Abstract
Solanum xanthocarpum (SX) has been used to treat a variety of diseases, including skin disorders like psoriasis (PSO). SX possesses many pharmacological activities of anti-inflammatory, anti-cancer, immunosuppressive, and healing qualities. However, the multi-target mechanism of SX on PSO still needs clarity. Materials and methods: The Indian Medicinal Plants, Phytochemicals and Therapeutics (IMPPAT) database and the Swiss Target Prediction online tool were used to find the active phytochemical components and their associated target proteins. OMIM and GeneCards databases were used to extract PSO-related targets. A Venn diagram analysis determined the common targets of SX against PSO. Subsequently, the protein–protein interaction (PPI) network and core PPI target analysis were carried out using the STRING network and Cytoscape software. Also, utilising the online Metascape and bioinformatics platform tool, a pathway enrichment analysis of common targets using the Kyoto Encyclopaedia of Genes and Genome (KEGG) and Gene Ontology (GO) databases was conducted to verify the role of targets in biological processes, cellular components and molecular functions with respect to KEGG pathways. Lastly, molecular docking simulations were performed to validate the strong affinity between components of SX and key target receptors. Results: According to the IMPPAT Database information, 8 active SX against PSO components were active. According to the PPI network and core targets study, the main targets against PSO were EGFR, SRC, STAT3, ERBB2, PTK2, SYK, EP300, CBL, TP53, and AR. Moreover, molecular docking simulations verified the binding interaction of phytochemical SX components with their PSO targets. Last but not least, enrichment analysis showed that SX is involved in several biological processes, including peptidyl-tyrosine phosphorylation, peptidyl-tyrosine modification, and peptidyl-serine modification. The relevant KEGG signalling pathways are the PI3K-AKT signalling pathway, the EGFR tyrosine kinase inhibitor resistance pathway, and the MAPK signalling pathway. Conclusion: The network pharmacology technique, which is based on data interpretation and molecular docking simulation techniques, has proven the multi-target function of SX phytoconstituents.
Keywords: Solanum xanthocarpum, Psoriasis, Network pharmacology, Molecular docking, IMPPAT database
1. Introduction
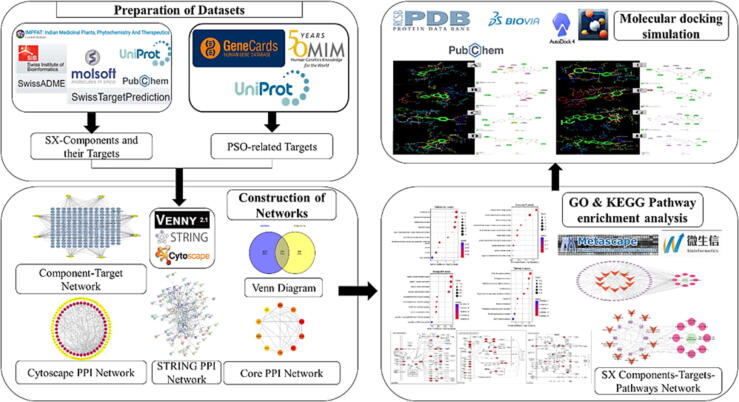
Psoriasis (PSO) is an immune-mediated chronic and recurring inflammatory skin or joint condition. The global prevalence of PSO is around 2% (Wang et al., 2022b). Although gender does not affect the occurrence of PSO, genetic predisposition and age are known to enhance its occurrence. PSO imposes a significant psychological burden because of the deformity and teratogenicity associated with this condition, which harms the quality of life. The physiological load is associated with recurring bouts of itching and several associated disorders (Sakai et al., 2016). More than 90% of cases are caused by Plaque PSO. Clinical signs include well-defined erythema and silvery scaly skin that can appear anywhere on the body (Amat-Samaranch and Puig, 2020). Topical glucocorticoids, vitamin D derivatives, calcineurin inhibitors, corticosteroids, phototherapy, and systemic therapy are common treatments for PSO. The use of topical medications is limited by practicability (the time necessary to apply therapy), convenience, and adverse responses (e.g., skin irritation), whereas systemic treatment is limited by drug interactions and cumulative organ damage (Martins et al., 2020). PSO is mediated by dendritic and T cells in a complex feedback loop involving antigen-presenting cells, neutrophils, keratinocytes, vascular endothelial cells, and the skin’s nervous system. The primary pathophysiology of PSO is resident immune cell interaction dysfunction in the skin, which is mediated by tumour necrosis factor-α, interferon-γ, interleukin-1β, and other cytokines (Christophers and van de Kerkhof, 2019). Hence, blocking the PSO-related immunological pathway with targeted therapy is effective; nevertheless, the high cost of biological agents makes it challenging to spread. Implementing the usage of natural remedies against the PSO produced superior outcomes because there were no negative side effects. Solanum xanthocarpum (SX) is an ancient medicinal plant used to cure various diseases, including skin disorders such as PSO (Joghee et al., 2019). One of the in-vivo trials revealed that SX targeting a phytoconstituent chlorogenic acid was more effective against PSO (Parmar et al., 2017), yet, none of the phytoconstituents of SX has a comprehensive understanding of the treatment’s molecular mechanism. To lay the theoretical groundwork for future research into the potential targets and signal pathways of SX in the treatment of PSO, this study uses network pharmacology to predict the potential targets and signal pathways of SX and combines it with molecular docking technology to aid verification. Network pharmacology approaches can be used to confirm the multiple-targeting effects of potential phytoconstituents of SX in the treatment of PSO. These methodologies can also be taken into further consideration to examine the effect against PSO. This study will clearly describe all of the pathophysiological changes that could be brought about by using SX to treat PSO. A flowchart of this study is shown in Fig. 1.
Fig. 1.
Workflow of the network pharmacological and molecular docking investigation strategy of SX in the treatment of PSO.
2. Materials and Methods
2.1. Active components and correlated target database establishment
SX active components screening on the IMPPAT (Indian Medicinal Plants, Phytochemicals and Therapeutics) (https://cb.imsc.res.in/imppat/home) database (Mohanraj et al., 2018), the plant name “S. xanthocarpum / virginianum” was entered sequentially to retrieve the matching phytochemical substances and related information (Murali et al., 2022). The screening conditions for obtaining the effective components and corresponding targets of each drug of SX were set according to the principle of pharmacokinetics (ADME), bioavailability score (BS) ≥ 0.3 and drug-likeness (DL) ≥ 0.18 (Chen et al., 2022a, Chen et al., 2022b) obtained from Swiss ADME (https://www.swissadme.ch/) (Ranjith and Ravikumar, 2019) and Molsoft L.L.C. (Drug-likeness prediction, https://molsoft.com/mprop/) (Ahsan et al., 2011) respectively. The chemical formulas of components and the canonical SMILES were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) to check the final components of SX (Butz et al., 2017). Based on the Swiss Target Prediction (https://www.swisstargetprediction.ch/), the targets linked with SX components were further determined, which is a webserver to accurately identify the targets of active molecules (Daina et al., 2019) and deduplicated to remove repeated targets.
2.2. Network construction of active Components-Potential targets
To reflect the complicated interaction between active chemicals and probable targets, a complete network was built using Cytoscape 3.9.1 software (https://www.cytoscape.org/) (Jin et al., 2021). Edges indicate the relationships between components and targets, whereas nodes represent the components and targets.
2.3. Collection of potential PSO associated targets
To confirm the data’s completeness and quality, the probable PSO targets in the GeneCards database (Shen et al., 2022) (https://www.genecards.org/), OMIM database (Amberger and Hamosh, 2018) (https://omim.org/), were searched with “psoriasis” as keywords. The score in the GeneCards database reflects the proximity between the target and the illness, thus the target with a score larger than the median was chosen as a prospective PSO target. The chemical components that did not have corresponding targets were eliminated, the repetitive targets were deleted, and the UniProt database (https://www.uniprot.org/uploadlists/) was used to standardize the target names through the above methods (Xia et al., 2021).
2.4. Screening Component-Disease overlapping targets
The Venn diagram (https://bioinfogp.cnb.csic.es/tools/venny/) was filled with the screened SX phytoconstituent targets and PSO disease targets (Chen et al., 2022a, Chen et al., 2022b) and as prospective objectives for additional investigation, the intersection targets of active components in SX and PSO were gathered.
2.5. Network construction of Component-Disease common targets
The STRING (https://string-db.org/) platform was used to generate a protein–protein interaction (PPI) network (Doncheva et al., 2019), this includes almost all functional relationships among the produced proteins. The protein type was set to Homo sapiens, full STRING network type, the meaning of network edges was set to confidence, active interaction sources were experiments, not more than 10 interactors and the minimum required interaction source was set to medium confidence (0.400) (Cui et al., 2021). Target interaction data produced from the STRING database was transferred into the Cytoscape version 3.9.1 software, where it was integrated and analysed (Tang et al., 2015). The top ten target protein scores in network string interactions were produced and rated using the MCC technique using the CytoHubba plug-in (Zhang et al., 2021). The major targets were chosen based on the components-targets-pathways network and the results of molecular docking.
2.6. Gene Ontology (GO) and Kyoto Encyclopaedia of genes and Genomes (KEGG) pathway enrichment analyses
The GO and KEGG enrichment were performed on the Database for Metascape (https://metascape.org) (Zhou et al., 2019). GO functionally annotates key genes into 3 main terms, i.e., cellular components (CCs), molecular functions (MFs), and biological processes (BPs) (Wang et al., 2022b). The KEGG enrichment analysis reveals potential biological processes with critical targets. The words of considerably enriched CCs, MFs, and BPs (P value < 0.05, minimum count of 3, and enrichment factor of greater than 1.5) were shown (Kanehisa et al., 2022). The bioinformatics platform (https://www.bioinformatics.com.cn/) was used to generate the bubble chart of GO and KEGG enrichment analyses (Chen et al., 2023).
