promote CPC KT dissociation in metaphase.
Fin1-PP1 promotes CPC translocation in cells lacking active Cdc14 phosphatase
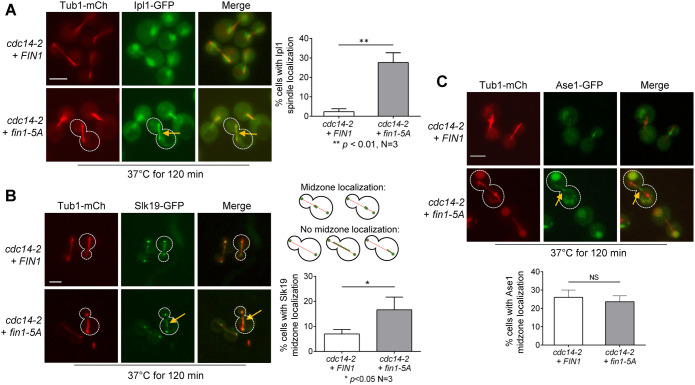
During anaphase, CPC dissociates from the KT and binds to the elongated spindle. Previous works indicate that Cdc14-dependent dephosphorylation of Sli15 is required for CPC translocation to the spindle (Pereira and Schiebel, 2003). In cdc14-2 mutant cells, mitotic exit is blocked, but spindle elongation still occurs, albeit slowly (Visintin et al., 1998; Pereira and Schiebel, 2003; Yellman and Roeder, 2015). Nevertheless, CPC shows no spindle association in this mutant. Because Fin1-5A, but not Fin1, shows KT localization in cdc14-2 mutant cells (Bokros et al., 2021), we tested whether Fin1-PP1 KT localization was sufficient for CPC translocation to the spindle in the absence of Cdc14 activity. For this purpose, we introduced WT FIN1 and phospho-deficient fin1-5A plasmids into cdc14-2 cells with Ipl1-GFP and Tub1-mCherry (spindle). As shown in Figure 3A, after 120 min incubation at 37°C, we observed few FIN1 cells that showed Ipl1 spindle localization (3%). However, 27% of cells with fin1-5A showed Ipl1 spindle localization, and the difference between these two strains is significant. This result indicates that the KT localization of Fin1-PP1 leads to CPC translocation to the spindle, at least partially, in the absence of Cdc14 activity.
FIGURE 3:
Fin1-PP1 promotes CPC translocation in cells with inactive Cdc14. (A) Premature KT localization of Fin1-PP1 promotes Ipl1 translocation to the spindle in cdc14-2 mutants. cdc14-2 IPL1-3GFP TUB1-mCherry cells (4052-9-2) containing either FIN1 (pMB6) or fin1-5A (pMB7) plasmids were grown to log phase in synthetic dropout medium at 25°C then shifted to 37°C for 2 h to inactivate Cdc14. Colocalization of Ipl1 with spindle (Tub1-mCherry) was quantified. Yellow arrows represent Ipl1 spindle localization in the cell outlined with white-dotted line. Scale bar, 5 μm. The pictures are representative of three experimental repeats (N = 3) where 100 cells were counted for each experiment. The bars represent mean values ± SD. p values were obtained by using a two-tailed unpaired t test, with asterisks indicating *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. (B) fin1-5A restores Slk19 midzone localization in cdc14-2 cells. cdc14-2 SLK19-GFP TUB1-mCherry cells (4210-1-2) containing either FIN1 (pMB6) or phospho-deficient fin1-5A (pMB7) plasmids were grown to log-phase in YPD at 25°C then shifted to 37°C for 2 h to inactivate Cdc14. The percentage of cells with Slk19 midzone localization was quantified. The guide for counting midzone localization is shown in the schematic. Representative cells are outlined in white, with yellow arrows representing Slk19 midzone localization. Scale bar, 5 μm. The pictures are representative of three experimental repeats (N = 3) where 100 cells were counted for each experiment. The bars represent mean values ± SD. p values were obtained by using a two-tailed unpaired t test, with asterisks indicating *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. (C) fin1-5A does not restore Ase1 midzone localization in cdc14-2 cells. cdc14-2 ASE1-GFP TUB1-mCherry cells (4209-1-1) containing either FIN1 (pMB6) or fin1-5A (pMB7) plasmids were subjected to the same experiment as in panel B. The percentage of Ase1 midzone localization was quantified. The guide for counting midzone localization is shown in the schematic in panel B. Yellow arrows represent Ase1 midzone localization in the cell outlined with white dotted line. Scale bar, 5 μm. The pictures are representative of three experimental repeats. The statistical analysis was performed as described above.
The spindle binding of CPC regulates the localization of spindle midzone proteins. Slk19 was originally identified as a KT protein that promotes KT capture and clustering in cells with disrupted spindles (Richmond et al., 2013; Norell et al., 2021). However, in anaphase Slk19 also localizes at spindle midzones for spindle stability (Sullivan et al., 2001). Slk19 midzone localization is dependent on CPC spindle association, which is triggered by Cdc14 activation. In support of this notion, phospho-deficient sli15-6A mutant that lacks the CDK phosphorylation sites can restore Slk19 midzone localization in the absence of Cdc14 activity (Pereira and Schiebel, 2003). To test whether Fin1-PP1 mediated Sli15 dephosphorylation can also induce Slk19 spindle midzone binding in the absence of Cdc14 activity, we constructed cdc14-2 strains containing Slk19-GFP and Tub1-mCherry and introduced WT FIN1 and fin1-5A plasmids into these strains. After 2-h incubation at 37°C to inactivate Cdc14 (Figure 3B), we found that 7% of cells with WT FIN1 showed Slk19 midzone localization, but this number increased to 17% in fin1-5A cells, and the difference is significant. Therefore, fin1-5A mutant partially restores the spindle midzone localization of Slk19 in the absence of Cdc14 activity. It is likely that the KT localization of Fin1-PP1 in cdc14-2 cells promotes CPC spindle localization and the subsequent recruitment of Slk19 to the spindle midzone.
