Abstract

Research on siRNA delivery has seen tremendous growth over the past few decades. As one of the major delivery strategies, siRNA bioconjugates offer the potential to enhance and extend the pharmacological properties of siRNAs while minimizing toxicity. In this paper, we suggest the development of a siRNA conjugate platform with peptides and proteins that are ligands of target receptors for cancer treatment. The siRNA bioconjugates target and block the receptor membrane proteins, enter the cells through receptor-mediated endocytosis, and inhibit the expression of that same target membrane receptor, thereby doubly controlling the function of the membrane proteins. The three kinds of bioconjugates targeting CD47, PD-L1, and EGFR were synthesized via two different copper-free click chemistry reactions. Results showed the cellular uptake of each conjugate, reduction of target gene expression, and efficient functional control of receptor proteins. This platform provides an effective approach for regulating membrane proteins in various diseases beyond cancer.
1. Introduction
Signaling between cancer and stromal cells in the tumor microenvironment (TME) is known to drive oncogenesis and is mediated by ligand–receptor interactions.1,2 For example, the epidermal growth factor receptor (EGFR) is a major factor in malignant epithelial tumors, and its function is stimulated by an EGFR ligand (e.g., TGFα and EGF).3 The EGFR in cancer is often constantly stimulated because of the sustained production of EGFR ligands in the TME or as a result of a mutation in the EGFR itself. In addition, the representative immune checkpoints programmed death-ligand 1 (PD-L1) and CD47 enable cancer-immune crosstalk across different tumor types.4 The PD-L1 immune checkpoint is well known to attenuate T cell-mediated immune responses by binding to its partner, PD-1, expressed on the surface of T cells.5,6 CD47, a cell surface receptor in the immunoglobulin superfamily, is another established target that is highly expressed in various malignant cells.7 When CD47 binds to its receptor, signal regulatory protein alpha (SIRPα), on various myeloid cells, CD47 functions as an innate inhibitory immune checkpoint by interrupting the tumor cell phagocytosis.8,9
Not surprisingly, these membrane receptors have emerged as a principal target for therapeutic intervention in cancer therapy. For example, antibodies targeting overexpressed membrane receptors on cancer cells have been designed to block the interaction between tumor and stromal cells, thereby inhibiting tumor development, progression, and metastasis.10 To date, six monoclonal antibodies (mAbs) targeting PD-1 and PD-L1 and four targeting the EGFR have been approved for several types of cancer.11,12 However, despite their high specificity and affinity for target membranes, mAbs are associated with inherent limitations that include complex synthesis or purification steps due to their large molecular size as well as insufficient cytotoxic efficacy for achieving complete tumor regression. Furthermore, the fragment crystallizable region (Fc region) may contribute to hyperprogression and adverse reactions during immune checkpoint inhibitor therapy.13,14
On the other hand, small interfering RNA (siRNA) enables one to downregulate target gene expression by degrading mRNA in a highly sequence-specific manner. siRNA is a type of macromolecular drug composed of 21–25 nucleotides that can bind to and degrade the expressed mRNA.15 Once siRNA molecules are internalized into the cell and incorporated into the RNA-induced silencing complex (RISC), they enable the complex to target complementary mRNA transcripts for degradation, leading to a decrease in the expression of the targeted protein.16 siRNA therapeutics are considered a key player in gene therapy to treat various diseases including cancer.17 Despite the enormous therapeutic potential of siRNAs that directly target mRNAs, their clinical application remains challenging, mainly due to the lack of efficient delivery systems.
In this article, we suggest siRNA bioconjugates for the effective control of membrane receptors for cancer therapy. The bioconjugates include one part that specifically targets the tumor cell via a membrane receptor overexpressed on its surface and another part that downregulates the gene encoding the same receptor. Herein, we synthesized three types of siRNA bioconjugates targeting CD47, PD-L1, or EGFR via one-step copper-free click chemistry. We aimed to investigate the efficacy of a drug delivery platform that targets the same receptors via two distinct mechanisms of action, with the primary objective of evaluating the potential of this platform as an anticancer therapeutic. Our results demonstrated that this approach enhanced the cellular binding, uptake, and gene silencing efficiency of siRNA, resulting in target protein downregulation at the cellular level. By targeting overexpressed membrane receptors, our approach represents a promising strategy for cancer therapy that effectively controls oncogenic signaling and inhibits tumor development.
2. Experimental Section
2.1. Materials
The CD47-targeting protein was purified from E. coli BL21(DE3) in our lab as previously mentioned.18 Peptide binding to either PD-L1 or the EGFR was synthesized and provided after terminal modification with an azide group from Peptron (Korea). The azide-PEG3-maleimide kit was obtained from Alfa Aesar and Thermo Scientific (USA). Maleimide PEG2-succinimidyl ester (NHS ester) was obtained from the Tokyo Chemical Industry (TCI, Japan). siRNA was ordered from two companies depending on the terminal modification. Diarylcyclooctyne (DBCO)-labeled double-stranded siRNA was provided by Bioneer (Korea), while amine-modified siRNA and negative siRNA were purchased from Integrated DNA Technologies (IDT, USA). The siRNA sequences were obtained from the TriFECTa Kit by Integrated DNA Technologies for selected receptors (CD47, PD-L1, and EGFR).
Primers were designed and purchased from CosmoGenetech and Bioneer. The RNeasy Plus mini kit from Qiagen (Germany) was used to extract RNA from the cell. For qRT-PCR, the SYBR green master mix from Enzynomics (Korea) has been employed.
The nucleic acid stain dye YOYO-1 iodide was acquired from Thermo Fisher Scientific (USA). Hoechst 33342, which was used to stain the cell nucleus, was purchased from Invitrogen (MA, USA). The dyes that have been used for the phagocytosis assay, CellTracker Green CMFDA and pHrodo Red, were also obtained from Thermo Fisher.
The primary antibody targeting β-actin was purchased from Santa Cruz Biotechnology (TX, USA) in an HRP-conjugated form (sc-47778 hp). The following antibodies were used for western blot: CD47 (Abcam ab175388; MA, USA), PD-L1 (Abcam ab213480), and EGFR (Cell Signaling Technology no. 4267; MA, USA). The secondary antibody of goat antirabbit IgG (H+L)-HRP conjugate from Bio-Rad (1706515; CA, USA) was used for the visualization of the western blot band.
2.2. Cell Cultures
Murine colon carcinoma cells (CT26.CL25), murine mammary carcinoma cells (4T1), and human lung carcinoma cells (A549) were all cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS; Gibco, USA) and 1% antibiotic antimycotic acid (AA; Welgene, Taiwan). All cell lines were incubated at 37 °C, 5% CO2 condition.
2.3. Preparation of siRNA Conjugates
Peptide and protein sequences were designed as follows: CD47 binding protein (EEELQIIQPDKSVLVAAGETATLRCTITSLFPVGPIQWFRGAGPGRVLIYNQRQGPFPRVTTVSDTTKRNNMDFSIRIGNITPADAGTYYCIKFRKGSPDDVEFKSGAGTELSVRAKP),9,18 PD-L1 targeting peptide (D-peptide NYSKPTDRQYHF),19,20 and EGFR binding protein (YHWYGYTPQNVI).21 The sequences were obtained and slightly modified from the previous reports of binding proteins and peptides of tumor-overexpressed receptors. The additional cysteine group was added on the C terminal of the CD47 binding protein, while PD-L1 and EGFR peptides were modified with an azide group on the N terminal. siRNA sequences were tested in six-well plate cells seeded to reach 80% confluency on the next day. The commercial transfection agent Lipofectamine RNAiMAX (Thermo Fisher, USA) was used for testing the individual siRNA sequence silencing efficacy. After following the manufacturer’s protocol of forward transfection, the cells were collected and analyzed by using RT-PCR and western blot.
For the CD47 targeting conjugates, siRNA modified with a 5′-amino-modifier C6 linker was used. The maleimide-PEG2-NHS ester (250 nmol, 106 μg) was mixed with siRNA (100 μM, 5 nmol) at 4 °C overnight with vigorous shaking. After the incubation, the unbound maleimide-PEG2-NHS ester was removed using a 3 kDa Amicon ultra-0.5 centrifugal filter. After purification, the siRNA-maleimide and CD47 protein were incubated at 4 °C overnight, resulting in a maleimide–thiol reaction between the siRNA and the protein. The conjugates for PD-L1 and the EGFR were formed with a copper-free click chemistry reaction. The azide-functionalized peptide (10 μg/μL, 2 nmol) and DBCO-attached siRNA (100 μM, 1 nmol) were mixed at 37 °C at 1200 rpm for 2 h. The reaction was incubated in a two-to-one molar ratio, and the synthesis was confirmed with 3% agarose TBE gel and 16–20% SDS-PAGE gel. Agarose TBE gels were prestained using the SYBR-safe DNA gel (Invitrogen, USA), while the SDS-PAGE gel was poststained using the ethidium bromide (Invitrogen). The gel image was analyzed by an iBright 750 Imaging System (Thermo Fisher Scientific).
