SUMMARY
Ligation of Retinoic acid receptor alpha (RARα) by RA promotes varied transcriptional programs associated with immune activation and tolerance, but genetic deletion approaches suggest impact of RARα in TCR signaling. Here we examined whether RARα would exert roles beyond transcriptional regulation. Specific deletion of the nuclear isoform of RARα revealed an RARα isoform in the cytoplasm of T cells. Extranuclear RARα was rapidly phosphorylated upon TCR stimulation and recruited to the TCR signalosome. RA interfered with extranuclear RARα signaling, causing suboptimal TCR activation while enhancing FOXP3+ regulatory T cell conversion. TCR activation induced expression of CRABP2, which translocates RA to the nucleus. Deletion of Crabp2 led to increased RA in the cytoplasm and interfered with signalosome-RARα resulting in impaired anti-pathogen immunity and suppressed autoimmune disease. Our findings underscore the significance of subcellular RA/RARα signaling in T cells and identify extranuclear RARα as a component of the TCR signalosome and a determinant of immune responses.
Keywords: Retinoic acid receptor alpha, Retinoic acid, extranuclear, T cell receptor, ZAP-70, signal transduction, proliferation, effector differentiation, FOXP3+Regulatory T cells, cellular retinoic acid binding protein, nuclear receptor, autoimmune disease, anti-pathogen immunity
eTOC blurb:
Nuclear retinoic acid receptor alpha (RARα) regulates gene transcription upon binding of retinoic acid (RA). Larange, Takazawa, Kakugawa, Thiault et al., identify an RARα isoform in the cytoplasm of T cells (cRARα), and reveal that cRARα binds the TCR-ZAP-70 complex thereby regulating T cell proliferation and effector differentiation. RA counteracts TCR-engaged RARα resulting in suboptimal TCR activation but enhanced Treg cell differentiation.
Graphical Abstract
INTRODUCTION
Retinoic acid receptor (RAR) signaling greatly impacts immune responses, ranging from protective immunity to immune tolerance, tissue homing and lymph node organogenesis(1). Defects in RAR functions weaken immunity against infections and cancers and lead to immune deregulation and inflammatory diseases(1–4).
RARs consist of three isotypes (RARα, RARβ and RARγ) encoded by separate genes, and each can give rise to several transcripts and alternative protein isoforms. Similar to other nuclear receptors, RARs exhibit a modular organization, comprising a conserved DNA-binding domain (DBD) and C-terminal ligand-binding domain (LBD), whereas the N-terminal region and F-domain vary greatly between different RAR isoforms(1).
At steady state, nuclear RARα participates in transcription factor complexes that silence target genes. Upon ligation with the vitamin A metabolite, retinoic acid (RA), and in response to post-translational modulations, RARα undergoes conformational changes of its LBD. These changes result in the release of corepressors and recruitment of coregulators that alter the activity of the transcription complex(5). RA is present in blood and tissues and as a small molecule it can freely enter cells, but requires active transportation by cellular retinoic acid binding protein 2 (CRABP2) to translocate to the nucleus(6). CRABP2 not only transports RA, but it interacts with RARα and specifically directs and facilitates RA ligation(6). This transactivates RARα bound to retinoic acid responsive elements (RARE) in regulatory regions of RA target genes and promotes transcription(6, 7).
In T cells, RA ligation of RARα activates transcription of signature genes, including Ccr9 and a4b7 that direct T cell migration to the intestine(8, 9). RA/RARα signaling greatly affects proliferation and CD4+ T helper polarization(3, 10) especially influencing the reciprocal regulation of TGF-β-driven polarization by counteracting the generation of inflammatory T helper 17 (Th17) cells while promoting the differentiation of suppressive FOXP3-expressing regulatory T (Treg) cells(11–16). Although inhibition of pro-inflammatory cytokines produced by activated bystander T cells may indirectly contribute to enhanced FOXP3+Treg cell conversion(17), RA/RARα signaling also promotes FOXP3 expression of purified naïve T cells activated in an antigen presenting cell (APC)-free system(18). Similar to interleukin (IL)-2-mediated control of Treg/Th17 cell polarization, the influence of RA/RARα on FOXP3 expression coincides with activation of the transcription factor STAT5 and suppression of STAT3-driven IL-17 expression(19–22). However, the enhancing effect of RA on FOXP3 induction persists even in the absence of IL-2, STAT5 or STAT3(23). Moreover, while the interaction of RA with RARα amplified TGFβ-dependent phosphorylated SMAD3-binding to the enhancer I in the Foxp3 locus(24), absence of SMAD3 expression does not prevent the promoting effect of RA/RARα signaling on FOXP3+ Treg cell conversion(25). All together, these observations suggest that the main mode of action by RA/RARα is not solely explained by the transcriptional control of Foxp3 expression.
TCR signal strength also influences the induction of Treg cells. Absence of or low CD28 co-stimulation or transient reduction in the phosphatidyl-inositol-3-kinase (PI3K)/AKT pathway, favors Treg cell over T effector differentiation(25, 26). Notably, conditional deletion or overexpression of RARα, impacts PI3K/AKT signaling in response to TCR stimulation and causes important downstream consequences for T cell activation, proliferation and differentiation(3, 27). These immediate effects downstream of the TCR suggest that RARα signaling during T cell activation might involve non-genomic and extranuclear actions.
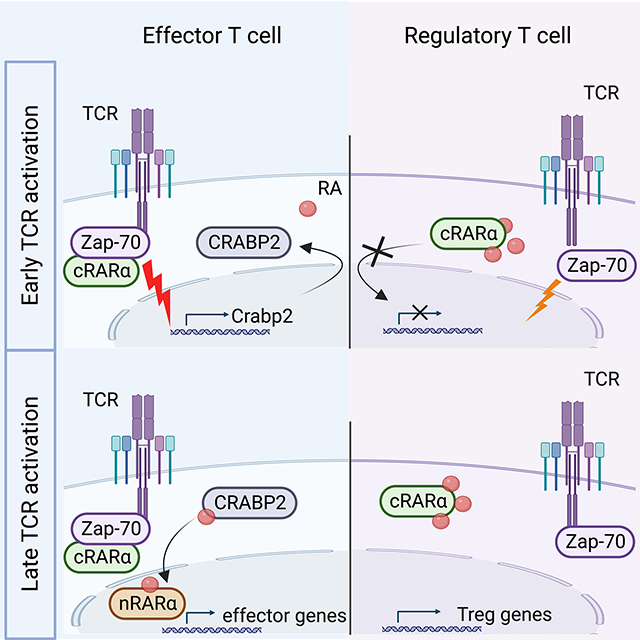
Here we examined whether RARα would exert roles beyond transcriptional regulation. Specific deletion of the nuclear isoform of RARα revealed the presence of RARα in the cytoplasm. TCR stimulation activated extranuclear RARα and promoted its recruitment to the immunological synapse (IS). Participation of extranuclear RARα in the signalosome was critical for signal transduction at the plasma membrane and to promote downstream signaling cascades that subsequently also affected nuclear RARα-controlled gene transcription. Effective extranuclear RARα signaling was also required for TCR-induced CRABP2 expression, thereby constituting a circuit that induces RA responsiveness and RARα-controlled gene transcription. In contrast to RA-mediated transactivation of nuclear RARα, RA negatively impacted non-genomic signaling by RARα in the cytoplasm, leading to sub-optimal activation, reduced proliferation and impaired effector differentiation, but enhanced FOXP3+Treg cell conversion. These data identify extranuclear RARα as a critical participant in the TCR signalosome and a determinant of T cell fate that ultimately controls the inflammatory or regulatory nature of the immune response.
RESULTS
RA enhances Treg cell conversion independently of nuclear RARα
T cells with a global deletion of Rara, which eliminates all RARα isoforms, do not enhance FOXP3 expression in the presence of exogenously added RA(3, 25). In contrast, T cells with a specific deletion of nuclear RARα1 (nRARα-deficient) isoform responded with enhanced FOXP3 expression (Figure 1A and Figure S1A). These data suggest that transcriptional control by nuclear RARα might not be the main mechanism that regulates RA-mediated enhanced FOXP3 expression. To address this possibility, we analyzed T cells that overexpressed a dominant negative (DN) isoform of RARα (RAR403CD4-Cre) that blocks the transcriptional function of RARα(28). As expected, the presence of DNRARα interfered with the transcription of Ccr9 (Figure S1B). In contrast, it did not affect the RA-mediated increase in TGF-β-induced FOXP3 expression in naïve T cells activated in vitro in an APC-free system (Figure 1B). Notably, when the addition of RA was delayed by 24 hours, RA no longer enhanced FOXP3 expression (Figure 1C). Taken together the results suggest that RA targets an RARα-function during early TCR stimulation independently of transactivation of nuclear RARs.
FIGURE 1: RA enhances Treg cell conversion independently of nuclear RARα. (Related data shown in Figure S1).
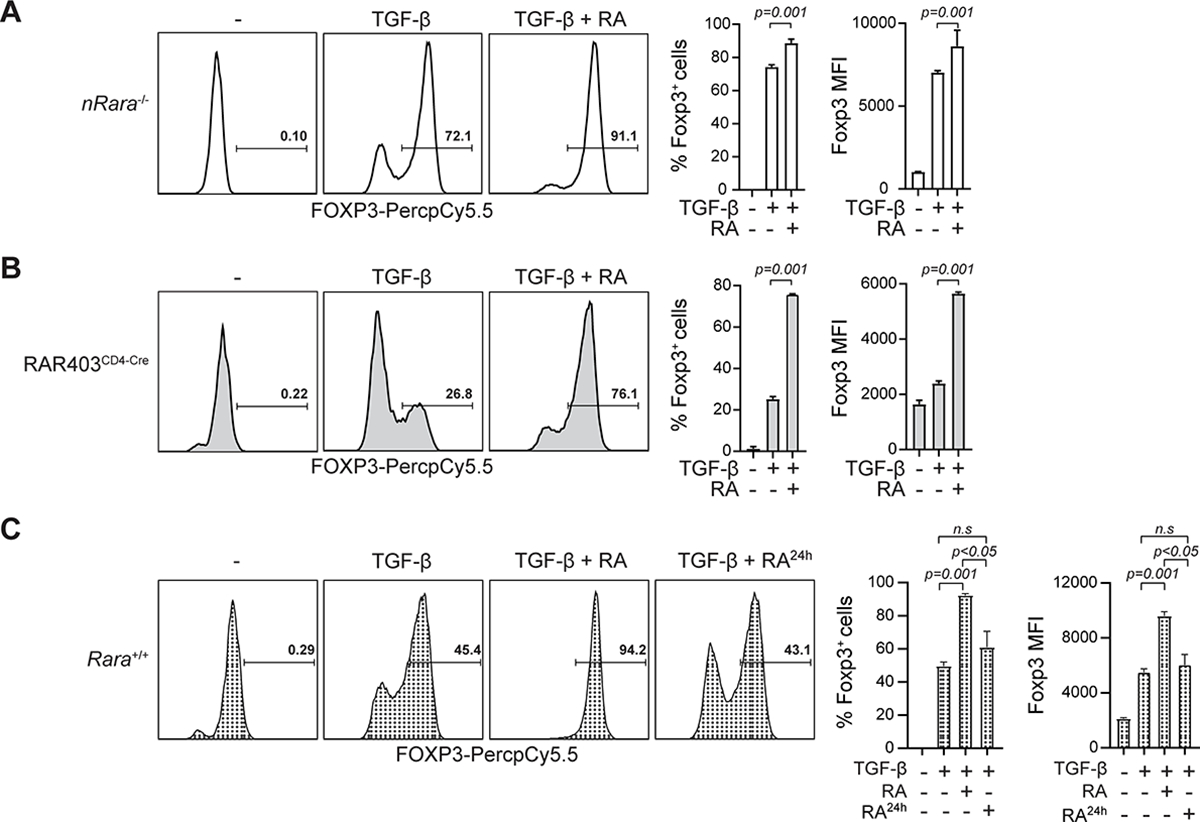
(A-B) FOXP3 intracellular staining of (A) nRARα-deficient or (B) DNRARα (RAR403CD4-Cre) spleen CD4 T cells stimulated for 96hrs with α-CD3/CD28 with or without TGF-β or RA (10 nM). (C) Naïve WT spleen CD4 T cells stimulated with α-CD3/CD28 with or without TGF-β. After 24hrs RA (10 nM) was added or not and cells were cultured for another 72hrs and stained for intracellular FOXP3. Shown representative histograms of 3 independent experiments (left) and statistical analyses of FOXP3+ T cell frequency and mean fluorescence intensity (MFI) (right). Statistical significance calculated with two-tailed Student t-test. Shown mean +/− SEM.
T cells express an extranuclear isoform of RARα
To investigate physical participation of RARα in early T cell activation, we used confocal imaging of ovalbumin peptide (OVAp) (SIINFEKL)-responsive OT-I TCR transgenic CD8αβ T cells activated in vitro with OVAp-loaded DCs and stained with RARα-specific antibodies together with anti-CD3ζ and anti-ZAP70 antibodies. Consistent with an extranuclear presence, RARα was detected at the IS in close proximity to CD3ε and ZAP-70 (Figure 2A). Colocalization of RARα and CD3ε alone, was not observed, suggesting an indirect link between RARα and the TCR itself. FLAG-tagged constructs for nuclear Rara1 and Rara2 transduced in primary T cells that were activated with anti-CD3/CD28 and subsequently analyzed by western blotting, showed that RARα1 and -α2 remained in the nucleus upon TCR stimulation (Figure 2B), and thus that there must be another Rara gene expressed in T cells, which encodes this cytoplasmic molecule. High-throughput sequencing and analysis of double positive (DP) thymocyte and CD4 and CD8αβ spleen T cell mRNA (RNA-seq) revealed that in addition to Rara1 and -2 transcripts, there were other Rara sequences with different 5’ splice junctions (Figure 2C). Three transcripts (J3, J4 and J5) aligned with predicted sequences in the RefSeq database (www.ncbi.nlm.nih.gov/refseq)(29). Rara transcripts that predominate in thymocytes were distinct from those in mature T cells (Figure S2A). All alternative transcript shared the same 3’ end sequence with Rara1 and -2 but each one had a unique 5’end sequence (Figure 2D), indicating a transcriptional start site (TSS) distinct from that of Rara1 or -2 (Figure S2B). This was supported by data from the Functional ANnoTation Of the Mammalian genome (FANTOM)5 database(30), which indicated a TSS downstream of the Rara2 TSS and upstream of the 5’ end of the variant transcripts (Figure 2E). Complete sequence analysis specified a GC-rich 5’ end with one or two variable non-translated exons (Figure S2C) and conserved exons 9 to 15 shared with Rara1 and -2 (Figure 2E). All transcript variants encoded the same protein with a putative AUG translational start in exon 10 (Figures 2E and S2D). The alternative RARα differed from nuclear RARα by truncation with a predicted molecular weight of approximately 38kDa. There were no typical transcriptional features like a functional DNA binding domain (DBD) or nuclear translocation sequences (NLS). A complete ligand binding domain (LBD) and an intact F-domain however, were present suggesting that, similar to nuclear RARα, this isoform potentially interacts with binding partners to form dynamic active (open) and inactive (closed) protein complexes. Gfp-tagged constructs of alternative Rara, referred to as Rara3 (RARα3-eGFP) and Rara1 (RARα1-eGFP) as control were transduced into mouse MCC-T CD4 hybridoma T cells(31) and analyzed by imaging for protein expression. As expected GFP-tagged RARα1 localized almost exclusively to the nucleus (Figure 2F). In contrast, a major fraction of GFP-tagged RARα3 was detected in the cytoplasm (Figure 2F), indicating that Rara3 encodes an RARα isoform that can reside outside the nucleus. Analysis of the cytoplasmic and nuclear fractions of human T cells confirmed RARα in the cytoplasm of a similar size as mouse extranuclear RARα (Figure 2G and 2H). Overall, the data indicate that T cells express a splice isoform of RARα that is expressed in the cytoplasm.
FIGURE 2: T cells express an extranuclear isoform of RARα. (Related data shown in Figure S2).