Slk19 works with other midzone proteins to stabilize the spindle during anaphase. Ase1 becomes dephosphorylated by Cdc14 during early anaphase to allow its MT midzone binding (Khmelinskii et al., 2007). However, mutated sli15-6A that lacks CDK phosphorylation sites is unable to restore Ase1 midzone localization in the absence of Cdc14, indicating that the midzone localization of Ase1 is controlled directly by Cdc14 and is independent of CPC spindle interaction (Pereira and Schiebel, 2003; Khmelinskii et al., 2007). Thus, we performed a similar experiment to examine spindle midzone localization of Ase1-GFP in cdc14-2 cells expressing FIN1 or fin1-5A (Figure 3C). After 2-h incubation at 37°C, similar numbers of cells showed spindle midzone localization of Ase1 in cells expressing either FIN1 (26%) or fin1-5A (24%), indicating that Fin1 KT association does not affect Ase1 midzone location. Therefore, only CPC-dependent spindle midzone localization of Slk19 is enhanced by the expression of fin1-5A.
Loss of Fin1 delocalizes Slk19 from the spindle midzone and yields unstable spindles in cells arrested in telophase
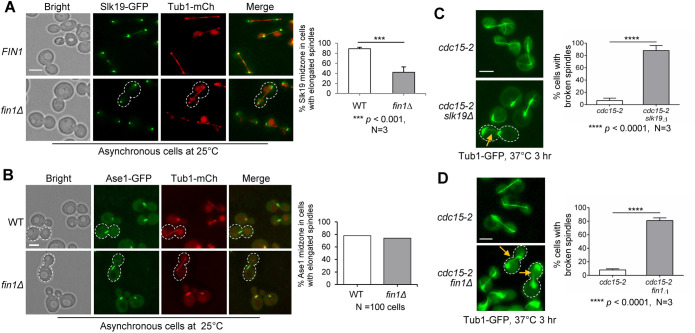
If premature Fin1 KT association partially restores Slk19 midzone localization when Cdc14 is absent, then Slk19 midzone localization might be compromised in fin1Δ cells. We checked Slk19 spindle midzone localization in asynchronous WT and fin1Δ cells. After scoring large-budded cells with elongated spindles (Figure 4A), we found that 89% of WT cells showed Slk19 midzone colocalization. Interestingly, only 42% of fin1Δ cells showed Slk19 midzone colocalization, and the drop is significant. This is likely because of compromised CPC spindle translocation in fin1Δ cells, although we failed to detect obvious CPC translocation defect in fin1Δ cells. In contrast, we did not detect significant difference in Ase1 midzone localization in WT (63%) or fin1Δ cells (65%) with elongated spindles (Figure 4B). Thus, the midzone localization of Slk19, but not Ase1, is regulated by Fin1.
FIGURE 4:
Loss of Fin1 impairs Slk19 spindle midzone localization and causes broken spindles in telophase. (A) Slk19 spindle midzone localization is reduced in fin1Δ cells. WT (4207-1-3) and fin1Δ (4207-4-1) cells containing Slk19-GFP and Tub1-mCherry were cultured overnight at 25°C in YPD medium. Asynchronous cells were collected for fluorescence microscopy. The percentage of cells with elongated spindles harboring Slk19 midzone localization was quantified. White dotted boundary represents cell lacking Slk19 midzone localization. Scale bar, 5 μm. The pictures are representative of three experimental repeats (N = 3) where 100 cells were counted for each experiment. The bars represent mean values ± SD. p values were obtained by using a two-tailed unpaired t test, with asterisks indicating ***p < 0.001. (B) Ase1 spindle midzone localization does not change in fin1Δ mutant cells. WT (2567-2-1) and fin1Δ (4205-5-1) cells containing Ase1-GFP and Tub1-mCherry were subjected to the same experiment as in panel A. White dotted boundary represents Ase1 midzone localization in cells with elongated spindles. Scale bar, 5 μm. The pictures and graph represent the quantification of counting 100 cells with elongated spindles for the WT and mutant, respectively. (C) Loss of Slk19 results in broken spindles in cells arrested at telophase. cdc15-2 (870-1-2) and cdc15-2 slk19Δ (870-2-2) cells containing Tub1-GFP were grown to log phase in YPD at 25°C then shifted to 37°C for 3 h to inactivate Cdc15. The percentage of cells with broken spindles was quantified. The yellow arrow points to a broken spindle in a representative cell outlined with white dotted line. Scale bar, 5 μm. The pictures are representative of three experimental repeats (N = 3) where 100 cells were counted for each experiment. The bars represent mean values ± SD. p values were obtained by using a two-tailed unpaired t test, with asterisks indicating **** p < 0.0001. (D) Loss of Fin1 results in broken spindles in cells arrested at telophase. cdc15-2 (4370-4-3) and cdc15-2 fin1Δ (4370-1-2) cells containing Tub1-GFP were subjected to the same experiment as in panel C. Yellow arrows point to broken spindles in representative cells outlined with white dotted lines. Scale bar, 5 μm. The pictures are representative of three experimental repeats. The same method was used for statistical analysis as described in panel C.
Slk19 midzone localization has been implicated in spindle stability (Sullivan et al., 2001; Havens et al., 2010; Norell et al., 2021). We previously showed broken spindles in slk19Δ cells arrested at telophase by cdc15-2 (Jin et al., 2008). Although cdc15-2 single mutant cells had very few broken spindles after 3 h at 37°C (7%), 88% of cdc15-2 slk19Δ cells exhibited broken spindles (Figure 4C). Because fin1Δ cells show reduced Slk19 midzone localization, fin1Δ cells may also exhibit spindle instability in telophase. Strikingly, 82% of fin1Δ cdc15-2 cells showed broken spindles compared with cdc15-2 (8%) after telophase arrest at 37°C (Figure 4D). This contradicts a previous report stating that cdc15-2 fin1Δ has no discernable spindle defects (Woodbury and Morgan, 2007a). We confirmed this phenotype with more than one strain, and one possible explanation for this difference is that strains with different genetic backgrounds were used. Surprisingly, fin1Δ cells showed no obvious spindle defects in normal cell cycle, so it is possible that the robust Cdc14 activity during anaphase can bypass the requirement of Fin1 for spindle stability, but the role of Fin1 becomes critical when Cdc14 activity is compromised in cdc15-2 mutants. Although we observed strong spindle instability in telophase arrested fin1Δ cells, Fin1-dependent CPC translocation may contribute only partially to this phenotype. Because Fin1 itself has been shown to localize to the spindle in anaphase and fin1Δ ase1Δ cells are synthetically lethal (Tong et al., 2004), Fin1 may also regulate spindle stability directly.
