Abstract
Purpose
To investigate the potential causal associations between the use of sun/ultraviolet (UV) protection and ease of skin tanning and the risk of pseudoexfoliation glaucoma (PXG) in European populations.
Methods
Single nucleotide polymorphisms (SNPs) associated with the use of sun/UV protection and ease of skin tanning were selected from the UK Biobank genome-wide association study database consisting of 498,751 European participants. SNPs of PXG were obtained from the FinnGen study including 3424 PXG cases and 326,434 controls. Two-sample Mendelian randomization (MR) analyses were performed to assess the association between the use of sun/UV protection and ease of skin tanning and risk of PXG.
Results
Inverse variance weighted regression of genetic susceptibility predicted that both use of sun/UV protection and ease of skin tanning were potentially positively associated with the decreased risk of PXG in the European ancestry (use of sun/UV protection: odds ratio [OR] = 0.47; 95% confidence interval [CI], 0.24-0.92; P = 0.028; ease of skin tanning: OR = 0.81; 95% CI, 0.67-0.97; P = 0.025).
Conclusions
We found genetic evidence supporting a potential causal association between UV protection and a decreased risk of PXG in European population. Further research will help elucidate the underlying mechanisms and promote UV protection for eyes, especially in people with a high risk of PXG.
Keywords: pseudoexfoliation glaucoma, Mendelian randomization, ultraviolet protection, skin tanning
Pseudoexfoliation glaucoma (PXG), recognized as a disease within the heterogeneous spectrum of glaucoma, is a common and severe subtype of open-angle glaucoma that accounts for a substantial risk of blindness. It affects all populations worldwide, with prevalence rates varying between 5% and 30% in individuals aged over 60 years.1 It is believed to be brought on by aggregation and deposition of the pathologic extracellular matrix product in pseudoexfoliation (PEX), which obstructs the aqueous humor outflow pathway, raising intraocular pressure (IOP) and causing glaucomatous optic nerve damage.2 In its early stage, the condition is referred to as pseudoexfoliation syndrome, characterized by the absence of damage to retinal ganglion cells. As it progresses into a more severe form, it is termed pseudoexfoliation glaucoma (PXG), associated with the degeneration of cells in the optic nerve head region.3 Initially, PXG is usually treated using ocular pharmaceuticals aiming to lower and control IOP. Unfortunately, after a period of treatment, most patients cease to respond, and doctors have to resort to laser therapy or surgical management.4,5 Therefore it is important and urgent to reduce the development of the disease in the early stages according to the prevention of risk factors.
Recently, exposure to ultraviolet (UV) radiation from sunlight and artificial sources has been reported to increase the risk of PXG in different ancestries.6–8 Studies have also found that UV-related cancers may be a risk factor for PXG.9–11 However, there has been no research on the relationship between the protection of UV radiation and the reduction of PXG risk, and whether these associations are causal remains unclear. Using the Mendelian randomization (MR) technique, we sought to assess the causality between the use of sun/UV protection and ease of skin tanning and the risk of PXG. Because the skin tanning reaction is considered a skin protective response from UV-related DNA photodamage, we included it in the analysis as a protective factor.12
MR is increasingly being applied to infer credible causal relationships between the risk factors and disease outcomes.13 Using genetic variants (generally single nucleotide polymorphisms [SNPs]) associated with the risk factor of interest as instrumental variables (IVs) and estimating the effect of the risk factor on the outcomes, MR serves as a useful tool for determining causation, particularly when it is impractical to perform randomized controlled trials.14 MR analyses are similar to randomized controlled trials in that they are randomized, blinded, and less susceptible to bias due to the random distribution of genetic variants during meiosis.15 They are less susceptible to bias brought by reverse causality and confounding factors that commonly hamper traditional observational studies.16
In this study, we used MR analyses to investigate the impact of sun/UV protection and ease of skin tanning on the risk of PXG in European populations. Our research not only contributes to understanding the potential protective roles of these factors but also provides valuable insights for developing preventive intervention strategies aimed at reducing the risk of PXG.
Methods
Study Design
Using genome-wide association study (GWAS) summary statistics, we conducted a two-sample Mendelian randomization analysis to investigate the causal effect of sun/UV protection and ease of skin tanning on the risk of PXG.
