Abstract
Cutavirus (CuV) is associated with cutaneous T-cell lymphoma (CTCL), of which parapsoriasis is a precursor. Our study reveals a significantly higher CuV-DNA prevalence in skin swabs of parapsoriasis patients (6/13; 46.2%) versus those of healthy adults (1/51; 1.96%). Eight patients (8/12; 66.7%) had CuV DNA in biopsied skin, and 4 developed CTCL.
Keywords: protoparvovirus, disease association, cutaneous T-cell lymphoma, parapsoriasis, in situ hybridization
Cutaneous T-cell lymphomas (CTCLs) are rare skin malignancies of mature T cells that are more often diagnosed in men than in women, and usually in middle-age. The most common subtype of CTCL is mycosis fungoides (MF), accounting for 60% of CTCL cases [1]. Although MF can occur de novo, many patients with MF first present with longstanding reactive inflammatory conditions such as parapsoriasis en plaques (PP) [2]. Parapsoriasis en plaques is divided into 2 clinically different entities: small plaque parapsoriasis (SPP) and large plaque parapsoriasis (LPP). The exact cause of CTCL remains unknown, but it has been speculated that chronic antigen stimulation with long-lasting inflammatory responses possibly could induce T-cell proliferation, which would develop into malignancy. A viral etiology for CTCL has therefore long been sought but not found [3].
In 2012–2016, metagenomic analysis of pediatric stools revealed 3 novel parvoviruses of the Protoparvovirus genus named bufavirus (BuV), tusavirus (TuV), and cutavirus (CuV) [4]. CuV was also found in skin biopsies exclusively from 4 patients with CTCL/MF. Soon thereafter, our group and others detected CuV DNA in CTCL lesions, whereas the skin biopsies of other skin lymphomas and of healthy subjects were CuV-DNA negative, indicating a significant association of CuV with CTCL [5, 6]. CuV immunoglobulin G (IgG) was further detected in serum sampled up to 20 years prior to the biopsy, revealing that CuV infection is persistent [6]. If CuV could induce a persistent antigenic T-cell stimulation, it would be present in the pre-CTCL PP stage. Hence, the aim of our current study was to elucidate the prevalence of CuV DNA in the skin of patients with nonmalignant PP, as compared with healthy controls.
METHODS
Participants and Skin Samples
Skin samples from 13 study subjects with the rare PP (6 with SPP and 7 with LPP), collected in 2014 and studied for the skin bacterial microbiome [7], were analyzed for CuV DNA: 8 men and 5 women, with an age range of 37–86 years (mean: 64.3 years). Six patients were between 37 and 65 years of age (mean: 58.8 years). None of the patients showed any signs of systemic immune suppression and they had not received any ultraviolet phototherapy within the previous 12 months [7]. The diagnoses of the patients were confirmed histopathologically at the Dermatopathology Unit of the Department of Dermatology and Allergology, Helsinki University Hospital, Finland. A total of 52 swabs, 2 from patch/plaque and 2 from healthy skin areas, were collected from all 13 patients with PP, and respective paired formalin-fixed paraffin-embedded (FFPE) skin biopsy samples, were available from 12 patients and analyzed in a blinded fashion (Supplementary Materials and Methods).
Altogether, 171 skin swabs of 4 skin regions—forearm, elbow bend, forehead, and behind the ear— from 51 healthy volunteers working at the Helsinki University or Hospital were collected in 2016 (15 males and 36 females; age range: 18–65 years; mean: 41.3 years) (Supplementary Materials and Methods). Twenty-eight patients were in the 37–65-year age group (mean: 52.8 years).
Analysis
DNA of the 3 human protoparvoviruses—CuV, BuV, and TuV—was detected and quantified by a multiplex real-time quantitative polymerase chain reaction (qPCR), and all positive results were re-analyzed by single-plex qPCR, as previously described [6]. Non-template controls were included in all PCR runs. The CuV-PCR amplicons were further verified by cloning and sequencing, and longer sequences (∼230 and ∼570 bp) were obtained from samples with high viral loads. We also searched for DNA of parvovirus B19, Merkel-cell (MCPyV) and BK polyomaviruses (BKV), and all 9 human herpesviruses. In addition, RNAscope in situ hybridization (RISH), with CuV probes targeting the virus protein (VP) region, was performed on 6 CuV PCR-positive PP FFPE-skin biopsies and, as controls, 2 CuV PCR-negative PP FFPE-skin biopsies. Probes targeting the human gene PPIB and the bacterial gene DapB (ACDbio, USA) served as positive and negative technical controls, respectively, and insect cells infected and noninfected with CuV VP-bearing baculovirus as positive and negative CuV-probe controls, respectively. Detailed methods are described in the Supplementary Materials and Methods.
