Abstract
Due primarily to the excellent soft tissue contrast depictions provided by MRI, the widespread application of head and neck MRI in clinical practice serves to assess various diseases. Artificial intelligence (AI)-based methodologies, particularly deep learning analyses using convolutional neural networks, have recently gained global recognition and have been extensively investigated in clinical research for their applicability across a range of categories within medical imaging, including head and neck MRI. Analytical approaches using AI have shown potential for addressing the clinical limitations associated with head and neck MRI. In this review, we focus primarily on the technical advancements in deep-learning-based methodologies and their clinical utility within the field of head and neck MRI, encompassing aspects such as image acquisition and reconstruction, lesion segmentation, disease classification and diagnosis, and prognostic prediction for patients presenting with head and neck diseases. We then discuss the limitations of current deep-learning-based approaches and offer insights regarding future challenges in this field.
Keywords: artificial intelligence, deep learning, head and neck, magnetic resonance imaging
Introduction
MRI is one of the important modalities for assessing patients with head and neck diseases in routine clinical practice. The region of the head and neck is characterized by a highly complex anatomy comprised of numerous organs within a narrow space, including the skull base, temporal bone, orbit, laryngopharynx, oral cavity, nasal/sinonasal cavity, major salivary glands, and many other organs/spaces. A major advantage of MRI in assessing head and neck diseases is its higher soft-tissue contrast in representative imaging sequences (e.g., T1-weighted imaging [T1WI] and T2-weighted imaging [T2WI]) compared to CT, thus making MRI highly useful to assess diseases in the head and neck region without radiation exposure.1 In addition, several advanced MR techniques can provide functional information (e.g., diffusion-weighted imaging [DWI] and perfusion/permeability weighted imaging); these additional techniques provide useful information for assessments of head and neck diseases.2–4 However, MRI of the head and neck still presents challenges in various clinical applications: (1) In image acquisition, due to the complex anatomical structure of the head and neck, images with higher spatial resolution are often required, leading to longer acquisition times, which can be problematic. The complex morphology of the head and neck can also result in B0 and B1 field inhomogeneity, which can make it difficult to obtain sufficient image quality. The presence of air in the nasal/sinonasal cavity or laryngopharynx and metallic substances in the oral cavity can cause magnetic susceptibility artifacts, which are also an important problem in image quality assessment.2,5 (2) In lesion detection and segmentation, it can be difficult to detect lesions and to accurately segment lesions because of the anatomical complexity of the head and neck. Segmenting a lesion and normal anatomical structures around the lesion is crucial for radiotherapy treatment planning, but performing this task manually is very time-consuming.6 (3) In lesion diagnosis and differentiation, the signal intensity in the lesion is often nonspecific (e.g., low signal intensity on T1WI and high signal intensity on T2WI), which can be a source of difficulty for radiologists. Related to the disease diagnosis, it is also difficult to predict prognoses (particularly in cancer patients) because various factors such as lesion size, surface irregularity, intralesional heterogeneity, and several other factors affect the treatment outcome. Therefore, although MRI is indispensable for assessing head and neck lesions, these limitations should be resolved.
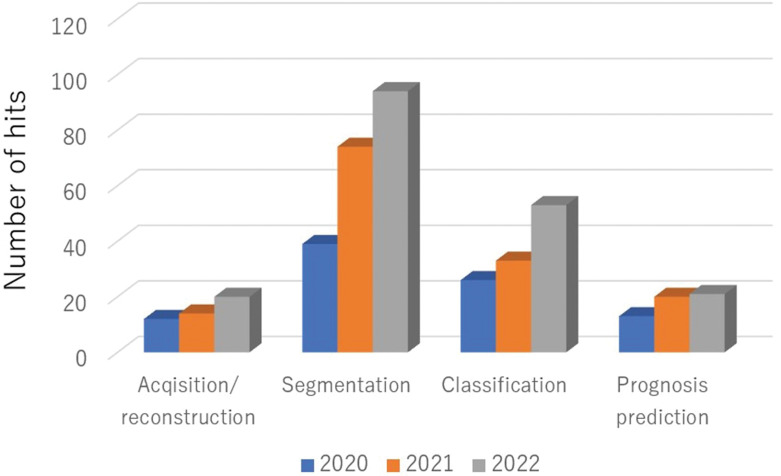
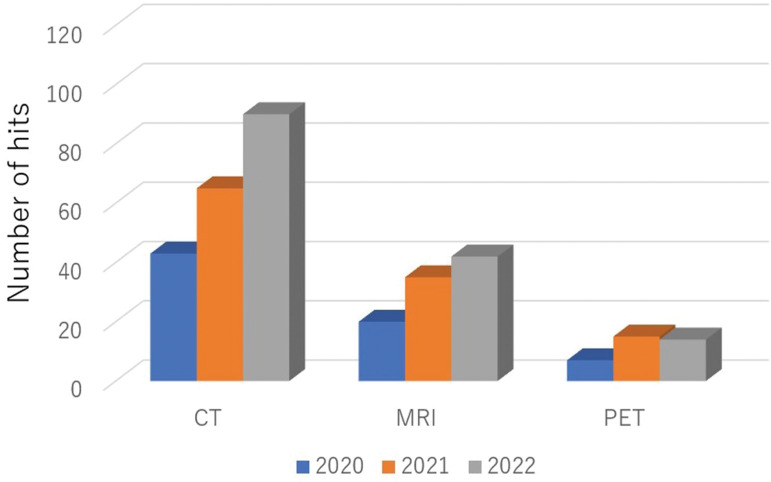
Over the past decade, advanced analytical methods such as texture analyses and radiomics have gained attention in the research field of radiology.7 Compared to the interpretation of conventional imaging findings, these advanced methods are capable of more accurately quantifying the information contained within the images at a higher level of detail, thus providing high diagnostic performance.8–28 More recently, remarkable advances have been made in the development and clinical application of artificial intelligence (AI) in the field of radiology.29–32 In particular, the analysis of deep learning using convolutional neural networks (CNNs) has become mainstream, and extensive clinical research on its application continues for various categories in medical imaging, including that of the head and neck (Fig. 1).33–51 First, there has been remarkable progress in the development of deep-learning reconstruction techniques in image acquisition, focusing on both the reduction of image noise and reconstructions to high-resolution imaging (i.e., super-resolution); these techniques provide high-quality images and/or short scanning time-derived images. Second, in the segmentation of targets, the application of semantic segmentation with deep-learning algorithms has progressed, with reports of high-precision delineations of ROIs in various targets, including lesions such as malignant tumors and normal organs. Third, in the diagnosis or the differentiation of lesions, classifications using a deep-learning method have achieved very high diagnostic accuracy rates in various disease differentiations, suggesting that deep-learning methods may achieve diagnostic ability that is comparable to that of board-certified radiologists and to become useful as diagnostic support tools for all physicians. Fourth, as another classification task using a deep-learning approach, prognosis prediction (especially in patients with cancer) will be beneficial for the appropriate patient care. In the investigations that focused on the head and neck, the number of segmentation-related studies has been the highest compared to other categories, followed by disease classification, prognosis prediction, and image acquisition in recent studies using a deep-learning analysis (Fig. 2). In addition, although the utility of deep-learning analyses in the head and neck has been investigated mainly using CT, the number of reports investigating deep learning with other imaging modalities, including MRI, have been increasing (along with those of CT) (Fig. 3).
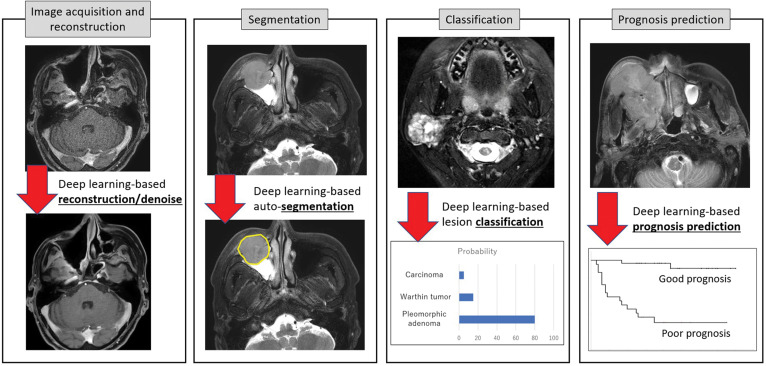
Fig. 1.
Representative artificial intelligence (AI)-based clinical appreciation in head and neck MRI. Representative clinical applications using AI-based methods are shown: image reconstruction with denoising, lesion segmentation, disease classification, and prognosis prediction from an imaging dataset. AI, artificial intelligence.
Fig. 2.