Exposure Data Source: Use of Sun/UV Protection and Ease of Skin Tanning
GWAS data for exposure (use of sun/UV protection, ease of skin tanning), which were adjusted for covariates including age and sex, were obtained from the UK Biobank (http://www.nealelab.is/uk-biobank/), a longitudinal population-based study exploring the effects of lifestyle and genetics on health. The dataset included over 500,000 participants aged 56.53 ± 8.09 years recruited from the UK Biobank between 2006 and 2010.17 In the latest dataset version released in July 2018, age, age2, inferred_sex, age × inferred_sex, and age2 × inferred_sex were included as covariates in the GWAS analysis model. All participants analyzed in this study were of European ancestry and had completed a touchscreen questionnaire regarding their use of sun/UV protection (sunscreen) and ease of tanning. The GWAS data for sun/UV protection included 498,790 participants, with a female ratio of 53.62%. Participants were categorized into five groups based on their sun/UV protection habits: never/rarely, sometimes, most of the time, always, and those who do not go out in the sunshine. The GWAS data for ease of skin tanning comprised 498,271 participants, with a female ratio of 53.74%. Participants were classified into four categories based on their skin tanning responses: never tan, only burn, get mildly or occasionally tanned, get moderately tanned, and get very tanned.
Outcome Data Source: Pseudoexfoliation Glaucoma
GWAS statistics of PXG, consisting of 3424 European ancestry patients and 326,434 European ancestry controls, were extracted from the eighth release of the data on PXG from the FinnGen Study, which included genotype and health registry data from 412,000 Finnish individuals from 22 research institutes in Finland as of August 2020.18 PXG cases had a female ratio of 55.20%, with a median age of 72.00 years for females and 72.75 years for males. Participants with PXG were identified based on International Classification of Diseases (ICD)-8, ICD-9, and ICD-10 diagnoses. PXG was characterized by the obstruction of aqueous humor outflow pathways because of the deposition of extracellular fibrillar aggregates, leading to elevated IOP and subsequent glaucomatous optic nerve damage.19 Other forms of glaucoma were excluded in the control group.
Selection of Genetic Instruments
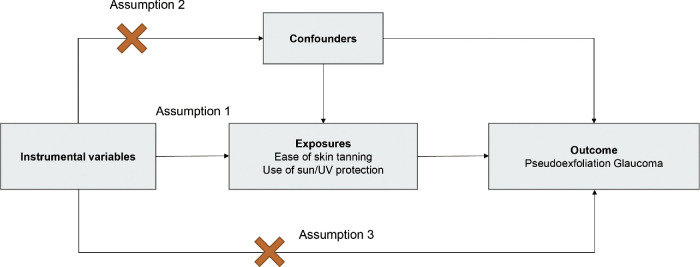
To ensure the validity of the MR analysis, we adhered to three crucial conditions: (1) the IVs must be significantly related to the exposure, (2) the IVs must be independent, and (3) the IVs should not introduce any effects on the results except through the exposure. Figure 1 depicts the general study design.
Figure 1.
Three general conditions that must be met for MR analysis. Assumption 1: This assumption reflects the causal link between IVs and the exposure of interest. Assumption 2: This assumption represents the causal connection between IVs and potential confounding factors that influence both the exposure and the outcome. Assumption 3: This assumption signifies the causal relationship between IVs and the outcome being studied.
The IVs for exposure (use of sun/UV protection, ease of skin tanning) were identified using several criteria. First, we selected SNPs associated with exposure at a genome-wide significance level (P < 5 × 10−8). Second, SNP clumping was conducted using the PLINK algorithm with stringent criteria (LD cutoff r2 < 0.001, 10,000 kb). Third, SNPs with an F-statistic < 10 were excluded from the MR analyses to avoid bias due to weak IVs.20 The F-statistic was calculated using the formula: F = R2(N − 2)/(1 − R2), where R2 is the proportion of variance in the exposures explained by the IVs, and N is the sample size. To calculate R2 for the extended 10 IVs, the following formula was used: R2 = 2 × β2 × MAF × (1 − MAF)/[2 × β2 × MAF × (1 − MAF) + 2 × N × se2 × MAF × (1 − MAF)],21,22 where β is the beta coefficient effect size, se is the standard error of β, and MAF is the minor allele frequency of the IVs. After harmonizing the IVs of the exposure and PXG to exclude palindromic and incompatible SNPs, MR analysis was performed using reserved SNPs to determine the causal effects of the use of sun/UV protection and ease of skin tanning on PXG.