2.7. Construction of SX Components-Targets-Pathways network
Using the Cytoscape version 3.9.1 software, a network of components-targets-pathways was built to describe the treatment mechanisms of SX for PSO (Patwardhan et al., 2005). The nodes in the network with varied colours and forms indicate components, targets, or disease-related pathways, respectively, and the “edge” connecting the nodes represents the link.
2.8. Molecular docking
The main components of SX from the component-target network and the core targets from the PPI involved in the components-targets-pathways network were docked using molecular docking. The main target’s protein structure was obtained from the RCSB PDB database (Jiménez-García et al., 2021) (https://www.rcsb.org/). The selection criteria (Hsin et al., 2013) are as follows: (a) X-ray structures with a resolution of 2.5 Å or better were included, if available; (b) if two or more structures were available, that with the best resolution was selected; (c) a structure with a ligand bound to its nucleotide-binding site was selected; (d) non-modified and non-phosphorylated residues found in the binding site were selected with priority; (e) the organism was human. The PubChem (https://pubchem.ncbi.nlm.nih.gov/) database was used to obtain the tiny molecular structure of essential components (Kim et al., 2021) as 3D Conformer SDF. The PyMOL 2.4.0 (Mooers, 2020) software was used to dewater, hydrogenate, and separate the original ligand of the core target protein. The molecular docking was completed in the AutoDock Tools 1.5.6 (Trott and Olson, 2010) and AutoDock Vina 4.2 (Afolabi et al., 2022) software. Then, using AutoDock Tools 1.5.6, the docking grid box was built for each target protein’s active site and stored in the pdbqt format. Molecular docking of potential targets and active substances and the evaluation of free binding energies were done using AutoDock Vina 4.2 (Wang et al., 2022a). When the binding energy was negative, it suggested that the receptor and ligand might bind spontaneously; when the binding energy was less than −5 kcal mol−1, it showed that the receptor and ligand had excellent binding activity (Hsin et al., 2013). The results with higher activity were visualized by PyMOL 2.4.0 (https://pymol.org/2/) and Discovery Studio BIOVIA 2021 (https://discover.3ds.com/discovery-studio-visualizer-download) (Ru et al., 2014).
3. Results
3.1. Active components of SX
The IMPPAT database was used to select the active phytochemical components of SX, and 23 components were obtained. Finally, seven components were chosen for further investigation based on BS ≥ 0.3 and DL ≥ 0.18 property thresholds. One more phytochemical component, chlorogenic acid, was included due to its skin permeation ability of −8.76 cm/s and DL of 0.79 with potential activity in PSO as determined by a literature review (Parmar et al., 2017) (Table 1).
Table 1.
General information of phytoconstituents of SX.
| Identifier | Phytochemical name | Canonical SMILES | Drug likeness | Bioavailability score | MW |
|---|---|---|---|---|---|
| 5,280,443 | Apigenin | C1 = CC(=CC = C1C2 = CC(=O)C3 = C(C = C(C = C3O2)O)O)O | 0.39 | 0.55 | 270.24 |
| 222,284 | Beta-Sitosterol | CCC(CCC(C)C1CCC2C1(CCC3C2CC = C4C3(CCC(C4)O)C)C)C(C)C | 0.78 | 0.55 | 414.71 |
| 5997 | Cholesterol | CC(C)CCCC(C)C1CCC2C1(CCC3C2CC = C4C3(CCC(C4)O)C)C | 0.49 | 0.55 | 386.65 |
| 173,183 | Campesterol | CC(C)C(C)CCC(C)C1CCC2C1(CCC3C2CC = C4C3(CCC(C4)O)C)C | 0.59 | 0.55 | 400.68 |
| 5,280,794 | Stigmasterol | CCC(C = CC(C)C1CCC2C1(CCC3C2CC = C4C3(CCC(C4)O)C)C)C(C)C | 0.62 | 0.55 | 412.69 |
| 70,699,351 | Stigmast-5-en-3beta-yl beta-D-glucopyranoside | CCC(CCC(C)C1CCC2C1(CCC3C2CC = C4C3(CCC(C4)OC5C(C(C(C(O5)CO)O)O)O)C)C)C(C)C | 0.5 | 0.55 | 576.85 |
| 70,699,355 | Stigmasteryl glucoside | CCC(C = CC(C)C1CCC2C1(CCC3C2CC = C4C3(CCC(C4)OC5C(C(C(C(O5)CO)O)O)O)C)C)C(C)C | 0.33 | 0.55 | 574.83 |
| 1,794,427 | Chlorogenic acid | C1C(C(C(CC1(C(=O)O)O)OC(=O)C = CC2 = CC(=C(C = C2)O)O)O)O | 0.79 | 0.11 | 354.31 |
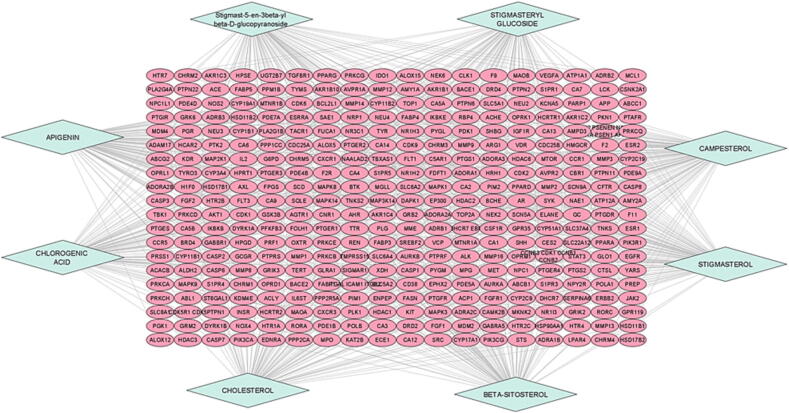
3.2. Component-Target network construction
A component-target network was created to show the interaction connection between the SX components and their respective targets (Fig. 2). By mapping 8 components to 800 possible targets, the network consists of 368 nodes and 800 edges, with blue circles representing putative targets and yellow circles representing SX components. Apigenin, Beta-sitosterol, Campesterol, Chlorogenic acid, Cholesterol, Stigmast-5-en-3beta-yl beta-D-glucopyranoside, Stigmasterol, and Stigmasteryl glucoside are phytochemical components that correspond to 51, 55, 58, 51, 57, 56, 54, and 51 targets, respectively. The findings indicate that these eight components are likely to play important therapeutic roles in PSO.
Fig. 2.
Network constructed for Active Components-Potential Targets, the blue diamond shape nodes represent the selected phytoconstituents of SX and the pink ellipse shape nodes represent the possible targets of components.
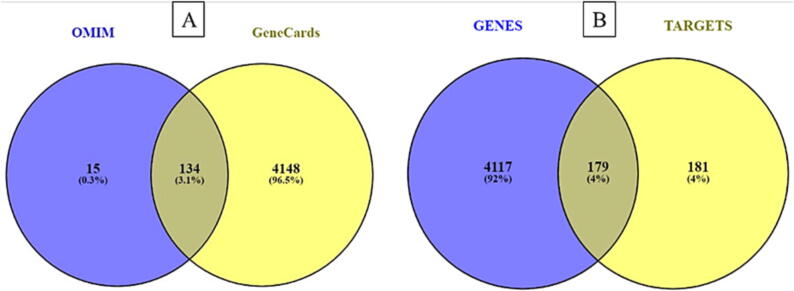
3.3. Predicting PSO-Related targets
The findings were combined by extracting the PSO-associated targets from the GeneCards and OMIM databases. The probable target genes collected from the GeneCards and OMIM databases are 4282 and 149, respectively, as shown in Fig. 3(A). Following deduplication, 4297 genes were acquired for future investigation. Fig. 3(B) depicts 179 putative target genes identified by the application Venn diagram screening of 360 possible targets of SX active components and 4296 genes of PSO.
Fig. 3.
(A) Venn diagram of OMIM & GeneCards targets, blue and yellow portion represents the genes collected from OMIM and GeneCards databases respectively, the intersection portion of 134 gene genes are common genes between these two databases; (B) Venn diagram of disease and drug targets, the blue and yellow portion represents the genes obtained from the above databases and SX components associated targets, 179 intersection genes obtained the intersection portion.