The numbers of head and neck imaging studies in various clinical applications using deep-learning approach in recent years. The search results in PubMed (https://pubmed.ncbi.nlm.nih.gov/) with ‘deep learning neck imaging (acquisition or reconstruction)’ were categorized as acquisition/reconstruction, ‘deep learning neck imaging segmentation’ as segmentation, ‘deep learning neck imaging classification’ as classification, and ‘deep learning neck imaging prognosis prediction’ as prognosis prediction. The data from 2020 to 2022 are presented.
Fig. 3.
The numbers of head and neck imaging studies in various modalities by a deep-learning approach in recent years. The search results in PubMed (https://pubmed.ncbi.nlm.nih.gov/) with ‘deep learning neck imaging CT’, ‘deep learning neck imaging MRI’, and ‘deep learning neck imaging PET’ were, respectively, categorized as CT-, MRI-, and PET-related investigations using deep learning. The data from 2020 to 2022 are shown. PET, positron emission tomography.
As mentioned above, there has been an increasing number of studies of technological innovations utilizing AI in the field of head and neck MRI, particularly with a focus on deep learning, and the studies have established the utility of AI in various applications such as image acquisition, lesion segmentation, disease diagnosis, and prognosis prediction. In this review, we comprehensively describe the usefulness of AI, especially deep learning, in the field of head and neck MRI, and we provide an overview of the demonstrated benefits and limitations as well as future perspectives.
Image Acquisition and Reconstruction
Numerous important anatomical structures are present within the small FOV of the head and neck, and high-spatial-resolution imaging is thus generally required for the sufficient quality of image reading. Scanning with a short acquisition time is also required because imaging of the head and neck region always suffers from the patient’s motion by, for example, respiration and swallowing. Susceptibility artifacts in the region located around the air space of the laryngopharynx and metallic items in the oral cavity are also problematic in the efforts to maintain good image quality. To address these problems, the designs of MRI sequences and acquisition methods have evolved and have been applied to the head and neck over the last decade or so. A short scanning time without a reduction of the SNR is considered a crucial task for head and neck MRI acquisition.
The methods for image acceleration that are most frequently used in daily clinical practice include parallel imaging, half-scan (half Fourier), and the zero filling interpolation, among others. A compressed sensing technique was introduced in 2015 in the main field of fast MRI acquisition.52 This technique is an image reconstruction method that uses the undersampling of k-space data with sparse image signals and a repeated denoising cycle performed by wavelet transformation. There are several studies in which a compressed sensing technique was applied in head and neck MRI.53–55 Takumi et al. performed a comparative study of the quality of contrast-enhanced (CE) fat-suppressed 3D gradient-echo imaging with both quantitative and qualitative assessments between images with and without compressed sensing, using a dataset of patients with pharyngolaryngeal squamous cell carcinoma (SCC).53 They reported that the images obtained with compressed sensing demonstrated better image quality compared to those without, and they concluded that compressed sensing improves the image quality of 3D-based CE-T1WI for the evaluation of pharyngolaryngeal SCC, without requiring additional acquisition time. Kami et al. also observed improved image quality of compressed sensing-based CE-3D-T1WI compared to the 2D multi-slice spin echo sequence in patients with maxillofacial lesions.54 The superiority of a T2WI sequence with compressed sensing compared to the conventional parallel imaging-based sequence for the evaluation of normal structures in the oral cavity was demonstrated by Tomita et al.55
More recently, deep-learning-based reconstruction algorithms have been reported worldwide.56,57 In most of the assessments of the image quality of deep-learning-based reconstruction, the novel reconstruction method using a deep-learning-based reconstruction algorithm was observed to be the best method to obtain superb image quality in various image-reconstruction techniques; even the above-mentioned compressed-sensing technique tended to provide inferior image quality compared to the deep-learning-based reconstruction. The number of investigations that performed deep-learning-based MRI reconstruction for the evaluation of head and neck disorders is very limited. Naganawa et al. evaluated the contrast-to-noise ratio (CNR) of endolymph to perilymph in MRI examinations of endolymphatic hydrops based on 3D fluid-attenuated inversion recovery (FLAIR) imaging performed 4 hours after the intravenous administration of a contrast agent.40 They assessed the hybrid of (i) the reversed image of the positive endolymph signal and (ii) the native image of the perilymph signal multiplied with the heavily T2-weighted MR cisternography images (named ‘HYDROPS-Mi2’) with a deep-learning-based denoising algorithm. Their results revealed that the CNR of the images obtained with deep-learning-based denoising (7738.6 ± 5149.2) was increased by more than fourfold compared to the images obtained without denoising (1681.8 ± 845.2).40 In another study, Naganawa et al. performed the fast scanning of endolymphatic hydrops imaging with the use of HYDROPS2-Mi2 and a deep-learning-based denoising algorithm. They obtained sufficient image quality within a 5-minute imaging protocol with the help of deep-learning reconstruction, whereas the conventional imaging method with HYDROPS2-Mi2 required 12 minutes for image acquisition.47
Deep-learning-based super-resolution is another featured technology in the field of deep-learning reconstruction. Koktzoglou et al. applied the deep-learning-based super-resolution technique to head and neck MR angiography (MRA).58 This super-resolution model was created with the training for the estimation of difference between the high- and low-resolution images based on image domain deep learning method. They reported that deep-learning-based super-resolution (up to fourfold) as well as twofold lower-resolution input volumes provided the same quality of images obtained from high-resolution ground-truth volumes with a long scanning time, based on their evaluation with the Dice similarity coefficient (DSC), structural similarity (SSIM), arterial diameters, and arterial sharpness. They concluded that head and neck MRA with deep-learning-based super-resolution has the potential for an up to fourfold reduction in acquisition time.58 Some of the structures that comprise the head and neck are especially small and anatomically complex. It has been difficult to achieve the sufficient depiction of several small normal structures (e.g., the inner ear, the parathyroid) and lesions (e.g., T1-stage cancers) by routine imaging sequences because of their smallness and/or the similarity of the signal intensity located near the target.59 Super-resolution techniques could thus contribute significantly to the acquisition of good-quality and high-spatial-resolution images without increasing the acquisition time.
Artifact reduction is an important issue in the field of MRI. Head and neck DWI is challenging and often results in poor image quality with severe image distortion and susceptibility artifact due to the complex anatomical shapes and the presence of air and/or metallic substances. A study published in 2023 investigated the new technique of susceptibility artifact correction in an echo planar imaging sequence using an unsupervised deep neural model.60 This technique can be effective for improving the image quality of the head and neck DWI.
Synthetic imaging between modalities (i.e., image-to-image translation) is another notable deep-learning-based reconstruction strategy. More specifically, bone MR imaging to synthetic CT techniques enables the visualization of cortical bone without the need for radiation exposure, which is particularly useful for pediatric patients. Although acquisition-based bone MRI technique was previously investigated, the zero TE imaging is one of the representative methods of bone MRI to visualize short-T2 materials (i.e., cortical bone);61 deep-learning-based bone MRI is also considered as an interesting technique for the bone MRI. Bambach et al. demonstrated the optimization that can be used to create a robust deep-learning model for the conversion of bone MR imaging to synthetic CT with Light U-Net architecture, and they suggested the possibility of using this technique in clinical practice.50 Including the abovementioned deep-learning-based super-resolution technique and the synthetic imaging, the potential utility of these techniques is highly expected; however, derived images are created by incorporating data that were not originally acquired from the target and may contain artificially reconstructed false information. This is considered one of the potential important limitations of these types of deep-learning-based image processing. Such images should be carefully reviewed under the close supervision of a radiologist.
Segmentation
Target delineation and segmentation is important to accurately understand the main localization of lesions and the range of disease. Several research groups have suggested the utility of lesion delineation or detection of the lesion extent and segmentation using several modalities such as CE-CT, non-CE- or CE-MRI, and 18F-fluorodeoxyglucose positron-emission tomography/CT (FDG-PET/CT).62,63 The accurate and rapid accomplishment of this procedure clinically is thus desired. More specifically, target segmentation is crucial in head and neck imaging, especially for successful radiotherapy procedures. For treatment planning in radiotherapy, accurate delineation of the target lesion and the organs at risk is mandatory, and the delineation procedure directly influences the quality of the radiotherapy.64,65 Because various types of soft tissue are present in the head and neck, MRI is an effective modality due to its excellent contrast of soft tissue compared to other modalities (e.g., CT) for delineating the target lesion and organs at risk, and it has thus become a more common imaging technique for the delineation in radiotherapy treatment planning.66 However, the delineation of target lesions and organs at risk in the head and neck region is a very labor-intensive, time-consuming, and observer-dependent procedure. The delineation process is typically performed by radiologists or radiation oncologists, and it takes an average of ~3 hours to delineate a full set of tumor volumes and organs at risk, mainly because of the complex anatomy of the head and neck.6 To address this limitation, AI-based segmentation methods, particularly deep-learning-based techniques, have been widely introduced as novel supportive tools and are investigated with excellent initial results, as follows.