Statistical Analysis
The causal effects of the use of sun/UV protection and ease of skin tanning on PXG were assessed using five MR analytical approaches to address the possible pleiotropic impacts of genetic variations. We applied an inverse variance weighted (IVW) fixed- effects estimate for the main analysis, which combined the Wald ratio of each SNP on the outcome and obtained a pooled causal estimate. Furthermore, as complements to IVW, the maximum likelihood, weighted median, and MR-Egger regression methods were used because they can deliver more reliable estimates across a larger variety of scenarios. Subsequently, we performed the following tests to determine the robustness of the MR results: (1) heterogeneity of the IVW estimations was detected by the Cochran Q test and the MR-PRESSO, and heterogeneity and invalid instruments were proved if the P value of the Cochran Q test was less than 0.05, with heterogeneity proved, MR-PRESSO was performed to eliminate outliers, and residual SNPs was reperformed to evaluate the robustness; (2) horizontal pleiotropy was test using the MR Egger intercept test. If the intercept from the MR-Egger analysis is not equal to zero (P < 0.05), there could be a potential pleiotropy; and (3) Rucker framework was used through goodness-of-fit heterogeneity statistics to decide whether the IVW or MR-Egger regression model is better.23 R 4.2.2 (https://www.R-project.org/) was used to conduct all the analyses. The R packages "TwoSampleMR" and "MRPRESSO" were both curated from the MR-Base platform (https://www.mrbase.org/) to conduct the MR analysis.
Results
Causal Association Between the Use of Sun/UV Protection and PXG
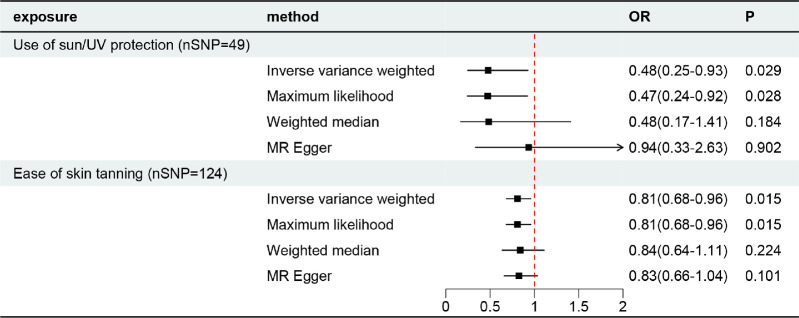
Fifty-one independent SNPs were selected as IVs for the use of sun/UV protection. After excluding palindromic and incompatible SNPs (no outliers were tested using MR-PRESSO), 49 SNPs were retained to explore the causal effect. Among the tested phenotypes, IVW analysis indicated that the use of sun/UV protection decreased the risk of PXG (odds ratio [OR] = 0.48; 95% confidence interval [CI], 0.25-0.93; P = 0.029). Maximum likelihood also showed a similar result (OR = 0.47; 95% CI, 0.24-0.92; P = 0.028). Weighted median (OR = 0.48; 95% CI, 0.17-1.41; P = 0.184) and MR Egger (OR = 0.94; 95% CI, 0.33-2.63; P = 0.902) results showed a consistent but nonsignificant direction (Figs. 2, 3). Forest plots of the MR for each SNP are provided in the Appendix.
Figure 2.
Causal effect of sun/UV protection and ease of skin tanning on PXG.
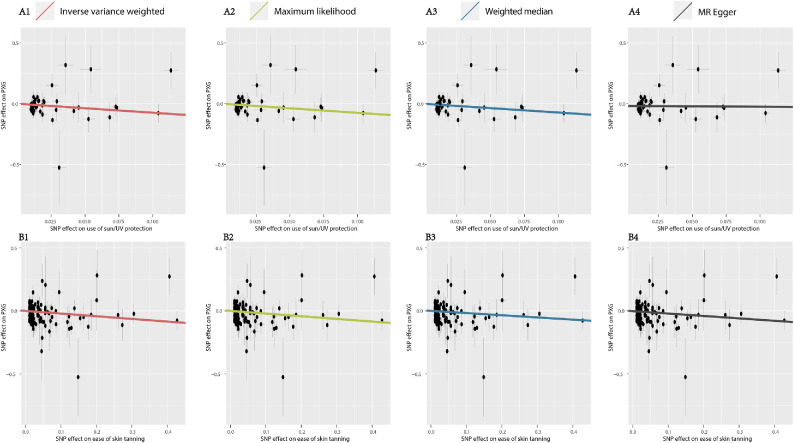
Figure 3.