Abolishment of Ipl1-dependent phosphorylation in Sli15 results in sensitivity to syntelic attachments
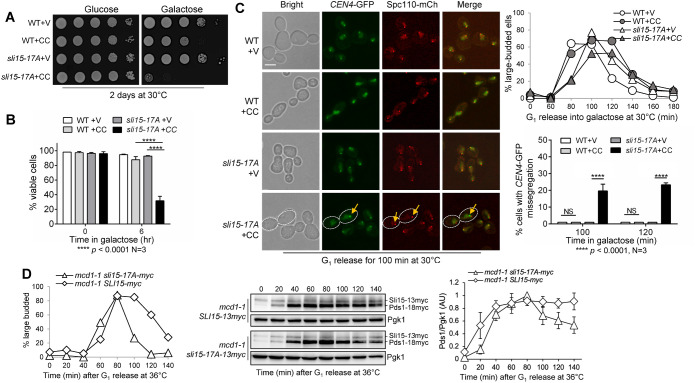
We previously showed that premature targeting of Fin1-PP1 to KTs in fin1-5A cells results in a defective checkpoint response to tensionless syntelic attachment (Bokros et al., 2016; Bokros et al., 2021). We speculated that premature KT localization of Fin1-5A-PP1 in fin1-5A cells delocalizes CPC from the KT and compromises the checkpoint. In that case, phospho-deficient sli15 mutants may also show a checkpoint defect in response to tensionless attachment. Thus, we examined the sensitivity of sli15-17A mutant cells (Figure 5A), which lack the Ipl1 phosphorylation sites, to the overexpression of the coiled–coiled domain of CIK1 (CIK1-CC). CIK1-CC overexpression disrupts the formation of kinesin dimer Cik1/Kar3 and induces tensionless syntelic attachment (Jin et al., 2012). On galactose plates, which induce CIK1-CC overexpression, WT cells showed mild slow growth compared with control cells, as CIK1-CC overexpression delays the cell cycle due to SAC activation by syntelic attachments (Jin et al., 2012). Strikingly, sli15-17A mutant cells showed dramatic growth defects on galactose plates. Consistently, sli15-17A mutants exhibited significant viability loss after CIK1-CC overexpression (Figure 5B). A high percentage (88%) of WT cells overexpressing CIK1-CC remained viable after 6-h incubation in galactose medium, whereas only 32% of sli15-17A cells were viable. The compromised checkpoint in sli15-17A cells likely contributes to the viability loss.
FIGURE 5:
Phospho-deficient sli15-17A mutant cells show defective checkpoint in response to syntelic attachment induced by CIK1-CC overexpression. (A) Phospho-deficient sli15-17A cells show sick growth when CIK1-CC is overexpressed. WT (Y300) and sli15-17A (4368-3-1) cells with either an empty vector control (V, p1218) or PGALCIK1-CC (CC, pHL002) were grown to saturation then 10fold serially diluted and spotted onto glucose and galactose plates. The plates were incubated at 30°C for 2 d before imaging. (B) sli15-17A mutant cells show severe viability loss after CIK1-CC overexpression. The same strains used in panel A were grown to log-phase in raffinose media at 25°C. Galactose was added to a final concentration of 2% to induce CIK1-CC overexpression. Samples were taken at time 0 and after 6 h, and then spread on a YPD plate. The plating efficiency (viability) was examined after overnight incubation at 25°C. The experiment was repeated three times (N = 3) where 300 cells were counted for each experiment. The bars represent mean values ± SD. p values were obtained by using a two-tailed unpaired t test, with asterisks indicating **** p < 0.0001. (C) sli15-17A mutant cells show increased chromosome missegregation when syntelic attachment is induced. WT (4379-3-3) and sli15-17A (4368-3-1) strains containing CEN4-GFP and Spc110-mCherry with either vector control (V, p1218) or PGALCIK1-CC (CC, pHL002) were arrested in G1 in raffinose media at 30°C. Cells were released into 2% galactose media to induce CIK1-CC overexpression, and samples were collected for budding index (N = 100 cells, top, right). Pictures were taken at 100 and 120 min to determine the rate of chromosome missegregation for 100 cells. The yellow arrow points to chromosome missegregation in a representative cell with white dotted boundary. Scale bar, 5 μm. The pictures are representative of three experimental repeats (N = 3) where 100 cells were counted for each experiment. The bars represent mean values ± SD. p values were obtained by using a two-tailed unpaired t test, with asterisks indicating **** p < 0.0001. (D) Abolishment of Ipl1-dependent Sli15 phosphorylation in sli15-17A cells causes a tension checkpoint defect. mcd1-1 sli15-13myc (4517-1-2) and mcd1-1 sli15-17A-13myc (4505-4-3) cells with Pds1-18myc were arrested in G1 in YPD medium at 25°C, and then released into the cell cycle at 36°C. Cells were collected every 20 min for budding index (N = 100 cells; left), and for the detection of Pds1 protein level with anti-myc antibody (center). Pgk1, loading control. We used ImageJ to acquire the intensity of each Pds1 band from western blotting images from three independent time courses and mean values ± SD are plotted (right). Pds1 levels were normalized to the loading control (Pgk1) and are shown relative to 80 min following G1 release, where Pds1 levels peak.
To further examine the checkpoint response to CIK1-CC overexpression in sli15-17A mutants, we followed cell-cycle progression and sister chromosome segregation using strains with CEN4-GFP (GFP-marked centromere of chromosome IV) and Spc110-mCherry (marks spindle pole bodies; Sherwin et al., 2022). G1-arrested WT and sli15-17A cells carrying vector or PGALCIK1-CC plasmids were released into galactose media to overexpress CIK1-CC. WT cells overexpressing CIK1-CC presented with more large-budded cells at later time points, indicating delayed cell cycle, which is consistent with previous observations (Jin et al., 2012). However, this cell-cycle delay was abolished in sli15-17A cells, indicating a defective checkpoint response to tensionless attachments. After G1 release for 100 and 120 min, we examined the CEN4-GFP signal in cells with elongated spindles (Figure 5C). WT cells showed no chromosome missegregation with or without CIK1-CC overexpression, indicating an intact checkpoint response. In contrast, sli15-17A displayed an increased frequency of CEN4-GFP missegregation. At 100 and 120 min after CIK1-CC induction, 20% and 23% of cells showed CEN4-GFP missegregation, respectively.