Investigations of MR-based ROI delineation using deep-learning methods for head and neck cancers or organs at risk have been performed in patients with nasopharyngeal cancer67–70 and oropharyngeal cancer.71,72 To delineate the appropriate ROIs, most of these investigations used the deep-learning architecture of an Encoder-Decoder-type CNN, and the 2D U-net or 3D U-net was the most popular type of this model. Automatic MRI-based delineation for the primary lesion of nasopharyngeal cancer using CNNs has been applied in several studies.38,67–70 For example, Lin et al. used U-net architecture for the delineation of the primary lesion of nasopharyngeal cancer on CE-T1WI.67 They reported the DSC of 0.79 with the ground truth by an expert radiation oncologist or radiologist’s manual assessment, whereas the DSCs by qualified radiation oncologists ranged from 0.71 to 0.74. The use of their CNN model as an assistive tool for radiologists also reduced the intraobserver variation (the interquartile deviation of DSC) by 36.4%, and it reduced the contouring time by 39.4% (from 30.2 to 18.3 minutes).67 Chen et al. proposed a novel multi-modality MRI fusion network (MMFNet) in which T1WI, T2WI, and CE-T1WI sequences were well integrated to provide a complete accurate segmentation of nasopharyngeal cancer. Their model consists of a multi-encoder-based network and one decoder to capture modality-specific features. Their study achieved the DSC of 0.72, which was higher than that obtained in a U-net-based analysis with a single sequence of T1WI, T2WI, or CE-T1WI.68 Although Chen et al.’s study was performed with a multi-modal evaluation, the DSC value they reported is lower than that obtained in the above-cited study by Lin et al. This might be due to the variability in the two studies’ patient cohorts. In contrast, Wong et al. demonstrated the performance a U-Net-based CNN using only the non-CE-sequence (i.e., fat-suppressed T2WI), and their results showed no significant difference in the DSC (0.71) compared to the CE-T1WI sequence.38
Investigations of the segmentation of oropharyngeal cancer have also been reported.71,72 Rodríguez Outeiral et al. obtained the best DSC values with the combination of multiple MRI sequences (T1WI, T2WI, and CE-T1WI) as input using 3D U-Net. Notably, their DSC results suggested that manually reducing the context around the tumor, which they defined as a semi-automatic method, results in better segmentations than those given by a completely automatic method; with the semi-automatic method the DSC value was 0.74, whereas with the completely automatic method the DSC value was 0.55.71 Wahid et al. also demonstrated improved delineation of oropharyngeal cancer lesions with the combination use of conventional sequences (T1WI and T2WI) and quantitative functional image sequences; they used the apparent diffusion coefficient (ADC) from DWI and the volume transfer coefficient (Ktrans) and the volume of the extravascular extracellular space (Ve) from dynamic CE (DCE) perfusion as input channels to a 3D Residual U-Net.72
The automatic delineation of organs at risk is also an important task in patients with head and neck cancer who need radiotherapy planning — especially the planning of intensity-modulated radiation therapy (IMRT). Dai et al. used the datasets of both CT for bony-structure contrast and MRI for soft-tissue contrast in model training for the delineation of multiple organs at risk (totally 18 organs) with the CNN, resulting in high DSC values (approximately 0.8–0.9).73 Interestingly, their study acquired the MRI data not directly; rather, it was acquired from the CT dataset by a pre-trained cycle-consistent generative adversarial network (GAN).73 In contrast, Korte et al. investigated the delineations of the parotid gland, submandibular gland, and neck lymph nodes by using a T2WI dataset with multiple 3D-U-Net systems, resulting in DSC values of approximately 0.8.74
The segmentation of other lesions and anatomical structures such as vestibular schwannoma in the cerebellopontine angle,75,76 the inner ear and its related structures (e.g., cochlea, vestibule),77–79 and venous malformations of the neck80 has been described. Segmentation of the inner ear and its related structures necessary for the diagnosis of endolymphatic hydrops has also been a concern (see the section below titled ‘Disease classification and diagnosis’) and would be valuable for clinical practice.
Due to the excellent soft-tissue contrast obtained from MRI, MRI-based segmentation could contribute to the highly accurate segmentation of various lesions and organs at risk. The currently used segmentation process is usually performed for treatment-planning CT, but the combination use of multiple modalities, including MRI, is expected to be established in the future as a more useful method. Unfortunately, most of the studies mentioned above did not use external validation; they used internal validation with a hold-out or cross-validation scheme for the test set when assessing the created model’s accuracy. The performance of each new deep-learning model must therefore be interpreted carefully before its use in clinical practice.
Disease Classification and Diagnosis
There is a wide variety of differential diagnoses in head and neck imaging depending on the various anatomical origins. The ability to accurately determine the final diagnosis with high diagnostic confidence using images can help reduce unnecessary invasive examinations and surgical procedures. Numerous radiological investigations have described lesion enhancement patterns, lesion morphological shapes, and morphological changes in lesions over a given follow-up period, and several additional parameters obtained from functional imaging techniques are valuable for the diagnostic prediction of the histological type or differentiation of benign and malignant lesions in various head and neck diseases.81–83 However, there are still many nonspecific imaging findings in head and neck MRI, and standard imaging sequences such as T1WI and T2WI can be limited in their ability to reliably differentiate, for example, benign from malignant lesions, resulting in difficulty reaching a diagnosis even among experienced radiologists. Advanced analytical methods such as a texture analysis or radiomics approach have been reported to provide mostly equivalent diagnostic accuracy (or sometimes better accuracy) in classifying head and neck diseases compared to classifications by expert radiologists who specialize in head and neck imaging.84 There is a great need for a supplementary tool, and AI-based methods have the potential to make a significant contribution to this application. We next describe the aspects of the usefulness of AI-based techniques, especially deep-learning methods, for the diagnoses and classification of head and neck diseases.
Major salivary gland tumors; parotid tumors
The origin of approximately 80% of salivary gland tumors is a parotid gland. Pleomorphic adenoma is the most common histological subtype of parotid tumors, followed by Warthin tumor. Mucoepidermoid carcinoma is the most common subtype in malignant tumors. MRI is often used to investigate a parotid tumor’s diagnosis based on various imaging findings (mainly T2WI and CE-T1WI).85 However, the imaging findings of parotid gland tumors are sometimes unspecific and often difficult to diagnose even by radiologists who specialize in the head and neck. Chang et al. demonstrated the differentiation of parotid tumors among pleomorphic adenoma, Warthin tumor, and malignant tumors with the use of 2D U-Net and five types of MRI sequences (T1WI, T2WI, CE-T1WI, DWI with b-value of 0 s/mm2 [DWIb0], DW with b-value of 1000 s/mm2 [DWIb1000], and ADC) with a fully automated method. They achieved high overall diagnostic performance with a DWI-based model consisting of DWIb0, DWIb1000, and ADC (0.71–0.81 accuracy).86 Interestingly, combining DWI and other sequences did not improve the prediction accuracy. Matsuo et al. performed the integration of T1WI and T2WI into single pseudo-color images and then conducted an analysis with the combination use of a VGG16-based deep learning-model and the L2-constrained softmax loss function. The highest area under the curve (AUC) value to differentiate between benign and malignant parotid gland tumors (0.86) was obtained with this method, higher than that obtained visually by a board-certified radiologist (0.74).87 More recently, the ‘transformer’ method has been applied to computer vision, including disease classifications in medical imaging; this technique uses the self-attention function which enables early information collection and combining of the total global information by dividing images into several parts.88 Dai et al. used an MRI dataset and the combination use of a CNN and the transformer method for the classification of parotid gland tumors.89 In their created model, named as TransMed, the CNN was used to extract a low-level feature from multi-modal imaging dataset; the output of CNN was conducted by a linear projection layer to create a low-level feature-based map. This low-level feature map was analyzed by the transformer layer to provide the final classification. Their proposed model achieved the diagnostic accuracy of 0.89 in differentiating the parotid gland tumors, which was superior compared to a CNN model’s performance.89 Liu et al. performed a two-step approach using Res-Net deep-learning architecture and then the Transformer network. This two-step method employed aligns with the technique utilized in the aforementioned study by Dai, et al. First, a vector describing image features was produced using image dataset through the Resnet-18. Subsequently, a sequence classification network by transformer model identified the subtype of tumors using the extracted image by Resnet-18 as an input. Their results demonstrated that the multiple sequences-based model with T2WI, CE-T1WI, and DWI produced the best results with 0.85 accuracy to differentiate between malignant and benign parotid tumors.90
Head and neck squamous cell carcinoma
Head and neck SCC is the most common malignancy arising from various regions of the head and neck. The main anatomy includes the pharyngolarynx and the nasal/sinonasal cavity. The correct diagnosis of this pathology is crucial for prompt decision-making regarding treatment planning in appropriate patient care. Deep-learning-based approaches have been assessed as a potential supportive tool for the rapid and precise diagnosis of these head and neck cancers. Wong et al. performed a discrimination analysis between early-stage (i.e., T1 stage) nasopharyngeal cancer and benign hyperplasia, using fat-suppressed T2WI with the Residual Attention Network and 3D volume input and multiple 2D acquired slices.91 They achieved high diagnostic accuracy (0.92) with a CNN-based algorithm, and this value was not significantly different from the diagnostic performance of an experienced radiologist. In another study, deep-learning-based T-staging in patients with nasopharyngeal cancer was investigated using imaging datasets of T1WI, T2WI, and CE-T1WI with the deep-learning architecture of ResNet.92 Those authors used the ‘weakly supervised learning’ method in which slice-by-slice analyses of multiple slices are conducted, after which the slice with the highest score (named the T-score) was used in the model’s training procedure. Their created model achieved 0.76 accuracy in the determination of T-staging. In patients with nasal or sinonasal cavity tumors, the differentiation between benign inverted papillomas and those with malignant transformation to SCC is important but challenging, particularly by imaging findings alone. Liu et al. investigated an MRI-based deep-learning approach to accomplish this challenge.93 They performed a 3D-CNN analysis using T1WI with or without a contrast agent and/or T2WI, resulting in the highest diagnostic accuracy (0.78) with their developed CNN model, named ‘All-Net,’ a 3D CNN model with four convolutional layers followed by rectified linear unit activation functions and max pooling layers.