Scatter plot of respective effect size of each SNP for the use of sun/UV protection (A) and ease of skin tanning (B) on PXG. (1) Inverse variance weighted (A1 & B1), a primary regression with Wald ratios estimated for each SNP, meta-analyzed under a fixed-effects model; (2) maximum likelihood (A2 & B2), which uses a maximum likelihood method where the genetic effects on the exposure and outcome are modeled directly as a bivariate normal distribution; (3) weighted median (A3 & B3), which provides robust point estimates even when up to 50% of the IVs are invalid instruments; (4) MR Egger (A4 & B4), which accounts for directional pleiotropy.
Causal Association Between the Ease of Skin Tanning and PXG
A total of 135 independent SNPs associated with the ease of skin tanning were selected as IVs. After excluding palindromic SNPs, incompatible SNPs, and outliers (rs75223371, tested by MR-PRESSO), 124 SNPs were retained to explore the causal effects. Among the tested phenotypes, IVW analysis indicated that ease of skin tanning decreased the risk of PXG (OR = 0.81; 95% CI, 0.67-0.97; P = 0.025). Maximum likelihood also showed a similar result (OR = 0.81; 95% CI, 0.68-0.96; P = 0.015). Weighted median (OR = 0.84; 95% CI, 0.64-1.11; P = 0.224) and MR Egger (OR = 0.83; 95% CI, 0.66-1.04; P = 0.101) results showed a consistent but nonsignificant direction (Figs. 2, 3).
Heterogeneity, Horizontal Pleiotropy, and Rucker Framework
The Table presents the results of heterogeneity, horizontal pleiotropy, and the Rucker framework. All P values of the Cochran Q and MR-Egger intercept tests were > 0.05, indicating the absence of heterogeneity or horizontal pleiotropy. Additionally, we employed the MR Rucker framework to determine the superiority between the IVW and MR-Egger regression models. As shown in the Table, the P values of the Rucker Framework test were > 0.05, indicating that IVW is the preferred method.
Table.
Heterogeneity, Horizontal Pleiotropy, and Rucker Framework Test of MR Analyses
| Exposure | P of Cochran Q Test | P of Intercept Test | P of Rucker Framework |
|---|---|---|---|
| Use of sun/UV protection | 0.485 | 0.101 | 0.096 |
| Ease of skin tanning | 0.091 | 0.761 | 0.739 |
Discussion
This study, based on extensive GWAS data from the UK Biobank and FinnGen Study, represents the first attempt to estimate the causal effect of sun/UV protection and ease of skin tanning on the risk of PXG using multiple MR approaches. Our findings indicate that both genetic predispositions to using sun/UV protection and having an ease of skin tanning response are associated with a decreased risk of PXG in the European population.
Previous research has established that UV exposure from sunlight and artifacts sources can contribute to the risk of various diseases through DNA damage response signaling pathways.24–26 Recently, studies have also linked UV radiation exposure to PEX and PXG.6,7,9,10,19,27,28 Pasquale et al.7 revealed that ocular exposure to light from reflective surfaces could be a significant factor in PEX development, as evidenced by correlations with labor over snow or water and a lack of correlation with wearing brimmed hats in the United States and Israel. Using conjunctival UV autofluorescence photography, Sureshkumar et al.6 demonstrated a significant association between ocular UV exposure and PXG risk. Studies involving patients with UV-associated dermatological carcinomas (basal cell and squamous cell carcinoma) in the head and neck region further support the notion that UV exposure may be a risk factor for PXG.10,11 Greater time spent outdoors, which increases UV exposure, was found to be associated with the risk of PXG or suspected PXG in young adults.6,8 However, because of the absence of GWAS data directly related to UV exposure, we cannot definitively establish a causal relationship between UV exposure and PXG in this study.
Skin tanning response to sun exposure is a process involving melanin pigmentation, which can reduce DNA damage resulting from UV radiation and restrict the damage to the upper layer of the skin.12 Ease of skin tanning represents the skin's ability to resist the damaging effects of UV radiation and serves as a photoprotective factor, similar to the use of sun/UV protection. Although we did not directly measure UV exposure, our study underscores the significance of UV protection and ease of skin tanning as potentially modifiable factors that could influence PXG risk.