Ethics Approvals
The study was approved by the HUS ethics review board (approval 12/13/03/01/2012 for the patients with parapsoriasis en plaques (PP) and approval 553/E6/01, §106/2014 for the healthy subjects). Written informed consent was obtained from all individuals in the study. The studies were conducted in accordance with the relevant guidelines and regulations.
RESULTS AND DISCUSSION
The prevalence of CuV DNA among the skin swabs from the 13 patients with parapsoriasis was 45% (6/13) and from the healthy controls it was 1.96% (1/51), the difference of which is statistically significant (RD: 44; 95% CI: .174, .706; P = .0001, Fisher's exact test). This difference remained significant when comparing only those aged 37–65 years old in both groups: 50% (3/6) versus 3.5% (1/28) (P = .0124) (Supplementary Table 1). Our results thus indicate that cutaneous CuV is associated with plaque-type parapsoriasis, a precursor of CTCL/MF to which CuV has been associated [4–6]. CuV DNA has previously been found to be significantly more frequent in skin swabs of HIV-positive men than those of healthy controls (17.1% vs 3.8%; P < .001) [8]. The high prevalence in HIV-positive individuals could possibly be attributed to their immunocompromised state. However, our patients with PP were not immunosuppressed. Moreover, in previous studies of skin biopsies, only 4 of 136 (2.9%) immunosuppressed solid-organ transplant recipients and 1 of 25 (4%) highly immunocompromised hematopoietic stem-cell transplant recipients harbored CuV DNA in their skin [6, 9], indicating that immunodeficiency is not a major factor affecting the presence of CuV. Likewise, all skin biopsies of 13 individuals with atopic dermatitis [6] and of 85 individuals with non-CTCL/MF skin lymphomas [5] were CuV negative, indicating that other lymphoproliferative skin diseases would not be prone to CuV-DNA carriage.
In the current study, CuV DNA was also detected in 15 biopsied FFPE skins of 8 of 12 (66.7%) patients with PP, 4 of whom had SPP and 4 LPP (Supplementary Table 2). Interestingly, all 6 patients with CuV DNA in their skin swabs also had CuV DNA in their skin biopsies, which were analyzed in a blinded fashion (Supplementary Table 2). Of note, during a median follow-up time of 14 years (range: 10–22 years), 4 patients with PP, 3 with LPP, and 1 with SPP, of whom 2 were CuV DNA-positive, progressed to histologically confirmed CTCL/MF (Supplementary Table 2). That only 1 of our 6 patients with SPP developed CTCL/MF is in accordance with our previous observation in which 35% of the LPPs and only 10% of SPPs progressed to CTCL/MF [2]. It is not known whether the previously determined association of CuV with CTCL/MF is causal or if CuV, using cellular polymerases, merely favors rapidly dividing cancer cells for its replication. However, we have now observed an even higher prevalence of CuV in a nonmalignant skin disease, PP, which indicates that it does not merely target cancer cells but instead persists in PP skin, perhaps contributing to the development of CTCL/MF cancer. Parvoviruses in general have not been shown to be oncogenic, but they are known to persist in host tissues for years [10, 11], which could result in long-lasting inflammatory responses possibly leading to malignancy. The persistence of CuV in skin is corroborated by the previously observed long CuV seropositivity for up to 20 years before skin biopsy [6].
The high prevalence (66.7%) of CuV DNA in our PP skins contrasts to the 0% prevalence in 8 PP skins reported previously [4]. This may be due to our shorter PCR targets (91 vs >400 bp), leading to increased sensitivity of PCR (Supplementary Materials and Methods). Upon comparison of the PCR amplicons from different individuals, nucleotide mismatches were observed, confirming that the CuV-PCR positives did not result from cross-contamination or from the plasmid control (Supplementary Figures 1 and 2).
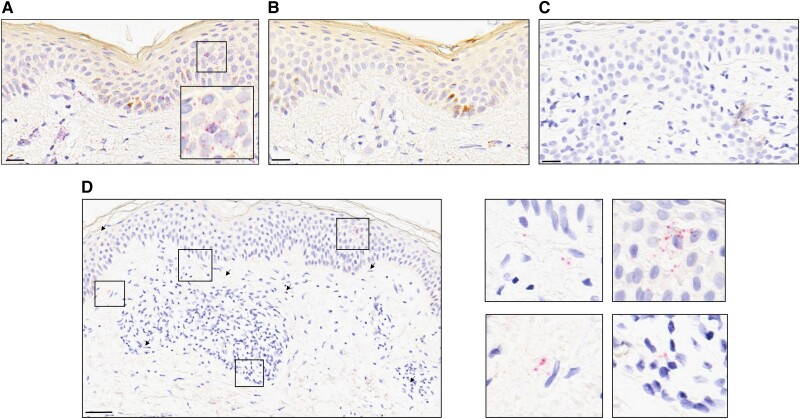
In RISH experiments of the 6 high-load skin samples, CuV-specific signals were observed in the nucleus or cytoplasm of cells localized in both the epidermis and dermis (Figure 1). Both technical control probes worked as expected, and CuV probes were specific (Figure 1, Supplementary Figure 3).