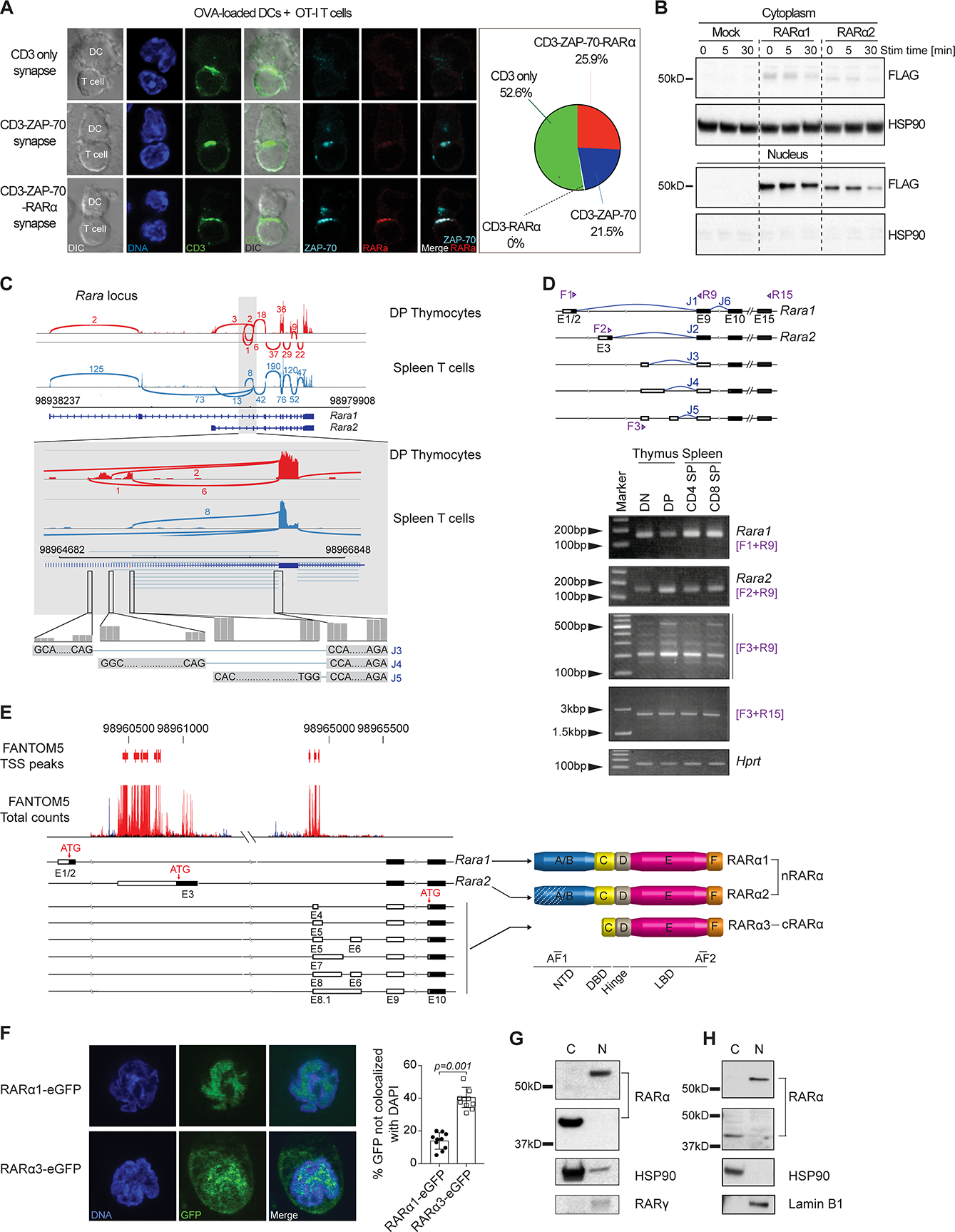
(A) CD3, ZAP-70 and RARα staining of OT-I T cells co-cultured with OVA-loaded DCs for 40min. DAPI marks DNA. Representative cells are shown. Frequencies of synapse types from 1 representative experiment out of 2. (B) Naive spleen T cells transduced with FLAG-tagged RARα1 or RARα2 activated with α-CD3/CD28 for 5 or 30min. Localization of FLAG-tagged RARα1 and RARα2 assessed by western blotting. HSP90 as control. Data are representative of 3 experiments. (C) Mouse DP thymocytes and spleen T cells RNA-seq data aligned on UCSC mm10. Rara-mapped short-reads around exon 9 (gray background) shown. Numbers indicate short-reads of exons. J3, J4 and J5 are new splice junctions. (D) Junction usage with exon 9 in common. Forward (F) and reverse (R) primers indicated. RT-PCR of DN and DP thymocytes and CD4 and CD8 T cells with primers as shown. F1+R9 and F2+R9 primers detect Rara1 and -2. F3 primer is of 5’ end of exon E6 of XM_006532597.2. (E) Transcription start site(s) (TSS) of new Rara transcripts using FANTOM5 database. Forward and reverse transcription direction marked by red and blue peaks. New exon sequences shown. New RARα protein sequences with internal ATG translational starts. Domains in RARα1, -α2 and -α3 are shown. (F) CD4 T hybridoma cells transduced with RARα1-GFP or RARα3-GFP analyzed by confocal microscopy and localization of GFP assessed. Images show 1 focal plane of representative cells. Graph represents mean +/− SEM from 150 or more cells from 1 representative experiment out of 2 (each dot represents 15–20 cells). (G-H) RARα expression in cytoplasm (C) and nucleus (N) of human (Jurkat) (G) and primary mouse T cells (H). Expression of RARγ, Lamin B1 and HSP90 as control for N or C fractions, respectively. Data are representative from 2 (h) and 3 (m) independent experiments.
Extranuclear RARα controls TCR signal transduction
Primary T cells transduced with either Rara1 or Rara3 constructs were stimulated in vitro with anti-CD3+CD28 and proximal TCR signaling events were assessed. T cells, which overexpressed RARα3 but not RARα1, displayed markedly enhanced phosphorylation of ZAP-70 and phospholipase C gamma-1 (PLCγ1) relative to the mock control (Figures 3A and 3B). Notably, in vitro stimulation of OT-I TCR transgenic T cells with OVAp-loaded DCs followed by immunoprecipitation with RARα-specific antibody and western blotting for ZAP-70, indicated that RARα antibody precipitated ZAP-70 from lysates of activated but not resting OT-I T cells (Figure 3C). These data showed that extranuclear RARα influenced TCR/ZAP-70 activation and moreover that activated ZAP-70 directly or indirectly interacted with RARα.
FIGURE 3: Extranuclear RARα controls TCR signal transduction. (Related data shown in Figure S3).
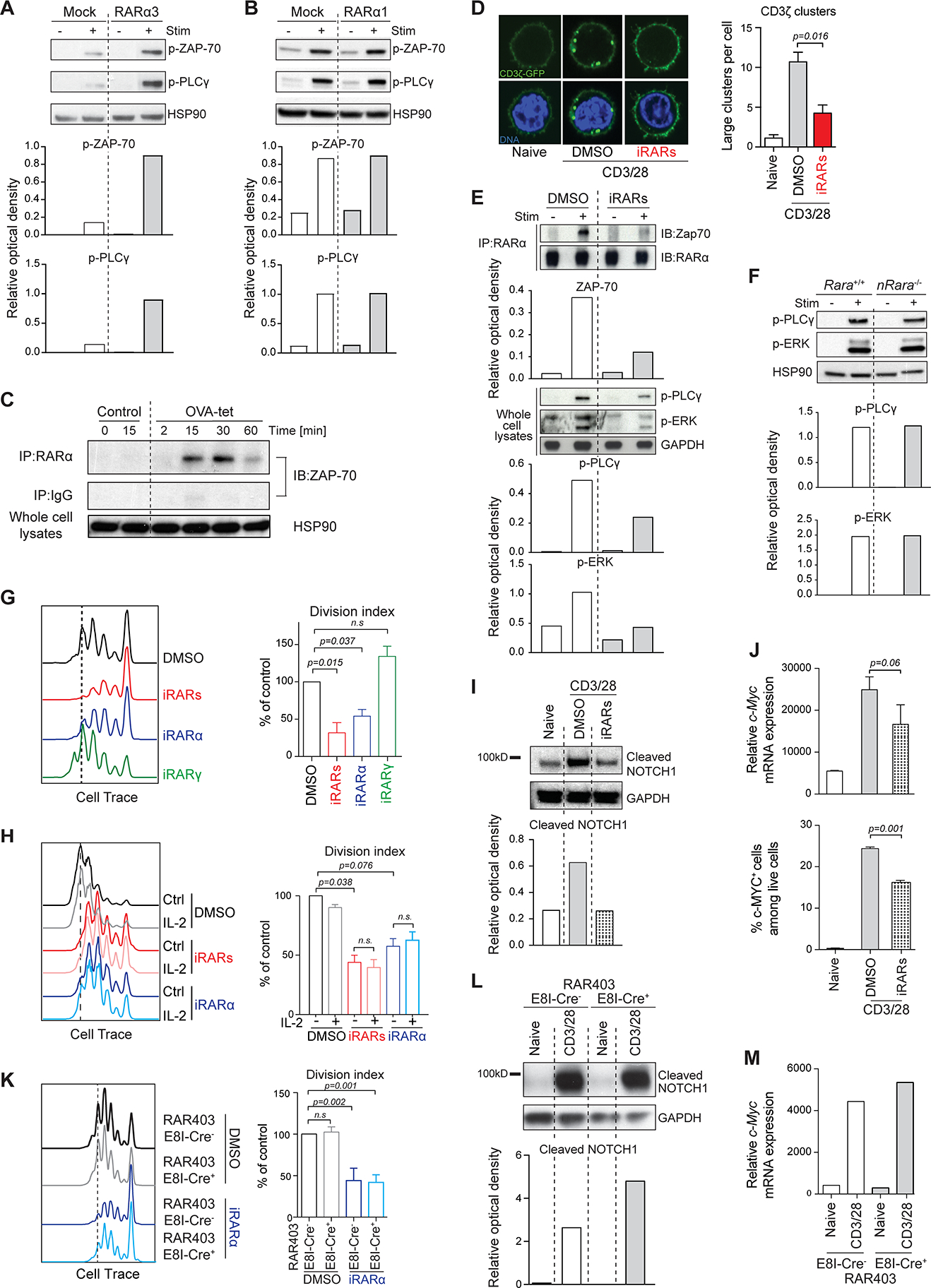
(A-B) Primary mouse spleen T cells transduced with (A) Rara3- or (B) Rara1 cDNA, were activated with α-CD3/CD28 for 5min. Phosphorylation of ZAP-70 and PLCγ assessed by western blotting. HSP90 used as control. Representative of 2 experiments. (C) OT-I CD8 T cells stimulated with H2Kb-OVA tetramers. Cell-lysates immunoprecipitated with α-RARα. Precipitates were assessed for ZAP-70 by western blotting. HSP90 in whole cell lysates as control. Shown 1 representative out of 3 experiments. (D) Spleen T cells transduced with CD3ζ-GFP activated with α-CD3/CD28 for 30min. CD3ζ-GFP clustering was quantified. Representative cells shown. Graph is mean +/− SEM from > 150 cells from 3 experiments. A large cluster was empirically defined. (E) OT-I T cells stimulated with H2Kb-OVA tetramers with or without RARα inhibitor (Ro41–5253). Cell-lysates were immunoprecipitated with α-RARα and blotted for ZAP-70 or RARα. (F) WT or RARα1-deficient spleen T cells activated with α-CD3/CD28 for 5min. Phosphorylation of PLCγ and ERK was assessed. HSP90 as control. Shown representative western blot out of 3. (G-H) Cell trace-labeled spleen T cells treated with DMSO, a pan-RAR antagonist LE540 (iRARs), an RARα-specific antagonist Ro 41–5253 (iRARα) or an RARγ antagonist MM11253 (iRARγ), activated with α-CD3/CD28 without (G) or with rIL-2 (H). Proliferation assessed after 72hrs. Shown representative data. Graph means +/− SEM of division index calculated from 3 experiments expressed as percentage of control. (I) Primary spleen T cells treated with DMSO or RAR antagonist were activated with α-CD3/CD28 for 4hrs. Cleaved NOTCH1-intracellular domain assessed by western blotting. Shown 1 representative out of 3. (J) Primary spleen T cells with DMSO or inhibitor activated with α-CD3/CD28 for 4hrs and analyzed for c-Myc mRNA (upper panel) and c-MYC protein (lower panel). Graph represents means +/− SEM from 3 experiments. (K) Cell trace-labeled dnRara cre− or cre+ T cells with DMSO or iRARα and activated with α-CD3/CD28. Proliferation assessed after 72hrs. Shown representative experiment. Graph represents means +/− SEM of division index from 3 experiments expressed as a percentage of control. (L) dnRara cre− or cre+ spleen T cells activated with α-CD3/CD28 for 4hrs. Cleaved NOTCH1 was assessed. Shown 1 representative western blot out of 3. (M) dnRara cre− or cre+ spleen T cells activated with α-CD3/CD28 for 4hrs. c-Myc mRNA assessed. Graph shows means +/− SEM from 2 experiments. Where appropriate band intensities were quantified by Relative Optical Density (ROD) relative to control.
TCR downstream signal transduction depends on cytoskeletal remodeling and micro-domain scaffolds that recruit signaling molecules including kinases and adaptors(32–35). To further assess the influence of RARα on the TCR-ZAP-70 activation complex, we added RARα inhibitors to CD3ζ-GFP transduced primary T cells stimulated in vitro with anti-CD3+CD28. In addition to the reduced Ccr9 gene transcription (Figure S3A), RARα inhibitor also reduced CD3ζ-GFP clustering efficiency at the plasma membrane (Figure 3D) and notably disrupted the RARα/ZAP-70 activation complex resulting in weakened TCR signal transduction (Figure 3E). Conversely, specific deletion of nuclear RARα1, which eliminated Ccr9 transcription (Figure S1), had no effect on early TCR activation (Figure 3F). Moreover, transient blocking with specific RARα-, but not RARγ-inhibitor, reduced T cell proliferation (Figure 3G), and addition of IL-2 did not restore this defect (Figure 3H). IL-2-independent proliferation of activated T cells is mediated by TCR-induced activation of the NOTCH/cMYC pathway(34, 36). Accordingly, inhibition of RARα signaling decreased TCR-dependent cleavage of NOTCH (Figure 3I), to a similar degree as γ-secretase inhibitor (Figure S3B) and comprised upregulation of c-Myc mRNA and c-MYC protein expression (Figure 3J and Figure S3C). Overexpression of DNRARα did not affect TCR-driven proliferation, which was only blocked when also the inhibitor was added (Figure 3K). Also, overexpression of DNRARα affected neither TCR-mediated NOTCH activation (Figure 3L) nor upregulation of cMyc expression (Figure 3M).
All together, these results demonstrate that RARα participates in TCR signal transduction, independently of its classical genomic function as a transcription factor.
Extranuclear and nuclear RARα are activated by distinct mechanisms
RARs are phosphoproteins that alter their affinity for interacting binding partners through conformational changes induced by phosphorylation of specific sites(37). To assess phosphorylation of RARα following TCR stimulation, we performed a large-scale quantitative phosphoproteomic analysis of Jurkat E6.1 T cells activated short term with anti-CD3 and anti-CD28 antibodies. Mass spectrometric analysis of whole lysates identified TCR-induced phosphorylation of a serine residue (S445) located within the F-domain of RARα (Figure 4A). Phosphorylation of RARα S445 (pS445) immediately increased roughly four-fold upon TCR stimulation but returned to basal levels after 10 minutes and continued to decline for some time after the initial stimulation (Figure 4B). Serine 445 is followed by a proline, indicating a proline directed serine kinase is likely responsible for the TCR-induced phosphorylation of the F domain (38). Thus, TCR stimulation leads to rapid phosphorylation of RARα. Moreover, the fast kinetics of this activation process underscore the proximity and participation of RARα in the TCR-ZAP-70 signalosome.
FIGURE 4: Extranuclear and nuclear RARα are activated by distinct mechanisms.
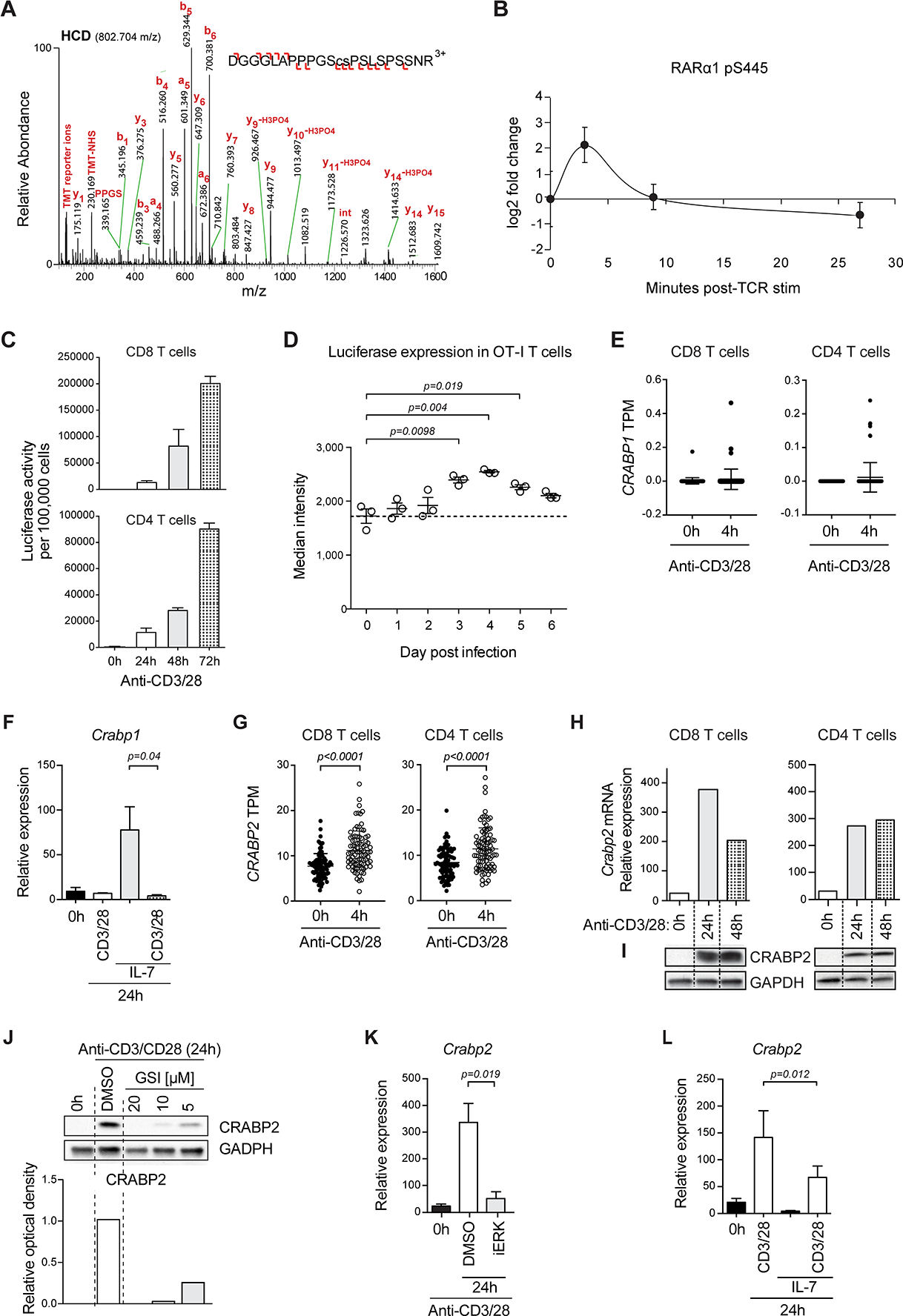
(A) Higher-energy C-trap dissociation (HCD) MS2 mass spectrum identifying TCR-induced phosphorylation of RARα at S445. Sequence of a 22 amino acid F domain peptide with phosphorylated serine is in lower case. Difference between measured and theoretical m/z expressed as parts per million (ppm). Product ions leading to the peptide sequence are shown, with b- (N-terminal) and y-series (C-terminal) ions indicated with left and right facing signs. (B) Phosphorylation dynamics of RARα pS445 in the first 30min of T cell stimulation, relative to unstimulated, determined by tandem mass tag (TMT)-based quantification. Error bars show standard deviation for double or triple biological replicates. (C) RARE-Luc CD8 or CD4 T cells activated with α-CD3/CD28 for 24hrs, 48hrs and 72hrs. Luciferase activity was measured. The means +/− SEM of 2 experiments are shown. (D) Three OT-I TCR transgenic RARE-Luc mice infected i.v. with ActA− Lm-OVA/day analyzed until 6 days post-infection for luciferase expression in spleen cells. Median fluorescence intensity of luciferase expression per mouse is shown. Data are from 1 experiment out of 2. (E) RNA-Seq data from human naïve CD8 or CD4 (CD3+ CD45RA+ CD127+ CCR7+) peripheral blood T cells stimulated with α-CD3/CD28 for 4hrs analyzed for CRABP1 expression. CRABP1 mRNA in transcripts per million (TPM) for each donor (n=88). P value calculated by Mann-Whitney U test. Datasets are from Database of Immune Cell Expression, expression quantitative trait loci (eQTLs) and epigenomics (DICE); https://dice-database.org/. (F) Mouse spleen CD8 T cells activated with α-CD3/CD28 or IL-7 or both. Crabp1 mRNA was quantified. Data are means +/− SEM of 4 experiments. (G) Expression of CRABP2 mRNA in activated human T cells assessed as in (E). (H-I) CD8 or CD4 mouse spleen T cells activated with α-CD3/CD28 and Crabp2 mRNA (H) and protein (I) were evaluated. Relative expression (mRNA) or western blot (Protein) of 1 of 3 experiments. (J) Expression of CRABP2 in CD4 mouse spleen T cells activated for 24hrs with α-CD3/CD28 alone or together with various doses of γ-secretase inhibitor (GSI). Quantification of the optical band density relative to control HSP90. Representative western blot of two independent experiments. (K) Spleen T cells activated with α-CD3/CD28 for 24hrs in DMSO or with MEK/ERK inhibitor U0126 (iERK). Crabp2 mRNA was evaluated. Means +/− SEM of Crabp2 mRNA are shown of 3 experiments. (L) Mouse spleen T cells activated with α-CD3/CD28 or IL-7 or both. mRNA of Crabp2 was quantified. Data are means +/− SEM of 4 experiments.