The exact underlying pathways connecting UV radiation and the risk of PXG remain enigmatic, and various possible mechanisms have been proposed. One possible explanation is that UV radiation may influence the expression of nonpigmented ciliary epithelial cells (NPE) in humans through an aryl hydrocarbon receptor (AHR)-associated pathway, thereby contributing to the development of PXG. Notably, we identified SNP rs117132860 in our single SNP MR analysis (See Supplementary files SingleMR1 and SingleMR2) of both two exposures (use of sun/UV protection and ease of skin tanning). This SNP has been recognized as a functional variant within a UVB-responsive element located at chromosome band 7p21.1, and it is associated with the allelic expression of AHR.29 Importantly, the AHR-mediated pathway has been implicated in the regulation of mRNA and protein expression in NPE.30 Given that clusterin, produced by NPE, serves as an effective extracellular chaperone, its deficiency in the anterior segment can promote stress-induced aggregation and the stable deposition of pathologic extracellular matrix products—hallmarks of PXG.31 Furthermore, Zenkel et al.31 have observed an oxidative milieu in the anterior chamber of PEX eyes, potentially leading to stress-induced protein modifications and misfolding. However, whether oxidation of the UV radiation contributes to this process remains uncertain. Consequently, future investigations should explore the relationship between UV exposure and the expression of PXG-related proteins.
Another possible mechanism involves the lysyl oxidase like 1 (LOXL1) enzyme, which may serve as a key mediator linking UV radiation to an increased risk of PXG. LOXL1, encoded by the LOXL1 gene on chromosome 15q24.1, plays a crucial role in the cross-linking of collagen and elastin.32 The amalgamation of LOXL1 with the elastin matrix during elastin deposition has been proposed to contribute to increased matrix accumulation.32 Genetic variants within the LOXL1 gene can cause abnormal accumulation of these fibrillar materials, which could potentially obstruct trabecular drainage, elevate IOP, and increase the susceptibility to PXG development.33 Additionally, studies have reported downregulation of LOXL1 in the lamina cribrosa of PXG cases, leading to decreased stiffness in the lamina cribrosa and peripapillary sclera. This structural weakness renders these tissues vulnerable to IOP-induced optic nerve damage.34 Moreover, LOXL1 has been identified as a target of the autophagy pathway. Defects in LOXL1 protein folding might lead to autophagy dysfunction, subsequently facilitating the degradation of weakened exfoliation.35 Previous research has demonstrated that TGF-β1, oxidation and UV radiation can induce a significant upregulation in LOXL1 expression in human tenon fibroblasts and PEX.32,36 UVB has been associated with the increased levels of TGF-beta 1 mRNA, and UVA exposure can induce oxidative damage.37 Nevertheless, it remains uncertain whether UV radiation affects LOXL1 through direct DNA damage, TGF-β1 mediation, or oxidative stress. Two protein-coding SNPs (rs1048661 and rs3825942) in exon 1 of LOXL1 have been identified, showing an increased risk of PXG in various populations, including European (Germans and Italians), Middle Eastern, and Latin/Central American cohorts.38–40 Individuals with the high-risk haplotype (G-G) for these two SNPs exhibited an approximately 700-fold higher risk of PXG compared to those with the low-risk haplotype.41 Several noncoding variants (rs16958477, rs12914489, rs11638944, and rs7173049) have also been reported to influence LOXL1 and contribute to PXG development in conjunction with exonic variants.36,42–44 However, these SNPs were not identified among our significant IVs sourced from the UK biobank, potentially because of differences in participant ancestry. The precise mutation within LOXL1 induced by UV radiation warrant further exploration.
Within the scope of our MR analysis, a notable distinction emerged, highlighting a more robust association between the use of sun/UV protection and the risk of PXG when contrasted with the relationship involving ease of skin tanning. This distinction may stem from the direct impact of sun/UV protection on regulating the extent of UV radiation exposure to the eyes, effectively shielding ocular structures from potential damage.7 In contrast, although the degree of ease of skin tanning might signify heightened melanin production, it does not inherently govern the precise quantity of UV radiation that reaches the eyes. Extended exposure to UV radiation could still contribute to ocular damage despite the skin's adaptive response. Additionally, prolonged UV exposure can induce skin damage and alterations in melanin distribution.45,46 These alterations might, in turn, influence the complex interplay between skin tanning and the risk of PXG. The intricacies underpinning the divergent associations of UV protection and ease of skin tanning with PXG risk beckon for comprehensive exploration through dedicated research endeavors and population-based investigations.
