Abstract
Purpose of Review
Clonal hematopoiesis (CH) is a prevalent condition that results from the acquisition of somatic mutations in hematopoietic stem cells. When these mutations occur in “driver” genes, they can potentially confer fitness advantages to the cell, leading to a clonal expansion. While most clonal expansions of mutant cells are generally considered to be asymptomatic since they do not impact overall blood cell numbers, CH carriers display long-term risks of all-cause mortality and age-associated diseases including cardiovascular disease (CVD). This review summarizes recent findings in CH related to aging, atherosclerotic CVD, and inflammation, emphasizing epidemiological and mechanistic studies, and potential therapeutic options to treat CVDs that are promoted by CH.
Recent Findings
Epidemiological studies have revealed associations between CH and CVDs. Experimental studies with CH models employing the Tet2- and Jak2-mutant mouse lines display inflammasome activation and a chronic inflammatory state that leads to accelerated atherosclerotic lesion growth.
Summary
A body of evidence suggests that CH represents a new causal risk factor for CVD. Studies also indicate that understanding an individual’s CH status could provide guidance for personalized approaches to treat atherosclerosis and other CVDs with anti-inflammatory drugs.
Keywords: Age-related clonal hematopoiesis, Clonal hematopoiesis of indeterminate potential, CANTOS, Inflammasome
Introduction
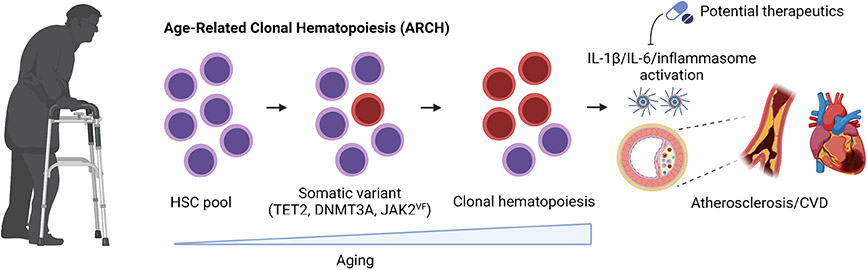
Multiple lineages of blood cells are produced by hematopoietic stem cells (HSCs) in the bone marrow. Over time, HSCs randomly acquire somatic mutations and thus become increasingly genetically distinct as the organism ages. While most of these mutations produce few if any biological effects, some can alter cell fate decisions toward self-renewal, decreased cell death or differentiation, or some combination of these fates, leading to their positive selection through a process referred to as clonal hematopoiesis (CH) (Fig. 1). These somatic mutations are also passed to progeny leukocytes, and can alter the inflammatory properties of immune cells and contribute to disease states [1].
Fig. 1.
Scheme of age-related clonal hematopoiesis. As individuals age, their hematopoietic stem cells can acquire somatic mutations in their hematopoietic stem cells that confer a fitness advantage in the bone marrow niche. As a result of this process, clones harboring mutations in TET2, DNMT3A, JAK2, or other driver genes increasingly expand, promoting the risk of all-cause mortality, cardiovascular disease, and less often hematological malignancies. Created with BioRender.com
Age-Related Clonal Hematopoiesis
The development of CH is best appreciated in the context of aging, and hence it can be referred to as age-related CH (ARCH). Alternatively, this phenomenon has been referred to as clonal hematopoiesis of indeterminate potential (CHIP). As individuals age, they will inevitably accumulate somatic mutations that include (1) single nucleotide variants (SNVs), including the spontaneous cytosine deamination that is a mutational signature of aging; (2) small insertions and deletions (indels); and (3) chromosomal alterations that include large chromosome insertions and deletions, inversions, loss of heterozygosity, and aneuploidy [2, 3]. While CH will increase the risk of hematological neoplasia, in most cases this condition does not progress to malignancy. CH is distinguished from hematological malignancies because it is believed that the clones are composed of fewer driver gene mutations, and the individual does not present with cytopenia, dysplasia, and abnormal blood cell counts [4]. CH is receiving increasing scientific interest because it has been recognized that this condition can produce physiological consequences beyond a neoplastic phenotype. In particular, the clonal expansions of blood cells can increase the risk of cardiovascular disease (CVD) and other non-neoplastic disorders.