Lesions in the temporal bone
Diseases in the temporal bone for which an MRI-based diagnosis is useful include cholesteatoma, Meniere’s disease (i.e., endolymphatic hydrops), vestibular and facial schwannoma, and many others. In particular, the diagnosis of endolymphatic hydrops in medical imaging can be attained exclusively through the utilization of MRI incorporating specific techniques. In the past, an intratympanic injection of a gadolinium-based contrast agent was required to obtain an SNR that was sufficient to visualize the endolymphatic hydrops. However, with the widespread use of high-field MRI and the optimization of imaging protocols, endolymphatic hydrops can be well detected with an intravenous (not intratympanic) injection of a contrast agent and image acquisition 4 hours later, and this imaging sequence has been well developed for clinical use.94–97 A deep-learning-based supportive tool for the assessment of endolymphatic hydrops was reported in 2020. Cho et al. described the combination use of automatically segmented images of the cochlea and vestibule by a deep-learning method with VGG19-based architecture and post-CE-3D-FLAIR images to obtain diagnostic information that indicates the degree of endolymphatic hydrops. This deep-learning-based output was highly consistent with the results obtained manually by experts, with an intraclass correlation coefficient of 0.971.77 Park et al. further improved this deep-learning-based model by developing it with the use of the architectures of the Inception-v3 and U-Net combination use; the newly developed system successfully achieved the automatic selection of representative images from a full MRI dataset with a very short total analysis time: 3.5 seconds.79
In disease classification and diagnoses using image datasets, deep-learning techniques are considered a promising method and have provided high overall diagnostic performance. However, most of the above-cited studies did not use images with the whole FOV for preparing the input used to train the deep-learning model. Rather, they used segmented images that include the lesion and exclude the area outside the lesion obtained by a manual procedure. A fully automated approach would be in great demand for easier clinical use because approaches using manual methods require high amounts of time and human effort. This methodological background may impede the worldwide use of deep-learning-based applications in clinical practice. Fully automated procedures with lesion detection, delineation, and classification abilities are desired for practical use in the field of diagnostic imaging. As in the studies of segmentation, most of the investigations of disease classification and diagnoses used internal (and not external) validation for the test set in order to assess the model’s performance. The diagnostic performance described in these reports might thus be higher than the real-world accuracy.98 This limitation should be addressed by future studies.
Prognosis Prediction
Challenges in predicting the prognoses of patients with head and neck lesions, for example, predictions of the initial treatment outcome and long-term predictions of the disease control, are still difficult. Prognosis prediction contributes to the optimization of treatment strategies, including the selection of initial therapy and the indication for adjuvant therapies, and the prediction is also considered useful to validate the treatment plan for each patient with head or neck cancer. Over the past few decades, imaging methods have been well developed and used together with advanced imaging techniques such as DWI and DCE,99–110 or a high-dimensional analytical method such as a texture analysis or radiomics method14,22,24,111,112 in various diseases as well as head and neck lesions. However, achieving accurate predictions is still challenging. Recently developed deep-learning-based techniques can make significant contributions to the prediction of patient prognoses as effective support tools with higher accuracy compared to MR imaging biomarkers and older analytical techniques.
Among the prognosis prediction studies using deep-learning-based methods with an MRI dataset, the number of studies of nasopharyngeal cancer is relatively high, and each of these studies assessed the prognosis in patients receiving chemoradiotherapy. As an initial result, Qiang et al. reported the utility of deep-learning-based MRI scores using T1WI, T2WI, and CE-T1WI.113 Their analysis was performed using MR images fed into 3D-DenseNet, and the low-dimensional features were then analyzed using DeepSurvivalNet and outputted as an MRI score. A combination diagnostic model of the clinical features and the deep-learning-based MRI score by a conventional machine learning algorithm of XGBoost were finally created. The model described by Qiang et al. demonstrated a statistically significant improvement in prognosis prediction with a Concordance Index (C-index) around 0.7–0.8 compared to the conventional tumor, node, metastasis (TNM) staging system. Notably, they used whole FOV images rather than manually segmented images, and they used external validation for the test set with the C-index of 0.72–0.76; the results of this analysis might be valuable toward the use of deep-learning-based prognosis prediction in other institutions.113 Zhong et al. investigated a deep-learning-radiomics model to predict the prognoses of patients with T3-stage nasopharyngeal cancer.114 They performed deep-learning-based feature map extraction by SE Res-Net using manually segmented T1WI, T2WI, and CE-T1WI. These deep-learning-based features were finally integrated by radiomics signature building. The created model showed excellent prognostic ability for disease-free survival, with the C-index of 0.8–0.9.114 Li et al. reported the utility of an ensemble learning method that effectively combined two sets of single deep-learning-based results for the prediction of the prognosis of patients with nasopharyngeal carcinoma at an advanced clinical stage.115 They used the Resnet-V2 deep-learning architecture with pre-treatment and post-treatment MRI, finally combining the datasets into one deep-learning-based diagnostic model. The ensemble learning-based diagnostic model exhibited the best performance compared to a single deep-learning-based model and conventional clinical staging, probably because the combined use of two datasets effectively improved the model’s performance. Li et al. also performed an analysis of gradient-weighted class activation mapping (Grad-CAM) images.115 Grad-CAM is used for visualization when a deep-learning diagnostic model is used to look at a target image. Interestingly, the deep-learning model suggested that the areas around the tumor and some cervical lymph nodes were strongly related to the prognosis of the tumor, whereas the relationship between the signal of the primary tumor area and the prognosis was not as strong as expected in many cases.115 In another study, Li et al. investigated deep-learning-based prognosis predictions and conducted a Grad-CAM analysis using several sizes of FOVs fed into the deep-learning model, and they observed a similar tendency: the area around the tumor is the most important for predicting the prognosis of patients with nasopharyngeal carcinoma.116
Regarding other primary sites of head and neck cancers, Tomita et al. investigated the utility of DWI with a deep-learning analysis in patients with hypopharyngeal or laryngeal SCC treated by definitive chemoradiotherapy. They trained the deep learning of Xception architecture by using anatomically masked DWI including the primary lesion at two time points (pre-treatment and intra-treatment). The results revealed that a deep-learning model trained with intra-treatment DWI provided the highest diagnostic performance to predict local recurrence, with the accuracy value of 0.767.117
In addition to directly estimating the prognosis, it is important to predict the genetic information that is well known as an independent prognostic factor, such as human papillomavirus (HPV) in oropharyngeal cancer and Epstein-Barr virus (EBV) in nasopharyngeal cancer. Image-based parameter integration and calculation for the prediction of the genomic information is recognized as ‘radiogenomics.’118 Research using deep-learning-based radiogenomics is still in progress, but the use of radiogenomics in clinical practice may be realized in the near future.
Duan et al. investigated the MRI findings of patients with large vestibular aqueduct syndrome and performed a deep-learning analysis to predict the patients’ hearing prognosis. They used images from a segmented T2-weighted sequence to include the target anatomical structure for the evaluation. The GoogLeNet-trained diagnostic model provided the best diagnostic accuracy (0.98) to differentiate between patients with stable versus fluctuating hearing loss.119
Prognosis prediction is essential for the determination of the best treatment planning and the appropriate post-treatment strategy, particularly in patients with cancer of the head and neck.120 Few studies have investigated this, but more precise patient classification might be feasible in the near future. Because MRI provides excellent contrast of various tissues and a large amount of information about the target lesion, combinations of a deep-learning method and the information derived from MRI will bring the diagnostic performance to a higher stage.