Our MR analysis is bolstered by several notable strengths. Primarily, we harnessed the power of extensive sample sizes gleaned from GWAS databases, affording us the capability to scrutinize the causal relationship between UV protection and PXG in European populations. Furthermore, the MR approach inherently mitigates the sway of confounding factors and the potential for reverse causality, enhancing the robustness of our findings. The horizontal pleiotropy test, buttressing our outcomes, effectively dispelled concerns regarding genetic variants employed as IVs influencing PXG through pathways distinct from UV radiation's impact. Moreover, MR analysis mitigates biases, such as the systematic selection of individuals with specific traits. This approach helps counteract any tendencies towards differential UV exposure that might arise, for instance, from disparities in outdoor activities between PXG cases and controls. In particular, our utilization of the IVW analysis,47 known for its heightened statistical power compared to other MR methods, fortifies the reliability of our results, with significant outcomes emerging for the causal effect of UV protection and ease of skin tanning on PXG. This robustness is corroborated by ancillary methods probing IVW's validity, namely the Cochran Q test and Rucker framework.
However, it is essential to acknowledge the study's limitations. First, the absence of GWAS data specifically linked to UV exposure impedes a direct MR-based validation of the UV radiation-PXG relationship. Second, the absence of additional mediator analyses leaves the metabolic mechanisms underpinning the causal connection between UV exposure and PXG risk unexplored. To fully comprehend these pathways, future investigations are warranted. Third, the study's confinement to individuals of European ancestry underscores the need for more extensive exploration across diverse ethnic backgrounds, given the notable role ethnicity plays in PXG.27 Fourth, because of data access limitations, we couldn't delve into the phenotypic associations between UV protection and ease of skin tanning. Last, the unavailability of stratified gender and age-related PXG data within the publicly accessible FinnGen Study database constrained our ability to perform gender- or age-specific analyses, leading to potential confounding factors persisting in our analysis.
In conclusion, our findings lend support to the potential protective roles of sun/UV protection and ease of skin tanning against PXG. Nonetheless, our study falls short of conclusively establishing the mechanisms through which safeguarding against UV radiation damage may contribute to PXG. This limitation arises from the absence of direct investigations into potential intermediaries, such as AHR and LOXL1. Therefore further research is imperative to definitively elucidate the underlying pathways that connect UV damage and its protection with PXG. This exploration holds the promise of not only unveiling potential genetic screening tools but also spotlighting behaviors linked to UV exposure, particularly in individuals at heightened risk of PXG.
Supplementary Material
Acknowledgments
The authors thank all the participants and investigators involved in the UK Biobank and FinnGen study.
Supported by the Young Scientists Fund of the National Natural Science Foundation of China (grant no. 82201180), and Peking University Medicine Fund of Fostering Young Scholars’ Scientific & Technological Innovation (BMU2022PYB018).
Disclosure: J. Dai, None; L. Suo, None; H. Xian, None; Z. Pan, None; C. Zhang, None
References
- 1. Schlotzer-Schrehardt U, Naumann GOH.. Ocular and systemic pseudoexfoliation syndrome. Am J Ophthalmol. 2006; 141: 921–937. [DOI] [PubMed] [Google Scholar]
- 2. Ritch R, Schlotzer-Schrehardt U. Exfoliation syndrome. Surv Ophthalmol. 2001; 45: 265–315. [DOI] [PubMed] [Google Scholar]
- 3. Ritch R. Exfoliation syndrome-the most common identifiable cause of open-angle glaucoma. J Glaucoma. 1994; 3: 176–177. [PubMed] [Google Scholar]
- 4. Plateroti P, Plateroti AM, Abdolrahimzadeh S, Scuderi G.. Pseudoexfoliation syndrome and pseudoexfoliation glaucoma: a review of the literature with updates on surgical management. J Ophthalmol. 2015; 2015: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mastronikolis S, Pagkalou M, Baroutas G, Kyriakopoulou K, Makri OE, Georgakopoulos CD.. Pseudoexfoliation syndrome: the critical role of the extracellular matrix in pathogenesis and treatment. Iubmb Life. 2022; 74: 995–1002. [DOI] [PubMed] [Google Scholar]
