Abstract
Isoniazid (INH) activation in vitro is associated with reduction of the mycobacterial ferric KatG catalase-peroxidase by hydrazine and reaction with O2 to form an oxyferrous enzyme complex. Since this complex could also form directly via reaction of ferric KatG with superoxide, intracellular activation might be responsive to superoxide concentration. When Mycobacterium smegmatis carrying the M. bovis katG gene was treated with nontoxic levels of plumbagin, a generator of superoxide, the bacteriostatic activity of INH increased unless a plasmid-borne superoxide dismutase gene was also present. Thus, endogenous superoxide probably contributes to intracellular activation of INH.
Isoniazid (isonicotinic acid hydrazide [INH]) has been a first-line antitubercular agent for decades, but only recently has an understanding of its action against Mycobacterium tuberculosis emerged. One of the key observations was that defects in katG, the gene encoding the catalase-peroxidase of M. tuberculosis, lead to INH resistance (25). Resistance can also be obtained by mutation of a locus called inhA (1, 7). In M. smegmatis, the InhA protein is an NADH-dependent fatty acyl enoyl acyl carrier protein reductase thought to participate in the synthesis of mycolic acid (3). Since INH interferes with mycolic acid synthesis (16, 21), there emerged the idea that INH enters the cell as a prodrug, is activated by the KatG protein, and then inactivates the InhA protein. Supporting biochemical studies subsequently showed that INH inactivates purified InhA protein in the presence of Mn2+ through a reaction stimulated by the KatG catalase-peroxidase (10, 23). For M. tuberculosis, it has been argued that InhA is not the primary target (15), and the AcpM-KasA complex, which is also involved in mycolic acid synthesis, has been found bound to activated INH (1a). Activation of INH may also affect DNA, proteins, and other macromolecules through formation of reactive oxygen species (9, 11). The latter idea is supported by three related observations (reviewed in reference [2]): (i) the KatG-dependent modification of INH generates reactive oxygen species; (ii) M. tuberculosis is hypersensitive to INH and lacks a functional copy of oxyR, a positive regulator of the oxygen defense response; and (iii) low-level INH resistance is conferred by upregulation of ahpC, whose product, alkyl hydroperoxide reductase, detoxifies organic peroxides (24). Moreover, inactivation of ahpC or addition of hydrogen peroxide leads to an increase in the INH sensitivity of M. smegmatis (18, 24). Thus, oxidative activation of INH can have both a specific effect on mycolic acid synthesis and a more general toxicity for proteins and nucleic acids. These general ideas are sketched in Fig. 1.
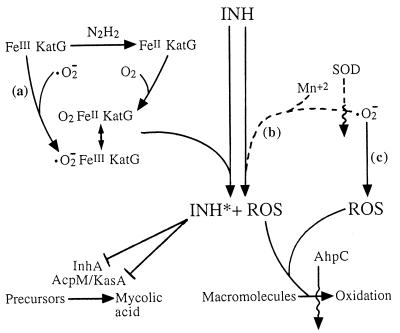
FIG. 1.
Potential involvement of superoxide in INH action. INH is shown as a prodrug activated by KatG protein (catalase-peroxidase) or Mn2+ action. At the left of the figure is a schema showing the resting form of KatG (FeIII KatG) being converted to an active form by two pathways, one of which (a) requires superoxide (·O2−). On the right is a schema showing activation of INH by an Mn2+-dependent pathway that also involves superoxide (b), as indicated by inhibition by superoxide dismutase (SOD). This pathway is shown with a dashed line because it is unlikely to be significant in vivo (see text); in vivo, there may be a comparable katG-independent, peroxide-dependent activation (18). Activated INH (INH*) blocks mycolic acid synthesis by inactivating InhA in M. smegmatis and AcpM-KasA in M. tuberculosis. Reactive oxygen species (ROS) arise during activation of INH (19, 20) or from the presence of superoxide (c). The AhpC protein appears to limit accumulation of the macromolecular oxidative damage that is expected to arise from activation of INH or the presence of superoxide. Wavy lines indicate inhibition of a pathway; lines with a perpendicular bar indicate inhibition of enzymes.