As the reverse phase HPLC to confirm the conjugation between ligand and siRNA, 5–50% acetonitrile with 0.1% trifluoroacetic acid versus 3DW with 0.1% trifluoroacetic acid was run over 30 min at a flow rate of 0.5 mL/min. The detection of the peak was made at a UV wavelength of 280 nm using HPLC (Agilent, USA).
2.4. Assessment of Cytotoxicity
To analyze the cellular toxicity of the conjugate, the CT26.CL25 and 4T1 cells were seeded in 96-well plates at 5 × 103 cells/well. After 24 h of stabilization, either lig(p)-siR or an equivalent dose of Lipofectamine-encapsulated siRNA was added accordingly and treated for 24 or 48 h. Using the cell counting kit-8 (Dojindo, Japan), the viability of each well was measured at an absorbance of 450 nm.
2.5. Assessment of In Vitro Binding and Cellular Uptake
To appraise the binding efficacy of the protein and peptide, the CT26.CL25 cells and A549 cells were seeded in a 35 mm glass-bottom dish at a density of 2 × 105 cells/well. After 24 h of incubation, the cells were treated with 200 pmol of conjugates or siRNA in a 1 mL serum-free medium. For the antibody preblocked group in CT26.CL25, the murine PD-L1 antibody (BE0101, Bioxcell, USA) was treated at 1 mg/mL 1 h before the addition of the conjugates. For 4T1, cells were added with 1 μM PD-L1 antibody or PD-L1 ligand 30 min before the lig(p)-siR treatment. Before being added to the cell medium, unstained siRNA and conjugates were stained with the YOYO-1 dye. Either siRNA or conjugates were mixed with YOYO-1 dye in a one-to-five molar ratio and incubated at room temperature for 30 min. The equivalent volume of siRNA and conjugates was used during the incubation with the dye, and if not, then it was matched with RNase-free water. In contrast to CD47 siRNA and EGFR siRNA, Cy5-tagged PD-L1 siRNA was used for the binding and uptake assay; therefore, PD-L1 siRNA was not poststained with the YOYO-1 dye. For the binding assay, cells were fixated with 4% formaldehyde (Biosesang, Korea) after 30 min of treatment at 4 °C. For the staining of nuclei, PBS-diluted Hoechst 33342 (1:2000) was added to the solution and treated at RT for 10 min.
For the cellular uptake image, CT26.CL25 cells and A549 cells were seeded in a 35 mm glass-bottom dish with 1 × 105 cells/well. The YOYO-1 dye-stained siRNA and siRNA conjugates were also used for the cellular uptake assay. siRNA or the conjugates were added to the cell medium at a final concentration of 200 nM in a serum-free medium. The cells were fixated using 4% formaldehyde according to the varying time points and room temperature incubation for 10 min. Hoechst 33342 (1:2000) was used for nuclei staining. A TCS SP8 confocal laser microscope (Leica, Germany) was used for the images of binding and uptake assay, which applied CD47 siRNA and PD-L1 siRNA on CT26.CL25 cells. For A549 cells, the binding and uptake images were taken by using an EVOS M5000 imaging system (Thermo Fisher Scientific).
2.6. Evaluation of mRNA Silencing Efficacy
To evaluate the relative mRNA level of a target gene, reverse transcription PCR (RT-PCR) gel electrophoresis or real-time quantitative reverse transcription PCR (qRT-PCR) was performed. CT26.CL25 cells and A549 cells were seeded in a six-well plate at a density of 2 × 105 cells/well. The cells were then treated according to the assigned treatment group and annotated treatment dose. After being treated for 24 h, cells were then collected after being trypsinized. From the cell pellets, total RNA was isolated using the RNeasy Plus mini kit by following its instructions. The RNA levels were measured using a NanoDrop Lite spectrophotometer (Thermo Fisher) from the eluted solution. With the calculation, 600 ng of RNA was used for each group for cDNA. The cDNA was synthesized using the AccuPower premix from Bioneer and the Oligo(dT) 20 primer. For the gel electrophoresis-based RT-PCR detection method, 1% TBE agarose gel was used to visualize the PCR band. The detailed PCR cycle for PD-L1 was as follows: initial denaturation at 95 °C 5 min, denaturation at 95 °C 10 s, annealing at 60 °C 15 s, elongation at 72 °C 15 s, and followed by elongation at 72 °C 30 s. For qRT-PCR, SYBR-green from Enzynomics was used with six duplicates for each sample. Using a StepOne Real-Time PCR system (Applied Biosystems, Thermo Fisher Scientific), qRT-PCR was performed. The relative level of the target gene has been measured using the 2–ΔΔCt method. For the comparison, the control housekeeping gene of GAPDH has been used. The specific sequences for the primers are listed in Table 1.
Table 1. Primer Sequences.
| gene | primer sequences | ref |
|---|---|---|
| HPRT | F: 5′ GGCTATAAGTTCTTTGCTGACCTG 3′ | (22) |
| R: 5′ GCTTGCAACCTTAACCATTTTGGG 3′ | ||
| GAPDH | F: 5′ CCACCCAGAAGACTGTGGAT 3′ | (23) |
| R: 5′ CACATTGGGGGTAGGAACAC 3′ | ||
| CD47 | F: 5′ AGGAGAAAAGCCCGTGAAG 3′ | (24) |
| R: 5′ TGGCAATGGTGAAAGAGGTC 3′ | ||
| PD-L1 | F: 5′ GCTCCAAAGGACTTGTACGTG 3′ | (25) |
| R: 5′ TGATCTGAAGGGCAGCATTTC 3′ | ||
| GAPDH (human) | F: 5′ ACAACTTTGGTATCGTGGAAGG 3′ | (26) |
| R: 5′ GCCATCACGCCACAGTTTC 3′ | ||
| EGFR (human) | F: 5′ TCCCTCAGCCACCCATATGTAC 3′ | (27) |
| R: 5′ GTCTCGGGCCATTTTGGAGAATCC 3′ |
2.7. Evaluation of siRNA Conjugate-Mediated Protein Expression
CT26.CL25 cells and A549 cells were seeded in six-well plates at 2 × 105 cell/well density. The cells were treated with DPBS, siRNA, or a conjugate in a final concentration of 200 nM. The incubation times were different for the receptors (CD47, PD-L1, and EGFR). For PD-L1 and the EGFR, the siRNA conjugates were treated for 48 h, while for CD47 receptors, the conjugates were treated for 5 days. After the incubation, the cells were lysed using RIPA buffer (Biosesang) that was added with a 1% Halt protease inhibitor cocktail (Thermo Fisher). The lysis process was continued at 4 °C for 30 min. The cells are then centrifuged at 12,000 x g for 20 min, and the supernatants are collected for protein quantification via the Pierce BCA assay kit (Thermo Fisher). In a 10% SDS-PAGE gel, the proteins were separated. The gels were then transferred to a 0.2 μm nitrocellulose membrane. Using the blocking buffer of 5% skimmed milk in a TBS-T buffer, the membrane was incubated at room temperature for 90 min. The primary antibody of the target gene (CD47, PD-L1, and EGFR) and the housekeeping gene (GAPDH and β-actin) was detected and developed using an iBright 750 instrument (Thermo Fisher).