Unlike modulation of RARα in response to TCR stimulation, transactivation of nuclear RARs is a direct consequence of RA ligation(1). T cell activation also leads to transcriptional activation of nuclear RARα(3, 10). Accordingly, CD8αβ- and CD4 T cells from transgenic mice that expressed a luciferase transgene (Luc) transcriptionally controlled by a retinoic acid responsive element (RARE)(39), showed a gradual increase of RAR-controlled luciferase expression when activated with anti-CD3/CD28 in an APC-free system (Figure 4C). Also, spleen T cells from OT-I/RARE-Luc double transgenic mice induced increasing levels of luciferase after infection with OVA antigen-expressing Listeria monocytogenes (Lm-OVA) (Figure 4D). In contrast to the rapid activation of RARα in the TCR/ZAP-70 complex, RA-dependent transactivation of nuclear RARα was a late event measurable after 24 hours and peaked several days later (Figure 4C and 4D). RA is present at low physiological concentrations (<1nM) in blood and in most tissues and freely enters the cytoplasmic space of cells. Subcellular distribution and local concentration of RA are regulated by cellular retinoic acid binding proteins (CRABPs). CRABP1 targets RA for degradation via the CYP26 pathway(6, 40), whereas binding of RA to CRABP2 induces translocation of the CRABP2/RA complex to the nucleus and results in transactivation of nuclear RARs and RAR-controlled gene transcription(41, 42). Analysis of naïve or anti-CD3+CD28 stimulated T cells showed no CRABP1 expression (Figure 4E and 4F), but Crabp1 mRNA was rapidly induced when triggered by IL-7 cytokine (Figure 4F). Expression of Crabp2 mRNA was also insignificant at steady state but strongly induced by TCR stimulation (Figure 4G–4I), whereas blocking of proximal TCR signaling at the plasma membrane with γ-secretase inhibitor (Figure 4J) or further downstream with ERK inhibitor (Figure 4K), interfered with CRABP2 induction. Both Crabp2 mRNA and CRABP2 protein (Figure 4H and 4I), remained highly expressed during later stages of activation indicating that activation of the TCR promotes constant translocation of RA from the cytoplasm to the nucleus. TCR signals counteracted RA degradation by suppressing IL-7-induced CRABP1 expression (Figure 4F) whereas addition of IL-7 suppressed TCR-induced CRABP2 expression (Figure 4L). Altogether, the dose and subcellular location of RA is highly regulated in T cells. Furthermore, TCR activation induces CRABP2 expression, which removes RA from the cytoplasm and facilitates ligation of RA with nuclear RARα.
RA counteracts extranuclear RARα signaling in T cells
T cells with a genetic deletion for Crabp2 failed to efficiently induce RA responsiveness, as indicated by reduced Ccr9 expression when activated in the presence of a pharmacological dose (10 nM) of RA (Figure 5A and S4A). Also, RARE-Luc T cells on a Crabp2-deficient background showed inefficient luciferase gene transcription when stimulated with anti-CD3 and -CD28 in regular culture medium containing a physiological dose of RA (Figure 5B). TCRγδ and TCRαβ thymocytes and Foxp3+ natural Treg cell develop normally in CRABP2-deficient mice (Figure S4B), indicating that CRABP2 function influences mature T cells and enables RA-responsiveness associated with TCRαβ stimulation. Accordingly, blocking of proximal TCR signaling diminished CRABP2/RA-dependent luciferase gene transcription (Figure 5C). Notably, RARα inhibitors interfered with TCR-induced CRABP2 expression (Figure 5D), thereby linking the TCR-mediated activation of extranuclear RARα with RA-responsiveness and the RA-dependent transactivation of nuclear RARα.
FIGURE 5: RA counteracts extranuclear RARα signaling in T cells. (Related data shown in Figure S4).
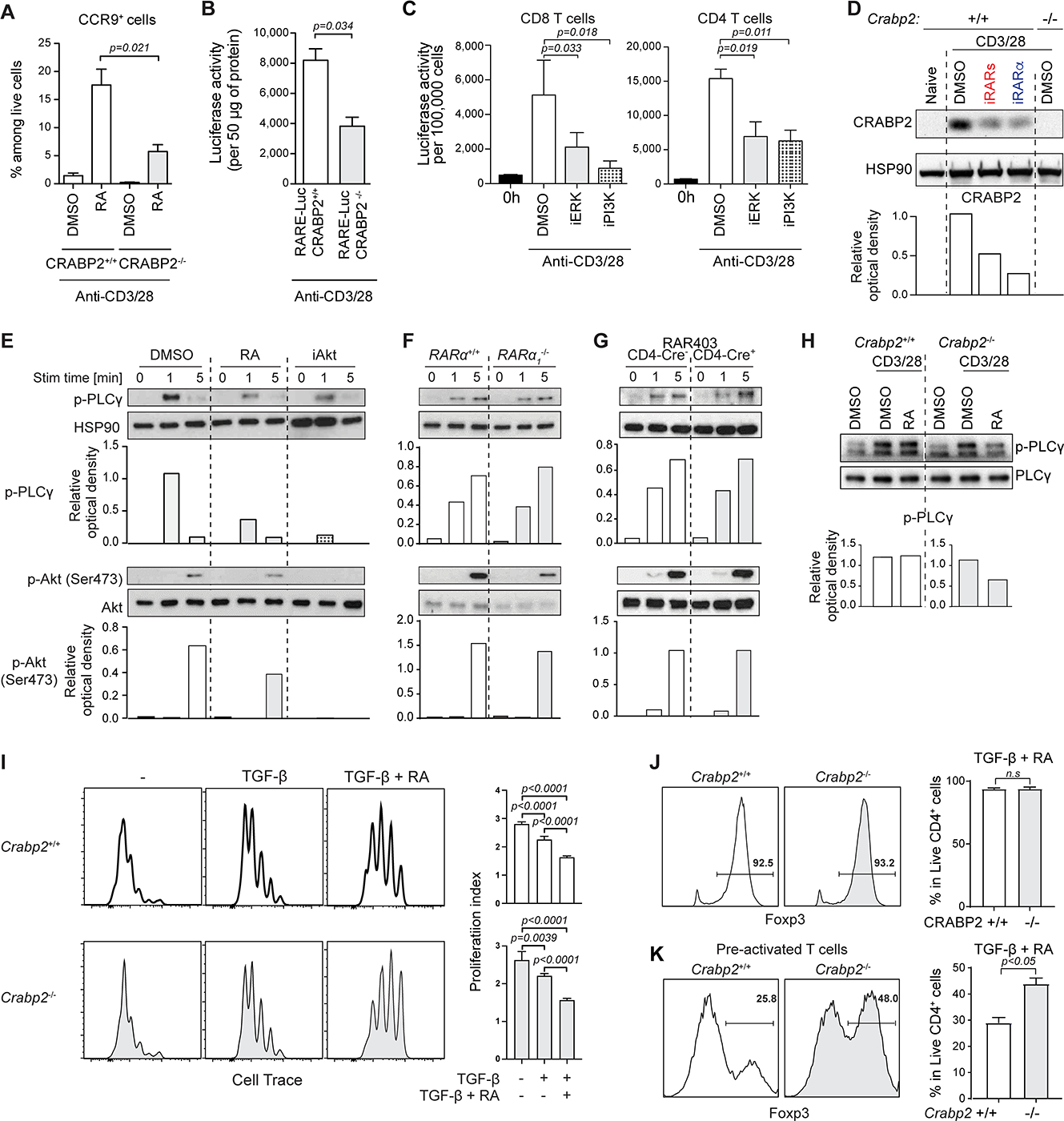
(A) WT or CRABP2-deficient spleen T cells activated with α-CD3/CD28 and RA for 72hrs and assessed for CCR9 expression. Graphs show means +/− SEM of CCR9+ T cells from 3 experiments. (B) WT- or CRABP2-deficient-RARE-Luc spleen T cells activated with α-CD3/CD28 for 48hrs and assessed for luciferase activity. Shown are means +/− SEM of luciferase activity per 50μg of protein for 3 experiments. (C) RARE-Luc CD8 (left) or CD4 (right) T cells activated with α-CD3/CD28 for 24hrs in DMSO, or a MEK/ERK inhibitor U0126 (iERK) or a PI3K inhibitor LY294002 (iPI3K). Luciferase activity was measured. Data are shown as means +/− SEM of 3 experiments. (D) WT or CRABP2-deficient spleen T cells activated with α-CD3/CD28 for 24hrs in DMSO and for WT T cells also with pan-RAR antagonist (iRARs) or RARα antagonist (iRARα). CRABP2 expression is evaluated. Representative western blot of 2 independent experiments. (E) Naïve CD4 T cells in DMSO, Retinoic Acid (RA) or an AKT inhibitor (AKT Inhibitor VIII) and activated with α-CD3/CD28 for indicated times. Phosphorylation of PLCγ and AKT was assessed. (F-G) Naïve CD4 T cells from nRARα-deficient (F) or DNRARα mice (G), activated with α-CD3/CD28 for indicated times were assessed for phosphorylation of PLCγ and AKT. (H) WT or CRABP2-deficient spleen T cells activated with α-CD3/CD28 for 72hrs were sorted and re-activated for 1min in DMSO or DMSO + (10nM) RA and PLCγ phosphorylation evaluated. Total PLCγ was measured as control. Data in (E-H) are representative from 3 experiments. (I) Proliferation of cell trace-labeled WT or CRABP2-deficient CD4 T cells activated with α-CD3/CD28 alone or with TGF-β or TGF-β + RA for 72hrs was evaluated. Histograms show a representative experiment and graphs show means +/− SEM of the division index from 3 experiments, expressed as a percentage of control. (J) Naïve WT or CRABP2-deficient CD4 T cells activated with α-CD3/CD28 alone or with TGF-β or TGF-β + RA for 72hrs were assessed for FOXP3. Frequency of FOXP3+ cells is shown. A representative histogram and_means +/− SEM of 3 experiments are shown. (K) Naïve WT or CRABP2-deficient CD4 T cells activated with α-CD3/CD28 alone or with TGF-β or TGF-β + RA for 72hrs after 24hrs pre-activation without TGF-β + RA. FOXP3 was assessed and frequency of FOXP3+ cells assessed. A representative histogram and means +/− SEM of 3 experiments are shown. Student t-test was used for the statistical analysis. Where appropriate band intensity was quantified by ROD relative to control.
To examine if RA also influences RARα in the signalosome, we analyzed TCR signal transduction events in WT T cells activated in vitro with anti-CD3 and -CD28 together with a pharmacological dose (10nM) of RA. Contrary to its promoting effect on gene transcription, RA interfered with signaling events at the plasma membrane, including phosphorylation of phospholipase C gamma-1 (PLCγ1) and AKT S473 (Figure 5E). Conversely, the absence or impaired transactivation of nuclear RARα in Rara1-deletion mutant (Figure 5F) or dnRara T cells (Figure 5G), respectively, had no measurable effect on proximal TCR/CD28 signaling. Addition of RA after 24 hours of stimulation, when CRABP2 was induced, no longer interfered with proximal signaling (Figure 5H), indicating that by translocating RA to the nucleus, CRABP2 eliminated the inhibitory effect of RA on early TCR signaling. Accordingly, in CRABP2-deficient T cells, RA added after 24 hours of stimulation continued to interfere with TCR signal transduction (Figure 5H).
Sub-optimal TCR activation promotes Treg cell conversion(25, 26). It is thus conceivable that the mechanism that drives the RA-enhanced FOXP3 Treg cell differentiation of naïve T cells might be due to the negative influence of RA on proximal TCR signaling. In agreement with this, RA-enhanced Treg cell conversion coincided with decreased activation-induced proliferation (Figure 5J). This was even more pronounced when RA accumulated in the cytoplasm of activated CRABP2-deficient T cells (Figure 5I). Notably, with inefficient nuclear translocation and impaired transactivation of nuclear RARα, RA still enhanced Treg cell conversion of CRABP2-deficient T cells (Figure 5J), even when added 24 hours after the initial stimulation (Figure 5K). Overall, these results indicated that the absence of CRABP2 expression or a large dose of RA interfere with the function of RARα in the TCR signalosome resulting in suboptimal activation and reduced proliferation but enhanced Treg cell conversion in vitro.
Extranuclear RARα signaling controls TCR-induced proliferation in vivo
To examine the significance of extranuclear RARα signaling and the impact of RA interference during T cell responses in vivo, we compared the response of CRABP2-deficient and WT T cells in a pathogen infection. Naïve OT-I TCR transgenic T cells on a CRABP2-deficient background and WT OT-I T cells were co-transferred into C57BL/6 recipient mice that were subsequently infected with Lm-OVA (Figure 6A). When analyzed 7 days later, the frequency of CRABP2-deficient OT-I T cells was much reduced compared to WT OT-I cells (Figure 6B), indicating a cell-intrinsic defect consistent with impaired TCR-driven proliferation and or less survival. To test if the activation defect was due to the accumulation of RA in the cytoplasm in the absence of TCR-induced CRABP2 expression, we co-transferred WT and CRABP2-deficient naïve OT-I T cells into vitamin A-depleted (Vit A−) recipient animals, which had not received vitamin A since before birth. Vit A− recipient mice were subsequently infected with Lm-OVA and analyzed 7 days later. As expected, without RA, homing of WT OT-I T cells to the intestinal epithelium was impaired (Figure 6C). Conversely, in the absence of RA, Lm-OVA responding CRABP2-deficient OT-I T cells in the spleen expanded vigorously, even to a greater extent than WT OT-I cells (Figure 6D). The frequency of recipient-derived OVA-reactive CD8 T cells did not differ between Vitamin A-sufficient or -depleted conditions (Figure S5), indicating that the reversed frequency ratio of donor WT and CRABP2-deficient OT-I T cells in VitA− mice was not due to excessive cell death of the WT cells in the absence of RA. Re-supplementing RA restored intestinal homing of the OT-I T cells (Figure 6C) and prevented again the relative increase in number of Lm-OVA responding CRABP2-deficient OT-I T cells compared to WT OT-I cells (Figure 6E). The results underscored the significance of activation induced CRABP2 to prevent accumulation of RA in the cytoplasm and interference of RA with RARα signaling in the TCR signalosome. The impaired proliferation and/or survival of the Lm-responding CRABP2-deficient T cells is in contrast to the vigorous expansion of Lm-responding dnRara T cells(43) and underscores the significance of participation of extranuclear RARα in the TCR signalosome as a critical determinant of the magnitude of the T cell response in vivo.
Figure 6: Extranuclear RARα signaling controls TCR-induced proliferation in vivo. (Related data shown in Figure S5).