To better understand INH activation, several studies have focussed on the role of the KatG catalase-peroxidase in cell-free systems. Surprisingly, activation of INH is not simply a direct peroxidation, as evidenced by the following: the enzyme catalyzes INH oxidation in the absence of peroxide (10); oxidation of INH requires a reducing agent such as hydrazine, a spontaneous decomposition product appearing in INH solutions (14); and INH is activated only under aerobic conditions (14). These observations led to the idea (14) that a reduced form of the enzyme, containing ferrous heme, rather than the ferric “resting” enzyme reacts with O2 to produce an active oxyferrous enzyme (Fig. 1). Whether such a reaction occurs in vivo is not known. The oxyferrous enzyme, analogous to peroxidase compound III, is a resonance equivalent of the superoxyferric enzyme; consequently, a direct reaction between the ferric resting enzyme and endogenous superoxide anion could also activate the enzyme (Fig. 1, pathway a). It has also been reported that a KatG-independent pathway for INH attack on the InhA protein can be prevented by the presence of superoxide dismutase (23) (Fig. 1, pathway b). If either the direct, superoxy route for activation of the KatG catalase-peroxidase or the KatG-independent pathway is important inside cells, INH cytotoxicity should depend on the availability of superoxide anion. Below we describe experiments with M. smegmatis in which INH activity increased when cultures were treated with plumbagin, a redox cycling agent expected to increase the intracellular superoxide concentrations (5). The enhancing action of plumbagin was reversed by introducing into M. smegmatis a plasmid that allows overexpression of superoxide dismutase, a procedure expected to lower the superoxide concentration. These observations led to the conclusion that superoxide stimulates the intracellular action of INH.
M. smegmatis was selected for this study rather than M. tuberculosis because the former exhibits a higher level of expression of ahpC (4). As pointed out above, the product of this gene detoxifies organic peroxides, and so treatments that increase the superoxide concentration are less likely to have a confounding toxic effect in M. smegmatis (plumbagin was expected to stimulate pathway c in Fig. 1). To minimize the effects of differences between the catalase-peroxidases of M. smegmatis and those of members of the M. tuberculosis complex, we introduced the katG gene from M. bovis BCG into an INH-resistant mutant of M. smegmatis that expressed low levels of catalase (8). For this construction, transformation of strain BH1 (8) was carried out with a derivative of the integration vector pYUB295 (provided by A. Brown, Albert Einstein School of Medicine) into which the M. bovis BCG katG gene had been subcloned from pYUB318 (provided by E. Dubnau, Public Health Research Institute). The resulting plasmid, pJYW1333, lacks a mycobacterial origin of replication and has an att site for integration; consequently, transformants were expected to carry chromosomal insertions of katG. Southern transfer hybridization confirmed the presence of M. bovis BCG katG in the chromosome of M. smegmatis (data not shown). The resulting strain, KD1337, was 30-fold more susceptible to INH than the parental strain (BH1) when assayed for colony-forming ability on 7H10 agar containing INH.
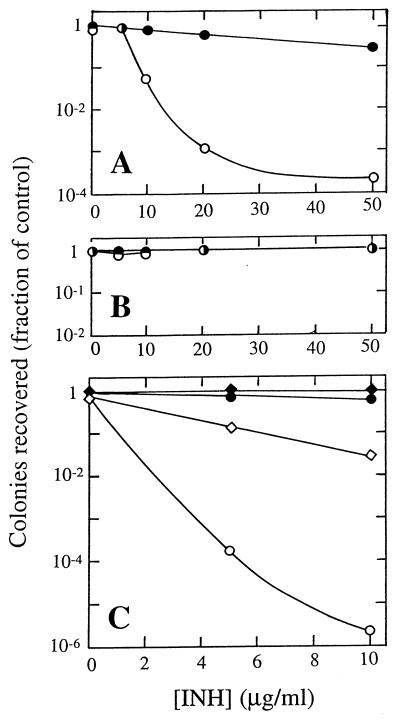
The effect of elevation of the superoxide concentration on INH activity was observed by plating strain KD1337 (katG+) on 7H10 agar containing combinations of INH and plumbagin (technical grade; Sigma Chemical Co., St. Louis, Mo.; 20 μM). As shown in Fig. 2A, the presence of plumbagin made INH considerably more effective at blocking colony formation. Under these conditions, plumbagin by itself reduced colony formation by less than 20%. As a control, the same experiment was carried out with the parental strain (BH1), which is deficient in catalase-peroxidase. In this case, cultures were quite resistant to INH, and 20 μM plumbagin had no stimulatory effect (Fig. 2B). Similar results were obtained when the two strains were grown in 7H9 liquid medium and growth was determined by measuring changes in culture turbidity (data not shown). The inability of plumbagin to enhance INH potency in the katG mutant (strain BH1) indicates that a KatG-independent pathway for INH activation (pathway b in Fig. 1) plays at most a minor role in living cells.
FIG. 2.