2.8. In Vitro Phagocytosis Assay
For the phagocytosis assay, bone marrow-derived macrophages (BMDMs) were used. Six-week-old BALB/c mice were purchased from Orient Bio (Korea). The mice were sacrificed, and bone marrow cells were isolated from both of the hind legs. The bone marrow cells from tibias and femurs were differentiated in RPMI media with the macrophage colony-stimulating factor (Peprotech, USA) in a 20 ng/mL final concentration. The cytokines were added on days 1, 2, 3, 4, and 6, while the media were changed on days 2, 4, and 6. On the seventh day since the initial seeding, the BMDM was seeded to a 35 mm glass plate dish at 2 × 105 cells/well. On the eighth day, the BMDM cells were stained using the CellTracker Green dye for 30 min at room temperature. After the staining and washing with DPBS, BMDM cells were cocultured with different groups of cancer cells for 2 h in a 37 °C CO2 incubator. The cancer cells were pretreated with DPBS, protein, or protein–siRNA conjugates in a final concentration of 200 nM. The cancer cells were labeled using pHrodo Deep Red for 30 min at room temperature. The cancer cells were resuspended in a serum-free medium after being washed and added to the BMDM for the coculture. After 2 h of incubation, the dishes were washed softly using DPBS and were imaged using a fluorescence microscope (Nikon, Japan). The phagocytosis index was measured using the taken images and calculated based on colocalized red signals with the green. A detailed explanation of the phagocytosis assay was published previously.28
2.9. Membrane Receptor Expression Analysis Using Flow Cytometry
The 4T1 cells were seeded in a density of 0.5 × 105 cells in 2 mL per well in a six-well plate. After stabilization overnight, the cells were treated with 500 pmol/mL for both ligand and ligand-siRNA. The cells were treated for 48 h in a 37 °C incubator. For the 30 min treated group, cells were treated 30 min before trypsinization. The cells were then detached using trypsin-EDTA and washed with DPBS. Cells were resuspended at 106 cells/100 μL. To measure the expression of PD-L1 on the cell membrane, a single-cell suspension of 4T1 was stained with the APC-conjugated PD-L1 antibody (124312; BioLegend, USA) at a 1:5000 ratio for 30 min at 4 °C. The cells were then centrifuged and resuspended to wash with DPBS using a CytoFLEX flow cytometer (Beckman Coulter, USA).
2.10. In Vitro Scratch Migration Assay
The A549 cells were seeded in six-well plates with 3 × 105 cells/well. As the cells reached a confluency of 80%, they were scratched using 10 μL sterile pipet tips and thoroughly washed using DPBS to remove the detached cells. The cells were then incubated in a serum-free medium and treated with an equivalent volume of either DPBS, siRNA, peptide, or siRNA conjugate. One nmol of siRNA and siRNA conjugate were each treated in the wells in a 2 mL medium, and 2 nmol of the peptide was added to the 2 mL medium in the wells to account for the one-to-two molar ratio during the synthesis of the siRNA–peptide conjugate. The medium was changed to a fresh serum-free medium after 3 days. The images were taken every 24 h to analyze the distance between the scratch using a tabletop optical microscope (CK40; Olympus, Japan). The width of the scratch was measured and analyzed using the ImageJ program.
2.11. Statistical Analysis
The statistical analysis was performed using the Prism 8.0 (GraphPad) program. Statistical significance was determined by the Student’s t-test or the one-way analysis of variance (ANOVA) test depending on the number of the group. A p-value less than 0.05 was considered statistically significant, and the statistical significance was marked as * p < 0.05, ** p < 0.01, *** p < 0.001, and ****p < 0.0001.
3. Results
3.1. Preparation of the Ligand–siRNA Conjugate
In this article, we developed a therapeutic modality that targets cancer receptors at both the protein and RNA levels (Figure 1). This approach employs ligand–siRNA conjugates, which are designed to enhance tumor-specific targeting efficiency and therapeutic outcomes by blocking the receptors with peptides or protein ligands and by selectively silencing the target gene with siRNA. To synthesize ligand–siRNA conjugates, we utilized two types of copper-free click chemistry: the DBCO-azide and the maleimide-thiol reaction29 (Figures 1A and 2A–C). CD47-targeting conjugates utilized a thiol-maleimide reaction, and the protein sequence was manipulated to contain extra cysteine on the C-terminus along with several glycine residues to facilitate the conjugation process (Figure 2A). In contrast, the DBCO-azide reaction was adopted for conjugating N-terminal azide-modified PD-L1 and EGFR-binding peptides with DBCO-functionalized siRNA on the 3′ sense strand (Figure 2B,C). The ligand–siRNA conjugates can then be internalized into the cell through PD-L1, EGFR, or CD47 receptor-mediated endocytosis, wherein the attached siRNA enters the endosome and executes RNA interference, ultimately leading to downregulation of the receptors on the surface membrane (Figure 1B). This dual-targeting approach could potentially enhance the cytotoxic effects on cancer cells while reducing the risk of off-targeting effects.
Figure 1.
Overall schematic diagram of the ligand–siRNA conjugates and their double targeting mechanism of action. (A) CD47-ligand, PD-L1-ligand, and EGFR-ligand are conjugated with siRNA via a one-step copper-free click reaction, enhancing receptor-mediated endocytosis. (B) Dual function of ligand–siRNA conjugates is targeting and blocking the receptor, as well as silencing the receptor pathway via RNAi-mediated gene regulation. As the ligand interacts with the receptor, it functions as the blocking agent that inhibits downward signaling (1). The receptor-mediated uptake occurs accompanying conjugate into the cells (2). Among the endocytosed conjugates, part of the siRNA escapes and leaks into the cytosols before the endosome matures into a lysosome (3). Escaped siRNA functions by binding to the target mRNA (4, 5) and silences the receptor expression, which further downregulates the targeted pathway (6).
Figure 2.
Chemical structure of the conjugate used in the study. (A) C′ terminal amino acids were displayed, and the thiol used for the conjugation has been marked in red. The maleimide tagged at the 3′ sense strand of siRNA is also denoted in red. (B) PD-L1 binding peptide and linked PD-L1 siRNA were partially illustrated in their chemical structure. The azide and DBCO groups are shown in red. (C) EGFR binding peptide and part of the EGFR siRNA were shown, with a chemical group associated with the conjugation displayed in red.
To determine the most effective siRNA sequences for the intervening CD47, PD-L1, and EGFR expression, we screened several candidate sequences (Figure 3A). We assessed the silencing efficacy of the candidates by delivering them to cancer cells using the commercial transfection agent Lipofectamine RNAiMAX. We utilized the murine colon carcinoma cell line CT26.CL25, in which high expression levels of these receptors were reported, for screening CD47 and PD-L1-siRNA sequences. The RT-PCR data revealed that the CD47 mRNA expression level was reduced in the first and third sequence groups. In contrast, DPBS, scrambled siRNA, and the second candidate showed an insignificant reduction in the CD47 mRNA level (Figure 3B). When the PD-L1 mRNA expression was analyzed using the same methodology, the second and third candidates of PD-L1 targeting siRNA showed the most reduced PD-L1 mRNA signals (Figure 3C). Consistent with the RT-PCR results, the western blot image also showed a reduced level of PD-L1 receptors in the second and third siRNA-treated groups when compared to that in other groups (Figure 3D). For the selection of the EGFR-siRNA sequence, we used the human lung cancer cell line A549 to represent the EGFR wild type. After EGFR siRNA candidates were added to A549 cells using Lipofectamine, the target gene (Figure 3E) and protein expression (Figure 3F) were analyzed, revealing that only the third siRNA sequence showed reduced EGFR expression in both gene and protein levels. Given the variable response level of siRNA silencing, we selected the most efficient sequences for the upcoming experiments, namely, the first sequence of CD47 and the third sequence of both PD-L1 and the EGFR.
Figure 3.
siRNA sequence selection for ligand–siRNA conjugates. (A) Sequences for siRNA candidates with their expected binding sites and mass. Lower case indicates DNA nucleotides; uppercase represents the RNA nucleotides. (B) RT-PCR gel electrophoresis results and quantified band intensity using ImageJ software. (C) RT-PCR gel electrophoresis (D) and quantified western blot gel results for each siRNA candidate for PD-L1. (E) RT-PCR gel and (F) quantified western blot images after treatment of EGFR siRNA with a commercial transfection agent. The siRNA sequences that have been used for the following experiments are bolded and marked in blue. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; one-way ANOVA with the Holm–Sidak posthoc test. Data are presented as the mean ± s.d.
3.2. CD47 Ligand–siRNA Conjugates
The conjugation of CD47 binding ligands and CD47 siRNA (lig(c)-siR) via a thiol-maleimide reaction was confirmed by observing a band shift in gel electrophoresis. (Figure 4A). The SDS-PAGE and agarose gel images showed that overnight incubation of the CD47-ligand with CD47 siRNA resulted in a conjugation efficiency of up to 90%,
Figure 4.