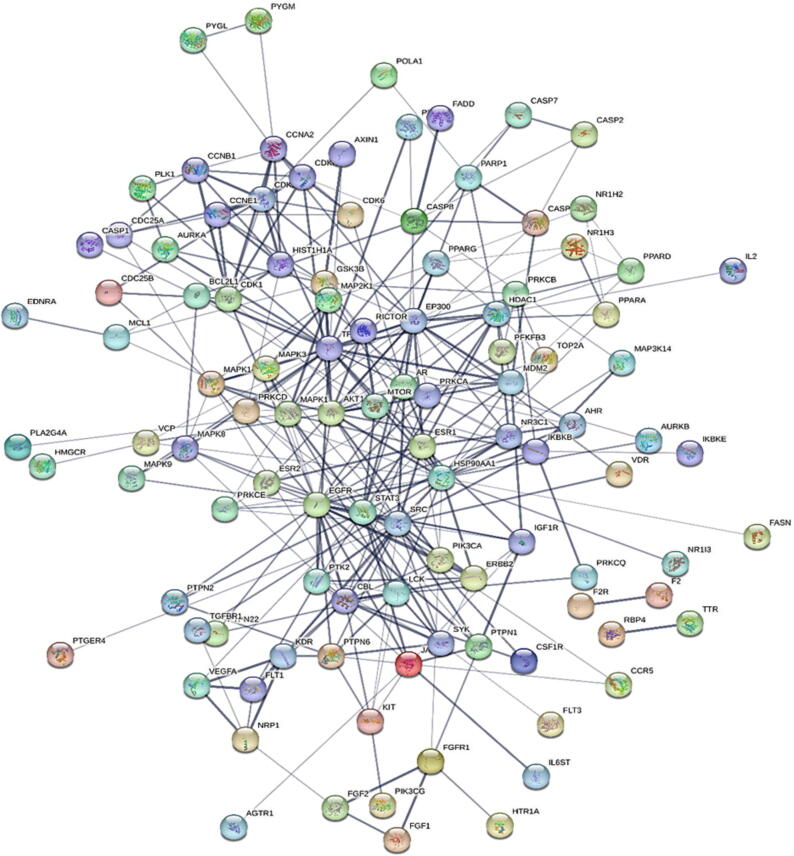
3.4. Common target PPI network
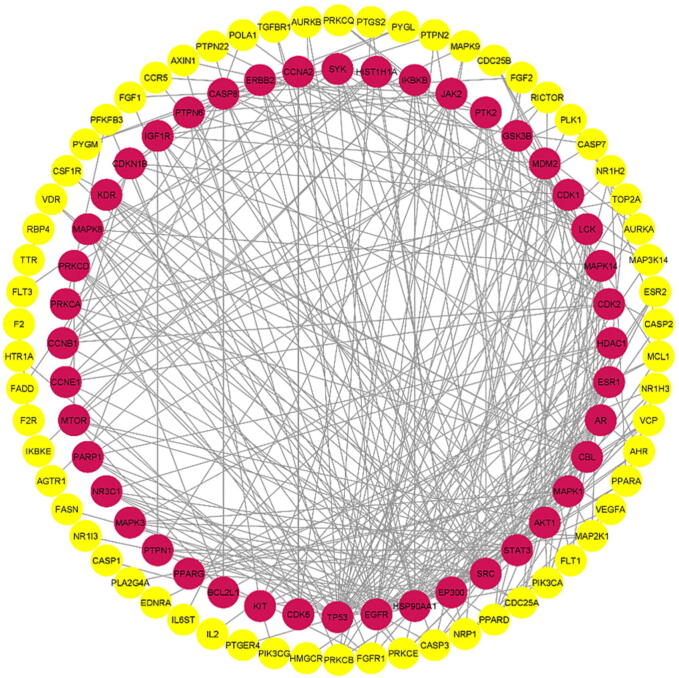
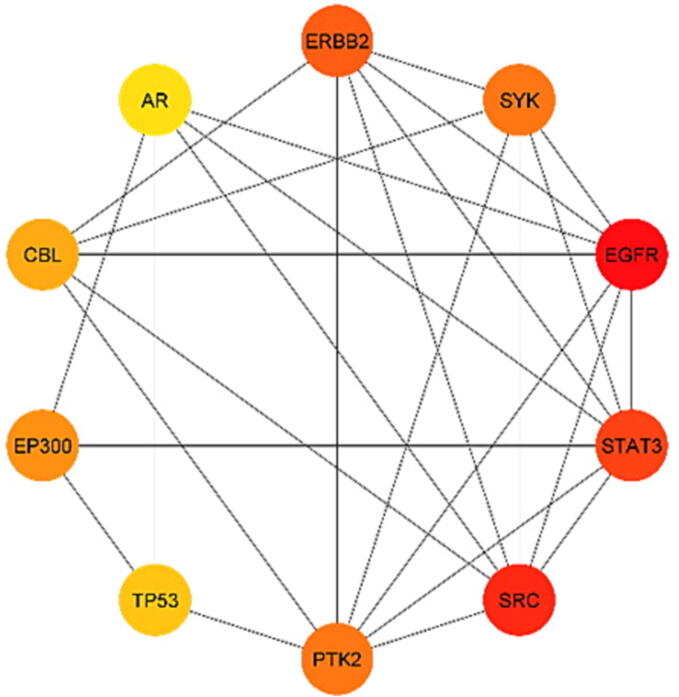
The STRING database was used to import 179 potential target genes associated with PSO for analysis and network formation. The interaction network was based on the selected targets with confidence network edges, only active experiments for active interaction sources, a medium confidence score of 0.400, maximum of 10 interactors (Fig. 4). After analysis, 189 nodes and 323 edges were embodied, with the average node degree being 3.42. The data were imported into Cytoscape for additional analysis and network creation (Fig. 5). The edges in this picture reflect the interaction between two prospective targets, the nodes represent the targets, and the degree value represents the strength of the target interaction. Then, CytoHubba was put in to extract the core PPI network by calculating the top 10 nodes with the shortest path using the MCC approach (Fig. 6). In descending order, the top ten nodes represented EGFR, SRC, STAT3, ERBB2, PTK2, SYK, EP300, CBL, TP53, and AR targets were chosen as the core targets for components-targets-pathways network construction and selected targets for molecular docking with the core components of SX for PSO treatment. All 77 target genes obtained by PPI were gathered for pathway enrichment analysis.
Fig. 4.
PPI Network obtained from STRING, nodes represent the proteins and the edges represent the interaction between proteins.
Fig. 5.
PPI Network obtained from Cytoscape software, nodes represent the proteins and the edges represent the interaction between proteins.
Fig. 6.
Core PPI network from CytoHubba by MCC method, nodes represent the 10 proteins following the shortest path and the edges represent the interaction between proteins.
3.5. GO and KEGG enrichment analyses
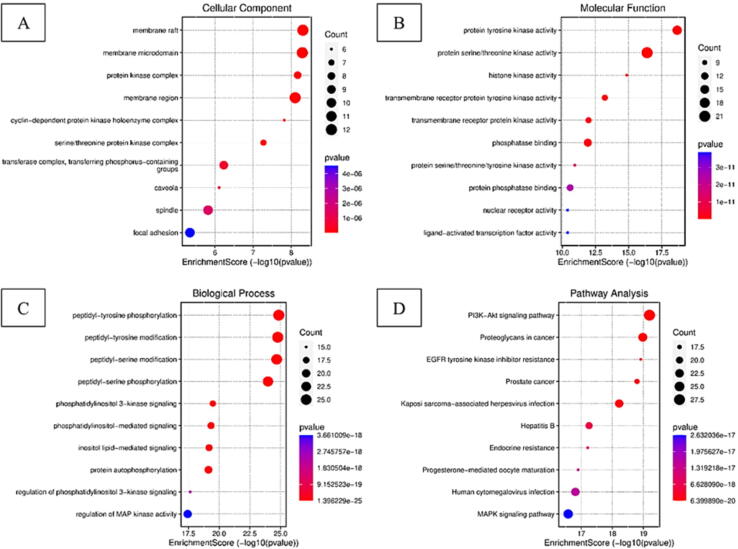
The top ten substantially enriched CCs, MFs, and BPs terms (p < 0.05, minimum count of 3, and enrichment factor of greater than 1.5) were examined. The size of the dot in the bubble chart represents the number of target genes in the associated function pathway, and the enrichment is the ratio of target genes belonging to all annotated genes found in the pathway. Enriched were 208 cellular components, 324 molecular activities, and 3323 biological processes. For CCs, the top 3 significant enrichment with the key targets were GO:0045121 membrane raft, GO:0098857 membrane microdomain, and GO:1902911 protein kinase complex [Fig. 7(A)]. For MFs, the top 3 significant enrichment were GO:0004713 protein tyrosine kinase activity, GO:0004674 protein serine/threonine kinase activity, and GO:0035173 histone kinase activity [Fig. 7(B)], while GO:0018108 peptidyl-tyrosine phosphorylation, GO:0018212 peptidyl-tyrosine modification, and GO:0018209 peptidyl-serine modification were the top 3 significant enrichments in BPs [Fig. 7(C)]. These mechanisms are crucial in understanding the therapeutic mechanism of SX in the treatment of PSO. The KEGG enrichment revealed how SX interacts with the pathway, performing a therapeutic effect in PSO. As shown in Fig. 7(D) and Table 2, the top 10 significant signalling pathways (p < 0.05) according to their enrichment score [-log10 (p-value)] were picked out for further analysis based on the 77 key target genes, including hsa04151 PI3K-AKT signalling pathway, hsa05205 Proteoglycans in cancer, hsa01521 EGFR tyrosine kinase inhibitor resistance as the top ones and the total number of 202 pathways were obtained. The results showed that SX may treat PSO by regulating the PI3K-AKT signalling pathway, EGFR tyrosine kinase inhibitor resistance pathway and MAPK signalling pathway.
Fig. 7.
(A) Dot-plot diagram for CCs enrichment (The pathway shorted by Fold enrichment, dots show the number of genes counts x-axis shows -logP and colour show the p-value); (B) Dot-plot diagram for MFs enrichment (The pathway shorted by Fold enrichment, dots show the number of gene count x-axis shows -logP and colour show the p-value); (C) Dot-plot diagram for BPs enrichment (The pathway shorted by Fold enrichment, dots show the number gene count, the x-axis shows -logP and colour show the p-value); (D) Dot plot diagram for KEGG Pathway enrichment analysis (The pathway shorted by Fold enrichment, dots show the number of gene count, the x-axis shows -logP and colour show the p-value).