Epidemiology: CH, Mortality, and CVD
Busque et al. [5••] reported the first association between CH and mutations in the driver gene ten-eleven translocation 2 (TET2). Later studies detected candidate driver gene clones by employing large-scale, next-generation sequencing (NGS) on peripheral blood cells from individuals with no hematological disorders [6••, 7••]. These studies reported common features including a strong association between age and the prevalence of CH. Both studies also reported clonal expansions predominantly associated with mutations in epigenetic regulator genes, specifically DNA methyltransferase three alpha (DNMT3A), TET2, and additional sex combs-like 1 (ASXL1). To a lesser extent, distinct clones also exhibited mutations in Janus kinase 2 (JAK2), tumor protein 53 (TP53), protein phosphatase, Mg2+/Mn2+-dependent 1D (PPM1D), Splicing Factor 3b Subunit 1 (SF3B1), and other genes. Based on low-coverage DNA sequence analysis, Jaiswal et al. [7••] reported that individuals under 40 years of age rarely exhibited CH, but showed an increase in CH frequency with age. A commonly quoted finding from this study is that clones arising from mutations in CH driver genes occurred in approximately 10% of individuals older than 70 years. However, studies employing different methodology for clone identification reveal that this underestimates the extent of CH, and this will be discussed below. As noted previously, Genovese et al. [6••] also reported the age-dependent prevalence of hematopoietic clones and similar frequencies of mutated driver genes. Notably, this study assessed the full exome rather than candidate driver genes, and they reported that approximately half of the clonal events found in blood could be attributed to mutations in known driver genes.
Since these initial reports, other methods of analysis have been applied to assess clonality in the blood. An early study by Zink et al. [8••] employed a non-biased, barcoded whole-genome DNA sequence analysis. They could detect clonal events in 50% of individuals by 85 years of age. These authors also noted that clonal hematopoiesis was probably inevitable with advanced age and that less than 15% of the clonal events could be attributed to the known clonal hematopoiesis driver genes identified previously [6••, 7••]. Others have reached similar conclusions [9-12]. In addition to these nonbiased analyses of clonal hematopoiesis, error-corrected ultra-deep DNA sequencing of a subset of driver gene candidates has revealed that small variant allele fraction (VAF) clones are essentially ubiquitous by middle age [13, 14].
Loh et al. [2] has examined clonal events in blood that are associated with age-related mosaic chromosomal alterations (mCAs). In a large cohort of individuals, mCAs occurred at a frequency of 5.6%, where most of the detected alterations are gains, deletions, or copy-number-neutral loss of heterozygosity. Similar to analyses of SNVs and small indels, the frequency of mCA-derived clones was found increased with age. Terao et al. [15] also found specific patterns of genomic mutations and clonal selection in hematopoietic cells in an analysis of autosomal mCAs in a Japanese cohort. In this study, mCAs were detected in more than 35% of individuals older than 90 years. Finally, mosaic loss of the Y chromosome (mLOY), another form of CH, represents the most prevalent human post-zygotic mutation. Like SNVs, indels, and other mCAs, the prevalence of mLOY is strongly associated with age, and mLOY can be detected in 40% of men who are 70 years of age [16].
Many studies have associated CH with mortality. In earlier studies, Genovese et al. [6••] and Jaiswal et al. [7••] found that CH carriers exhibited significantly reduced overall survival during a follow-up period. Zink et al. [8••] also reported that CH carriers had substantially higher rates of all-cause mortality. Notably, both Zink et al. and Genovese et al. reported increased mortality associated with CH regardless of whether the clone could be attributed to a known candidate driver gene or was the result of an unknown mechanism. The association between CH and mortality was further observed in carriers of large mCAs. Loh et al. [2] showed that those with an mCA had a 2-fold increased risk for all-cause mortality. Supporting these findings, analysis of the Japanese cohort also revealed that mCAs contribute to an increase in the risk of death [15]. Finally, mLOY has also been associated with an increased risk of all-cause mortality [17-19••].