INH activity during perturbation of the intracellular superoxide concentration. (A) Enhancement by plumbagin of INH activity against M. smegmatis containing a katG gene from M. tuberculosis. An exponentially growing culture of strain KD1337 (107 cells/ml) was serially diluted and plated on 7H10 agar containing the indicated concentration of INH plus 0 μM (solid symbols) or 20 μM (open symbols) plumbagin. Numbers of colonies are expressed as fractions of the number of colonies obtained on control plates lacking INH or plumbagin. (B) katG is required to observe plumbagin enhancement of INH activity. M. smegmatis BH1 was plated on agar containing the indicated concentrations of INH in the presence or absence of plumbagin, as in panel A. (C) Antagonism of INH activity by expression of plasmid-borne superoxide dismutatase. Strains KD1337 (parental) (circles) and KD1593 (pSOD3) (diamonds) were plated on 7H10 agar containing the indicated concentrations of INH with no addition (solid symbols) or 40 μM plumbagin (open symbols). Numbers of colonies are expressed as fractions of the number of colonies obtained on control plates lacking INH or plumbagin.
We also performed the same plating experiment in cells transformed with plasmid pSOD3, which expresses superoxide dismutase (6). Strain KD1337 was transformed with pSOD3, and the effect of plumbagin on INH activity was measured as described above. As shown in Fig. 2C, the presence of plasmid pSOD3 limited the enhancement by plumbagin of INH toxicity. A vector identical to pSOD3 except for the presence of the katG gene and associated linking sequences had no effect on the ability of plumbagin to enhance INH activity (data not shown). These results argue that the effect of plumbagin is via production of superoxide anion.
As an additional control, we examined the ability of plumbagin treatment to elevate expression of KatG, since that would provide an explanation for increased INH activity unrelated to activation of the catalase-peroxidase. Catalase activity was assayed (8) in extracts prepared from KD1337 grown in 7H9 medium with or without INH (50 μg/ml) or plumbagin (30 μM). INH treatment caused a 50% increase in catalase activity, from 0.1 to 0.15 U/mg of total protein; plumbagin reduced activity by 20%, to 0.08 U/mg of total protein.
In summary, chemical considerations led to the expectation that superoxide stimulates INH activity in mycobacteria. We showed that plumbagin, a superoxide generator, enhanced INH action except when a plasmid-borne superoxide dismutase gene was present. It should be possible to improve INH therapy for mycobacterial diseases by supplementing INH treatments with INH and agents such as clofazimine. This superoxide-producing compound (22) is sometimes used for treatment of leprosy (12, 13) and shows activity against M. tuberculosis (17).
Acknowledgments
We thank D. Wu, Y. Dong, and B.-Y. Zhao for construction and testing of KD1337; S. Cole, E. Dubnau, and V. Escuyer for providing strains or plasmids; V. Deretic for stimulating discussions; and M. Gennaro and I. Smith for critical comments on the manuscript.
This work was supported by NIH grant AI35257.
REFERENCES
- 1.Banerjee A, Dubnau E, Quémard A, Balasubramanian V, Um K S, Wilson T, Collins D, de Lisle G, Jacobs W R. inhA, a gene encoding a target for isoniazid and ethionamide resistance in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 1a.Barry, C. Personal communication.
- 2.Deretic V, Pagan-Ramos E, Zhang Y, Dhandayuthapani S, Via L. The extreme sensitivity of Mycobacterium tuberculosis to the front-line antituberculosis drug isoniazid. Nat Biotechnol. 1996;14:1557–1561. doi: 10.1038/nbt1196-1557. [DOI] [PubMed] [Google Scholar]
- 3.Dessen A, Quémard A, Blanchard J, Jacobs W, Sacchettini J C. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science. 1995;267:1638–1641. doi: 10.1126/science.7886450. [DOI] [PubMed] [Google Scholar]
- 4.Dhandayuthapani S, Zhang Y, Mudd M H, Deretic V. Oxidative stress response and its role in sensitivity to isoniazid in mycobacteria: characterization and inducibility of ahpC by peroxides in Mycobacterium smegmatis and lack of expression in M. aurum and M. tuberculosis. J Bacteriol. 1996;178:3641–3649. doi: 10.1128/jb.178.12.3641-3649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiGuiseppe J, Fridovich I. Oxygen toxicity in Streptococcus sanguis: the relative importance of superoxide and hydroxyl radicals. J Biol Chem. 1982;257:4046–4051. [PubMed] [Google Scholar]