Table 2.
Top 10 highly enriched pathways from KEGG pathways enrichment analysis with ID, P-value, genes involved, gene count.
| Pathway ID | Pathway Name | P-value | Gene IDs | Gene Count |
|---|---|---|---|---|
| hsa04151 | PI3K-Akt signalling pathway | 6.39989E-20 | MAP2K1/FLT3/FGFR1/FGF2/FGF1/IGF1R/MAPK3/CCNE1/CDK2/ERBB2/CSF1R/FLT1/KIT/PTK2/PRKCA/KDR/BCL2L1/TP53/CDK6/MAPK1/IKBKB/MTOR/GSK3B/EGFR/MDM2/AKT1/HSP90AA1/JAK2 | 28 |
| hsa05205 | Proteoglycans in cancer | 1.04844E-19 | MAP2K1/FGFR1/FGF2/PTPN6/IGF1R/MAPK3/ERBB2/PTK2/PRKCA/KDR/CBL/TP53/STAT3/MAPK1/SRC/MTOR/CASP3/PRKCB/EGFR/MDM2/MAPK14/AKT1/ESR1 | 23 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 1.22578E-19 | MAP2K1/FGF2/IGF1R/MAPK3/ERBB2/PRKCA/KDR/BCL2L1/STAT3/MAPK1/SRC/MTOR/GSK3B/PRKCB/EGFR/AKT1/JAK2 | 17 |
| hsa05215 | Prostate cancer | 1.61055E-19 | MAP2K1/FGFR1/IGF1R/MAPK3/CCNE1/CDK2/ERBB2/TP53/MAPK1/IKBKB/MTOR/GSK3B/EGFR/MDM2/AKT1/HSP90AA1/AR/EP300 | 18 |
| hsa05167 | Kaposi sarcoma-associated herpesvirus infection | 6.01722E-19 | MAP2K1/IL6ST/FGF2/PTGS2/MAPK9/MAPK3/CCR5/CASP8/MAPK8/TP53/CDK6/STAT3/MAPK1/IKBKB/SRC/MTOR/GSK3B/CASP3/MAPK14/AKT1/EP300/JAK2 | 22 |
| hsa05161 | Hepatitis B | 5.59834E-18 | MAP2K1/MAPK9/MAPK3/CCNE1/CDK2/CCNA2/PRKCA/CASP8/MAPK8/TP53/STAT3/MAPK1/IKBKB/SRC/CASP3/PRKCB/MAPK14/AKT1/EP300/JAK2 | 20 |
| hsa01522 | Endocrine resistance | 6.18228E-18 | MAP2K1/ESR2/MAPK9/IGF1R/MAPK3/ERBB2/PTK2/MAPK8/TP53/MAPK1/SRC/MTOR/EGFR/MDM2/MAPK14/AKT1/ESR1 | 17 |
| hsa04914 | Progesterone-mediated oocyte maturation | 1.26355E-17 | MAP2K1/MAPK9/IGF1R/MAPK3/CDC25B/CDC25A/CDK1/CDK2/CCNB1/CCNA2/MAPK8/PLK1/AURKA/MAPK1/MAPK14/AKT1/HSP90AA1 | 17 |
| hsa05163 | Human cytomegalovirus infection | 1.54453E-17 | PTGER4/MAP2K1/PTGS2/MAPK3/CCR5/PTK2/PRKCA/CASP8/TP53/CDK6/STAT3/MAPK1/IKBKB/SRC/MTOR/GSK3B/CASP3/PRKCB/EGFR/MDM2/MAPK14/AKT1 | 22 |
| hsa04010 | MAPK signalling pathway | 2.63204E-17 | MAP2K1/FLT3/FGFR1/FGF2/FGF1/MAPK9/IGF1R/MAPK3/CDC25B/ERBB2/CSF1R/FLT1/KIT/PRKCA/KDR/MAPK8/TP53/MAPK1/IKBKB/CASP3/PRKCB/EGFR/MAPK14/AKT1 | 24 |
3.6. SX Components-Targets-Pathways network construction
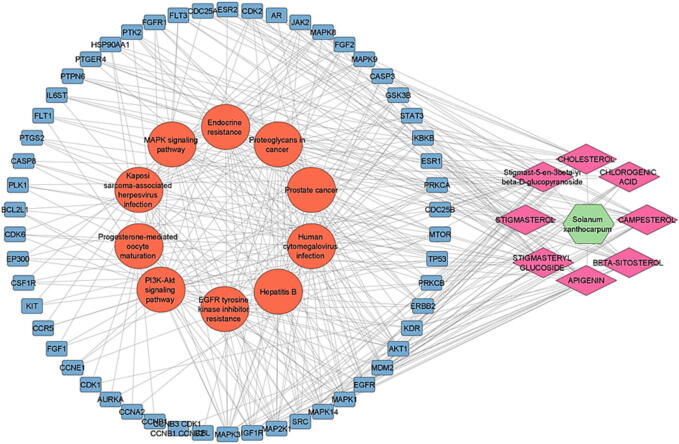
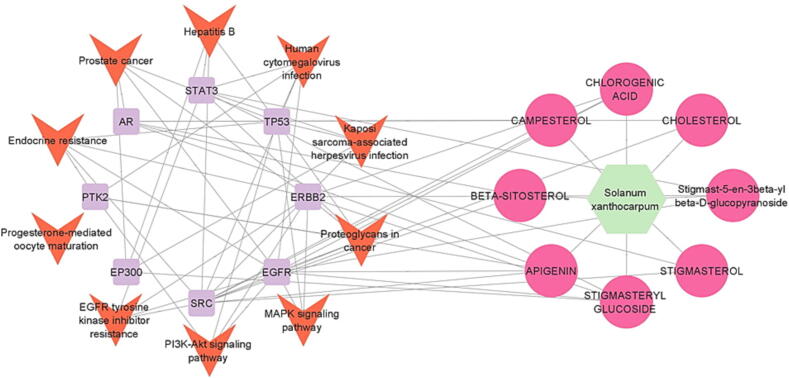
The target pathway data from KEGG analysis were imported into Cytoscape software to create a network graph of components, targets, and pathways to more easily show which component is engaged in which targets and which pathway each target is involved in. Fig. 8 depicted the SX components-targets-paths network, which included 73 nodes (1 SX, 8 components, 54 targets, and 10 pathways) and 342 edges. Multiple SX components, according to the network study, act on at least ten target genes. Fig. 9 depicted the network created by keeping the top eight targeted genes (excluding CBL and SYK since they play no role in these pathways) from the core PPI network. This network has 74 edges and 27 nodes (1 SX, 8 components, 8 targets, and 10 routes). Furthermore, the majority of the target genes were regulated by at least one active component, and 5 genes were possibly engaged in PSO-related targeted pathways. This network analysis revealed the properties of numerous SX components and targets in the treatment of PSO.
Fig. 8.
SX Components-Targets-Pathways Network (Green hexagon node shows SX, pink diamond nodes show the components, blue rectangle nodes show target genes, orange ellipse-shape nodes show pathways and edges show the interaction between two nodes).
Fig. 9.
SX Components-top 8 Targets-Pathways Network (Green hexagon node shows SX, pink circle nodes show the components, purple rectangle nodes show target genes, orange V-shape nodes show pathways and edges show the interaction between two nodes).