In vitro effect of the CD47-ligand–siRNA conjugate (lig(c)-siR). (A) Gel electrophoresis of siRNA and lig(c)-siR in both 16% SDS-PAGE gel (left) and 3% agarose gel (right). (B) Representative images of CT26.CL25 cellular binding when treated with 200 nM of either siRNA or the conjugate for 30 min in 4 °C. (C) Representative confocal images of cellular uptake in CT26.CL25 after 1, 3, and 6 h of 200 nM treatment. (D) qRT-PCR results of relative CD47 mRNA levels in CT26.CL25 cells after incubating the cells with varying final concentrations of the conjugates (0, 100, and 200 nM) for 24 h. All were normalized by their GAPDH level. (E) Western blot analysis of the CD47 protein expression level of cells treated with varying final concentrations (0, 100, and 200 nM) of lig(c)-siR for 5 days. Data were presented as the mean ± s.d. *p < 0.05, ***p < 0.001, and ****p < 0.0001; one-way ANOVA with the Tukey–Kramer posthoc test.
To assess the ability of the CD47 targeting moiety, we examined in vitro cellular binding and uptake of the protein in CT26.CL25 cancer cells. The CD47 siRNA was labeled with the YOYO-1 dye and conjugated with the CD47-ligand. Naked siRNA without the binding moiety showed no cellular binding activity, whereas high binding efficiency was observed in lig(c)-siR-treated cells (Figure 4B). Consistent with this binding tendency, we also observed the cellular uptake of siRNA by a time-dependent increase in lig(c)-siR-treated CT26.CL25 (Figure 4C). Notably, we detected little fluorescence signal of membrane-bound and internalized siRNAs without the CD47-ligand, highlighting the importance of the receptor-targeting ligand. To examine the effectiveness of internalized siRNA in inducing gene silencing, the cellular expression level of CD47 mRNA was analyzed using qRT-PCR assay. The results demonstrated a significant dose-dependent reduction in the level of CD47 mRNA expression (Figure 4D). Similar results were also observed for the CD47 expression at the protein level using a western blot assay under the same experimental conditions (Figure 4E). Taken together, these data suggest that lig(c)-siR not only enhances the cellular binding and uptake efficiency via the CD47-ligand but also intervenes in the CD47 protein synthesis pathway via CD47-siRNA.
CD47 overexpression in cancer cells plays a role in evading immune surveillance, allowing them to continue growing and spreading without being attacked by immune cells.30 When CD47 binds to SIRPα, primarily expressed in immune cells such as monocytes, granulocytes, and macrophages, it sends a “don’t eat me” signal that prevents macrophages from engulfing and destroying cancer cells.31 Thus, the downregulation of CD47 receptors can be an efficient approach for inhibiting tumor development by stimulating the phagocytic activity of macrophages. To evaluate the functional effects of lig(c)-siR, phagocytic activity was assessed in vitro using differentiated BMDMs. As expected, the engulfment of cancer cells (red) by BMDMs (green) was greatly enhanced in CT26.CL25 treated with lig(c)-siR (Figure 5). The phagocytic index (%) was also analyzed for both time points (30 min and 48 h). The phagocytic index at 30 min treatment was similar in the CD47-ligand and lig(c)-siR treated group, indicating receptors and inhibitory ligands were sufficiently bound at 30 min and blocking the CD47/SIRPα axis and increasing phagocytosis. However, the phagocytic index for 48 h treatment showed a different pattern for the groups; for the CD47-ligand treated group, 48 h treatment showed similar results to the 30 min treatment, whereas the phagocytic index significantly increased after 48 h in the conjugate-treated group. The results of this study suggest that lig(c)-siR can enhance therapeutic efficacy due to the RNA interference mechanism in addition to the effects induced from protein alone, which overall inhibits the expression of CD47 and promotes macrophage phagocytosis and extends the effects of inhibiting the CD47/SIRPα axis.
Figure 5.
Representative images of phagocytosis assay for the 48 h treated group for the protein and the conjugate. The quantification of the phagocytosis index was measured and presented in a box and whisker plot. Within each box, the dashed lines indicate the mean value, while solid lines indicate the median values. The boxes are extended from the 25th to the 75th percent of each group. The symbols denote all values for the group.
3.3. PD-L1 Ligand–siRNA Conjugates
Next, we synthesized a PD-L1 ligand–siRNA conjugate (lig(p)-siR) via a DBCO-azide click reaction. The overexpression of PD-L1 on cancer cells is believed to be one of the mechanisms that enable tumors to evade the immune system via the PD-1/PD-L1 checkpoint signaling pathway.32 When PD-L1 binds to PD-1, primarily expressed in the T cell membrane, it sends an inhibitory signal to the T cells, preventing them from attacking cancer cells.33 Thus, blocking the PD-1/PD-L1 interaction by reducing the PD-L1 expression level in cancer cells can be a reasonable approach for cancer immunotherapy.
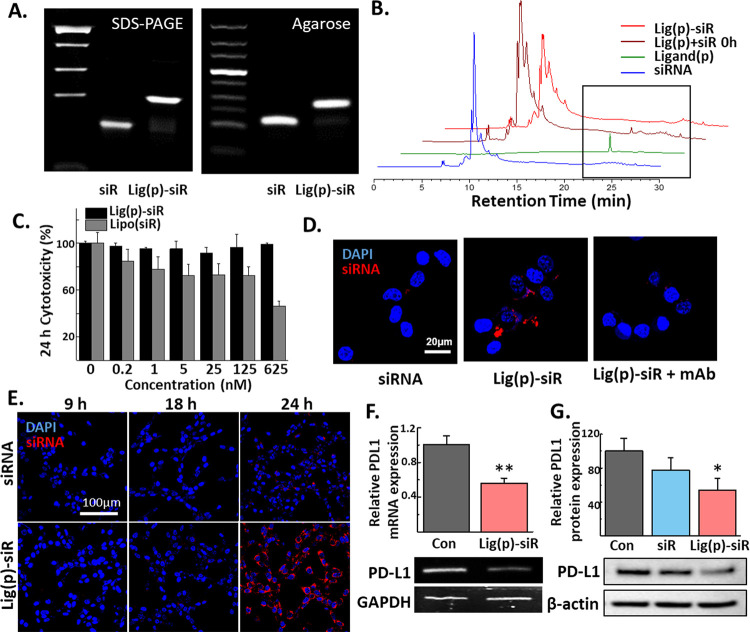
To address the PD1/PD-L1 pathway, we adjoined PD-L1 binding peptides with anti-PD-L1 siRNA. The successful conjugation was confirmed by detecting a band shift in 16% SDS-PAGE and 3% agarose gel (Figure 6A). The conjugation efficiency was found to be over 80% only after a 2 h reaction, highlighting the facile and reproducible synthesis steps of this conjugate. The conjugation of siRNA and the ligand is further supported via HPLC analysis (Figure 6B). With the distinct ligand peak at 280 nm around 25.6–25.7 min, it is clear that the ligand signal decreases as the reaction time passes. As the reaction was held in a one-to-one molar ratio between the ligand and siRNA, it indicates that the ligand peak disappears as the ligand is conjugated to siRNA. The peak eventually is plateaued, which signifies that almost all ligands are conjugated within 1 h of incubation. In addition to its rapid and uncomplicated synthesis process, lig(p)-siR demonstrated a very favorable toxicity profile. Even at a high dose of 625 nM, lig(p)-siR treatment resulted in a nonsignificant difference in cell cytotoxicity for CT26.CL25, while the group treated with a commercial transfection agent, Lipofectamine, has shown a significant decrease with the increased dose (Figure 6C).
Figure 6.