To further assess the checkpoint efficiency in sli15-17A cells, the degradation kinetics of Pds1 were analyzed in cohesion-defective cells using mcd1-1 mutants. Lack of cohesion leads to tensionless KT attachment that triggers Ipl1-dependent anaphase entry delay (Biggins and Murray, 2001; Jin and Wang, 2013). The degradation of anaphase inhibitor Pds1 marks anaphase entry (Cohen-Fix et al., 1996). mcd1-1 SLI15-13myc and mcd1-1 sli15-17A-13myc strains expressing Pds1-18myc were used for this assay. Cells were synchronized in G1 and released at 36°C to inactivate cohesin, resulting in tensionless attachments. Pds1 levels were quantified by normalization to the loading control and are relative to timepoint 80 min following G1 release, where Pds1 levels peak (Figure 5D). mcd1-1 SLI15-13myc cells exhibited relatively persistent Pds1 level and delayed disappearance of large-budded cells. However, mcd1-1 sli15-17A-13myc cells showed a clear drop in Pds1 level beginning at 100 min after G1 release, and the number of large-budded cells also reduced faster compared with the control cells, suggesting a tension-checkpoint defect. Taken together, we conclude that abolishment of Ipl1-dependent Sli15 phosphorylation in sli15-17A cells causes defective tension checkpoint as well as increased chromosome missegregation in response to syntelic attachments, which is likely due to premature CPC delocalization from KTs.
Abolishment of CDK-dependent phosphorylation in Sli15 also results in sensitivity to syntelic attachments
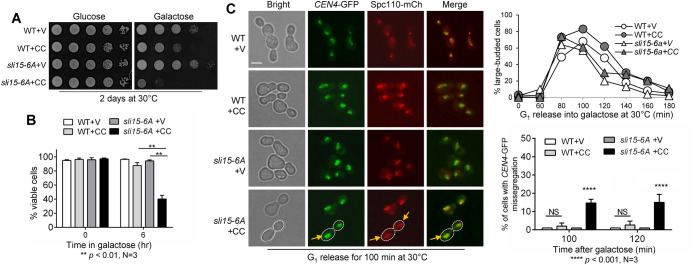
The sli15-6A mutant allele has six of the CDK consensus sites mutated to alanine, and CPC prematurely binds the spindle in this mutant (Pereira and Schiebel, 2003). We hypothesized that sli15-6A mutant would have a similar checkpoint defect in response to syntelic attachment as sli15-17A. As expected, sli15-6A cells showed a dramatic growth defect on galactose plates compared with WT cells after CIK1-CC induction as shown in Figure 6A. Consistently, sli15-6A mutant also showed more significant viability loss (35% viable cells) after 6-h growth in galactose medium for CIK1-CC overexpression compared with WT cells (88% viable), indicating a compromised checkpoint (Figure 6B). We also examined CEN4-GFP segregation in these cells and found that sli15-6A mutant exhibited 15% chromosome missegregation after G1 release for 100 and 120 min when CIK1-CC was overexpressed (Figure 6C), whereas less chromosome missegregation was observed in WT cells. sli15-6A also abolished the anaphase entry delay caused by CIK1-CC overexpression, as indicated by the drop in large-budded cells at later time points. This result is comparable with that of sli15-17A mutant, suggesting that premature Sli15 dephosphorylation by either Cdc14 or PP1 is able to cause a checkpoint defect in response to tensionless attachments.
FIGURE 6:
Phospho-deficient sli15-6A mutant cells are sensitive to syntelic attachment induced by CIK1-CC overexpression. (A) sli15-6A mutant cells show sick growth when syntelic attachment is induced by CIK1-CC overexpression. WT (4379-3-3) and sli15-6A (4388-1-1) cells with either vector control (V, p1218) or PGALCIK1-CC (CC, pHL002) were grown to saturation then 10fold serially diluted and spotted onto glucose and galactose plates. The plates were incubated at 30°C for 2 d for imaging. (B) sli15-6A mutants show viability loss when syntelic attachment is induced by CIK1-CC overexpression. The same strains used in panel A were grown to log-phase in raffinose media at 25°C. Galactose was added to a final concentration of 2% to induce CIK1-CC overexpression. Samples were taken at time 0 and after 6 h and spread on a YPD plate. Platting efficiency was counted for viability after overnight incubation at 25°C. The percentage of viable colonies was quantified from three independent experiments (N = 3) where 300 cells were counted for each experiment. The bars represent mean values ± SD. p values were obtained by using a two-tailed unpaired t test, with asterisks indicating **p < 0.01. (C) sli15-6A mutant cells show increased chromosome missegregation when syntelic attachment is induced. WT (4379-3-3) and sli15-6A (4388-1-1) strains containing CEN4-GFP and Spc110-mCherry with either vector control (V, p1218) or PGALCIK1-CC (CC, pHL002) were arrested in G1 in raffinose media at 30°C. Cells were released into 2% galactose media, and samples were collected for budding index (N = 100 cells, top, right). Pictures were taken at 100 and 120 min to determine the rate of chromosome missegregation for 100 cells. Yellow arrows point to chromosome missegregation in a representative cell with white dotted boundary. Scale bar, 5 μm. The pictures are representative of three experimental repeats (N = 3) where 100 cells were counted for each experiment. The bars represent mean values ± SD. p values were obtained by using a two-tailed unpaired t test, with asterisks indicating **** p < 0.0001.