3.7. Molecular docking simulation of components and targets
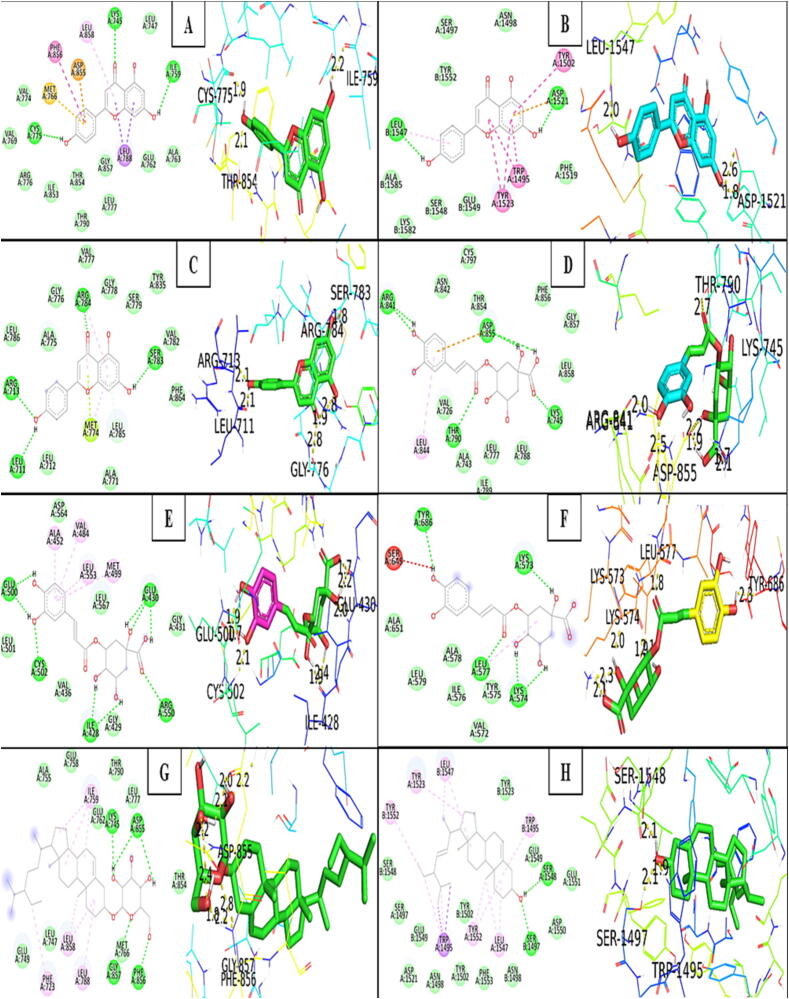
To validate the findings from network pharmacology, we employed molecular docking to evaluate screened active components and targets. The active components and key targets involved in the components-targets-pathways network analysis were selected for molecular docking. The absorption, distribution, metabolism and excretion properties of 8 selected components were determined by SwissADME (https://www.swissadme.ch/) web-based tool. Targets-related information like PDB IDs, grid box dimensions, centre coordinates and components name with their PubChem IDs are mentioned in Table 3. Table 4 contains the information on the free binding energies (kcal/mol) of key targets and active components after performing the molecular docking respectively. Molecular docking simulation results were visualized by PyMOL 2.4.0 and Discovery studio BIOVIA 2021, the amino acid residues involved in the hydrogen bond with their bond lengths in Å given in Table 5 for respective targets and active components. The stronger the bond between the ligand and receptor, the lesser the binding energy. When affinity is lesser than −4.25 kcal/mol, the ligand binds to the receptor with certainty. Strong binding activity is indicated by an affinity of less than −7.0 kcal/mol and good binding activity is indicated by an affinity of less than −5.0 kcal/mol. All the components showed good interaction of free binding energy ≤ -5.0 kcal/mol with all the key targets selected for molecular docking, except SRC. Apigenin and chlorogenic acid, surprisingly, had better binding affinities, making more hydrogen bonds to the binding pocket regions of their respective targets. Furthermore, Stigmasteryl glucoside interacted strongly with all of the targets, establishing at least one hydrogen bond to the binding pocket region except SRC. Except for STAT3 and AR, Stigmast-5-en-3beta-yl beta-D-glucopyranoside binds to the key targets into at least one hydrogen bond. In decreasing order, the best binding energy score attained by Beta-sitosterol, Stigmasterol, Campesterol, and Cholesterol to the target SRC was (-12.3, −12.58, −12.38, and −12.03) kcal/mol. Apigenin has the best binding energy score of −8.36 kcal/mol with EGFR by 3 amino acid residues bound with 3 hydrogen bonds, but it also creates 6 hydrogen bonds with 5 amino acid residues implicated by the target ERBB2 with a binding energy score of −7.54 kcal/mol. Chlorogenic acid formed the highest binding energy score of −7.98 kcal/mol with EGFR by binding 4 amino acid residues to 7 hydrogen bonds, 6 hydrogen bonds to 6 amino acid residues to the target SRC of −6.45 kcal/mol, 8 hydrogen bonds to 5 amino acid residues to the target TP53 of −6.23 kcal/mol, and 8 hydrogen bonds to 4 residues to the target PTK2 of −7.18 kcal/mol. Beta-sitosterol, Campesterol, Cholesterol, Stigmasterol, and Stigmasteryl glucoside bind to TP53 more readily than other targets due to the presence of hydrogen bonds. The data suggested that EGFR, STAT3, ERBB2, and TP53 were the major targets for achieving this effect, whereas Apigenin, Chlorogenic acid, and Stigmasteryl glucoside might be predicted as the active components of SX for PSO. In Fig. 10, the primary targets and active SX components were represented by the findings of molecular docking.
Table 3.
Information on targets and components for molecular docking.
| Target | PDB ID | Grid box(X, Y, Z–dimension) | Centre coordinates(x, y, z centre) | Component name | PubChem ID |
|---|---|---|---|---|---|
| EGFR | 8A27 | 126, 126, 126 | 10.004, −6.509, −16.145 | Apigenin | 5,280,443 |
| SRC | 2HBH | 74, 90, 68 | 2.722, 36.101, 40.392 | Beta-Sitosterol | 222,284 |
| STAT3 | 6NJS | 126, 126, 126 | 5.717, 51.338, 9.433 | Cholesterol | 5997 |
| ERBB2 | 7PCD | 126, 126, 126 | 2.245, −13.102, −16.917 | Campesterol | 173,183 |
| PTK2 | 4C7T | 70, 86, 70 | −9.472, 3.619, 6.75 | Stigmasterol | 5,280,794 |
| EP300 | 6GYT | 52, 64, 68 | –33.943, 106.875, 187.261 | Stigmast-5-en-3beta-yl beta-D-glucopyranoside | 70,699,351 |
| TP53 | 8EOM | 126, 126, 126 | 8.061, 0.0, 5.911 | Stigmasteryl glucoside | 70,699,355 |
| AR | 2PIV | 126, 126, 126 | 18.791, 5.461, 11.638 | Chlorogenic acid | 1,794,427 |
Table 4.
Molecular docking score; free binding energy (kcal/mol) of key targets and active components.
| Phytochemical name | EGFR | SRC | STAT3 | ERBB2 | PTK2 | EP300 | TP53 | AR |
|---|---|---|---|---|---|---|---|---|
| Apigenin | −8.36 | −7.53 | −6.53 | −7.54 | −7.59 | −7.09 | −8.14 | −8.03 |
| Beta-Sitosterol | −10.87 | −12.6 | −7.94 | −10.68 | −10.62 | −8.29 | 9.95 | −8.82 |
| Campesterol | −11.06 | −12.38 | −8.1 | −10.6 | −9.69 | −8.42 | −10.09 | −10.02 |
| Chlorogenic acid | −7.98 | −6.45 | −5.7 | −7.18 | −7.18 | −6.05 | −6.23 | −5.82 |
| Cholesterol | −11.11 | −12.03 | −7.75 | −10.11 | −10.16 | −8.26 | −9.61 | −8.19 |
| Stigmast-5-en-3beta-yl beta-D-glucopyranoside | −9.57 | −6.14 | −6.55 | −9.24 | −8.26 | −6.67 | −8.5 | −7.7 |
| Stigmasterol | −11.75 | −12.58 | −7.96 | −11.25 | −10.8 | −8.2 | −10.01 | −8.76 |
| Stigmasteryl glucoside | −9.17 | −4.96 | −7.54 | −8.76 | −9.15 | −6.65 | −10.51 | −6.45 |
Table 5.
Docking simulation results of key targets and active components with their residues involved in hydrogen bonding and bond length in Å.
| Target | Apigenin | Beta-Sitosterol | Campesterol | Chlorogenic acid | Cholesterol | Stigmast-5-en-3beta-yl beta-D-glucopyranoside | Stigmasterol | Stigmasteryl glucoside |
|---|---|---|---|---|---|---|---|---|
| EGFR | CYS-775: 1.9 Å; ILE-759: 2.2 Å; THR-854: 2.1 Å | GLU-758: 2.0 Å | ASP-800: 2.8 Å | LYS-745: 1.7 Å, 2.1 Å; THR-790: 2.7 Å; ARG-841: 2.0 Å, 2.5 Å; ASP-855: 1.9 Å, 2.9 Å | _ | LYS-745: 2.2 Å; ASP-855: 2.0 Å, 2.1 Å, 2.2 Å, 2.4 Å; PHE-856: 1.8 Å; GLY-857: 2.2 Å, 2.8 Å | LUE-718: 2.0 Å | PRO-794: 1.8 Å, 1.8 Å, 1.9 Å |
| SRC | THR-174: 2.5 Å; ASP-176: 1.8 Å; SER-265: 2.7 Å; SER-306: 1.7 Å | _ | _ | THR-174: 2.8 Å; ASP-176: 1.9 Å; TYR- 264: 2.1 Å; SER-265: 2.0 Å; ARG- 302: 1.9 Å; SER-303: 2.2 Å | _ | ASP-177: 2.1 Å; SER-178: 2.1 Å, 2.2 Å | SER-306: 1.7 Å | _ |
| STAT3 | SER-636: 1.9 Å; GLN-644: 2.1 Å, 2.2 Å | ASP-334: 2.1 Å | _ | LYS-573: 2.0 Å, 2.1 Å, 2.3 Å; LYS-574: 1.9 Å, 2.1 Å; LEU-577: 1.8 Å; TYR-686: 2.3 Å | ASP-334: 2.1 Å, 2.2 Å | _ | ASP-334: 3.1 Å | GLU-638: 1.8 Å |
| ERBB2 | LEU-711: 2.1 Å; ARG-713: 2.1 Å; GLY-776: 2.8 Å; SER-783: 1.8 Å; ARG-784: 1.9 Å, 2.8 Å | GLU-770: 2.1 Å | ILE-767: 2.1 Å | LYS-724: 1.9 Å; LYS-736: 1.9 Å, 1.9 Å; PRO-802: 1.8 Å, 2.0 Å | ILE-767: 1.8 Å | ASP-808: 1.8 Å, 2.1 Å, 2.6 Å | SER-783: 2.3 Å; LEU-785: 1.9 Å | LYS-724: 2.2 Å |
| PTK2 | GLU-500: 2.0 Å; CYS-502: 2.2 Å, 2.2 Å, 2.6 Å; GLU-506: 1.8 Å; ASP-564: 2.1 Å | GLU-500: 1.9 Å | ARG-500: 2.3 Å, 2.5 Å | ILE-428: 1.9 Å, 2.4 Å; GLU-430: 1.7 Å, 2.0 Å, 2.2 Å; GLU-500: 1.7 Å, 1.9 Å; CYS-502: 2.1 Å | GLU-500: 2.0 Å | ARG-426: 2.7 Å; GLN-438: 2.1 Å, 2.2 Å | GLU-500: 1.9 Å | ARG-426: 2.1 Å, 2.4 Å; GLN-438: 2.2 Å |
| EP300 | VAL-1259: 2.1 Å; GLY-1161:2.1 Å; TYR-1162:2.2 Å; ASP-1273: 2.1 Å; GLY-1274: 2.8 Å | _ | _ | ARG-1252: 2.1 Å; ASP-1273: 2.0 Å, 2.3 Å; LYS-1278: 1.7 Å, 1.9 Å | CYS-1258: 2.2 Å | LYS-1278: 2.2 Å | LYS-1278: 2.2 Å | CYS-1163: 1.9 Å, 2.5 Å |
| TP53 | ASP-1521: 1.8 Å, 2.6 Å; LEU-1547: 2.0 Å | SER-1497: 2.0 Å; SER-1548: 1.6 Å | TRP-1495: 2.3 Å; SER-1548: 2.2 Å | LYS-1563:1.7 Å; GLY-1566: 1.9 Å, 2.2 Å; GLU-1567: 2.8 Å; LEU-1568: 1.9 Å; ARG-1583: 1.9 Å, 2.5 Å, 2.6 Å | TRP-1495: 1.8 Å; SER-1497: 1.7 Å; SER-1548: 1.9 Å | TYR-1552: 1.9 Å | TRP-1497: 1.9 Å; SER-1497: 2.1 Å; SER-1548: 2.1 Å | GLU-1575: 1.9 Å, 1.9 Å, 2.1 Å, 2.2 Å, 2.2 Å, 2.3 Å, 2.8 Å |
| AR | LEU-704: 3.3 Å; ASN-705: 2.1 Å, 3.5 Å; MET-745: 2.1 Å; ARG-752: 1.7 Å; LEU-873: 2.2 Å; THR-877: 2.7 Å | _ | ASN-756: 2.3 Å | ILE-816: 1.8 Å, 1.8 Å, 2.5 Å; LYS-912: 1.8 Å, 2.6 Å; TYR-915: 2.3 Å | _ | _ | _ | GLU-793: 2.0 Å; SER-865: 2.2 Å |
Fig. 10.
