Abstract
Genomic diversity of mutation in the mecI gene or mecA promoter/operator region was analyzed for clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA) and Staphylococcus epidermidis (MRSE). In most MRSA strains, a single base substitution was detected in either the mecI (three different positions) or the mecA promoter (two different positions), while a 28-base deletion in mecI was found in one strain. In contrast, no mutation was detected in these gene sequences of MRSE strains.
Methicillin-resistant Staphylococcus aureus (MRSA) is defined by the production of a specific penicillin-binding protein (PBP), PBP-2a, that has a reduced binding affinity for beta-lactam compounds (4, 25). PBP-2a functions as a transpeptidase in cell wall synthesis in MRSA at high concentrations of beta-lactam antibiotics that inhibit the growth of methicillin-susceptible strains with normal PBPs. This additional PBP is encoded by the structural gene mecA on the chromosome (18), which has also been detected in methicillin-resistant strains of other staphylococcal species (10, 16, 19, 24). The mecA gene is a component of a large DNA fragment designated mec DNA, which is located at the specific site of the S. aureus chromosome and has been suggested to be transmitted from other bacterial species (1–3, 7, 27). The acquisition of mec DNA is considered to be the first genetic requisite for methicillin resistance of staphylococci.
Expression of PBP-2a is controlled by two regulator genes on mec DNA, mecI and mecR1, located upstream of mecA, which encode mecA repressor protein and signal transducer protein, respectively (5, 14, 21). An MRSA carrying intact mecI and mecR1 together with mecA has been called pre-MRSA, which is represented by prototype S. aureus strain N315 (6). Since intact mecI product strongly represses the expression of PBP-2a, the pre-MRSA is apparently methicillin susceptible (6, 14). Hence, it is hypothesized that removal of the repressor function for mecA is a prerequisite for constitutive expression of methicillin resistance in S. aureus with mec DNA. Indeed, the deletion of mecI or point mutations in the mecI gene has been found in a number of methicillin-resistant staphylococcal isolates (6, 8, 12, 20). In some strains, point mutations were detected in the mecA promoter region corresponding to a presumptive operator of mecA, i.e., the binding site of the repressor protein. Furthermore, genetic alteration on the chromosome which causes high methicillin resistance was presented as another mechanism of evolution of MRSA, although the details are not known (6).
We previously studied the presence of mec regulator genes in a number of clinical isolates of MRSA and methicillin-resistant Staphylococcus epidermidis (MRSE). Most strains were found to possess mecI and mecR1 genes, and the possibility of mutation in the mecI or mecA promoter region was suggested (12). In the present study, we analyzed the genetic diversity of mutations in the mecI and mecA promoters of 20 MRSA and 11 MRSE strains isolated at Sapporo Medical University Hospital in Japan between 1993 and 1995. The presence of the mecI and mecR1 genes in these strains has been confirmed previously (12). Oxacillin MICs for the MRSA and MRSE strains ranged from 256 to 1,024 μg/ml and from 16 to 256 μg/ml, respectively. Table 1 lists some characteristics of MRSA strains. Most of the MRSA strains belonged to coagulase type II.
TABLE 1.
Properties of MRSA strains analyzed in this study
| Year of isolation | Strain | Coagulase typea | RFLP pattern of coagulase geneb | MIC (μg/ml) of oxacillinc | RFLP pattern of mecI gened |
|---|---|---|---|---|---|
| 1993 | SH1 | II | A | 256 | 1 |
| SH12 | II | A | 256 | 1 | |
| SH13 | II | A | 256 | 3 | |
| SH19651 | IV | C | 256 | 2 | |
| SH15 | IV | C | 1,024 | 2 | |
| SH20 | II | A | 1,024 | 1 | |
| SH22 | VII | B | 1,024 | 1 | |
| SH24 | II | A | 256 | 1 | |
| SH27 | II | A | 256 | 1 | |
| 1994 | SH153 | II | A | 256 | 1 |
| SH155 | II | A | 512 | 1 | |
| SH158 | II | A | 256 | 1 | |
| SH165 | II | A | 512 | 1 | |
| SH212 | II | A | 512 | 2 | |
| 1995 | SH219 | II | A | 256 | 1 |
| SH320 | II | A | 256 | 1 | |
| SH321 | II | A | 256 | 1 | |
| SH324 | II | A | 512 | 1 | |
| SH326 | II | A | 512 | 1 | |
| SH327 | II | A | 512 | 1 |
Coagulase type (I to VIII) was determined with coagulase type-specific antisera.