Future Perspective
Analyses of deep-learning techniques using MRI datasets are expected to be useful for a variety of purposes, including image acquisition/reconstruction, target segmentation, disease classification/diagnosis, and prognosis prediction. Such innovative methods will bring about a higher level of diagnostic performance compared to conventional methods. However, several issues remain. First, almost all of the studies related to head and neck MRI described above were performed with a transfer learning technique, whereas the CNN architecture used in the studies were quite different. In addition, some of the studies used a single CNN network, but other studies combined multiple architectures into one diagnostic model. Moreover, a wide range of methods for setting hyperparameters was present. The majority of the investigations cited above applied various deep-learning architectures and hyperparameters. With this background, it is challenging to establish a universally applicable model setting method, and it might be necessary to optimize the technical issues in model development based on each institution. A recent report by Sellergren et al. described a solution to address this limitation. Their general pretrained model followed by additional pretraining by medical images achieved higher model accuracy, and this technique may therefore contribute to the generalization of the pretraining procedure.121 It might also be helpful to build a basic model using large-scale medical images before the individual use; it could be effective to first build a fundamental deep-learning diagnostic model using a large sample size as the basis to accommodate image-quality differences due to multiple vendors and varying parameter settings, and then conduct fine-tuning of the model for individual uses for various purposes. Such an approach may help unify the processes of deep-learning techniques. In addition, the use of eXplainable AI (i.e., XAI) is an important method that can improve the utility of deep-learning algorithms for routine clinical practice. This function will aid the human interpretation of deep-learning-based diagnoses more objectively. A Grad-CAM approach as a type of XAI has been used frequently in recent investigations to identify the site at which a deep-learning model focuses (Fig. 4). Second, instead of using the entire images (those including the full FOV) as the input in model training, most of the prior analyses have been of the images after a manual segmentation of the lesion by a physician. Particularly, as described above in the ‘Disease classification and diagnosis’ section, most of the studies were performed using the segmented images for model training and testing. A full FOV image might be difficult to analyze for appropriate lesion detection/segmentation or diagnosis, probably because the complex anatomical structure of the head and neck includes a large amount of information. However, even with localized input data as well as a full FOV input, AI might output data by focusing on different parts of the target areas as the basis for decision-making.115 Such findings can also be revealed by using the above-mentioned function of Grad-CAM. Moreover, only a few studies have used external validation for the test set to estimate their diagnostic models. The deep-learning model created in such a situation might result in very limited use; the created model is likely to be used only in its developers’ institution. A review published in 2022 also indicated that among the AI-related studies focused on the head and neck, the number of investigations with the use of external validation in the test session is relatively small.122 For an appropriate external-validation procedure, the use of large amounts of data from public academic institutions (e.g., J-MID from the Japan Radiology Society) may be one of the steps to solve this problem.123 Further research is necessary to address these limitations, as are discussions to integrate the existing knowledge and future aspects. From this perspective, we speculate that the incorporation of deep-learning-based clinical tools into daily clinical practice may require more time. However, we believe that with the further development and careful monitoring of appropriate approaches and basic/clinical investigations, clinically significant achievements can be made.
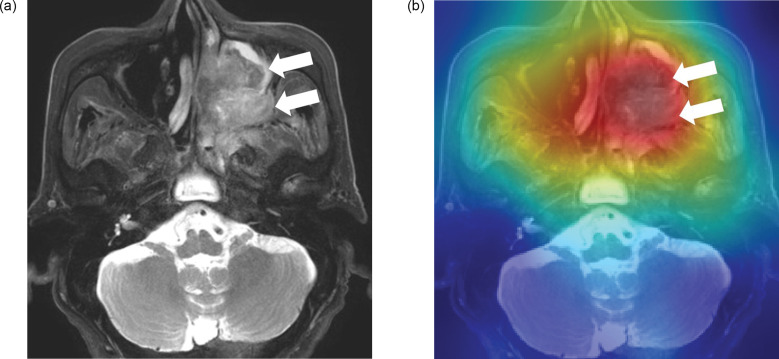
Fig. 4.
An example of gradient-weighted class activation mapping (Grad-CAM) for a deep-learning analysis targeting head and neck cancer. (a) Left maxillary cancer is observed on fat-suppressed T2WI (arrows). (b) The image shows the Grad-CAM displaying where the deep-learning model will be looking; this colormap represents a mathematical computation of the degree to which each pixel influences the final diagnosis; pixels with a stronger degree of influence are represented with a more intense red on the red-green-blue (RGB) color scale. In this case, the model analyzed the presence or absence of head and neck cancers on the MRI. Hot spots can be easily identified on the tumor lesion in this map (arrows). Grad-CAM, gradient-weighted class activation mapping; RGB, red-green-blue; T2WI, T2-weighted imaging.
Conclusion
AI-related techniques, particularly deep learning, are considered promising for the assessment of head and neck MRI, including image acquisition/reconstruction, segmentation, classification/diagnosis, and prognosis prediction. However, several limitations must be addressed before the deep-learning techniques can be applied in general medical practice in the field of radiology, and all of the future development of AI-based techniques should be closely monitored.
Footnotes
Conflicts of interest
The authors declare that they have no conflicts of interest.
References
- 1.Tshering Vogel DW, Thoeny HC. Cross-sectional imaging in cancers of the head and neck: how we review and report. Cancer Imaging 2016; 16:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatnagar P, Subesinghe M, Patel C, Prestwich R, Scarsbrook AF. Functional imaging for radiation treatment planning, response assessment, and adaptive therapy in head and neck cancer. Radiographics 2013; 33:1909–1929. [DOI] [PubMed] [Google Scholar]
- 3.King AD, Thoeny HC. Functional MRI for the prediction of treatment response in head and neck squamous cell carcinoma: potential and limitations. Cancer Imaging 2016; 16:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albano D, Bruno F, Agostini A, et al. Dynamic contrast-enhanced (DCE) imaging: state of the art and applications in whole-body imaging. Jpn J Radiol 2022; 40:341–366. [DOI] [PubMed] [Google Scholar]
- 5.Touska P, Connor SEJ. Recent advances in MRI of the head and neck, skull base and cranial nerves: new and evolving sequences, analyses and clinical applications. Br J Radiol 2019; 92:20190513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosmin M, Ledsam J, Romera-Paredes B, et al. Rapid advances in auto-segmentation of organs at risk and target volumes in head and neck cancer. Radiother Oncol 2019; 135:130–140. [DOI] [PubMed] [Google Scholar]
- 7.Fusco R, Granata V, Grazzini G, et al. Radiomics in medical imaging: pitfalls and challenges in clinical management. Jpn J Radiol 2022; 40:919–929. [DOI] [PubMed] [Google Scholar]
- 8.Aerts HJWL, Velazquez ER, Leijenaar RTH, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014; 5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y-Q, Liang C-H, He L, et al. Dvelopment and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 2016; 34:2157–2164. [DOI] [PubMed] [Google Scholar]
- 10.Lian C, Ruan S, Denœux T, Jardin F, Vera P. Selecting radiomic features from FDG-PET images for cancer treatment outcome prediction. Med Image Anal 2016; 32:257–268. [DOI] [PubMed] [Google Scholar]
- 11.Kickingereder P, Burth S, Wick A, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 2016; 280:880–889. [DOI] [PubMed] [Google Scholar]
- 12.Nie K, Shi L, Chen Q, et al. Rectal Cancer: Asessment of neoadjuvant chemoradiation outcome based on radiomics of multiparametric MRI. Clin Cancer Res 2016; 22:5256–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohki K, Igarashi T, Ashida H, et al. Usefulness of texture analysis for grading pancreatic neuroendocrine tumors on contrast-enhanced computed tomography and apparent diffusion coefficient maps. Jpn J Radiol 2021; 39:66–75. [DOI] [PubMed] [Google Scholar]
- 14.Bos P, van den Brekel MWM, Gouw ZAR, et al. Improved outcome prediction of oropharyngeal cancer by combining clinical and MRI features in machine learning models. Eur J Radiol 2021; 139:109701. [DOI] [PubMed] [Google Scholar]