Docking simulation results obtained from PyMOL for 3D images and Discovery Studio BIOVIA for 2D images); (A: EGFR- Apigenin; B: TP53- Apigenin; C: ERBB2- Apigenin; D: EGFR-Chlorogenic acid; E: PTK2- Chlorogenic acid; F: STAT3- Chlorogenic acid; G: EGFR - Stigmast-5-en-3beta-yl-beta-D-glucopyranoside; H: TP53- Stigmasterol).
4. Discussion
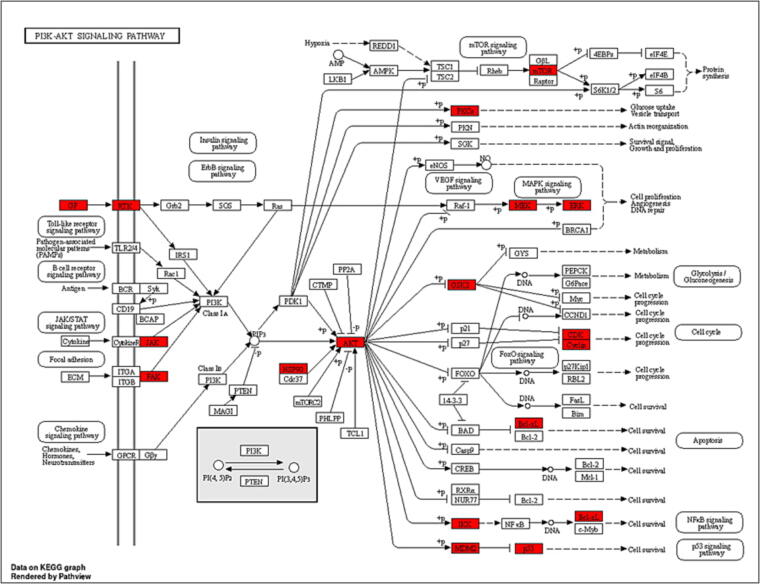
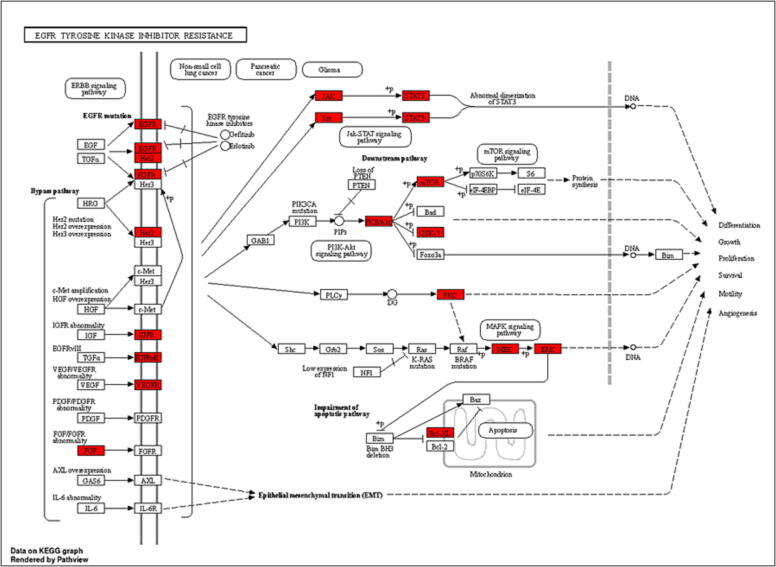
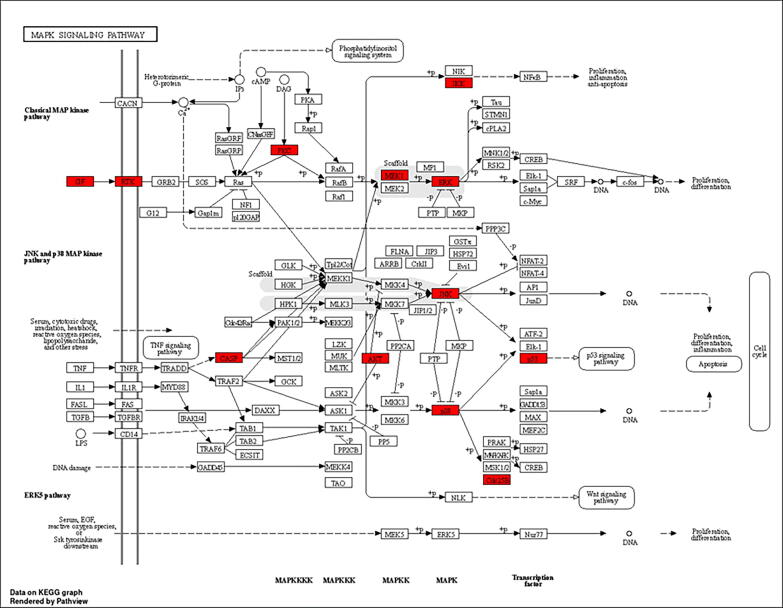
PSO is characterised by areas of thickened, inflamed, itchy, red skin that are commonly covered in silvery scales. PSO is a chronic inflammatory autoimmune skin disorder. Its etiology is genetic material, which also results in epidermal keratinocyte dysregulation and proliferation. Traditional Indian medical systems such as Ayurveda and Unani natural medicines have been utilised for disease prevention and treatment based on methodical multi-target and multi-component techniques for thousands of years (Byard et al., 2010). Ayurveda and Unani natural therapies in particular have been utilised for many years as a treatment alternative for disorders associated with PSO (Ramanunny et al., 2019). SX is frequently used to treat autoimmune and inflammatory diseases because of its potent anti-inflammatory and immunosuppressive characteristics (Joghee, 2019, Srivastava et al., 2022). SX consists of a variety of active phytochemical components that have an impact on several targets and pathways in the formation of PSO (Parmar et al., 2017). Eight active ingredients from SX were selected for this study based on the BS and DL, including Apigenin, Beta-sitosterol, Cholesterol, Campesterol, Stigmasterol, Stigmast-5-en-3-beta-yl beta-D-glucopyranoside, Stigmasteryl glucoside, and Chlorogenic acid. This work used network pharmacology and molecular docking approaches to comprehend the molecular biology of SX for the treatment of PSO. Eight SX components suggested 360 potential targets using Swiss Target Prediction, and 4297 PSO-related targets were acquired from the GeneCards and OMIM databases. The Venn diagram eliminated 179 potential SX-PSO targets. According to the components-targets-pathways network analysis, all of the phytoconstituents in SX have the potential to act on various targets and follow associated pathways for the treatment of PSO. Apigenin plays a major significance in the MAPK signalling pathway by acting on the EGFR target and downregulating keratinocyte differentiation (Yoon et al., 2023). To cure PSO, beta-sitosterol may potentially impact the EGFR target via processing through the EGFR tyrosine kinase inhibitor resistance pathway (Abbas et al., 2019). Chlorogenic acid acts on EGFR and MAPK, which was predicted by components-targets-pathways network and molecular docking study. Additionally, a study has demonstrated that chlorogenic acid can treat PSO by following MAPK and STAT3 signalling pathways to produce an antiproliferative effect in keratinocytes (Nguyen et al., 2020). In PSO, the active components in SX have well-demonstrated therapeutic benefits. PSO and other inflammatory skin disorders are characterised by EGFR and its ligands. EGFR is expressed in epithelial cells such as keratinocytes and hair follicles and is essential for healthy skin development and immune system function (Pastore et al., 2014). To perform GO enrichment and forecast the mechanism of SX in treating PSO, 179 common targets between the components of SX and PSO were chosen. These objectives included CCs, MFs, and BPs. It was discovered that the potential targets are connected to many CCs, including membrane raft, membrane microdomain, and protein kinase complex. It serves as an indirect illustration of the heterogeneity of PSO’s etiology and the harm it does to several cellular components (Balato and Gaspari, 2009, Laude and Prior, 2004, McFadden et al., 2012). SX phytoconstituents may have a role in controlling these CCs, thereby reducing the severity of PSO. Moreover, PSO is directly associated with tyrosine kinase, protein serine/threonine kinase, and histone kinase activities that are mainly enriched in MFs (Nibbering et al., 1997, Zhang and Zhang, 2019). SX phytoconstituents might play a possible role by targeting these MFs and treating PSO. Lastly, peptidyl-tyrosine phosphorylation, peptidyl-tyrosine modification, and peptidyl-serine modification are possible targets that are associated with many BPs. The majority of the BPs involve active targets such as ERBB2, PTK2, TP53, STAT3, SRC, and EGFR. As a result, the etiology and mechanism of PSO are generally compatible with the BPs of a multitude of targets. 