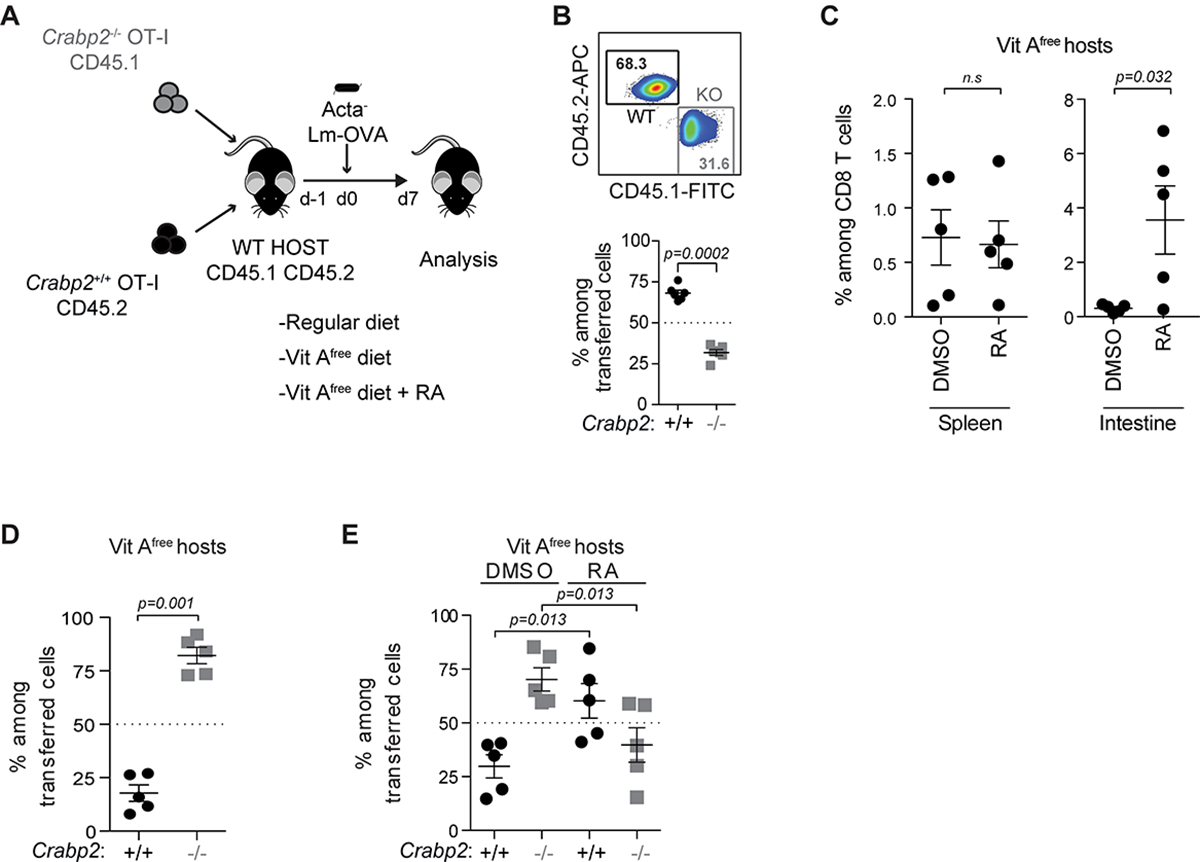
(A) Experimental strategy. (B) Tracking of adoptively transferred naïve CD45.2+ WT and CD45.1+ CRABP2-deficient OT-I cells in spleen of CD45.1+CD45.2+ recipient mice 7 days after intravenous (i.v.) infection with ActA− Lm-OVA. Percentage of WT(CD45.2+) and CRABP2-deficient (CD45.1+) OT-I cells among donor cells is shown. (C) Tracking of adoptively transferred naïve CD45.2+ WT OT-I cells in spleen and small intestine epithelium of CD45.1+CD45.2+ vitamin A-free recipient mice treated with DMSO or RA, 7 days after infected i.v. with ActA− Lm-OVA. Percentage is shown of WT OT-I T cells among total CD8 T cells. (D) Frequency of donor WT and CRABP2-deficient OT-I cells in the spleen of vitamin A-free CD45.1+CD45.2+ recipient mice infected i.v. with ActA− Lm-OVA and assessed 7 days later. Percentage of WT and CRABP2-deficient OT-I cells among donor cells is shown. (E) Frequency of donor WT and CRABP2-deficient OT-I cells in spleen of DMSO or RA treated vitamin A-free CD45.1+CD45.2+ recipient mice infected i.v. with ActA− Lm-OVA and assessed 7 days later. Percentage of WT and CRABP2-deficient OT-I cells among donor cells is shown. Data are representative of 2 experiments.
Extranuclear RARα signaling controls effector differentiation in vivo
To investigate the impact of cell intrinsic RARα signaling during T cell activation and effector differentiation in vivo, we induced EAE in mice with a T cell-specific deletion of Crabp2, using distal Lck-Cre for specific deletion of Crabp2 in mature T cells. Mice with a conditional deletion of Crabp2 and WT mice were immunized subcutaneously with myelin oligodendrocyte glycoprotein (MOG35–55) peptide in complete Freund’s adjuvant and pertussis toxin on day 0 and day 2. Onset of disease was similar for Crabp2 conditional deletion mutant mice and WT mice however disease did not progress in Crabp2 conditional deletion mutant animals (Figure 7A and Figure S6A). In contrast by day 25, some of the WT mice had developed severe symptoms, including paralyzed hind limbs (Figures 7B and 7C). Analysis of the mice at the peak of disease in the WT animals, showed more cell infiltration, including more T cells, in the spinal cord (Figure 7D and Figure S6B) and an overall higher histological disease score (Figure 7E) compared to Crabp2 conditional deletion mutant mice. Moreover, the proportion of FOXP3+RORγt− Treg cells was increased in several of the Crabp2 conditional deletion mutant mice compared to the WT mice (Figure S6C), whereas in contrast, the absolute number (Figure 7F) and proportion of IFNγ-producing cells (Figure S6D) was reduced in the Crabp2 conditional deletion mutant mice. Unlike Lm-responding dnRara effector cells, however, which showed conversion of IFNγ-expressing cells to RORγt and IL-17 expressing cells(43), Crabp2 conditional deletion mutant CD4 T cells in this EAE model did not convert more to inflammatory Th17 cells (Figure S6C). Instead comparison of the ratios of effector cells (IFNγ+) over non-effector cells (IFNγ− and IL-17−) at day 25 of disease induction, indicated a significant overall decrease in inflammatory effector differentiation of the CRABP2 Crabp2 conditional deletion mutant cells compared to WT cells (Figure 7G).
FIGURE 7: Extranuclear RARα signaling controls effector differentiation in vivo. (Related data shown in Figure S6).
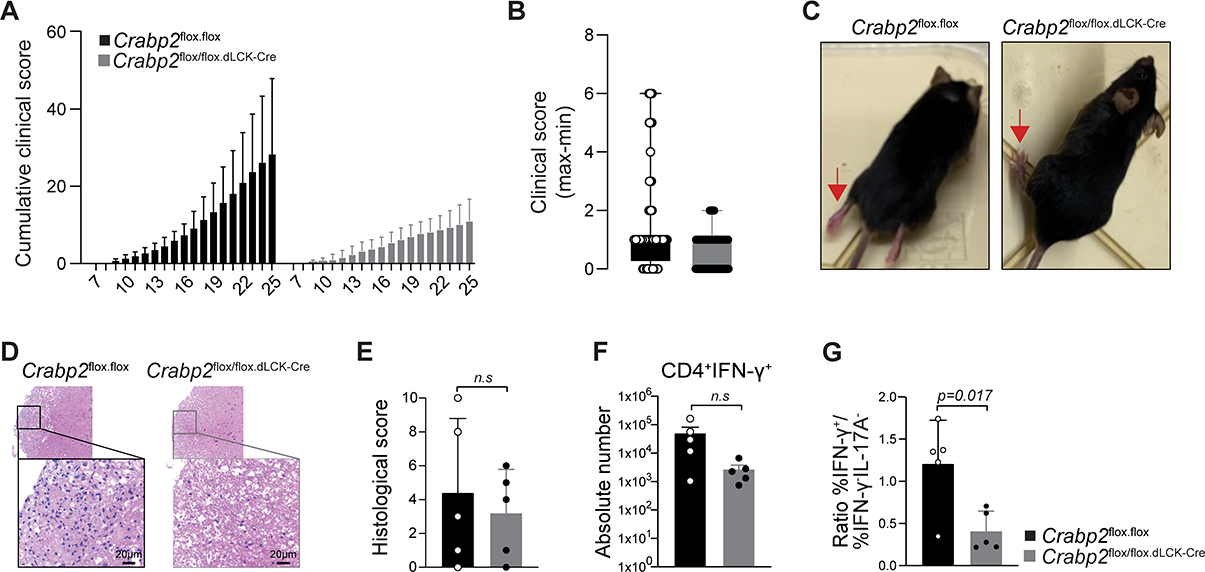
(A-C) EAE was induced and the clinical score was evaluated daily. (A) Cumulative clinical score, (B) maximum and minimum clinical scores and (C) images of WT and Crabp2 conditional deletion mutant mice at day 22. Red arrows point to the hind limb position. (D-G) Histopathology analyses at day25 after EAE induction. (D) H&E staining of spinal cords from WT or Crabp2 conditional deletion mutant mice, (E) Histological score comprising inflammation severity and axon dilation, (F) IFNγ expression of FOXP3−CD45+TGF-β+CD4 T cells from the spinal cord, (G) Ratio of IFNγ+ cells over IFNγ− and IL-17− cells. P value was calculated by Student t-test. Five female mice of 8 to 12 weeks old were analyzed per condition in 2 EAE experiments. Shown mean +/− SD.
The global weakened effector response in Crabp2 conditional deletion mutant mice highlights the essential role of extranuclear RARα as a critical TCR-induced component of the signalosome required for T cell activation, expansion and optimal effector differentiation in vivo.
DISCUSSION
Here we identified an isoform of RAR that plays decisive roles in the mechanisms that control T cell activation in vitro and in vivo. Alternative RARα is expressed in the cytoplasm and rapidly phosphorylated upon TCR stimulation. Extranuclear RARα interacts with a ZAP-70-activation complex and participates in signal transduction during TCR-driven proliferation and effector differentiation. In contrast to RA-dependent transcriptional activation of nuclear RARs, RA negatively influences RARα at the plasma membrane, which results in suboptimal activation and enhanced Treg cell differentiation. Transactivation of nuclear RARα by RA, is important to control effector gene expression programs and depends on nuclear translocation of RA by TCR-induced CRABP2 expression. The non-genomic and genomic functions of RARα have distinct kinetics, with actions of cytoplasmic RARα occurring much faster. The decisive effects of extranuclear and nuclear RARα signaling in T cells underscore the importance of their spatial and temporal bipartite regulation in controlling appropriate T cell responses to immune challenges.
Blocking of RARα function with inhibitors known to interfere with structural dynamics of nuclear RARs also disrupted the RARα/ZAP-70 complex, impaired phosphorylation of AKT and other signaling components downstream of the TCR and interfered with rapidly-induced actin-based events at the plasma membrane, including TCR/CD3-microdomain clustering(44, 45) and non-canonical ligand-independent cleavage of NOTCH(34, 36, 46, 47). These findings suggest that, similar to transcription-complex-building by nuclear RARα, the mode of action of signalosome RARα might also consist of a structural interactome that coordinates functions of various cytoplasmic binding partners downstream of the TCR. These non-genomic roles of extranuclear RARα are consistent with previously described findings showing that early T cell activation was influenced by the absence or overexpression of RARα(3, 4, 27) and negatively impacted by RA(27).
Cytoplasmic RARα in T cells is encoded by multiple transcripts with variable 5’ non-translated ends. Each transcript encodes the same truncated RARα isoform, which lacks typical transcriptional features, including a complete DBD and a functional NLS. Conversely, the conserved LBD, which interacts with binding partners and the regulatory F-domain are fully present. The F domain is unique and highly variable in length and sequence in different RAR proteins(5). Although its function is not fully understood, post-translational modifications of the F domain have been reported to counteract the transcriptional activity of nuclear RARα and RA(5). Several putative phosphorylation sites for proline-directed kinases (Pdks) were previously identified in the F domain of RARα(48). Phosphorylation of these sites is independent of RA ligation, and instead RA prevents the phosphorylation of the F region(48). Signals that lead to F domain phosphorylation had not been identified, but here we showed that phosphorylation of a predicted Pdk phosphorylation site (serine 445) in the F domain occurred within minutes of TCR stimulation. Phosphorylation of the F domain coincided with the interaction of RARα with activated ZAP-70, suggesting that TCR-induced modulation of the F domain might facilitate the dynamic interaction of RARα LBD with a ZAP-70-containing activation complex.
The current findings expand the repertoire of non-genomic actions by nuclear receptor superfamily members(49) to include participation of extranuclear RARα in the TCR signalosome. Rapid, non-transcriptional functions for RARα in actin remodeling were described in human platelets(50) and in neurons during the cleavage of amyloid preprotein by secretases(51), a process that parallels that of TCR-mediated NOTCH cleavage. In mouse embryonic fibroblasts and human mammary cancer cells, RARα cooperates with G protein alpha Q (Gαq) and directs Rho-GTPases and p38MAPK signaling(52), and in neuroblastoma cells extranuclear RARα interacts with phosphatidylinositol 3-kinase subunit, p85 and recruits the catalytic subunit, p110 to the plasma membrane resulting in AKT and ERK1/2 phosphorylation(53). All these observations are consistent with participation of extranuclear RARα in protein complexes that affect cytoskeleton remodeling and downstream signaling cascades in various cell types, similar to its role in the TCR activation process.
RA targets cytoplasmic RARα resulting in diminished TCR signal strength and enhanced Treg cell induction, consistent with other studies linking reduced TCR signals with Treg cell differentiation(25, 54–56).
In contrast to the rapid TCR-mediated activation of extranuclear RARα, transactivation of nuclear RARα is much later and requires ligation by RA(1). Translocation and ligation are promoted by CRABP2(6, 41). We show here that CRABP2 is not expressed in resting T cells, but rapidly induced upon activation. TCR-induced CRABP2 not only established RA-responsiveness but it also prevented accumulation and interference of RA with extranuclear RARα signaling. In the absence of CRABP2 induction, RA translocation and transactivation of nuclear RARα is greatly impaired. However, accumulation of RA in the cytoplasm of CRABP2-deficient T cells did not impact the RA- enhanced TGFβ-induced Treg cell conversion, underscoring that transcriptional control by nuclear RARs is not the main mechanism for RA-mediated enhanced Foxp3 expression. Instead the data are consistent with a negative impact of a high dose of RA in the cytoplasm on the non-genomic function of extranuclear RARα in the TCR signalosome, which led to suboptimal activation and impaired effector differentiation and survival but favored RA-enhanced Treg cell conversion. In WT T cells, TCR stimulation resulted in CRABP2 induction, which transports RA to the nucleus and away from the TCR signalosome. Addition of RA after CRABP2 induction no longer led to reduced signaling and no longer enhanced Treg cell conversion. In contrast, in pre-activated CRABP2-deficient T cells, RA accumulation in the cytoplasm counteracted extranuclear RARα signaling and continued to promote Treg cell conversion, even when added after 24hrs of pre-activation. Interference with extranuclear RARα signaling not only favored Treg cell conversion by weakening TCR signaling, but also hampered TCR-induced CRABP2 expression and consequently reduced transactivation of nuclear RARα. Lack of TCR-induced CRABP2 expression significantly compromised the extranuclear RARα action in TCR-induced activation and effector differentiation which caused a global suppression of EAE in Crabp2 conditional deletion mutant mice.
The non-genomic role of RARα increases the scope of its biological functions in T cells beyond its role as a regulator of gene transcription. A paradigm unfolds by which extranuclear RARα signaling integrates early TCR-induced signaling events at the plasma membrane with subsequent nuclear events, coordinated by CRABP2. In transporting RA from the cytoplasm to the nucleus, TCR-induced CRABP2 functions as an intrinsic trait of the spatial and temporal regulation of RARα signaling during T cell activation, enabling continuing participation of extranuclear RARα in the TCR signalosome and promoting RA-dependent transactivation of nuclear RARα. The findings reveal a fundamental process for extranuclear and nuclear RARα signaling in control of both regulatory and effector arms of adaptive T cell immunity.
Limitation of the Study:
The cRARα sequence is imbedded in the common sequence shared by all RARα isoforms. Because deletion of a common exon is lethal, we attempted alternative strategies to specifically interfere with cRARα expression. We targeted the putative promoter for Rara3 located in the intron that separates the unique Rara1 and -2 N-terminal sequences from the common C-portion. All mutant mice however still expressed cRARα encoded by Rara3 transcripts initiated from the Rara2 promoter. Next, we altered the internal translational start codon of cRARα to a neutral amino acid codon. The alternative codon remained active when only one nucleotide was replaced, whereas the exchange of two nucleotides led to expression of cRARα starting from an alternative CUG start codon upstream of the internal methionine codon. Therefore, the results suggested that specific targeting of cRARα was not feasible without also affecting expression or function of nRARα and/or viability of the mutant mice.
STAR Method
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hilde Cheroutre (hilde@lji.org).
Materials availability
Mice and embryonic stem cells used in this study are commercially available. Newly generated plasmids in this study are available from the lead contact with a completed material transfer agreement.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table.