Two distinct Sli15 dephosphorylation pathways have additive effects on CPC translocation
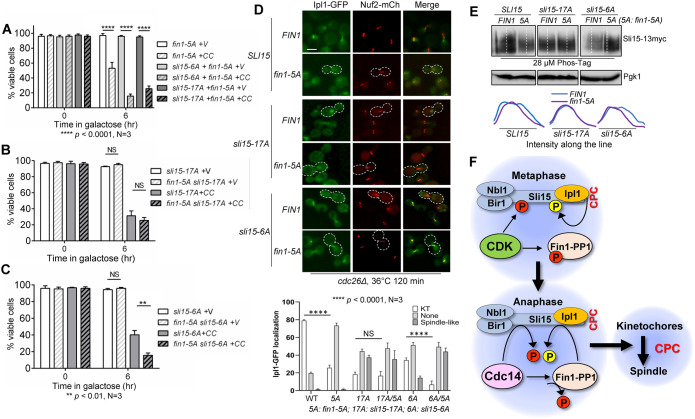
Sli15 is phosphorylated by both CDK and Ipl1, whereas Cdc14 and PP1 reverse the CDK and Ipl1-mediated phosphorylation, respectively. Thus, we speculated that combined dephosphorylation by both phosphatases could exacerbate viability loss in cells with syntelic attachment. We therefore introduced fin1-5A into sli15-6A and sli15-17A strains containing CIK1-CC plasmid. Both fin1-5A sli15-6A and fin1-5A sli15-17A double mutants showed dramatic growth defect on galactose plates after CIK1-CC induction (Supplemental Figure S4). After CIK1-CC overexpression for 6 h in galactose medium, cells with either fin1-5A sli15-6A or fin1-5A sli15-17A exhibited low viability (16% and 26%, respectively; Figure 7A). We also compared the viability with sli15-6A and sli15-17A single mutants after CIK1-CC overexpression (Figure 7B). We found that sli15-17A (32%) and fin1-5A sli15-17A (26%) had no significant difference in their viability loss in response to CIK1-CC overexpression. This is likely due to the fact that these two mutants act in the same pathway to reverse Ipl1-dependent Sli15 phosphorylation. Contrastingly, in response to CIK1-CC overexpression, fin1-5A sli15-6A suffered more viability loss (16%) than sli15-6A alone (35%), a statistical difference (Figure 7C). We were unable to analyze CEN4-GFP segregation in these mutants because of strain-construction technicalities.
FIGURE 7:
Combined Sli15 dephosphorylation by Cdc14 and Fin1-PP1 has an additive effect on CPC translocation and checkpoint response to syntelic attachment. (A) Viability loss in strains with the combination of phospho-deficient sli15 mutants (sli15-6A and sli15-17A) with fin1-5A after CIK1-CC overexpression. WT (Y300), sli15-6A (Y3594), and sli15-17A (4053-4-1) cells containing fin1-5A (pMB7) plasmid with either vector control (V, p1218) or PGALCIK1-CC (CC, pHL002) were grown to log-phase in raffinose media at 25°C. Galactose was added to a final concentration of 2% to induce CIK1-CC overexpression. Samples were taken at time 0 and after 6 h and spread on a YPD plate for overnight incubation at 25°C. The percentage of viable colonies was quantified. The experiments were repeated three times (N = 3) and 300 cells were counted for viability. The bars represent mean values ± SD. p values were obtained by using a two-tailed unpaired t test, with asterisks indicating **** p < 0.0001. (B) The combination of sli15-17A with fin1-5A does not have an additive effect on the sensitivity to syntelic attachment. The same protocol was to analyze the effect of the combination of sli1-17A with fin1-5A on the viability loss after syntelic attachment induction. Statistical analysis was performed as described in panel A. (C) The combination of sli15-6A with fin1-5A shows an additive effect on the sensitivity to syntelic attachment. The same method was used to determine the effect of the combination of sli15-6A with fin1-5A on the viability loss after syntelic attachment induction. The experiments were repeated three times (N = 3) and 300 cells were counted for each experiment. See above for statistical analysis. (D) The combination of sli15-6A with fin1-5A shows an additive effect on CPC dissociation from KTs in metaphase cells. cdc26∆ IPL1-3GFP NUF2-mCherry (4421-3-2, WT), sli15-17A cdc26∆ IPL1-3GFP NUF2-mCherry (4437-1-3), and sli15-6A cdc26∆ IPL1-3GFP NUF2-mCherry (4426-5-3) cells containing either FIN1 (pMB6) or phospho-deficient fin1-5A (pMB7) plasmids were grown to log-phase in YPD at 25°C then shifted to 36°C for 2 h to inactivate Cdc26 for metaphase arrest. Samples were collected for imaging. Ipl1 localization was categorized as no KT localization, KT localization, or spindle-like localization. Cells representing loss of Ipl1 KT localization are outlined with white dotted line. Scale bar, 5 μm. The pictures are representative of three experimental repeats (N = 3) where 100 cells were counted for each experiment. The bars represent mean values ± SD. p values were obtained by using a two-tailed unpaired t test, with asterisks indicating ****p < 0.0001. 17A: sli15-17A; 6A: sli15-6A; 5A: fin1-5A. (F) A schematic model describing two distinct pathways of Sli15 dephosphorylation by Cdc14 and Fin1-PP1 that promote CPC translocation in anaphase.
We further investigated whether the combined Sli15 dephosphorylation by both phosphatases, Cdc14 and PP1, would show an enhanced CPC removal from the KT in cells arrested at metaphase with cdc26∆. Consistent with our previous experiments, introducing fin1-5A significantly reduced KT localization of Ipl1 (24%) as compared with cells expressing FIN1 (78%; Figures 7D). The KT localization of Ipl1 in sli15-17A (18%) and fin1-5A sli15-17A (16%) showed no significant difference. In clear contrast, loss KT localization of Ipl1 in sli15-6A (37%) was significantly reduced when coupled with fin1-5A (8%). Additionally, we observed a significantly higher proportion of fin1-5A sli15-6A cells (43%) with premature spindle association of Ipl1 compared with sli15-6A (14%). We next examined the effect of Fin1-5A expression on the phosphorylation of WT and phospho-deficient Sli15 mutants, Sli15-6A, and Sli15-17A. The WT and mutated SLI15 genes were tagged with 13myc and we used Phos-tag SDS–PAGE to visualize Sli15 protein phosphorylation in asynchronized cells (Figure 7E). We noticed an appreciable decrease in Sli15-6A phosphorylation in fin1-5A cells as compared with FIN1 cells, but no visible difference in Sli15-17A phosphorylation was detected in fin1-5A and FIN1 cells. This result indicates that mutations of the Ipl1 phosphorylation sites (Sli15-17A), but not the CDK sites (Sli15-6A), in Sli15 abolishes its further dephosphorylation by Fin1-PP1. Together, our results support the conclusion that the combination of dephosphorylation of Sli15 by Cdc14 and PP1 contributes to the additive defect on CPC translocation and the checkpoint response to syntelic attachments (Figure 7F).