179 common targets being used for analysing the KEGG pathways enrichment which predicts the pathways might be followed by the targets in the pathogenesis of PSO. The studies have been conducted previously on PSO show the top ten pathways obtained from the KEGG pathways enrichment analysis, the pathways like PI3K-AKT signalling pathway (Zhao et al., 2022), EGFR tyrosine kinase inhibitor resistance pathway (Furue et al., n.d.), and MAPK signalling pathway (Xu et al., 2019) are potentially involved in the pathogenesis of PSO. PI3K-AKT signalling pathway involves targets like EGFR, ERBB2, JAK and p53 (Abraham and O’Neill, 2014, Lu et al., 2008), which play a vital role in the inflammation causing PSO (Fig. 11). Then it stimulates signalling to the p53 signalling and NF-κB signalling pathway (Subhan et al., 2017). The pathophysiological function achieved by the PI3K-AKT signalling pathway mainly focuses on apoptosis, differentiation, and angiogenesis, which is eventually the reason for PSO (Karar and Maity, 2011). The ERBB2, STAT3, SRC, and EGFR targets of the EGFR tyrosine kinase inhibitor resistance pathway promote differentiation, proliferation, and angiogenesis (Fig. 12), which results in PSO by upregulating inflammatory cytokines (Furue et al., 2020). MAPK signalling pathway involves ERBB2, TP53, and EGFR, which are the reason for the development of inflammation, proliferation and differentiation, and also lead to stimulating the p53 signalling pathway (Fig. 13) (Subhan et al., 2017, Xu et al., 2019). Subsequently, molecular docking was employed to analyse eight key target proteins (EGFR, SRC, STAT3, ERBB2, PTK2, EP300, TP53, and AR) and active components. Mostly all components and targets bind with a good binding energy of less than −5 kcal/mol. EGFR, ERBB2, PTK2 and TP53 targets showed the highest number of hydrogen bond involvement the with lowest binding energy compared to all 8 targets.
Fig. 11.
KEGG pathway diagram for PI3K-AKT signalling pathway.
Fig. 12.
KEGG pathway diagram for EGFR tyrosine kinase inhibitor resistance pathway.
Fig. 13.
KEGG pathway diagram for MAPK signalling pathway.
5. Conclusion
In closing, the current study is the first to in-depth examine the pharmacological and molecular mechanism of action of SX in the treatment of PSO utilising bioinformatics tools like network pharmacology and molecular docking simulations. In all, 8 active components were detected in SX, and 800 active component targets as well as 4297 PSO-related targets were accumulated. PPI analysis screened out a total of 10 significant targets, and signalling pathways such as the PI3K-AKT signalling pathway, the EGFR tyrosine kinase inhibitor resistance pathway, and the MAPK signalling pathway were demonstrated to be crucial in the mechanism of SX for the treatment of PSO. The outcomes of network pharmacology revealed a strong connection between these pathways and PSO, whereas SX can inhibit these pathways. Moreover, the most significant target proteins were identified to be TP53, EGFR, ERBB2, PTK2, and STAT3. The multi-component and multi-pathway characteristics of phytoconstituents of SX and its mechanism of action were the main themes of the current investigation. These results are anticipated to direct the use of SX and its continued development in the management of PSO. There were still significant restrictions, though, because this study mostly depended on the network database and molecular docking. Further in-vivo and in-vitro experimental verification of the exact therapeutic action mechanism is required.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The Authors are thankful to the Researchers Supporting Project number (RSP2023R430), King Saud University, Riyadh, Saudi Arabia
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Perwez Alam, Email: aperwez@ksu.edu.sa.
Shikha Saxena, Email: skataria@amity.edu.
References
- Abbas M., Al-Rawi N., Abbas M., Al-Khateeb I. Naringenin potentiated β-sitosterol healing effect on the scratch wound assay. Res. Pharm. Sci. 2019;14:566–573. doi: 10.4103/1735-5362.272565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham A.G., O’Neill E. PI3K/Akt-mediated regulation of p53 in cancer. Biochem. Soc. Trans. 2014;42:798–803. doi: 10.1042/BST20140070. [DOI] [PubMed] [Google Scholar]
- Afolabi R., Chinedu S., Ajamma Y., Adam Y., Koenig R., Adebiyi E. Computational identification of Plasmodium falciparum RNA pseudouridylate synthase as a viable drug target, its physicochemical properties, 3D structure prediction and prediction of potential inhibitors. Infect. Genet. Evol. 2022;97 doi: 10.1016/j.meegid.2021.105194. [DOI] [PubMed] [Google Scholar]
- Ahsan M.J., Samy J.G., Khalilullah H., Nomani M.S., Saraswat P., Gaur R., Singh A. Molecular properties prediction and synthesis of novel 1,3,4-oxadiazole analogues as potent antimicrobial and antitubercular agents. Bioorg. Med. Chem. Lett. 2011;21:7246–7250. doi: 10.1016/j.bmcl.2011.10.057. [DOI] [PubMed] [Google Scholar]
- Amat-Samaranch V., Puig L. Safety of calcipotriene and betamethasone dipropionate foam for the treatment of psoriasis. Expert Opin. Drug Saf. 2020;19:423–432. doi: 10.1080/14740338.2020.1749594. [DOI] [PubMed] [Google Scholar]
- Amberger J.S., Hamosh A. Knowledgebase of human genes and genetic phenotypes. Curr. Protoc. Bioinformatics. 2018;58:1–20. doi: 10.1002/cpbi.27.Searching. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balato, A., Gaspari, A.A., 2009. NK cells and NKT cells in cutaneous disorders, Natural Killer Cells. Basic Science and Clinical Application. Elsevier Ltd. https://doi.org/10.1016/B978-0-12-370454-2.00023-5.