In vitro effect of the PD-L1 ligand–siRNA conjugate (lig(p)-siR). (A) Gel electrophoresis of siRNA and the conjugate in both 16% SDS-PAGE gel (left) and 3% agarose gel (right). (B) Reverse-phase HPLC peak analysis of siRNA, ligand, and lig(p)-siR. The peak of lig(p)-siR incubated for 0 and 1 h was analyzed. The ligand peak was marked with a black rectangle. (C) Cytotoxicity assay on CT26.CL25 for 24 h. (D) Representative images of CT26.CL25 cellular binding when treated with 200 nM of either siRNA or the conjugate for 30 min in 4 °C. (E) Representative confocal images of cellular uptake in CT26.CL25 after 9, 18, and 24 h of either siRNA or lig(p)-siR (200 nM). (F) qRT-PCR results of relative PD-L1 mRNA levels in CT26.CL25 cells after incubating the cells with DPBS or lig(p)-siR for 24 h. All were normalized by their GAPDH level. (G) Western blot gel image and quantified results of the PD-L1 protein expression level of CT26.CL25 cells treated with DPBS, siRNA, or lig(p)-siR for 2 days. Data were presented as the mean ± s.d.; the unpaired Student t-test and one-way ANOVA were used. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
To evaluate the capability of the PD-L1 targeting moiety to improve the binding to cell membranes and cytosolic delivery of siRNA, in vitro cellular binding and uptake assays were performed. After 30 min of treatment of free-siRNA and lig(p)-siR, the binding efficiency was observed using confocal microscopy (Figure 6D). Our results indicated that the use of PD-L1-binding ligands resulted in a substantial improvement of siRNA binding compared to the free-siRNA treated group, which exhibited minimal binding to the cell membrane. Importantly, the PD-L1 receptor-specific binding was confirmed through the use of a PD-L1 antibody pretreatment. As a result, the observed signals were significantly reduced almost to the level of free siRNA-treated groups. Moreover, the cellular uptake of lig(p)-siR was observed in CT26.CL25 cells. The siRNA fluorescence intensity in the cytosol is increased for 24 h, indicating that siRNA was internalized via PD-L1-mediated endocytosis (Figure 6E). In contrast, siRNA fluorescence of the free-siRNA treated group was not detected throughout the experiment, even after incubating with siRNA for 24 h. Lig(p)-siR displayed a superior uptake profile and 4.7-fold higher fluorescence intensity when compared to the siRNA-treated group at 24 h post treatment. To more precisely assay the gene silencing ability of lig(p)-siR, we confirmed the cellular PD-L1 expression level in the mRNA and protein. Consistent with the cellular uptake image results, lig(p)-siR decreased PD-L1 mRNA expression to 56% compared to the DPBS-treated group (Figure 6F). Furthermore, western blot analysis demonstrated that lig(p)-siR downregulates PD-L1 expression at both the RNA and protein levels (Figure 6G). When added to the CT26.CL25 cell, lig(p)-siR displayed a similar binding pattern to 4T1 cells, another cell line known to overexpress PD-L1 receptors. As in the previous assay with antibody pretreatment, the blocking with excess ligands resulted in the silencing of the signal, indicating that lig(p)-siR has bound with cells via PD-L1 receptors (Figure 7A). Similar to CT26.CL25, the limited cellular toxicity was noted for both 24 and 48 h treatment, which is further highlighted when compared to the Lipofectamine-treated group (Figure 7B,C). The dose-dependent reduction of PD-L1 mRNA and protein signifies that even at a high concentration, lig(p)-siR maintained a high level of cellular efficacy without the risk of cellular toxicity (Figure 7D,E). Although the amount of response varied between CT26.CL25 and 4T1 as the percentage of relative mRNA and protein levels slightly differed between the two cells, the overall response as the reduction of mRNA and protein levels signifies the RNAi effects of lig(p)-siR on both cells.
Figure 7.
Lig(p)-siR in vitro efficacy. (A) Representative images of cellular binding when treated with 200 nM conjugate for 30 min at 4 °C on 4T1 cells. For the competitive assay, two groups were pretreated with excess ligands or antibodies (2 nmol/mL) 1 h before the lig(p)-siR treatment. Cytotoxicity assay on 4T1 for (B) 24 h and (C) 48 h. The equivalent dose of siRNA encapsulated with Lipofectamine RNAiMAX was also tested on 4T1 for cellular toxicity. qRT-PCR (D) and western blot analysis (E) with varying doses of lig(p)-siR or Lipofectamine-encapsulated siRNA for 24 h on 4T1. (F) Representative histograms indicating the PD-L1 expression level in 4T1 cells. Cells were treated with DPBS, peptide, or lig(p)-siR (500 nM). The data of percentage for positive cells are presented in a box and whisker plot. Within each box, solid lines indicate median values. The boxes are extended from the 25th to the 75th percent of each group. The symbol denotes all values for the group. Data are presented as the mean ± s.d.; one-way ANOVA was used. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Next, to evaluate the additional benefits of attached PDL1 siRNA for extending the efficacy, we compared the expression level of PD-L1 receptors in 4T1 cells at two different time points, 1 and 48 h (Figure 7F). From the rate of the bound-fluorescence-tagged PD-L1 antibody to the 4T1 cells following the conjugate treatment, the PD-L1 receptor expression level was analyzed. PD-L1-ligand treatment was able to successfully obstruct the antibody from binding to the cells in 1 h (3.29%), but the signal was restored to the level of the control group after 48 h (7.82%). However, lig(p)-siR not only blocked the binding of the antibody to the cells in 1 h (3.32%) but also continued to reduce the expression of the PD-L1 receptor by the action of siRNA even after 48 h (3.28%). These results indicate that efficient and sustainable downregulation of PD-L1 membrane expression could be obtained from the PD-L1 gene silencing ability of siRNA.
3.4. EGFR Ligand–siRNA Conjugates
Aberrant overexpression of the EGFR in cancer cells has been identified as a key driver of cancer cell tumor growth, invasion, and metastasis.3 With an activated EGFR signaling pathway, cancer cell migration is enhanced by cytoskeletal rearrangement, extracellular matrix degradation, and cell adhesion. In addition, EGFR signaling has also been linked to the induction of epithelial-mesenchymal transition (EMT), a process by which epithelial cells lose their characteristic cell–cell adhesion and acquire a migratory and invasive phenotype.34 For these reasons, blocking the EGFR has been explored as a potential approach for impeding the migration of cancer cells, ultimately inhibiting tumor development and metastasis.35
Similar to PD-L1, for addressing EGFR signaling, the EGFR peptide and siRNA were conjugated. First, to validate the successful synthesis of the EGFR ligand–siRNA conjugate (lig(e)-siR) via the DBCO-azide click reaction, we utilized gel electrophoresis in 20% SDS-PAGE gel and 3% agarose gel (Figure 8A). The resulting gel images revealed a band shift in lig(e)-siR, indicating an 85% efficiency of conjugation between the EGFR peptide. Subsequently, we assessed the cellular binding ability of lig(e)-siR by incubating A549 cancer cells for 30 min (Figure 8B). The binding of green fluorescence-labeled siRNA on the A549 cell membrane is detected in the lig(e)-siR treated group, which is consistent with lig(c)-siR and lig(p)-siR. However, no fluorescence signals were detected when DPBS and free-siRNA were treated, emphasizing the importance of the EGFR ligand in binding to the cell membrane. Upon further incubation to assess the cellular uptake of lig (e)-siR, higher green fluorescence intensity was detected, compared to the free siRNA-treated group (Figure 8C). In all conjugates including lig (e)-siR, the noticeable cellular uptake signal was found even before 12 h, and for PD-L1- and EGFR-targeting conjugates, the cellular uptake was shown to increase time-dependently up to 24 h (Figures 4C, 5E, and 8C). Followed by the results of cellular uptake, the conjugate induced successful gene silencing, significantly downregulating the expression of target mRNA and protein levels (Figure 8D,E).
Figure 8.
In vitro effect of the EGFR ligand–siRNA conjugate (lig(e)-siR). (A) Gel electrophoresis of siRNA and the conjugate in both 20% SDS-PAGE gel (left) and 3% agarose gel (right). (B) Representative images of A549 cellular binding when treated with 200 nM of either siRNA or the conjugate for 30 min at 4 °C. (C) Representative fluorescence images of cellular uptake in A549 after 1, 12, and 24 h of either siRNA or lig(e)-siR. (D) qRT-PCR results of relative EGFR mRNA levels in A549 cells after incubating the cells with DPBS, siRNA, or lig(e)-siR (200 nM) for 24 h. All were normalized with their GAPDH level. Data were presented as the mean ± s.d. (E) Western blot gel images and quantified results of the EGFR protein expression level of cells treated with DPBS, siRNA, or lig(e)-siR for 2 days. Data presented as mean ± s.d. (*p < 0.05, **p < 0.01, and ****p < 0.0001; one-way ANOVA with the Tukey–Kramer posthoc test).
To examine the functional effects of lig(e)-siR in cell migration, a scratch migration assay, measuring how fast the cells would close the gap after scratching the confluent cells, was conducted (Figure 9). As expected, the treatment of the EGFR-ligand showed some efficacy in slowing cell migration. However, lig(e)-siR exhibited the greatest effect in delaying migration in vitro. The control group showed approximately 25% closure of the gap after 48 h, whereas the lig(e)-siR treated group had little closure of the gap from scratching. Parallel with previous results showing increased phagocytosis mediated by the CD47-targeting siRNA conjugate and decreased expression of the receptor by the PD-L1-targeting siRNA conjugate, the scratch migration assay also demonstrated superior functionality of ligand–siRNA conjugates over simply blocking target receptors.
Figure 9.
Representative images of scratch migration assays in A549 cells at 1 and 48 h of incubation. The data were presented in a bar graph after the width of the scratch was quantified by using the ImageJ program. The percentage was calculated by the relative width when compared to that of 48 h to the corresponding width of a 1 h time point. Data presented as mean ± s.d. (*p < 0.05, ****p < 0.0001; Brown–Forsythe and Welch ANOVA with Dunnett’s T3 posthoc test).