DISCUSSION
The SAC becomes active in response to tensionless or incorrect KT–MT attachments. The KT localized CPC corrects errors in KT–MT attachment and prevents premature SAC silencing (Wang et al., 2014). CPC component Sli15 is phosphorylated by both Ipl1 and CDK, which retains CPC localization at the KT. At early anaphase, a small amount of Cdc14 phosphatase is released from the nucleolus via the FEAR pathway (Stegmeier et al., 2002). This release is responsible for reversing CDK-imposed phosphorylation on Sli15 as well as CPC translocation (Pereira and Schiebel, 2003; Jin et al., 2008). Although mutation of the Ipl1 sites in Sli15 to alanine is sufficient for CPC translocation (Nakajima et al., 2011), it is currently unclear which phosphatase is responsible for reversing Ipl1-dependent Sli15 phosphorylation. Here we report that Cdc14-dependent dephosphorylation of Fin1 triggers KT recruitment of Fin1-PP1 and the subsequent reversal of Ipl1-imposed Sli15 phosphorylation, which promotes CPC translocation from the KT to the spindle as well. Therefore, our results uncovered a new layer of regulation for CPC translocation.
Recent work from our lab revealed the role of Fin1-PP1 in anaphase progression. As a PP1 regulatory subunit, Fin1 is a CDK substrate, and this phosphorylation prevents its KT localization. After FEAR activation in early anaphase, Fin1 is dephosphorylated by Cdc14 to enable the association of Fin1-PP1 with the KT (Akiyoshi et al., 2009). In this study, we further show that KT-localized Fin1-PP1 also promotes dephosphorylation of Ipl1 substrate Sli15, resulting in CPC translocation even when Cdc14 is inactive. Consistently, premature Fin1 KT localization in cdc14-2 mutant can partially restore spindle midzone localization of Slk19, which depends on CPC-spindle interaction. Moreover, premature Fin1 KT localization or mutation of the Ipl1/CDK consensus sites in Sli15 not only triggers CPC translocation but also sensitizes cells to syntelic chromosome attachments, as indicated by increased viability loss and chromosome missegregation. Notably, the combination of premature KT localization of Fin1 (fin1-5A) and mutation of CDK phosphorylation sites in Sli15 (sli15-6A) shows an additive effect on CPC translocation and sensitivity to syntelic attachment. Although previous studies have demonstrated the role of Cdc14 in CPC translocation by reversing Sli15 phosphorylation imposed by CDK, this study exposes Fin1-PP1 as a secondary layer of regulation for CPC translocation by reversing Sli15 phosphorylation imposed by Ipl1.
One open question is why cells require two separate pathways to regulate CPC translocation. We showed that reversal of either CDK- or Ipl1-mediated phosphorylation of Sli15 is sufficient for CPC translocation even in cells arrested at metaphase. One explanation is that two layers of dephosphorylation allow for robust CPC translocation after anaphase entry. Once Cdc14 is activated, it not only reverses CDK-mediated Sli15 phosphorylation but also dephosphorylates Fin1 and triggers KT recruitment of Fin1-PP1 to reverse Ipl1-mediated Sli15 phosphorylation. This positive feedback likely ensures robust CPC translocation after anaphase entry. This also fits our current understanding that multiple dephosphorylation events promote SAC silencing during the metaphase–anaphase transition. First, dephosphorylation of Sli15 by Cdc14 and Fin1-PP1 promotes the removal of CPC from the KT (Pereira and Schiebel, 2003; Nakajima et al., 2011). Second, the dephosphorylation of Ipl1 KT substrate Dam1 by PP1 stabilizes KT–MT interaction and facilitates SAC silencing (Jin and Wang, 2013). Moreover, the reversal of Ipl1-imposed Ndc80 phosphorylation helps clear Bub1/Bub3 SAC proteins from the KT, which results in SAC disassembly at the KT (Bokros et al., 2016; Bokros et al., 2021). Together, these concurrent dephosphorylation events ensure timely anaphase entry by stabilizing KT–MT interaction and preventing SAC reactivation.
Previous studies have uncovered how Sli15 dephosphorylation promotes CPC spindle binding in anaphase. Dephosphorylation of Sli15 within its MT-binding domain facilitates CPC MT binding (Pereira and Schiebel, 2003; Nakajima et al., 2011; Makrantoni et al., 2014). We showed that premature Fin1-PP1 KT localization as well as phospho-deficient mutations, sli15-6A and sli15-17A, all promote CPC KT delocalization in metaphase. However, only phospho-deficient Sli15 mutants but not fin1-5A mutants exhibited significant spindle-bound CPC in cells arrested at metaphase (Figure 2). Our explanation is that PP1-dependent dephosphorylation of Sli15 is likely partial in metaphase arrested cells expressing fin1-5A. This partial dephosphorylation may be sufficient for KT delocalization but not for spindle binding of CPC. In support of this idea, we observed that the combination of premature Fin1-PP1 KT localization and phospho-deficient mutant sli15-6A promoted CPC spindle localization in metaphase-arrested cells in Figure 7D. Furthermore, KT dissociation of CPC, but not spindle binding, was observed in cdc14-2 arrested cells. It is likely that the trace Cdc14 activity in cdc14-2 mutant cells is sufficient to induce CPC KT delocalization but is insufficient for spindle binding (Figure 3). In addition, recent works show several pools of Ipl1/Sli15 that localize to the centromere and KT-proximal regions (Fischbock-Halwachs et al., 2019; Garcia-Rodriguez et al., 2019). More work is needed to distinguish functions of Sli15 dephosphorylation by Cdc14 and PP1 in the regulation of centromere and KT-localized CPC.
Altogether, our results uncovered an additional pathway that regulates Sli15 dephosphorylation and CPC localization. The recruitment of Fin1-PP1 to the KT after anaphase onset promotes CPC translocation from the KT to the spindle during anaphase (Figure 7F). This translocation not only stabilizes KT-MT interaction and anaphase spindle structure but also facilitates SAC clearance from KTs. Because we showed that dysregulation of this process causes chromosome missegregation in response to tensionless chromosome attachment, this pathway is therefore critical for genome integrity.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Yeast strains, growth, and media
The relevant genotypes and sources of the yeast strains used in this study are listed in Supplemental Table S1. All the strains listed are isogenic to Y300, a W303 derivative, and they were constructed by tetrad dissection. Yeast cell growth conditions and synchronization were performed as described previously (Bokros et al., 2021). The plasmids used in this study are listed in Supplemental Table S2.