Original western blot images, raw flow cytometry data, (RT)-PCR, and microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESSOURCE TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| T cell stimulation anti-CD3e | Thermo Fisher Scientific | 16-0031-86 |
| T cell stimulation anti-CD28 | Thermo Fisher Scientific | 16-0281-86 |
| WB: anti-ZAP-70 (cl. D1C10E) | Cell Signaling Tech. | 3165T |
| WB: anti-GAPDH (cl. 14C10) | Cell Signaling Tech. | 85925SF |
| WB: anti-Lamin B1 (cl. D9V6H) | Cell Signaling Tech. | 13435S |
| WB: anti-NOTCH1 (cl. D1E11) | Cell Signaling Tech. | 3608T |
| WB: anti-RARα | Cell Signaling Tech. | 2554 |
| WB: anti-RARγ1 (cl. D3A4) | Cell Signaling Tech. | 8965T |
| WB: anti-CRABP2 | One World Lab | 11250 |
| WB: anti-RARa | Assay Biotechnology Company | R12-3424 |
| WB: anti-HSP90 (cl. AC16) | Santa Cruz | sc-101494 |
| WB: anti-Flag (cl. M2) | Millipore Sigma | A2220 |
| WB: anti-AKT | Cell Signaling Tech. | 9272 |
| WB: anti-Phospho-AKT | Cell Signaling Tech. | 9271T |
| WB: anti-Phospho- PLCγ1 | Cell Signaling Tech. | 2821 |
| WB: anti-Phospho-ZAP-70 (cl. 65E4) | Cell Signaling Tech. | 2717 |
| WB: anti-Phospho-ERK1/2 (cl. D13.14.4E) | Cell Signaling Tech. | 4370 |
| Activation: H-2Kb-SIINFEKL tetramer | In house | N/A |
| IP: anti-RARα | Santa Cruz | sc-551 |
| Flow /Conf. Im.: anti-CD16-CD32 (cl. 2.4G2) | In house | N/A |
| Flow: anti-CD4-APC (cl. RM4-5) | BD | 553051 |
| Flow: anti-TCRgd-FITC (cl. GL3) | BD | 553177 |
| Flow: anti-CD45.1-FITC (cl. A20) | BD | 553775 |
| Flow: anti-CD44-V450 (cl. IM7) | BD | 560452 |
| Flow: anti-CD8a-PercpCy5.5 (cl. 53-6.7) | BD | 551162 |
| Flow: anti- CD8a-PeCy7 (cl. 53-6.7) | BD | 552877 |
| Flow: anti-IFNg-BV711 (cl. XMG1.2) | Biolegend | 505835 |
| Flow: anti-Il-17-PeCy7 (cl. TC11-18H10.1) | Biolegend | 506922 |
| Flow: anti-CD8a-AF700 (cl. 53-6.7) | Biolegend | 100730 |
| Flow: anti-CD4-AF700 (cl. GK1.5) | Biolegend | 100430 |
| Flow: anti-FOXP3-PercpCy5.5 (cl. FJK-16s) | Thermo Fisher Scientific | 45-5773-82 |
| Flow: anti-CCR9-FITC (cl. eBioCW-1.2) | Thermo Fisher Scientific | 11-1991-85 |
| Flow: anti-TCRb-APCeF780 (cl. H57-597) | Thermo Fisher Scientific | 47-5961-82 |
| Flow: anti-CD45.2-APC (cl.104) | Thermo Fisher Scientific | 17-0454-82 |
| Flow: anti-RORgt-Pe (cl. AFKJS-9) | Thermo Fisher Scientific | 12-6988-80 |
| Flow: anti-CD45-eF450 (cl. 30-F11) | Thermo Fisher Scientific | 48-0451-82 |
| Flow: anti-FOXP3-AF488 (cl. FJK-16s) | Thermo Fisher Scientific | 53-5773-82 |
| Flow: anti-FOXP3-Pe (cl. FJK-16s) | Thermo Fisher Scientific | 12-5773-82 |
| Flow: anti-CD25-APC (cl. PC61.5) | Thermo Fisher Scientific | 17-0251-82 |
| Flow: anti-CD4-eF450 (cl. RM4-5) | Thermo Fisher Scientific | 48-0042-82 |
| Flow: anti-c-MYC-FITC (cl. 9E10) | Thermo Fisher Scientific | 13-2511 |
| Conf. Im.: anti- CD3ε-biotin (cl. 2C11) | Biolegend | 100304 |
| Conf. Im.: Goat anti-mouse IgG-DyLight™ 488 | Biolegend | 405310 |
| Conf. Im.: Rabbit anti-Goat IgG-Alexa Fluor 647 | Life Technology | A21446 |
| Conf. Im.: anti-ZAP70 (cl. M20) | Santa Cruz | sc-1526 |
| Conf. Im.: anti-RARa (c. Ralpha10) | Thermo Fisher Scientific | MA1810A |
| Bacterial and virus strains | ||
| Listeria monocytogenes expressing OVA (ActA− Lm-OVA) | (64) | Gift |
| Biological samples | ||
| N/A | ||
| Chemicals, peptides, and recombinant proteins | ||
| LIVE/DEAD™ Fixable Yellow Dead Cell Stain | Thermo Fisher Scientific | L34959 |
| EDTA | Thermo Fisher Scientific | 15575020 |
| Collagenase D | Millipore Sigma | 11088858001 |
| DNAse I | Millipore Sigma | 10104159001 |
| Human recombinant TGF-β | R&D System | 240-B-002 |
| Trans retinoic acid | Millipore Sigma | R2625 |
| RAR antagonist LE540 | FUJIFILM Wako Pure Chemical Corporation | Distributor 123-04521 Barcode No 4987481365261 |
| RARα-selective antagonist | Enzo lifescience | Ro 41-5253 |
| RARγ-selective antagonist MM 11253 | Tocris bioscience | 3822 |
| MEK1/2 inhibitor U0126 | Invivogen | tlrl-u0126 |
| PI3K inhibitor LY294002 | Invivogen | tlrl-ly29 |
| γ-secretase inhibitor GSI | Tocris bioscience | 2870 |
| Human recombinant IL-2 | Peprotech | 200-02 |
| AKT Inhibitor VIII | Cayman Chemical Company, Inc | 14870 |
| DMSO | Millipore Sigma | D2438-5X |
| TRIzol reagent | Thermo Fisher Scientific | 15596026 |
| TGX FastCast Acrylamide Solutions for SDS-PAGE gel | Biorad | 1610171 |
| PVDF membrane | Santa Cruz | sc-3723 |
| NaCl | Millipore Sigma | S9888 |
| Tris base | Santa Cruz | sc-362305 |
| Glycerol | Milliopore Sigma | G5516 |
| Triton X-100 | Milliopore Sigma | X100 |
| Protease and phosphatase inhibitor mixture | Milliopore Sigma | 11697498001 |
| ECL solution | Santa Cruz | sc-2048 |
| Polybrene | Milliopore Sigma | TR-1003-G |
| Poly I:C (HMW) | InvivoGen | tlrl-pic |
| OVA257-264 peptide | AnaSpec Inc. | AS-62572 |
| Poly-L-lysine | R&D System | 3438-100-01 |
| NH4Cl | Milliopore Sigma | A4514 |
| Saponin | Millipore Sigma | 47036 |
| Fish gelatin | Milliopore Sigma | G7041 |
| ProLong™ Gold Antifade Mountant with DNA Stain DAPI | Thermo Fisher Scientific | P36935 |
| Dithiothreitol | Milliopore Sigma | D2438 |
| Percoll | Cytiva | 17089109 |
| Freund’s adjuvant | BD | 263910 |
| M Tuberculosis H37Ra Dessicated | BD | 231141 |
| Pertussis toxin | List Biological Laboratories Inc | 180 |
| Zinc formalin | Anatech Lt | 175 |
| Pro-Par Clearant | Anatech Ltd | 510 |
| Conf. Im.: Streptavidine-Cyanine3 | Biolegend | 405215 |
| Critical commercial assays | ||
| MACS CD8α+ T cell isolation kit | Miltenyi Biotec | 130-096-543 |
| MACS CD4+ T cell isolation kit | Miltenyi Biotec | 130-104-453 |
| CD11c-Microbeads Ultrapure | Miltenyi Biotec | 130-125-835 |
| SureSelectXT RNA Direct Library Preparation kit | Agilent Technologies | G7564A |
| Pierce™ NE-PER® Nuclear and Cytoplasmic Extraction Reagent Kit | Thermo Fisher Scientific | 78833 |
| JetPrime transfection reagent | Polyplus transfection | 101000027 |
| Tandem mass tags (TMT) 11-plex reagents | Thermo Fisher Scientific | A34808 |
| Cell Trace Violet | Thermo Fisher Scientific | C34557 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE112609 |
| Database of Immune Cell Expression, Expression quantitative trait loci (eQTLs) and Epigenomics (DICE) | (66) | https://dice-database.org |
| UCSC mouse genome assembly (GRCm38/mm10) | UCSC Genome Browser Group | https://genome.ucsc.edu/cgi-bin/hgGateway |
| Experimental models: Cell lines | ||
| Platinum-E | Cell Biolabs | RV-101 |
| Jurkat, Clone E6-1 | ATCC | TIB-152 |
| MCC-T CD4 T cell hybridoma cells | (31) | Gift |
| Mutant ES cell (LoxP sites flanking Crabp2 exons) | EUCOMM | HEPD0679_8_F03 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Jackson Lab. | 000664 |
| Mouse: Crabp2−/−(10 generation backcrossed to B6 background) | Fawcett et al., 1995 | Gift |
| Mouse: RARE-Luciferase (DR5) | (39) | Gift |
| Mouse: RAR403 (DOMINANT NEGATIVE) | (28) | Gift |
| Mouse: CD4-Cre | (58) | Gift |
| Mouse: E8I-cre | (59) | Gift |
| Mouse: OT-I TCRM | Jackson Lab. | 003831 |
| Mouse: Act-mOVA | (60) | Gift |
| Mouse: RARα1−/− | Jackson Lab. | 023845 |
| Mouse: Crabp2flox/flox Crabp2tm1e(EUCOMM)Hmgu | This paper | N/A |
| Mouse: Distal LCK-Cre | Jackson Lab. | Jackson Lab. |
| Oligonucleotides | ||
| RT-RARa F1: CAGTCAGTGGTTACAGCACACCGTC | IDT DNA | N/A |
| RT-RARa F2: GGACTCCGCTTTGGAATGGCTC | IDT DNA | N/A |
| RT-RARa F3: GATGTCCAGGCCCAAGTAGAAGCCAG | IDT DNA | N/A |
| RT-RARa R1: GCTGCAATCTGCTGCTCATGCCTACAC | IDT DNA | N/A |
| RT-RARa R2: GGACTGACCTGCTGCAATCTGCTG | IDT DNA | N/A |
| RT-RARa R3: CTGGCTTCTACTTGGGCCTGGACATC | IDT DNA | N/A |
| RT-RARa R4: CAGGCTGTGAAAGACTCCTGCGGCT | IDT DNA | N/A |
| RT-RARa R5: CAGCATGTGTTATGCCAGGCTGTGAAAG | IDT DNA | N/A |
| RT-RARa R6: CCAACAGCATGTGTTATGCCAGGCTG | IDT DNA | N/A |
| RT-RARa R7: TTCACCTCACTTCCTTCTCGGGAG | IDT DNA | N/A |
| RT-RARa R9: CCCCATAGTGGTAGCCGGATGATTTG | IDT DNA | N/A |
| RT-RARa R15: TCAGTGGAAACCCAGCAGGAACAGGTG | IDT DNA | N/A |
| RT-Hprt F: GTCGTGATTAGCGATGATGAACC | IDT DNA | N/A |
| RT-Hprt R: ATGACATCTCGAGCAAGTCTTTCAG | IDT DNA | N/A |
| qRT-Crabp2 F: CCCAGAGCTTTTAGCATTTCC | IDT DNA | N/A |
| qRT-Crabp2 R: GAAGATCTAAAGAGAAAGCCACCT | IDT DNA | N/A |
| qRT-cMYC F: TTGAAGGCTGGATTTCCTTTGGGC | IDT DNA | N/A |
| qRT-cMYC R: TCGTCGCAGATGAAATAGGGCTGT | IDT DNA | N/A |
| qRT-GADPH F: CAGATGCCTGCTTCACCA | IDT DNA | N/A |
| qRT-GADPH R: ATGGCCTTCCGTGTTCCT | IDT DNA | N/A |
| qRT-bACTIN F: CCAGAAGGACTGTTATGTGGGA | IDT DNA | N/A |
| qRT-bACTIN R: GACTCCGTGTTCAATGGGATAC | IDT DNA | N/A |
| qRT-HPRT F: GTCGTGATTAGCGATGATGAACC | IDT DNA | N/A |
| qRT-HPRT R: ATGACATCTCGAGCAAGTCTTTCAG | IDT DNA | N/A |
| Recombinant DNA | ||
| Plasmid: MSCV-IRES-GFP | Addgene | 20672 |
| Plasmid: MSCV-IRES-Puromycin | Addgene | 68469 |
| Plasmid: CD3ζ-GFP | Katz et al., 2016 | Gift |
| Software and algorithms | ||
| FlowJo software v10.8.0 | Becton Dickinson & Company | https://www.flowjo.com/solutions/flowjo/downloads |
| HISAT2 | (67) | https://github.com/DaehwanKimLab/hisat2 |
| Tools (written in C using htslib) for manipulating next-generation sequencing data | Samtools | https://github.com/samtools/samtools |
| Integrative Genomics Viewer | Broad Institute | https://software.broadinstitute.org/software/igv/ |
| FACSDIVA | Becton Dickinson & Company | https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software |
| Spectrum Mill | Agilent/Broad Institute | https://proteomics.broadinstitute.org/millhome.htm |
| ZEN software | ZEISS | https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html |
| Prism 8.4.2 | GraphPad | https://www.graphpad.com/features |
| Other | ||
| Penicillin, streptomycin | Thermo Fisher Scientific | 15140122 |
| Puromycin | Invivogen | ant-pr-1 |
| Balsticidin | Invivogen | ant-bl-05 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
In vivo Experimental Models
All in vivo models used in this study were performed in mice.
Age matched male and female mice were both analyzed separately and compared except where specifically indicated otherwise.
Gender- and age-matched male and female animals were used between the ages of 2 months to 4 months.
Mice were maintained and bred under specific pathogen-free conditions in the animal facility of the La Jolla Institute for Immunology. Animal care and experimentation were consistent with the guidelines of the US National Institutes of Health and were approved by the Institutional Animal Care and Use Committee of the La Jolla Institute for Immunology. All experiments were performed in accordance with the approved protocols.
In vivo T cell activation model.
RARE-Luc transgenic mice were crossed to OVA-responsive OT-I TCR transgenic mice to generate OT-I/RARE-Luc double transgenic mice.
In one case age-matched male and female mice were separately infected intravenous with ActA− Lm-OVA and analyzed until 6 days post-infection for luciferase expression in harvested spleen cells.
In another case, naïve CD45.2+ WT and CD45.1+ CRABP2-deficient OT-I cells isolated from CD45.2+ OT-I WT or from CD45.1+ CRABP2-deficient OT-I male or female age-matched donor mice were adoptively transferred to gender- and age-matched normal or vitamin A-free CD45.1+CD45.2+ C57BL/6 recipient mice that were infected intravenous with ActA− Lm-OVA.
Donor and endogenous spleen and intestinal T cells isolated from the recipient mice were analyzed 7 days later. In some cases, vitamin A-free recipient mice were treated with DMSO or RA.
Experimental Autoimmune Encephalomyelitis (EAE) experiment
Age-matched female C57BL/6 control mice or conditional Crabp2 deletion mice between the ages of 2 months to 4 months, were analyzed per condition and per experiment. Littermates of 2 to 5 litters were randomly assigned to experimental groups.
In vitro Experimental Models
Primary cells and cell lines
-
T cell hybridoma/cell lines
Jurkat (cl. E6–1, purchased from ATCC) and MCC-T CD4 T cell hybridoma cells were cultured in RPMI complemented with 10% FBS, penicillin, streptomycin and sodium pyruvate. Only low passage cells (<20) were used.
-
Primary cells
Naïve sorted primary CD8αβ+or CD4+ spleen T cells or total or sorted thymocytes or spleen dendritic cells isolated from male or female age-matched mice were harvested and used or analyzed in this study.
In vitro culture and activation of T cells
For activation of naïve primary CD4+ or CD8αβ+ T cells isolated from the spleen of either female or male age-matched donor mice, 105 cells were cultured in 96-well plates coated with anti-CD3ε (1 μg/ml, cl. 145–2C11, #16–0031-86, eBioscience) and soluble anti-CD28 (0.5 μg/ml, cl. 37.51, #16–0281-85, eBioscience).
For Treg cell differentiation, naïve primary CD4+ spleen T cells isolated from age-matched male or female donor mice were activated with anti-mouse CD3ε and anti-mouse CD28 in the presence of human recombinant TGF-β 5 ng/ml and all trans retinoic acid (10 nM).
For the short-term activation followed by the analysis of proximal signaling events, up to 106 naïve primary CD4+ or CD8αβ+ T cells isolated from the spleen of either female or male age-matched donor mice were kept on ice for 10min in serum-free HBSS, then loaded with anti-CD3ε (10 μg/ml) and anti-CD28 (10 μg/ml) on ice for another 10–15min. Anti-hamster crosslinker (18 μg/ml) was added and the cells were immediately placed in a water-bath (37°C).
For stimulation of pre-activated T cells, 105 naïve WT or Crabp2 deficient spleen T cells isolated from male or female age-matched donor mice were cultured in 96-well plates coated with anti-CD3ε (1 μg/ml) and soluble anti-CD28 (0.5 μg/ml) for 72h. Live activated T cells were sorted according to their FSC/SSC parameters. Equal numbers of pre-activated T cells were further activated in vitro according to the short-term activation protocol described hereinbefore.
METHOD DETAILS
Cell lines
Platinum-E (PLAT-E) (Cell Biolabs, San Diego, CA, USA) cells were cultured in DMEM supplemented with 10% FBS, penicillin, streptomycin, puromycin 1μg/ml and Balsticidin 10μg/ml. Jurkat (cl. E6–1, purchased from ATCC) and MCC-T CD4 T cell hybridoma cells(31) were cultured in RPMI complemented with 10% FBS, penicillin, streptomycin and sodium pyruvate. Only low passage cells (<20) were used.