4. Discussion and Conclusions
Thanks to the success of cholesterol–siRNA conjugates (chol-siRNA) and GalNAc–siRNA conjugates, the delivery strategy for siRNA therapeutics has recently shifted from complex polymeric nanoparticles to chemically well-defined, homogeneous siRNA bioconjugates.36 Bioconjugation has evolved into a powerful delivery strategy that imparts multiple functions to siRNAs for efficient delivery, ranging from liver diseases such as hepatic porphyria and hepatitis to muscle and brain diseases.37 Compared to unconjugated siRNA, these siRNA bioconjugates have significantly higher tissue biocompatibility, longer biological half-lives, targetability, and better pharmacological properties with increased delivery efficiency, while maintaining gene silencing efficacy.
siRNAs can be linked with targeting ligands for cell type-specific delivery. Compared with antibodies, peptides or proteins are relatively small in size and can be easily prepared by synthesis or purification. The wide application of phage display technology has identified a large number of peptide ligands targeting a broad spectrum of receptors and proteins. In addition to the well-established strategy to screen the protein and peptide of cell surface display systems using phages and yeasts,38 the recent development of in-silico techniques has further facilitated the discovery of the ligand.39 Thus, peptide/protein ligands are one of the most commonly applied agents for drug conjugates and delivery due to their high specificity and affinity for aimed receptors.
In this article, we synthesized ligand–siRNA conjugates targeting CD47, PD-L1, and EGFR using a copper-free click chemistry reaction. Our results indicated that these conjugates exhibited enhanced cellular binding capability and internalization through receptor-mediated endocytosis facilitated by the attached targeting ligand. Furthermore, all three conjugates could significantly downregulate gene expression. A treatment concentration of 200 nM was sufficient for all three conjugates to induce significant gene silencing. The receptor expression levels are greatly reduced with the conjugate treatment, but the time taken for the decrease in protein expression may vary depending on the stability of the receptors. For example, the EGFR-targeting conjugate downregulated the protein expression after 48 h of treatment, whereas the CD47-targeting conjugate was shown to take more than 120 h to reduce the expression of receptors at the protein level.
Yet, there were still limitations that we have not addressed fully. As we only conducted in vitro assays, the necessity of chemical modification to protect siRNA against the nucleases was not as imminent. However, as the chemical modification of the siRNA in both sugar and backbone modification has been known to increase efficacy as well as stability,40 the optimization of the siRNA in terms of chemical modification can lead to an increase in silencing effects and a longer duration of action. In the in vivo assays to assay the efficacy of these siRNA bioconjugates, consideration of siRNA metabolic stability and an attempt to increase it will be made before the experiments. The safety and immune response against the nonmodified siRNA has been noted in previous studies,41 which further urges the optimization, and evaluating the safety profile of all components of the conjugate would be required as well.
Many previous studies have shown promising results that the use of peptides/proteins delayed tumor growth by inhibiting interactions between receptors and ligands in the tumor microenvironments.20,42 One of the relevant published studies was conducted in our lab using the same PD-L1 peptide sequence with the same “click chemistry” design.20 The cargo was miRNA in contrast to current siRNA. Although in that study the PD-L1 peptide-binding ability was fully utilized, the focus was to discover the synergistic effects of miRNA and PD-L1 blocking. However, in this current study, we attempted to fully suppress the specific receptor for antitumoral efficacy. In addition to the different foci of the studies, the previously published study contained animal data that supported the tumor growth suppression activity using lig(p). Although this study lacked in vivo experiments, even with the limited scope of in vitro experiments, our conjugate platform demonstrated that it can not only competitively bind the membrane receptors but also be sequentially uptaken by cells via endocytosis, efficiently reducing the expression of target receptors through the action of RNA interference. Therefore, we believe that the use of conjugates rather than protein/peptide or siRNA alone provides additional benefits for the functional control of receptors. Therefore, in addition to anticancer therapy, this platform is expected to be utilized in various disease-targeting membrane receptors.
Acknowledgments
This work was supported by the Mid-Career Researcher Program (NRF-2022R1A2C2006861), the KU-KIST Graduate School of Converging Science and Technology (Korea University), and the Intramural Research Program of Korea Institute of Science and Technology (KIST).
Glossary
ABBREVIATIONS
- TME
tumor microenvironment
- EGFR
epidermal growth factor receptor
- PD-L1
programmed death-ligand 1
- SIRPα
signal regulatory protein alpha
- mAb
monoclonal antibodies
- Fc region
fragment crystallizable region
- siRNA
small interfering RNA
- nt
nucleotides
- DBCO
diarylcyclooctyne
- BMDMs
bone marrow-derived macrophages
- EMT
epithelial-mesenchymal transition
The authors declare no competing financial interest.
References
- Baghban R.; Roshangar L.; Jahanban-Esfahlan R.; Seidi K.; Ebrahimi-Kalan A.; Jaymand M.; Kolahian S.; Javaheri T.; Zare P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal 2020, 18 (1), 59. 10.1186/s12964-020-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun J. Y.; Leem S. H.; Lee J. H.; Kim H. S. Dual Relationship Between Stromal Cells and Immune Cells in the Tumor Microenvironment. Front. Immunol. 2022, 13, 864739. 10.3389/fimmu.2022.864739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar S.; Emdad L.; Das S. K.; Fisher P. B. EGFR: An essential receptor tyrosine kinase-regulator of cancer stem cells. Adv. Cancer Res. 2020, 147, 161–188. 10.1016/bs.acr.2020.04.003. [DOI] [PubMed] [Google Scholar]
- Akinleye A.; Rasool Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J. Hematol. Oncol. 2019, 12 (1), 92. 10.1186/s13045-019-0779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Casey S. C.; Tong L.; Li Y.; Do R.; Walz S.; Fitzgerald K. N.; Gouw A. M.; Baylot V.; Gutgemann I.; Eilers M.; et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016, 352 (6282), 227–231. 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang J.; Liu F.; Li C.; Liang X.; Li C.; Liu Y.; Yi Z.; Zhang L.; Fu S.; Zeng Y. Role of CD47 in tumor immunity: a potential target for combination therapy. Sci. Rep. 2022, 12 (1), 9803. 10.1038/s41598-022-13764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lian S.; Xie R.; Ye Y.; Lu Y.; Cheng Y.; Xie X.; Li S.; Jia L. Dual blockage of both PD-L1 and CD47 enhances immunotherapy against circulating tumor cells. Sci. Rep. 2019, 9 (1), 4532. 10.1038/s41598-019-40241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q.; Qiu X.; Zhang Z.; Zhang S.; Zhang Y.; Liang Y.; Guo J.; Peng H.; Chen M.; Fu Y. X.; et al. PD-L1 on dendritic cells attenuates T cell activation and regulates response to immune checkpoint blockade. Nat. Commun. 2020, 11 (1), 4835. 10.1038/s41467-020-18570-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Lee C. K.; Lin C. H.; Gassen R. B.; Xu X.; Huang Z.; Xiao C.; Bonorino C.; Lu L. F.; Bui J. D.; et al. PD-L1:CD80 Cis-Heterodimer Triggers the Co-stimulatory Receptor CD28 While Repressing the Inhibitory PD-1 and CTLA-4 Pathways. Immunity 2019, 51 (6), 1059–1073.e9. 