What is driving the accelerated mortality in individuals with CH? An obvious explanation would be an increase in death from hematologic malignancies, as this would make sense from the standpoint that CH is an early step in the progression to blood cancer. While the incidence of blood cancer is elevated in individuals with CH [2, 6••, 7••], deaths from hematologic malignancies cannot explain these large increases in mortality associated with CH. Instead, it appears that increased CVD-associated death is a major factor in accounting for the increased mortality in CH carriers. This was first noted by a secondary analysis performed by Jaiswal et al. [7••] where it was found that CH carriers had a higher risk of coronary heart disease and ischemic stroke. Following this initial study, the same group performed prospective and retrospective studies to illustrate the association between CH and CVD [20•]. It was found that CH carriers had 1.9-fold increased risk of coronary heart disease and a 4.0-fold increased risk of myocardial infarction. In a separate cohort, coronary-artery calcification was 3 times higher in CH carriers than in non-carriers [20•]. They also reported that the coronary-artery calcification score was 12 times higher in carriers of large CH clones (VAF ≥ 10%), whereas carriers of small CH clones showed no difference [20•]. A large cohort study of the UK Biobank has provided further evidence for the association between CH and the increased risk of CVD, and this study also reported that large clone size was associated with greater CVD risk [21]. Finally, it has been reported that there is an association between CH and incident coronary artery disease events in postmenopausal middle-aged women [22].
In addition to the studies mentioned above, it has been reported that carriers of DNMT3A and TET2 who undergo aortic valve implantation demonstrate an increase in mortality during follow-up [23]. Yu et al. [24] examined whether CH predisposes to hospitalized HF incidence in a large population. They found that mutations in TET2, JAK2, and ASXL1 were strongly associated with an increased risk of HF, whereas DNMT3A sequence variants were not. They also found a strong association between ASXL1-mediated CH and reduced left ventricular ejection fraction. In HF patients, CH was found to be associated with morbidity and mortality combined with rehospitalization [25•-27]. Dorsheimer et al. [27] analyzed bone marrow and peripheral blood mononuclear cells of 419 patients with chronic ischemic HF using deep error-corrected sequencing. A total of 223 mutations were found in 154 of the 419 patients with a 0.5% VAF detection threshold. As expected, most mutations occurred in the DNMT3A and TET2 genes, and the carriers of these mutant clones had worse prognosis than the non-carriers. In a subsequent study, this group analyzed the VAF threshold for prognostic significance of CH in HF [25•]. They determined a VAF cut-off value of 0.73% for TET2 and 1.15% for DNMT3A CH mutations. Pascual-Figal et al. [28] have extended these studies and reported that DNMT3A- and TET2-mediated CH contributes to HF progression, independently of its ischemic or nonischemic etiology.
In view of findings discussed above, it should be emphasized that multiple approaches are employed to assess CH in patient populations. On one hand, some investigations have employed relatively shallow, whole-exome sequence analysis to assess clonal expansions in blood. The advantage of this approach is that it is relatively unbiased and it can employ existing DNA sequencing data from large biobanks (such as the UK Biobank). A disadvantage of this approach is the low sensitivity of variant detection such that only very large clonal events can be detected. On the other hand, some investigations have employed ultra-deep, error-corrected sequencing on relatively small, but more highly phenotyped, cohorts [29]. The advantage of this approach is that it can detect associations between smaller clones and disease conditions. The disadvantage with this approach is that the analysis is biased because it only focuses on a panel of pre-selected, candidate driver genes to contain costs.