RFLP patterns based on the staphylocoagulase gene were classified as described previously (11).
Susceptibility to oxacillin was measured by a broth microdilution assay with cation-supplemented Mueller-Hinton broth (BBL) containing 2% NaCl as recommended by the National Committee for Clinical Laboratory Standards (15).
Differentiation of the RFLP patterns to detect mutation in the mecI gene was performed as described previously (12; see text).
Two point mutations have been most frequently detected in the mecI gene (base substitutions C to T at position 202 and T to A at position 260) (6, 20), yielding an additional MseI site (5′T↓TAA3′) in the mecI sequence. Digestion of PCR product containing the complete open reading frame of the mecI gene with MseI enabled us to differentiate three patterns of restriction fragment length polymorphism (RFLP) described in the previous study (12), with pattern 1 representing a prototype of the mecI gene, pattern 2 indicating the presence of mutation at position 202, and pattern 3 having a smaller fragment than that in pattern 1, suggesting a deletion of nucleotides. As shown in Table 1, the mecI gene of most MRSA isolates was assigned to RFLP pattern 1, while three strains and one strain were classified as RFLP patterns 2 and 3, respectively. All 11 MRSE strains analyzed in the present study exhibited mecI RFLP pattern 1.
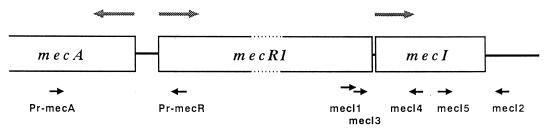
DNA samples for PCR were prepared with achromopeptidase as described previously (10). Either of two primer pairs, mecI1 and mecI2 or mecI3 and mecI2, was used to amplify DNA fragments containing the mecI gene, and another primer pair, Pr-mecA and Pr-mecR, was used to amplify the promoter regions of mecA and mecR1. Sequences of these primers are shown in Table 2, and the locations of the primers in mec DNA are depicted in Fig. 1. DNA amplification was performed with a thermal cycler as described previously (10). The presence of amplified PCR product (481 or 469 bp for the mecI gene and 748 bp for the mecA promoter region) was ascertained by electrophoresis on a 1% agarose gel and staining with ethidium bromide. With the PCR-amplified DNA fragments as templates, nucleotide sequences of the mecI gene (369 bases) and the promoter regions of mecA and mecR1 (99 bases) were determined by the dideoxynucleotide chain termination method with the Sequenase kit, version 2.0 (United States Biochemical Corp., Cleveland, Ohio). Primers listed in Table 2 were used for DNA sequencing.
TABLE 2.
Oligonucleotide primers
| Primer name | Sequencea |
|---|---|
| mecI1 | 5′-AATGGCGAAAAAGCACAACA-3′ |
| mecI2 | 5′-GACTTGATTGTTTCCTCTGTT-3′ |
| mecI3 | 5′-GCACAACAAATTTCTGAGCG-3′ |
| mecI4 | 5′-TGGACTCCAGTCCTTTTGC-3′ |
| mecI5 | 5′-CTTGTAGAAGAAAGTGATAT-3′ |
| Pr-mecA | 5′-CCTGTATTGGCCAATTCCAC-3′ |
| Pr-mecR | 5′-AATGGAATTAACGTGGAGAC-3′ |
The location of each primer on mec DNA is shown in Fig. 1.