- 15.Geng Z, Zhang Y, Wang S, et al. Radiomics analysis of susceptibility weighted imaging for hepatocellular carcinoma: exploring the correlation between histopathology and radiomics features. Magn Reson Med Sci 2021; 20:253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Yu S, Yin X, et al. An AI-based auxiliary empirical antibiotic therapy model for children with bacterial pneumonia using low-dose chest CT images. Jpn J Radiol 2021; 39:973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cay N, Mendi BAR, Batur H, Erdogan F. Discrimination of lipoma from atypical lipomatous tumor/well-differentiated liposarcoma using magnetic resonance imaging radiomics combined with machine learning. Jpn J Radiol 2022; 40:951–960. [DOI] [PubMed] [Google Scholar]
- 18.Hu P, Chen L, Zhong Y, et al. Effects of slice thickness on CT radiomics features and models for staging liver fibrosis caused by chronic liver disease. Jpn J Radiol 2022; 40:1061–1068. [DOI] [PubMed] [Google Scholar]
- 19.Tsuneta S, Oyama-Manabe N, Hirata K, et al. Texture analysis of delayed contrast-enhanced computed tomography to diagnose cardiac sarcoidosis. Jpn J Radiol 2021; 39:442–450. [DOI] [PubMed] [Google Scholar]
- 20.Valletta R, Faccioli N, Bonatti M, et al. Role of CT colonography in differentiating sigmoid cancer from chronic diverticular disease. Jpn J Radiol 2022; 40:48–55. [DOI] [PubMed] [Google Scholar]
- 21.Zeydanli T, Kilic HK. Performance of quantitative CT texture analysis in differentiation of gastric tumors. Jpn J Radiol 2022; 40:56–65. [DOI] [PubMed] [Google Scholar]
- 22.Kunimatsu A, Yasaka K, Akai H, Sugawara H, Kunimatsu N, Abe O. Texture analysis in brain tumor mr imaging. Magn Reson Med Sci 2022; 21:95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin L-Y, Zhang F, Yu Y, et al. Noninvasive evaluation of hypoxia in rabbit VX2 lung transplant tumors using spectral CT parameters and texture analysis. Jpn J Radiol 2022; 40:289–297. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Lu S, Mao Y, et al. An MRI-based radiomics-clinical nomogram for the overall survival prediction in patients with hypopharyngeal squamous cell carcinoma: a multi-cohort study. Eur Radiol 2022; 32:1548–1557. [DOI] [PubMed] [Google Scholar]
- 25.Ohno Y, Aoyagi K, Arakita K, et al. Newly developed artificial intelligence algorithm for COVID-19 pneumonia: utility of quantitative CT texture analysis for prediction of favipiravir treatment effect. Jpn J Radiol 2022; 40:800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anai K, Hayashida Y, Ueda I, et al. The effect of CT texture-based analysis using machine learning approaches on radiologists’ performance in differentiating focal-type autoimmune pancreatitis and pancreatic duct carcinoma. Jpn J Radiol 2022; 40:1156–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Chai W, Sun K, Fu C, Yan F. The value of whole-tumor histogram and texture analysis based on apparent diffusion coefficient (ADC) maps for the discrimination of breast fibroepithelial lesions: corresponds to clinical management decisions. Jpn J Radiol 2022; 40:1263–1271. [DOI] [PubMed] [Google Scholar]
- 28.Kuno H, Qureshi MM, Chapman MN, et al. CT Texture analysis potentially predicts local failure in head and neck squamous cell carcinoma treated with chemoradiotherapy. AJNR Am J Neuroradiol 2017; 38:2334–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueda D, Shimazaki A, Miki Y. Technical and clinical overview of deep learning in radiology. Jpn J Radiol 2019; 37:15–33. [DOI] [PubMed] [Google Scholar]
- 30.Nakata N. Recent technical development of artificial intelligence for diagnostic medical imaging. Jpn J Radiol 2019; 37:103–108. [DOI] [PubMed] [Google Scholar]
- 31.Barat M, Chassagnon G, Dohan A, et al. Artificial intelligence: a critical review of current applications in pancreatic imaging. Jpn J Radiol 2021; 39:514–523. [DOI] [PubMed] [Google Scholar]
- 32.Chassagnon G, De Margerie-Mellon C, Vakalopoulou M, et al. Artificial intelligence in lung cancer: current applications and perspectives. Jpn J Radiol 2023; 41:235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakhani P, Sundaram B. Deep learning at chest radiography: Automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 2017; 284:574–582. [DOI] [PubMed] [Google Scholar]
- 34.Matsukiyo R, Ohno Y, Matsuyama T, et al. Deep learning-based and hybrid-type iterative reconstructions for CT: comparison of capability for quantitative and qualitative image quality improvements and small vessel evaluation at dynamic CE-abdominal CT with ultra-high and standard resolutions. Jpn J Radiol 2021; 39:186–197. [DOI] [PubMed] [Google Scholar]
- 35.Kawamura M, Tamada D, Funayama S, et al. Accelerated acquisition of high-resolution diffusion-weighted imaging of the brain with a multi-shot echo-planar sequence: Deep-learning-based denoising. Magn Reson Med Sci 2021; 20:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueda D, Yamamoto A, Takashima T, et al. Visualizing “featureless” regions on mammograms classified as invasive ductal carcinomas by a deep learning algorithm: the promise of AI support in radiology. Jpn J Radiol 2021; 39:333–340. [DOI] [PubMed] [Google Scholar]
- 37.Ichikawa Y, Kanii Y, Yamazaki A, et al. Deep learning image reconstruction for improvement of image quality of abdominal computed tomography: comparison with hybrid iterative reconstruction. Jpn J Radiol 2021; 39:598–604. [DOI] [PubMed] [Google Scholar]
- 38.Wong LM, Ai QYH, Mo FKF, Poon DMC, King AD. Convolutional neural network in nasopharyngeal carcinoma: how good is automatic delineation for primary tumor on a non-contrast-enhanced fat-suppressed T2-weighted MRI?. Jpn J Radiol 2021; 39:571–579. [DOI] [PubMed] [Google Scholar]
- 39.Nakai H, Fujimoto K, Yamashita R, et al. Convolutional neural network for classifying primary liver cancer based on triple-phase CT and tumor marker information: a pilot study. Jpn J Radiol 2021; 39:690–702. [DOI] [PubMed] [Google Scholar]
- 40.Naganawa S, Nakamichi R, Ichikawa K, et al. MR imaging of endolymphatic hydrops: utility of iHYDROPS-Mi2 combined with deep learning reconstruction denoising. Magn Reson Med Sci 2021; 20:272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okuma T, Hamamoto S, Maebayashi T, et al. Quantitative evaluation of COVID-19 pneumonia severity by CT pneumonia analysis algorithm using deep learning technology and blood test results. Jpn J Radiol 2021; 39:956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagawa H, Fushimi Y, Nakajima S, et al. Deep learning-based noise reduction for fast volume diffusion tensor imaging: assessing the noise reduction effect and reliability of diffusion metrics. Magn Reson Med Sci 2021; 20:450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitahara H, Nagatani Y, Otani H, et al. A novel strategy to develop deep learning for image super-resolution using original ultra-high-resolution computed tomography images of lung as training dataset. Jpn J Radiol 2022; 40:38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishii T, Funama Y, Kato S, et al. Comparison of visibility of in-stent restenosis between conventional- and ultra-high spatial resolution computed tomography: coronary arterial phantom study. Jpn J Radiol 2022; 40:279–288. [DOI] [PubMed] [Google Scholar]
- 45.Yasaka K, Akai H, Sugawara H, et al. Impact of deep learning reconstruction on intracranial 1.5T magnetic resonance angiography. Jpn J Radiol 2022; 40:476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakao T, Hanaoka S, Nomura Y, Hayashi N, Abe O. Anomaly detection in chest 18F-FDG PET/CT by Bayesian deep learning. Jpn J Radiol 2022; 40:730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naganawa S, Ito R, Kawai H, et al. MR imaging of endolymphatic hydrops in five minutes. Magn Reson Med Sci 2022; 21:401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ozaki J, Fujioka T, Yamaga E, et al. Deep learning method with a convolutional neural network for image classification of normal and metastatic axillary lymph nodes on breast ultrasonography. Jpn J Radiol 2022; 40:814–822. [DOI] [PubMed] [Google Scholar]
- 49.Kaga T, Noda Y, Mori T, et al. Unenhanced abdominal low-dose CT reconstructed with deep learning-based image reconstruction: image quality and anatomical structure depiction. Jpn J Radiol 2022; 40:703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bambach S, Ho M-L. Deep learning for synthetic CT from bone MRI in the head and neck. AJNR Am J Neuroradiol 2022; 43:1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nai Y-H, Loi HY, O’Doherty S, Tan TH, Reilhac A. Comparison of the performances of machine learning and deep learning in improving the quality of low dose lung cancer PET images. Jpn J Radiol 2022; 40:1290–1299. [DOI] [PubMed] [Google Scholar]