- 6. Sureshkumar I, Gunalan V, Nareshkumar RN, et al.. Evaluating the impact of ocular UV exposure for the development for pseudoexfoliation syndrome in a South Indian population. Clin Exp Optom. 2023; 106: 734–740. [DOI] [PubMed] [Google Scholar]
- 7. Pasquale LR, Jiwani AZ, Zehavi-Dorin T, et al.. Solar exposure and residential geographic history in relation to exfoliation syndrome in the United States and Israel. JAMA Ophthalmol. 2014; 132: 1439–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kang JH, Wiggs JL, Pasquale LR.. Relation between time spent outdoors and exfoliation glaucoma or exfoliation glaucoma suspect. Am J Ophthalmol. 2014; 158: 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kang JH, VoPham T, Laden F, et al.. Cohort study of nonmelanoma skin cancer and the risk of exfoliation glaucoma. J Glaucoma. 2020; 29: 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang J, Geduldig JE, Jacobs EB, et al.. Dermatologic UV-related cancers in the head and neck region are associated with exfoliation syndrome in a New York City-based clinic population. Invest Ophthalmol Vis Sci. 2022; 63: 4. [Google Scholar]
- 11. Huang JJ, Geduldig JE, Jacobs EB, et al.. Head and neck region dermatological ultraviolet-related cancers are associated with exfoliation syndrome in a clinic-based population. Ophthalmol Glaucoma. 2022; 5: 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miyamura Y, Coelho SG, Wolber R, et al.. Regulation of human skin pigmentation and responses to ultraviolet radiation. Pigment Cell Res. 2007; 20: 2–13. [DOI] [PubMed] [Google Scholar]
- 13. Richmond RC, Smith GD.. Mendelian randomization: concepts and scope. Cold Spring Harbor Perspect Med. 2022; 12: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sekula P, Del Greco M F, Pattaro C, Kottgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. 2016; 27: 3253–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thanassoulis G, O'Donnell CJ. Mendelian randomization nature's randomized trial in the post-genome era. JAMA. 2009; 301: 2386–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burgess S, Butterworth A, Thompson SG.. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013; 37: 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sudlow C, Gallacher J, Allen N, et al.. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015; 12: e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurki MI, Karjalainen J, Palta P, et al.. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023; 613: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schlotzer-Schrehardt U, Khor CC.. Pseudoexfoliation syndrome and glaucoma: from genes to disease mechanisms. Curr Opin Ophthalmol. 2021; 32: 118–128. [DOI] [PubMed] [Google Scholar]
- 20. Burgess S, Thompson SG.. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011; 40: 755–764. [DOI] [PubMed] [Google Scholar]
- 21. Shim H, Chasman DI, Smith JD, et al.. A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. Plos One. 2015; 10: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Papadimitriou N, Dimou N, Tsilidis KK, et al.. Physical activity and risks of breast and colorectal cancer: a Mendelian randomisation analysis. Nat Commun. 2020; 11: 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brion MJ, Shakhbazov K, Visscher PM.. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013; 42: 1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sample A, He YY.. Autophagy in UV damage response. Photochem Photobiol. 2017; 93: 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Britt AB. Repair of DNA damage induced by solar UV. Photosynthesis Res. 2004; 81: 105–112. [Google Scholar]
- 26. Afaq F, Mukhtar H.. Effects of solar radiation on cutaneous detoxification pathways. J Photochem Photobiol B. 2001; 63: 61–69. [DOI] [PubMed] [Google Scholar]
- 27. Schweitzer C. Pseudoexfoliation syndrome and pseudoexfoliation glaucoma. J Fr Ophthalmol. 2018; 41: 78–90. [DOI] [PubMed] [Google Scholar]
- 28. Kozobolis VP, Detorakis ET, Sourvinos G, Pallikaris IG, Spandidos DA.. Loss of heterozygosity in pseudoexfoliation syndrome. Invest Ophthalmol Vis Sci. 1999; 40: 1255–1260. [PubMed] [Google Scholar]