- Butz A.M., von Christopher S., Bartheld J.B. Getting the most out of pubchem for virtual screening HHS public access. Physiol. Behav. 2017;176:139–148. doi: 10.1080/17460441.2016.1216967.Getting. [DOI] [Google Scholar]
- Byard R.W. A review of the potential forensic significance of traditional herbal medicines. J. Forensic Sci. 2010;55:89–92. doi: 10.1111/j.1556-4029.2009.01252.x. [DOI] [PubMed] [Google Scholar]
- Chen W., Kan H., Qin M., Yang J., Tao W. Investigation of the active compounds and important pathways of Huaiqihuang granule for the treatment of immune thrombocytopenia using network pharmacology and molecular docking. Biomed Res. Int. 2023;2023:5984361. doi: 10.1155/2023/5984361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.Y., Luo J., Liu Y., Yu X.B., Liu X.Y., Tao Q.W. Network pharmacology analysis and experimental validation to investigate the mechanism of total flavonoids of rhizoma drynariae in treating rheumatoid arthritis. Drug Des. Devel. Ther. 2022;16:1743–1766. doi: 10.2147/DDDT.S354946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang K., Yang J., Zhang A., Dong X., Zhou Z., Li T., Fan R. Mechanism of ferroptosis in hypertensive nephropathy. Transl. Androl. Urol. 2022;11(5):617–626. doi: 10.21037/tau-22-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophers E., van de Kerkhof P.C.M. Severity, heterogeneity and systemic inflammation in psoriasis. J. Eur. Acad. Dermatol. Venereol. 2019;33(4):643–647. doi: 10.1111/jdv.15339. [DOI] [PubMed] [Google Scholar]
- Cui Y., Wang H., Wang D., Mi J., Chen G., Li F., Wang Y., Zhang Y. Network pharmacology analysis on the mechanism of Huangqi Sijunzi decoction in treating cancer-related fatigue. J. Healthcare Eng. 2021;18:9780677. doi: 10.1155/2021/9780677. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Daina A., Michielin O., Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357–W3664. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncheva N.T., Morris J.H., Gorodkin J., Jensen L.J. Cytoscape StringApp: network analysis and visualization of proteomics data. J. Proteome Res. 2019;18:623–632. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furue K., Ito T., Tanaka Y., Hashimoto-Hachiya A., Takemura M., Murata M., Kido-Nakahara M., Tsuji G., Nakahara T., Furue M. The EGFR-ERK/JNK-CCL20 pathway in scratched keratinocytes may underpin koebnerization in psoriasis patients. Int. J. Mol. Sci. 2020;21(2):434. doi: 10.3390/ijms21020434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin K.Y., Ghosh S., Kitano H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS One. 2013;8(12):e83922. doi: 10.1371/journal.pone.0083922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-García B., Teixeira J.M.C., Trellet M., Rodrigues J.P.G.L.M., Bonvin A.M.J.J. PDB-tools web: a user-friendly interface for the manipulation of PDB files. Proteins. 2021;89(3):330–335. doi: 10.1002/prot.26018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D., An X., Zhang Y., Zhao S., Duan L., Duan Y., Lian F., Tong X. Potential mechanism prediction of herbal medicine for pulmonary fibrosis associated with SARS-CoV-2 infection based on network analysis and molecular docking. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.602218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joghee S. Solanum Xanthocarpum: a review. IPCM. 2019;3:1–7. doi: 10.23880/ipcm-16000177. [DOI] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022;31:47–53. doi: 10.1002/pro.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karar J., Maity A. PI3K/AKT/mTOR pathway in angiogenesis. Front. Mol. Neurosci. 2011;4:51. doi: 10.3389/fnmol.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., Zaslavsky L., Zhang J., Bolton E.E. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude A.J., Prior I.A. Plasma membrane microdomains: organization, function and trafficking (Review) Mol. Membr. Biol. 2004;21:193–205. doi: 10.1080/09687680410001700517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Zhou J., Xu C., Lin H., Xiao J., Wang Z., Yang B. JAK/STAT and PI3K/AKT pathways form a mutual transactivation loop and afford resistance to oxidative stress-induced apoptosis in cardiomyocytes. Cell. Physiol. Biochem. 2008;21:305–314. doi: 10.1159/000129389. [DOI] [PubMed] [Google Scholar]
- Martins A.M., Ascenso A., Ribeiro H.M., Marto J. Current and future therapies for psoriasis with a focus on serotonergic drugs. Mol. Neurobiol. 2020;57:2391–2419. doi: 10.1007/s12035-020-01889-3. [DOI] [PubMed] [Google Scholar]
- McFadden J., Fry L., Powles A.V., Kimber I. Concepts in psoriasis: psoriasis and the extracellular matrix. Br. J. Dermatol. 2012;167:980–986. doi: 10.1111/j.1365-2133.2012.11149.x. [DOI] [PubMed] [Google Scholar]
- Mohanraj K., Karthikeyan B.S., Vivek-Ananth R.P., Chand R.P.B., Aparna S.R., Mangalapandi P., Samal A. IMPPAT: a curated database of Indian medicinal plants. Phytochem. Therapeutics. Sci. Rep. 2018;8(1):4329. doi: 10.1038/s41598-018-22631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers B.H.M. Shortcuts for faster image creation in PyMOL. Protein Sci. 2020;29:268–276. doi: 10.1002/pro.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali M., Nair B., Vishnu V.R., Aneesh T.P., Nath L.R. 2,4-Dihydroxycinnamic acid as spike ACE2 inhibitor and apigenin as RdRp inhibitor in Nimbamritadi Panchatiktam Kashayam against COVID-19: an in silico and in vitro approach. Mol. Divers. 2022;2022:1–11. doi: 10.1007/s11030-022-10552-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen U.T., Nguyen L.T.H., Kim B.A., Choi M.J., Yang I.J., Shin H.M. Natural compound mixture, containing Emodin, Genipin, Chlorogenic Acid, Cimigenoside, and Ginsenoside Rb1, ameliorates psoriasis-like skin lesions by suppressing inflammation and proliferation in keratinocytes. Evid. Based Complement. Alternat. Med. 2020;2020:9416962. doi: 10.1155/2020/9416962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibbering P.H., Thio B., Bezemer A.C., Beijersbergen R.L., Zomerdijk T.P. Intracellular signalling by binding sites for the antipsoriatic agent monomethylfumarate on human granulocytes. Br. J. Dermatol. 1997;137:65–75. [PubMed] [Google Scholar]
- Parmar K.M., Itankar P.R., Joshi A., Prasad S.K. Anti-psoriatic potential of Solanum xanthocarpum stem in Imiquimod-induced psoriatic mice model. J. Ethnopharmacol. 2017;198:158–166. doi: 10.1016/j.jep.2016.12.046. [DOI] [PubMed] [Google Scholar]
- Pastore S., Lulli D., Girolomoni G. Epidermal growth factor receptor signalling in keratinocyte biology: Implications for skin toxicity of tyrosine kinase inhibitors. Arch. Toxikol. 2014;88:1189–1203. doi: 10.1007/s00204-014-1244-4. [DOI] [PubMed] [Google Scholar]
- Patwardhan B., Warude D., Pushpangadan P., Bhatt N. Ayurveda and traditional Chinese medicine: a comparative overview. Evid. Based Complement. Alternat. Med. 2005;2:465–473. doi: 10.1093/ecam/neh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanunny A.K., Wadhwa S., Singh S.K., Sharma D.S., Khursheed R., Awasthi A. Treatment strategies against psoriasis: principle, perspectives and practices. Curr. Drug Deliv. 2019;17:52–73. doi: 10.2174/1567201816666191120120551. [DOI] [PubMed] [Google Scholar]
- Ranjith D., Ravikumar C. SwissADME predictions of pharmacokinetics and drug-likeness properties of small molecules present in Ipomoea mauritiana Jacq. J. Pharmacogn. Phytochem. 2019;8:2063–2073. [Google Scholar]
- Ru J., Li P., Wang J., Zhou W., Li B., Huang C., Li P., Guo Z., Tao W., Yang Y., Xu X., Li Y., Wang Y., Yang L. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K., Sanders K.M., Youssef M.R., Yanushefski K.M., Jensen L., Yosipovitch G., Akiyama T. Mouse model of imiquimod-induced psoriatic itch. Pain. 2016;157:2536–2543. doi: 10.1097/j.pain.0000000000000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Lu J., Wang G., Wang C., Li Z. Molecular mechanism of YuPingFeng in the treatment of asthma based on network pharmacology and molecular docking technology. Comput. Math. Methods Med. 2022;2022:7364126. doi: 10.1155/2022/7364126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V., Navabharath M., Gupta S., Singh S.V., Ahmad S. Exploration of Solanum xanthocarpum Schrad. & Wendl. against Mycobacterium avium Subspecies paratuberculosis and assessment of its immunomodulatory and anti-inflammatory potential. Pharmaceuticals (Basel) 2022;15(11):1367. doi: 10.3390/ph15111367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subhan F., Kang H.Y., Lim Y., Ikram M., Baek S.Y., Jin S., Jeong Y.H., Kwak J.Y., Yoon S. Fish scale collagen peptides protect against CoCl2/TNF- α-Induced cytotoxicity and inflammation via inhibition of ROS, MAPK, and NF-κ B pathways in HaCaT cells. Oxid. Med. Cell. Longev. 2017;2017:9703609. doi: 10.1155/2017/9703609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Li M., Wang J., Pan Y., Wu F.X. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yuan Y., Wang W., He Y., Zhong H., Zhou X., Chen Y., Cai X.J., Liu L. Mechanisms underlying the therapeutic effects of Qingfeiyin in treating acute lung injury based on GEO datasets, network pharmacology and molecular docking. Comput. Biol. Med. 2022;145 doi: 10.1016/j.compbiomed.2022.105454. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhang H.M., Guo Y.R., Li L.L. Molecular mechanisms of Biyu decoction as treatment for psoriasis: a network pharmacology and molecular docking study. World J. Clin. Cases. 2022;10:7224–7241. doi: 10.12998/wjcc.v10.i21.7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M., Liu D., Liu H., Zhao J., Tang C., Chen G., Liu Y., Liu H. Based on network pharmacology tools to investigate the mechanism of tripterygium wilfordii against IgA nephropathy. Front Med (Lausanne) 2021;8 doi: 10.3389/fmed.2021.794962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Xu J., Xiong X., Deng Y. Salidroside inhibits MAPK, NF-κB, and STAT3 pathways in psoriasis-associated oxidative stress via SIRT1 activation. Redox Rep. 2019;24:70–74. doi: 10.1080/13510002.2019.1658377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.H., Kim M.Y., Cho J.Y. Apigenin: a therapeutic agent for treatment of skin inflammatory diseases and cancer. Int. J. Mol. Sci. 2023;24(2):1498. doi: 10.3390/ijms24021498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhang X. The role of PI3K/AKT/FOXO signaling in psoriasis. Arch. Dermatol. Res. 2019;311:83–91. doi: 10.1007/s00403-018-1879-8. [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhou Y., Ma Z. Multi-target mechanism of Tripteryguim wilfordii Hook for treatment of ankylosing spondylitis based on network pharmacology and molecular docking. Ann. Med. 2021;53:1090–1098. doi: 10.1080/07853890.2021.1918345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Xie Y., Li X., Song J., Guo M., Xian D., Zhong J. The protective effect of proanthocyanidins on the psoriasis-like cell models via PI3K/AKT and HO-1. Redox Rep. 2022;27:200–211. doi: 10.1080/13510002.2022.2123841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]