FIG. 1.
Schematic representation of the mecA, mecR1, and mecI genes and locations of primers (Table 1) used in this study. The direction of the nucleotide extension reaction (5′ to 3′) of each primer is shown by a solid arrow. The shaded arrows indicate the directions of transcription of the structural genes.
The nucleotide sequences of the mecI gene and the promoter regions of mecA and mecR1 of MRSA strains employed in this study were compared with those of S. aureus N315 (5), a prototype strain possessing intact mec regulator genes. In all the MRSA strains, a point mutation or a deletion was detected in one of the two gene sequences, except in one strain (SH212) which had mutations in both sequences. As shown in Table 3, the mutations detected in MRSA strains were classified into seven groups (M1 to M7); five of these mutations, M1, M2, M4, M5, and M7, were identified for the first time in the present study. In contrast, no mutation was found in the mecI or mecA promoter region of the MRSE strains or in the mecR1 promoter regions of the MRSA or MRSE strains.
TABLE 3.
Mutations detected in mecI gene and mecA promoter region of MRSA strains
| Mutation group | MRSA strain(s) | Mutation in mecI gene
|
Mutation in mecA promoter regiona | |
|---|---|---|---|---|
| Nucleotide position change | Codon change | |||
| M1 | SH20 | 43 (G→T) | Val15→Phe | None |
| M2 | SH22 | 163 (A→T) | Lys55→stop codon | None |
| M3 | SH15, SH19651 | 202 (C→T) | Gln68→stop codon | None |
| M4 | SH212 | 202 (C→T) | Gln68→stop codon | C→A |
| M5 | SH13 | Deletion of 28 bases (28–55 or 29–56)b | Stop codon after the 11th amino acid | None |
| M6 | SH1, SH12, SH24, SH27, SH153, SH158, SH165, SH219, SH320, SH321, SH324, SH326, SH327 | None | None | C→A |
| M7 | SH155 | None | None | G→A |
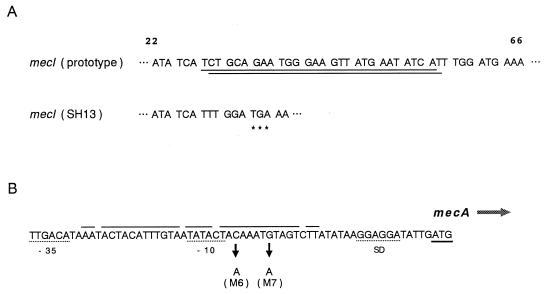
A nucleotide substitution at position 202 (M3) in mecI was detected in three strains (SH15, SH19651, and SH212) which had exhibited mecI RFLP pattern 2. Other base substitutions, M1 in strain SH20 and M2 in strain SH22, generated an amino acid change and a new termination codon, respectively. The deletion of 28 bases (M5) near the 5′ end of the mecI gene was found in MRSA strain SH13, which had shown mecI RFLP pattern 3. This base deletion, shown in Fig. 2A, caused a premature termination at position 33 on the mecI gene.
FIG. 2.
(A) Partial nucleotide sequences of the mecI gene from prototype S. aureus N315 (5) and MRSA SH13. Presumptive nucleotide sequences which were deleted in the SH13 mecI gene are underlined. Triple asterisks under the mecI gene sequence of strain SH13 denote a putative stop codon caused by the deletion. (B) Nucleotide sequence of the mecA promoter region. Positions of base substitution (M6 and M7) are shown by solid arrows. Putative −35 and −10 promoter sequences and Shine-Dalgarno (SD) sequences are shown by dotted lines, and a pair of palindrome sequences are indicated by solid lines above the sequence. The direction of mecA gene transcription is shown by a shaded arrow, with the initiation codon indicated by solid underlining.