Mechanistic Studies on CH and Atherosclerosis
Epidemiological studies have identified associations between CH and multiple disease processes [30]. However, these studies are generally descriptive in nature and they do not provide information about cause-and-effect relationships or the directionality of the association. Thus, to understand the causal and mechanistic relationships between CH and CVD in more detail, we and others have developed several mouse models of CH that recapitulate human CH [31-36]. Details on these mouse models have been presented elsewhere [37-39].
Tet2 and Atherosclerosis
To study TET2-mediated CH in a CVD model, our group used a competitive bone marrow transplantation (BMT) to mimic the hematopoietic clone expansion event in humans. Atherosclerosis-prone low-density lipoprotein receptor-deficient mice were engrafted with 10% Tet2+/+ or Tet2−/− bone marrow cells and treated with a high-cholesterol, Western diet. Over time, the Tet2−/− cells underwent expansion, with a slight myeloid bias, in bone marrow, spleen, peripheral blood, and the infiltrating pool of immune cells of the aorta [32••]. These conditions promoted atherosclerosis progression and increased plaque size. Mechanistically, it was found that Tet2-deficient macrophages display upregulated expression of IL-1β, its precursor pro-IL-1β, and components of NLRP3 inflammasome [32••]. Furthermore, treatment with an NLRP3 inhibitor reduced plaque size, suggesting that an inflammasome product, such as IL-1β, plays a crucial role in the Tet2-mediated acceleration of atherogenesis (Fig. 1). Subsequently, it was reported that a conventional BMT method, i.e., the transplantation of 100% Tet2+/+ or Tet2−/− mouse bone marrow cells, also led to an increase in atherosclerotic plaque size [20•]. While this model was confounded by the systemic effects of a complete BMT of Tet2−/− cells, this study provided further support for the notion that TET2-mediated CH accelerates atherosclerosis.
JAK2V617F and atherosclerosis
The JAK2V617F variant is a gain-of-function mutation that constitutively activates the STAT family of transcription factors. JAK2V617F was originally identified as a myeloproliferative neoplasm (MPN)–associated mutation, but it is increasingly recognized that hematopoietic cell clones with this mutation are also detected in non-symptomatic individuals with few if any indications of MPN [6••, 7••, 40]. Fidler et al. [31] utilized Cre/LoxP-mediated recombination and a BM transplantation strategy to achieve Jak2V617F expression specifically in myeloid cells to avoid the MPN phenotype. They found that hyperlipidemic recipient mice reconstituted with macrophage specific-Jak2V617F donor cells exhibited increased necrotic core sizes, despite lower levels of circulating LDL cholesterol. They attributed the enhanced atherogenesis to greater DNA oxidative damage, replication stress, AIM2 inflammasome activation, and IL-1β production [31].
Mechanistic Studies on CH and Other Types of CVD
The mechanistic relationships between CH and other CVD entities have also been examined by experimental studies in mice. Models of TET2, DNMT3A, PPM1D, and JAK2V617F CH have been found to promote pathological remodeling in models of HF that are induced by LAD ligation, angiotensin II infusion, and/or transverse aortic constriction [33-35, 39, 41•, 42•]. Furthermore, the adoptive transfer model of TET2-mediated CH in non-conditioned mice has been shown to accelerate age-associated cardiomyopathy [36], and the adoptive transfer model of TP53-mediated CH has been shown to promote anthracycline-induced cardiomyopathy [34]. In a model of TET2-mediated CH, increased insulin resistance was observed in aging mice and in young mice fed a high calorie diet [43]. In contrast to these findings with traditional driver gene-mediated clonal hematopoiesis, it has been reported that mLOY promotes profibrotic signaling and diminishes IL-1β-signaling [19••], indicating the complex actions of somatic mutations on immune cell function.