- 52.Jaspan ON, Fleysher R, Lipton ML. Compressed sensing MRI: a review of the clinical literature. Br J Radiol 2015; 88:20150487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takumi K, Nagano H, Nakanosono R, Kumagae Y, Fukukura Y, Yoshiura T. Combined signal averaging and compressed sensing: impact on quality of contrast-enhanced fat-suppressed 3D turbo field-echo imaging for pharyngolaryngeal squamous cell carcinoma. Neuroradiology 2020; 62:1293–1299. [DOI] [PubMed] [Google Scholar]
- 54.Kami Y, Chikui T, Togao O, Ooga M, Yoshiura K. Comparison of image quality of head and neck lesions between 3D gradient echo sequences with compressed sensing and the multi-slice spin echo sequence. Acta Radiol Open 2020; 9:2058460120956644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomita H, Deguchi Y, Fukuchi H, et al. Combination of compressed sensing and parallel imaging for T2-weighted imaging of the oral cavity in healthy volunteers: comparison with parallel imaging. Eur Radiol 2021; 31:6305–6311. [DOI] [PubMed] [Google Scholar]
- 56.Higaki T, Nakamura Y, Tatsugami F, Nakaura T, Awai K. Improvement of image quality at CT and MRI using deep learning. Jpn J Radiol 2019; 37:73–80. [DOI] [PubMed] [Google Scholar]
- 57.Lin DJ, Johnson PM, Knoll F, Lui YW. Artificial Intelligence for MR image reconstruction: An Overview for Clinicians. J Magn Reson Imaging 2021; 53:1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koktzoglou I, Huang R, Ankenbrandt WJ, Walker MT, Edelman RR. Super-resolution head and neck MRA using deep machine learning. Magn Reson Med 2021; 86:335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li S, Zhang S, Yu Z, Lin Y. MRI of the intraorbital ocular motor nerves on three-dimensional double-echo steady state with water excitation sequence at 3.0T. Jpn J Radiol 2021; 39:749–754. [DOI] [PubMed] [Google Scholar]
- 60.Bao Q, Xie W, Otikovs M, et al. Unsupervised cycle-consistent network using restricted subspace field map for removing susceptibility artifacts in EPI. Magn Reson Med 2023; 90:458–472. [DOI] [PubMed] [Google Scholar]
- 61.Lu A, Gorny KR, Ho M-L. Zero TE MRI for craniofacial bone imaging. AJNR Am J Neuroradiol 2019; 40:1562–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hiyama T, Kuno H, Sekiya K, Tsushima S, Oda S, Kobayashi T. Subtraction iodine imaging with area detector CT to improve tumor delineation and measurability of tumor size and depth of invasion in tongue squamous cell carcinoma. Jpn J Radiol 2022; 40:167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baba A, Hashimoto K, Kuno H, et al. Assessment of squamous cell carcinoma of the floor of the mouth with magnetic resonance imaging. Jpn J Radiol 2021; 39:1141–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aliotta E, Nourzadeh H, Siebers J. Quantifying the dosimetric impact of organ-at-risk delineation variability in head and neck radiation therapy in the context of patient setup uncertainty. Phys Med Biol 2019; 64:135020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai X, Lei Y, Wang T, et al. Multi-organ auto-delineation in head-and-neck MRI for radiation therapy using regional convolutional neural network. Phys Med Biol 2022; 67:025006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chandarana H, Wang H, Tijssen RHN, Das IJ. Emerging role of MRI in radiation therapy. J Magn Reson Imaging 2018; 48:1468–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin L, Dou Q, Jin Y-M, et al. Deep learning for automated contouring of primary tumor volumes by MRI for nasopharyngeal carcinoma. Radiology 2019; 291:677–686. [DOI] [PubMed] [Google Scholar]
- 68.Chen H, Qi Y, Yin Y, et al. MMFNet: A multi-modality MRI fusion network for segmentation of nasopharyngeal carcinoma. Neurocomputing 2020; 394:27–40. [Google Scholar]
- 69.Ma Z, Zhou S, Wu X, et al. Nasopharyngeal carcinoma segmentation based on enhanced convolutional neural networks using multi-modal metric learning. Phys Med Biol 2019; 64:025005. [DOI] [PubMed] [Google Scholar]
- 70.Li Q, Xu Y, Chen Z, et al. Tumor segmentation in contrast-enhanced magnetic resonance imaging for nasopharyngeal carcinoma: Deep learning with convolutional neural network. BioMed Res Int 2018; 2018:9128527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodríguez Outeiral R, Bos P, Al-Mamgani A, Jasperse B, Simões R, van der Heide UA. Oropharyngeal primary tumor segmentation for radiotherapy planning on magnetic resonance imaging using deep learning. Phys Imaging Radiat Oncol 2021; 19:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wahid KA, Ahmed S, He R, et al. Evaluation of deep learning-based multiparametric MRI oropharyngeal primary tumor auto-segmentation and investigation of input channel effects: Results from a prospective imaging registry. Clin Transl Radiat Oncol 2021; 32:6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dai X, Lei Y, Wang T, et al. Automated delineation of head and neck organs at risk using synthetic MRI-aided mask scoring regional convolutional neural network. Med Phys 2021; 48:5862–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Korte JC, Hardcastle N, Ng SP, Clark B, Kron T, Jackson P. Cascaded deep learning-based auto-segmentation for head and neck cancer patients: Organs at risk on T2-weighted magnetic resonance imaging. Med Phys 2021; 48:7757–7772. [DOI] [PubMed] [Google Scholar]
- 75.Neve OM, Chen Y, Tao Q, et al. Fully Automated 3D vestibular schwannoma segmentation with and without gadolinium-based contrast material: A multicenter, multivendor study. Radiol Artif Intell 2022; 4:e210300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao P, Shavit SS, Shin J, Selesnick S, Phillips CD, Strauss SB. Segmentation of vestibular schwannomas on postoperative gadolinium-enhanced T1-wighted and noncontrast T2-weighted magnetic resonance imaging using deep learning. Otol Neurotol 2022; 43:1227–1239. [DOI] [PubMed] [Google Scholar]
- 77.Cho YS, Cho K, Park CJ, et al. Automated measurement of hydrops ratio from MRI in patients with Ménière’s disease using CNN-based segmentation. Sci Rep 2020; 10:7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vaidyanathan A, van der Lubbe MFJA, Leijenaar RTH, et al. Deep learning for the fully automated segmentation of the inner ear on MRI. Sci Rep 2021; 11:2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park CJ, Cho YS, Chung MJ, et al. A fully automated analytic system for measuring endolymphatic hydrops ratios in patients with ménière disease via magnetic resonance imaging: Deep learning model development study. J Med Internet Res 2021; 23:e29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ryu JY, Hong HK, Cho HG, et al. Deep learning for the automatic segmentation of extracranial venous malformations of the head and neck from MR Images Using 3D U-Net. J Clin Med 2022; 11: 5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gumeler E, Kurtulan O, Arslan S, et al. Assessment of 4DCT imaging findings of parathyroid adenomas in correlation with biochemical and histopathological findings. Jpn J Radiol 2022; 40:484–491. [DOI] [PubMed] [Google Scholar]
- 82.Baba A, Matsushima S, Fukuda T, et al. Improved assessment of middle ear recurrent/residual cholesteatomas using temporal subtraction CT. Jpn J Radiol 2022; 40:271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang J-R, Song Y, Jia Y-L, Ruan L-T. Application of multimodal ultrasonography for differentiating benign and malignant cervical lymphadenopathy. Jpn J Radiol 2021; 39:938–945. [DOI] [PubMed] [Google Scholar]
- 84.Wang X, Dai S, Wang Q, Chai X, Xian J. Investigation of MRI-based radiomics model in differentiation between sinonasal primary lymphomas and squamous cell carcinomas. Jpn J Radiol 2021; 39:755–762. [DOI] [PubMed] [Google Scholar]
- 85.Suto T, Kato H, Kawaguchi M, et al. MRI findings of epithelial-myoepithelial carcinoma of the parotid gland with radiologic-pathologic correlation. Jpn J Radiol 2022; 40:578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chang Y-J, Huang T-Y, Liu Y-J, Chung H-W, Juan C-J. Classification of parotid gland tumors by using multimodal MRI and deep learning. NMR Biomed 2021; 34:e4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matsuo H, Nishio M, Kanda T, et al. Diagnostic accuracy of deep-learning with anomaly detection for a small amount of imbalanced data: discriminating malignant parotid tumors in MRI. Sci Rep 2020; 10:19388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raghu M, Unterthiner T, Kornblith S, Zhang C, Dosovitskiy A. Do vision transformers see like convolutional neural networks?. arXiv [csCV] 2021.