Point mutations in the mecA promoter region (M6 and M7) were detected in 15 strains, although M7 was found in only one strain (SH155). Both mutations M6 and M7 are located downstream of the mecA promoter sequence (−10) on a palindrome structure corresponding to the presumptive operator of the mecA gene (Fig. 2B) (6, 18). It was reported previously that a C-to-A substitution (corresponding to M6) caused a decrease in the stacking energy of the stem (6); this also seems to be the case with the G-to-A (M7) nucleotide substitution detected in the present study.
A mutation observed in strain SH212 was assigned to group M4, because a point mutation was found in the mecI and the mecA promoter regions, coinciding with M3 and M6, respectively. In spite of the presence of double mutations in SH212, no significant difference in the MIC of oxacillin was seen for SH212 and other MRSA strains with a single mutation. The emergence of this peculiar MRSA strain can be explained as follows. Although this strain originally possessed a point mutation only in the mecI gene, a mutation in the mecA promoter region occurred subsequently and the mutant was selected to escape from the blaI repressor protein, which is a plasmid factor controlling blaZ, a penicillinase gene (9, 13, 26, 28). This assumption is based on the findings that the mecA promoter sequence is quite similar to that of blaZ on the plasmid and that expression of PBP-2a is also regulated by the bla regulator in some MRSA strains (17, 22, 23).
The finding that all 11 MRSE strains harbored no mutations in the two gene sequences was unexpected. Although the expression of mecI and mecA has not been confirmed in these strains, some mechanisms may be considered as explanations of the methicillin resistance of these MRSE strains, e.g., genomic alteration in a mec regulator gene other than mecI or the presence of certain unknown genetic factors controlling mecA expression that may suppress regulation by mecI. In any case, methicillin resistance in MRSE strains is presumably mediated by a mechanism different from that observed in MRSA strains.
REFERENCES
- 1.Archer G L, Niemeyer D M. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 1994;2:343–347. doi: 10.1016/0966-842x(94)90608-4. [DOI] [PubMed] [Google Scholar]
- 2.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Couto I, de Lencastre H, Severina E, Kloos W, Webster J A, Hubner R J, Sanches I S, Tomasz A. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb Drug Resist. 1996;2:377–391. doi: 10.1089/mdr.1996.2.377. [DOI] [PubMed] [Google Scholar]
- 4.Hartman B J, Tomasz A. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA) FEBS Lett. 1992;298:133–136. doi: 10.1016/0014-5793(92)80039-j. [DOI] [PubMed] [Google Scholar]
- 6.Hiramatsu K. Molecular evolution of MRSA. Microbiol Immunol. 1995;39:531–543. doi: 10.1111/j.1348-0421.1995.tb02239.x. [DOI] [PubMed] [Google Scholar]
- 7.Hiramatsu K, Kondo N, Ito T. Genetic basis for molecular epidemiology of MRSA. J Infect Chemother. 1996;2:117–129. doi: 10.3412/jsb.52.417. [DOI] [PubMed] [Google Scholar]
- 8.Hürlimann-Dalel R L, Ryffel C, Kayser F H, Berger-Bächi B. Survey of the methicillin resistance-associated genes mecA, mecR1-mecI, and femA-femB in clinical isolates of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:2617–2621. doi: 10.1128/aac.36.12.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imanaka T, Himeno T, Aiba S. Cloning and nucleotide sequence of the penicillinase antirepressor gene penJ of Bacillus licheniformis. J Bacteriol. 1987;169:3867–3872. doi: 10.1128/jb.169.9.3867-3872.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi N, Wu H, Kojima K, Taniguchi K, Urasawa S, Uehara N, Omizu Y, Kishi Y, Yagihashi A, Kurokawa I. Detection of mecA, femA, and femB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiol Infect. 1994;113:259–266. doi: 10.1017/s0950268800051682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi N, Taniguchi K, Kojima K, Urasawa S, Uehara N, Omizu Y, Kishi Y, Yagihashi A, Kurokawa I. Analysis of methicillin-resistant and methicillin-susceptible Staphylococcus aureus by a molecular typing method based on coagulase gene polymorphisms. Epidemiol Infect. 