Figure 1.
RNAscope™ 2.5 HD Assay-RED on FFPE skin of patients with parapsoriasis, counterstained with hematoxylin. A, B Sample AS11 hybridized with (A) human PPIB mRNA probe and (B) bacterial DapB probe, as positive and negative technical controls. C, D CuV VP probe on (C) CuV PCR-negative sample AS8 and (D) the CuV PCR-positive sample AS11. Representative positive probe signals (red punctuated dots) are indicated by black arrows and as a zoomed-in box at the bottom-right corner of panel A and four zoomed-in boxes at the right side of panel D. Scale bars, 20 μm (A–C) and 100 μm (D). As a further probe control, CuV RISH on insect cells with and without CuV VP-mRNA expression is shown in Supplementary Figure 3. Abbreviations: CuV, cutavirus; FFPE, formalin-fixed paraffin-embedded; PCR, polymerase chain reaction; RISH, RNAscope in situ hybridization.
The CuV-DNA loads ranged from 2.91 × 104 to 2.86 × 107 copies per 1 million cells (cpm) in the PP skin swabs and from 2.49 × 104 to 5.88 × 107 cpm in the skin biopsies. All samples were human RNaseP-PCR positive, indicating that the DNA was intact. All patients, except for P6, further exhibited higher CuV-DNA loads in the parapsoriasis patch/plaque biopsies than their healthy counterparts, but this did not reach statistical significance (Supplementary Table 2). Also, there was no significant difference in the CuV-DNA loads between patch/plaque and healthy swabs of the patients with PP. The average numbers of cells for 1 μL of DNA eluate in CuV-DNA–positive plaque (5.9 × 101) versus healthy (2.2 × 101) and CuV-DNA–positive (4.14 × 101) and –negative (3.35 × 101) skin-swab extracts were similar. The single CuV-positive healthy individual was a 50-year-old female who had low loads (0.14 to 196 copies/μL of DNA eluate) of CuV DNA in 3 of her 4 skin swabs, which also were positive for MCPyV. RNaseP qPCR indicated that the DNA extractions of swabs from the healthy controls contained similar amounts of human DNA as those from the patients (Supplementary Materials and Methods).
All 24 paired skin biopsies available from the 12 patients with PP who underwent biopsy were negative for DNA of BuV, TuV, BKV, and all 9 herpesviruses (Supplementary Materials and Methods), thus corroborating the specificity of CuV PCR and lack of contamination. Parvovirus B19 (B19V) and MCPyV are known to persist in skin of both diseased and healthy individuals, even though MCPyV is associated with Merkel-cell cancer [12–14]. However, parapsoriasis skin has not previously been studied for other parvoviruses or polyomaviruses. In our study, the healthy biopsy sample of patient P3 harbored MCPyV DNA, and both biopsies of patient P2 harbored B19V DNA.
In conclusion, we report here the first observation of CuV DNA in the skin of patients with plaque-type parapsoriasis, a precursor of CTCL/MF. In contrast to the infrequent detection of other parvoviruses, herpesviruses, or polyomaviruses, CuV DNA was observed in high prevalence both in skin biopsies (66%) and in skin swabs (46%) of patients with PP, whereas the skin swabs of only a single healthy individual (1.96%) harbored CuV. This difference was statistically significant, indicating that human CuV was associated with plaque-type parapsoriasis. CuV-DNA positivity was further localized to both the epidermis and dermis. Prospective studies with more patients together with functional experiments are needed to assess whether this relationship between CuV, PP, and CTCL/MF is causal or consequential; perhaps instead of being caused by CuV, PP skin could merely provide a good habitat for the virus. Intriguingly, in both cases, this persistence could eventually lead to malignancy. The presence of CuV in parapsoriasis skin may serve as a biomarker for the development of CTCL/MF.
Supplementary Data
Supplementary Materials and Methods are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Ushanandini Mohanraj, Department of Virology, University of Helsinki, Helsinki, Finland.
Tuomas Konttinen, Department of Virology, University of Helsinki, Helsinki, Finland; Pediatric Research Center, Children's Hospital, University of Helsinki and Helsinki University Hospital, Helsinki, Finland; Research Program for Clinical and Molecular Metabolism, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Alexander Salava, Department of Dermatology, Allergology, and Venereology, University of Helsinki and Helsinki University Central Hospital, Helsinki, Finland.
Liisa Väkevä, Department of Dermatology, Allergology, and Venereology, University of Helsinki and Helsinki University Central Hospital, Helsinki, Finland.