10.1016/j.immuni.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu T.; Li B.; Wang Y. Targeting CD47/SIRPalpha as a therapeutic strategy, where we are and where we are headed. Biomark. Res. 2022, 10 (1), 20. 10.1186/s40364-022-00373-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bouwstra R.; van Meerten T.; Bremer E. CD47-SIRPalpha blocking-based immunotherapy: Current and prospective therapeutic strategies. Clin. Transl. Med. 2022, 12 (8), e943 10.1002/ctm2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jalil A. R.; Andrechak J. C.; Discher D. E. Macrophage checkpoint blockade: results from initial clinical trials, binding analyses, and CD47-SIRPalpha structure-function. Antib. Ther. 2020, 3 (2), 80–94. 10.1093/abt/tbaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf K.; Jahchan N. S.; Schnorr P. J.; Cristea S.; Ring A. M.; Maute R. L.; Volkmer A. K.; Volkmer J. P.; Liu J.; Lim J. S.; et al. CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J. Clin. Invest. 2016, 126 (7), 2610–2620. 10.1172/JCI81603. [DOI] [PMC free article] [PubMed] [Google Scholar]; Majeti R.; Chao M. P.; Alizadeh A. A.; Pang W. W.; Jaiswal S.; Gibbs K. D. Jr; van Rooijen N.; Weissman I. L. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 2009, 138 (2), 286–299. 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf K.; Ring A. M.; Ho C. C.; Volkmer J. P.; Levin A. M.; Volkmer A. K.; Ozkan E.; Fernhoff N. B.; van de Rijn M.; Weissman I. L.; et al. Engineered SIRPalpha variants as immunotherapeutic adjuvants to anticancer antibodies. Science 2013, 341 (6141), 88–91. 10.1126/science.1238856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon H.; Reichert J. M. Antibodies to watch in 2021. mAbs 2021, 13 (1), 1860476. 10.1080/19420862.2020.1860476. [DOI] [PMC free article] [PubMed] [Google Scholar]; Das M.; Mohanty C.; Sahoo S. K. Ligand-based targeted therapy for cancer tissue. Expert Opin. Drug Delivery 2009, 6 (3), 285–304. 10.1517/17425240902780166. [DOI] [PubMed] [Google Scholar]; Carter P. J.; Senter P. D. Antibody-drug conjugates for cancer therapy. Cancer J. 2008, 14 (3), 154–169. 10.1097/PPO.0b013e318172d704. [DOI] [PubMed] [Google Scholar]
- Cai W. Q.; Zeng L. S.; Wang L. F.; Wang Y. Y.; Cheng J. T.; Zhang Y.; Han Z. W.; Zhou Y.; Huang S. L.; Wang X. W.; et al. The Latest Battles Between EGFR Monoclonal Antibodies and Resistant Tumor Cells. Front. Oncol. 2020, 10, 1249. 10.3389/fonc.2020.01249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey J. D.; Zhang B. Cancer Immunotherapy Update: FDA-Approved Checkpoint Inhibitors and Companion Diagnostics. AAPS J. 2021, 23 (2), 39. 10.1208/s12248-021-00574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adashek J. J.; Subbiah I. M.; Matos I.; Garralda E.; Menta A. K.; Ganeshan D. M.; Subbiah V. Hyperprogression and Immunotherapy: Fact, Fiction, or Alternative Fact?. Trends Cancer 2020, 6 (3), 181–191. 10.1016/j.trecan.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camelliti S.; Le Noci V.; Bianchi F.; Moscheni C.; Arnaboldi F.; Gagliano N.; Balsari A.; Garassino M. C.; Tagliabue E.; Sfondrini L.; et al. Mechanisms of hyperprogressive disease after immune checkpoint inhibitor therapy: what we (don’t) know. J. Exp. Clin. Cancer Res. 2020, 39 (1), 236. 10.1186/s13046-020-01721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplen N. J.; Parrish S.; Imani F.; Fire A.; Morgan R. A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 9742. 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb G. B.; Brown K. M.; Khurana J.; Rana T. M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct Mol. Biol. 2005, 12 (2), 133–137. 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- Devi G. R. siRNA-based approaches in cancer therapy. Cancer Gene Ther. 2006, 13 (9), 819–829. 10.1038/sj.cgt.7700931. [DOI] [PubMed] [Google Scholar]; Deng Y.; Wang C. C.; Choy K. W.; Du Q.; Chen J.; Wang Q.; Li L.; Chung T. K.; Tang T. Therapeutic potentials of gene silencing by RNA interference: principles, challenges, and new strategies. Gene 2014, 538 (2), 217–227. 10.1016/j.gene.2013.12.019. [DOI] [PubMed] [Google Scholar]; Bora R. S.; Gupta D.; Mukkur T. K.; Saini K. S. RNA interference therapeutics for cancer: challenges and opportunities (review). Mol. Med. Rep. 2012, 6 (1), 9–15. 10.3892/mmr.2012.871. [DOI] [PubMed] [Google Scholar]
- Ko Y. J.; Lee J. W.; Kim H.; Cho E.; Yang Y.; Kim I.-S.; Kim S. H.; Kwon I. C. Versatile activatable vSIRPα-probe for cancer-targeted imaging and macrophage-mediated phagocytosis of cancer cells. J. Controlled Release 2020, 323, 376–386. 10.1016/j.jconrel.2020.04.037. [DOI] [PubMed] [Google Scholar]
- Chang H. N.; Liu B. Y.; Qi Y. K.; Zhou Y.; Chen Y. P.; Pan K. M.; Li W. W.; Zhou X. M.; Ma W. W.; Fu C. Y.; et al. Blocking of the PD-1/PD-L1 Interaction by a D-Peptide Antagonist for Cancer Immunotherapy. Angew. Chem., Int. Ed. Engl. 2015, 54 (40), 11760–11764. 10.1002/anie.201506225. [DOI] [PubMed] [Google Scholar]
- Kim E. H.; Lee J.; Kwak G.; Jang H.; Kim H.; Cho H.; Jang Y.; Choi J.; Chi S. G.; Kim K.; et al. PDL1-binding peptide/anti-miRNA21 conjugate as a therapeutic modality for PD-L1(high) tumors and TAMs. J. Controlled Release 2022, 345, 62–74. 10.1016/j.jconrel.2022.02.031. [DOI] [PubMed] [Google Scholar]
- Hossein-Nejad-Ariani H.; Althagafi E.; Kaur K. Small Peptide Ligands for Targeting EGFR in Triple Negative Breast Cancer Cells. Sci. Rep. 2019, 9 (1), 2723. 10.1038/s41598-019-38574-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath M. F.; Ogawa T.; Bold A. J. D. Ras dexamethasone-induced protein 1 is a modulator of hormone secretion in the volume overloaded heart. Am. J. Physiol.: Heart Circ. Physiol. 2012, 302 (9), H1826–H1837. 10.1152/ajpheart.01085.2011. [DOI] [PubMed] [Google Scholar]
- Seo S.; Takayama K.; Uno K.; Ohi K.; Hashimoto R.; Nishizawa D.; Ikeda K.; Ozaki N.; Nabeshima T.; Miyamoto Y.; et al. Functional analysis of deep intronic SNP rs13438494 in intron 24 of PCLO gene. PLoS One 2013, 8 (10), e76960 10.1371/journal.pone.0076960. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jeong A. K.; Sung Hak K.; In Sung K.; Da Yoon Y.; Gwang Il K.; Yang Soo M.; Sung Chan K.; Seung Ho L.; Sang Suk L.; Cheol Heui Y.; et al. Galectin-9 Induced by Dietary Prebiotics Regulates Immunomodulation to Reduce Atopic Dermatitis Symptoms in 1-Chloro-2,4-Dinitrobenzene (DNCB)-Treated NC/Nga Mice. J. Microbiol. Biotechnol. 2020, 30 (9), 1343–1354. 10.4014/jmb.2005.05017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Xie X.; Yeganeh P. N.; Lee D. J.; Valle-Garcia D.; Meza-Sosa K. F.; Junqueira C.; Su J.; Luo H. R.; Hide W.; et al. Immunotherapy for breast cancer using EpCAM aptamer tumor-targeted gene knockdown. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (9), e2022830118 10.1073/pnas.2022830118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Huang X.; Xu J.; Li E.; Lao M.; Tang T.; Zhang G.; Guo C.; Zhang X.; Chen W.; et al. NEK2 inhibition triggers anti-pancreatic cancer immunity by targeting PD-L1. Nat. Commun. 2021, 12 (1), 4536. 10.1038/s41467-021-24769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang S.; You X.; Xu T.; Chen Q.; Li H.; Dou L.; Sun Y.; Xiong X.; Meredith M. A.; Sun Y. PD-L1 induction via the MEK-JNK-AP1 axis by a neddylation inhibitor promotes cancer-associated immunosuppression. Cell Death Dis. 2022, 13 (10), 844. 10.1038/s41419-022-05292-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Chan S. T.; Kim J. Y.; Ou J. J. Hepatitis C Virus Induces the Ubiquitin-Editing Enzyme A20 via Depletion of the Transcription Factor Upstream Stimulatory Factor 1 To Support Its Replication. mBio 2019, 10 (4), e01660–19 10.1128/mBio.01660-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasson M.; Hedman H.; Guo D.; Ljungberg B.; Henriksson R. LRIG1 and epidermal growth factor receptor in renal cell carcinoma: a quantitative RT–RCR and immunohistochemical analysis. Br. J. Cancer 2004, 90 (12), 2425. 10.1038/sj.bjc.6601773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam G. H.; Hong Y.; Choi Y.; Kim G. B.; Kim Y. K.; Yang Y.; Kim I. S. An optimized protocol to determine the engulfment of cancer cells by phagocytes using flow cytometry and fluorescence microscopy. J. Immunol. Methods 2019, 470, 27–32. 10.1016/j.jim.2019.04.007. [DOI] [PubMed] [Google Scholar]