Mice
C57BL/6 mice (WT) were bred in house. Crabp2 deletion mutant mice(57) were a gift from V. Giguere (McGill University, Canada). Crabp2 deletion mutant mice were backcrossed to the C57BL/6 genetic background for at least 10 generations. RARE-Luciferase (DR5) transgenic mice(39) and DNRARα mice(28) were a gift from R. Noelle (Dartmouth-Hitchcock Medical College). The DNRARα mice, that express a dnRara transgene preceded by a floxed stop, were crossed with CD4cre(58) or E8Icre (also known as CD8cre) (59) transgenic mice in order to obtain littermates expressing or not the cre recombinase in mature CD4 and/or CD8 T cells. RARE-Luc transgenic mice were crossed to OVA-responsive OT-I TCR transgenic mice to generate OT-I/RARE-Luc double transgenic mice. S. Schoenberger provided the Act-mOVA mice(60). Rara1 deletion mutant mice(61) were purchased from the Jackson laboratory. Crabp2 -flox mice were generated with mutant ES cell clones with LoxP sites flanking Crabp2 exons, obtained from EUCOMM. Crabp2-flox mice were crossed with distal Lck Cre transgenic mice to generate mature T cell specific Crabp2 deletion mutant mice. Gender-matched and age-matched (from 2 to 4month-old) animals were used. Mice were maintained and bred under specific pathogen–free conditions in the animal facility of the La Jolla Institute for Immunology. Animal care and experimentation were consistent with the guidelines of the US National Institutes of Health and were approved by the Institutional Animal Care and Use Committee of the La Jolla Institute for Immunology.
T cell isolation, cell sorting and Cell Trace labeling
Splenic CD8 T cells or CD4 T cells were purified by magnetic negative selection with the MACS CD8α+ T cell isolation kit or naïve CD4 T cell isolation kit according to the manufacturer’s protocol (#130–104-453, Miltenyi Biotec). Naïve T cells were sorted with a FACSAria (Becton Dickinson). In some cases, sorted cells were labeled with Cell Trace reagent (Invitrogen) according to the manufacturer’s protocol. The division index, which is the average number of cell divisions that a cell in the original population has undergone, was calculated using the FlowJo software (Tristar). LIVE/DEAD™ Fixable Yellow Dead Cell Stain Kit, for 405 nm excitation, Thermo Fisher, L34968.
DC isolation
Spleens from naïve mice were perfused with 1 mg/ml collagenase D in the presence of DNAse I. After perfusion the spleens were cut into small fragments and then digested with frequent mixing for 25min at room temperature. To disrupt DC-T cell complexes, EDTA (0.1 M) was added, and mixing continued for 5min. Cells were enriched using CD11c-beads (Miltenyi).
In vitro stimulation of T cells
For activation of T cells, 105 naïve CD4 or CD8 T cells were cultured in 96-well plates coated with anti-CD3ε (1 μg/ml, cl. 145–2C11, #16–0031-86, eBioscience) in the presence of soluble anti-CD28 (0.5 μg/ml, cl. 37.51, #16–0281-85, eBioscience). For Treg cell differentiation, naïve CD4+ (APC Rat anti-mouse, #553051, BD) T cells were isolated with naive CD4 + T Cell Isolation Kit, mouse, #130–104-453, Miltenyi Biotec) were activated with anti-mouse CD3ε and anti-mouse CD28 in the presence of human recombinant TGF-β 5 ng/ml, (#240-B-002, R&D) and all trans retinoic acid (RA) (10 nM, #R2625, Sigma).
For the short-term activation followed by the analysis of the signaling events, up to 106 cells were rested on ice for 10min in serum-free HBSS, then loaded with anti-CD3ε (10 μg/ml) and anti-CD28 (10 μg/ml) on ice for 10–15min. Anti-hamster crosslinker (18 μg/ml, Jackson Immunoresearch) was added and the cells were immediately placed in a water-bath (37°C).
For stimulation of pre-activated T cells, 105 naïve WT or Crabp2 deletion T cells were cultured in 96-well plates coated with anti-CD3ε (1 μg/ml) in the presence of soluble anti-CD28 (0.5 μg/ml) for 72h. Live activated T cells were then sorted according to their FSC/SSC parameters. Equal numbers of T cells were then activated according to the short-term activation protocol described hereinbefore.
The RAR antagonist LE540 (iRARs, 5 μM, Waco chemicals), the RARα-selective antagonist Ro 41–5253 (iRARα, 5 μM, Enzo lifescience), the RARγ-selective antagonist MM11253 (iRARγ, 5 μM, Tocris bioscience), MEK1/2 inhibitor U0126 (iERK, 10 μM, InvivoGen), the PI3K inhibitor LY294002 (iPI3K, 20 μM, InvivoGen), the γ-secretase inhibitor GSI (Tocris bioscience), RA (10 nM, Sigma) or rhIL-2 (40 U/ml, Peprotech), AKT-inhibitor (AKT Inhibitor VIII, Cayman Chemical Company, Inc, # 14870, at 0.3 μM), were used in some cultures. When appropriate, DMSO (0.05 to 0.1%) was used as a control.
RNA-sequencing
After total cellular RNA extraction, 106 purified DP thymocytes or spleen T cells were collected to prepare total RNA using TRIzol reagent (Invitrogen). 200–300 ng total RNA was used for library construction with a SureSelect Strand Specific RNA-Seq Library Preparation kit (Agilent Technologies) according to the manufacturer’s protocol. Sequencing was performed by the genomics facility at RIKEN IMS with an Illumina HiSeq 1500. Sequences retrieved in RNA-seq experiments were aligned on the UCSC mouse genome assembly (GRCm38/mm10) using the HISAT2 (hierarchical indexing for spliced alignment of transcripts 2). And the obtained BAM files were then sorted by Samtools and visualized by Integrative Genome viewer (IGV). Raw fastq files for the RNA-seq libraries were deposited at the Gene Expression Omnibus (GEO) database under the accession number GEO: GSE112609.
RNA-seq data of human naïve CD8 (CD3+ CD8+ CD45RA+ CD127+ CCR7+) or CD4 (CD3+ CD4+ CD45RA+ CD127+ CCR7+) T cells sorted from peripheral blood mononuclear cells (PBMC) were obtained through the Database of Immune Cell Expression, Expression quantitative trait loci (eQTLs) and Epigenomics (DICE); https://dice-database.org/.
RT-PCR
Total RNA was obtained as described above. cDNA was synthesized using SuperScript IV (Thermo Fisher Scientific) and an oligo dT primer, then each Rara or control transcript was measured with combinations of the following primers.
F1: CAGTCAGTGGTTACAGCACACCGTC
F2: GGACTCCGCTTTGGAATGGCTC
F3: GATGTCCAGGCCCAAGTAGAAGCCAG
R1: GCTGCAATCTGCTGCTCATGCCTACAC
R2: GGACTGACCTGCTGCAATCTGCTG
R3: CTGGCTTCTACTTGGGCCTGGACATC
R4: CAGGCTGTGAAAGACTCCTGCGGCT
R5: CAGCATGTGTTATGCCAGGCTGTGAAAG
R6: CCAACAGCATGTGTTATGCCAGGCTG
R7: TTCACCTCACTTCCTTCTCGGGAG
R9: CCCCATAGTGGTAGCCGGATGATTTG
R15: TCAGTGGAAACCCAGCAGGAACAGGTG
Hprt F: GTCGTGATTAGCGATGATGAACC
Hprt R: ATGACATCTCGAGCAAGTCTTTCAG
Quantitative RT-PCR
RNA was extracted with TRIzol (Invitrogen) and cDNA was synthesized with the iScript cDNA Synthesis kit (Bio-Rad). A 480 Real-Time PCR System (Roche) was used for real-time PCR. Values were normalized to the amount of Gapdh, β-actin or Hprt in each sample and multiplied by 10,000. The primers for quantitative PCR were as follows:
Crabp2 F: CCCAGAGCTTTTAGCATTTCC
Crabp2 R: GAAGATCTAAAGAGAAAGCCACCT
c-Myc F: TTGAAGGCTGGATTTCCTTTGGGC
c-Myc R: TCGTCGCAGATGAAATAGGGCTGT
Gapdh F: CAGATGCCTGCTTCACCA
Gapdh R: ATGGCCTTCCGTGTTCCT
β-actin F: CCAGAAGGACTGTTATGTGGGA
β-actin R: GACTCCGTGTTCAATGGGATAC
Hprt F: GTCGTGATTAGCGATGATGAACC
Hprt R: ATGACATCTCGAGCAAGTCTTTCAG
Immunofluorescence staining and flow cytometry
Cells were pre-incubated with anti-CD16-CD32 (2.4G2 mAb prepared in-house) to block binding of antibodies to the Fc receptor, and then stained in cold PBS containing 0.5% (vol/vol) FBS and 0.05% (wt/vol) sodium azide with the relevant labeled antibodies and tetramers. The following antibodies were used: anti-CD4 (cl. RMA4–5), anti-TCRγδ (cl. GL3), anti CD45.1 (cl. A20), anti-CD44 (cl. IM7), anti-CD8α (cl. 53–6.7) all from BD; anti-IFNγ (cl. XMG1.2), anti-Il-17A (cl. TC11–18H10.1), anti-CD8α (cl. 53–6.7), anti-CD4 (cl. GK1.5) all from Biolegend; anti-FOXP3 (cl. FJK-16s), anti-CCR9 (cl. eBioCW-1.2), anti-CD45.2 (cl. 104), anti-RORγt (cl. AFKJS-9), anti-CD45 (cl. 30-F11), anti-CD25 (cl. PC61.5), anti-CD4 (cl. RMA4–5), anti-c-MYC (cl. 9E10) all from Thermo Fisher Scientific. A Fix/Perm kit was used according to the manufacturer’s directions (eBioscience) for intracellular staining of c-MYC or FOXP3, after surface staining. For detection of cell death, cells were stained with the Live/Dead fixable dead cell stain kit according to the manufacturer’s protocol (Invitrogen) and cells were analyzed immediately after staining. All stained cells were processed on an LSR-II (Becton Dickinson) and the data analyzed using FlowJo software (TreeStar). Dead cells were excluded from the analysis.
Western-blot analysis
Cells were lysed in cold triton lysis buffer (137 mM NaCl, 20 mM Tris base at pH 7.4, 10% glycerol and 1% Triton X-100) supplemented with a protease and phosphatase inhibitor mixture (Roche) for 20min on ice, and centrifuged at 15,000 rpm for 15min at 4°C. In some experiments, nuclear/cytoplasmic fractionation was performed according to the manufacturer’s protocol (Pierce). Equal amounts of denatured proteins were loaded onto an SDS-PAGE gel (Biorad) and transferred onto a PVDF membrane (Invitrogen). The membranes were then incubated with antibodies to ZAP-70 (cl. D1C10E), GAPDH (cl. 14C10), Lamin B1 (cl. D9V6H), NOTCH1 (cl. D1E11), RARα (#2554), RARγ1 (cl. D3A4) (all from Cell Signaling Technology), CRABP2 (#11250, One World Lab), RARα (R12–3424, Assay Biotechnology Company), HSP90 (cl. AC16, Santa Cruz), FLAG (cl. M2, Sigma Aldrich), AKT (#9272) or with antibodies to the phosphorylated forms of AKT (Ser473) (#9271), PLCγ1 (#2821), ZAP-70 (cl. 65E4) or ERK1/2 (cl. D13.14.4E) (all from Cell Signaling Technology). Immuno-reactive bands were detected by chemiluminescence (ECL solution, Santa Cruz).
Immunoprecipitation
OT-I CD8 T cells were activated with H-2Kb-SIINFEKL tetramers in order to avoid non-specific binding of anti-CD3 stimulating Abs to the beads used for immunoprecipitation. Cells were lysed in cold triton lysis buffer (137 mM NaCl, 20 mM Tris base at pH 7.4, 10% glycerol and 1% Triton X-100) supplemented with protease and phosphatase inhibitor mixture (Roche) for 20min on ice and centrifuged at 15,000 rpm for 15min at 4°C. Cell lysates were pre-cleared with A/G beads (Pierce) prior to immunoprecipitation. Equal amounts (at least three hundred micrograms) of protein were immunoprecipitated with 2 μg of antibodies to RARα (#sc-551, Santa Cruz) or with a rabbit IgG overnight at 4°C, followed by capture with 25 μl of protein A/G beads (Pierce). Denatured proteins were then loaded onto an SDS-PAGE gel (Biorad) and transferred onto a PVDF membrane (Invitrogen). Immunoblotting was performed with antibodies to ZAP-70 (cl. D1C10E) (Cell Signaling Technology). Immuno-reactive bands were detected by chemiluminescence (ECL solution, Santa Cruz).
Retroviral plasmids
Sequences of interest were inserted into an MSCV-IRES-GFP or into an MSCV-IRES-Puromycin (for GFP-tagged proteins) plasmid (Addgene, #20672 and #68469).
The sequences used for overexpression code for these protein sequences:
RARα1-FLAG:
MASNSSSCPTPGGGHLNGYPVPPYAFFFPPMLGGLSPPGALTSLQHQLPVSGYSTPSPATIETQSSSSEEIVPSPPSPPPLPRIYKPCFVCQDKSSGYHYGVSACEGCKGFFRRSIQKNMVYTCHRDKNCIINKVTRNRCQYCRLQKCFDVGMSKESVRNDRNKKKKEAPKPECSESYTLTPEVGELIEKVRKAHQETFPALCQLGKYTTNNSSEQRVSLDIDLWDKFSELSTKCIIKTVEFAKQLPGFTTLTIADQITLLKAACLDILILRICTRYTPEQDTMTFSDGLTLNRTQMHNAGFGPLTDLVFAFANQLLPLEMDDAETGLLSAICLICGDRQDLEQPDKVDMLQEPLLEALKVYVRKRRPSRPHMFPKMLMKITDLRSISAKGAERVITLKMEIPGSMPPLIQEMLENSEGLDTLSGQSGGGTRDGGGLAPPPGSCSPSLSPSSHRSSPATQSPDYKDDDDK
RARα2-FLAG:
MYESVEVGGLTPAPNPFLVVDFYNQNRACLLQEKGLPAPGPYSTPLRTPLWNGSNHSIETQSSSSEEIVPSPPSPPPLPRIYKPCFVCQDKSSGYHYGVSACEGCKGFFRRSIQKNMVYTCHRDKNCIINKVTRNRCQYCRLQKCFDVGMSKESVRNDRNKKKKEAPKPECSESYTLTPEVGELIEKVRKAHQETFPALCQLGKYTTNNSSEQRVSLDIDLWDKFSELSTKCIIKTVEFAKQLPGFTTLTIADQITLLKAACLDILILRICTRYTPEQDTMTFSDGLTLNRTQMHNAGFGPLTDLVFAFANQLLPLEMDDAETGLLSAICLICGDRQDLEQPDKVDMLQEPLLEALKVYVRKRRPSRPHMFPKMLMKITDLRSISAKGAERVITLKMEIPGSMPPLIQEMLENSEGLDTLSGQSGGGTRDGGGLAPPPGSCSPSLSPSSHRSSPATQSPDYKDDDDK
RARα3:
MVYTCHRDKNCIINKVTRNRCQYCRLQKCFDVGMSKESVRNDRNKKKKEAPKPECSESYTLTPEVGELIEKVRKAHQETFPALCQLGKYTTNNSSEQRVSLDIDLWDKFSELSTKCIIKTVEFAKQLPGFTTLTIADQITLLKAACLDILILRICTRYTPEQDTMTFSDGLTLNRTQMHNAGFGPLTDLVFAFANQLLPLEMDDAETGLLSAICLICGDRQDLEQPDKVDMLQEPLLEALKVYVRKRRPSRPHMFPKMLMKITDLRSISAKGAERVITLKMEIPGSMPPLIQEMLENSEGLDTLSGQSGGGTRDGGGLAPPPGSCSPSLSPSSHRSSPATQSP
RARα1-eGFP:
MASNSSSCPTPGGGHLNGYPVPPYAFFFPPMLGGLSPPGALTSLQHQLPVSGYSTPSPATIETQSSSSEEIVPSPPSPPPLPRIYKPCFVCQDKSSGYHYGVSACEGCKGFFRRSIQKNMVYTCHRDKNCIINKVTRNRCQYCRLQKCFDVGMSKESVRNDRNKKKKEAPKPECSESYTLTPEVGELIEKVRKAHQETFPALCQLGKYTTNNSSEQRVSLDIDLWDKFSELSTKCIIKTVEFAKQLPGFTTLTIADQITLLKAACLDILILRICTRYTPEQDTMTFSDGLTLNRTQMHNAGFGPLTDLVFAFANQLLPLEMDDAETGLLSAICLICGDRQDLEQPDKVDMLQEPLLEALKVYVRKRRPSRPHMFPKMLMKITDLRSISAKGAERVITLKMEIPGSMPPLIQEMLENSEGLDTLSGQSGGGTRDGGGLAPPPGSCSPSLSPSSHRSSPATQSPGASLEMVSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITLGMDELYK
RARα3-eGFP:
MVYTCHRDKNCIINKVTRNRCQYCRLQKCFDVGMSKESVRNDRNKKKKEAPKPECSESYTLTPEVGELIEKVRKAHQETFPALCQLGKYTTNNSSEQRVSLDIDLWDKFSELSTKCIIKTVEFAKQLPGFTTLTIADQITLLKAACLDILILRICTRYTPEQDTMTFSDGLTLNRTQMHNAGFGPLTDLVFAFANQLLPLEMDDAETGLLSAICLICGDRQDLEQPDKVDMLQEPLLEALKVYVRKRRPSRPHMFPKMLMKITDLRSISAKGAERVITLKMEIPGSMPPLIQEMLENSEGLDTLSGQSGGGTRDGGGLAPPPGSCSPSLSPSSHRSSPATQSPGASLEMVSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITLGMDELYK
Retroviral transduction
Retroviral plasmids were transfected into the Platinum-E Retroviral Packaging cell line (Cell Biolabs) using the JetPrime transfection reagent (Polyplus Transfection) according to the manufacturer’s instructions. Retrovirus-containing supernatants were collected 48hrs or 72hrs after transfection and filtered through a 0.45 μm filter. For infection, cells were spin-infected with retroviral supernatants containing 5 μg/ml polybrene (Sigma Aldrich) for 2h at room temperature at 2000 rpm. Supernatants were gently removed 4–5hrs later and replaced with adapted culture media.