Annamari Ranki, Department of Dermatology, Allergology, and Venereology, University of Helsinki and Helsinki University Central Hospital, Helsinki, Finland.
Maria Söderlund-Venermo, Department of Virology, University of Helsinki, Helsinki, Finland.
Notes
Author contributions. Conceptualization: U. M., M. S.-V. Data curation: U. M. Formal analysis: U. M. Funding acquisition; M. S.-V., . Investigation: U. M., T. K. Methodology: U. M. Resources: A. S., L. V., A. R. Software: U. M. Supervision: M. S.-V. Validation: U. M. Visualization: U. M. Writing—original draft preparation: U. M. Writing—review and editing: A. R., M. S.-V.
Acknowledgments. The authors thank Alli-Kaarina Tallqvist for technical help and Taina Härkönen for help with supervision of the sampling from the controls.
Financial support. This work was supported by the Finnish-Norwegian Medical Foundation (2022035; to U. M.), Ida Montin Foundation (20220151; to U. M.), Viral Disease Research Foundation (U. M.), the Sigrid Jusélius Foundation (M. S.-V.), the Life and Health Medical Support Association (M. S.-V.), and the Swedish Cultural Foundation (M. S.-V.).
Data availability. Cutavirus sequences are submitted to GenBank (OP751138—OP751146). Requests to access raw datasets should be directed to the corresponding author.
References
- 1. Willemze R, Cerroni L, Kempf W, et al. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood 2019; 133:1703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Väkevä L, Sarna S, Vaalasti A, Pukkala E, Kariniemi AL, Ranki A. A retrospective study of the probability of the evolution of parapsoriasis en plaques into mycosis fungoides. Acta Derm Venereol 2005; 85:318–23. [DOI] [PubMed] [Google Scholar]
- 3. Mirvish JJ, Pomerantz RG, Falo LD, Geskin LJ. Role of infectious agents in cutaneous T-cell lymphoma: facts and controversies. Clin Dermatol 2013; 31:423–31. [DOI] [PubMed] [Google Scholar]
- 4. Phan TG, Dreno B, da Costa AC, et al. A new protoparvovirus in human fecal samples and cutaneous T cell lymphomas (mycosis fungoides). Virology 2016; 496:299–305 [DOI] [PubMed] [Google Scholar]
- 5. Kreuter A, Nasserani N, Tigges C, et al. Cutavirus infection in primary cutaneous B- and T-cell lymphoma. JAMA Dermatol 2018; 154:965–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Väisänen E, Fu Y, Koskenmies S, et al. Cutavirus DNA in malignant and nonmalignant skin of cutaneous T-cell lymphoma and organ transplant patients but not of healthy adults. Clin Infect Dis 2019; 68:1904–10. [DOI] [PubMed] [Google Scholar]
- 7. Salava A, Pereira P, Aho V, et al. Skin microbiome in small- and large-plaque parapsoriasis. Acta Derm Venereol 2017; 97:685–91. [DOI] [PubMed] [Google Scholar]
- 8. Wieland U, Silling S, Hufbauer M, et al. No evidence for role of cutavirus in malignant melanoma. Emerg Infect Dis 2019; 25:1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zanella MC, Cordey S, Laubscher F, Docquier M, Vieille G, Van Delden C. Unmasking viral sequences by metagenomic next-generation sequencing in adult human blood samples during steroid-refractory/dependent graft-versus host disease. Microbiome 2021; 9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hokynar K, Norja P, Hedman K, Söderlund-Venermo M. Tissue persistence and prevalence of B19 virus types 1–3. Future Virol 2007; 2:377–88. [Google Scholar]
- 11. Xu M, Perdomo MF, Mattola S, et al. Persistence of human bocavirus 1 in tonsillar germinal centers and antibody-dependent enhancement of infection. mBio 2021; 12:e03132-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Santonja C, Santos-Briz A, Palmedo G, Kutzner H, Requena L. Detection of human parvovirus B19 DNA in 22% of 1815 cutaneous biopsies of a wide variety of dermatological conditions suggests viral persistence after primary infection and casts doubts on its pathogenic significance. Br J Dermatol 2017; 177:1060–5. [DOI] [PubMed] [Google Scholar]
- 13. Wang Y, Keinonen A, Koskenmies S, et al. Occurrence of newly discovered human polyomaviruses in skin of liver transplant recipients and their relation with squamous cell carcinoma in situ and actinic keratosis—a single-center cohort study. Transpl Int 2019; 32:516–22. [DOI] [PubMed] [Google Scholar]
- 14. Bopp L, Wieland U, Hellmich M, Kreuter A, Pfister H, Silling S. Natural history of cutaneous human polyomavirus infection in healthy individuals. Front Microbiol 2021; 12:740947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.