1995;115:419–426. doi: 10.1017/s095026880005857x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi N, Taniguchi K, Kojima K, Urasawa S, Uehara N, Omizu Y, Kishi Y, Yagihashi A, Kurokawa I, Watanabe N. Genomic diversity of mec regulator genes in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Epidemiol Infect. 1996;117:289–295. doi: 10.1017/s0950268800001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi T, Zhu Y F, Nicholls N J, Lampen J O. A second regulatory gene, blaR1, encoding a potential penicillin-binding protein required for induction of β-lactamase in Bacillus licheniformis. J Bacteriol. 1987;169:3873–3878. doi: 10.1128/jb.169.9.3873-3878.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuwahara-Arai K, Kondo N, Hori S, Tateda-Suzuki E, Hiramatsu K. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP 2′ production. Antimicrob Agents Chemother. 1996;40:2680–2685. doi: 10.1128/aac.40.12.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. Approved standard M7-A3. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 3rd ed. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 16.Ryffel C, Tesch W, Birch-Machin I, Reynolds P E, Barberis-Maino L, Kayser F H, Berger-Bächi B. Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Gene. 1990;94:137–138. doi: 10.1016/0378-1119(90)90481-6. [DOI] [PubMed] [Google Scholar]
- 17.Ryffel C, Kayser F H, Berger-Bächi B. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 1992;36:25–31. doi: 10.1128/aac.36.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song M D, Wachi M, Doi M, Ishino F, Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987;221:167–171. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki E, Hiramatsu K, Yokota T. Survey of methicillin-resistant clinical strains of coagulase-negative staphylococci for mecA gene distribution. Antimicrob Agents Chemother. 1992;36:429–434. doi: 10.1128/aac.36.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki E, Kuwahara-Arai K, Richardson J F, Hiramatsu K. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob Agents Chemother. 1993;37:1219–1226. doi: 10.1128/aac.37.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tesch W, Ryffel C, Strässle A, Kayser F H, Berger-Bächi B. Evidence of a novel staphylococcal mec-encoded element (mecR) controlling expression of penicillin-binding protein 2′. Antimicrob Agents Chemother. 1990;34:1703–1706. doi: 10.1128/aac.34.9.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ubukata K, Yamashita N, Konno M. Occurrence of a β-lactam-inducible penicillin-binding protein in methicillin-resistant staphylococci. Antimicrob Agents Chemother. 1985;27:851–857. doi: 10.1128/aac.27.5.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ubukata K, Nonoguchi R, Matsuhashi M, Konno M. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J Bacteriol. 1989;171:2882–2885. doi: 10.1128/jb.171.5.2882-2885.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ubukata K, Nonoguchi R, Song M D, Matsuhashi M, Konno M. Homology of mecA gene in methicillin-resistant Staphylococcus haemolyticus and Staphylococcus simulans to that of Staphylococcus aureus. Antimicrob Agents Chemother. 1990;34:170–172. doi: 10.1128/aac.34.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Utsui Y, Yokota T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1985;28:397–403. doi: 10.1128/aac.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittman V, Wong H C. Regulation of the penicillinase genes of Bacillus licheniformis: interaction of the pen repressor with its operators. J Bacteriol. 1988;170:3206–3212. doi: 10.1128/jb.170.7.3206-3212.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu S, Piscitelli C, de Lencastre H, Tomasz A. Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb Drug Resist. 1996;2:435–441. doi: 10.1089/mdr.1996.2.435. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y F, Curran I H A, Joris B, Ghuysen J-M, Lampen J O. Identification of BlaR, the signal transducer for β-lactamase production in Bacillus licheniformis, as a penicillin-binding protein with strong homology to the OXA-2 β-lactamase (class D) of Salmonella typhimurium. J Bacteriol. 1990;172:1137–1141. doi: 10.1128/jb.172.2.1137-1141.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]