Microscopy analysis
Purified DCs were activated with 1 μg/ml poly(I:C) (Invivogen) for 4hrs at 37°C and then pulsed with 0.25 μg/μl OVA257–264 peptide (AnaSpec Inc.) for 1hr at 37°C. Activated DCs and naïve OT-I cells were co-cultured for 40min at a 1:1 ratio at 37°C. During the last 10min, cells were transferred onto poly-L-lysine-coated microscopy slides. DC-T conjugates were fixed with 4% paraformaldehyde (PFA), quenched (50 mM NH4Cl), permeabilized (0.3% Triton-X-100) and blocked (0.01% saponin /0.25% fish gelatin /anti-CD16/32). Cells were then stained with biotinylated anti-CD3ε (Biolegend), anti-RARα (cl. Ralpha 10, Thermo Fisher) and anti-ZAP-70 (cl. M20, Santa Cruz) at 4°C overnight. Staining with secondary antibodies and streptavidin, listed in the key resources table, was carried out at room temperature for 1hr. Images were recorded using a FV1000 laser scanning confocal microscope (Olympus). Images of at least 100 synapses were recorded and the number of CD3 only, CD3-ZAP-70 and CD3-ZAP-70-RARα synapses was quantified. CD3-RARα synapses were not observed.
T cells transduced with GFP-tagged RARα1 or RARα3 were fixed (PBS1X PFA 2%). Staining with DAPI was then performed for 1hr at 4°C. Samples were imaged on an Olympus Fluoview FV10i confocal microscope with a 60x oil immersion objective and acquired using FV10i SW. Cytosolic/nuclear localization was based on DAPI staining and quantified using Imaris software. CD8 T cells transduced with CD3ζ-GFP were activated with anti-CD3/CD28 for 30min. Clustering of CD3ζ-GFP was quantified following microscopic (Olympus) imaging of entire cells.
Mass spectrometry-based phosphoproteomic analysis
Jurkat E6.1 cells (ATCC) were passaged in RPMI + 10% heat inactivated FBS and kept at cell densities between 2e5 and 1e6 cell per mL. Jurkat cells were activated using plate bound anti-CD3 and anti-CD28 antibodies at 3.33 μg/mL in 96 well plates. At select time points cells were transferred to cold PBS to stop signaling, washed twice with cold PBS and flash frozen. The zero timepoint was performed in biological duplicate, the others in triplicate. Cell pellets were prepared and were then analyzed for quantitative phosphoproteomics as previously described in (62, 63). Iron immobilized metal affinity chromatography (IMAC) was used to enrich for phosphopeptides, and tandem mass tags (TMT) 11-plex reagents (Thermo Scientific) were used for quantitation. Data was processed by Spectrum Mill (Agilent/Broad Institute). Mass spectra were manually annotated.
Bacterial infection
Acta deficient Listeria monocytogenes expressing OVA (ActA− Lm-OVA)(64) was prepared from cultures in brain-heart–infusion broth. Bacteria were washed and resuspended in 1X PBS before intravenous infection (2.5 × 105 CFU per mouse).
Diet Studies
At day 14.5 of gestation, pregnant females were fed with either vitamin A-deficient or -sufficient diet (Dyets Inc.) and maintained on the diet until weaning of the litters. Weanlings were maintained on either diet until the experiment. In some experiments, mice were administered DMSO or 250 μg all trans retinoic acid intraperitoneally every other day for 7 days prior to immunization and until euthanasia of the mice.
Preparation of intraepithelial T cells
Small intestines were removed and separated from Peyer’s patches and cut longitudinally and into pieces of 0.5 cm in length. Pieces were shaken for 30min in magnesium-free, calcium-free Hank’s balanced-salt solution supplemented with 1 mM dithiothreitol (SIGMA Aldrich), 5mM EDTA (SIGMA-Aldrich) and 5% (vol/vol) FCS. Cells were collected from the washes and were passed over a discontinuous 40–70% (vol/vol) gradient of Percoll (Pharmacia Biotech) for 20min at 1100g. Intraepithelial T cells were then isolated from the Percoll gradient interface and washed free of Percoll.
Experimental Autoimmune Encephalomyelitis (EAE) experiment
MOG35–55 peptide (200 μg/mouse, Anaspec) was mixed with an equal volume of incomplete Freund’s adjuvant (#263910, 200μl/mouse, BD) and M.Tuberculosis (#231141, 400μg/mouse, BD), then, subcutaneously injected to each mouse. Diluted pertussis toxin from B. pertussis (#180, 400 ηg/mouse, List Biological Laboratories Inc) was intraperitoneally injected at day 0 and day 2. Clinical score was evaluated on a daily basis from day 7(65). All experiments were performed in accordance with the approved protocol by the institutional animal care committee.
Histology
Spines were cut open and spinal cords were extracted and fixed with 4% zinc formalin and dehydrated with 70%, 80%, 90% and 100% isopropanol and Pro-Par Clearant (Anatech) by Tissue-Tek VIP (SAKURA), then, embedded in Paraffin and sectioned. Tissue sections were stained with Hematoxylin and Eosin, then, scanned by Axio Scan Z1 (ZEISS) and analyzed by ZEN software (ZEISS).
Other Software
Graphic abstract created with BioRender.com.
QUANTIFICATIONS AND STATISTICAL ANALYSIS
Quantifications
Quantitative phosphoproteomics analysis was performed as previously described in(62, 63). Iron immobilized metal affinity chromatography (IMAC) was used to enrich for phosphopeptides, and tandem mass tags (TMT) 11-plex reagents (Thermo Scientific) were used for quantitation. Data was processed by Spectrum Mill (Agilent/Broad Institute). Mass spectra were manually annotated.
Signaling intensity was measured by quantifying the optical density of immuno-reactive bands of proteins analyzed by western blot, detected by chemiluminescence (ECL solution, Santa Cruz). ImageJ software from NIH image (Version 1.51 23 April 2018) was used to quantify the intensity of each band in the negative picture after performing a gray scale 8-bit transformation on the scans and calibrating the threshold.
Statistics
Statistical comparisons were performed using a two-tailed unpaired Student t-test, Mann-Whitney test or, when appropriate, a two-tailed paired Student t-test, with the Prism software (Graphpad).
When applicable, variances were calculated and were similar between compared groups.
For in vivo experiments, animals were gender- and age-matched and divided in groups where n= >3. The specific number of animals used per group and per experiment is indicated in the figure and/or figure legend.
No statistical method was used to predetermine the number of animal sample size.
No sample or animal was excluded from analysis.
No randomization method was used to determine how samples/animals were allocated to experimental groups and processed.
For in vitro experiments, the minimal sample size chosen was n=3 independent repeats, each independent experiment was performed in triplicates (3 technical replicates), except for signaling studies where technical replicates are not feasible.
ADDITIONAL RESOURCES
Sequences retrieved in RNA-seq experiments were aligned on the UCSC mouse genome assembly (GRCm38/mm10) using the HISAT2 (hierarchical indexing for spliced alignment of transcripts 2). And the obtained BAM files were then sorted by Samtools and visualized by Integrative Genome viewer (IGV). Raw fastq files for the RNA-seq libraries were deposited at the Gene Expression Omnibus (GEO) database under the accession number GEO: GSE112609.
Analysis of high-throughput sequencing identified other Rara sequences with different 5’ splice junctions. The transcripts were then aligned with predicted sequences in the RefSeq database (www.ncbi.nlm.nih.gov/refseq).
All alternative transcript shared the same 3’ end sequence with Rara1 and -2 but each one had a unique 5’end sequence. To identify a putative Rara3 promoter region and transcriptional start site (TSS) data from the Functional ANnoTation Of the Mammalian genome (FANTOM)5 database was used, which indicated a TSS downstream of the Rara2 TSS and upstream of the 5’ end of the variant transcripts.
RNA-seq data of human naïve CD8αβ+ (CD3+ CD8+ CD45RA+ CD127+ CCR7+) or CD4+ (CD3+ CD4+ CD45RA+ CD127+ CCR7+) peripheral blood T cells were from the Database of Immune Cell Expression, Expression quantitative trait loci (eQTLs) and Epigenomics (DICE); https://dice-database.org/.
Supplementary Material
HIGHLIGHTS.
A cytoplasmic isoform of RARα (cRARα) is activated by the TCR and binds TCR-ZAP-70
RA counteracts cRARα for suboptimal T cell activation and enhanced Treg conversion
Effective cRARα-TCR signaling induces CRABP2 for RA translocation to the nucleus
Defects in CRABP2 expression reduce anti-pathogen immunity and autoimmune disease
ACKNOWLEDGMENTS
We thank Y. Wang-Zhu and Gabriela Aguila for help with breeding and typing of the mice; R. Noelle for providing the RARαDN mice; C. Kim, D. Hinz, L. Nosworthy, K. Van Gunst and A. Jose for cell sorting. Z. Mikulski for help with microscopy. Sara McArdle and Kenneth Kim for help with the histology analysis and scoring. This work was supported by the US National Institutes of Health (U01 AI125957 and R01 AI106298 (HC). Support from the Philippe Foundation (AL), CCFA Fellowship 697275 (IT) and MK by the P01 DK46763.
This is manuscript 1634 from the La Jolla Institute for Immunology.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
MATERIALS AVAILABILITY
All unique/stable reagents generated in this study are available from the lead contact with a completed material transfer agreement.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Larange A, Cheroutre H. Retinoic Acid and Retinoic Acid Receptors as Pleiotropic Modulators of the Immune System. Annu Rev Immunol. 2016;34:369–94. [DOI] [PubMed] [Google Scholar]
- 2.di Masi A, Leboffe L, De Marinis E, Pagano F, Cicconi L, Rochette-Egly C, et al. Retinoic acid receptors: from molecular mechanisms to cancer therapy. Mol Aspects Med. 2015;41:1–115. [DOI] [PubMed] [Google Scholar]
- 3.Hall JA, Cannons JL, Grainger JR, Dos Santos LM, Hand TW, Naik S, et al. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity. 2011;34(3):435–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall JA, Grainger JR, Spencer SP, Belkaid Y. The role of retinoic acid in tolerance and immunity. Immunity. 2011;35(1):13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farboud B, Privalsky ML. Retinoic acid receptor-alpha is stabilized in a repressive state by its C-terminal, isotype-specific F domain. Mol Endocrinol. 2004;18(12):2839–53. [DOI] [PubMed] [Google Scholar]
- 6.Dong D, Ruuska SE, Levinthal DJ, Noy N. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J Biol Chem. 1999;274(34):23695–8. [DOI] [PubMed] [Google Scholar]
- 7.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129(4):723–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21(4):527–38. [DOI] [PubMed] [Google Scholar]
- 9.Ohoka Y, Yokota A, Takeuchi H, Maeda N, Iwata M. Retinoic acid-induced CCR9 expression requires transient TCR stimulation and cooperativity between NFATc2 and the retinoic acid receptor/retinoid X receptor complex. J Immunol. 2011;186(2):733–44. [DOI] [PubMed] [Google Scholar]
- 10.Pino-Lagos K, Guo Y, Brown C, Alexander MP, Elgueta R, Bennett KA, et al. A retinoic acid-dependent checkpoint in the development of CD4+ T cell-mediated immunity. J Exp Med. 2011;208(9):1767–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007;204(8):1765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204(8):1757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317(5835):256–60. [DOI] [PubMed] [Google Scholar]
- 14.Mucida D, Cheroutre H. TGFbeta and retinoic acid intersect in immune-regulation. Cell Adh Migr. 2007;1(3):142–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schambach F, Schupp M, Lazar MA, Reiner SL. Activation of retinoic acid receptor-alpha favours regulatory T cell induction at the expense of IL-17-secreting T helper cell differentiation. Eur J Immunol. 2007;37(9):2396–9. [DOI] [PubMed] [Google Scholar]
- 16.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204(8):1775–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill JA, Hall JA, Sun CM, Cai Q, Ghyselinck N, Chambon P, et al. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008;29(5):758–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mucida D, Pino-Lagos K, Kim G, Nowak E, Benson MJ, Kronenberg M, et al. Retinoic acid can directly promote TGF-beta-mediated Foxp3(+) Treg cell conversion of naive T cells. Immunity. 2009;30(4):471–2; author reply 2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26(3):371–81. [DOI] [PubMed] [Google Scholar]
- 20.Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, O’Malley JT, et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178(8):4901–7. [DOI] [PubMed] [Google Scholar]
- 21.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282(13):9358–63. [DOI] [PubMed] [Google Scholar]
- 22.Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109(10):4368–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elias KM, Laurence A, Davidson TS, Stephens G, Kanno Y, Shevach EM, et al. Retinoic acid inhibits Th17 polarization and enhances FoxP3 expression through a Stat-3/Stat-5 independent signaling pathway. Blood. 2008;111(3):1013–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, Kitani A, Strober W. Molecular mechanisms regulating TGF-beta-induced Foxp3 expression. Mucosal Immunol. 2010;3(3):230–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nolting J, Daniel C, Reuter S, Stuelten C, Li P, Sucov H, et al. Retinoic acid can enhance conversion of naive into regulatory T cells independently of secreted cytokines. J Exp Med. 2009;206(10):2131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Etemire E, Krull M, Hasenberg M, Reichardt P, Gunzer M. Transiently reduced PI3K/Akt activity drives the development of regulatory function in antigen-stimulated Naive T-cells. PLoS One. 2013;8(7):e68378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friesen LR, Gu B, Kim CH. A ligand-independent fast function of RARalpha promotes exit from metabolic quiescence upon T cell activation and controls T cell differentiation. Mucosal Immunol. 2021;14(1):100–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajaii F, Bitzer ZT, Xu Q, Sockanathan S. Expression of the dominant negative retinoid receptor, RAR403, alters telencephalic progenitor proliferation, survival, and cell fate specification. Dev Biol. 2008;316(2):371–82. [DOI] [PubMed] [Google Scholar]
- 29.Leroy P, Krust A, Zelent A, Mendelsohn C, Garnier JM, Kastner P, et al. Multiple isoforms of the mouse retinoic acid receptor alpha are generated by alternative splicing and differential induction by retinoic acid. EMBO J. 1991;10(1):59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.So T, Soroosh P, Eun SY, Altman A, Croft M. Antigen-independent signalosome of CARMA1, PKCtheta, and TNF receptor-associated factor 2 (TRAF2) determines NF-kappaB signaling in T cells. Proc Natl Acad Sci U S A. 2011;108(7):2903–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Britton GJ, Ambler R, Clark DJ, Hill EV, Tunbridge HM, McNally KE, et al. PKCtheta links proximal T cell and Notch signaling through localized regulation of the actin cytoskeleton. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dustin ML, Choudhuri K. Signaling and Polarized Communication Across the T Cell Immunological Synapse. Annu Rev Cell Dev Biol. 2016;32:303–25. [DOI] [PubMed] [Google Scholar]
- 34.Guy CS, Vignali KM, Temirov J, Bettini ML, Overacre AE, Smeltzer M, et al. Distinct TCR signaling pathways drive proliferation and cytokine production in T cells. Nat Immunol. 2013;14(3):262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palaga T, Miele L, Golde TE, Osborne BA. TCR-mediated Notch signaling regulates proliferation and IFN-gamma production in peripheral T cells. J Immunol. 2003;171(6):3019–24. [DOI] [PubMed] [Google Scholar]
- 37.Chebaro Y, Amal I, Rochel N, Rochette-Egly C, Stote RH, Dejaegere A. Phosphorylation of the retinoic acid receptor alpha induces a mechanical allosteric regulation and changes in internal dynamics. PLoS Comput Biol. 2013;9(4):e1003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu KP, Liou YC, Zhou XZ. Pinning down proline-directed phosphorylation signaling. Trends Cell Biol. 2002;12(4):164–72. [DOI] [PubMed] [Google Scholar]
- 39.Svensson RU, Shey MR, Ballas ZK, Dorkin JR, Goldberg M, Akinc A, et al. Assessing siRNA pharmacodynamics in a luciferase-expressing mouse. Mol Ther. 2008;16(12):1995–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Won JY, Nam EC, Yoo SJ, Kwon HJ, Um SJ, Han HS, et al. The effect of cellular retinoic acid binding protein-I expression on the CYP26-mediated catabolism of all-trans retinoic acid and cell proliferation in head and neck squamous cell carcinoma. Metabolism. 2004;53(8):1007–12. [DOI] [PubMed] [Google Scholar]
- 41.Budhu AS, Noy N. Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol Cell Biol. 2002;22(8):2632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sessler RJ, Noy N. A ligand-activated nuclear localization signal in cellular retinoic acid binding protein-II. Mol Cell. 2005;18(3):343–53. [DOI] [PubMed] [Google Scholar]
- 43.Brown CC, Esterhazy D, Sarde A, London M, Pullabhatla V, Osma-Garcia I, et al. Retinoic acid is essential for Th1 cell lineage stability and prevents transition to a Th17 cell program. Immunity. 2015;42(3):499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Billadeau DD, Nolz JC, Gomez TS. Regulation of T-cell activation by the cytoskeleton. Nat Rev Immunol. 2007;7(2):131–43. [DOI] [PubMed] [Google Scholar]
- 45.Yokosuka T, Sakata-Sogawa K, Kobayashi W, Hiroshima M, Hashimoto-Tane A, Tokunaga M, et al. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat Immunol. 2005;6(12):1253–62. [DOI] [PubMed] [Google Scholar]
- 46.Dongre A, Surampudi L, Lawlor RG, Fauq AH, Miele L, Golde TE, et al. Non-Canonical Notch Signaling Drives Activation and Differentiation of Peripheral CD4(+) T Cells. Front Immunol. 2014;5:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinbuck MP, Arakcheeva K, Winandy S. Novel TCR-Mediated Mechanisms of Notch Activation and Signaling. J Immunol. 2018;200(3):997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rochette-Egly C, Adam S, Rossignol M, Egly JM, Chambon P. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 1997;90(1):97–107. [DOI] [PubMed] [Google Scholar]
- 49.Unsworth AJ, Flora GD, Gibbins JM. Non-genomic effects of nuclear receptors: insights from the anucleate platelet. Cardiovasc Res. 2018;114(5):645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rondina MT, Freitag M, Pluthero FG, Kahr WH, Rowley JW, Kraiss LW, et al. Non-genomic activities of retinoic acid receptor alpha control actin cytoskeletal events in human platelets. J Thromb Haemost. 2016;14(5):1082–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarvis CI, Goncalves MB, Clarke E, Dogruel M, Kalindjian SB, Thomas SA, et al. Retinoic acid receptor-alpha signalling antagonizes both intracellular and extracellular amyloid-beta production and prevents neuronal cell death caused by amyloid-beta. Eur J Neurosci. 2010;32(8):1246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piskunov A, Rochette-Egly C. A retinoic acid receptor RARalpha pool present in membrane lipid rafts forms complexes with G protein alphaQ to activate p38MAPK. Oncogene. 2012;31(28):3333–45. [DOI] [PubMed] [Google Scholar]
- 53.Masia S, Alvarez S, de Lera AR, Barettino D. Rapid, nongenomic actions of retinoic acid on phosphatidylinositol-3-kinase signaling pathway mediated by the retinoic acid receptor. Mol Endocrinol. 2007;21(10):2391–402. [DOI] [PubMed] [Google Scholar]
- 54.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205(3):565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merkenschlager M, von Boehmer H. PI3 kinase signalling blocks Foxp3 expression by sequestering Foxo factors. J Exp Med. 2010;207(7):1347–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A. 2008;105(22):7797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fawcett D, Pasceri P, Fraser R, Colbert M, Rossant J, Giguere V. Postaxial polydactyly in forelimbs of CRABP-II mutant mice. Development. 1995;121(3):671–9. [DOI] [PubMed] [Google Scholar]
- 58.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–74. [DOI] [PubMed] [Google Scholar]
- 59.Maekawa Y, Minato Y, Ishifune C, Kurihara T, Kitamura A, Kojima H, et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat Immunol. 2008;9(10):1140–7. [DOI] [PubMed] [Google Scholar]
- 60.Ehst BD, Ingulli E, Jenkins MK. Development of a novel transgenic mouse for the study of interactions between CD4 and CD8 T cells during graft rejection. Am J Transplant. 2003;3(11):1355–62. [DOI] [PubMed] [Google Scholar]
- 61.Li E, Sucov HM, Lee KF, Evans RM, Jaenisch R. Normal development and growth of mice carrying a targeted disruption of the alpha 1 retinoic acid receptor gene. Proc Natl Acad Sci U S A. 1993;90(4):1590–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katrancha SM, Shaw JE, Zhao AY, Myers SA, Cocco AR, Jeng AT, et al. Trio Haploinsufficiency Causes Neurodevelopmental Disease-Associated Deficits. Cell Rep. 2019;26(10):2805–17 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat Protoc. 2018;13(7):1632–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4(10):812–23. [DOI] [PubMed] [Google Scholar]
- 65.Bittner S, Afzali AM, Wiendl H, Meuth SG. Myelin oligodendrocyte glycoprotein (MOG35–55) induced experimental autoimmune encephalomyelitis (EAE) in C57BL/6 mice. J Vis Exp. 2014(86). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell. 2018;175(6):1701–15 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table.