- Kim E.; Koo H. Biomedical applications of copper-free click chemistry: in vitro, in vivo, and ex vivo. Chem. Sci. 2019, 10 (34), 7835–7851. 10.1039/c9sc03368h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eladl E.; Tremblay-LeMay R.; Rastgoo N.; Musani R.; Chen W.; Liu A.; Chang H. Role of CD47 in Hematological Malignancies. J. Hematol. Oncol. 2020, 13 (1), 96. 10.1186/s13045-020-00930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha B.; Lv Z.; Bian Z.; Zhang X.; Mishra A.; Liu Y. ’Clustering’ SIRPalpha into the plasma membrane lipid microdomains is required for activated monocytes and macrophages to mediate effective cell surface interactions with CD47. PLoS One 2013, 8 (10), e77615 10.1371/journal.pone.0077615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H.; Hsu J. M.; Yang W. H.; Hung M. C. Mechanisms regulating PD-L1 expression in cancers and associated opportunities for novel small-molecule therapeutics. Nat. Rev. Clin. Oncol. 2022, 19 (5), 287–305. 10.1038/s41571-022-00601-9. [DOI] [PubMed] [Google Scholar]
- Dong H.; Zhu G.; Tamada K.; Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 1999, 5 (12), 1365–1369. 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- Uramoto H.; Iwata T.; Onitsuka T.; Shimokawa H.; Hanagiri T.; Oyama T. Epithelial-mesenchymal transition in EGFR-TKI acquired resistant lung adenocarcinoma. Anticancer Res. 2010, 30 (7), 2513–2517. [PubMed] [Google Scholar]
- Cascone T.; Martinelli E.; Morelli M. P.; Morgillo F.; Troiani T.; Ciardiello F. Epidermal growth factor receptor inhibitors in non-small-cell lung cancer. Expert Opin. Drug Discovery 2007, 2 (3), 335–348. 10.1517/17460441.2.3.335. [DOI] [PubMed] [Google Scholar]
- Soutschek J.; Akinc A.; Bramlage B.; Charisse K.; Constien R.; Donoghue M.; Elbashir S.; Geick A.; Hadwiger P.; Harborth J.; et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004, 432 (7014), 173–178. 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]; Shmushkovich T.; Monopoli K. R.; Homsy D.; Leyfer D.; Betancur-Boissel M.; Khvorova A.; Wolfson A. D. Functional features defining the efficacy of cholesterol-conjugated, self-deliverable, chemically modified siRNAs. Nucleic Acids Res. 2018, 46 (20), 10905–10916. 10.1093/nar/gky745. [DOI] [PMC free article] [PubMed] [Google Scholar]; Foster D. J.; Brown C. R.; Shaikh S.; Trapp C.; Schlegel M. K.; Qian K.; Sehgal A.; Rajeev K. G.; Jadhav V.; Manoharan M.; et al. Advanced siRNA Designs Further Improve In Vivo Performance of GalNAc-siRNA Conjugates. Mol. Ther. 2018, 26 (3), 708–717. 10.1016/j.ymthe.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Janas M. M.; Schlegel M. K.; Harbison C. E.; Yilmaz V. O.; Jiang Y. F.; Parmar R.; Zlatev I.; Castoreno A.; Xu H. L.; Shulga-Morskaya S.; et al. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 2018, 9, 723. 10.1038/s41467-018-02989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W.; Choi J.; Choi Y.; Kim K.; Yang Y.; Kim S. H.; Yoon H. Y.; Kwon I. C. Molecularly engineered siRNA conjugates for tumor-targeted RNAi therapy. J. Controlled Release 2022, 351, 713–726. 10.1016/j.jconrel.2022.09.040. [DOI] [PubMed] [Google Scholar]; Chernikov I. V.; Vlassov V. V.; Chernolovskaya E. L. Current Development of siRNA Bioconjugates: From Research to the Clinic. Front. Pharmacol. 2019, 10, 444. 10.3389/fphar.2019.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tai W. Current Aspects of siRNA Bioconjugate for In Vitro and In Vivo Delivery. Molecules 2019, 24 (12), 2211. 10.3390/molecules24122211. [DOI] [PMC free article] [PubMed] [Google Scholar]; Biscans A.; Coles A.; Haraszti R.; Echeverria D.; Hassler M.; Osborn M.; Khvorova A. Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo. Nucleic Acids Res. 2019, 47 (3), 1082–1096. 10.1093/nar/gky1239. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kuwahara H.; Nishina K.; Yoshida K.; Nishina T.; Yamamoto M.; Saito Y.; Piao W.; Yoshida M.; Mizusawa H.; Yokota T. Efficient in vivo delivery of siRNA into brain capillary endothelial cells along with endogenous lipoprotein. Mol. Ther. 2011, 19 (12), 2213–2221. 10.1038/mt.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittrup K. D. Protein engineering by cell-surface display. Curr. Opin. Biotechnol. 2001, 12 (4), 395–399. 10.1016/S0958-1669(00)00233-0. [DOI] [PubMed] [Google Scholar]; Ueda M. Establishment of cell surface engineering and its development. Biosci., Biotechnol., Biochem. 2016, 80 (7), 1243–1253. 10.1080/09168451.2016.1153953. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Rajendran V.; Sethumadhavan R.; Purohit R. In silico prediction of a disease-associated STIL mutant and its affect on the recruitment of centromere protein J (CENPJ). FEBS Open Bio. 2012, 2, 285–293. 10.1016/j.fob.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Purohit R.; Rajasekaran R.; Sudandiradoss C.; George Priya Doss C.; Ramanathan K.; Rao S. Studies on flexibility and binding affinity of Asp25 of HIV-1 protease mutants. Int. J. Biol. Macromol. 2008, 42 (4), 386–391. 10.1016/j.ijbiomac.2008.01.011. [DOI] [PubMed] [Google Scholar]; Tanwar G.; Mazumder A. G.; Bhardwaj V.; Kumari S.; Bharti R.; Yamini; Singh D.; Das P.; Purohit R. Target identification, screening and in vivo evaluation of pyrrolone-fused benzosuberene compounds against human epilepsy using Zebrafish model of pentylenetetrazol-induced seizures. Sci. Rep. 2019, 9 (1), 7904. 10.1038/s41598-019-44264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud M.; El Madani M.; Massaad-Massade L. Small interfering RNA from the lab discovery to patients’ recovery. J. Controlled Release 2020, 321, 616–628. 10.1016/j.jconrel.2020.02.032. [DOI] [PubMed] [Google Scholar]; Chiu Y. L.; Rana T. M. siRNA function in RNAi: a chemical modification analysis. RNA 2003, 9, 1034. 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hu B.; Zhong L.; Weng Y.; Peng L.; Huang Y.; Zhao Y.; Liang X. J. Therapeutic siRNA: state of the art. Signal Transduction Targeted Ther. 2020, 5 (1), 101. 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid M. I.; Moazzam M.; Kato S.; Yeseom Cho K.; Tiwari R. K. Overcoming Barriers for siRNA Therapeutics: From Bench to Bedside. Pharmaceuticals 2020, 13 (10), 294. 10.3390/ph13100294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K.; Pang L.; Pan X.; Fan S.; Wang X.; Wang Q.; Dai P.; Gao W.; Gao J. GE11 Modified PLGA/TPGS Nanoparticles Targeting Delivery of Salinomycin to Breast Cancer Cells. Technol. Cancer Res. Treat. 2021, 20, 15330338211004954. 10.1177/15330338211004954. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang X.; Chen L.; Zhang Y.; Zhou S.; Cai H. H.; Li T.; Jin H.; Cai J.; Zhou H.; Pi J. GE11 Peptide Conjugated Liposomes for EGFR-Targeted and Chemophotothermal Combined Anticancer Therapy. Bioinorg. Chem. Appl. 2021, 2021, 5534870. 10.1155/2021/5534870. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang X.; Zhao M.; Wu Z.; Chen C.; Zhang Y.; Wang L.; Guo Q.; Wang Q.; Liang S.; Hu S.; et al. Nano-ultrasonic Contrast Agent for Chemoimmunotherapy of Breast Cancer by Immune Metabolism Reprogramming and Tumor Autophagy. ACS Nano 2022, 16 (2), 3417–3431. 10.1021/acsnano.2c00462. [DOI] [PubMed] [Google Scholar]