Budding index
For the indicated time points, samples were taken from the culture and fixed with 10% formaldehyde (3.7%). After sonication, cells were counted and categorized as single cell, small budded, and large budded cells based on the existence and size of the daughter cell. A cell was counted as large budded when the diameter of the daughter cell was greater than half of the diameter of the mother cell. The percentage of large-budded cells over time was plotted.
Plating efficiency/viability
A small aliquot of cells was 10fold diluted and then spread onto a YPD plate. After incubation at 25°C overnight, cell viability was examined under a microscope and cells were categorized as viable and nonviable. Cells that formed mini colonies were counted as viable, and nonviable cells were determined as single dead cells or a small cluster of sick cells. For each strain, more than 300 cells or cell colonies were counted to obtain the percentage of viable cells.
Western blotting
Yeast cells (1 ml) were collected by centrifugation and the cell pellets were resuspended in 200 μl of 0.1-M NaOH. After incubation at room temperature for 5 min, the sample was centrifuged, and the pellets were resuspended in 100-μl 1 × SDS protein loading buffer. The protein samples were boiled for 5 min and resolved by 8% SDS–PAGE. After probing for anti-myc (9E10, Covance Research Products) and anti-Pgk1 antibodies (Molecular Probes, Eugene, OR) followed by horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology), the protein levels were detected with enhanced chemiluminescence (ECL, PerkinElmer). A Bio-Rad ChemiDoc imaging system was used to image blots.
Sli15 phosphorylation was detected using Phos-tag SDS–PAGE. We added 28-μM Phos-tag and 150-μM MnCl2 to SDS–PAGE gel, which was run at 20 mA/gel for 120 min in the cold. The gel was washed with transfer buffer containing 10-mM EDTA for 10 min and then washed with transfer buffer alone for 10 min. The blots were then prepared as described above.
Cytological techniques
For fluorescence microscopy, collected yeast cells at indicated time points were resuspended in 1 × PBS (pH 7.2) for the examination of fluorescence signals using a microscope with a 60 × objective (BZ-X800 from Keyence). A Z-stack with 9–11 planes of 0.2 µm was acquired for each field and converted to a maximum projection using Keyence BZ-X800 software.
Statistical analysis
All statistical analyses were done using Graph Pad/Prism software. Results from fluorescence microscopy experiments were determined by counting 100 cells for each yeast strain. Experiments repeated three times are indicated as such. The mean values were calculated and shown with corresponding standard deviations. We performed two-tailed unpaired t tests to determine *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 and is denoted as such. For individual Ipl1 intensity at KTs, a defined area was used to determine a ratio of the intensity of Ipl1-GFP (AU) over the intensity of the Nuf2-mCherry signal (AU) for 100 KTs using ImageJ software and final ratios were plotted. For respective Ipl1-GFP localization in individual cells, a straight line of consistent length was drawn through the Nuf2-mCherry and Ipl1-GFP signals and the intensity (AU) along the line was plotted using ImageJ. For quantification of Sli15 protein bandshift, a straight line of consistent length was drawn through the Sli15 protein for the indicated timepoints and intensity of the protein signal (AU) across the line was plotted. When necessary, lines were shifted to the edge of each protein sample because the signal was often weaker in the middle of the sample due to western blotting inconsistencies.
Supplementary Material
Acknowledgments
We are grateful to the yeast community at Florida State University for reagents and helpful suggestions. We thank Drs. Sue Biggins, Georjana Barnes, and Frank Uhlmann for providing yeast strains and plasmids. We thank Dr. Terra Bradley for reading through this manuscript. This work was supported by R01GM121786 from the National Institutes of Health to Y.W.
Abbreviations used:
- CDK
cyclin-dependent kinase
- CPC
chromosomal passenger complex
- FEAR
Cdc14 early anaphase release
- KT
kinetochore
- MT
microtubule
- PP1
protein phosphatase 1
- SAC
spindle assembly checkpoint
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-02-0063) on July 5, 2023.
REFERENCES
- Akiyoshi B, Nelson CR, Ranish JA, Biggins S (2009). Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev 23, 2887–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A (2000). A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102, 21–31. [DOI] [PubMed] [Google Scholar]
- Biggins S, Bhalla N, Chang A, Smith DL, Murray AW (2001). Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics 159, 453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Murray AW (2001). The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev 15, 3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokros M, Gravenmier C, Jin F, Richmond D, Wang Y (2016). Fin1-PP1 helps clear spindle assembly checkpoint protein Bub1 from kinetochores in anaphase. Cell Rep 14, 1074–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokros M, Sherwin D, Kabbaj MH, Wang Y (2021). Yeast Fin1-PP1 dephosphorylates an Ipl1 substrate, Ndc80, to remove Bub1-Bub3 checkpoint proteins from the kinetochore during anaphase. PLoS Genet 17, e1009592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremmer SC, Hall H, Martinez JS, Eissler CL, Hinrichsen TH, Rossie S, Parker LL, Hall MC, Charbonneau H (2012). Cdc14 phosphatases preferentially dephosphorylate a subset of cyclin-dependent kinase (Cdk) sites containing phosphoserine. J Biol Chem 287, 1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad AJ, DeLuca JG (2020). The right place at the right time: aurora B kinase localization to centromeres and kinetochores. Essays Biochem 64, 299-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S, Tatsutani SY, Vermaak D, Biggins S (2003). The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J Cell Biol 160, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CS, Desai A (2013). Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature 497, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR 3rd, Chan CSM, Drubin DG, Barnes G (2002). Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D (1996). Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev 10, 3081–3093. [DOI] [PubMed] [Google Scholar]
- Edgerton H, Johansson M, Keifenheim D, Mukherjee S, Chacon JM, Bachant J, Gardner MK, Clarke DJ (2016). A noncatalytic function of the topoisomerase II CTD in Aurora B recruitment to inner centromeres during mitosis. J Cell Biol 213, 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbock-Halwachs J, Singh S, Potocnjak M, Hagemann G, Solis-Mezarino V, Woike S, Ghodgaonkar-Steger M, Weissmann F, Gallego LD, Rojas J, et al. (2019). The COMA complex interacts with Cse4 and positions Sli15/Ipl1 at the budding yeast inner kinetochore. Elife 8, e42879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodriguez LJ, Kasciukovic T, Denninger V, Tanaka TU (2019). Aurora B-INCENP localization at centromeres/inner kinetochores is required for chromosome bi-orientation in budding yeast. Curr Biol 29, 1536–1544.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW (1996). Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science 273, 953–956. [DOI] [PubMed] [Google Scholar]