CH and Inflammation
Experimental work on several different CH driver genes in multiple murine models of CVD have led to the general conclusion that CH promotes CVD risk through mechanisms involving inflammation. Notably, a number of these mechanistic studies have implicated the NLRP3 inflammasome in many of these processes [32••, 41•-43]. For example, in multiple CVD models in mice, we have shown that treatment with an inflammasome inhibitor MCC950 counteracted the effect of CH on atherosclerosis, HF, and insulin resistance [32••, 41•-43]. Consistent with the experimental work, studies have reported that CH carriers also exhibit elevated inflammatory markers [21, 44, 45]. For example, single-cell RNA sequencing analyses of circulating peripheral monocytes revealed that individuals with aortic valve stenosis who carried sequence variations in the DNMT3A driver gene displayed increased expression of IL-6, CXCL2, IL-1β, TNF, NLRP3, and genes involved in T-cell stimulation [44].
The Canakinumab Anti-inflammatory Thrombosis Outcomes Study (CANTOS) trial tested the hypothesis that blockade of IL-1β using a specific monoclonal antibody reduces the rate of major adverse cardiovascular events (MACEs) in patients with a previous myocardial infarction and evidenced for elevated systemic inflammation (Fig. 1) [46]. This large trial found that the middle dose of canakinumab led to a statistically significant 15% reduction in MACE. A subsequent exploratory analysis assessed the association of clinical outcome with CH in a subset of this cohort [47••]. Whereas individuals that harbored no detectable CH exhibited a 7% relative risk reduction of MACE in response to any dose of canakinumab, those with TET2-mediated CH displayed a 62% relative risk reduction in MACE. These findings are consistent with experimental studies of CVD showing that mice are superior responders to an inflammasome inhibitor if they also harbor an expansion of Tet2-deficient cells in the hematopoietic system [32••, 42•]. The beneficial effects among the population with a specific form of CH suggest an opportunity to implement personalized medicine based on the identity of mutations that have been acquired in the somatic cells of an individual.
Mosaic Loss of the Y Chromosome
Recent studies have provided evidence that hematopoietic mLOY in men can be causally linked to CVD. This condition is remarkably prevalent, affecting a larger portion of the population than CH mediated by any individual ARCH/CHIP driver gene or the combined prevalence of all known driver genes associated with ARCH/CHIP [30]. Sano et al. [19••] have reported that mLOY is associated with cardiovascular disease and heart failure-associated mortality in the UK Biobank and that mice reconstituted with bone marrow lacking the Y chromosome display accelerated cardiac dysfunction and other pathologies. In contrast to the pro-inflammation paradigm that has been developed from studies of ARCH/CHIP [48], mLOY leads to an over-activation of pro-fibrotic signaling in the immune system and a reduction in pro-inflammatory signaling. These data suggest that the age-associated somatic mosaicism that develops in the immune system can give rise to a complex interplay of pro- and anti-inflammatory processes. Collectively, these data indicate new avenues for experimental and clinical CH research.
Limitations of CH Research
It should be noted that studies of CH currently suffer from numerous limitations. First, it should be recognized that there is a high degree of observational bias in the epidemiological analyses. This bias largely results from an incomplete knowledge of the driver gene repertoire that give rise to these clonal events. Second, there is an overfocus on the large clones and insufficient appreciation of the potential cumulative effect of many smaller clones that are difficult to accurately detect [11, 26, 49]. Related to this point, there are technical limitations due to the high degree of error associated with base calling with current NGS procedures, masking the ability to detect small clones [13, 14]. Finally, most clinical analyses treat CH as a binary readout. Treating CH as a simple binary readout does not account for the distinct mechanistic features of the different driver genes. For example, some clones could be derived from driver genes that are highly pathological, whereas other driver gene clones might be benign, or even protective, as has been reported in other systems [50-52]. Thus, current conclusions about the association of CH with CVD are likely based on analyses of data that are rife with false-positive as well as false-negative signals, suggesting that there is great need for improving these analyses through a deeper understanding of the biology of CH.