- 6.Escuyer V, Haddod N, Frehel C, Berche P. Molecular characterization of a surface-exposed superoxide dismutase of Mycobacterium avium. Microb Pathog. 1996;20:41–55. doi: 10.1006/mpat.1996.0004. [DOI] [PubMed] [Google Scholar]
- 7.Heym B, Alzari P, Honore N, Cole S. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol Microbiol. 1995;15:235–245. doi: 10.1111/j.1365-2958.1995.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 8.Heym B, Cole S T. Isolation and characterization of isoniazid-resistant mutants of Mycobacterium smegmatis and M. aurum. Res Microbiol. 1992;143:721–730. doi: 10.1016/0923-2508(92)90067-x. [DOI] [PubMed] [Google Scholar]
- 9.Ito K, Yamamoto K, Kawanishi S. Manganese-mediated oxidative damage of cellular and isolated DNA by isoniazid and related hydrazines: non-Fenton-type hydroxyl radical formation. Biochemistry. 1992;31:11606–11613. doi: 10.1021/bi00161a046. [DOI] [PubMed] [Google Scholar]
- 10.Johnsson K, King D, Schultz P. Studies on the mechanism of action of isoniazid and ethionamide in the chemotherapy of tuberculosis. J Am Chem Soc. 1995;117:5009–5010. [Google Scholar]
- 11.Johnsson K, Schultz P. Mechanistic studies of the oxidation of isoniazid by the catalase peroxidase from Mycobacterium tuberculosis. J Am Chem Soc. 1994;116:7425–7426. [Google Scholar]
- 12.Kapoor V K. Clofazimine. Anal Profiles Drug Subst. 1989;18:91–120. [Google Scholar]
- 13.Karat A B, Jeevaratnam A, Karat S, Rao P S. Controlled clinical trial of clofazimine in untreated lepromatous leprosy. Br Med J. 1971;4:514–516. doi: 10.1136/bmj.4.5786.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magliozzo R S, Marcinkeviciene J A. Evidence for isoniazid oxidation to oxyferrous mycobacterial catalase-peroxidase. J Am Chem Soc. 1996;118:11303–11304. [Google Scholar]
- 15.Mdluli K, Sherman D, Hickey M, Kreiswirth B, Morris S, Stover C K, Barry C E. Biochemical and genetic data suggest that InhA is not the primary target for activated isoniazid in Mycobacterium tuberculosis. J Infect Dis. 1996;174:1085–1090. doi: 10.1093/infdis/174.5.1085. [DOI] [PubMed] [Google Scholar]
- 16.Quémard A, Lacave C, Lanéelle G. Isoniazid inhibition of mycolic acid synthesis by cell extracts of sensitive and resistant strains of Mycobacterium aurum. Antimicrob Agents Chemother. 1991;35:1035–1039. doi: 10.1128/aac.35.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy V M, Nadadhur G, Daneluzzi D, O’Sullivan J F, Gangadharam P R J. Antituberculosis activities of clofazimine and its new analogs B4154 and B4157. Antimicrob Agents Chemother. 1996;40:633–636. doi: 10.1128/aac.40.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosner J L, Storz G. Effects of peroxides on susceptibilities of Escherichia coli and Mycobacterium smegmatis to isoniazid. Antimicrob Agents Chemother. 1994;38:1829–1833. doi: 10.1128/aac.38.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoeb H A, Bowman B U, Jr, Ottolenghi A C, Merola A J. Enzymatic and nonenzymatic superoxide-generating reactions of isoniazid. Antimicrob Agents Chemother. 1985;27:408–412. doi: 10.1128/aac.27.3.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoeb H A, Bowman B U, Jr, Ottolenghi A C, Merola A J. Evidence for the generation of active oxygen by isoniazid treatment of extracts of Mycobacterium tuberculosis H37Ra. Antimicrob Agents Chemother. 1985;27:404–407. doi: 10.1128/aac.27.3.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takayama K, Wang L, David H L. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1972;2:29–35. doi: 10.1128/aac.2.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warek U, Falkinham J O., III Action of clofazimine on the Mycobacterium avium complex. Res Microbiol. 1996;147:43–48. doi: 10.1016/0923-2508(96)80202-6. [DOI] [PubMed] [Google Scholar]
- 23.Zabinski R F, Blanchard J S. The requirement for manganese and oxygen in the isoniazid-dependent inactivation of Mycobacterium tuberculosis enoyl reductase. J Am Chem Soc. 1997;119:2331–2332. [Google Scholar]
- 24.Zhang Y, Dhandayuthapani S, Deretic V. Molecular basis for the exquisite sensitivity of Mycobacterium tuberculosis to isoniazid. Proc Natl Acad Sci USA. 1996;93:13212–13216. doi: 10.1073/pnas.93.23.13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]