- 29. Xu M, Mehl L, Zhang TW, et al.. A UVB-responsive common variant at chromosome band 7p21.1 confers tanning response and melanoma risk via regulation of the aryl hydrocarbon receptor, AHR. Am J Hum Genet. 2021; 108: 1611–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Volotinen M, Maenpaa J, Kankuri E, et al.. Expression of cytochrome P450 (CYP) enzymes in human nonpigmented ciliary epithelial cells: induction of CYP1B1 expression by TCDD. Invest Ophthalmol Vis Sci. 2009; 50: 3099–3105. [DOI] [PubMed] [Google Scholar]
- 31. Zenkel M, Kruse FE, Juenemann AG, Naumann GOH, Schloetzer-Schrehardt U. Clusterin deficiency in eyes with pseudoexfoliation syndrome may be implicated in the aggregation and deposition of pseudoexfoliative material. Invest Ophthalmol Vis Sci. 2006; 47: 1982–1990. [DOI] [PubMed] [Google Scholar]
- 32. Zenkel M, Krysta A, Pasutto F, Juenemann A, Kruse FE, Schlotzer-Schrehardt U.. Regulation of lysyl oxidase-like 1 (LOXL1) and elastin-related genes by pathogenic factors associated with pseudoexfoliation syndrome. Invest Ophthalmol Vis Sci. 2011; 52: 8488–8495. [DOI] [PubMed] [Google Scholar]
- 33. Bernstein AM, Ritch R, Wolosin JM.. LOXL1 folding in exfoliation glaucoma. In: Donev R (ed.), Protein Misfolding . London: Academic Press Ltd–Elsevier Science Ltd; 2020: 273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Braunsmann C, Hammer CM, Rheinlaender J, Kruse FE, Schaffer TE, Schlotzer-Schrehardt U. Evaluation of lamina cribrosa and peripapillary sclera stiffness in pseudoexfoliation and normal eyes by atomic force microscopy. Invest Ophthalmol Vis Sci. 2012; 53: 2960–2967. [DOI] [PubMed] [Google Scholar]
- 35. Bernstein AM, Ritch R, Wolosin JM.. Exfoliation syndrome: a disease of autophagy and LOXL1 proteopathy. J Glaucoma. 2018; 27(Suppl 1): S44–S53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berner D, Zenkel M, Pasutto F, et al.. Posttranscriptional regulation of LOXL1 expression via alternative splicing and nonsense-mediated mRNA decay as an adaptive stress response. Invest Ophthalmol Vis Sci. 2017; 58: 5922–5932. [DOI] [PubMed] [Google Scholar]
- 37. Lee HST, Kooshesh F, Sauder DN, Kondo S.. Modulation of TGF-beta 1 production from human keratinocytes by UVB. Exp Dermatol. 1997; 6: 105–110. [DOI] [PubMed] [Google Scholar]
- 38. Yaz Y, Yildirim N, Yaz YA, Cilingir O, Yuksel Z, Mutlu F.. Three single nucleotide polymorphisms of LOXL1 in a Turkish population with pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Turk J Ophthalmol. 2018; 48: 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pasutto F, Krumbiegel M, Mardin CY, et al.. Association of LOXL1 common sequence variants in German and Italian patients with pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Invest Ophthalmol Vis Sci. 2008; 49: 1459–1463. [DOI] [PubMed] [Google Scholar]
- 40. Jaimes M, Rivera-Parra D, Miranda-Duarte A, Valdes G, Zenteno JC.. Prevalence of high-risk alleles in the LOXL1 gene and its association with pseudoexfoliation syndrome and exfoliation glaucoma in a Latin American population. Ophthalm Genet. 2012; 33: 12–17. [DOI] [PubMed] [Google Scholar]
- 41. Thorleifsson G, Magnusson KP, Sulem P, et al.. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007; 317: 1397–1400. [DOI] [PubMed] [Google Scholar]
- 42. Pasutto F, Zenkel M, Hoja U, et al.. Pseudoexfoliation syndrome-associated genetic variants affect transcription factor binding and alternative splicing of LOXL1. Nat Commun. 2017; 8: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fan BJ, Pasquale LR, Rhee D, Li TS, Haines JL, Wiggs JL.. LOXL1 promoter haplotypes are associated with exfoliation syndrome in a US Caucasian population. Invest Ophthalmol Vis Sci. 2011; 52: 2372–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ferrell G, Lu MY, Stoddard P, et al.. A single nucleotide polymorphism in the promoter of the LOXL1 gene and its relationship to pelvic organ prolapse and preterm premature rupture of membranes. Reprod Sci. 2009; 16: 438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Routaboul C, Denis A, Vinche A.. Immediate pigment darkening: description, kinetic and biological function. Eur J Dermatol. 1999; 9: 95–99. [PubMed] [Google Scholar]
- 46. Moyal D, Wichrowski K, Tricaud C.. In vivo persistent pigment darkening method: a demonstration of the reproducibility of the UVA protection factors results at several testing laboratories. Photodermatol Photoimmunol Photomed. 2006; 22: 124–128. [DOI] [PubMed] [Google Scholar]
- 47. Lin ZT, Deng YQ, Pan W.. Combining the strengths of inverse-variance weighting and Egger regression in Mendelian randomization using a mixture of regressions model. Plos Genet. 2021; 17: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.