Concluding Remarks
CH is increasingly recognized as a causal risk factor for CVD. CH results from the age-dependent acquisition of mutations in somatic cells, and it represents a new mechanism of CVD that occurs at the interface of aging and cancer (Fig. 1). Assessing CH in patient populations will likely provide opportunities for improving prognosis and guiding interventions. While CH can be determined by the analysis of a blood sample, currently this assessment is technically difficult and expensive. However, a better understanding of the pathophysiology of CH and technical improvements in DNA sequence analysis could make the analysis of CH a routine procedure by a clinical laboratory.
Regardless of the limitations discussed in the section above, considerable progress has been made in documenting the utility of CH measurements in assessing CVD risk and patient prognosis. Notably, the exploratory analysis of CH in the CANTOS trial provides a compelling indication that the analysis of CH could offer a new avenue in the development of personalized medicine approaches where therapies target the effects of specific somatic mutations [47••]. Thus, it is reasonable to expect that the associations between CH and disease prognosis will strengthen considerably with advances in DNA sequence methodology and a better understanding of how this condition mechanistically contributes to CVD.
Funding
This work was supported by US National Institutes of Health grants to KW (AG073249, AG072095, HL141256, HL139819, and HL142650).
Footnotes
Conflict of Interest The authors declare that they have no financial interests.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Evans MA, Sano S, Walsh K. Cardiovascular disease, aging, and clonal hematopoiesis. Annu Rev Pathol. 2020;15:419–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loh PR, Genovese G, Handsaker RE, Finucane HK, Reshef YA, Palamara PF, et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature. 2018;559:350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559:400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.••. Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–81 This work reported the first association between CH and TET2.
- 6.••. Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87 This paper showed the relationship between aging, CH, and mortality.
- 7.••. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98 This paper showed the relationship between aging, CH, and mortality.
- 8.••. Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130:742–52 This work used a non-biased DNA analysis to identify clonal expansions in the hematopoietic system.
- 9.Fabre MA, de Almeida JG, Fiorillo E, Mitchell E, Damaskou A, Rak J, et al. The longitudinal dynamics and natural history of clonal haematopoiesis. Nature. 2022;606:335–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKerrell T, Park N, Moreno T, Grove CS, Ponstingl H, Stephens J, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10:1239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell E, Spencer Chapman M, Williams N, Dawson KJ, Mende N, Calderbank EF, et al. Clonal dynamics of haematopoiesis across the human lifespan. Nature. 2022;606:343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poon GYP, Watson CJ, Fisher DS, Blundell JR. Synonymous mutations reveal genome-wide levels of positive selection in healthy tissues. Nat Genet. 2021;53:1597–605. [DOI] [PubMed] [Google Scholar]
- 13.Watson CJ, Papula AL, Poon GYP, Wong WH, Young AL, Druley TE, et al. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science. 2020;367:1449–54. [DOI] [PubMed] [Google Scholar]
- 14.Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7:12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terao C, Suzuki A, Momozawa Y, Akiyama M, Ishigaki K, Yamamoto K, et al. Chromosomal alterations among age-related haematopoietic clones in Japan. Nature. 2020;584:130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson DJ, Genovese G, Halvardson J, Ulirsch JC, Wright DJ, Terao C, et al. Genetic predisposition to mosaic Y chromosome loss in blood. Nature. 2019;575:652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsberg LA, Rasi C, Malmqvist N, Davies H, Pasupulati S, Pakalapati G, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet. 2014;46:624–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loftfield E, Zhou W, Graubard BI, Yeager M, Chanock SJ, Freedman ND, et al. Predictors of mosaic chromosome Y loss and associations with mortality in the UK Biobank. Sci Rep. 2018;8:12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.••. Sano S, Horitani K, Ogawa H, Halvardson J, Chavkin NW, Wang Y, et al. Hematopoietic loss of Y chromosome leads to cardiac fibrosis and heart failure mortality. Science. 2022;377:292–7 This experimental study revealed direct effects of losing the male chromosome on heart failure and mortality.