- 89.Dai Y, Gao Y, Liu F. TransMed: Transformers Advance multi-modal medical image classification. Diagnostics (Basel) 2021; 11: 1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu X, Pan Y, Zhang X, et al. A deep learning model for classification of parotid neoplasms based on multimodal magnetic resonance image sequences. Laryngoscope 2023; 133:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wong LM, King AD, Ai QYH, et al. Convolutional neural network for discriminating nasopharyngeal carcinoma and benign hyperplasia on MRI. Eur Radiol 2021; 31:3856–3863. [DOI] [PubMed] [Google Scholar]
- 92.Yang Q, Guo Y, Ou X, Wang J, Hu C. Automatic t staging using weakly supervised deep learning for nasopharyngeal carcinoma on MR images. J Magn Reson Imaging 2020; 52:1074–1082. [DOI] [PubMed] [Google Scholar]
- 93.Liu GS, Yang A, Kim D, et al. Deep learning classification of inverted papilloma malignant transformation using 3D convolutional neural networks and magnetic resonance imaging. Int Forum Allergy Rhinol 2022; 12:1025–1033. [DOI] [PubMed] [Google Scholar]
- 94.Naganawa S, Kawai H, Taoka T, Sone M. Improved HYDROPS: Imaging of endolymphatic hydrops after intravenous administration of gadolinium. Magn Reson Med Sci 2017; 16:357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Naganawa S, Kawai H, Taoka T, Sone M. Improved 3D-real Inversion Recovery: A Robust Imaging Technique for Endolymphatic Hydrops after Intravenous Administration of Gadolinium. Magn Reson Med Sci 2019; 18:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ohashi T, Naganawa S, Takeuchi A, Katagiri T, Kuno K. Quantification of endolymphatic space volume after intravenous administration of a single dose of gadolinium-based contrast agent: 3D-real Inversion Recovery versus HYDROPS-Mi2. Magn Reson Med Sci 2020; 19:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Naganawa S, Ito R, Kato Y, et al. Intracranial distribution of intravenously administered gadolinium-based contrast agent over a period of 24 hours: Evaluation with 3D-real IR Imaging and MR fingerprinting. Magn Reson Med Sci 2021; 20:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nomura Y, Hanaoka S, Nakao T, et al. Performance changes due to differences in training data for cerebral aneurysm detection in head MR angiography images. Jpn J Radiol 2021; 39:1039–1048. [DOI] [PubMed] [Google Scholar]
- 99.Yamaguchi K, Nakazono T, Egashira R, et al. Maximum slope of ultrafast dynamic contrast-enhanced MRI of the breast: Comparisons with prognostic factors of breast cancer. Jpn J Radiol 2021; 39:246–253. [DOI] [PubMed] [Google Scholar]
- 100.Kato E, Mori N, Mugikura S, Sato S, Ishida T, Takase K. Value of ultrafast and standard dynamic contrast-enhanced magnetic resonance imaging in the evaluation of the presence and extension of residual disease after neoadjuvant chemotherapy in breast cancer. Jpn J Radiol 2021; 39:791–801. [DOI] [PubMed] [Google Scholar]
- 101.Inoue A, Furukawa A, Takaki K, et al. Noncontrast MRI of acute abdominal pain caused by gastrointestinal lesions: indications, protocol, and image interpretation. Jpn J Radiol 2021; 39:209–224. [DOI] [PubMed] [Google Scholar]
- 102.Jia C, Liu G, Wang X, Zhao D, Li R, Li H. Hepatic sclerosed hemangioma and sclerosing cavernous hemangioma: a radiological study. Jpn J Radiol 2021; 39:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matsuura K, Inoue K, Hoshino E, et al. Utility of magnetic resonance imaging for differentiating malignant mesenchymal tumors of the uterus from T2-weighted hyperintense leiomyomas. Jpn J Radiol 2022; 40:385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xie Y, Zhang S, Liu X, et al. Minimal apparent diffusion coefficient in predicting the Ki-67 proliferation index of pancreatic neuroendocrine tumors. Jpn J Radiol 2022; 40:823–830. [DOI] [PubMed] [Google Scholar]
- 105.Takao S, Kaneda M, Sasahara M, et al. Diffusion tensor imaging (DTI) of human lower leg muscles: correlation between DTI parameters and muscle power with different ankle positions. Jpn J Radiol 2022; 40:939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Taoka T, Ito R, Nakamichi R, et al. Diffusion-weighted image analysis along the perivascular space (DWI-ALPS) for evaluating interstitial fluid status: age dependence in normal subjects. Jpn J Radiol 2022; 40:894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jiang J, Fu Y, Zhang L, et al. Volumetric analysis of intravoxel incoherent motion diffusion-weighted imaging in preoperative assessment of non-small cell lung cancer. Jpn J Radiol 2022; 40:903–913. [DOI] [PubMed] [Google Scholar]
- 108.Ichikawa K, Taoka T, Ozaki M, Sakai M, Yamaguchi H, Naganawa S. Impact of tissue properties on time-dependent alterations in apparent diffusion coefficient: a phantom study using oscillating-gradient spin-echo and pulsed-gradient spin-echo sequences. Jpn J Radiol 2022; 40:970–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Özer H, Yazol M, Erdoğan N, Emmez ÖH, Kurt G, Öner AY. Dynamic contrast-enhanced magnetic resonance imaging for evaluating early response to radiosurgery in patients with vestibular schwannoma. Jpn J Radiol 2022; 40:678–688. [DOI] [PubMed] [Google Scholar]
- 110.Taoka T, Kawai H, Nakane T, et al. Diffusion analysis of fluid dynamics with incremental strength of motion proving gradient (DANDYISM) to evaluate cerebrospinal fluid dynamics. Jpn J Radiol 2021; 39:315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mes SW, van Velden FHP, Peltenburg B, et al. Outcome prediction of head and neck squamous cell carcinoma by MRI radiomic signatures. Eur Radiol 2020; 30:6311–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Du G, Zeng Y, Chen D, Zhan W, Zhan Y. Application of radiomics in precision prediction of diagnosis and treatment of gastric cancer. Jpn J Radiol 2023; 41:245–257. [DOI] [PubMed] [Google Scholar]
- 113.Qiang M, Li C, Sun Y, et al. A prognostic predictive system based on deep learning for locoregionally advanced nasopharyngeal carcinoma. J Natl Cancer Inst 2021; 113:606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhong L, Dong D, Fang X, et al. A deep learning-based radiomic nomogram for prognosis and treatment decision in advanced nasopharyngeal carcinoma: A multicentre study. EBioMedicine 2021; 70:103522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li S, Deng Y-Q, Hua H-L, et al. Deep learning for locally advanced nasopharyngeal carcinoma prognostication based on pre- and post-treatment MRI. Comput Methods Programs Biomed 2022; 219:106785. [DOI] [PubMed] [Google Scholar]
- 116.Li S, Wan X, Deng Y-Q, et al. Predicting prognosis of nasopharyngeal carcinoma based on deep learning: peritumoral region should be valued. Cancer Imaging 2023; 23:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tomita H, Kobayashi T, Takaya E, et al. Deep learning approach of diffusion-weighted imaging as an outcome predictor in laryngeal and hypopharyngeal cancer patients with radiotherapy-related curative treatment: a preliminary study. Eur Radiol 2022; 32:5353–5361. [DOI] [PubMed] [Google Scholar]
- 118.Boot PA, Mes SW, de Bloeme CM, et al. Magnetic resonance imaging based radiomics prediction of Human Papillomavirus infection status and overall survival in oropharyngeal squamous cell carcinoma. Oral Oncol 2023; 137:106307. [DOI] [PubMed] [Google Scholar]
- 119.Duan B, Xu Z, Pan L, Chen W, Qiao Z. Prediction of hearing prognosis of large vestibular aqueduct syndrome based on the pytorch deep learning model. J Healthc Eng 2022; 2022:4814577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120.Zhang Y, Luo D, Guo W, Liu Z, Zhao X. Utility of mono-exponential, bi-exponential, and stretched exponential signal models of intravoxel incoherent motion (IVIM) to predict prognosis and survival risk in laryngeal and hypopharyngeal squamous cell carcinoma (LHSCC) patients after chemoradiotherapy. Jpn J Radiol 2023; 41:712–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sellergren AB, Chen C, Nabulsi Z, et al. Simplified transfer learning for chest radiography models using less data. Radiology 2022; 305:454–465. [DOI] [PubMed] [Google Scholar]
- 122.Adeoye J, Hui L, Su Y-X. Data-centric artificial intelligence in oncology: a systematic review assessing data quality in machine learning models for head and neck cancer. J Big Data 2023; 10: 28. [Google Scholar]
- 123.Toda N, Hashimoto M, Arita Y, et al. Deep learning algorithm for fully automated detection of small (≤4 cm) renal cell carcinoma in contrast-enhanced computed tomography using a multicenter database. Invest Radiol 2022; 57:327–333. [DOI] [PubMed] [Google Scholar]