Original western blot images, raw flow cytometry data, (RT)-PCR, and microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESSOURCE TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| T cell stimulation anti-CD3e | Thermo Fisher Scientific | 16-0031-86 |
| T cell stimulation anti-CD28 | Thermo Fisher Scientific | 16-0281-86 |
| WB: anti-ZAP-70 (cl. D1C10E) | Cell Signaling Tech. | 3165T |
| WB: anti-GAPDH (cl. 14C10) | Cell Signaling Tech. | 85925SF |
| WB: anti-Lamin B1 (cl. D9V6H) | Cell Signaling Tech. | 13435S |
| WB: anti-NOTCH1 (cl. D1E11) | Cell Signaling Tech. | 3608T |
| WB: anti-RARα | Cell Signaling Tech. | 2554 |
| WB: anti-RARγ1 (cl. D3A4) | Cell Signaling Tech. | 8965T |
| WB: anti-CRABP2 | One World Lab | 11250 |
| WB: anti-RARa | Assay Biotechnology Company | R12-3424 |
| WB: anti-HSP90 (cl. AC16) | Santa Cruz | sc-101494 |
| WB: anti-Flag (cl. M2) | Millipore Sigma | A2220 |
| WB: anti-AKT | Cell Signaling Tech. | 9272 |
| WB: anti-Phospho-AKT | Cell Signaling Tech. | 9271T |
| WB: anti-Phospho- PLCγ1 | Cell Signaling Tech. | 2821 |
| WB: anti-Phospho-ZAP-70 (cl. 65E4) | Cell Signaling Tech. | 2717 |
| WB: anti-Phospho-ERK1/2 (cl. D13.14.4E) | Cell Signaling Tech. | 4370 |
| Activation: H-2Kb-SIINFEKL tetramer | In house | N/A |
| IP: anti-RARα | Santa Cruz | sc-551 |
| Flow /Conf. Im.: anti-CD16-CD32 (cl. 2.4G2) | In house | N/A |
| Flow: anti-CD4-APC (cl. RM4-5) | BD | 553051 |
| Flow: anti-TCRgd-FITC (cl. GL3) | BD | 553177 |
| Flow: anti-CD45.1-FITC (cl. A20) | BD | 553775 |
| Flow: anti-CD44-V450 (cl. IM7) | BD | 560452 |
| Flow: anti-CD8a-PercpCy5.5 (cl. 53-6.7) | BD | 551162 |
| Flow: anti- CD8a-PeCy7 (cl. 53-6.7) | BD | 552877 |
| Flow: anti-IFNg-BV711 (cl. XMG1.2) | Biolegend | 505835 |
| Flow: anti-Il-17-PeCy7 (cl. TC11-18H10.1) | Biolegend | 506922 |
| Flow: anti-CD8a-AF700 (cl. 53-6.7) | Biolegend | 100730 |
| Flow: anti-CD4-AF700 (cl. GK1.5) | Biolegend | 100430 |
| Flow: anti-FOXP3-PercpCy5.5 (cl. FJK-16s) | Thermo Fisher Scientific | 45-5773-82 |
| Flow: anti-CCR9-FITC (cl. eBioCW-1.2) | Thermo Fisher Scientific | 11-1991-85 |
| Flow: anti-TCRb-APCeF780 (cl. H57-597) | Thermo Fisher Scientific | 47-5961-82 |
| Flow: anti-CD45.2-APC (cl.104) | Thermo Fisher Scientific | 17-0454-82 |
| Flow: anti-RORgt-Pe (cl. AFKJS-9) | Thermo Fisher Scientific | 12-6988-80 |
| Flow: anti-CD45-eF450 (cl. 30-F11) | Thermo Fisher Scientific | 48-0451-82 |
| Flow: anti-FOXP3-AF488 (cl. FJK-16s) | Thermo Fisher Scientific | 53-5773-82 |
| Flow: anti-FOXP3-Pe (cl. FJK-16s) | Thermo Fisher Scientific | 12-5773-82 |
| Flow: anti-CD25-APC (cl. PC61.5) | Thermo Fisher Scientific | 17-0251-82 |
| Flow: anti-CD4-eF450 (cl. RM4-5) | Thermo Fisher Scientific | 48-0042-82 |
| Flow: anti-c-MYC-FITC (cl. 9E10) | Thermo Fisher Scientific | 13-2511 |
| Conf. Im.: anti- CD3ε-biotin (cl. 2C11) | Biolegend | 100304 |
| Conf. Im.: Goat anti-mouse IgG-DyLight™ 488 | Biolegend | 405310 |
| Conf. Im.: Rabbit anti-Goat IgG-Alexa Fluor 647 | Life Technology | A21446 |
| Conf. Im.: anti-ZAP70 (cl. M20) | Santa Cruz | sc-1526 |
| Conf. Im.: anti-RARa (c. Ralpha10) | Thermo Fisher Scientific | MA1810A |
| Bacterial and virus strains | ||
| Listeria monocytogenes expressing OVA (ActA− Lm-OVA) | (64) | Gift |
| Biological samples | ||
| N/A | ||
| Chemicals, peptides, and recombinant proteins | ||
| LIVE/DEAD™ Fixable Yellow Dead Cell Stain | Thermo Fisher Scientific | L34959 |
| EDTA | Thermo Fisher Scientific | 15575020 |
| Collagenase D | Millipore Sigma | 11088858001 |
| DNAse I | Millipore Sigma | 10104159001 |
| Human recombinant TGF-β | R&D System | 240-B-002 |
| Trans retinoic acid | Millipore Sigma | R2625 |
| RAR antagonist LE540 | FUJIFILM Wako Pure Chemical Corporation | Distributor 123-04521 Barcode No 4987481365261 |
| RARα-selective antagonist | Enzo lifescience | Ro 41-5253 |
| RARγ-selective antagonist MM 11253 | Tocris bioscience | 3822 |
| MEK1/2 inhibitor U0126 | Invivogen | tlrl-u0126 |
| PI3K inhibitor LY294002 | Invivogen | tlrl-ly29 |
| γ-secretase inhibitor GSI | Tocris bioscience | 2870 |
| Human recombinant IL-2 | Peprotech | 200-02 |
| AKT Inhibitor VIII | Cayman Chemical Company, Inc | 14870 |
| DMSO | Millipore Sigma | D2438-5X |
| TRIzol reagent | Thermo Fisher Scientific | 15596026 |
| TGX FastCast Acrylamide Solutions for SDS-PAGE gel | Biorad | 1610171 |
| PVDF membrane | Santa Cruz | sc-3723 |
| NaCl | Millipore Sigma | S9888 |
| Tris base | Santa Cruz | sc-362305 |
| Glycerol | Milliopore Sigma | G5516 |
| Triton X-100 | Milliopore Sigma | X100 |
| Protease and phosphatase inhibitor mixture | Milliopore Sigma | 11697498001 |
| ECL solution | Santa Cruz | sc-2048 |
| Polybrene | Milliopore Sigma | TR-1003-G |
| Poly I:C (HMW) | InvivoGen | tlrl-pic |
| OVA257-264 peptide | AnaSpec Inc. | AS-62572 |
| Poly-L-lysine | R&D System | 3438-100-01 |
| NH4Cl | Milliopore Sigma | A4514 |
| Saponin | Millipore Sigma | 47036 |
| Fish gelatin | Milliopore Sigma | G7041 |
| ProLong™ Gold Antifade Mountant with DNA Stain DAPI | Thermo Fisher Scientific | P36935 |
| Dithiothreitol | Milliopore Sigma | D2438 |
| Percoll | Cytiva | 17089109 |
| Freund’s adjuvant | BD | 263910 |
| M Tuberculosis H37Ra Dessicated | BD | 231141 |
| Pertussis toxin | List Biological Laboratories Inc | 180 |
| Zinc formalin | Anatech Lt | 175 |
| Pro-Par Clearant | Anatech Ltd | 510 |
| Conf. Im.: Streptavidine-Cyanine3 | Biolegend | 405215 |
| Critical commercial assays | ||
| MACS CD8α+ T cell isolation kit | Miltenyi Biotec | 130-096-543 |
| MACS CD4+ T cell isolation kit | Miltenyi Biotec | 130-104-453 |
| CD11c-Microbeads Ultrapure | Miltenyi Biotec | 130-125-835 |
| SureSelectXT RNA Direct Library Preparation kit | Agilent Technologies | G7564A |
| Pierce™ NE-PER® Nuclear and Cytoplasmic Extraction Reagent Kit | Thermo Fisher Scientific | 78833 |
| JetPrime transfection reagent | Polyplus transfection | 101000027 |
| Tandem mass tags (TMT) 11-plex reagents | Thermo Fisher Scientific | A34808 |
| Cell Trace Violet | Thermo Fisher Scientific | C34557 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE112609 |
| Database of Immune Cell Expression, Expression quantitative trait loci (eQTLs) and Epigenomics (DICE) | (66) | https://dice-database.org |
| UCSC mouse genome assembly (GRCm38/mm10) | UCSC Genome Browser Group | https://genome.ucsc.edu/cgi-bin/hgGateway |
| Experimental models: Cell lines | ||
| Platinum-E | Cell Biolabs | RV-101 |
| Jurkat, Clone E6-1 | ATCC | TIB-152 |
| MCC-T CD4 T cell hybridoma cells | (31) | Gift |
| Mutant ES cell (LoxP sites flanking Crabp2 exons) | EUCOMM | HEPD0679_8_F03 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Jackson Lab. | 000664 |
| Mouse: Crabp2−/−(10 generation backcrossed to B6 background) | Fawcett et al., 1995 | Gift |
| Mouse: RARE-Luciferase (DR5) | (39) | Gift |
| Mouse: RAR403 (DOMINANT NEGATIVE) | (28) | Gift |
| Mouse: CD4-Cre | (58) | Gift |
| Mouse: E8I-cre | (59) | Gift |
| Mouse: OT-I TCRM | Jackson Lab. | 003831 |
| Mouse: Act-mOVA | (60) | Gift |
| Mouse: RARα1−/− | Jackson Lab. | 023845 |
| Mouse: Crabp2flox/flox Crabp2tm1e(EUCOMM)Hmgu | This paper | N/A |
| Mouse: Distal LCK-Cre | Jackson Lab. | Jackson Lab. |
| Oligonucleotides | ||
| RT-RARa F1: CAGTCAGTGGTTACAGCACACCGTC | IDT DNA | N/A |
| RT-RARa F2: GGACTCCGCTTTGGAATGGCTC | IDT DNA | N/A |
| RT-RARa F3: GATGTCCAGGCCCAAGTAGAAGCCAG | IDT DNA | N/A |
| RT-RARa R1: GCTGCAATCTGCTGCTCATGCCTACAC | IDT DNA | N/A |
| RT-RARa R2: GGACTGACCTGCTGCAATCTGCTG | IDT DNA | N/A |
| RT-RARa R3: CTGGCTTCTACTTGGGCCTGGACATC | IDT DNA | N/A |
| RT-RARa R4: CAGGCTGTGAAAGACTCCTGCGGCT | IDT DNA | N/A |
| RT-RARa R5: CAGCATGTGTTATGCCAGGCTGTGAAAG | IDT DNA | N/A |
| RT-RARa R6: CCAACAGCATGTGTTATGCCAGGCTG | IDT DNA | N/A |
| RT-RARa R7: TTCACCTCACTTCCTTCTCGGGAG | IDT DNA | N/A |
| RT-RARa R9: CCCCATAGTGGTAGCCGGATGATTTG | IDT DNA | N/A |
| RT-RARa R15: TCAGTGGAAACCCAGCAGGAACAGGTG | IDT DNA | N/A |
| RT-Hprt F: GTCGTGATTAGCGATGATGAACC | IDT DNA | N/A |
| RT-Hprt R: ATGACATCTCGAGCAAGTCTTTCAG | IDT DNA | N/A |
| qRT-Crabp2 F: CCCAGAGCTTTTAGCATTTCC | IDT DNA | N/A |
| qRT-Crabp2 R: GAAGATCTAAAGAGAAAGCCACCT | IDT DNA | N/A |
| qRT-cMYC F: TTGAAGGCTGGATTTCCTTTGGGC | IDT DNA | N/A |
| qRT-cMYC R: TCGTCGCAGATGAAATAGGGCTGT | IDT DNA | N/A |
| qRT-GADPH F: CAGATGCCTGCTTCACCA | IDT DNA | N/A |
| qRT-GADPH R: ATGGCCTTCCGTGTTCCT | IDT DNA | N/A |
| qRT-bACTIN F: CCAGAAGGACTGTTATGTGGGA | IDT DNA | N/A |
| qRT-bACTIN R: GACTCCGTGTTCAATGGGATAC | IDT DNA | N/A |
| qRT-HPRT F: GTCGTGATTAGCGATGATGAACC | IDT DNA | N/A |
| qRT-HPRT R: ATGACATCTCGAGCAAGTCTTTCAG | IDT DNA | N/A |
| Recombinant DNA | ||
| Plasmid: MSCV-IRES-GFP | Addgene | 20672 |
| Plasmid: MSCV-IRES-Puromycin | Addgene | 68469 |
| Plasmid: CD3ζ-GFP | Katz et al., 2016 | Gift |
| Software and algorithms | ||
| FlowJo software v10.8.0 | Becton Dickinson & Company | https://www.flowjo.com/solutions/flowjo/downloads |
| HISAT2 | (67) | https://github.com/DaehwanKimLab/hisat2 |
| Tools (written in C using htslib) for manipulating next-generation sequencing data | Samtools | https://github.com/samtools/samtools |
| Integrative Genomics Viewer | Broad Institute | https://software.broadinstitute.org/software/igv/ |
| FACSDIVA | Becton Dickinson & Company | https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software |
| Spectrum Mill | Agilent/Broad Institute | https://proteomics.broadinstitute.org/millhome.htm |
| ZEN software | ZEISS | https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html |
| Prism 8.4.2 | GraphPad | https://www.graphpad.com/features |
| Other | ||
| Penicillin, streptomycin | Thermo Fisher Scientific | 15140122 |
| Puromycin | Invivogen | ant-pr-1 |
| Balsticidin | Invivogen | ant-bl-05 |