- 20.•. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–21 Evidence of the relationship between CH and CVD in mice models and humans.
- 21.Bick AG, Pirruccello JP, Griffin GK, Gupta N, Gabriel S, Saleheen D, et al. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation. 2020;141:124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honigberg MC, Zekavat SM, Niroula A, Griffin GK, Bick AG, Pirruccello JP, et al. Premature menopause, clonal hematopoiesis, and coronary artery disease in postmenopausal women. Circulation. 2021;143:410–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mas-Peiro S, Hoffmann J, Fichtlscherer S, Dorsheimer L, Rieger MA, Dimmeler S, et al. Clonal haematopoiesis in patients with degenerative aortic valve stenosis undergoing transcatheter aortic valve implantation. Eur Heart J. 2020;41:933–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu B, Roberts MB, Raffield LM, Zekavat SM, Nguyen NQH, Biggs ML, et al. Supplemental association of clonal hematopoiesis with incident heart failure. J Am Coll Cardiol. 2021;78:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.•. Assmus B, Cremer S, Kirschbaum K, Culmann D, Kiefer K, Dorsheimer L, et al. Clonal haematopoiesis in chronic ischaemic heart failure: prognostic role of clone size for DNMT3A- and TET2-driver gene mutations. Eur Heart J. 2021;42:257–65 This study showed optimized VAF cut-off levels in clone size related to heart failure 5-year mortality incidence.
- 26.Cremer S, Kirschbaum K, Berkowitsch A, John D, Kiefer K, Dorsheimer L, et al. Multiple somatic mutations for clonal hematopoiesis are associated with increased mortality in patients with chronic heart failure. Circ Genom Precis Med. 2020;13:e003003. [DOI] [PubMed] [Google Scholar]
- 27.Dorsheimer L, Assmus B, Rasper T, Ortmann CA, Ecke A, Abou-El-Ardat K, et al. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol. 2019;4:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pascual-Figal DA, Bayes-Genis A, Diez-Diez M, Hernandez-Vicente A, Vazquez-Andres D, de la Barrera J, et al. Clonal hematopoiesis and risk of progression of heart failure with reduced left ventricular ejection fraction. J Am Coll Cardiol. 2021;77:1747–59. [DOI] [PubMed] [Google Scholar]
- 29.Crowgey EL, Mahajan N, Wong WH, Gopalakrishnapillai A, Barwe SP, Kolb EA, et al. Error-corrected sequencing strategies enable comprehensive detection of leukemic mutations relevant for diagnosis and minimal residual disease monitoring. BMC Med Genomics. 2020;13:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans MA, Walsh K. Clonal hematopoiesis, somatic mosaicism, and age-associated disease. Physiol Rev. 2023;103:649–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fidler TP, Xue C, Yalcinkaya M, Hardaway B, Abramowicz S, Xiao T, et al. The AIM2 inflammasome exacerbates atherosclerosis in clonal haematopoiesis. Nature. 2021;592:296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.••. Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355:842–7 The first experimental evidence that CH causally contributes to CVD.