- Havens KA, Gardner MK, Kamieniecki RJ, Dresser ME, Dawson DS (2010). Slk19p of Saccharomyces cerevisiae regulates anaphase spindle dynamics through two independent mechanisms. Genetics 186, 1247–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT (1991). S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66, 507–517. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Murray AW (1997). A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis. Mol Biol Cell 8, 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian VB, Stern BM, Murray AW (2005). The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science 307, 130–133. [DOI] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Lechner J, Shevchenko A, Shevchenko A, Magiera MM, Schramm C, Schiebel E (2001). The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J 20, 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Bokros M, Wang Y (2017). The phosphorylation of a kinetochore protein Dam1 by Aurora B/Ipl1 kinase promotes chromosome bipolar attachment in yeast. Sci Rep 7, 11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Liu H, Li P, Yu HG, Wang Y (2012). Loss of function of the cik1/kar3 motor complex results in chromosomes with syntelic attachment that are sensed by the tension checkpoint. PLoS Genet 8, e1002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Liu H, Liang F, Rizkallah R, Hurt MM, Wang Y (2008). Temporal control of the dephosphorylation of Cdk substrates by mitotic exit pathways in budding yeast. Proc Natl Acad Sci USA 105, 16177–16182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Wang Y (2013). The signaling network that silences the spindle assembly checkpoint upon the establishment of chromosome bipolar attachment. Proc Natl Acad Sci USA 110, 21036–21041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Cheeseman IM, Kallstrom G, Velmurugan S, Barnes G, Chan CS (2001). Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J Cell Biol 155, 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y (2007). Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev 21, 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Lawrence C, Roostalu J, Schiebel E (2007). Cdc14-regulated midzone assembly controls anaphase B. J Cell Biol 177, 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW (1991). Feedback control of mitosis in budding yeast. Cell 66, 519–531. [DOI] [PubMed] [Google Scholar]
- Loog M, Morgan DO (2005). Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434, 104–108. [DOI] [PubMed] [Google Scholar]
- Mackay AM, Eckley DM, Chue C, Earnshaw WC (1993). Molecular analysis of the INCENPs (inner centromere proteins): separate domains are required for association with microtubules during interphase and with the central spindle during anaphase. J Cell Biol 123, 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrantoni V, Corbishley SJ, Rachidi N, Morrice NA, Robinson DA, Stark MJR (2014). Phosphorylation of Sli15 by Ipl1 is important for proper CPC localization and chromosome stability in Saccharomyces cerevisiae. PLoS One 9, e89399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirchenko L, Uhlmann F (2010). Sli15(INCENP) dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr Biol 20, 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl DA, Huddleston MJ, Collingwood TS, Annan RS, Deshaies RJ (2009). Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J Cell Biol 184, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y, Cormier A, Tyers RG, Pigula A, Peng Y, Drubin DG, Barnes G (2011). Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP regulates CPC-spindle interaction to ensure proper microtubule dynamics. J Cell Biol 194, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norell S, Ortiz J, Lechner J (2021). Slk19 enhances cross-linking of microtubules by Ase1 and Stu1. Mol Biol Cell 32, ar22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Schiebel E (2003). Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302, 2120–2124. [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Kotwaliwale CV, Tatsutani SY, Breed CA, Biggins S (2006a). Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol Cell Biol 26, 2648–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Kung C, Shokat KM, Biggins S (2006b). The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol 8, 78–83. [DOI] [PubMed] [Google Scholar]
- Queralt E, Lehane C, Novak B, Uhlmann F (2006). Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125, 719–732. [DOI] [PubMed] [Google Scholar]
- Richmond D, Rizkallah R, Liang F, Hurt MM, Wang Y (2013). Slk19 clusters kinetochores and facilitates chromosome bipolar attachment. Mol Biol Cell 24, 566–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozelle DK, Hansen SD, Kaplan KB (2011). Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake. J Cell Biol 193, 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin D, Huetteman A, Wang Y (2022). Yeast Kinesin-5 motor protein CIN8 promotes accurate chromosome segregation. Cells 11, 2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin D, Wang Y (2019). The opposing functions of protein kinases and phosphatases in chromosome bipolar attachment. Int J Mol Sci 20, 6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ (1999). Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97, 233–244. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A (2002). Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108, 207–220. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Lehane C, Uhlmann F (2001). Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat Cell Biol 3, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJR, Nasmyth K (2002). Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108, 317–329. [DOI] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN (2010). Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol 189, 713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AHY, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. (2004). Global mapping of the yeast genetic interaction network. Science 303, 808–813. [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A (1998). The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell 2, 709–718. [DOI] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A (1999). Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398, 818–823. [DOI] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JMG (2010). Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jin F, Higgins R, McKnight K (2014). The current view for the silencing of the spindle assembly checkpoint. Cell Cycle 13, 1694–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ng TY (2006). Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell 17, 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples WG, Chahwan C, Ciechonska M, Lavoie BD (2009). Putting the brake on FEAR: Tof2 promotes the biphasic release of Cdc14 phosphatase during mitotic exit. Mol Biol Cell 20, 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury EL, Morgan DO (2007). Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol 9, 106–112. [DOI] [PubMed] [Google Scholar]
- Woodbury EL, Morgan DO (2007). The role of self-association in Fin1 function on the mitotic spindle. J Biol Chem 282, 32138–32143. [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y (2010). Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330, 239–243. [DOI] [PubMed] [Google Scholar]
- Yellman CM, Roeder GS (2015). Cdc14 early anaphase release, FEAR, is limited to the nucleus and dispensable for efficient mitotic exit. PLoS One 10, e0128604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Shin TH, Galova M, Obermaier B, Nasmyth K (1996). Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science 274, 1201–1204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.