- 33.Sano S, Oshima K, Wang Y, Katanasaka Y, Sano M, Walsh K. CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. Circ Res. 2018;123:335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sano S, Wang Y, Ogawa H, Horitani K, Sano M, Polizio AH, et al. TP53-mediated therapy-related clonal hematopoiesis contributes to doxorubicin-induced cardiomyopathy by augmenting a neutrophil-mediated cytotoxic response. JCI Insight. 2021:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sano S, Wang Y, Yura Y, Sano M, Oshima K, Yang Y, et al. JAK2 (V617F) -Mediated clonal hematopoiesis accelerates pathological remodeling in murine heart failure. JACC Basic Transl Sci. 2019;4:684–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Sano S, Yura Y, Ke Z, Sano M, Oshima K, et al. Tet2-mediated clonal hematopoiesis in nonconditioned mice accelerates age-associated cardiac dysfunction. JCI Insight. 2020:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park E, Evans MA, Doviak H, Horitani K, Ogawa H, Yura Y, et al. Bone marrow transplantation procedures in mice to study clonal hematopoiesis. J Vis Exp. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sano S, Wang Y, Evans MA, Yura Y, Sano M, Ogawa H, et al. Lentiviral CRISPR/Cas9-mediated genome editing for the study of hematopoietic cells in disease models. J Vis Exp. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Sano S, Ogawa H, Horitani K, Evans MA, Yura Y, et al. Murine models of clonal haematopoiesis to assess mechanisms of cardiovascular disease. Cardiovasc Res. 2022;118:1413–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cordua S, Kjaer L, Skov V, Pallisgaard N, Hasselbalch HC, Ellervik C. Prevalence and phenotypes of JAK2 V617F and calreticulin mutations in a Danish general population. Blood. 2019;134:469–79. [DOI] [PubMed] [Google Scholar]
- 41.•. Yura Y, Miura-Yura E, Katanasaka Y, Min KD, Chavkin N, Polizio AH, et al. The cancer therapy-related clonal hematopoiesis driver gene Ppm1d promotes inflammation and non-ischemic heart failure in mice. Circ Res. 2021;129:684–98 This experimental study provides a mechanistic framework that links therapy-related CH and heart failure.
- 42.•. Sano S, Oshima K, Wang Y, MacLauchlan S, Katanasaka Y, Sano M, et al. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1beta/NLRP3 inflammasome. J Am Coll Cardiol. 2018;71:875–86 This experimental study provides a mechanistic framework that links age-related CH and heart failure.
- 43.Fuster JJ, Zuriaga MA, Zorita V, MacLauchlan S, Polackal MN, Viana-Huete V, et al. TET2-Loss-of-function-driven clonal hematopoiesis exacerbates experimental insulin resistance in aging and obesity. Cell Rep. 2020;33:108326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abplanalp WT, Cremer S, John D, Hoffmann J, Schuhmacher B, Merten M, et al. Clonal hematopoiesis-driver DNMT3A mutations alter immune cells in heart failure. Circ Res. 2021;128:216–28. [DOI] [PubMed] [Google Scholar]
- 45.Bick AG, Weinstock JS, Nandakumar SK, Fulco CP, Bao EL, Zekavat SM, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 2020;586:763–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377:1119–31. [DOI] [PubMed] [Google Scholar]
- 47.••. Svensson EC, Madar A, Campbell CD, He Y, Sultan M, Healey ML, et al. TET2-Driven clonal hematopoiesis and response to canakinumab: an exploratory analysis of the CANTOS randomized clinical trial. JAMA Cardiol. 2022;7:521–8 Provides clinical evidence indicating that the analysis of CH could provide guidance for the treatment of CVD.
- 48.Chavkin NW, Min KD, Walsh K. Importance of clonal hematopoiesis in heart failure. Trends Cardiovasc Med. 2022;32:198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiefer KC, Cremer S, Pardali E, Assmus B, Abou-El-Ardat K, Kirschbaum K, et al. Full spectrum of clonal haematopoiesis-driver mutations in chronic heart failure and their associations with mortality. ESC Heart Fail. 2021;8:1873–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colom B, Herms A, Hall MWJ, Dentro SC, King C, Sood RK, et al. Mutant clones in normal epithelium outcompete and eliminate emerging tumours. Nature. 2021;598:510–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng SWK, Rouhani FJ, Brunner SF, Brzozowska N, Aitken SJ, Yang M, et al. Convergent somatic mutations in metabolism genes in chronic liver disease. Nature. 2021;598:473–8. [DOI] [PubMed] [Google Scholar]
- 52.Zhu M, Lu T, Jia Y, Luo X, Gopal P, Li L, et al. Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell. 2019;177(608–621):e612. [DOI] [PMC free article] [PubMed] [Google Scholar]